Abstract
The scavenger receptor C-type lectin (SRCL) is a glycan-binding receptor that has the capacity to mediate endocytosis of glycoproteins carrying terminal Lewisx groups (Galβ1–4(Fucα1–3)GlcNAc). A screen for glycoprotein ligands for SRCL using affinity chromatography on immobilized SRCL followed by mass spectrometry-based proteomic analysis revealed that soluble glycoproteins from secondary granules of neutrophils, including lactoferrin and matrix metalloproteinases 8 and 9, are major ligands. Binding competition and surface plasmon resonance analysis showed affinities in the low micromolar range. Comparison of SRCL binding to neutrophil and milk lactoferrin indicates that the binding is dependent on cell-specific glycosylation in the neutrophils, as the milk form of the glycoprotein is a much poorer ligand. Binding to neutrophil glycoproteins is fucose-dependent, and mass spectrometry-based glycomic analysis of neutrophil and milk lactoferrin was used to establish a correlation between high affinity binding to SRCL and the presence of multiple clustered terminal Lewisx groups on a heterogeneous mixture of branched glycans, some with poly N-acetyllactosamine extensions. The ability of SRCL to mediate uptake of neutrophil lactoferrin was confirmed using fibroblasts transfected with SRCL. The common presence of Lewisx groups in granule protein glycans can thus target granule proteins for clearance by SRCL. PCR and immunohistochemical analysis confirm that SRCL is widely expressed on endothelial cells and thus represents a distributed system that could scavenge released neutrophil glycoproteins both locally at sites of inflammation or systemically when they are released in the circulation.
Keywords: Endocytosis, Glycoprotein, Lectin, Neutrophil, Receptors, Glycomics, Lactoferrin
Introduction
The scavenger receptor C-type lectin (SRCL,2 also designated collectin P1) is an endothelial cell surface receptor able to mediate endocytosis of several classes of ligands (1–3). The extracellular portion of the receptor consists of three distinct regions as follows: a coiled coil of α-helices ∼450 Å in length; a collagen-like region of a similar length; and a globular C-terminal C-type carbohydrate-recognition domain (CRD). These regions are predicted to be linked by flexible joints (4). Although the first two types of domains are found in other class A scavenger receptors, SRCL is the only example of a scavenger-type receptor in this class that contains a C-type CRD (5).
In functional terms, the α-helical region in SRCL probably forms a stalk projecting from the cell surface, and studies with truncated forms of the receptor indicate that, as with other scavenger receptors, yeast and other microbes bind to the collagen-like domains, probably through regions containing clusters of positively charged amino acid residues (1). The CRD shows high selectivity for glycans terminating with the Lewisx trisaccharide (Galβ1–4(Fucα1–3)GlcNAc and recognizes Lewisx-containing glycoconjugates (6), but no physiological ligands for the receptor have been reported. The endothelial expression of SRCL suggests that the receptor could be involved in mediating adhesion of circulating cells to the endothelium during leukocyte extravasation, a function analogous to that of the selectin family of C-type lectins (7). Consistent with a role for SRCL in cell adhesion, the Lewisx structure is found on the surfaces of leukocytes and has been implicated in neutrophil adhesion (8). It has also been proposed that SRCL mediates adhesion of Lewisx-expressing cancer cells to endothelium (9). However, a role of SRCL in cell adhesion remains to be established definitively.
The endocytic activity of SRCL supports the alternative suggestion that this receptor may be involved in uptake and degradation of glycoproteins (6). Several other receptors that contain C-type CRDs as well as other types of sugar-binding domains have been implicated in glycoprotein turnover (10). The roles of the mannose receptor on macrophages in the clearance of glycoprotein hormones tagged with GalNAc-4-SO4 and released lysosomal enzymes bearing high mannose oligosaccharides have been particularly well characterized (11, 12), and the importance of the hepatic asialoglycoprotein receptor in clearance of specific subclasses of serum glycoproteins has been extensively studied (13, 14).
In the studies reported here, the function of SRCL was probed by identifying specific glycoprotein ligands associated with cells in the blood. The predominant SRCL ligands in neutrophils were found to be soluble granule proteins rather than cell surface proteins, suggesting that a major role for SRCL is likely to be in the clearance of glycoproteins released at sites of inflammation, which may be tagged with Lewisx epitopes to facilitate their uptake after release.
EXPERIMENTAL PROCEDURES
Generation of Biotin-tagged CRD of SRCL
The coding sequence for the biotinylation tag Gly-Leu-Asn-Asp-Ile-Phe-Glu-Ala-Gln-Lys-Ile-Glu-Trp-His-Glu, in which the lysine residue is a target for biotinylation (15), was added to the C terminus of the CRD in the expression vector pT5T (16). Escherichia coli strain BL21/DE3 cells containing this expression plasmid as well as a plasmid containing the chloramphenicol resistance gene and the birA biotin ligase gene (17) were grown in 1-liter batches, and inclusion bodies were isolated and dissolved in guanidine as described previously for preparation of other biotin-tagged CRDs (18). Following renaturation by dilution into 4 volumes of loading buffer (0.5 m NaCl, 25 mm Tris-Cl, pH 7.8, 25 mm CaCl2) and dialysis against three changes of the same buffer, the tagged CRD was isolated following the procedure developed for the untagged CRD (6).
