Abstract
The exocyst is a protein complex that ensures spatial targeting of exocytotic vesicles to the plasma membrane. We present microarray data obtained from differentiating mouse embryonic stem cell cultures that identify an up-regulation of exocyst complex component 3-like 2 (exoc3l2) mRNA in sprouting blood vessels. Vascular expression of exoc3l2 is confirmed by qPCR analysis of different mouse tissues and immunofluorescence analyses of mouse brain sections. We detect an up-regulation of exoc3l2 mRNA synthesis in primary human endothelial cells in response to VEGFA, and this response is enhanced when the cells are grown on a three-dimensional collagen I matrix. Myc-tagged EXOC3L2 co-precipitates with the exocyst protein EXOC4, and immunofluorescence detection of EXOC3L2 shows partial subcellular colocalization with EXOC4 and EXOC7. Finally, we show that exoc3l2 silencing inhibits VEGF receptor 2 phosphorylation and VEGFA-directed migration of cultured endothelial cells.
Keywords: Cell Migration, Chemotaxis, Exocytosis, Microarray, Receptor Recycling, Angiogenesis, VEGFA
Introduction
The vascular system is an intricate, well defined structure that forms in every fetus with striking accuracy and then has the ability to remodel itself to ensure proper delivery of oxygen and nutrients in every corner of the body. The formation and regulation of the vascular network have to be precise or the organism will face severe consequences such as inadequate oxygen and nutrient delivery. Aberrant vessel growth is a common feature in proliferative retinopathies, psoriasis, atherosclerosis, and tumor growth. The formation of new blood vessels depends on growth factors and morphogens produced by cells surrounding the vessels and by the vascular endothelial cells themselves (1). Members of the vascular endothelial growth factor (VEGF)2 family, several fibroblast growth factors, and the angiopoetins are important regulators of the vascular system as are many ephrins, netrins, and semaphorins (2). It has been shown on numerous occasions that these growth factors can be presented as gradients in the tissue to which the endothelial cells respond, and that they can act as attractants, repellents, or sometimes both (3, 4). The process causing a cell to migrate in response to a morphogen gradient is called chemotaxis and occurs frequently in many cell systems (5). In eukaryotes, the immune cells are probably the most studied in this context due to their extreme mobility, but also endothelial cells have been shown to be sensitive to molecular gradients (6).
Angiogenesis describes the formation of new blood vessels from existing ones. To date there are three different models describing this process: sprouting angiogenesis, intussusceptive angiogenesis, and looping angiogenesis. In sprouting angiogenesis the endothelial cell at the tip of the sprout is responsible for navigation, and thus migrates toward or away from a morphogen source (2). Several receptor-ligand pairs have been identified to participate in this process, most importantly perhaps VEGFR2/VEGFA and NOTCH/DLL4, but many other signaling systems have also been implicated (7, 8).
One of the important in vitro models for sprouting angiogenesis is the embryoid body (EB) model where embryonic stem cells are aggregated to spheroids and stimulated with pro-angiogenic factors to form vascular structures (9). In the present study we have used this model in combination with the microarray technique to identify potential new genes involved in sprouting angiogenesis or cell migration. One gene displaying a very interesting expression pattern was exocyst complex component 3-like 2 (exoc3l2). The name of the gene reflects its structural similarity to other members of the exocyst complex.
The exocyst complex consists of eight subunits (EXOC1–8) and its structure and function seems to be well conserved in species ranging from yeast to mammals. The major function of the exocyst complex is to ensure proper localization of vesicles to the plasma membrane immediately before fusion, which in turn is mediated by the SNARE proteins (10). This makes the exocyst important for processes such as cell polarization and migration, cytokinesis, ciliogenesis, tumor invasion, cell-cell communication, and cell growth (11). The exocyst is also likely to control the assembly of the SNARE complex (12). EXOC3L was discovered as a member of the exocyst complex in 2008 and proposed to be involved in insulin secretion by Saito et al. (13). When it comes to exoc3l2 not much is known about the gene or the corresponding protein. A recent publication associates the exoc3l2 gene locus with late onset Alzheimer disease (14), but this association is probably explained by correlation of exoc3l2 with the Alzheimer risk factor gene APOE (15). A microarray study done by Wallgard et al. (16) suggests an up-regulation of the mouse exoc3l2 homologue in glomerulus and in the vasculature of the brain.
In this paper we describe exoc3l2 in terms of expression pattern and function. We show that endothelial cells in developing blood vessels express elevated levels of exoc3l2, and that the EXOC3L2 protein associates with components of the exocyst complex. Finally, we provide evidence that exoc3l2 function is required for VEGFR2 activation and directional migration of endothelial cells in response to gradients of VEGFA.
EXPERIMENTAL PROCEDURES
Preparation and Culture of EBs and Microarray Study
Murine embryonic stem cells (line R1) (17) were cultured on growth-arrested mouse embryonic fibroblasts in DMEM-Glutamax (Invitrogen) supplemented with leukemia inhibitory factor (1000 units ml−1), 15% fetal bovine serum (Invitrogen), 1.2 mm sodium pyruvate, 25 mm HEPES (pH 7.4), and 19 mm monothiolglycerol. At day 0, stem cells were trypsinized and resuspended in cell medium without leukemia inhibitory factor and thereafter allowed to differentiate in drops hanging from the lid of a non-adherent culture dish (1200 cells/drop) (9). After 4 days the EBs were collected and seeded between two layers of 1.5 mg/ml of collagen I gel (three-dimensional culture), or on a glass slide (two-dimensional culture). Medium was added to give a final concentration of 30 ng of VEGFA/ml of culture and was replaced after 4 days and then every other day for the rest of the culture time. Three-dimensional cultured EBs were harvested for RNA extraction on day 14. Only EBs with clearly visible sprouting (VEGF treated) or outgrowth of cells (untreated) were selected. The entire core region of the EB was isolated using the tip of a Pasteur pipette and the peripheral area was collected afterward. Approximately 20–25 EBs were harvested from each condition. RNA was extracted using the RNeasy quick spin kit (Qiagen). The cDNA synthesis and amplification was carried out according to the two-cycle cDNA synthesis protocol (Affymetrix) using a two-cycle cDNA synthesis kit (Invitrogen) at the Uppsala array platform. A total amount of 100 ng of RNA was used for each reaction. Samples were labeled and hybridized to a GeneChip Mouse Expression 430 array (Affymetrix) according to the manufacturer's instructions. Hybridization was performed automatically using a Fluidics Station 450 (Affymetrix) and scanned using the GeneChip® Scanner 3000 7G (Affymetrix). Analysis of the gene expression data were conducted in the statistical computing language R using packages available from the Bioconductor project. Normalization of raw data were done using the robust multiarray average (18).
