Abstract
Metabolomics can map the large metabolic diversity in species, organs, or cell types. In addition to gains in enzyme specificity, many enzymes have retained substrate and reaction promiscuity. Enzyme promiscuity and the large number of enzymes with unknown enzyme function may explain the presence of a plethora of unidentified compounds in metabolomic studies. Cataloguing the identity and differential abundance of all detectable metabolites in metabolomic repositories may detail which compounds and pathways contribute to vital biological functions. The current status in metabolic databases is reviewed concomitant with tools to map and visualize the metabolome.
Keywords: Drug Metabolism, Enzyme Mutation, Evolution, Mass Spectrometry (MS), Metabolomics, Databases, Pathway Reconstruction
Introduction
Biological databases are indispensable for comparing genomes, proteins, and biological regulation. GenBankTM, Protein Data Bank (PDB), and Gene Expression Omnibus (GEO) are prime examples of how collecting biological information in a coherent manner enables novel insights into evolution and to derive testable hypotheses for gene function, yet biochemical databases on substrate-product relationships and organism-specific metabolic networks have lagged behind. Much of this lag is due to the inherent complexity of enzymology. Small changes in protein folding or in mutations in catalytic sites not only may change reaction kinetics but also have large impact on substrate specificity. Enzymes may have much broader substrate specificity than usually considered. Moreover, many enzymes exert reaction promiscuity (1), which is exploited in bioengineering but which also complicates the reconstruction of metabolic networks. Low-abundant enzymatic side reactions have likely not been reported in favor of the dominant and apparently biologically relevant functions and are consequently lacking in biochemical databases, yet such side reactions may become the major enzyme function through evolutionary pressure. Hence, the number of metabolites per species (or per cell type in multicellular organisms) is hard to predict except for the most conserved metabolic pathways.
Accordingly, a surprisingly small fraction of detected metabolites can be readily identified by sensitive screening tools such as HPLC- or GC-coupled MS (Fig. 1). It appears that the metabolome is much larger than anticipated. Phenotypes of species need to be determined by their individual metabolic capacities, defined by the plasticity and flexibility of their metabolic networks. Metabolites can act as intracellular and extracellular signals at very low concentrations and enable communication between organs, as well as serve multiple and vital roles for species in their ecological niches, e.g. for defense or reproductive purposes.
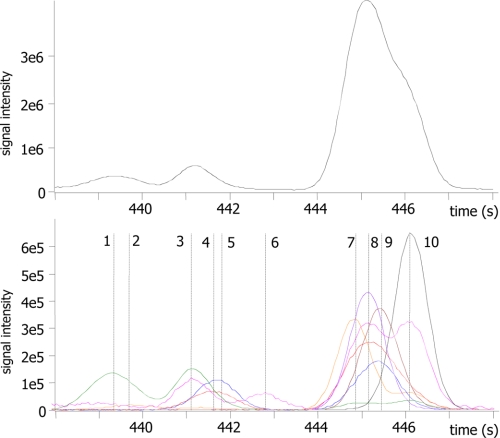
FIGURE 1.
Metabolome diversity observed by MS. Shown are the results from cold injection GC/TOF MS of human ileal effluent (70). Upper panel, total ion chromatogram for a 10-s retention time window out of a 20-min chromatogram. Lower panel, extracted ion chromatogram for the same 10-s retention time window. As each ion trace can be deconvoluted into individual peaks with resolved mass spectra, many co-eluting compounds can be separated and identified. Novel compounds of unknown structure are detected along with known “primary” metabolites. Compound 1, unknown; compound 2, unknown; compound 3, serine; compound 4, unknown; compound 5, benzoic acid; compound 6, unknown; compound 7, glycerol; compound 8, ethanolamine; compound 9, phosphate; compound 10, isoleucine.
Metabolome Diversity Originates from Enzyme Substrate and Reaction Promiscuity
Enzyme evolution has progressed to ever more biochemical specificity. Hence, scientific reports and consequently biochemical databases emphasize specificity over diversity. On the other hand, it is well known that substrate specificity can still be broad (e.g. lipases (2)), and the exact substrate preferences often remain unclear or untested. Enzymes may use a broad range of substrates yet remain high stereospecificity (3). Different ligands may induce large conformational changes in the cytochrome P450 enzymes, leading to different kinetic parameters, e.g. in metabolizing exogenous compounds in humans (4). In fact, many enzymes still lack rigorous functional characterizations and are only broadly classified as “cytochrome P450” or “oxidoreductases.” Such classifications are too vague to deduce enzyme functions in genome-based metabolic reconstructions.
Enzymes may also exert specific and promiscuous compartments in the same catalytic site, as shown for human carboxylesterase 1, which can even bind two different molecules simultaneously (5). Moreover, substrate ambiguity may exert large survival benefits for microorganisms, e.g. for detoxifying a range of different exogenous compounds simultaneously (6).