Detection of SRCL Expression by PCR
PCR-ready first strand cDNAs for a range of tissues were obtained from BioChain. PCR was performed using the Advantage 2 polymerase kit (Takara) using the standard conditions with the following primers: gtgcccctggccctgcagaatgagccaacc (forward) and aatttgctcatgtgatcccatcacagtccg (reverse). After the initial incubation at 95 °C for 1 min, cycles of 30 s at 95 °C and 1 min at 65 °C were performed. Products were resolved on a 2% agarose gel containing SYBR-Safe (Invitrogen) to visualize DNA.
Gels and Blotting
An aliquot (100 μg) of the extracellular domain of SRCL purified from transfected Chinese hamster ovary cells (6) was resuspended in 500 μl of 25 mm Bicine, pH 8.5, 0.5 m NaCl, 25 mm CaCl2 and incubated for 20 min at room temperature with 250 μCi of 125I-Bolton-Hunter Reagent (PerkinElmer Life Sciences). Labeled protein was applied to a 1-ml column of galactose-Sepharose that was washed with four 1-ml aliquots of loading buffer (0.5 m NaCl, 25 mm Tris-Cl, pH 7.8, 25 mm CaCl2) and eluted with five 0.5-ml aliquots of eluting buffer (0.5 m NaCl, 25 mm Tris-Cl, pH 7.8, 2.5 mm EDTA). Streptavidin-conjugated bacterial alkaline phosphatase (100 μg, Sigma) was incubated with a large excess of purified biotinylated CRD of SRCL in loading buffer at 4 °C overnight with rotary agitation. The resulting complexes were repurified on a 1-ml column of galactose-Sepharose in the same way. Elution fractions containing SRCL-streptavidin complexes (SRCL conjugated to alkaline phosphatase) were identified by SDS-PAGE.
SDS-polyacrylamide gels were run in the buffer system of Laemmli. All gels contained 17.5% acrylamide unless otherwise indicated, and blotting was performed on nitrocellulose (19). Blots were blocked with 2% bovine hemoglobin in low salt loading buffer (150 mm NaCl, 25 mm Tris-Cl, pH 7.8, 25 mm CaCl2) for 60 min at room temperature with agitation, followed by radiolabeled or alkaline phosphatase-tagged receptor in this same hemoglobin solution for 90 min. After four 10-min washes with low salt loading buffer, radioactivity on blots was detected with a phosphorimager from GE Healthcare, and alkaline phosphate was detected with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium substrate (Calbiochem). Immunoblotting was performed using rabbit polyclonal antibodies to pan-CEACAM (Abcam) at a 1:500 dilution followed by visualization using alkaline phosphatase-conjugated protein A.
Purification of Ligands from Human Granulocytes by Affinity Chromatography
An SRCL affinity column was created by passing ∼5 mg of biotinylated CRD of SRCL in eluting buffer over a 2-ml column of avidin-agarose (Pierce) pre-rinsed with the same buffer. The column was washed extensively with eluting buffer followed by loading buffer. A sample of the washed resin was analyzed by SDS-PAGE to confirm binding of the CRD to the resin.
Leukocytes were isolated from ∼50 ml of fresh human blood containing 1.5 mg/ml EDTA as an anticoagulant. Granulocytes were isolated by centrifugation through Ficoll-Paque PLUS (d = 1.077 g/cm3; Amersham Biosciences) followed by lysis of erythrocytes in ammonium bicarbonate (20). Granulocytes were lysed by sonication in low salt loading buffer containing 1% Triton X-100 and a protease inhibitor mixture (Calbiochem protease inhibitor mixture set 1), incubated for 30 min on ice, and centrifuged at 4,000 rpm for 10 min in an Eppendorf 5810R centrifuge. The supernatant was passed over the SRCL affinity column, which was washed with five 1-ml aliquots of low salt loading buffer containing 0.1% Triton X-100 and eluted with five 1-ml aliquots of low salt eluting buffer containing 0.1% Triton X-100. For repurification of ligands, the elution fractions were pooled, adjusted to 25 mm CaCl2, and applied to the column again. The column was washed and eluted as before.