Isolation of Endothelial Cells from Mouse Organs
Embryos were dissected at days 11.5 or 14.5 and adult organs were taken from male mice not more than 2 years of age. Lungs, kidneys, and brain were removed and cut to pieces with a scalpel. The organs were then incubated end-over-end in Hank's balanced salt solution, 1% BSA, 1× glucose, 100 units of DNase, 0.7 mg/ml of hyaluronidase for 15 min at 37 °C. Samples were homogenized with a 21-gauge syringe and passed through 100- and 40-μm filters consecutively and then centrifuged at 16,000 × g for 5 min at 4 °C. 100 μl of 1:1 Dynabead slurry (goat anti-rat preincubated with rat anti-PECAM antibody) was added to each sample and incubated end-over-end for 30 min at 4 °C. The beads were collected and washed 5× 5 min in PBS and the supernatant was centrifuged for 20,000 × g for 5 min in 4 °C. Beads and supernatant were lysed using 350 μl of lysis buffer (Qiagen) and RNA was prepared with RNeasy quick spin kit (Qiagen) according to the manufacturer's instructions.
Cell Culture Conditions
Primary endothelial cell cultures (HMVEC and HUVEC) were purchased from Promocell and maintained in EGM-2-MV (full medium) (Lonza) until passage six. Human fetal lung fibroblasts (HFL1) (ECACCI) and immortalized human fibroblasts (IMR-90) (19) were maintained in modified Eagle's medium supplemented with 10% FBS, 2 mm Glutamax, 1 mm sodium pyruvate, 25 mm HEPES, and non-essential amino acids. The murine fibroblast cell line NIH-3T3 was maintained in Dulbecco's modified Eagle's medium (high glucose, Invitrogen) supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Sigma). The human T-lymphocyte cell line Jurkat (ATCC) was maintained in RPMI 1640 + Glutamax (Invitrogen), supplemented with 10% FBS and 1% penicillin/streptomycin (Sigma). The embryonic stem cell line E14/T was cultured in the absence of feeder cells on gelatin-coated (Sigma) cell culture plastic (Corning). Cells were maintained in Glasgow modified Eagle's medium (Sigma) supplemented with 5% fetal calf serum (Invitrogen), 5% KnockOutTM serum replacement (Invitrogen), 2 mm glutamine (Invitrogen), 1× nonessential amino acids (Invitrogen), 1 mm sodium pyruvate (Invitrogen), 0.1 mm 2-mercaptoethanol (Sigma), and 1000 units/ml of leukemia inhibitory factor (Millipore). Cells were grown in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. Medium was changed every day and cells were split using 0.05% trypsin/EDTA (Invitrogen) every 2 days to prevent a too high confluence.
qPCR
Cells were lysed on the plate using Qiagen lysis buffer and RNA was prepared with the RNeasy quick spin kit (Qiagen) according to the manufacturer's instructions. cDNA synthesis was performed with SuperScript (Invitrogen) reversed transcriptase. SYBR Green (Bio-Rad) was used for the qPCR. The following primers were used: ExoC3l2 human, forward, ACCCAGACATCAGGCAGAAG, reverse, AGGGATGTCTGCAAAGAAGG; ExoC3l2 mouse, forward, TCTGAAAGTGGTGCCCTACC, reverse, AGGAGGGTCTATCAGCAGCA; ExoC3, forward, TCAAAGACATCCAGCAGTCG, reverse, GGGTCTCCCTCACAATCTCA; ExoC3l, forward, TCCAGGCAAGTGTGTCTCAG, reverse, GGCCACACGAATGTTCTCTT; ExoC6, forward, GGGAAAGTTGCTCAGACAGC, reverse, CAGAGCTGGCAAACAATTCA; ExoC2, forward, TACCATGGACGAAGGTCACA, reverse, CATTTTTGGCTTGTCCCACT; ExoC1, forward, TCTGGCAGTCAAACAGCAAC, reverse, CTTGGCATATCGGAGCAAAT; ExoC7, forward, CATGGGTTATCAGGGGATTG, reverse, GAGGTCCAGGTGTGGGTAGA; PECAM, forward, CTCCAACAGAGCCAGCAGTA, reverse, GACCACTCCAATGACAACCA; β-actin human, forward, TCTACAATGAGCTGCGTGTG, reverse, AGCCTGGATAGCAACGTACA; β-actin mouse, forward, AAGAGCTATGAGCTGCCTGA, reverse, TACGGATGTCAACGTCACAC.
Immunofluorescence
Samples were rinsed in ice-cold PBS followed by a 10- (monolayers) or 15-min (EBs) incubation with 4% paraformaldehyde on ice. After this step all incubations were performed at room temperature if not stated otherwise. Samples were then briefly rinsed with PBS, incubated for 5 min with 0.2% Triton in PBS, and washed 3× 5 min with 0.1% Triton in TBS. Before blocking for 1 h in TNB blocking buffer (PerkinElmer Life Sciences) samples were rinsed with TBS. Primary antibody was diluted as indicated below in TNB and incubated overnight at 4 °C. Following incubation the samples were washed 2× 5 min in 0.1% Triton TBS, rinsed with TBS, and incubated with secondary antibody for 1 h. The samples were washed 3× 5 min in 0.1% Triton TBS followed by a 5-min incubation in TBS with Hoechst and finally mounted in fluoromount and stored at 4 °C. The following antibodies were used for immunofluorescence stainings: anti-EXOC4; Abcam (ab13254) mouse monoclonal (1:50), anti-EXOC7; Abcam (57402) mouse monoclonal (1:200), anti-EXOC3L2 (central part); Santa Cruz (sc-136672) goat polyclonal (1:100), anti-EXOC3L2 (N-terminal); Storkbio (raised against peptide QEDEEHWGSLEDQPSSLA) rabbit polyclonal (10 μg/ml), anti-PECAM; BD Pharmingen (553370) rat monoclonal (1:1000), anti-GFAP; Sigma (G3893) mouse monoclonal (1:200), anti-VEGFR2; Santa Cruz (sc-505) rabbit polyclonal (1:50), anti-pVEGFR2(Tyr-1175); Cell Signaling Technology (number 2478) rabbit monoclonal (1:1000), anti-GAPDH; Santa Cruz (sc-25778) rabbit polyclonal (1:1000), anti-β-actin; Abcam (ab6276) mouse monoclonal (1:5000), and anti-Myc tag (9B11); Cell Signaling Technology (number 2276) mouse monoclonal (1:1000). Alexa Fluor secondary antibodies were used diluted 1:500. TRITC-phalloidin was used to stain F-actin. Images were captured with an inverted confocal microscope (LSM 700, Zeiss) using a Plan-Apochromat ×20/0.16 or a C-Apochromat ×63/1.2 objective. Zen 2009 software (Zeiss) was used for acquisition and image processing.
SDS-PAGE and Western Blotting
Cells were lysed on the plate using 0.1% Nonidet P-40 in Tris buffer (pH 7.4), passed 3 times through a 27-guage needle, and centrifuged at 15,000 × g for 30 min at 4 °C. The lysates were heated to 95 °C with SDS sample buffer containing β-mercaptoethanol, separated on a 10% acrylamide gel (1.5 mm), and transferred using wet transfer at 100 V for 2 h at 4 °C. The membrane was blocked for 1 h in Odyssey blocking buffer (LiCor) and incubated with primary antibody overnight at 4 °C. The primary antibodies used are described under “Immunofluorescence.” Secondary antibodies (Alexa Fluor 680 (Invitrogen) or IR-dye 800 (Rockland)) were incubated at RT for 1 h. Membranes were developed using the Odyssey Imaging System (LiCor) and the Odyssey 2.1 software was used to quantify the intensity of the bands.
