Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers worldwide. Despite significant progresses in the last decades, the origin of this cancer remains unclear and no efficient therapy exists. PDAC does not arise de novo: three remarkable different types of pancreatic lesions can evolve towards pancreatic cancer. These precursor lesions include: Pancreatic intraepithelial neoplasia (PanIN) that are microscopic lesions of the pancreas, Intraductal Papillary Mucinous Neoplasms (IPMN) and Mucinous Cystic Neoplasms (MCN) that are both macroscopic lesions. However, the cellular origin of these lesions is still a matter of debate.
Classically, neoplasm initiation or progression is driven by several genetic and epigenetic alterations. The aim of this review is to assemble the current information on genetic mutations and epigenetic disorders that affect genes during pancreatic carcinogenesis. We will further discuss the interest of the genetic and epigenetic alterations for the diagnosis and prognosis of PDAC. Large genetic alterations (chromosomal deletion/amplification) and single point mutations are well described for carcinogenesis inducers. Mutations classically occur within key regions of the genome. Consequences are various and include activation of mitogenic pathways or silencing of apoptotic processes. Alterations of K-RAS, P16 and DPC4 genes are frequently observed in PDAC samples and have been described to arise gradually during carcinogenesis. DNA methylation is an epigenetic process involved in imprinting and X chromosome inactivation. Alteration of DNA methylation patterns leads to deregulation of gene expression, in the absence of mutation. Both genetic and epigenetic events influence genes and non-coding RNA expression, with dramatic effects on proliferation, survival and invasion. Besides improvement in our fundamental understanding of PDAC development, highlighting the molecular alterations that occur in pancreatic carcinogenesis could provide new clinical tools for early diagnosis of PDAC and the molecular basis for the development of new effective therapies.
Keywords: Pancreas, Cancer, Preneoplastic lesions, DNA methylation, Genetic alteration.
1. INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the fourth cause of death by cancer in Western countries, while representing only 3% of the new cancer cases each year [1]. Tobacco, diabetes, obesity and chronic pancreatitis are described as risk factors for PDAC [2]. However, with the exception of familial PDAC, no high risk population is defined. Consequently, screening for PDAC with current imaging techniques seems possible only for patients presenting a familial history of PDAC or already managed for others pancreatic diseases. Hence, PDAC is diagnosed at late stage due to the absence of specific symptoms (abdominal discomfort and abdominal mass due to compression of pancreatic duct) [2, 3]. Up to 60% of patients have advanced PDAC at the time of diagnosis and are not eligible for surgery; their median survival is only 3 to 6 months [1]. Resection relies on the general condition of the patient, the location, size and tumor local invasion. Because of its poor prognosis, improvement of PDAC prognostic requires detecting and managing this cancer before regional invasion and metastasis [3]. For a better clinical care and a better understanding of PDAC, remarkable works have been done to elucidate the early events driving pancreatic carcinogenesis. We will first describe the main preneoplastic lesions leading to PDAC, to further introduce the molecular alterations associated with PDAC and their lesion-stage dependent appearance. We will finally discuss the molecular marker potential and the clinical interest of such alterations.
2. PRENEOPLASTIC LESIONS OF THE PANCREAS
To date, three types of neoplasic lesion precursor to PDAC are described and characterized. The study of these lesions is of prime importance to:
detect and manage PDAC before it becomes invasive and incurable.
better understand PDAC carcinogenesis to elaborate new therapies.
Development of such lesions is most of the time asymptomatic, and their discovery occurs together with unresecable carcinoma. They all follow a multistep progression to invasive cancer classified upon morphological and cytological features [2]. The sequential evolution from precursor lesion to cancer is still a matter of debate because of the absence of early detection and follow-up of these lesions. Main obstacles for precursor detection and follow-up using current diagnostic technologies are their small size and the absence of high risk population to screen. Consequently, studies are usually conducted on fixed resected tissue. In other words, the following observations are in favor of a multistep carcinogenesis:
duct lesions are far more common in pancreas with infiltrating carcinoma [4, 5].
there is an increase in the grade of lesions surrounding infiltrating carcinoma [5].
patients diagnosed with intraductal mucinous neoplasms that are not resected eventually develop infiltrating carcinoma [4, 5].
2.1. Pancreatic Intraepithelial Neoplasia (PanIN)
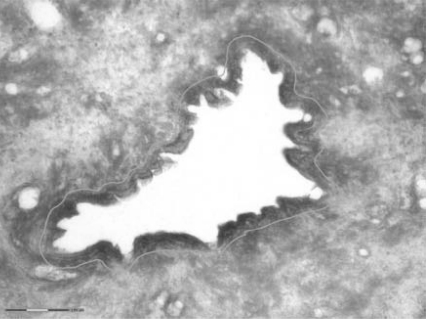
PanINs are microscopic ductal lesions usually measuring less than 1 cm, generally located in the head of the pancreas. The small size of PanINs renders illusive their detection by MRI (Magnetic Resonance Imagery) or CT (Computed Tomography). Observation of these lesions is usually fortuitous and concomitant to tumor resection. PanINs are classified into 3 grades: PanIN-1 (which are subdivided into PanIN-1A and PanIN-1B), PanIN-2 and PanIN-3/ in situ carcinoma (Fig. 1). This classification depends on the degree of architectural and cytological atypia. Briefly, flat and normal epithelia of secondary duct branches show increasing papillary architecture and finally adopt a severe cellular atypia and frequent mitosis, preceding invasive carcinoma [2]. PanINs are the most frequent preneoplastic lesions of the pancreas and are observed in 82% of pancreas with pancreatic cancer [5]. Low grade PanINs appear to be common lesions found in 16-80% of normal pancreas in the absence of neoplasia, while high grade PanINs are nearly exclusively associated with adenocarcinoma [5-8]. Besides, it is suggested that only 1% of PanINs evolve into PDAC [9]. As a result, it is tempting to consider low grade PanINs as benign lesions in absence of key point mutations. Key genetic events would enhance PanIN-1 to PanIN-2 transition, leading to a “no turning back” development into PDAC. A mouse model expressing the constitutively activated K-RAS G12D proto-oncogene recapitulates PanIN lesions and is the common in vivo model for PanINs study [10].
Fig. (1).
Microscopic view of Pancreatic intraepithelial neoplasia.