Besides accepting diverse substrates, enzymes may also catalyze more than one biochemical reaction, called enzyme promiscuity. It has been shown that enzymes may perform novel catalytic reactions elsewhere than in the previously identified catalytic site (7), as has been shown, for example, for phosphotriesterase (8). This phenomenon can explain how infectious microbes may quickly develop resistance against drug therapies but can also be experimentally verified in forced evolution experiments (9). Indeed, even structurally unrelated proteins may perform the same reactions (on a primitive scale) (10), thus explaining metabolic diversity as well as the limited impact of knock-out mutations that is often observed. The ability of recovering and magnifying promiscuous catalytic reactions is even useful for synthesis of new chemicals (11) or for metabolic engineering, which strives to advance metabolic capacities in organisms (12, 13). Enzyme evolution and retention of promiscuity may thus explain the size of the metabolome (Fig. 2).
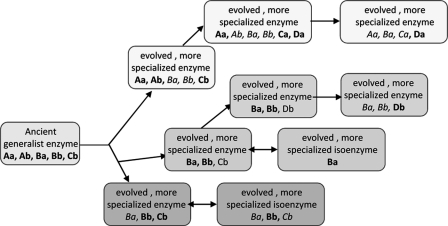
FIGURE 2.
Enzyme evolution toward higher specificity, retaining some substrate and reaction ambiguity. Shown is a schematic diagram of the origin of metabolome diversity between species and within species (adapted from Ref. 1). Given the currently accepted model of enzyme evolution by gene duplication and subsequent specialization, a generalist progenitor enzyme may have performed catalytic reactions (a and b) on substrates (A, B, and C). The phylogenetic tree for this enzyme may have led to isoenzymes that accept only substrate B for reaction a, whereas others retained some level of substrate and reaction ambiguity, leading to higher metabolome diversity (e.g. adding reaction a to substrate C or accepting the novel substrate D for reactions a and b).
Exploring the array of potential substrates in a high-throughput manner and simultaneously testing reaction promiscuity call for unbiased reliable assays such as metabolomic techniques. Metabolomic technology has matured to enrich biochemical databases by hypothesis-driven biochemical research and could also be employed for broad analysis of mutant collections and metabolic diversity of species or to fill gaps in metabolic networks. Current biochemical and metabolomic databases can be largely distinguished by the respective input data in repositories that focus on genes, pathways, and enzymes and libraries that are focused on compound-centric data (Table 1).
TABLE 1.
Overview of selected biochemical and chemical databases for metabolomics
| Pathway- and enzyme-centric databases and tools | Compound-centric databases |
|
|---|---|---|
| Chemical databases | Spectral libraries | |
| >400 MetaCyc databases (18) | PubChem (31) | MassBank (38) |
| KEGG (19) | ChemSpider (61) | PlantMetabolomics (45) |
| BRENDA (24) | ChEBI (33) | BinBase/SetupX (47) |
| Reactome (25) | HMDB (35) | GMD (39) |
| YeastNet (17) | KNApSAcK (36) | Kazusa OMICS |
| SMPDB (65) | CAS (29) | NMRShiftDB (53) |
| MetPA (64) | NIST MS (51) | |
| Ingenuity Systems IPA | METLIN (25) | |
| Ariadne Genomics Pathway Studio | ||
| GeneGo MetaCore | ||
Reconstructing Genomic Information toward Enzymes and Pathways
Genomes of ever more species are being sequenced. Open reading frames are first tentatively annotated and later enriched, curated, and complemented by the research community interested in that species, including highlighting pathway gaps. For this work, a range of tools have been developed, compiled in the BioCyc pathway tool collections (14). Genome-reconstructed metabolic databases are now available for hundreds of species, most of which are automatically annotated. Curation of these databases depends heavily on the input of users from the biochemical community as exemplified for mammalian systems (HumanCyc and MouseCyc) to plants (AraCyc (15), MedicCyc, and RiceCyc) and microbial databases, e.g. Escherichia coli (16) and yeast (17). The umbrella databases MetaCyc and BioCyc (18) today cover more than 1500 organisms and 1100 metabolic pathways. Interestingly, newly sequenced organisms often do not convey many novel predicted enzymes or pathways, pointing to the relatively poor annotation of substrate specificity for non-primary metabolism, e.g. for the P450 enzyme superfamily. By evaluating the number of enzymes, reactions, and chemical compounds deposited in MetaCyc, it becomes apparent that the number of biochemical entities increases more rapidly than the number of pathways, indicating that many of the newly added enzymatic reactions are not yet connected into the broader metabolic network. Here, gap filling by verifying missing links through metabolome analysis might be highly fruitful.
A similar approach for genomic reconstruction has been presented by the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database, which can either be used as a generic pathway tool or be restricted to specific organisms (19). The LIGAND repository within the KEGG Database is one of the best known and most often used reference databases of substrate-product reaction pairs (KEGG RPAIR). Within the past year, the KEGG LIGAND Database has increased from 15,217 metabolites to 16,746 compounds that are assigned to 5317 enzymes and 12,457 reaction pairs. Unfortunately, the LIGAND Database comprised many erroneous structures, often concerning stereochemistry (20). KEGG covers 366 reference pathways that are assigned to 1550 taxonomic species, mostly automatically annotated without extensive curation. Nevertheless, this automatic annotation can be used to constrain KEGG to species-related information (18, 21–23). Of a total of 3575 EC numbers stored in the KEGG Pathway Database, 1269 have more than one reaction annotation. Interestingly, ∼1200 reactions in KEGG do not have EC annotations, and about half of all metabolites deposited in KEGG do not have any reaction annotation. These compounds might be formed by chemical rather than biochemical reactions; however, it seems more likely that there are many metabolites included in KEGG that are not yet linked to enzymes and genes. For example, the plant hormone methyl salicylate is present as a compound in LIGAND (C12305) but is not associated with the enzyme salicylate methyltransferase, which can be retrieved from the better curated AraCyc Database as AtBSMT1 (At3g11480). A number of tools guide KEGG queries, e.g. the KEGG BRITE collection of hierarchical classifications and links to a range of further genomics databases such as the National Center for Biotechnology Information (NCBI), UniProt, Swiss-Prot, and BRENDA, the classic enzyme database that has recently extended its functionality (24).