Preparation of Brain Sections from Adult Mice
Adult C57bl6 mice were put under deep anesthesia and transcardially perfused with 4% paraformaldehyde. Brains were extracted and postfixed in 4% paraformaldehyde (overnight, 4 °C). After fixation the brains were cryopreserved in 30% sucrose in PBS (overnight, 4 °C) and then snap frozen in isopentane. The brains were sectioned in 14-μm coronal sections with a cryostat.
Transfection of the EXOC3L2-ORF Containing Plasmid and Immunoprecipitation
The exoc3l2-ORF plasmid was purchased from Origene (RC211213) and was propagated in Escherichia coli. HMVECS (p3–5) were transfected when they were ∼75% confluent. 9 μg of plasmid DNA (exoc3l2-ORF or pmaxGFP (Lonza)) and 18 μl of a non-lipid polymer transfection reagent (MegaTran, Origene) were used to transfect one 10-cm plate containing 7.2 ml of fresh EGM-2-MV. The cells were incubated for 24 h and then lysed with 0.1% Nonidet P-40 in Tris buffer (pH 7.4), passed 3 times through a 27-guage needle, and centrifuged at 15,000 × g for 30 min at 4 °C. The lysates were incubated end-over-end with or without primary antibody overnight at 4 °C. 50 μl of 1:1 bead slurry (sheep anti-mouse preincubated with mouse anti-Myc antibody) was added to each sample and incubated end-over-end for 1 h in 4 °C. The supernatant was removed and stored at −20 °C. The beads were washed 3× 5 min plus 2× 30 min in PBS at 4 °C and then collected and lysed in SDS sample buffer and stored at −20 °C.
siRNA Transfection
For each reaction 4 μl of siRNA (50 μm) was transfected to ∼300,000 cells with the Amaxa electroporation system. The following siRNAs were used: siexoc3l2-1 (Ambion, 42659) targeting exon 8,9, siexoc3l2-2 (Ambion, s40336) targeting exon 1,2 and negative control (Ambion, AM611).
Microfluidic Cell Migration Assay and Stimulations with VEGFA
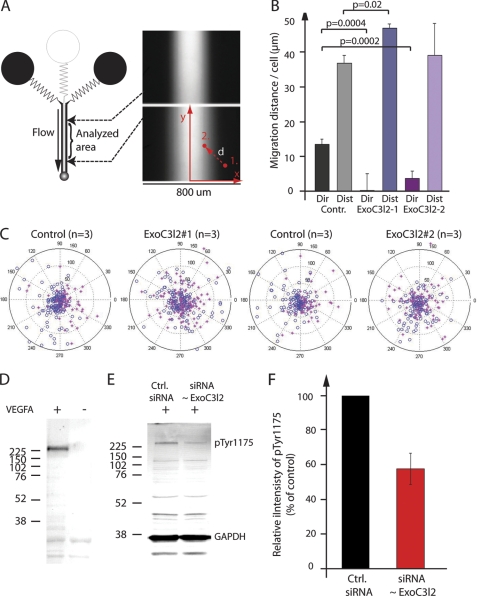
HMVECs were transfected as described above and seeded on a 3.5-cm culture dish precoated with 1% gelatin in PBS. After 24 h, the cells were starved in 0.2% BSA in endothelial basal medium (Lonza) for 16–20 h after which the cells were stimulated for 5 min with 50 ng/ml of VEGFA (for measurement of VEGFR2 phosphorylation) or exposed to a gradient ranging from 0 to 20 ng/ml of VEGFA in endothelial basal medium (for measurement of directed cell migration), as previously described (6). The gradient was produced by a continuous flow of medium from three different channels of which the middle one contains 20 ng/ml of VEGFA. When the three channels fuse into one big channel a hill-shaped gradient is formed by diffusion (see Fig. 7A). If the cells are attracted by the gradient they will migrate toward the center of this channel. The individual trajectory of each cell is recorded as described in Fig. 7A. Cell migration was observed with a Zeiss Axiovert 200 microscope equipped with temperature and CO2 controls, keeping the cells at 5% CO2 and 37 °C. Software for time-lapse imaging and cell tracking was from AxioVision (Zeiss). Phase-contrast images of cells were taken every 5 min through a ×10 objective (Plan-Neofluar, NA 0.3, Zeiss). Migration in response to the gradient was scored in three independent experiments. In each experiment, data were collected from at least 5 different non-overlapping fields of cells growing in the chamber over a period of 2.5 h.
FIGURE 7.
Exoc3l2 is required for directional endothelial cell migration in response to a gradient of VEGFA. A, graphic overview of the microfluidic system used to generate VEGFA gradients (right panels). Cells that are attracted by the growth factor will migrate toward the center of the channel. A cell that moves from position 1 to 2 will record a distance (length of the vector d) and a direction (angle of d). B, two siRNA were used to reduce exoc3l2 expression. Cells were exposed to a VEGFA gradient ranging from 0 to 20 ng/ml, and the average directionality (dir) and distance (dist) of the cells were measured for each experiment. Error bars represent S.D. by Student's t test (p < 0.05). C, polar plots of the distance and direction of each cell. Cells on the left side of the chamber are represented by red stars and cells on the right side by blue circles. A separation of the stars from the circles indicates that the cells receive directional information from the gradient. D, Western blot of pVEGFR2 (Tyr-1175) after VEGFA stimulation for 5 min. Total cell lysates from HMVECs were used for all experiments. E, VEGFA-induced VEGFR2 phosphorylation following siRNA transfection to suppress EXOC3L2 expression. F, quantification of the results in E. Error bars represent S.D. (n = 3).
RESULTS
Gene Expression Profiling of EB-derived Sprouts
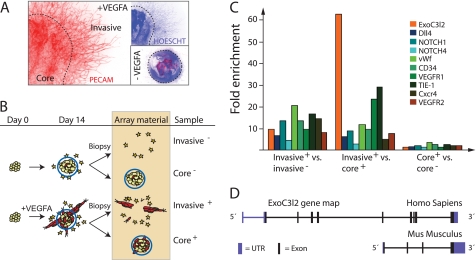
To analyze the gene expression profile of growing and actively invading vascular sprouts, we performed a microarray study where we compared different fractions of VEGF-stimulated and control EB cultures. The following four fractions were analyzed: Invasive+ was defined as the fraction of VEGFA-stimulated EB cultures that contained blood vessels and cells migrating out from the EB core. Stationary cells of the EB core were defined as the core+ fraction. Unstimulated EBs were used as controls, in which the fractions were labeled invasive− and core−, respectively (Fig. 1, A and B). Genes such as Tie1, Pecam, Cxcr4, Vwf, CD43, Notch4, Notch1, Dll4, Vegfr1, and Vegfr2 were all up-regulated in the invasive+ fraction and highly ranked in terms of p value. These genes have previously been implicated in angiogenesis, which lends credibility to the present array strategy as a method to predict genes involved in vessel formation (Fig. 1C). Of note, a similar microarray experiment where tube-forming endothelial cells harvested from three-dimensional collagen cultures were compared with proliferating endothelial cells cultured on fibronectin, recently identified tie1, vegfr1, and notch4 as differentiation-regulated genes (20). A list of the top 1000 genes from the present study, ranked by p value, is available under supplemental Table S1.
