Abstract
The development of intratumoral hypoxia, a hallmark of rapidly progressing solid tumors, renders tumor cells resistant to chemotherapy and radiation therapy. We have recently shown that inhibition of aldose reductase (AR), an enzyme that catalyzes the reduction of lipid aldehydes and their glutathione conjugates, prevents human colon cancer cell growth in culture as well as in nude mouse xenografts by inhibiting the NF-κB-dependent activation of oxidative stress-mediated inflammatory and carcinogenic markers. However, the role of AR in mediating hypoxic stress signals is not known. We therefore investigated the molecular mechanisms by which AR inhibition prevents the hypoxia-induced human colon cancer cells growth and invasion. Our results indicate that AR inhibition by the pharmacological inhibitor fidarestat or ablation by AR-specific siRNA prevents hypoxia-induced proliferation of HT29, SW480, and Caco-2 colon cancer cells. Furthermore, hypoxia-induced increase in the level of HIF-1α in colon cancer cells was significantly decreased by AR inhibition. During hypoxic conditions, treatment of HT29 cells with the AR inhibitor fidarestat significantly decreased the expression of vascular endothelial growth factor, a down target of HIF-1α, at both mRNA and protein levels and also prevented the activation of PI3K/AKT, GSK3β, Snail, and lysyl oxidase. Furthermore, inhibition of hypoxia-induced HIF-1α protein accumulation by AR inhibition was abolished in the presence of MG132, a potent inhibitor of the 26 S proteasome. In addition, AR inhibition also prevented the hypoxia-induced inflammatory molecules such as Cox-2 and PGE2 and expression of extracellular matrix proteins such as MMP2, vimentin, uPAR, and lysyl oxidase 2. In conclusion, our results indicate that AR mediates hypoxic signals, leading to tumor progression and invasion.
Keywords: Colon Cancer, Hypoxia, Proteasome, Signal Transduction, Tumor, HIF-1, Aldose Reductase
Introduction
Hypoxia, a condition of low oxygen tension, is a characteristic feature of solid tumors (>1 cm3) due to inadequate blood supply (1). In cancer patients, the extent of tumor hypoxia directly correlates with metastasis, resistance to radiation, and chemotherapeutic drugs (2). During hypoxia stress, tumor cells express transcriptional activator hypoxia-inducible factor-1 (HIF-1),4 which plays an important role in promoting cancer angiogenesis and metastasis. During normoxia, oxygen-dependent degradation domain of HIF-1α interacts with the von Hippel-Lindau protein, a recognition component of an E3 ubiquitin-protein ligase complex, and causes ubiquitinylation and degradation of HIF-1α protein via the 26 S proteasome. Under hypoxic conditions, the blockade of prolyl hydroxylation, ubiquitinylation, and degradation leads to accumulation of HIF-1α protein in the cytoplasm and translocation to the nucleus, where it forms an active complex with HIF-1β, an aryl hydrocarbon receptor nuclear translocator. This heterodimeric complex binds to consensus promoter region of target genes such as VEGF and activates their transcription (3, 4).
Another hypoxia-mediated regulatory pathway reported to be critical for controlling invasion and metastasis of colon cancer cells is involvement of lysyl oxidase (LOX) proteins (5). LOX proteins such as LOX2 and LOX3 interact with Snail to stabilize the protein in a manner that is dependent on two Snail lysine residues (5). Snail acts as direct repressor of E-cadherin expression in epithelial cells and activator of transcription of genes associated with mesenchymal differentiation such as vimentin and fibronectin (6). Phosphorylation of the Snail protein by GSK-3β leads to ubiquitination and proteasomal degradation by β-transducin repeat containing protein (6). Furthermore, during growth factor- and/or hypoxia-induced conditions, PI3K/AKT-mediated phosphorylation of GSK-3β renders activation of Snail protein and promotes angiogenesis and metastasis (7, 8).
Various reports show that hypoxia causes stabilization of HIF-1 and increased transcription of various inflammatory growth factors, cytokines, and chemokines. The increased cytokines and growth factors generate reactive oxygen species via NADPH oxidase by an autocrine and paracrine fashion (9–12). Among them, VEGF plays an important role in many physiological and pathological processes, including tumor growth, proliferation, and angiogenesis (11, 12). Indeed, numerous reports show that antioxidants, which are available in fruits, vegetables, and beverages derived from plants and in many dietary supplements or herbal remedies, could prevent hypoxia-mediated tumor cell invasion migration and metastasis in cancer cells (11, 13–17). Previous studies have shown that antioxidants such as quercetin inhibit HIF-1α protein accumulation in cytoplasm and translocation into the nucleus during hypoxic conditions via inhibiting the PI3K/AKT signaling pathway (11, 18–20). Therefore, use of antioxidants represents one of the most promising approaches to control tumor growth. Indeed, several antioxidants are under drug development program and clinical trials to identify novel anti-angiogneic and anti-cancer agents.
Our recent studies (21–24) with human colon cancer Caco-2, HT29, SW480 cells, and vascular umbilical endothelial cells suggest that the polyol pathway enzyme aldose reductase (AR), a member of the aldo-keto reductase superfamily, is a regulator of activation of NF-κB and expression of inflammatory cytokines, chemokines, and growth factors, which are induced by reactive oxygen species via the PI3K/AKT pathway. In addition to reducing aldo sugars, AR efficiently catalyzes the reduction of lipid aldehydes such as 4-hydroxy-trans-2-nonenal (HNE) and their glutathione conjugates such as glutathione-4-hydroxy-trans-2-nonenal (GS-HNE) to 1,4-dihydroxynonene (DHN) and GS-DHN, respectively with low (μm) Km compared with glucose with Km in mm range. Furthermore, inhibition of AR prevents PKC, NF-κB, and AP-1 activation and the increase in cell growth caused by HNE and GS-HNE but not by GS-DHN, suggesting that the already reduced form of glutathione lipid aldehyde, GS-DHN, is insensitive to AR inhibition and could be the main mediator of oxidative stress-induced NF-κB activation (21, 23, 24). Most remarkably, in the nude mouse xenograft model, we have shown that inhibition of AR by AR siRNA as well as by the AR inhibitor fidarestat completely prevented progression of tumor growth of human colon cancer cells (SW480 and HT29) implanted in nude mice subcutaneously (21). Furthermore, recent results with azoxymethane model in male BALB/c mice showed that inhibition of AR by pharmacological inhibitor of AR as well as AR gene knock-out in mice significantly prevented aberrant crypt foci formation and azoxymethane-induced expression of inflammatory markers, inducible nitric oxide synthase and cyclooxygenase-2 (Cox-2) and preneoplastic marker proteins, cyclin D1 and β-catenin and activation of NF-κB in mouse colons (25). However, the involvement of AR in the hypoxia-mediated carcinogenesis is unknown. Because the PI3K/AKT pathway plays a key role in the activation of HIF-1α, which regulates tumor growth during hypoxic conditions, we hypothesized that inhibition of AR could inhibit HIF-1α-mediated carcinogenesis.