KEGG, BioCyc, and Reactome (25) are now stored within the umbrella database NCBI BioSystems to link pathway information to the well established Entrez (22). The NCBI BioSystems Database is accessible via web services and Entrez query tools. Similar query functionalities are presented by the BioMart, Pathway Commons, and MetaCyc “advanced search” tools. MetaCyc provides an extensive description of its pathways with literature references; such description is lacking in the KEGG Database. To improve and accelerate the process of curating metabolic pathways, the WikiPathways Database has been developed recently (21). WikiPathways enables the community at large to construct, curate, and submit pathway maps. The maps are provided with descriptions, hyperlinks, and literature. Within the past 2 years, the WikiPathways Database has compiled ∼1300 pathways. These pathway maps can be visualized online (26) or can be downloaded and imported in Cytoscape or PathVisio software for visualization (27). Data sets of metabolome or gene expressions can be visualized on these pathways using an Atlas mapper.
Metabolome Databases
Cataloging the metabolome itself by experimental data and by literature information can complement genomic reconstructions of metabolism. Just like for pathways, information can be generated by information mining, as demonstrated for the Flavonoid Viewer through MediaWiki (28). Metabolome repositories are compound-centric databases that may be enriched by mass spectral libraries or links to pathway databases. They link the chemical identity of metabolites to presence and potentially to concentrations in a species, organ, or cell type. It is of paramount importance that the underlying database information is based on the chemical structure, not on names or “identifiers.” Metabolite names are not unambiguous identifiers, as metabolites may be naturally occurring in different chiral forms, e.g. d- and l-amino acids. Hence, the best annotation for a metabolite is its chemical structure. Encoding the structure in a string of letters in an open access format was standardized by the International Union of Pure and Applied Chemistry (IUPAC) in 2004 by introducing the International Chemical Identifier (InChI) code. This code has been abbreviated as InChI hash key to be readily used in tables or publications.
Linking Compounds to Chemical and Biological Information
A wealth of information is available through published literature, some of which can be accessed through generic chemistry databases. CAS, the Chemical Abstracts Service Database, is a fee-based service that compiles published literature on a compound-by-compound basis (29), comprising >50 million unique chemical substances. CAS does not distinguish biochemical metabolites from man-made small molecules. CAS also restricts batch downloads for chemical structures, compound names, or other metadata to be used for retrieving the complement of all chemicals that had been previously reported for a given species. Compound annotation by CAS numbers may change over time. For example, a variety of CAS numbers can be found for a single structure such as ribosylnicotinamide, which is annotated as 19131-72-7, 20299-13-2, 954368-04-8, and 1341-23-7 (30). CAS entries are also not linked to biochemical pathways databases.
Alternatively, public databases have been constructed. Most importantly, PubChem (31) presents a very versatile, open access database for small molecules. It is maintained at NCBI as part of the Entrez information retrieval system (32). Records for PubChem compound identifiers have increased to >31 million unique structures. All PubChem contents can be freely downloaded in batch mode, including compound properties such as lipophilicity, proton donor number, synonyms, and pharmacology and toxicology information. PubChem compound identifiers are linked to many other biochemical databases, from PDB to KEGG. Unlike CAS, PubChem is not a literature-curated database but depends on depositor information. Hence, PubChem compounds cannot be queried for presence or concentrations in biological species, biofluids, or tissues. A third example of a chemistry-focused database is Chemical Entities of Biological Interest (ChEBI) (33, 34). ChEBI compounds can be queried using chemical ontologies, enabling searches by chemical class information such as “d-aldohexose.” At this point, ChEBI does not store concentration data for species, organs, or cells.
Because of the lack of species-related metabolome information in chemistry resources, researchers have started collecting information from literature. Most prominently, the Human Metabolome Database (HMDB Version 2.5) has been constructed over the past 5 years to detail information for almost 8000 metabolites that are present in human organs or reported in conjunction with human health (35). Related databases that compile information about exogenous human metabolites, e.g. phytochemical components from food or metabolites of xenobiotics, are stored in accessory repositories (DrugBank and FooDB). HMDB stores concentrations for >4000 metabolites for a variety of tissues and body fluids and can thus now serve as a reference repository to compare metabolome data. HMDB can serve as a leading example of how further metabolome literature databases might be compiled.