FIGURE 1.
Microarray analysis of differentiating EBs identifies genes that are highly expressed in invading vascular sprouts. A, the samples were collected from EBs after 14 days in culture. The core was separated from the invading cells as indicated by the dashed line. B, schematic drawing of a sample preparation for the microarray study. C, summary of the relative expression of a number of genes previously implicated in angiogenesis along with the expression pattern of exoc3l2. Invasive+ versus invasive− compares the sprouts and invading cells from the VEGFA-stimulated cultures to the invading cells of the unstimulated control. Invasive+ versus core+ compares the gene expression of the sprout fraction to that of the core fraction in VEGFA-stimulated cultures, and core+ versus core− compares the two core fractions (n = 3 for all samples). D, map of the exoc3l2 gene in human (chromosome 19, 9 exons, ENST00000252482) and mouse (chromosome 7, 4 exons, ENSMUSG00000011263). The mouse gene lacks the N-terminal equivalent of the human gene, but in the C-terminal region the similarity between the human and mouse sequences is around 85%.
A largely uncharacterized gene that showed a 60-fold up-regulation when comparing the invasive+ fraction to the core+ fraction, and a 10-fold up-regulation when comparing the two invasive fractions, was exoc3l2 (Fig. 1C). This gene, which is located on chromosome 19 in humans, is also known as HBV X-transactivated gene 7. It is predicted to produce a 409-amino acid long protein that is related to exocyst complex component 3 (EXOC3, also known as SEC6). In the mouse exoc3l2 is located on chromosome 7 and the predicted protein is much shorter than the human homolog; around 250 amino acids (Fig. 1D). The structural relationship between EXOC3, EXOC3L, and EXOC3L2 is obvious from the tertiary structure prediction shown in supplemental Fig. S1, but in contrast to EXOC3L2, EXOC3 showed no differential expression in the microarray analysis. This was also the case for the other exocyst complex components suggesting a specialized function for EXOC3L2 in developing blood vessels (supplemental Table S1).
Expression of exoc3l2 mRNA and Protein in Endothelial Cells
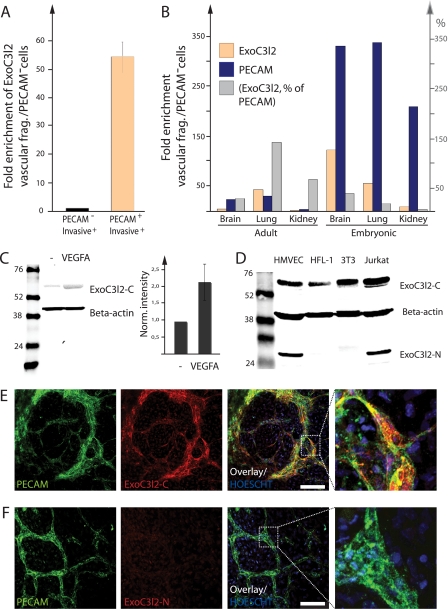
Next we wanted to analyze exoc3l2 expression in the invasive+ fraction of the EBs as well as to evaluate the expression of exoc3l2 in the vasculature in vivo. We used magnetic beads coated with antibodies against the vascular marker PECAM to split the invasive+ fraction into two subfractions: the PECAM positive cells (endothelial cells) and the PECAM negative cells (PECAM−). exoc3l2 mRNA was shown to be up-regulated more than 50-fold in endothelial cells compared with PECAM− cells (Fig. 2A). The elevated exoc3l2 mRNA expression in the endothelial cell population of the EB was observed already after 6 days in culture (day 2 in collagen, see “Experimental Procedures”) at which point no sprouts are visible (supplemental Fig. S2, A and B). The PECAM-coupled magnetic beads were also used to isolate endothelial cells from adult and embryonic mouse lung, kidney, and brain. exoc3l2 mRNA was enriched in the endothelial cell fraction compared with the PECAM− fraction in all tissues examined (Fig. 2B). The level of Pecam is shown as an indicator of isolation efficiency and the gray bars represent exoc3l2 mRNA expression in percent of Pecam mRNA expression.
FIGURE 2.
Expression pattern of exoc3l2 mRNA and protein. A, vascular fragments were extracted from the invasive+ fraction using magnetic beads coated with PECAM antibodies to show that endothelial cells were responsible for most of the exoc3l2 expression. Bars represent S.D. (n = 3). B, the expression levels of exoc3l2 in vascular fragments isolated from adult and embryonic mouse organs as measured by qPCR (orange bars, the results are normalized against PECAM−). PECAM expression was analyzed in parallel to assess the purity of the extraction (blue bars). The gray bars represent the expression levels of exoc3l2 in percent of PECAM expression. C, total cell lysates from feeder-free mouse embryonic stem cells (E14/T) were analyzed for EXOC3L2 protein expression by Western blot (left). The expression level in undifferentiated cells was low but increased upon differentiation for 5 days in the presence of 50 ng/ml of VEGFA (right). D, EXOC3L2 protein expression was analyzed in human endothelial cells (HMVEC), human fibroblasts (HFL1), mouse embryonic fibroblasts (3T3), and human T-cells (Jurkat). The larger EXOC3L2 protein band was detected in all cell lysates analyzed, whereas the smaller version was only detected in human endothelial cells and human T-cells. E, murine EBs were grown on glass slides in VEGFA-supplemented medium and analyzed for PECAM and EXOC3L2 expression. Co-staining for ExoC3L2-C and PECAM demonstrated that endothelial cells expressed high levels of EXOC3L2. F, ExoC3L2-N raised against the N-terminal region of human EXOC3L2 failed to recognize the mouse homologue, which lacks the N-terminal equivalent of the human gene.
The expression of EXOC3L2 protein was analyzed by Western blot using two different antibodies, one directed against the N-terminal domain of the human protein, denoted ExoC3l2-N, and one antibody directed against the central part, denoted ExoC3l2-C. The exoc3l2 mRNA is expected to produce a protein of 409 amino acids (∼45 kDa). However, when used for Western blotting of total cell lysate from human microvascular cells, neither of the antibodies could detect a band of that size. Instead there was one band with a molecular mass of ∼65 kDa detected by ExoC3l2-C, and one band with a molecular mass of ∼28 kDa detected by ExoC3l2-N (supplemental Fig. S3A). Both antibodies, however, recognized EXOC3L2 when it was overexpressed as a Myc-tagged fusion protein (supplemental Fig. S3A). When human microvascular endothelial cells (HMVECs) were transfected with siRNA against exoc3l2 the intensities of both the larger and smaller band were reduced compared with the control (supplemental Fig. S3, B and D), but further studies are needed to identify all the endogenous variants (sizes) of the EXOC3L2 protein.
In mouse ES cells, the expression of the larger EXOC3l2 protein was induced by differentiation in the presence of VEGFA (Fig. 2C). The smaller protein could not be detected because the mouse protein lacks the N-terminal domain of the human homologue (recognized by ExoC3l2-N). The larger EXOC3l2 protein was detected in all cell types investigated, whereas the shorter was expressed only in human endothelial cells and T-cells (Fig. 2D). Western blot analysis also revealed an increase in EXOC3l2 protein expression in the invasive+ fraction of the mouse EBs (supplemental Fig. S3, E and F).