2.2. Intraductal Papillary Mucinous Neoplasms (IPMN)
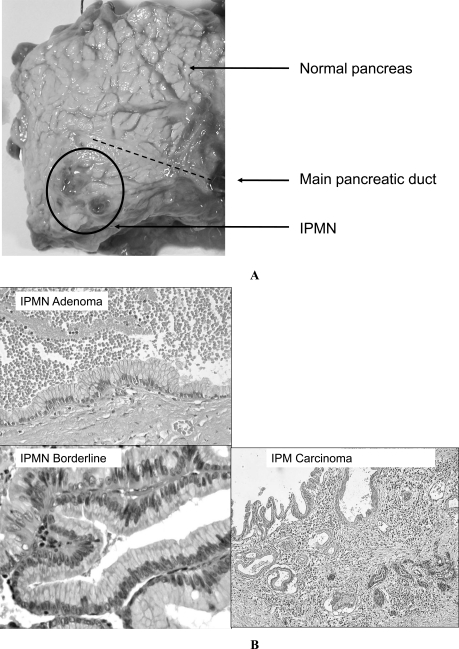
IPMNs are far less common than PanIN lesions and account for 3-5% of pancreatic tumors [11]. IPMNs are divided into adenoma, borderline and intraductal papillary mucinous carcinoma (IPMC) according to the highest degree of dysplasia detected (Fig. 2A and 2B) [12]. They comprise large lesions with long finger-like papillary architecture, larger than 1 cm and delimited by a columnar mucin-secreting epithelium [13, 14]. IPMNs communicate with pancreatic main duct. Invasive carcinoma occurs in 20-50% of IPMNs and are located on the main pancreatic duct or in the secondary branches of the main duct [11]. Additionally, further anatomical criteria depending on the extension of the disease in the organ are taken into consideration. In fact, IPMNs are classified into three groups: main pancreatic duct IPMN (MPD-IPMN), branch duct IPMN (BD-IPMN), and mixed IPMN [15]. Current knowledge on IPMN indicates that approximately 70% of MPD-IPMNs may progress into invasive cancer [15]. Resection represents the only curative treatment for IPMNs [15]. The long-term follow-up of resected patients shows 5-year survival rates of 88% for benign and non-invasive lesions and between 36 and 60% for IPMC [15, 16].
Fig. (2).
A: Macroscopic view of an Intraductal Papillary Mucinous Neoplasm. B: Microscopic view of an Intraductal Papillary Mucinous Neoplasm.
PanINs and IPMNs lesions exhibit similar histological and molecular features and the distinction between those two types of lesions is based on their size and location. Depending on the lesion origin (main duct or peripheral ductules), PanINs and IPMNs can be considered as similar lesions with different clinical outcome. Moreover, a mouse model develops PanIN lesions in addition to cystic lesions of the pancreas resembling human IPMNs following TGF-α overexpression in mutant K-RAS(G12D) transgenic mice [17]. This observation is in favor of a common origin of both types of lesions.
2.3. Mucinous Cystic Neoplasms (MCN)
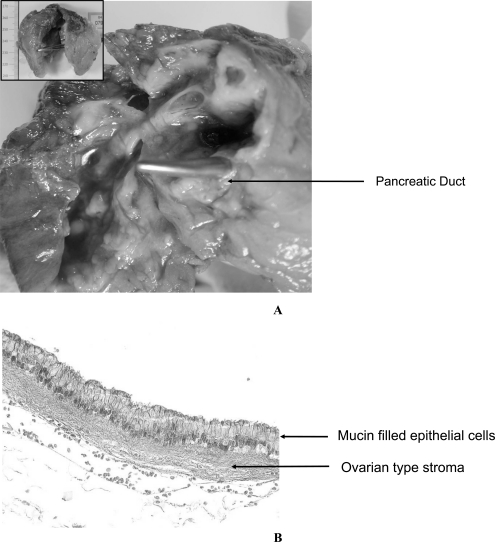
MCNs are rare and large (1 to 3 cm) mucin secreting neoplasms [13]. This type of lesions is lined by a columnar, mucin-filled epithelium and shows a characteristic ovarian-type stroma beneath the epithelial layer, separating the lesion from the pancreatic main duct (Fig. 3A and 3B). MCNs present a higher incidence in women as compared to men (ratio 20 to 1) for unknown reasons. MCNs are classified according to their degree of epithelial dysplasia: Mucinous cystadenoma, borderline mucinous cystic neoplasms and mucinous cystic neoplasms with in situ carcinoma [18]. Because of their size and their location, they are often associated with non-specific symptoms (abdominal discomfort and abdominal mass due to compression of pancreatic duct). About 20% of MCNs are associated with PDAC [11]. All MCNs may progress to mucinous cystadenocarcinoma, which has a very low resectability and a very poor prognosis [35, 36, 15]. The 5 year-survival rate for patient with a surgically resected MCNs in the absence of PDAC is close to 100%; such survival rate falls to 60% when an adjacent neoplasia is diagnosed [19]. Izeradjene and co-workers demonstrated that mice expressing activated K-RAS(G12D) and deleted for Smad4/Dpc4 develop pancreatic cystic neoplasms in the body and the tail of the pancreas in addition to low-grade PanINs [20]. According to the size of these lesions, IPMNs and MCNs can be detected using CT and MRI and present a better prognostic as compared to microscopic lesions PanIN.
Fig. (3).
A: Macroscopic view of Mucinous Cystic Neoplasm. B: Microscopic view of Mucinous Cystic Neoplasm.
3. CELLULAR ORIGIN OF PRENEOPLASTIC LESIONS
All three known precursor lesions bear ductal epithelial cells characteristics, but the precise cellular origin of these lesions is still highly debated.
In mice, induction of short term pancreatic injuries using caerulein leads to transient acinar cells dedifferentiation into mesenchymal cells. These proliferating cells differentiate into acinar cells to partially repopulate the wounded organ [21]. In mice carrying activating K-RAS mutation, this process seems altered and mesenchymal cells differentiation is impaired, leading to acinar-to-ductal metaplasia, and then to PDAC precursor lesions such as PanINs [21]. The contribution of immature pancreatic precursors to PDAC progression raises the possibility of the presence of pancreatic cancer stem cells (CSC). Classically, stem cells constitute a rare cellular population characterized by its self-renewal, specific cell surface markers, and cellular efflux through multidrug resistance channels. Furthermore, these cells give rise to an heterogeneous lineage [22-25]. Cancer stem cells may derive from this particular population, to acquire a slightly transformed phenotype, known as “minimal deviation” [23].