In this study, we have shown for the first time that inhibition of AR effectively suppresses hypoxia- induced expression of HIF-1α, VEGF, MMP2, MET, LOX2, vimentin, and E-cadherin and proliferation of human colon cancer cells. Our results also show that AR inhibition prevents hypoxia-induced and/or bFGF- or serum-induced invasion and migration of HT29 cells. The mechanisms by which AR inhibition prevents hypoxia-induced and/or bFGF- or serum-induced HIF-1α protein expression seemed to involve an increase in invasion of HT29 cells via the PI3K/AKT, GSK3β, LOX, and Snail signaling pathways. These unique findings provided further understanding of the molecular mechanisms underlying the hypoxia and AR inhibition, which help to delineate further targets of therapeutic intervention and chemoprevention of human cancers.
EXPERIMENTAL PROCEDURES
Materials
DMEM, McCoy's medium, RPMI 1640, PBS, penicillin/streptomycin, trypsin, and FBS were purchased from Invitrogen. Antibodies against AKT, p65, membrane type 1 matrix metalloproteinase (MT1-MMP), MMP2, MMP9, LOX, Snail, and GAPDH were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Fidarestat was obtained as a gift from Sanwa Kagaku Kenkyusho Co., Ltd. (Nagoya, Japan). The cell invasion assay kit was obtained from Chemicon International Inc. (Billerica, MA). FGFs and other reagents used in Western blot analysis were obtained from Sigma. HIF-1α- and uPAR-overexpressing plasmids were obtained from Genecopeia, Inc. (Rockville, MD). All other reagents used were of analytical grade.
Cell Culture and Hypoxic Culture Conditions
Human colon cancer HT29, SW480, and Caco-2 cells were obtained from ATCC and grown in McCoy's 5A medium, RPMI 1640, and DMEM supplemented with 10% FBS and 1% penicillin/streptomycin, respectively. For hypoxic conditions, cells were cultured to 80% confluence at normoxia conditions and transferred to hypoxia chamber (catalogue no. 27310; StemCell Technologies, Vancouver, BC, Canada) at 37 °C in a 1% O2, 5% CO2, and 94% N2 atmosphere for various periods of time.
Measurement of Cytotoxicity
HT29, SW480, and Caco-2 cells were grown to confluence in McCoy's medium and DMEM, respectively, and trypsinized. Cells were plated in a 96-well plate at 2500 per well. Subconfluent cells were growth-arrested in 0.1% FBS with or without the AR inhibitor fidarestat (5 μm) or transfected with AR siRNA or control siRNA using RNAiFect reagent (Qiagen). After 24 h, cells were treated with bFGF (10 ng/ml) or serum (5%) and incubated at normoxia or hypoxia conditions for 24 h. Cells incubated with the AR inhibitor or transfection reagent alone served as control. Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay as described previously (21).
Invasive Assay
An invasive assay was performed using basement membrane extract. Fifty microliters of basement membrane extract solution were added on top of 8-micron polyethylene terephthalate membrane to each well of 96-well plates and incubated at 37 °C for 4 h to allow gel formation. Human colon cancer (20,000 cells/well) cells in basal medium with or without bFGF (10 ng/ml) or serum (5%) and/or AR inhibitor (fidarestat (5 μm)) were plated on Matrigel and incubated under hypoxic conditions. After 24 h of incubation, the invasion of cells toward bottom side of the well was measured using calcein-AM fluorescent dye.
Western Blot Analysis
To examine the expression/phosphorylation of MT1-MMP, MMP2, MMP9, AKT, p65, and GAPDH proteins, Western blot analysis was carried out. Equal amounts of protein from cell lysates were subjected to 12% SDS-PAGE followed by transfer of proteins to nitrocellulose filters and probing with the indicated antibodies. The antigen-antibody complex was detected by enhanced chemiluminescence (Pierce). All blots were probed with either GAPDH or actin as a loading control, and densitometry analysis was performed using Kodak Image station 2000R (Eastman Kodak Co., Rochester, NY).
Determination of HIF-1α by ELISA
Total HIF-1α was determined by ELISA as recommended by manufacturer (R&D Systems, Minneapolis, MN). Briefly, growth-arrested HT29 cells were treated with hypoxia and/or bFGF (10 ng/ml) or serum in the presence and absence of fidarestat (5 μm) for 24 h. The total HIF-1α protein in cell lysate was determined by sandwich ELISA method in 96-well plates, and the development of color against TMB was measured at 450 nm using ELISA reader.
Determination of PI3K Activity
PI3K activity in human umbilical vascular endothelial cells (HUVEC) was determined using a competitive ELISA kit (Echelon Biosciences, Inc., Salt Lake City, UT) as described previously (26).
Reverse Transcription-PCR Analysis
Total RNA was isolated from cell lysates by using RNeasy mini isolation kit (Qiagen, Valencia, CA). Total RNA (1.0 μg) sample was reverse-transcribed with Omniscript and Sensiscript reverse transcriptase one-step RT-PCR system with HotStarTaq DNA polymerase (Qiagen) at 55 °C for 30 min followed by PCR amplification. The oligonucleotide primer sequences were as follows: 5′-TCACCACAGGACAGTACAGGATGC-3′ (sense) and 5′-CCAGCAAAGTTAAAGCATCAGGTTCC-3′ (antisense) for HIF-1α, 5′-AGGAGGGCAGAATCATCACG-3′ (sense) and 5′-CAAGGCCCACAGGGATTTTCT-3′ (antisense) for VEGF, and 5′-GTTTGAGACCTTCAACACCCC-3′ (sense) and 5′-GTGGCCATCTCCTGCTCGAAGTC-3′ (antisense) for β-actin. PCR was carried out in a GeneAmp 2700 thermocycler (Applied Biosystems, Foster City, CA) under the following conditions: initial denaturation at 95 °C for 15 min and 35 cycles of 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 1 min and then 72 °C for 5 min for final extension. PCR products were electrophoresed in 2% agarose-1× Tris-acetate-EDTA gels containing 0.5 μg/ml ethidium bromide. Bands were quantified using Kodak Image Station 2000R.
Data Analysis
Data are presented as mean ± S.E., and the p values were determined using the unpaired Student's t test, with significant differences determined as p < 0.05.
RESULTS
Inhibition of AR Prevents Hypoxia-induced Proliferation of Human Colon Cancer Cells
At first, we examined the efficacy of AR inhibition in suppressing the growth promoting activity of hypoxia in three different human colon cancer cells lines, HT29, SW480, and Caco-2. The results shown in Fig. 1A demonstrate that during normoxic conditions, AR inhibition did not affect the growth, whereas exposure of HT29 or Caco-2 cells to hypoxic conditions for 24 h significantly increased the proliferation. Supplementation of bFGF or (5%) complete FBS to hypoxic cells further increased the proliferation of HT29 or Caco-2 compared with hypoxic cells alone. Treatment of HT29, SW480, or Caco-2 cells with AR inhibitor, fidarestat significantly prevented the hypoxia alone and hypoxia+bFGF or serum- induced cell growth. Furthermore, to rule out the nonspecific effects of pharmacological AR inhibitor, we also examined hypoxia-induced cell growth in the AR-ablated cancer cells by using AR-specific siRNA. Ablation of AR significantly prevented the hypoxia alone and hypoxia+bFGF- or serum-induced proliferation of HT29, Caco-2, and SW480 cells (Fig. 1B). Transfection of colon cancer cells with AR-SiRNA decreased AR protein expression by >95% (Fig. 1B, insets). These results suggest that AR mediates hypoxia-induced proliferation of human colon cancer cells.