Vascular structures in two-dimensional EBs (see “Experimental Procedures”) were identified by immunofluorescence analysis using an antibody against the vascular marker PECAM and the EBs were counterstained for EXOC3L2. ExoC3l2-C staining was enriched in the vascular structures (Fig. 2E), whereas ExoC3l2-N did not recognize the mouse homologue of EXOC3L2 (Fig. 2F) as expected. The specificity of the antibodies was tested in a competition experiment with the peptides that were used for immunization (supplemental Fig. S3G).
VEGFA and Collagen I Stimulates exoc3l2 Expression
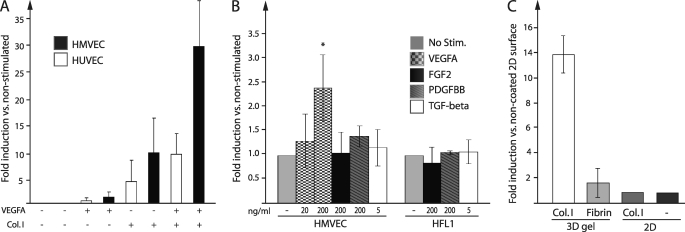
To further characterize the expression of exoc3l2 we used primary endothelial cells from human foreskin (HMVECs) or from the umbilical cord (HUVECs), which were grown in monolayers. Initially, we found that exoc3l2 mRNA was not highly expressed in the cultured endothelial cells when grown under standard conditions. In fact, the expression levels of exoc3l2 in endothelial cells were comparable with the levels of human fibroblast cell lines IMR-90 and HFL1 (supplemental Fig. S4A). This was rather puzzling considering the high expression levels detected in the endothelial cell fraction of the EBs, why we again measured the expression trying to mimic the conditions in the EB culture. In both HMVECs and HUVEC there was an increased expression of exoc3l2 mRNA upon stimulation with VEGFA and/or when cells were cultured on a gel composed of collagen I, effects that were clearly additive (Fig. 3A). The cells were seeded on top of the collagen I gel but after 24 h most of the cells had adopted a morphology suggesting that they had infiltrated the gel. The induction of exoc3l2 expression by collagen I culture was not seen in the two fibroblast cell lines (supplemental Fig. S4B) and FGF2, PDGF-BB, and TGF-β could not induce a significant increase of exoc3l2 mRNA in any of the cell lines investigated (Fig. 3B).
FIGURE 3.
Exoc3l2 mRNA expression is induced by collagen I and VEGFA. A, induction of exoc3l2 mRNA expression in primary endothelial cells (HMVEC and HUVEC) as measured by qPCR. Cells were seeded on cell culture-treated plastic or onto a collagen I gel and incubated for 24 h in full medium with or without addition of 200 ng/ml of VEGFA. Error bars represent S.D. (n = 3). B, the induction of exoc3l2 mRNA expression following 24 h stimulation with different growth factors. The fibroblast cell line HFL1 was used for comparison. The significance of the results was assessed by Student's t test (p < 0.05) (n = 3). C, the effect of different substrates on exoc3l2 expression in HMVECs as measured by qPCR (n = 3).
To test if any type of three-dimensional matrix could trigger the expression we seeded the cells on a fibrin gel. This caused a small up-regulation of exoc3l2 mRNA but it was not comparable with the strong response seen when the cells were seeded on collagen I. Growing the cells on a two-dimensional surface coated with collagen I was also not enough to cause up-regulation of exoc3l2 mRNA synthesis (Fig. 3C). qPCR analysis of the expression levels of the other exocyst complex components upon collagen I and VEGFA stimulation revealed no significant differences save an about 2-fold induction of exoc3l mRNA (supplemental Fig. S4C).
Immunofluorescence Analysis Indicates Association between EXOC3L2 and the Exocyst Complex
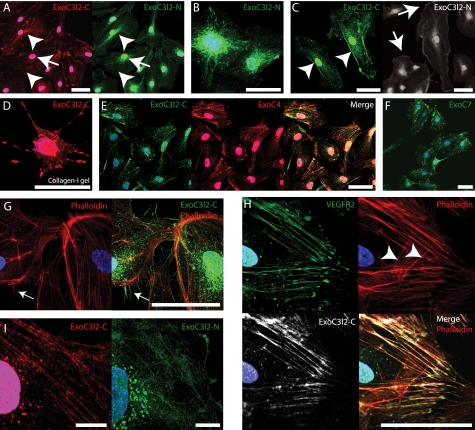
To analyze the subcellular distribution of EXOC3L2 the antibodies against EXOC3L2 were used for immunofluorescence analysis. The antibodies raised against the N-terminal and the central part of EXOC3L2, respectively, both stained HMVECs, and ExoC3l2-C could also be used to visualize the protein in human and mouse fibroblasts. ExoC3l2-N on the other hand failed to stain fibroblasts from mouse and only stained human fibroblasts quite weakly (supplemental Fig. S5). In HMVECs both antibodies stained stress fibers (Fig. 4A, arrowheads), vesicles (arrows), and nuclei. However, the staining patterns varied between cells and some cells lacked membrane staining but instead had a dot-like pattern around the nucleus (Fig. 4B). Altered distribution of the EXOC3L2 protein was also obvious during cell division (supplemental Fig. S6A). The ExoC3l2-N antibody mainly stained the protruding parts of the endothelial cell membranes (Fig. 4C, arrows), whereas the ExoC3l2-C antibody also stained the lateral cell membranes (arrowheads).
FIGURE 4.
The EXOC3L2 protein partially co-localizes with EXOC4 and VEGFR2. A–C, HMVECs grown on a two-dimensional surface and stained with antibodies directed against the central part (ExoC3l2-C) or the N-terminal part (ExoC3l2-N) of the EXOC3L2 protein sequence. Both antibodies stain nuclei, perinuclear vesicles (A, arrowheads), and stress fibers (A, arrows), but have a slightly different appearance in the peripheral regions. ExoC3l2-C stains the lateral part of the cell membrane (C, arrowheads), whereas anti-ExoC3l2-N stains the more dynamic, expanding or retracting, regions (C, arrows). D, HMVEC grown in collagen I gel stained for ExoC3l2-C. E and F, ExoC3l2-C co-localizes to a large extent with EXOC4 (Overlap coefficient = 0.75 ± 0.07) (E), which is also found predominantly associated with stress fibers and vesicles, whereas EXOC7 (F) shows a different staining pattern. G, filopodia stained with ExoC3l2-N (arrow). Actin fibers are visualized with phalloidin. H, ExoC3l2-C and VEGFR2 antibodies stain longitudinal actin fibers but not latitudinal (arrowheads) (overlap coefficient = 0.73 ± 0.04). Scale bars = 50 μm. I, ExoC3l2-C and ExoC3l2-N staining in ×63 magnification. Scale bars = 10 μm.
Because the expression of exoc3l2 was dramatically increased when the cells were cultured on collagen, we also wanted to study how the subcellular localization of the protein changed under these conditions. When the cells were cultured in a collagen I gel the expression pattern of EXOC3L2 was indeed altered, featuring a patchy staining with accumulation in the protruding structures (Fig. 4D).