CSCs have been identified in breast cancer, myeloid leukemia, colon and prostate cancer [26]. The identification of such CSCs in the pancreas only relies on the presence of cell surface markers and the up-regulation of Sonic Hedgehog pathway (SHH), participating in self renewal mechanisms [27]. Zhou and co-workers reported dye efflux capacities of PANC-1 cells side population in vitro but not in vivo [22, 28]. If the capacity of efflux, the location and the rarity of these cells within the tumor would contribute to explain their resistance against conventional therapies, this evidence only suggests the “stem” nature of a subpopulation of pancreatic cancer cells and further studies will be required to conclude on their status. Furthermore, the existence of pancreatic cancer stem cells implies the existence of adult pancreatic stem cells to deviate from. To date, existence of such cell type is still a matter of debate [21, 29]. On the other hand, one could hypothesize that a so-called stem cell population would rather arise from a subpopulation of de-differentiated acinar cell, unable to fully differentiate into epithelial ductal cells. Additionally, adult stem cell performing minimal deviation and leading irremediably to PDAC is contradictory because of the frequent observation of low grade PanINs in normal pancreas. “Cancer stem cell” would then originate from dedifferentiation failure during pancreas regeneration. The status of mesenchymal cell would allow them to migrate, colonize new niche and then, enter dormancy. Consequently, residual undifferentiated pancreatic cell in resection margin would explain frequent relapse of patients after surgery, together with distant, microscopic metastasis.
4. DNA ALTERATIONS IN PRENEOPLASTIC LESIONS
Classically, sequential evolution from precursor lesion to PDAC involves the accumulation of diverse molecular changes. Such alterations provide precious evidence for understanding PDAC development, and devising new detection and diagnostic tools. However, little is known about the kinetic of appearance of these molecular events. To date improvement in high throughput sequencing technologies provides whole genomic methylation patterns and single point mutation analysis. Moreover, recent design of mouse models recapitulating preneoplastic lesions of the pancreas allowed great advances in our understanding of PDAC related molecular alterations.
4.1. Genetic Alterations
4.1.1. Large Chromosomal Alterations in PDAC
Genetic alterations in pancreatic cancer have been described for decades. In the late 80’s, Korc and coworkers reported that Epidermal Growth Factor Receptor (EGFR) altered expression is linked with qualitative and quantitative chromosomal defects such as reciprocal translocation and chromosomal deletion of chromosome 7 [30]. EGFR belongs to the cell surface receptor family with tyrosine kinase activity. EGFR activation by its ligand EGF leads to signal transduction through three main pathways: Mitogen-activated protein (MAP) kinase cascade, phosphotidylinositol-3 kinase (PI3K) pathway and the signal transducer and activator of transcription (STAT) pathway, promoting cell division and survival [31]. In PDAC as in other cancers, EGFR is frequently subjected to gene amplification (65% of cases) [32-34]. Intragenic mutations are far less common (3.6% of PDAC) and lead to EGFR over-activation [35]. Overexpression of EGFR is an early event in pancreatic carcinogenesis and occurs from PanIN-1 stage to invasive PDAC [36]. Besides, 61.2% of malignant MCNs exhibit increased expression of EGFR, whereas EGFR is not detected in any of benign MCN [37].
Recent studies based on high throughput sequencing approaches described numerous original regions prone to frequent large genetic alterations during carcinogenesis. One hundred and forty four minimal regions identified in 119 independent loci are subjected to such changes and play a potential role in tumor progression, with loci encoding for p16INK4A, TP53, MYC, K-RAS2, and AKT2 previously described as duplicated or deleted in PDAC [38]. Using Representational Oligonucleotide Microarray (ROMA), Lucito et al. identified 31 amplifications and 25 deletions involving more than 500 genes in familial pancreatic cancer [39]. Campbell et al. recently analyzed patterns of genomic instability in PDAC and pancreatic metastasis [40]. Resulting evidence indicates that PDAC bears specific pattern of genomic alterations (deletion, fold-back inversion, tandem duplication), as compared to other type of cancer. Moreover, these alterations are sequentially detectable from primary tumor to metastasic disease. However, the status of such alterations in preneoplastic lesions is unknown. Additionally to tumor suppressor genes, genetic instability occurs in specific regions encoding microRNAs [41]. MicroRNAs (miRNA) are small non coding RNAs (20-22nt) involved in the negative regulation of mRNAs translation [42]. miRNAs may regulate about 60% of cellular processes and their deregulation is a critical event in carcinogenesis. Alteration in miRNAs expression is a hallmark of cancer and leads to oncogene loss of repression or direct tumor suppressor gene downregulation [42]. Such deregulation leads to a wide range of consequences, influencing tumor progression including cell cycle regulation, epithelial to mesenchymal transition, cellular migration and angiogenesis. Over 130 microRNAs are documented as deregulated in PDAC and pancreatic cancer cell lines but few data exist on the precise mechanisms of such alteration [43, 44]. Similarly to tumor suppressor genes, miRNAs expression alteration can occur anywhere from early to late precursor lesions during PDAC progression. In fact 52.5% of miR genes are in cancer-associated genomic regions or in fragile sites, subjected to loss of heterozygosity, minimal regions of amplification, or common breakpoint regions [45].