FIGURE 1.
Inhibition or ablation of AR prevents hypoxia-induced and/or FGF- or serum-induced growth in colon cancer cells. Growth-arrested HT29 or SW480 or Caco-2 cells were preincubated with fidarestat (Fid) or carrier (A) or transfected wth AR siRNA or scrambled (Scram) AR siRNA for 24 h (B), followed by stimulation with hypoxia and/or FGF or serum (5%) for another 24 h, and cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The insets in B shows the Western blot analysis for AR protein in untransfected (C), scrambled (S), and AR siRNA-transfected (R) cell extracts. Fold values of normalized intensity measured by densitometric scanning. Bars represent mean ± S.E. (n = 4). *, p < 0.001 compared with control and #, p < 0.001; ##, p < 0.01 compared with cells treated with hypoxia and FGF or serum. Con, control.
Inhibition of AR Prevents Hypoxia-induced Stabilization of HIF-1α Protein in HT29 Cells
Because HIF-1α has been shown to be the main protein responsible for hypoxia-induced pathologies (9, 11), we next determined the effect of AR inhibition by fidarestat or ablation by AR siRNA on the stabilization of HIF-1α protein during hypoxia conditions at different time intervals. Immunoblot analysis showed that HIF-1α protein rapidly increased in a time-dependent manner when HT29 cells were exposed to hypoxic conditions (Fig. 2A). Similar results were observed with the SW480 cells (data not shown). Pretreatment of HT29 cells with AR inhibitor (fidarestat) significantly prevented the accumulation of HIF-1α protein, which suggests that AR could mediate HIF-1α stabilization during hypoxic conditions.
FIGURE 2.
Effect of AR inhibition on hypoxia-induced and/or FGF- or serum-induced expression/degradation of HIF-1α protein in HT29 cells. A, growth-arrested HT29 cells were preincubated with fidarestat (Fid) or carrier for 24 h followed by incubation under hypoxia for different time intervals. Western blots were developed using antibodies against HIF-1α and GAPDH. B, HT29 cells were exposed to hypoxia for 18 h followed by treatment with cycloheximide (CHX; 10 μg/ml) in the presence or absence of fidarestat (5 μm) for different time periods. In another experiment, HT29 cells were treated with MG132 (20 μm) for 30 min and cultured in the presence and absence of fidarestat (5 μm) for 6 h under normoxic or hypoxic conditions. C, growth-arrested HT29 cells were treated with hypoxia and/or FGF or serum for 24 h and Western blot analysis and ELISA were performed. D, RT-PCR analysis of HIF-1α and β-actin mRNA. Bars represent mean ± S.E. (n = 4); *, p < 0.001 compared with control and #, p < 0.001 compared with cells treated with hypoxia and/or FGF or serum. Con, control.
Inhibition of AR Prevents Hypoxia-induced HIF-1α Protein Accumulation by Promoting Protein Degradation in HT29 Cells
Various studies demonstrate that hypoxia induces HIF-1α protein accumulation mainly by promoting its stability instead of increasing de novo synthesis (9–11, 16). To investigate how AR inhibition prevents HIF-1α protein accumulation/stabilization, we explored the effects of AR inhibitor on the stability of HIF-1α protein. HT29 cells at 70–80% confluence were exposed to hypoxia for 6 h, followed by treatment with cycloheximide to block de novo protein synthesis, in the presence or absence of AR inhibitor for different time periods. Our results showed that in HT29 cells treated with fidarestat, the hypoxia-induced HIF-1α protein accumulation gradually decreased in 4 h, following exposure to cycloheximide, whereas without AR inhibition, the HIF-1α protein level remained stable under hypoxic conditions (Fig. 2B). These findings suggest that AR inhibition exerts its inhibitory effect by promoting the degradation of hypoxia-induced HIF-1α protein. To further explore whether the degradation of hypoxia-induced HIF-1α protein promoted by AR inhibition is mediated by the proteasome degradation pathway, HT29 cells were treated with a potent and specific 26 S proteasome inhibitor, MG132, in the presence and absence of fidarestat. As shown in the Fig. 2B, treatment of HT29 cells under normoxic conditions with MG132 increased the total HIF-1α protein levels. However, the increased HIF-1α protein level induced by MG132 could not be attenuated by AR inhibition. As expected, under hypoxic conditions, MG132 treatment further increased total HIF-1α protein accumulation, whereas the inhibitory effects of AR inhibition on hypoxia-induced HIF-1α protein levels did not decrease in the presence of MG132 (Fig. 2B). Taken together, these results suggest that AR inhibition promoted the degradation of hypoxia-induced HIF-1α protein possibly via the proteasome degradation pathway in HT29 cells.
Effect of AR Inhibition on Hypoxia-induced and/or Growth Factor- or Serum-induced Expression of HIF-1α/β Proteins in HT29 Cells
Accumulating evidence suggests that some growth factors as well as serum could increase HIF-1α protein during hypoxia in various cancer cell lines (11). To understand the role of AR in hypoxia-induced and/or bFGF- or serum-induced expression of HIF-1α protein, serum-starved (for 24 h) HT29 cells were exposed to hypoxia and/or bFGF or serum with and without the AR inhibitor fidarestat for another 24 h. As shown in the Fig. 2C, hypoxia and/or bFGF or serum induced the expression of HIF-1α protein and inhibition of AR by fidarestat robustly inhibited expression, as determined using immunoblot analysis. AR inhibition did not affect the constitutively expressed HIF-1β protein levels. Furthermore, these results were confirmed with ELISA in total cell lysates of HT29 cells exposed to hypoxia and/or bFGF or serum (Fig. 2C), suggesting that inhibition of AR could prevent hypoxia-induced and/or bFGF- or serum-induced expression of HIF-1α protein in HT29 cells. Next, to determine whether the reduction of hypoxia-induced HIF-1α protein accumulation by AR inhibition was the result of transcriptional inhibition, HIF-1α levels were determined by RT-PCR. As shown in Fig. 2D, no apparent changes in HIF-1α mRNA levels were observed in HT29 cells after exposure to hypoxia and/or FGF or 5% serum for 8 h. These results suggest that AR inhibition decreased the hypoxia-induced HIF-1α protein accumulation via a post-transcriptional mechanism.