Most of the exocyst complex components are associated with exocytotic vesicles and depend on actin cables to reach the sites of exocytosis. The polarization of EXOC7, however, is partially actin-independent and instead of being transported with the exocytotic vesicle this molecule rather sits at the plasma membrane waiting for the cargo to arrive (21, 22). In agreement with this model the antibody recognizing EXOC4 stained actin fibers and vesicles, whereas the expression pattern of EXOC7 was more restricted to hot spots in the peripheral regions of the cell (Fig. 4, E and F, supplemental Fig. 6, B and C). The fact that EXOC3L2 colocalized to a large extent with EXOC4, suggests that EXOC3L2 is somehow associated with the core of the exocyst complex. It has previously been shown by Sugihara et al. (23) that the exocyst is involved in filopodia formation and staining using the ExoC3l2-N antibody identified EXOC3L2 protein throughout filopodia-like structures (Fig. 4G). When phalloidin was used to visualize actin filaments it was obvious that ExoC3l2-C recognized actin filaments oriented predominantly in one direction (Fig. 4H); this pattern was also seen when costaining for VEGFR2. ExoC3l2-C and ExoC3l2-N stainings in ×63 magnification are shown in Fig. 4I.
EXOC3L2 Is Expressed in Larger Vessels in the Mouse Brain
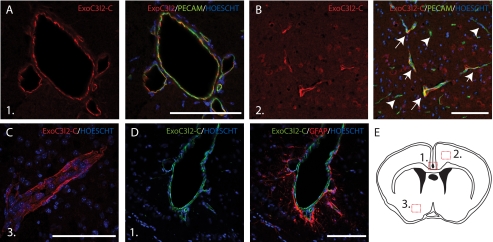
To investigate the expression pattern of EXOC3l2 in vivo we performed immunofluorescence analyses on coronal cryosections from adult mouse brains. The brain sections were stained for EXOC3l2 together with PECAM and expression of EXOC3l2 was detected in endothelial cells, particularly in the larger vessels (Fig. 5, A and B). The staining seemed to be evenly distributed in the vessel wall (Fig. 5C). The perivascular astrocytes on the other hand did not express EXOC3l2 (Fig. 5D).
FIGURE 5.
EXOC3L2 is expressed in the vasculature of the mouse brains. Adult mouse coronal brains were fixed in 4% paraformaldehyde, cryopreserved in 30% sucrose in PBS, and then snap frozen. 14-μm sections were stained for EXOC3L2 and PECAM and analyzed with confocal microscopy. The images show EXOC3L2 expression predominantly in the large vessels (A), but also in some of the smaller vessels (B, arrows), whereas others express PECAM only (B, arrowheads). EXOC3L2 seems to be distributed evenly in the vessel wall (C). Astrocytes (GFAP positive) surrounding the vessels do not express EXOC3L2 (D). E, schematic of a coronal cross-section, showing the location of the investigated areas in panels A–D.
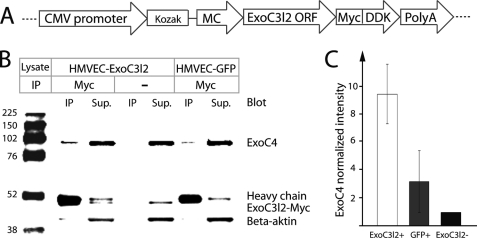
EXOC4 Co-precipitates with Myc-tagged EXOC3L2 Protein
The subcellular localization and the colocalization with EXOC4 indicate an association between EXOC3l2 and the exocyst complex. To verify this we overexpressed Myc-tagged human EXOC3L2 in HMVECs and used an antibody against the Myc tag for immunoprecipitation (Fig. 6A). The precipitates were analyzed by Western blot, and EXOC4 was found to co-precipitate with the EXOC3L2 fusion protein (Fig. 6, B and C). As negative control we used cells overexpressing GFP. These data support the hypothesis that the human EXOC3L2 protein interacts with the exocyst complex.
FIGURE 6.
EXOC3L2 physically interacts with EXOC4. A, the construct used to express Myc DDK-tagged human EXOC3L2 in endothelial cells (HMVEC). B, Western blot of co-immunoprecipitation (IP) of EXOC4 using the Myc-tagged EXOC3L2 as bait. The IP was also performed with no antibody or with HMVECs transfected with a GFP expressing plasmid (HMVEC-GFP) as negative controls. C, quantification of the data shown in B. The intensity of EXOC4 was normalized against intensity of β-actin in the supernatant and to the normalized intensity of the negative control. Error bars represent S.D. (n = 3).
EXOC3L2 Is Required in HMVECs for Migration in Response to a VEGFA Gradient
The exocyst plays a role in cell polarization and we therefore investigated the effect of exoc3l2 silencing on endothelial cell chemotaxis using siRNA-mediated gene silencing and a microfluidic cell migration assay (6) (Fig. 7A). In this assay it is possible to monitor and measure directed migration of endothelial cells in stable growth factor gradients. siRNA against exoc3l2 was shown to down-regulate transcript levels in endothelial cells by 30–70% after 24 h (supplemental Fig. S3C). After 48 h a 50–80% down-regulation of the EXOC3L2 protein level was observed (supplemental Fig. S3D). We detected no obvious phenotype in endothelial cell morphology, proliferation, or apoptosis, but there was a significant difference in chemotaxis in response to a gradient of VEGFA (Fig. 7, B and C). Two different siRNAs directed against different parts of the exoc3l2 mRNA were tested; both siRNAs were shown to reduce directional cell migration, but not to have an impact on the total distance migrated per cell. Transfection with one of the siRNAs even resulted in a slight increase of the average total migrated distance and this siRNA also gave the most severe phenotype in terms of reduced directional cell migration.
Cell migration in response to VEGFA is dependent on localized activation, internalization, and recycling of VEGFR2. We hypothesized that EXOC3L2 may be involved in the exocytosis of VEGFR2, and that a down-regulation of EXOC3L2 expression thus could have an effect on VEGFR2 phosphorylation. Indeed, siRNA silencing of ExoC3l2 was shown to reduce phosphorylation of VEGFR2 at tyrosine residue 1175 by ∼40% (Fig. 7, D–F).
DISCUSSION
In this paper we present a microarray study that identifies exoc3l2 as possibly involved in sprouting angiogenesis. exoc3l2 displays an expression pattern that resembles the profile of several genes already implicated in angiogenesis, and taking this observation as a starting point we have performed a series of experiments investigating exoc3l2 in terms of expression, regulation, and function. Although there are structural differences between the human and mouse exoc3l2 homologues, the vascular expression pattern seems conserved indicating that exoc3l2 is indeed important for vessel formation and/or function.
The subcellular localization of EXOC3L2, the effect on VEGFR2 phosphorylation, and the co-precipitation with EXOC4 all support the hypothesis that EXOC3L2 is associated with the exocyst. EXOC3L2 is predicted to be very similar to EXOC3 structurally and could be expected to have similar interaction partners (supplemental Fig. S1). However, EXOC3L2 lacks the N-terminal equivalent of EXOC3 and it has been shown by Sivaram et al. (24) that removing this part has an effect on some of the binding properties of EXOC3. Our data also suggest that the endogenous form of EXOC3L2 might be different from the protein produced from the cDNA ORF used to overexpress the protein.