Mammals have only limited anabolic capacities due to their adaptation to the diversity of micronutrients in their food, e.g. vitamins. Hence, natural products have been listed in the KNApSAcK repository (36). KNApSAcK is a species/metabolite database focused mostly on complex plant metabolites. Its size has doubled from 25,000 metabolites in 2008 to >50,000 entries in 2010. An alternative source, extending from plant metabolites to small molecules from microorganisms and fungi, is the commercial Dictionary of Natural Products (37). This library boasts 200,000 literature-based secondary plant, fungal, and microbial metabolites, including taxonomic reference species for most of the structures. Batch downloads of up to 100 search results are permitted.
Databases Linking Metabolomic Data to Compound Information
Finally, metabolome data repositories exist that are geared toward comprehensive identification of small molecules in biological samples, with the aim to unravel biochemical relationships, gene function annotation, and potentially biological function via statistical comparison of data sets. Several public databases store and disseminate spectral information for metabolites, and some databases exist for which experimental profiles can be downloaded, including raw annotated metabolite tables and raw files.
MassBank denotes a very ambitious project: the collection of a very large range of mass spectra from different instrument platforms and currently 15 collaborating institutions (38). MassBank entries span many metabolite classes. At current, >27,000 spectra have been collected from >13,000 compounds. Almost half of the spectra account for electron ionization mass spectra used in GC/MS analysis. However, it is unclear how many redundant spectra are housed and to what extent spectra are freely available, e.g. to develop new fragmentation algorithms. An alternative accurate mass database is the Kazusa OMICS Database, which resulted from a landmark paper on use of mass spectral data processing for compound identification. A process for metabolite annotations was outlined that was based on high-resolution accurate mass analysis by HPLC/electrospray/Fourier transform ion cyclotron resonance MS (40). Accurate masses for isotope clusters above a signal/noise ratio of 3:1 were averaged, and potential elemental formulas were calculated within 1-ppm mass windows. These formulas were constrained by heuristic rules similar to the those published in “Seven Golden Rules” (41) and compared between positive and negative electrospray ionization. A total of 869 metabolite peaks were detected in tomato, albeit still lacking metabolites that are very difficult to ionize by electrospray, e.g. carotenoids. A thorough comparison with previous work and published tomato metabolites (42, 43) led to the conclusion that at least 494 novel metabolites were detected (40). However, only 3.6% of all peaks were identifiable using authentic standards due to the vast complexity of plant natural products and the limited availability of pure reference chemicals. Annotation plausibility was further categorized using a novel approach (40): first, MS/MS spectra were interrogated for shared ions among metabolites and whether identical mass differences were observed for these compounds. Approximately 37% of the detected metabolites were assigned as “biologically relevant” using this schema. Aglycone backbones were found to be supplemented by additions of caffeic acid, amino groups, hydroxyl groups, hexoses, deoxyhexoses, malonic acid, or coumaric acid. Interestingly, a novel modification was detected as the addition of C3H7NO2S, which points to cysteine addition. Such modification had never been reported before for chalcone and flavonoid aglycones. Analysis of mutant plants led the authors to suggest a novel pathway from α-tomatine to esculoside, exemplifying how metabolomics can generate novel biochemical hypotheses to be tested in follow-up studies (40).
A less elaborate protocol using data from HPLC/quadrupole/TOF MS was used to suggest novel metabolites in seeds of Arabidopsis thaliana (44). Data sets of extracts of mutant and wild-type seeds were manually investigated for ion pairs formed by molecular ions and accompanying in-source fragmentation ions. These parent ion clusters were subsequently further investigated by MS/MS or in-source MS3 mass spectral acquisitions. The information obtained from MS/MS and MS3 spectra was applied as neutral loss substructure constraint into calculations of element formulas. Subsequently, the KNApSAcK (36), CAS, and PubChem (31) databases were queried to enumerate potential candidate structures. Comparative analysis of chalcone synthase and chalcone isomerase mutants eventually assigned novel phenolic choline esters that were suggested to indicate additional branch points in phenylpropanoid metabolic pathways (44).
Complementing HPLC/MS metabolomic investigations are databases for GC/electron ionization MS. BinBase/SetupX is a combined resource for metabolomic GC/TOF MS profiles using TOF MS (46). Data annotations employ the BinBase algorithm (47), which identifies metabolites based on mass spectral and retention index library matching using >2000 spectra of authentic reference compounds (48). BinBase maintains data for >25,000 samples that were acquired from >420 studies of >70 biological species. Each sample is associated with detailed information on taxonomy, genotypes, organs, cells, and experimental treatment data (biotic or abiotic treatments; time courses). A similar resource for freely downloadable GC/electron ionization mass spectra is available through the Golm Metabolome Database (GMD) (49). GMD has been recently updated by added functionalities such as substructure annotation for unidentified metabolites (50), similar to the well known substructure features given in the NIST MS software (51). The library details compounds by referencing to other databases such as KEGG as well as InChI codes but does not comprise taxonomic or biological metadata. GMD has a very useful associated web service that allows automatic programmatic access (Representational State Transfer (REST)).