4.1.2. Single Point Mutation
Single point mutations correspond to single base substitution in DNA sequence. Such replacement can have various outcomes depending on the genomic region targeted and the new codon generated by this modification. Many tumor suppressor genes and proto-oncogenes are associated with activating or inhibiting mutation in PDAC. If many of these mutations are known for decades, the dynamic of their appearence in preneoplastic lesions of the pancreas have long remained unclear. Here, we present a non-exhaustive list of genes for which single point mutation have been identified in preneoplastic grades. The K-RAS gene (Kirsten-RAS) encodes a RAS homolog that belongs to the cytosolic GTPase family. A single amino acid substitution from G to D at codon 12 is responsible for a constitutive activation observed in up to 95% (60-95%) of PDAC and therefore represents the most common mutation for this cancer [46]. Oncogenic K-RAS activates the MAP Kinase, and/or the PI3K pathways, leading to increased mitogenic activity. K-RAS mutation is an early event during pancreatic carcinogenesis and occurs in 35% of PanIN-1 lesions and reaches 75% in PanIN-3 [6, 46]. The low frequency of K-RAS mutation in PanIN-1 suggests that K-RAS mutation facilitates the progression from low-grade to high-grade PanINs but is not a sine qua non condition to PanIN-1 initiation. On the other hand, K-RAS G12D mutation specifically expressed in the whole mice pancreas at the embryonic stage is sufficient to develop PanIN lesions [10, 47]. K-RAS mutation is observed in a third of IPMN adenomas and its mutation frequency increases with lesion grade (50 % of mutated K-RAS in IPMN borderline and IPMC with invasion) [12, 48]. Similarly, MCNs harbor K-RAS mutation in 20% of benign MCNs, 33% of borderline lesions, and 89% of malignant MCNs [49]. B-RAF is a member of the RAF family, belonging to the MAP Kinase pathway. B-RAF mutation is observed in 7%-15% of PDAC [12, 50]. Activating intragenic mutation of B-RAF leads to activation of RAF effector MEK. Previous reports proposed that K-RAS and BRAF mutation are exclusive in PDAC [2, 51] and that oncogenic B-RAF would represent an alternative for MAP Kinase pathway activation in the absence of K-RAS mutation [50]. Ishimura and co-workers reported concomitant K-RAS and BRAF activating mutations in tumor samples [50], with no clinicopathological incidence as compared to K-RAS mutation alone, suggesting that there is no cumulative effect and that the addition of the two mutations is not an advantage for tumor progression. When compared to the very high K-RAS mutation frequency in preneoplastic lesions, mutation of B-Raf only occurs in 2.7% of IPMTs [12]. The study of B-RAF mutation in PanINs and MCNs is still to be done. As mentioned above, Schonleben et al reported that EGFR mutations are very infrequent in IPMNs (no mutation found in 36 IPMT) whereas another group reported EGFR point mutation in 7.5% of Intestine Type IPMNs [48, 52]. To date, no mutational analysis of EGFR concerning PanIN and MCN lesions are available. DKN2A is a tumor suppressor gene which encodes the P16 tumor suppressor protein. P16 indirectly provokes Rb protein phosphorylation and its loss of function leads to entry in cell cycle and acute cell proliferation. P16 is the most frequently inactivated tumor suppressor gene in PDAC and is observed in up to 95% of patients [13]. Interestingly, different mechanisms are responsible of its inactivation including homozygous deletion, intragenic mutation and epigenetic silencing by promoter methylation (40%, 40% and 15% of PDAC respectively) (see paragraph below) [2]. All of these invalidating modifications arise at late stages of pancreatic carcinogenesis, in both PanINs and IPMNs (adenoma 45%, borderline 50%, IPMC 53%) with a greater incidence in PanINs (PanIN-1 30-44%, PanIN-2 80%, PanIN-3 92-100%) [53]. However, these data are based on protein expression studies using immunohistochemistry detection and does not identify the alterations leading to p16 loss of function. Cell line derived from non invasive MCNs harbors p16 mutation at codon 58, indicating that such genetic alteration is an early event in MCNs multistep development [54]. SMAD4/DPC4 (Mothers against decapentaplegic homolog 4/Deleted in Pancreatic Cancer 4) is subjected to frequent deletion or intragenic mutation in PDAC (55%) [55]. SMAD4 is a key actor in TGF beta signaling pathway, and activates transcription of cell cycle inhibitory factors, particularly p21 [56]. Thus, loss of SMAD4 leads to relapse of growth inhibition and uncontrolled proliferation [53]. SMAD4 expression is normal in low grade PanINs (PanIN-1 and -2) and inactivation occurs at late stage in 31% of PanIN-3 [55]. Similarly, SMAD4 expression is normal in adenoma and borderline IPMNs and occurs in 38% of IPMCs [53]. During MCNs development, DPC4 loss of expression is a late event, as the protein is not detectable at invasive stage [57].
Mutation in Tumor Protein 53 (TP53) locus is the most common single point mutation in human cancer. TP53 acts as a transcription factor and binds to DNA to activate transcription of genes regulating DNA repair, cell cycle and apoptosis [56]. Inactivating mutations in TP53 are observed in up to 75% of pancreatic cancers [13]. Nonetheless, mutational inactivation of TP53 is a late event during PanIN multistep progression to PDAC (0% in PanIN-1 to 12% in PanIN-3) and seems to be more frequent in IPMN (30% in IPMN adenoma, 30% in IPMN borderline and in IPMC 50-63%) [2, 13, 53, 56, 58]. Cell line derived from non invasive MCN harbors p53 mutation at codon 132, indicating that such genetic alteration takes place before invasive MCN grade [54]. Whole genome analysis approach represent the next step for the discovery of new actors that have not previously been linked to cancer progression or initiation. The genome-wide study of twelve prospective cohorts of PDAC patients identified SNPs located in three novel genomic regions associated with the risk of PDAC [59]. Two of these regions are coding region while the third locus on chromosome 13q22.1 maps to a large non coding region that requires further characterization.
4.2. Epigenetic Alterations