It has previously been shown that the exocyst plays an important role in receptor trafficking and membrane shuffling (25). In endothelial cells, recycling and localization of VEGFR2 are important for VEGFA-directed angiogenesis and it is likely that EXOC3L2 is involved in this process. Supporting this hypothesis is the reduction in VEGFR2 phosphorylation and gradient responsiveness observed after siRNA-mediated silencing of exoc3l2. Loss of exocyst function has previously been shown to trap internalized cargoes in recycling endosomes (26, 27). The fact that the overall movement was not reduced after exoc3l2 silencing indicates that the cells were having trouble reading the gradient but did not experience a global migration problem, as they were still able to produce a front and a rear. Previous studies at the molecular level involving Rho family proteins indeed separate directed movement and motility. For example, it is possible for cells lacking functional Cdc42 or Rac to move about but not to sense direction (28).
Data presented in supplemental Fig. S6 indicate that the expression pattern of EXOC3L2 is altered during cytokinesis. This is in agreement with data presented by Murthy et al. (29). EXOC3L2 is clearly redistributed during the process of cell division, supporting an involvement of the protein in polarization, but does not appear to localize to the newly formed membrane between the daughter cells as was observed for EXOC4 by Chen et al. (30). Considering also that EXOC3L2 is found in larger, mature vessels it is possible that the protein plays a role in regulating, e.g. apical-basal polarity of endothelial cells.
As opposed to the other growth factors tested, VEGFA was able to increase the expression of exoc3l2 mRNA about 2-fold in cultured endothelial cells. However, this up-regulation was significantly higher when the cells were cultured on a three-dimensional collagen I gel or when the two treatments were combined. One explanation for this could be that the cells up-regulate exoc3l2 when they switch from two- to three-dimensional migratory mode, but this hypothesis was ruled out as the sole explanation when the cells failed to respond in a similar way when infiltrating a fibrin matrix. If, on the other hand, the increase in exoc3l2 expression was due to interactions with collagen-specific motifs, these have to be presented to the cell in three-dimensional fashion because the mRNA levels of exoc3l2 did not change when the cells were shifted to a collagen I coated two-dimensional surface. Another possible explanation is that exoc3l2 expression depends on matrix stiffness (31, 32). The staining pattern of EXOC3L2 was also altered when the cells were shifted from plastic to collagen gel. This shift was probably not caused by the differences in expression levels as this pattern has been observed for other components of the exocyst. In cells with no extensions, the staining is mostly located in the perinuclear region and in the plasma membrane and in cells with protrusions or extensions, the complex is found perinuclearly as well as in specific locations in the extensions (11). However, none of the other exocyst complex components showed signs of regulation upon stimulation with collagen and VEGFA.
Finally we need to address the specificity of the EXOC3L2 antibodies used in this study, and the apparent difference in size between the detected proteins. Both antibodies raised against EXOC3L2 detect one strong band when used for Western blotting of total endothelial cell lysate, but the bands differ greatly in size. When cells were expressing the Myc-tagged exoc3l2 ORF both antibodies recognized the Myc-tagged protein as well as the endogenous band (supplemental Fig. S4A). The artificial protein was slightly smaller than the protein recognized by ExoC3l2-C but about twice the size of the protein recognized by ExoC3l2-N. The identity of the bands was confirmed with siRNA, but the effect was larger on the smaller band (supplemental Fig. S3). Although both degradation and alternative splicing could produce these differences between the endogenous proteins and the Myc-tagged EXOC3L2, it is puzzling that one of the detected proteins is larger than the artificial construct. It is also of some concern that the large protein seems to have the same size in mouse and human samples. Worth mentioning is that the exoc3l2 ORF construct used to overexpress EXOC3L2 only contains the exons and thus does not allow for alternative splicing or post-translational modifications encoded by the introns. The fact that the smaller protein is not present in mouse cells is expected. It is, however, interesting that this protein is expressed in human endothelial cells and T-cells, which both express VEGFR2, but not in human fibroblasts. When used to stain paraformaldehyde-fixed cells, the antibodies produce similar staining patterns of expected structures; however, there are some distinct differences that could be explained by the existence of different splice variants. It remains to be determined if and to what extent different isoforms of EXOC3L2 exist, and whether the different isoforms may have diverse functions.
Supplementary Material
This work was supported by grants from the Swedish Research Council, Swedish Cancer Society, Swedish Childhood Cancer Foundation, Swedish Foundation for Strategic Research, and Uppsala University (to J. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- VEGFA
- vascular endothelial growth factor A
- VEGFR
- vascular endothelial growth factor receptor 2
- EB
- embryoid body
- EXOC3L2
- exocyst complex component 3-like 2
- EXOC4
- exocyst complex component 4
- EXOC7
- exocyst complex component 7
- PECAM
- platelet endothelial cell adhesion molecule
- HUVEC
- human umbilical vein endothelial cell
- HMVEC
- human microvascular endothelial cell
- HFL1
- human fetal lung fibroblast
- qPCR
- quantitative PCR
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Lundkvist A., Lee S., Iruela-Arispe L., Betsholtz C., Gerhardt H. (2007) Novartis Found. Symp. 283, 194–201 [DOI] [PubMed] [Google Scholar]
- 2. Adams R. H., Alitalo K. (2007) Nat. Rev. Mol. Cell Biol. 8, 464–478 [DOI] [PubMed] [Google Scholar]
- 3. Larrivée B., Freitas C., Trombe M., Lv X., Delafarge B., Yuan L., Bouvrée K., Bréant C., Del Toro R., Bréchot N., Germain S., Bono F., Dol F., Claes F., Fischer C., Autiero M., Thomas J. L., Carmeliet P., Tessier-Lavigne M., Eichmann A. (2007) Genes Dev. 21, 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Navankasattusas S., Whitehead K. J., Suli A., Sorensen L. K., Lim A. H., Zhao J., Park K. W., Wythe J. D., Thomas K. R., Chien C. B., Li D. Y. (2008) Development 135, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Haastert P. J., Devreotes P. N. (2004) Nat. Rev. Mol. Cell Biol. 5, 626–634 [DOI] [PubMed] [Google Scholar]