For metabolome analysis by NMR, two repositories are prominent. First, the Madison Metabolomics Consortium Database (MMCD) reports >700 experimental carbon, proton, and two-dimensional NMR spectra of a diverse set of metabolites (52). The compounds are well annotated using KEGG and PubChem identifiers to enable comparisons with other resources. Raw data are freely downloadable for academic purposes and comprise different types of NMR spectra for each compound such as one-dimensional 1H and 13C files as well as two-dimensional 1H-1H total correlation spectroscopic spectra and 1H-13C heteronuclear single quantum coherence data. An alternative open NMR database, NMRShiftDB (53), is maintained at the European Bioinformatics Institute.
Mapping and Visualization of Metabolomic Data to Biochemical Pathways
Once metabolome data are annotated by hundreds of identified metabolites, one might be tempted to map these compounds according to their location on pathway overview charts. A range of studies and reviews have been published (54–57) to demonstrate how a database can be used to integrate, query, and visualize pathway results (58, 59). A straightforward solution is presented by the KEGG “pathway mapping” tool, which directly displays metabolites on metabolic network graphs. More advanced variants first employ statistics assessments using the open R Project and subsequently use the Bioconductor tool (60). It has been shown that multiple pathway maps can be combined to construct a global pathway overview (19, 62). In addition, the KEGG Atlas mapping (61) and iPath (63) tools map data on global pathway graphs. More often, metabolic profiling studies include multiple class experiments such as wild-type/genotype or drug/non-drug treatments with multiple time points. Publicly available database services such as MetPA (64) and the Small Molecule Pathway Database (SMPDB) (65) can be used to visualize the coverage of submitted metabolites on different pathways. Commercial tools, including GeneGo MetaCore, Ingenuity Systems IPA, and Ariadne Genomics Pathway Studio, are oriented mostly toward microarray gene expression and proteomics data but recently also increased the coverage for small molecules and metabolic pathway analysis by actively analyzing the published literature.
The more metabolite nodes are visualized in a single pathway map, the more difficult it is to see details and obtain information on both larger biochemical modules and reactions. A web-based zoomable Pathway Projector has recently been proposed using the KEGG Atlas and Google Map technologies for magnifying details on global metabolic maps (66). However, such approaches necessarily fail when input metabolites (or enzymes) are not included in KEGG Atlas. Indeed, metabolomic surveys often detect novel metabolites for which no enzymatic reaction has been established. Moreover, biochemical pathway maps detail all intermediary steps, whereas in a biological sample, only some of the metabolites in a pathway might accumulate enough to be detectable. Hence, direct mapping approaches yield sparsely populated pathway charts. More abstract approaches have been used following the well known modular organization of biological systems (67). MapMan leaves out many low-abundant intermediates and summarizes metabolites into predefined sets of biochemical modules, e.g. TCA cycle, carbohydrates, amino acid biosynthesis, and glycolysis (68). These preset boxes are then superimposed with statistical analysis of metabolite expression to display differential regulation and hence enable highlighting the most prominent changes in biochemistry when looking at large data sets. As an alternative, network graphs may be used (69). A reconstruction of metabolic networks has been proposed (70) that employs biochemical and chemical similarity distances to visualize metabolic relationships and differential regulation in the open source Cytoscape tool (Fig. 3) (71). Metabolites are first mapped to presence in the KEGG RPAIR Database to provide a core biochemical structure, to which all identified metabolites are linked via their chemical similarity index. Chemical similarities are calculated via matrices that are obtained by decomposing all compounds into sets of substructures in the PubChem tool. Last, unknown metabolites can be mapped by calculating scores for mass spectral similarities to known compounds. As an example, a published data file on metabolic regulation of the human ileal effluent was downloaded from BinBase/SetupX (72) and used for network construction in Fig. 3.
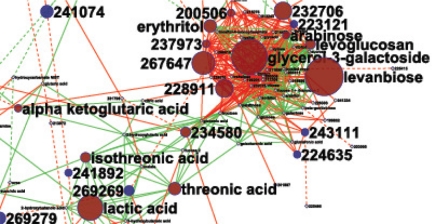
FIGURE 3.
Mapping metabolome regulation on biochemical networks. Shown is the regulation of the human ileal effluent metabolome after versus before ileostomy surgery (70), magnified for the carbohydrate cluster. Identified metabolites are mapped to the biochemical KEGG RPAIR Database and chemical similarity (green edges, dashed if <600 similarity), spanning a network displayed in Cytoscape. Unknown metabolites (BinBase Metabolome Database numbers (48)) are added by mass spectral similarity (red edges, dashed if <600 similarity). Red node metabolites are significantly increased in concentration (p < 0.05), blue nodes mark decreased compounds, and yellow nodes (small print) are not regulated. Node size indicates magnitude of change.
Analysis of the overall topology of biochemical networks can lead to novel insights into metabolic capacities of cells (73). Large-scale metabolic interactions have to be founded on the actual biochemical transformations that are performed. Earlier focus in computational analysis of metabolism had been geared toward mere node/edge topology analysis (74), but seemingly tight connectivities in topology networks are based mostly on hub metabolites like ATP and water and do not convey actual modifications of carbons and functional groups. Stoichiometric analysis of metabolism has been performed with great success for mapping microbial pathways in flux-constraint models (75, 76), and a combination of metabolome analysis of accumulating metabolites with genomic and fluxomic investigations appears to be most promising.