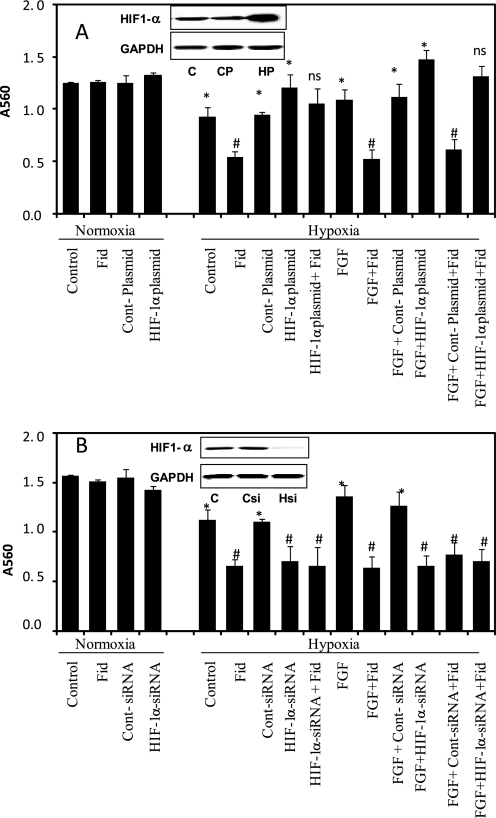
Overexpression of HIF-1α Rescues AR Inhibition
To validate the causal relationship between AR and HIF-1α, we next examined whether overexpression of HIF-1α rescues AR inhibition effect in colon cancer cells. Transient transfection of HT29 cells with the HIF-1α plasmid (GeneCopeia) increased HIF-1α levels by >8-fold as compared with control (Fig. 3A, inset). The HIF-1α-overexpressing cells were used to determine the efficacy of AR inhibitors in the prevention of hypoxia-induced cell proliferation. Our results shown in Fig. 3A indicate that under hypoxia, HT29 cells overexpressing HIF-1α in the absence and presence of FGF showed significantly increased cell proliferation as compared with control or FGF-treated cells alone, respectively. AR inhibitor prevented the proliferation of control and FGF-induced cells but not in HIF-1α-overexpressing cells, indicating that HIF-1α overexpression rescues AR inhibition-initiated effects. We next examined the efficacy of AR inhibition in the prevention of hypoxia-induced cancer cell growth in HIF-1α-ablated HT29 cells by using specific HIF-1α siRNA (Dharmacon). Transient transfection of HIF-1α in HT29 cells leads to a significant >90% decrease in the expression of HIF-1α as compared with scrambled siRNA-transfected cells (Fig. 3B, inset). Our results shown in the Fig. 3B indicate that the cells ablated with HIF-1α alone in the absence and presence of FGF showed significantly decreased proliferation of HT29 cells as compared with control and FGF-treated. Incubation of HT-29 cells with the AR inhibitor prevented the proliferation of control and FGF-induced cells but not in cells transfected with HIF-1α siRNA. These results indicate that AR regulates hypoxia-induced cell growth by modulating HIF-1α in colon cancer cells.
FIGURE 3.
Effect of AR inhibition on proliferation of HIF-1α plasmid and siRNA-transfected HT29 cells. A, HIF-1α-overexpressing and HIF-1α-ablated HT29 cells were incubated with fidarestat (Fid) or carrier subsequently stimulated with hypoxia and/or FGF for another 24 h, and cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Insets show the Western blot analysis for HIF-1α protein in untransfected, control plasmid and HIF-1α overexpressing plasmid transfected cell extracts. Bars represent mean ± S.E. (n = 4); *, p < 0.001 compared with control and #, p < 0.001 compared with cells treated with hypoxia and/or FGF.
Inhibition of AR Prevents Hypoxia-induced VEGF Expression and Secretion in HT29 Cells
The proangiogenic growth factor, VEGF is an immediate downstream target gene of HIF-1α, which plays an important role in tumor angiogenesis, especially during intratumoral hypoxia (4, 9). Because AR inhibition prevented the bFGF-induced increase of HIF-1α in HT29 cells, we hypothesized that AR inhibition would prevent VEGF expression. To determine whether AR inhibition can prevent hypoxia-induced VEGF expression in HT29 cells, VEGF protein expression and secretion levels were determined by Western blot and ELISA, respectively. Our results showed that treatment of HT29 cells with fidarestat resulted in a significant decrease in hypoxia-induced and/or bFGF- or serum-induced VEGF expression/secretion (Fig. 4, A and B) at 24 h. Similarly ablation of AR by using AR siRNA caused significant decrease in hypoxia-induced and/or bFGF- or serum-induced VEGF expression/secretion (Fig. 4, D and E) at 24 h. To further confirm the effects of AR inhibition on VEGF transcriptional activation, HT29 cells were exposed to hypoxia and/or bFGF or serum for 6 h, and the mRNA levels were determined by RT-PCR. As shown in the Fig. 4C treatment of HT29 cells with hypoxia and/or bFGF or serum strikingly increased (∼60%) the expression of VEGF mRNA, and AR inhibition prevented it. These results indicated that inhibition of AR prevented the hypoxia-induced and/or bFGF- or serum-induced VEGF protein accumulation through a translational and/or transcriptional mechanism.
FIGURE 4.
Inhibition of AR prevents hypoxia-induced and/or FGF- induced VEGF expression in HT29 cells. Growth-arrested HT29 cells were preincubated with fidarestat (Fid) or carrier for 24 h, followed by stimulation with hypoxia and/or FGF for another 24 h. A, Western blot analysis was performed using antibodies against VEGF and GAPDH. B, VEGF secretion in cell supernatants was measured. C, VEGF mRNA expression was measured after 6 h of hypoxia and/or FGF treatment. Bars represent mean ± S.E. (n = 4). *, p < 0.001 compared with control (Con) and #, p < 0.001 compared with cells treated with hypoxia and/or FGF. Scram, scrambled.
Inhibition of AR Prevents Hypoxia-induced Cox-2 Expression and PGE2 Production in HT29 Cells
Because Cox-2 is up-regulated during colorectal tumor progression and hypoxia, and also HIF-1α directly binds to Cox-2 promoter region, we next systematically investigated the effect of AR on hypoxia-mediated Cox-2 expression. As shown in Fig. 5A, treatment of HT29 cells with hypoxia and/or bFGF or serum caused a significant increase in the expression of HIF-1α protein, and inhibition of AR prevented it. To confirm the inducible Cox-2 expression during hypoxia, we next determined the Cox activity in total cell lysate of HT29 cells. Exposure of HT29 cells to hypoxia and/or bFGF or serum increased Cox activity (Fig. 5B), and preincubation with fidarestat abolished hypoxia-induced and/or bFGF- or serum-induced Cox activity. Because PGE2 is a primary product of arachidonic metabolism and is synthesized via the COX and prostaglandin synthase pathways, we next examined whether the inhibition of AR prevents hypoxia-induced PGE2 production in both HT29 cells (Fig. 5C). During normoxia, inhibition of AR did not affect the basal levels of PGE2 production, whereas inhibition of AR during hypoxia and/or bFGF or serum significantly prevented the production of PGE2. These results establish that AR could mediate the up-regulation of Cox-2 and PGE2 via HIF-1α regulation during hypoxia.
FIGURE 5.
Inhibition of AR prevents hypoxia-induced and/or FGF- or serum-induced Cox-2 expression, Cox activity, and PGE2 production in HT29 cells. Growth-arrested HT29 cells were preincubated with fidarestat (Fid) or carrier for 24 h, followed by stimulation under hypoxia and/or by FGF or serum for another 24 h. A, Western blot analysis using antibodies against Cox-2 and GAPDH. Fold values of normalized intensity measured by densitometric scanning from at least three to four independent analyses. Cox activity (B) and PGE2 production (C) in cell supernatants was measured. Bars represent mean ± S.E. (n = 4). *, p < 0.001 compared with control (Con) and #, p < 0.001 compared with cells treated with hypoxia and/or FGF or serum.