DNA methylation consists of the addition of a methyl (CH3) residue on cytosine preceding a guanosine, known as CpG dinucleotides (p stands for phosphate). This modification is catalyzed by a DNA methyltransferase family (DNMT1, 3a and 3b). CpG dinucleotides are not equally interspaced in the genome and are frequently grouped in CpG island. Sixty percent of gene in human genome display one or more CpG islands in their promoter and therefore can be potentially inactivated by methylation. Only 5% of these promoters are methylated. They promote transcription of genes involved in development , or subject to X chromosome inactivation and genomic imprinting [60]. Alteration in DNA methylation pattern is a hallmark of cancer and is associated with an overexpression of DNMTs [61, 62]. Interestingly, whole genome analysis indicates global hypomethylation and only local hypermethylation [63, 64]. Global hypomethylation phenomenon despite DNMTs overexpression may involve targeted DNA methylation [65]. However, the molecular mechanisms involved in such regulation remain unclear to date. Frequent over expression of proto-oncogene in cancer may be linked to their genomic sequence hypomethylation. Understanding global methylation patterns has long been limited by technological concerns limiting the number of genes analyzed. Using cell lines and tumor samples, Sato and co-workers analyzed the methylation status of a subset of 18 genes previously identified as over expressed in PDAC as compared to normal pancreas [66]. The authors describe that Mesothelin and Claudin 4 genes are frequently hypomethylated in PDAC (92% and 89%, respectively) and their methylation status correlates with their expression pattern. On the other hand, hypermethylation occurs during cancer, and may lead to gene silencing. Hypermethylation is responsible for the inactivation of numerous tumor suppressor genes and can occur independently or additionally to intragenic mutation [66-68]. As an example, P16 is equally subjected to mutation or hypermethylation, whereas RASSF1A, Cyclin D2, SOCS-1, APC are tumor suppressor genes silenced only following hypermethylation of their promoting sequence in PDAC [69-71]. By comparing the whole methylome of PDAC with healthy tissue using high throughput technologies, Omura et al. determined that MDF1, miR-9-1, ZNF415, CNTNAP2 and EVOLV-4 were the most frequently methylated loci among 88000 probes tested [68]. Interestingly, only miR-9-1 and CNTNAP2 have previously been linked to cancer progression or initiation. In PanINs, eight genes (CDH3, reprimo, CLDN5, LHX1, NPTX2, SARP2, SPARC and ST14) were reported to be differentially methylated between normal epithelia and PanIN lesions [72]. Interestingly the pattern of methylation during the progression of PanINs is variable among genes. This work proves that CpG island hypermethylation is an early event in PDAC initiation, and its prevalence progressively increases during neoplastic progression. Similar results were found in IPMNs concerning another subset of genes including P16, ppENK, P14, P15, P17, APC, hMLH, E-Cadherin and MGMT [73]. Gene methylation status have not been studied yet in MCN. Taken together, DNA aberrant hypermethylation correlates with DNMT1 over expression in PanINs, and strongly suggests that early methylation may induce precursor lesion progression, and is probably not a consequence of deregulated cellular processes [62]. As stated before, miRNAs have been linked to cancer initiation or progression but miR-148a, 107, 34a and Mir 9-3 are the only miRNA genes hypermethylated in cancer [41, 74-76]. Few miRNAs are described as early actors of pancreatic carcinogenesis. MiR-148a is described to be down regulated in numerous cancers (colon, melanoma, lung cancer) [41]. Our recent works demonstrated mir-148a down regulation by DNA hypermethylation in PanIN-1 and -2 [74]. To date, no data are available concerning miR-148a expression in IPMNs or MCNs. On the other hand, miR-200 and miR-34a alteration of expression are late events in PDAC. Indeed, miR-200 overexpression takes place in invasive PDAC consequently to its genomic sequence hypomethylation and indirectly suppresses E-cadherin expression and contributes to epithelial to mesenchymal transition [77]. Inversely, miR-34a is down-regulated by hypermethylation of its DNA coding sequence and its re-expression induces senescence and cell cycle arrest [76, 78]. Consistent work has been done on molecular characteristics of invasive cancer. Further efforts will be needed to improve our understanding of molecular alterations in preneoplactic lesion during their multistep evolution. In fact, most of the studies are performed on PanIN lesions, due to their higher frequency as compared to IPMNs and MCNs. Further work is needed to better understand IPMN and MCN carcinogenesis. As described above, alteration in miRNAs expression caused by aberrant DNA methylation is an early event in PDAC carcinogenesis, but the consequences of such modification is not clearly defined. Because miRNAs are fine regulator of multiple cellular pathways and because their alteration occurs very early in the oncogenic cascade, it is tempting to speculate that miRNAs may induce carcinogenesis. Transgenic mouse models for miR-17−92, miR-21, miR-29 or miR-155 miRNAs were recently published [79-82]. In these models, expression of miR specifically targeted to the hematopoietic compartment induced lymphopathies. To date, no PDAC transgenic mouse model with impaired miRNAs expression is available.
5. GENERAL INTEREST OF GENETIC MUTATION AND METHYLATED GENE AS BIOMARKERS FOR PDAC
For routine clinical use, molecular markers must be highly sensitive, highly specific and easy to access. To date, current molecular markers mainly rely on variations of proteins/mRNAs expression or histological features. However, the use of such markers presents several issues: i: proteins and mRNAs are not stable and are subject to extensive degradation following collection and ii: histological and protein analysis require consequent amount of tissue. PDAC grading is currently based on CA 19-9, mucin expression or histological study from pancreatic fluids or biopsy. Determination of new molecular markers to improve early diagnosis of this malignancy is impeded by the low frequency of PDAC (4% of new cases of cancer) and its late diagnosis [1]. Genetic and epigenetic alterations both represent an important source of biomarkers for PDAC early detection. In comparison to standard molecular markers, DNA mutation and DNA methylation patterns confer the advantage to be stable and quantified from very low amounts of material from diverse biological samples (biopsies, urines, stool, blood…).
5.1. Diagnostic Marker
Recent studies considered K-RAS mutation analysis in Fine Needle Aspiration (FNA) for PDAC diagnosis, and achieved respectable sensitivity and specificity [83-85]. Our work indicates that miR-148a genomic sequence hypermethylation also discriminate PDAC from chronic pancreatitis [74]. Moreover, 289 methylation patterns in 2016 genes have been demonstrated to successfully discriminate healthy tissue from PDAC [67]. However, this pattern is not specific for PDAC among other types of cancer and requires biopsy so is not suitable for large scale screening of PDAC. Finally, recent works proved that measuring methylation patterns specific for PDAC in blood samples is possible with a sensitivity of 81.7%, and can be a first step towards a sensitive, specific and non-invasive diagnosis tool [86].
5.1.1. Predictive Marker
Another promising application of genetic mutation and methylated gene derived biomarkers is to predict patients resistance to therapy. According to Tan and co-workers, methylation status of GSTM1 and ONECUT2_E96_F is predictive of tumor chemosensitivity [67]. Determination by high throughput technologies of global methylation patterns in patients diagnosed for PDAC and correlation with response to gemcitabine treatment after follow up could identify predictive and surrogate markers for chemotherapeutic response [67].