Perspective
This minireview has focused on enzyme pathways and metabolome databases that are deemed critical for more in-depth understanding of metabolic architectures. Quantitative considerations have been left out here but will be critical for using such databases, e.g. as constraints in flux-balance analysis. A range of metabolic pathway databases and public chemistry repositories can be used today as a backbone to understand metabolomic surveys. Lack of data on substrate specificity for large enzyme classes (ligases, P450, and oxidoreductases) explains the difficulty in identifying the plethora of detected signals and their biochemical relationships. Even for the best studied organisms such as yeast and E. coli, we cannot accurately enumerate the size of the minor-abundant compound metabolome, although a consensus exists for the major metabolic network in yeast (17).
Novel tools that suggest enzymatic relationships between novel metabolites may be exploited in the future (77). As of today, metabolomics can cover large parts of genome-reconstructed networks and is highly useful in targeted fluxomic investigations (78). However, novel pathways and novel enzymatic actions are more likely to be discovered in the vast array of species-specific metabolism (79), including lipid transformations.
Footnotes
This is the third article in the Thematic Minireview Series on Computational Systems Biology. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012
REFERENCES
- 1. Khersonsky O., Tawfik D. S. (2010) Annu. Rev. Biochem. 79, 471–505 [DOI] [PubMed] [Google Scholar]
- 2. Todd A. E., Orengo C. A., Thornton J. M. (2001) J. Mol. Biol. 307, 1113–1143 [DOI] [PubMed] [Google Scholar]
- 3. Bornscheuer U. T., Kazlauskas R. J. (2004) Angew. Chem. Int. Ed. Engl. 43, 6032–6040 [DOI] [PubMed] [Google Scholar]
- 4. Ekroos M., Sjögren T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bencharit S., Morton C., Xue Y., Potter P., Redinbo M. (2003) Nat. Struct. Mol. Biol. 10, 349–356 [DOI] [PubMed] [Google Scholar]
- 6. Fong D. H., Berghuis A. M. (2002) EMBO J. 21, 2323–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taglieber A., Höbenreich H., Carballeira J., Mondière R., Reetz M. (2007) Angew. Chem. 119, 8751–8754 [DOI] [PubMed] [Google Scholar]
- 8. Afriat L., Roodveldt C., Manco G., Tawfik D. S. (2006) Biochemistry 45, 13677–13686 [DOI] [PubMed] [Google Scholar]
- 9. Aharoni A., Gaidukov L., Khersonsky O., McQ Gould S., Roodveldt C., Tawfik D. S. (2005) Nat. Genet. 37, 73–76 [DOI] [PubMed] [Google Scholar]
- 10. James L. C., Tawfik D. S. (2001) Protein Sci. 10, 2600–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hult K., Berglund P. (2007) Trends Biotechnol. 25, 231–238 [DOI] [PubMed] [Google Scholar]
- 12. Yoshikuni Y., Ferrin T. E., Keasling J. D. (2006) Nature 440, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 13. Nobeli I., Favia A. D., Thornton J. M. (2009) Nat. Biotechnol. 27, 157–167 [DOI] [PubMed] [Google Scholar]
- 14. Karp P. D., Paley S. M., Krummenacker M., Latendresse M., Dale J. M., Lee T. J., Kaipa P., Gilham F., Spaulding A., Popescu L., Altman T., Paulsen I., Keseler I. M., Caspi R. (2010) Brief. Bioinform. 11, 40–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang P., Foerster H., Tissier C. P., Mueller L., Paley S., Karp P. D., Rhee S. Y. (2005) Plant Physiol. 138, 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feist A. M., Henry C. S., Reed J. L., Krummenacker M., Joyce A. R., Karp P. D., Broadbelt L. J., Hatzimanikatis V., Palsson B. Ø. (2007) Mol. Syst. Biol. 3, 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrgård M. J., Swainston N., Dobson P., Dunn W. B., Arga K. Y., Arvas M., Blüthgen N., Borger S., Costenoble R., Heinemann M., Hucka M., Le Novère N., Li P., Liebermeister W., Mo M. L., Oliveira A. P., Petranovic D., Pettifer S., Simeonidis E., Smallbone K., Spasiã I., Weichart D., Brent R., Broomhead D. S., Westerhoff H. V., Kirdar B., Penttilä M., Klipp E., Palsson B., Sauer U., Oliver S. G., Mendes P., Nielsen J., Kell D. B. (2008) Nat. Biotechnol. 26, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caspi R., Foerster H., Fulcher C. A., Kaipa P., Krummenacker M., Latendresse M., Paley S., Rhee S. Y., Shearer A. G., Tissier C., Walk T. C., Zhang P., Karp P. D. (2008) Nucleic Acids Res. 36, D623–D631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okuda S., Yamada T., Hamajima M., Itoh M., Katayama T., Bork P., Goto S., Kanehisa M. (2008) Nucleic Acids Res. 36, W423–W426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ott M. A., Vriend G. (2006) BMC Bioinformatics 7, 517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pico A. R., Kelder T., van Iersel M. P., Hanspers K., Conklin B. R., Evelo C. (2008) PLoS Biol. 6, e184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geer L. Y., Marchler-Bauer A., Geer R. C., Han L., He J., He S., Liu C., Shi W., Bryant S. H. (2010) Nucleic Acids Res. 38, D492–D496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. (2008) Nucleic Acids Res. 36, D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang A., Scheer M., Grote A., Schomburg I., Schomburg D. (2009) Nucleic Acids Res. 37, D588–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matthews L., Gopinath G., Gillespie M., Caudy M., Croft D., de Bono B., Garapati P., Hemish J., Hermjakob H., Jassal B., Kanapin A., Lewis S., Mahajan S., May B., Schmidt E., Vastrik I., Wu G., Birney E., Stein L., D'Eustachio P. (2009) Nucleic Acids Res. 37, D619–D622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelder T., Pico A. R., Hanspers K., van Iersel M. P., Evelo C., Conklin B. R. (2009) PLoS ONE 4, e6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Iersel M. P., Kelder T., Pico A. R., Hanspers K., Coort S., Conklin B. R., Evelo C. (2008) BMC Bioinformatics 9, 399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arita M., Suwa K. (2008) BioData Mining 1, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitley K. (2002) J. Am. Soc. Information Sci. Technol. 53, 1210–1215 [Google Scholar]
- 30. Kind T., Scholz M., Fiehn O. (2009) PLoS ONE 4, e5440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y., Xiao J., Suzek T. O., Zhang J., Wang J., Bryant S. H. (2009) Nucleic Acids Res. 37, W623–W633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S., Geer L. Y., Kapustin Y., Khovayko O., Landsman D., Lipman D. J., Madden T. L., Maglott D. R., Ostell J., Miller V., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Sirotkin K., Souvorov A., Starchenko G., Tatusov R. L., Tatusova T. A., Wagner L., Yaschenko E. (2007) Nucleic Acids Res. 35, D5–D12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Degtyarenko K., de Matos P., Ennis M., Hastings J., Zbinden M., McNaught A., Alcántara R., Darsow M., Guedj M., Ashburner M. (2008) Nucleic Acids Res. 36, D344–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Degtyarenko K., Hastings J., de Matos P., Ennis M. (2009) Curr. Protoc. Bioinformatics Unit 14.9 [DOI] [PubMed] [Google Scholar]
- 35. Wishart D. S., Knox C., Guo A. C., Eisner R., Young N., Gautam B., Hau D. D., Psychogios N., Dong E., Bouatra S., Mandal R., Sinelnikov I., Xia J., Jia L., Cruz J. A., Lim E., Sobsey C. A., Shrivastava S., Huang P., Liu P., Fang L., Peng J., Fradette R., Cheng D., Tzur D., Clements M., Lewis A., De Souza A., Zuniga A., Dawe M., Xiong Y., Clive D., Greiner R., Nazyrova A., Shaykhutdinov R., Li L., Vogel H. J., Forsythe I. (2009) Nucleic Acids Res. 37, D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shinbo Y., Nakamura Y., Altaf-Ul-Amin M., Asahi H., Kurokawa K., Arita M., Saito K., Ohta D., Shibata D., Kanaya S. (2006) in Plant Metabolomics (Nagata T., Lörz H., Widholm J. M. eds) Vol. 57, pp. 165–181, Springer-Verlag, Berlin [Google Scholar]
- 37. Buckingham J. (ed) (2010) Dictionary of Natural Products, Chapman & Hall/CRC, Boca Raton, FL [Google Scholar]
- 38. Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., Ojima Y., Tanaka K., Tanaka S., Aoshima K., Oda Y., Kakazu Y., Kusano M., Tohge T., Matsuda F., Sawada Y., Hirai M. Y., Nakanishi H., Ikeda K., Akimoto N., Maoka T., Takahashi H., Ara T., Sakurai N., Suzuki H., Shibata D., Neumann S., Iida T., Tanaka K., Funatsu K., Matsuura F., Soga T., Taguchi R., Saito K., Nishioka T.(2010) J. Mass Spectrom. 45, 703–714 [DOI] [PubMed] [Google Scholar]
- 39. Hummel J., Selbig J., Walther D., Kopka J. (2007) Top. Curr. Genet. 18, 75–95 [Google Scholar]
- 40. Iijima Y., Nakamura Y., Ogata Y., Tanaka K., Sakurai N., Suda K., Suzuki T., Suzuki H., Okazaki K., Kitayama M., Kanaya S., Aoki K., Shibata D. (2008) Plant J. 54, 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kind T., Fiehn O. (2007) BMC Bioinformatics 8, 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moco S., Bino R. J., Vorst O., Verhoeven H. A., de Groot J., van Beek T. A., Vervoort J., de Vos C. H. R. (2006) Plant Physiol. 141, 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grennan A. K. (2009) Plant Physiol. 151, 1701–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Böttcher C., von Roepenack-Lahaye E., Schmidt J., Schmotz C., Neumann S., Scheel D., Clemens S. (2008) Plant Physiol. 147, 2107–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bais P., Moon S. M., He K., Leitao R., Dreher K., Walk T., Sucaet Y., Barkan L., Wohlgemuth G., Roth M. R., Wurtele E. S., Dixon P., Fiehn O., Lange B. M., Shulaev V., Sumner L. W., Welti R., Nikolau B. J., Rhee S. Y., Dickerson J. A. (2010) Plant Physiol. 152, 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fiehn O., Wohlgemuth G., Scholz M. (2005) in Data Integration in the Life Sciences (Ludascher B., Raschid L. eds) pp. 224–239, Springer-Verlag, Berlin [Google Scholar]