Inhibition of AR Prevents Hypoxia-induced Activation of PI3K/AKT in HT29 Cells
Various studies indicate that multiple signaling pathways, particularly PI3K/AKT, are involved in hypoxia-induced HIF-1α protein accumulation and expression of its downstream target genes to drive invasion and migration of hypoxic tumor cells (11, 18). Furthermore, we have shown that inhibition of AR prevents the growth factors such as EGF- and FGF-induced activation of PI3K and AKT in HT-29 cells. In this study, to investigate how AR inhibition could prevent hypoxia-mediated activation of PI3K/AKT, HT29 cells were treated with hypoxia and/or FGF or serum for 2 h in the presence and absence of the AR inhibitor fidarestat and determined PI3K/AKT activation. Our results showed that AR inhibition by fidarestat significantly attenuated the phosphorylated PI3K and AKT levels (Fig. 6A). Furthermore, AR-mediated PI3K activation during hypoxia and/or bFGF or serum was confirmed by using a competitive ELISA. As shown in Fig. 6B, hypoxia and/or bFGF or serum treatment stimulated PI3K activity 2.0–2.5-fold as compared with control, and inhibition of AR significantly (∼65%) prevented it. Taken together, these results suggest that AR inhibition prevents hypoxia-induced HIF-1α protein accumulation and VEGF expression through the inhibition of PI3K/AKT in HT29 cells.
FIGURE 6.
Effect of AR inhibition on hypoxia-mediated and/or FGF- or serum-mediated PI3K/AKT, GSK3β, and Snail activities and invasion of HT29 cells. Growth-arrested HT29 cells were preincubated with fidarestat (Fid) or carrier for 24 h, followed by stimulation under hypoxia and/or FGF for another 24 h. A, Western blot analysis performed using antibodies against phospho-p85α, phospho-AKT, phospho-GSK3β, phospho-Snail, and GAPDH. Fold values of normalized intensity measured by densitometric scanning. B, PI3K activity was measured after 6 h of hypoxia and/or FGF or serum treatment. C, growth-arrested HT29 cells were plated at 20,000 cells/well in a 96-well plate containing Matrigel and treated with hypoxia and/or FGF (10 ng/ml) or serum with or without fidarestat (5 μm). After an overnight incubation, the invasion of cells toward the bottom side of the well was measured in situ using calcein-AM florescent dye at 480/520 nm. Bars represent mean ± S.E. (n = 4). *, p < 0.001 compared with control (Con) and #, p < 0.001 compared with cells treated with hypoxia and/or FGF or serum.
AR Mediates Phosphorylation of GSK3β and Expression of Snail and LOX during hypoxia in HT29 Cells
Because GSK3β is a well characterized downstream substrate of AKT, and phosphorylation of GSK3β at Ser-9 by AKT results in the inhibition of GSK activity (7, 11), we investigated the effect of AR inhibition on the phosphorylation of GSK3β during hypoxia. Hypoxia and/or bFGF or serum treatment resulted in an increase in the phosphorylation of GSK3β in HT29 cells, and AR inhibition prevented it (Fig. 6A). Next, we investigated the levels of Snail phosphorylation because Snail is a downstream substrate of GSK3β, and Snail phosphorylation by GSK3β undergoes proteasomal degradation (7, 8). Our results showed that inhibition of AR increased hypoxia and/or bFGF or serum down-regulated phosphorylation of Snail (i.e. inactivation of Snail protein) (Fig. 6A). Because Snail protein is regulated by LOX proteins during hypoxia, expression of LOX-2 protein was determined. Treatment of HT29 cells with hypoxia and/or bFGF or serum induced the expression of LOX-2 and AR inhibition prevented it. Taken together, these results strongly suggest that AR regulates the hypoxia-mediated signaling pathway via AKT/GSK3β/LOX/Snail.
Inhibition of AR Prevents Hypoxia-induced Invasion and Expression of Structural Proteins in HT29 Cells
Because hypoxia, serum, and growth factors are known to have stimulatory effects on cancer cell invasion and migration (9, 27), we performed in vitro cell migration assay to investigate whether AR inhibition could suppress the hypoxia-induced cancer cell invasion. Results shown in Fig. 6C indicate that invasion of HT29 cells was increased in the hypoxic conditions in absence and presence of FGF. No significant invasion of HT29 cells was observed in normoxic conditions. The hypoxia-induced and/or bFGF- or serum-induced invasion of HT29 cells was significantly decreased in the presence of the AR inhibitor fidarestat (5 μm). To further understand how AR inhibition could prevent invasion/migration during hypoxia, we next determined the expression of important molecules such as uPAR, MT1-MMP, active-MMP2, c-MET, vimentin, E-cadherin, and etc. that play an important role in invasion and migration. As shown in Fig. 7A, inhibition of AR by fidarestat prevented the invasion of HT29 cells by inhibiting the expression of structural proteins.
FIGURE 7.
Effect of AR inhibition on hypoxia-mediated expression of structural proteins and AR-catalyzed reaction products and hypoxia-mediated HIF-1α expression in HT29 cells. A and C, the growth-arrested HT29 cells were treated with hypoxia and/or FGF, serum, HNE, GS-HNE esters, or GS-DHN esters for 24 h, and Western blot analysis was performed in total cell lysates using antibodies against LOX2, uPAR, MT1-MMP, MMP2, c-MET, vimentin, HIF-1α, and GAPDH. Fold values of normalized intensity measured by densitometric scanning. B, uPAR overexpressing HT29 cells were incubated with fidarestat (Fid), or the carrier was subsequently stimulated with hypoxia and/or FGF for another 24 h, and cell viability was measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Insets show the Western blot analysis for uPAR protein in untransfected, control plasmid (Cont-Plasmid), and uPAR overexpressing plasmid transfected cell extracts. Bars represent mean ± S.E. (n = 4). *, p < 0.001 compared with control (Con) and #, p < 0.001 compared with cells treated with hypoxia and/or FGF or serum. C, control; CP, control plasmid; UP, uPAR plasmid.
Overexpression of uPAR Rescues AR Inhibition
Because uPAR plays a significant role in hypoxia-mediated migration, we next examined whether overexpression of uPAR rescues effects initiated by AR inhibition. Transient transfection of HT29 cells with uPAR plasmid (GeneCopeia) in HT-29 cells significantly increased the expression of uPAR by 6-fold (Fig. 7B, inset). The uPAR-overexpressing cells were used to determine the efficacy of AR inhibitors in the prevention of hypoxia-induced cell proliferation. Our results shown in Fig. 7B indicate that under hypoxia, HT29 cells overexpressing uPAR in the absence and presence of FGF showed significantly increased cell proliferation as compared with control or FGF-treated cells alone, respectively. The AR inhibitor prevented the proliferation of control and FGF-induced cells but not in uPAR-overexpressing cells, indicating that uPAR overexpression rescues AR inhibition-initiated effects.
Effect of AR Inhibition on Lipid Aldehyde-induced HIF-1α Expression during Hypoxia in HT29 Cells
We have shown previously that AR-catalyzed reaction products of GS-HNE could mediate mitogenicity in Caco-2 cells. Therefore, we examined whether GS conjugates of lipid aldehydes could be involved in hypoxia. Treatment of HT29 cells with HNE or cell-permeable esters of GS-HNE or GS-DHN resulted in an increased expression of HIF-1α (Fig. 7C). Inhibition of AR by fidarestat significantly (∼80%) prevented the HNE- and GS-HNE-induced HIF-1α expression but had no effect on GS-DHN-induced expression of HIF-1α. These results indicate that hypoxia-induced and/or bFGF- or serum-induced tumor cell survival/invasion in HT29 cells could be mediated by the reduced form of lipid aldehyde-glutathione conjugates catalyzed by AR.