6. Summary and future directions
Despite recent progress in understanding the multistep progression from precursor lesion to invasive cancer and metastasis, PDAC is still detected too late for surgical treatment. As a consequence, early diagnosis of this disease is urgently needed. Detecting alterations in DNA methylation patterns is one most promising avenue for biomarker discovery for PDAC. It is our belief that the future use of unbiased high throughput sequencing will allow the characterization of whole methylation patterns to identify new biomarkers helpful for PDAC diagnosis and prognosis.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun M J. Cancer statistics, 2009. CA Cancer J. Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Koorstra J B, Hustinx S R, Offerhaus G J, Maitra A. Pancreatic carcinogenesis. Pancreatology. 2008;8:110–125. doi: 10.1159/000123838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghaneh P, Costello E, Neoptolemos J P. Biology and management of pancreatic cancer. Gut. 2007;56:1134–1152. doi: 10.1136/gut.2006.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brat D J, Lillemoe K D, Yeo C J, Warfield P B, Hruban R H. Progression of pancreatic intraductal neoplasias to infiltrating adenocarcinoma of the pancreas. Am. J. Surg. Pathol. 1998;22:163–169. doi: 10.1097/00000478-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Hruban R H, Maitra A, Goggins M. Update on pancreatic intraepithelial neoplasia. Int. J. Clin. Exp. Pathol. 2008;1:306–316. [PMC free article] [PubMed] [Google Scholar]
- 6.Sipos B, Frank S, Gress T, Hahn S, Kloppel G. Pancreatic intraepithelial neoplasia revisited and updated. Pancreatology. 2009;9:45–54. doi: 10.1159/000178874. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi K, Yokohata K, Noshiro H, Chijiiwa K, Tanaka M. Mucinous cystic neoplasm of the pancreas or intraductal papillary-mucinous tumour of the pancreas. Eur. J. Surg. 2000;166:141–148. doi: 10.1080/110241500750009492. [DOI] [PubMed] [Google Scholar]
- 8.Andea A, Sarkar F, Adsay V N. Clinicopathological correlates of pancreatic intraepithelial neoplasia: a comparative analysis of 82 cases with and 152 cases without pancreatic ductal adenocarcinoma. Mod. Pathol. 2003;16:996–1006. doi: 10.1097/01.MP.0000087422.24733.62. [DOI] [PubMed] [Google Scholar]
- 9.Terhune P G, Phifer D M, Tosteson T D, Longnecker D S. K-ras mutation in focal proliferative lesions of human pancreas. Cancer Epidemiol. Biomarkers Prev. 1998;7:515–521. [PubMed] [Google Scholar]
- 10.Hingorani S R, Petricoin E F, Maitra A, Rajapakse V, King C, Jacobetz M A, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 11.Roggin K K, Chennat J, Oto A, Noffsinger A, Briggs A, Matthews J B. Pancreatic cystic neoplasm. Curr. Probl. Surg. 0000;47:459–510. doi: 10.1067/j.cpsurg.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Schonleben F, Qiu W, Bruckman K C, Ciau N T, Li X, Lauerman M H, Frucht H, Chabot JA, Allendorf JD, Re-motti HE, Su GH. BRAF and KRAS gene mutations in intraductal papillary mucinous neoplasm/carcinoma (IPMN/IPMC) of the pancreas. Cancer Lett. 2007;249:242–248. doi: 10.1016/j.canlet.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonezawa S, Higashi M, Yamada N, Goto M. Precursor lesions of pancreatic cancer. Gut Liver. 2008;2:137–154. doi: 10.5009/gnl.2008.2.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hruban R H, Adsay N V. Molecular classification of neoplasms of the pancreas. Hum. Pathol. 2009;40:612–623. doi: 10.1016/j.humpath.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Chari S, Adsay V, Fernandez-del Castillo C, Falconi M, Shimizu M, Yamaguchi K, Yamao K, Matsuno S International Association of Pancreatology. International consensus guidelines for management of intraductal papillary mucinous neoplasms and mucinous cystic neoplasms of the pancreas. Pancreatology. 2006;6:17–32. doi: 10.1159/000090023. [DOI] [PubMed] [Google Scholar]
- 16.Maire F, Hammel P, Terris B, Paye F, Scoazec J Y, Cellier C, Barthet M, O'Toole D, Rufat P, Partensky C, Cuillerier E, Lévy P, Belghiti J, Ruszniewski P. Prognosis of malignant intraductal papillary mucinous tumours of the pancreas after surgical resection. Comparison with pancreatic ductal adenocarcinoma. Gut. 2002;51:717–722. doi: 10.1136/gut.51.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siveke J T, Einwachter H, Sipos B, Lubeseder-Martellato C, Kloppel G, Schmid R M. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer Cell. 2007;12:266–279. doi: 10.1016/j.ccr.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Wilentz R E, Albores-Saavedra J, Hruban R H. Mucinous cystic neoplasms of the pancreas. Semin. Diagn. Pathol. 2000;17:31–42. [PubMed] [Google Scholar]
- 19.Hruban R H, Maitra A, Kern S E, Goggins M. Precursors to pancreatic cancer. Gastroenterol. Clin. North Am. 2007;36:831–849. doi: 10.1016/j.gtc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, Deng CX, Hruban RH, Adsay NV, Tuveson DA, Hingorani SR. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–243. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Jensen J N, Cameron E, Garay M V, Starkey T W, Gianani R, Jensen J. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Sergeant G, Vankelecom H, Gremeaux L, Topal B. Role of cancer stem cells in pancreatic ductal adenocarcinoma. Nat. Rev. Clin. Oncol. 2009;6:580–586. doi: 10.1038/nrclinonc.2009.127. [DOI] [PubMed] [Google Scholar]
- 23.Bhagwandin V J, Shay J W. Pancreatic cancer stem cells: fact or fiction? Biochim. Biophys. Acta. 2009;1792:248–259. doi: 10.1016/j.bbadis.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C J, Dosch J, Simeone D M. Pancreatic cancer stem cells. J. Clin. Oncol. 2008;26:2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 25.Hermann P C, Mueller M T, Heeschen C. Pancreatic cancer stem cells--insights and perspectives. Expert Opin. Biol. Ther. 2009;9:1271–1278. doi: 10.1517/14712590903246362. [DOI] [PubMed] [Google Scholar]
- 26.Simeone D M. Pancreatic cancer stem cells: implications for the treatment of pancreatic cancer. Clin. Cancer Res. 2008;14:5646–5648. doi: 10.1158/1078-0432.CCR-08-0584. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J, Wang CY, Liu T, Wu B, Zhou F, Xiong JX, Wu HS, Tao J, Zhao G, Yang M, Gou SM. Persistence of side population cells with high drug efflux capacity in pancreatic cancer. World J. Gastroenterol. 2008;14:925–930. doi: 10.3748/wjg.14.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iovanna J L, Lechene de la Porte P, Dagorn J C. Expression of genes associated with dedifferentiation and cell proliferation during pancreatic regeneration following acute pancreatitis. Pancreas. 1992;7:712–718. doi: 10.1097/00006676-199211000-00013. [DOI] [PubMed] [Google Scholar]