- 6. Barkefors I., Le Jan S., Jakobsson L., Hejll E., Carlson G., Johansson H., Jarvius J., Park J. W., Li Jeon N., Kreuger J. (2008) J. Biol. Chem. 283, 13905–13912 [DOI] [PubMed] [Google Scholar]
- 7. Hellström M., Phng L. K., Hofmann J. J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A. K., Karlsson L., Gaiano N., Yoon K., Rossant J., Iruela-Arispe M. L., Kalén M., Gerhardt H., Betsholtz C. (2007) Nature 445, 776–780 [DOI] [PubMed] [Google Scholar]
- 8. De Smet F., Segura I., De Bock K., Hohensinner P. J., Carmeliet P. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 639–649 [DOI] [PubMed] [Google Scholar]
- 9. Jakobsson L., Kreuger J., Claesson-Welsh L. (2007) J. Cell Biol. 177, 751–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sugita S. (2008) Acta Physiol. 192, 185–193 [DOI] [PubMed] [Google Scholar]
- 11. Hsu S. C., TerBush D., Abraham M., Guo W. (2004) Int. Rev. Cytol. 233, 243–265 [DOI] [PubMed] [Google Scholar]
- 12. He B., Guo W. (2009) Curr. Opin. Cell Biol. 21, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saito T., Shibasaki T., Seino S. (2008) Biomed. Res. 29, 85–91 [DOI] [PubMed] [Google Scholar]
- 14. Seshadri S., Fitzpatrick A. L., Ikram M. A., DeStefano A. L., Gudnason V., Boada M., Bis J. C., Smith A. V., Carassquillo M. M., Lambert J. C., Harold D., Schrijvers E. M., Ramirez-Lorca R., Debette S., Longstreth W. T., Jr., Janssens A. C., Pankratz V. S., Dartigues J. F., Hollingworth P., Aspelund T., Hernandez I., Beiser A., Kuller L. H., Koudstaal P. J., Dickson D. W., Tzourio C., Abraham R., Antunez C., Du Y., Rotter J. I., Aulchenko Y. S., Harris T. B., Petersen R. C., Berr C., Owen M. J., Lopez-Arrieta J., Varadarajan B. N., Becker J. T., Rivadeneira F., Nalls M. A., Graff-Radford N. R., Campion D., Auerbach S., Rice K., Hofman A., Jonsson P. V., Schmidt H., Lathrop M., Mosley T. H., Au R., Psaty B. M., Uitterlinden A. G., Farrer L. A., Lumley T., Ruiz A., Williams J., Amouyel P., Younkin S. G., Wolf P. A., Launer L. J., Lopez O. L., van Duijn C. M., Breteler M. M. (2010) JAMA 303, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naj A. C., Jun G., Beecham G. W., Wang L. S., Vardarajan B. N., Buros J., Gallins P. J., Buxbaum J. D., Jarvik G. P., Crane P. K., Larson E. B., Bird T. D., Boeve B. F., Graff-Radford N. R., De Jager P. L., Evans D., Schneider J. A., Carrasquillo M. M., Ertekin-Taner N., Younkin S. G., Cruchaga C., Kauwe J. S., Nowotny P., Kramer P., Hardy J., Huentelman M. J., Myers A. J., Barmada M. M., Demirci F. Y., Baldwin C. T., Green R. C., Rogaeva E., George-Hyslop P. S., Arnold S. E., Barber R., Beach T., Bigio E. H., Bowen J. D., Boxer A., Burke J. R., Cairns N. J., Carlson C. S., Carney R. M., Carroll S. L., Chui H. C., Clark D. G., Corneveaux J., Cotman C. W., Cummings J. L., Decarli C., Dekosky S. T., Diaz-Arrastia R., Dick M., Dickson D. W., Ellis W. G., Faber K. M., Fallon K. B., Farlow M. R., Ferris S., Frosch M. P., Galasko D. R., Ganguli M., Gearing M., Geschwind D. H., Ghetti B., Gilbert J. R., Gilman S., Giordani B., Glass J. D., Growdon J. H., Hamilton R. L., Harrell L. E., Head E., Honig L. S., Hulette C. M., Hyman B. T., Jicha G. A., Jin L. W., Johnson N., Karlawish J., Karydas A., Kaye J. A., Kim R., Koo E. H., Kowall N. W., Lah J. J., Levey A. I., Lieberman A. P., Lopez O. L., Mack W. J., Marson D. C., Martiniuk F., Mash D. C., Masliah E., McCormick W. C., McCurry S. M., McDavid A. N., McKee A. C., Mesulam M., Miller B. L., Miller C. A., Miller J. W., Parisi J. E., Perl D. P., Peskind E., Petersen R. C., Poon W. W., Quinn J. F., Rajbhandary R. A., Raskind M., Reisberg B., Ringman J. M., Roberson E. D., Rosenberg R. N., Sano M., Schneider L. S., Seeley W., Shelanski M. L., Slifer M. A., Smith C. D., Sonnen J. A., Spina S., Stern R. A., Tanzi R. E., Trojanowski J. Q., Troncoso J. C., Van Deerlin V. M., Vinters H. V., Vonsattel J. P., Weintraub S., Welsh-Bohmer K. A., Williamson J., Woltjer R. L., Cantwell L. B., Dombroski B. A., Beekly D., Lunetta K. L., Martin E. R., Kamboh M. I., Saykin A. J., Reiman E. M., Bennett D. A., Morris J. C., Montine T. J., Goate A. M., Blacker D., Tsuang D. W., Hakonarson H., Kukull W. A., Foroud T. M., Haines J. L., Mayeux R., Pericak-Vance M. A., Farrer L. A., Schellenberg G. D. (2011) Nat. Genet. 43, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wallgard E., Larsson E., He L., Hellström M., Armulik A., Nisancioglu M. H., Genove G., Lindahl P., Betsholtz C. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1469–1476 [DOI] [PubMed] [Google Scholar]
- 17. Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 19. Nichols W. W., Murphy D. G., Cristofalo V. J., Toji L. H., Greene A. E., Dwight S. A. (1977) Science 196, 60–63 [DOI] [PubMed] [Google Scholar]
- 20. Mellberg S., Dimberg A., Bahram F., Hayashi M., Rennel E., Ameur A., Westholm J. O., Larsson E., Lindahl P., Cross M. J., Claesson-Welsh L. (2009) FASEB J. 23, 1490–1502 [DOI] [PubMed] [Google Scholar]
- 21. Boyd C., Hughes T., Pypaert M., Novick P. (2004) J. Cell Biol. 167, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zajac A., Sun X., Zhang J., Guo W. (2005) Mol. Biol. Cell 16, 1500–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sugihara K., Asano S., Tanaka K., Iwamatsu A., Okawa K., Ohta Y. (2002) Nat. Cell Biol. 4, 73–78 [DOI] [PubMed] [Google Scholar]
- 24. Sivaram M. V., Furgason M. L., Brewer D. N., Munson M. (2006) Nat. Struct. Mol. Biol. 13, 555–556 [DOI] [PubMed] [Google Scholar]
- 25. Sans N., Prybylowski K., Petralia R. S., Chang K., Wang Y. X., Racca C., Vicini S., Wenthold R. J. (2003) Nat. Cell Biol. 5, 520–530 [DOI] [PubMed] [Google Scholar]
- 26. Oztan A., Silvis M., Weisz O. A., Bradbury N. A., Hsu S. C., Goldenring J. R., Yeaman C., Apodaca G. (2007) Mol. Biol. Cell 18, 3978–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prigent M., Dubois T., Raposo G., Derrien V., Tenza D., Rossé C., Camonis J., Chavrier P. (2003) J. Cell Biol. 163, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allen W. E., Zicha D., Ridley A. J., Jones G. E. (1998) J. Cell Biol. 141, 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murthy M., Teodoro R. O., Miller T. P., Schwarz T. L. (2010) Development 137, 2773–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen X. W., Inoue M., Hsu S. C., Saltiel A. R. (2006) J. Biol. Chem. 281, 38609–38616 [DOI] [PubMed] [Google Scholar]
- 31. Califano J. P., Reinhart-King C. A. (2010) J. Biomech. 43, 79–86 [DOI] [PubMed] [Google Scholar]
- 32. Yamamura N., Sudo R., Ikeda M., Tanishita K. (2007) Tissue Eng. 13, 1443–1453 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.