DISCUSSION
Hypoxia is known to be a hallmark of invasive solid tumors and increased angiogenesis and is strongly associated with the progression of malignant phenotypes, poor prognosis, and resistance to anticancer drugs and radiation therapy (1, 2). It has been shown that HIF-1 pathways are activated in many cancers and are amplified by a wide variety of oncogenic pathways and growth factors, cytokines, and chemokines (2, 9). Up-regulated HIF-1α is involved in tumor cell progressive steps such as cell survival, invasion, proliferation, migration, and metastasis (1, 9). Among the various growth factors secreted by tumor cells in response to hypoxia, VEGF and bFGF are known to play an important role in the up-regulation of HIF-1α expression mainly by enhancing HIF-1 translation as well as by inhibiting its degradation (9, 11). The hypoxia-mediated HIF-1α protein accumulation occurs mainly by inhibiting its degradation through the ubiquitin proteasomal pathway (9, 11). This leads to the establishment of an autocrine and paracrine loop between hypoxia and growth factors in tumors, thereby enhancing angiogenic capability, tumor growth, invasion, and metastases (2, 9). Because AR is involved in the oxidative stress-mediated inflammatory signaling induced by growth factors, cytokines, and chemokines in various pathological diseases such as congestive heart failure, cardiomyopathy, hypertension, atherosclerosis, wound healing, tissue remodeling, thrombosis, rheumatoid arthritis, diabetic retinopathy, and cancer (21–24, 28), we determined the possible role of AR in hypoxia. In addition, various reports show that AR is overexpressed during ischemia (29, 30), but they did not show the molecular mechanism of AR involvement during hypoxia. The present study reveals that inhibition of AR directly inhibits hypoxia-mediated HIF-1α protein accumulation by inhibiting its degradation via the proteasomal pathway in both human colon cancer HT29 and Caco-2 cells. These findings provide the first evidence supporting the anti-hypoxic effects of AR inhibition in the setting of in vitro hypoxia, which mimics the in vivo intratumoral hypoxia.
HIF-1α protein levels are tightly regulated by oxygen tension via the ubiquitination and 26 S proteasomal degradation system (31). During hypoxic conditions, the ubiquitination and degradation of HIF-1α protein is abolished. This led to stabilization and accumulation of HIF-1α protein (11). Wu et al. (16) showed that treatment of human colon cancer Lovo cells with antioxidant such as resveratrol prevented the hypoxia-induced HIF-1α protein expression and stabilization. In this direction, our results show that AR inhibition prevented the basal level of HIF-1α protein expression in HT29 cells and significantly shortened the half-life of hypoxia-induced HIF-1α protein (Fig. 2). Furthermore, we have showed that the inhibition of hypoxia-induced HIF-1α protein accumulation by AR inhibition was abolished in the presence of MG132, a potent inhibitor of the 26 S proteasome. This supports our hypothesis that AR inhibition prevents HIF-1α protein expression via regulating both protein translation and HIF-1α protein degradation.
Various growth factors, cytokines, chemokines, oncogenes, and tumor promoters up-regulate oxidative stress-mediated inflammation via Cox-2 expression by activating transcription factors such as NF-IL6, NF-κB, nuclear factor of activated T-cells, and PEA3 (23, 32–34). We have shown previously that inhibition of AR prevents growth factors such as bFGF, platelet-derived growth factor, and cytokine such as TNF-α-induced Cox-2 expression as well as PGE2 production in colon cancer Caco-2 cells (23). Furthermore, accumulating evidence suggests that up-regulation of Cox-2 is mandatory particularly in colon carcinogenesis and the subset of adenomas (35, 36). Various reports show that Cox-2 up-regulation by hypoxia has been described to be mediated by NF-κB and peroxisome proliferation-activated receptors, including HIF-1α (36, 37). Our results show that AR inhibitors are anti-inflammatory, and AR inhibition could prevent hypoxia-mediated Cox-2 expression and PGE2 production via inhibiting HIF-1α expression and stabilization. Recently, Kaidi et al. (38), using human colon cancer HT29 cells, showed that Cox-2 up-regulation is transcriptional and dependent on HIF-1α induction during hypoxia. Furthermore, they showed that inhibition of the Cox-2 enzyme by a non-steroidal anti-inflammatory drug such as NS-398 inhibited hypoxia-mediated PGE2 production.
It is well established that the progression of solid tumor completely depends on angiogenesis and VEGF, which is potently stimulated by major hypoxia-responsive transcription factor HIF-1α (4, 9, 11, 16). This indicates that HIF-1α/VEGF could be potential targets for the anti-angiogenic therapy of cancer. Accumulating evidences suggest that antioxidants such as reseveratrol, lycopene, and green tea have anti-angiogenic effect in several hypoxia-mediated tumor angiogenic models (11, 16). The anti-angiogenic mechanism(s) of these antioxidants seems to be associated with inhibition of VEGF production and/or VEGF receptor activity. Treatment of human colon cancer SW837 cells with epigallocatechin gallate decreased VEGF production and HIF-1α expression induced by hypoxia (39). Similarly, the results in accordance with AR showed that the AR inhibitor fidarestat prevented hyperglycemia-induced oxidative stress in retinal endothelial cells and the expression of VEGF in diabetic rats (40). Furthermore, Tang et al. (41) showed that inhibition of AR or sorbitol dehydrogenase (the second enzyme of the polyol pathway), both attenuated expression of HIF-1α, transferring and its receptor and intracellular iron content in the ischemia reperfusion of rat heart. These findings together with our results show that the AR inhibitor has anti-angiogenic effect via inhibition of HIF-1α and VEGF expression in HT29 cells.
Several studies have demonstrated increased expression of MMPs in various cancer cells, including colon cancer cells (42–44). MMPs are involved in a large number of physiological and pathological processes (45). Because HIF-1α also up-regulates MMPs, which are involved in tumor invasion and metastasis (46), we next examined the efficacy of AR inhibition on hypoxia-induced up-regulation of MMP-2 in colon cancer cells. Our results indicate that AR inhibition prevents hypoxia-induced MMP2 activation. MT1-MMP shown to be involved in the cancer cell invasion under normoxic and hypoxic conditions activates pro-MMP2 to active-MMP2 (47, 48). Furthermore, the MT1-MMP-mediated activation of MMP-2 is mainly observed following paracrine stimulation by stromal fibroblast-derived conditioned media or various other stimuli such as growth factors (47). Therefore, we next examined whether AR inhibition prevents MMP-2 activation via regulating the expression of MT1-MMP in colon cancer cells. Our results shown in Fig. 7 indicates that AR inhibition prevents hypoxia-induced MMP2 activation by suppressing the expression of MT1-MMP in colon cancer cells.