- 30.Korc M, Meltzer P, Trent J. Enhanced expression of epidermal growth factor receptor correlates with alterations of chromosome 7 in human pancreatic cancer. Proc. Natl. Acad. Sci. USA. 1986;83:5141–5144. doi: 10.1073/pnas.83.14.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papageorgio C, Perry M C. Epidermal growth factor receptor-targeted therapy for pancreatic cancer. Cancer Invest. 2007;25:647–657. doi: 10.1080/07357900701522653. [DOI] [PubMed] [Google Scholar]
- 32.Dancer J, Takei H, Ro J Y, Lowery-Nordberg M. Coexpression of EGFR and HER-2 in pancreatic ductal adenocarcinoma: a comparative study using immunohistochemistry correlated with gene amplification by fluorescencent in situ hybridization. Oncol. Rep. 2007;18:151–155. [PubMed] [Google Scholar]
- 33.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gal-linger S, Au HJ, Murawa P, Walde D, Wolff RA, Campos D, Lim R, Ding K, Clark G, Voskoglou-Nomikos T, Ptasynski M, Parulekar W National Cancer Institute of Canada Clinical Trials Group. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 34.Xiong HQ, Rosenberg A, LoBuglio A, Schmidt W, Wolff RA, Deutsch J, Needle M, Abbruzzese JL. Cetuximab, a monoclonal antibody targeting the epidermal growth factor receptor, in combination with gemcitabine for advanced pancreatic cancer: a multicenter phase II Trial. J. Clin. Oncol. 2004;22:2610–2616. doi: 10.1200/JCO.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 35.Kwak EL, Jankowski J, Thayer SP, Lauwers GY, Brannigan BW, Harris PL, Okimoto RA, Haserlat SM, Driscoll DR, Ferry D, Muir B, Settleman J, Fuchs CS, Kulke MH, Ryan DP, Clark JW, Sgroi DC, Haber DA, Bell DW. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin. Cancer Res. 2006;12:4283–4287. doi: 10.1158/1078-0432.CCR-06-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozaki N, Ohmuraya M, Hirota M, Ida S, Wang J, Takamori H, Higashiyama S, Baba H, Yamamura K. Serine protease inhibitor Kazal type 1 promotes proliferation of pancreatic cancer cells through the epidermal growth factor receptor. Mol. Cancer Res. 2009;7:1572–1581. doi: 10.1158/1541-7786.MCR-08-0567. [DOI] [PubMed] [Google Scholar]
- 37.Kirby R E, Lewandrowski K B, Southern J F, Compton C C, Warshaw A L. Relation of epidermal growth factor receptor and estrogen receptor-independent pS2 protein to the malignant transformation of mucinous cystic neoplasms of the pancreas. Arch. Surg. 1995;130:69–72. doi: 10.1001/archsurg.1995.01430010071014. [DOI] [PubMed] [Google Scholar]
- 38.Aguirre A J, Brennan C, Bailey G, Sinha R, Feng B, Leo C, Zhang Y, Zhang J, Gans JD, Bardeesy N, Cauwels C, Cordon-Cardo C, Redston MS, DePinho RA, Chin L. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc. Natl. Acad. Sci. USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucito R, Suresh S, Walter K, Pandey A, Lakshmi B, Kras-nitz A, Sebat J, Wigler M, Klein AP, Brune K, Palmisano E, Maitra A, Goggins M, Hruban RH. Copy-number variants in patients with a strong family history of pancreatic cancer. Cancer Biol. Ther. 2007;6:1592–1599. doi: 10.4161/cbt.6.10.4725. [DOI] [PubMed] [Google Scholar]
- 40.Campbell P J, Yachida S, Mudie L J, Stephens P J, Pleas-ance E D, Stebbings L A, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lujambio A, Calin G A, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartel D P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 43.Rachagani S, Kumar S, Batra S K. MicroRNA in pancreatic cancer: pathological, diagnostic and therapeutic implications. Cancer Lett. 2010;292:8–16. doi: 10.1016/j.canlet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Li M, Wang H, Fisher W E, Lin P H, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J. Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calin G A, Sevignani C, Dumitru C D, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feldmann G, Beaty R, Hruban R H, Maitra A. Molecular genetics of pancreatic intraepithelial neoplasia. J. Hepatobiliary Pancreat. Surg. 2007;14:224–232. doi: 10.1007/s00534-006-1166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habbe N, Shi G, Meguid R A, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, Maitra A. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc. Natl. Acad. Sci. USA. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schonleben F, Allendorf J D, Qiu W, Li X, Ho D J, Ciau N T, Fine RL, Chabot JA, Remotti HE, Su GH. Mutational analyses of multiple oncogenic pathways in intraductal papillary mucinous neoplasms of the pancreas. Pancreas. 2008;36:168–172. doi: 10.1097/MPA.0b013e318158a4d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimenez R E, Warshaw A L, Z'Graggen K, Hartwig W, Tay-lor D Z, Compton C C, Fernández-del Castillo C. Sequential accumulation of K-ras mutations and p53 overexpression in the progression of pancreatic mucinous cystic neoplasms to malignancy. Ann. Surg. 1999;230:501–509. doi: 10.1097/00000658-199910000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishimura N, Yamasawa K, Karim Rumi M A, Kadowaki Y, Ishihara S, Amano Y, Nio Y, Higami T, Kinoshita Y. BRAF and K-ras gene mutations in human pancreatic cancers. Cancer Lett. 2003;199:169–173. doi: 10.1016/s0304-3835(03)00384-7. [DOI] [PubMed] [Google Scholar]
- 51.Calhoun E S, Jones J B, Ashfaq R, Adsay V, Baker S J, Valentine V, Hempen PM, Hilgers W, Yeo CJ, Hruban RH, Kern SE. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am. J. Pathol. 2003;163:1255–1260. doi: 10.1016/S0002-9440(10)63485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chadwick B, Willmore-Payne C, Tripp S, Layfield L J, Hirschowitz S, Holden J. Histologic, immunohistochemical, and molecular classification of 52 IPMNs of the pancreas. Appl. Immunohistochem. Mol. Morphol. 2009;17:31–39. doi: 10.1097/PAI.0b013e31817c02c6. [DOI] [PubMed] [Google Scholar]
- 53.Biankin A V, Biankin S A, Kench J G, Morey A L, Lee C S, Head D R, Eckstein RP, Hugh TB, Henshall SM, Sutherland RL. Aberrant p16(INK4A) and DPC4/Smad4 expression in intraductal papillary mucinous tumours of the pancreas is associated with invasive ductal adenocarcinoma. Gut. 2002;50:861–868. doi: 10.1136/gut.50.6.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorio C, Capelli P, Lissandrini D, Moore P S, Balzarini P, Falconi M, Zamboni G, Scarpa A. Mucinous cystic carcinoma of the pancreas: a unique cell line and xenograft model of a preinvasive lesion. Virchows Arch. 2005;446:239–245. doi: 10.1007/s00428-004-1167-1. [DOI] [PubMed] [Google Scholar]