- 47. Fiehn O., Wohlgemuth G., Scholz M., Kind T., Lee D. Y., Lu Y., Moon S., Nikolau B. (2008) Plant J. 53, 691–704 [DOI] [PubMed] [Google Scholar]
- 48. Kind T., Wohlgemuth G., Lee D. Y., Lu Y., Palazoglu M., Shahbaz S., Fiehn O. (2009) Anal. Chem. 81, 10038–10048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmüller E., Dörmann P., Weckwerth W., Gibon Y., Stitt M., Willmitzer L., Fernie A. R., Steinhauser D. (2005) Bioinformatics 21, 1635–1638 [DOI] [PubMed] [Google Scholar]
- 50. Hummel J., Strehmel N., Selbig J., Walther D., Kopka J. (2010) Metabolomics 6, 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stein S. E. (1995) J. Am. Soc. Mass Spectrom. 6, 644–655 [DOI] [PubMed] [Google Scholar]
- 52. Cui Q., Lewis I. A., Hegeman A. D., Anderson M. E., Li J., Schulte C. F., Westler W. M., Eghbalnia H. R., Sussman M. R., Markley J. L. (2008) Nat. Biotechnol. 26, 162–164 [DOI] [PubMed] [Google Scholar]
- 53. Steinbeck C., Kuhn S. (2004) Phytochemistry 65, 2711–2717 [DOI] [PubMed] [Google Scholar]
- 54. Merico D., Gfeller D., Bader G. D. (2009) Nat. Biotechnol. 27, 921–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pavlopoulos G. A., Wegener A. L., Schneider R. R. (2008) BioData Mining 1, 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suderman M., Hallett M. (2007) Bioinformatics 23, 2651–2659 [DOI] [PubMed] [Google Scholar]
- 57. Atkinson H. J., Morris J. H., Ferrin T. E., Babbitt P. C. (2009) PLoS ONE 4, e4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bader G. D., Cary M. P., Sander C. (2006) Nucleic Acids Res. 34, D504–D506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bauer-Mehren A., Furlong L. I., Sanz F. (2009) Mol. Syst. Biol. 5, 290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang J. D., Wiemann S. (2009) Bioinformatics 25, 1470–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Williams A. J., Tkachenko V., Golotvin S., Kidd R., McCann G. (2010) J. Cheminform. 2, 1 20298528 [Google Scholar]
- 62. Antonov A. V., Dietmann S., Wong P., Mewes H. W. (2009) FEBS J. 276, 2084–2094 [DOI] [PubMed] [Google Scholar]
- 63. Letunic I., Yamada T., Kanehisa M., Bork P. (2008) Trends Biochem. Sci. 33, 101–103 [DOI] [PubMed] [Google Scholar]
- 64. Xia J., Wishart D. S. (2010) Bioinformatics 26, 2342–2344 [DOI] [PubMed] [Google Scholar]
- 65. Frolkis A., Knox C., Lim E., Jewison T., Law V., Hau D. D., Liu P., Gautam B., Ly S., Guo A. C., Xia J., Liang Y., Shrivastava S., Wishart D. S. (2010) Nucleic Acids Res. 38, D480–D487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kono N., Arakawa K., Ogawa R., Kido N., Oshita K., Ikegami K., Tamaki S., Tomita M. (2009) PLoS ONE 4, e7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bonneau R. (2008) Nat. Chem. Biol. 4, 658–664 [DOI] [PubMed] [Google Scholar]
- 68. Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L. A., Rhee S. Y., Stitt M. (2004) Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 69. Schnoes A. M., Brown S. D., Dodevski I., Babbitt P. C. (2009) PLoS Comput. Biol. 5, e1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hartman A. L., Lough D. M., Barupal D. K., Fiehn O., Fishbein T., Zasloff M., Eisen J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fiehn O. (2008) TrAC Trends Anal. Chem. 27, 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Arita M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1543–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jeong H., Tombor B., Albert R., Oltvai Z. N., Barabási A. L. (2000) Nature 407, 651–654 [DOI] [PubMed] [Google Scholar]
- 75. Fong S. S., Nanchen A., Palsson B. O., Sauer U. (2006) J. Biol. Chem. 281, 8024–8033 [DOI] [PubMed] [Google Scholar]
- 76. Pachkov M., Dandekar T., Korbel J., Bork P., Schuster S. (2007) Gene 396, 215–225 [DOI] [PubMed] [Google Scholar]
- 77. Yamanishi Y., Hattori M., Kotera M., Goto S., Kanehisa M. (2009) Bioinformatics 25, i179–i186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fischer E., Sauer U. (2003) Eur. J. Biochem. 270, 880–891 [DOI] [PubMed] [Google Scholar]
- 79. Sallaud C., Rontein D., Onillon S., Jabès F., Duffé P., Giacalone C., Thoraval S., Escoffier C., Herbette G., Leonhardt N., Causse M., Tissier A. (2009) Plant Cell 21, 301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]