Numerous studies show that multiple signaling pathways are involved in the regulation of hypoxia-induced HIF-1α protein stabilization and transactivation (11, 49, 50). Among various signaling pathways, the PI3K/AKT pathway is known to play a major role in the multiple signaling during hypoxia (3, 11). For example, activation of PI3K/AKT stabilizes/activates transcription factors such as HIF-1α and Snail via phosphorylation of both α and β subunits of HIF-1 and GSK3β (7, 8, 11, 50). Li et al. (51) reported that the HIF-1β subunit has a conserved AKT phosphorylation site, and phosphorylation of HIF-1β by AKT at Ser-271 increases the affinity between α- and β-subunits of HIF-1, which leads to increased HIF-1α expression and HIF-1 transcriptional activity. Our results show that AR inhibition could weaken the interaction between HIF-1α and β-subunits via PI3K/AKT inactivation, which renders HIF-1α to proteolytic degradation. AR inhibition-mediated suppression of AKT/PI3K activation is not restricted to the hypoxic condition because similar effects are evident during normoxia when HT29 cells are incubated with growth factors such as EGF and FGF. The AKT/GSK3 signaling pathway regulates HIF-1α protein stability (11, 50). Inhibition of GSK3β activation by LiCl or overexpression of dominant negative GSK3β mutant is known to increase of HIF-1α expression and activity (52, 53). Furthermore, various reports show that the transcription factor Snail is regulated by growth factors or integrins (6–8). Unphosphorylated form of Snail translocates to the nucleus to function as transcriptional repressor and activator to promote epithelial mesenchymal transition. However, to get progression of epithelial/mesenchymal transition GSK3β activity must be inhibited. Snail acts as a direct repressor of E-cadherin expression in epithelial cells, and the expression of Snail induces a full epithelial mesenchymal transition and increases migration/invasion in different physiological and pathological situations (8). Peinado et al. (54), showed that LOX gene family proteins (LOX, LOXL1, -2, -3, and -4) interact and cooperate with Snail to down-regulate E-cadherin expression. The individual function of the remaining LOX proteins remains unclear, although recent evidences suggest that overexpression of LOXL2 in epithelial cells induces an epithelial mesenchymal transition process, supporting their implication in tumor progression. Furthermore, RNA interference ablation of LOX2 caused apoptosis by decreasing the expression of mesenchymal, invasive and angiogenic markers in snail-overexpressing metastatic cancer cells (54). These studies provided the biological significance of LOX2 in cancer. Together, these results suggest that AR inhibition prevents hypoxia-mediated LOX2 and Snail protein expression in HT29 cells most likely by suppression of AKT/PI3K activation (Fig. 8).
FIGURE 8.
Schematic representation of the involvement of AR in hypoxia-mediated signals in colon cancer cells. ROS, reactive oxygen species.
In summary, we have shown that AR inhibition prevents hypoxia- and serum-induced HIF-1α protein accumulation and VEGF expression in HT29 cells by promoting HIF-1α protein degradation. AR inhibition also prevented the activation (phosphorylation) of PI3K/AKT as well as PI3K/AKT-mediated inactivation (phosphorylation) of GSK3β. Furthermore, inhibition of AR prevented the hypoxia-mediated expression of LOX2 and Snail proteins, which are important mediators in the epithelial mesenchymal transition of tumor cells. Collectively, these results for the first time show that AR mediates hypoxia signaling in human colon cancer HT29 cells via the PI3K/AKT/HIF-1α pathway, and AR inhibition could be used as a pharmacological intervention in HIF-1α-driven tumor progression/metastasis.
This work was supported in part by National Institutes of Health Grants GM71036 (to K. V. R.) and CA129383 and DK36118 (to S. K. S.).
- HIF-1
- hypoxia-inducible factor-1
- AR
- aldose reductase
- LOX
- lysyl oxidase
- VEGF
- vascular endothelial growth factor
- HNE
- 4-hydroxy-trans-2-nonenal
- GS-HNE
- glutathione HNE
- MT1-MMP
- membrane type 1 matrix metalloproteinase
- AKT
- also known as protein kinase B
- bF6F
- basic fibroblast growth factor
- uPAR
- urokinase-type plasminogen activator receptor.
REFERENCES
- 1. Cairns R. A., Khokha R., Hill R. P. (2003) Curr. Mol. Med. 3, 659–671 [DOI] [PubMed] [Google Scholar]
- 2. DeClerck K., Elble R. C. (2010) Front. Biosci. 15, 213–225 [DOI] [PubMed] [Google Scholar]
- 3. Gao N., Shen L., Zhang Z., Leonard S. S., He H., Zhang X. G., Shi X., Jiang B. H. (2004) Mol. Cell Biochem. 255, 33–45 [DOI] [PubMed] [Google Scholar]
- 4. Park S. Y., Jeong K. J., Lee J., Yoon D. S., Choi W. S., Kim Y. K., Han J. W., Kim Y. M., Kim B. K., Lee H. Y. (2007) Cancer Lett. 258, 63–69 [DOI] [PubMed] [Google Scholar]
- 5. Denko N. C., Fontana L. A., Hudson K. M., Sutphin P. D., Raychaudhuri S., Altman R., Giaccia A. J. (2003) Oncogene 22, 5907–5914 [DOI] [PubMed] [Google Scholar]
- 6. Yook J. I., Li X. Y., Ota I., Fearon E. R., Weiss S. J. (2005) J. Biol. Chem. 280, 11740–11748 [DOI] [PubMed] [Google Scholar]
- 7. Jope R. S., Yuskaitis C. J., Beurel E. (2007) Neurochem. Res. 32, 577–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., Hung M. C. (2004) Nat. Cell Biol. 6, 931–940 [DOI] [PubMed] [Google Scholar]
- 9. Rosmorduc O., Housset C. (2010) Semin. Liver Dis. 30, 258–270 [DOI] [PubMed] [Google Scholar]
- 10. Ushio-Fukai M., Nakamura Y. (2008) Cancer Lett. 266, 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Q., Tang X., Lu Q., Zhang Z., Rao J., Le A. D. (2006) Mol. Cancer Ther. 5, 1227–1238 [DOI] [PubMed] [Google Scholar]
- 12. Nam S. Y., Ko Y. S., Jung J., Yoon J., Kim Y. H., Choi Y. J., Park J. W., Chang M. S., Kim W. H., Lee B. L. (2011) Br. J. Cancer 104, 166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ichikawa H., Nakamura Y., Kashiwada Y., Aggarwal B. B. (2007) Curr. Pharm. Des. 13, 3400–3416 [PubMed] [Google Scholar]