- 55.Iacobuzio-Donahue C A, Klimstra D S, Adsay N V, Wilentz R E, Argani P, Sohn T A, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Dpc-4 protein is expressed in virtually all human intraductal papillary mucinous neoplasms of the pancreas: comparison with conventional ductal adenocarcinomas. Am. J. Pathol. 2000;157:755–761. doi: 10.1016/S0002-9440(10)64589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yachida S, Iacobuzio-Donahue C A. The pathology and genetics of metastatic pancreatic cancer. Arch. Pathol. Lab. Med. 2009;133:413–422. doi: 10.5858/133.3.413. [DOI] [PubMed] [Google Scholar]
- 57.Luttges J, Feyerabend B, Buchelt T, Pacena M, Kloppel G. The mucin profile of noninvasive and invasive mucinous cystic neoplasms of the pancreas. Am. J. Surg. Pathol. 2002;26:466–471. doi: 10.1097/00000478-200204000-00008. [DOI] [PubMed] [Google Scholar]
- 58.Flejou J F, Boulange B, Bernades P, Belghiti J, Henin D. p53 protein expression and DNA ploidy in cystic tumors of the pancreas. Pancreas. 1996;13:247–252. doi: 10.1097/00006676-199610000-00006. [DOI] [PubMed] [Google Scholar]
- 59.Petersen G M, Amundadottir L, Fuchs C S, Kraft P, Stolzenberg-Solomon R Z, Jacobs K B, Arslan AA, Bueno-de-Mesquita HB, Gallinger S, Gross M, Helzlsouer K, Holly EA, Jacobs EJ, Klein AP, LaCroix A, Li D, Mandelson MT, Olson SH, Risch HA, Zheng W, Albanes D, Bamlet WR, Berg CD, Boutron-Ruault MC, Buring JE, Bracci PM, Canzian F, Clipp S, Cotterchio M, de Andrade M, Duell EJ, Gaziano JM, Giovannucci EL, Goggins M, Hallmans G, Hankinson SE, Hassan M, Howard B, Hunter DJ, Hutchinson A, Jenab M, Kaaks R, Kooperberg C, Krogh V, Kurtz RC, Lynch SM, McWilliams RR, Mendelsohn JB, Michaud DS, Parikh H, Patel AV, Peeters PH, Rajkovic A, Riboli E, Rodriguez L, Seminara D, Shu XO, Thomas G, Tjønneland A, Tobias GS, Trichopoulos D, Van Den Eeden SK, Virtamo J, Wactawski-Wende J, Wang Z, Wolpin BM, Yu H, Yu K, Zeleniuch-Jacquotte A, Fraumeni JF, Hoover RN, Jr, Hartge P, Chanock SJ. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber M. [Profiles of DNA methylation in normal and cancer cells] Med Sci (Paris) 2008;24:731–734. doi: 10.1051/medsci/20082489731. [DOI] [PubMed] [Google Scholar]
- 61.Robertson K D, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales F A, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng D F, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kosuge T, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96:403–408. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueki T, Toyota M, Sohn T, Yeo C J, Issa J P, Hruban R H, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 64.Sato N, Maitra A, Fukushima N, van Heek N T, Matsubayashi H, Iacobuzio-Donahue C A, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 65.Hervouet E, Vallette F M, Cartron P F. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–499. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- 66.Ueki T, Toyota M, Skinner H, Walter K M, Yeo C J, Issa J P, Hruban RH, Goggins M. Identification and characterization of differentially methylated CpG islands in pancreatic carcinoma. Cancer Res. 2001;61:8540–8546. [PubMed] [Google Scholar]
- 67.Tan A C, Jimeno A, Lin S H, Wheelhouse J, Chan F, Solo-mon A, Rajeshkumar NV, Rubio-Viqueira B, Hidalgo M. Characterizing DNA methylation patterns in pancreatic cancer genome. Mol. Oncol. 2009;3:425–438. doi: 10.1016/j.molonc.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Omura N, Li C P, Li A, Hong S M, Walter K, Jimeno A, Hidalgo M, Goggins M. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol. Ther. 2008;7:1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klump B, Hsieh C J, Nehls O, Dette S, Holzmann K, Kiesslich R, Jung M, Sinn U, Otner M, Porschen R, Gregor M. Methylation status of p14ARF and p16INK4a as detected in pancreatic secretions. Br. J. Cancer. 2003;88:217–222. doi: 10.1038/sj.bjc.6600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, Boehm BO, Pfeifer GP, Hoang-Vu C. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene. 2003;22:3806–3812. doi: 10.1038/sj.onc.1206582. [DOI] [PubMed] [Google Scholar]
- 71.Kuroki T, Tajima Y, Kanematsu T. Role of hypermethylation on carcinogenesis in the pancreas. Surg. Today. 2004;34:981–986. doi: 10.1007/s00595-004-2858-6. [DOI] [PubMed] [Google Scholar]
- 72.Sato N, Fukushima N, Hruban R H, Goggins M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod. Pathol. 2008;21:238–244. doi: 10.1038/modpathol.3800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato N, Goggins M. Epigenetic alterations in intraductal papillary mucinous neoplasms of the pancreas. J. Hepatobiliary Pancreat. Surg. 2006;13:280–285. doi: 10.1007/s00534-005-1056-2. [DOI] [PubMed] [Google Scholar]
- 74.Hanoun N, Delpu Y, Suriawinata A A, Bournet B, Bureau C, Selves J, Tsongalis GJ, Dufresne M, Buscail L, Cordelier P, Torrisani J. The silencing of MicroRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin. Chem. 2010;56:1107–1118. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 75.Lee K H, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 77.Li A, Omura N, Hong S M, Vincent A, Walter K, Griffith M, Borges M, Goggins M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70:5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, Xu L. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mu P, Han Y C, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao C, Srinivasan L, Calado D P, Patterson H C, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphopro-liferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Medina P P, Nolde M, Slack F J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 82.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl. Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bournet B, Souque A, Senesse P, Assenat E, Barthet M, Lesavre N, Aubert A, O'Toole D, Hammel P, Levy P, Ruszniewski P, Bouisson M, Escourrou J, Cordelier P, Buscail L. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with KRAS mutation assay to distinguish pancreatic cancer from pseudotumoral chronic pancreatitis. Endoscopy. 2009;41:552–557. doi: 10.1055/s-0029-1214717. [DOI] [PubMed] [Google Scholar]
- 84.Hosoda W, Takagi T, Mizuno N, Shimizu Y, Sano T, Yamao K, Yatabe Y. Diagnostic approach to pancreatic tumors with the specimens of endoscopic ultrasound-guided fine needle aspiration. Pathol. Int. 2010;60:358–364. doi: 10.1111/j.1440-1827.2010.02527.x. [DOI] [PubMed] [Google Scholar]
- 85.Jang J Y, Park Y C, Song Y S, Lee S E, Hwang D W, Lim C S, Lee HE, Kim WH, Kim SW. Increased K-ras mutation and expression of S100A4 and MUC2 protein in the malignant intraductal papillary mucinous tumor of the pancreas. J. Hepatobiliary Pancreat. Surg. 2009;16:668–674. doi: 10.1007/s00534-009-0105-7. [DOI] [PubMed] [Google Scholar]
- 86.Liggett T, Melnikov A, Yi Q L, Replogle C, Brand R, Kaul K, Talamonti M, Abrams RA, Levenson V. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer. 2010;116:1674–1680. doi: 10.1002/cncr.24893. [DOI] [PubMed] [Google Scholar]