- 14. Nagle D. G., Zhou Y. D. (2006) Curr. Drug Targets 7, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aggarwal B. B., Ichikawa H., Garodia P., Weerasinghe P., Sethi G., Bhatt I. D., Pandey M. K., Shishodia S., Nair M. G. (2006) Expert Opin. Ther. Targets 10, 87–118 [DOI] [PubMed] [Google Scholar]
- 16. Wu H., Liang X., Fang Y., Qin X., Zhang Y., Liu J. (2008) Biomed. Pharmacother. 62, 613–621 [DOI] [PubMed] [Google Scholar]
- 17. Bae M. K., Kim S. H., Jeong J. W., Lee Y. M., Kim H. S., Kim S. R., Yun I., Bae S. K., Kim K. W. (2006) Oncol. Rep. 15, 1557–1562 [PubMed] [Google Scholar]
- 18. Oh S. J., Kim O., Lee J. S., Kim J. A., Kim M. R., Choi H. S., Shim J. H., Kang K. W., Kim Y. C. (2010) Food Chem. Toxicol. 48, 3227–3234 [DOI] [PubMed] [Google Scholar]
- 19. Gulati N., Laudet B., Zohrabian V. M., Murali R., Jhanwar-Uniyal M. (2006) Anticancer Res. 26, 1177–1181 [PubMed] [Google Scholar]
- 20. Hwang M. K., Song N. R., Kang N. J., Lee K. W., Lee H. J. (2009) Int. J. Biochem. Cell Biol. 41, 1592–1600 [DOI] [PubMed] [Google Scholar]
- 21. Tammali R., Ramana K. V., Singhal S. S., Awasthi S., Srivastava S. K. (2006) Cancer Res. 66, 9705–9713 [DOI] [PubMed] [Google Scholar]
- 22. Srivastava S. K., Ramana K. V., Bhatnagar A. (2005) Endocr. Rev. 26, 380–392 [DOI] [PubMed] [Google Scholar]
- 23. Tammali R., Ramana K. V., Srivastava S. K. (2007) Cancer Lett. 252, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramana K. V., Bhatnagar A., Srivastava S., Yadav U. C., Awasthi S., Awasthi Y. C., Srivastava S. K. (2006) J. Biol. Chem. 281, 17652–17660 [DOI] [PubMed] [Google Scholar]
- 25. Tammali R., Reddy A. B., Ramana K. V., Petrash J. M., Srivastava S. K. (2009) Carcinogenesis 30, 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramana K. V., Tammali R., Srivastava S. K. (2010) Mol. Cancer Ther. 9, 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krishnamachary B., Berg-Dixon S., Kelly B., Agani F., Feldser D., Ferreira G., Iyer N., LaRusch J., Pak B., Taghavi P., Semenza G. L. (2003) Cancer Res. 63, 1138–1143 [PubMed] [Google Scholar]
- 28. Ramana K. V., Srivastava S. K. (2010) Int. J. Biochem. Cell Biol. 42, 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reddy A. B., Ramana K. V. (2010) Recent Pat. Cardiovasc. Drug Discov. 5, 25–32 [DOI] [PubMed] [Google Scholar]
- 30. Ramasamy R., Goldberg I. J. (2010) Circ. Res. 106, 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang S., Liu Z., Wang L., Zhang X. (2009) Cell Mol. Immunol. 6, 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Celil Aydemir A. B., Minematsu H., Gardner T. R., Kim K. O., Ahn J. M., Lee F. Y. (2010) Bone 46, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mann J. R., Backlund M. G., DuBois R. N. (2005) Nat. Clin. Pract. Oncol. 2, 202–210 [DOI] [PubMed] [Google Scholar]
- 35. Yang Z., Li C., Wang X., Zhai C., Yi Z., Wang L., Liu B., Du B., Wu H., Guo X., Liu M., Li D., Luo J. (2010) J. Cell. Physiol. 225, 266–275 [DOI] [PubMed] [Google Scholar]
- 36. Plummer S. M., Holloway K. A., Manson M. M., Munks R. J., Kaptein A., Farrow S., Howells L. (1999) Oncogene 18, 6013–6020 [DOI] [PubMed] [Google Scholar]
- 37. Tafani M., Russo A., Di Vito M., Sale P., Pellegrini L., Schito L., Gentileschi S., Bracaglia R., Marandino F., Garaci E., Russo M. A. (2010) Cancer Sci. 101, 1014–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaidi A., Williams A. C., Paraskeva C. (2007) Nat. Cell Biol. 9, 210–217 [DOI] [PubMed] [Google Scholar]
- 39. Shimizu M., Shirakami Y., Sakai H., Yasuda Y., Kubota M., Adachi S., Tsurumi H., Hara Y., Moriwaki H. (2010) Chem. Biol. Interact. 185, 247–252 [DOI] [PubMed] [Google Scholar]
- 40. Obrosova I. G., Minchenko A. G., Vasupuram R., White L., Abatan O. I., Kumagai A. K., Frank R. N., Stevens M. J. (2003) Diabetes 52, 864–871 [DOI] [PubMed] [Google Scholar]
- 41. Tang W. H., Wu S., Wong T. M., Chung S. K., Chung S. S. (2008) Free Radic. Biol. Med. 45, 602–610 [DOI] [PubMed] [Google Scholar]
- 42. Fang J., Shing Y., Wiederschain D., Yan L., Butterfield C., Jackson G., Harper J., Tamvakopoulos G., Moses M. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3884–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kousidou O. C., Roussidis A. E., Theocharis A. D., Karamanos N. K. (2004) Anticancer Res. 24, 4025–4030 [PubMed] [Google Scholar]
- 44. Kousidou O. Ch., Berdiaki A., Kletsas D., Zafiropoulos A., Theocharis A. D., Tzanakakis G. N., Karamanos N. K. (2008) Mol Oncol. 2, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gialeli C., Theocharis A. D., Karamanos N. K. (2011) FEBS J. 278, 16–27 [DOI] [PubMed] [Google Scholar]
- 46. Ben-Yosef Y., Miller A., Shapiro S., Lahat N. (2005) Am. J. Physiol. Cell Physiol. 289, C1321–1331 [DOI] [PubMed] [Google Scholar]
- 47. Stahtea X. N., Roussidis A. E., Kanakis I., Tzanakakis G. N., Chalkiadakis G., Mavroudis D., Kletsas D., Karamanos N. K. (2007) Int. J. Cancer 121, 2808–2814 [DOI] [PubMed] [Google Scholar]
- 48. Muñoz-Nájar U. M., Neurath K. M., Vumbaca F., Claffey K. P. (2006) Oncogene. 25, 2379–2392 [DOI] [PubMed] [Google Scholar]
- 49. Semenza G. L. (2002) Biochem. Pharmacol. 64, 993–998 [DOI] [PubMed] [Google Scholar]
- 50. Cannito S., Novo E., Compagnone A., Valfrè di Bonzo L., Busletta C., Zamara E., Paternostro C., Povero D., Bandino A., Bozzo F., Cravanzola C., Bravoco V., Colombatto S., Parola M. (2008) Carcinogenesis 29, 2267–2278 [DOI] [PubMed] [Google Scholar]
- 51. Li Y. M., Zhou B. P., Deng J., Pan Y., Hay N., Hung M. C. (2005) Cancer Res. 65, 3257–3263 [DOI] [PubMed] [Google Scholar]
- 52. Schnitzer S. E., Schmid T., Zhou J., Eisenbrand G., Brüne B. (2005) FEBS Lett. 579, 529–533 [DOI] [PubMed] [Google Scholar]
- 53. Mottet D., Dumont V., Deccache Y., Demazy C., Ninane N., Raes M., Michiels C. (2003) J. Biol. Chem. 278, 31277–31285 [DOI] [PubMed] [Google Scholar]
- 54. Peinado H., Del Carmen Iglesias-de la Cruz M., Olmeda D., Csiszar K., Fong K. S., Vega S., Nieto M. A., Cano A., Portillo F. (2005) EMBO J. 24, 3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]