Proteomic Analysis by Mass Spectrometry
Affinity-purified proteins were resolved by SDS-PAGE and stained with Coomassie Blue. Bands for proteomic analysis were excised from the gel, reduced and carboxymethylated, digested with trypsin, and subjected to MALDI mass spectrometry (21). MS and MS/MS data were used to search 17,869 human entries in release 54.6 of the SwissProt data base with version 2.2 of the Mascot data base search algorithm. For the search, peptide masses were fixed as monoisotopic; the mass tolerance was set at 75 ppm for precursor ions and 0.1 Da for fragment ions; partial carboxymethylation of cysteine residues and partial oxidation of methionine residues were considered, and up to one missed trypsin cleavage site was allowed. Protein matches from both MS and MS/MS data were used to generate probability-based Mowse protein scores. Scores greater than 55 were considered significant (p < 0.05) (22). Additional information, such as agreement between molecular weight of the tentatively matched protein and mobility on the SDS-polyacrylamide gel, was used to evaluate lower confidence matches, where the number of peptides detected might be low because the protein is scarce or small, has few trypsin cleavage sites, or is post-translationally modified. For instances in which the mass spectrometry results suggested that a single band contained more than one protein, it was ensured that each protein match was derived from nonoverlapping sets of detected peptides.
Fucosidase Treatment of Neutrophil Glycoproteins
Lactoferrin from human neutrophils (Sigma) was dialyzed against water and lyophilized. An aliquot (5 μg) was digested with 1 unit/ml bovine kidney α-l-fucosidase (Sigma) at 37 °C for 48 h in 50 μl of 100 mm sodium citrate, pH 5.6. Samples equivalent to 1 μg of lactoferrin were analyzed by gel electrophoresis and blotting. Matrix metalloproteinase (MMP) 9 from human neutrophils (Calbiochem), at a concentration of 100 μg/ml in 50 μl in 50 mm Tris-Cl, pH 7.0, 200 mm NaCl, 5 mm CaCl2, 1 μm ZnCl2, 0.05% BRIJ 35 detergent, 0.05% sodium azide, was supplemented with 12.5 μl of 1 m sodium citrate, pH 5.6. A 25-μl aliquot of this solution (2 μg of protein) was incubated for 24 h at 37 °C with 5 μl of fucosidase solution.
Binding Assays
Solid phase binding assays were performed as described previously (6). Surface plasmon resonance measurements were made using a BIAcore 3000 instrument with BIAcore 3000 Control software and BIAevaluation software. Neutrophil lactoferrin was coupled to activated CM5 sensor chips (BIAcore) by flowing a 50 μg/ml solution of protein in 10 mm sodium acetate, pH 5.0, over the chip for 10 min at a flow rate of 10 μl/min. CRD of SRCL was resuspended at 1 mg/ml in 10 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm CaCl2, 0.005% P20 detergent, and serial dilutions were prepared using the same buffer, with 1 μg/ml as the lowest concentration. The sensor chip was regenerated between samples using 10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% P20 detergent. The response values after 80 s of binding were fitted to a direct binding equation using SigmaPlot software.
Glycan Analysis
Purified lactoferrin derived from milk or neutrophils (∼0.5 mg) was precipitated with trichloroacetic acid and resuspended in 25 μl of 8 m urea, 10 mm HEPES, pH 8.0. The protein was digested at 37 °C overnight with l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin from bovine pancreas (Sigma) at a final concentration of 50 μg/ml in a final volume of 250 μl of 50 mm ammonium bicarbonate, pH 8.4. Isolation of glycopeptides, release of glycans, and permethylation were performed as described previously (23). The data presented were obtained from the 50% acetonitrile fraction of permethylated glycans, except that data for the 35% acetonitrile fraction are shown in supplemental Fig. S6A. The fractionated glycans were dissolved in 10 μl of methanol, and a 1-μl aliquot of dissolved sample was mixed with 1 μl of 2,5-dihydroxybenzoic acid matrix solution (20 mg/ml in 70% methanol). Two aliquots of 0.5 μl were spotted onto a target plate and dried under vacuum. MALDI MS and MS/MS data were acquired using an Applied Biosystems 4800 MALDI-time-of-flight/time-of-flight mass spectrometer. The collision energy was set to 1 kV for MS/MS fragmentation, and argon was used as collision gas. The Calmix 4700 calibration standard kit (Applied Biosystems) was used as the external calibrant for the MS mode and [Glu1] fibrinopeptide B human (Sigma) was used as an external calibrant for the MS/MS mode. MS and MS/MS data were processed using Data Explorer 4.9 software (Applied Biosystems). The proposed assignments for the selected peaks were based on 12C isotopic composition together with knowledge of the biosynthetic pathways with the aid of the glycobioinformatics tool GlycoWorkbench (24). The proposed structures were then confirmed by MS/MS experiments.
Aliquots of underivatized N-glycans derived from 50 μg of neutrophil lactoferrin were subjected to chemical and enzymatic degradation. Sialidase digestion was performed in 200 μl of 50 mm sodium acetate, pH 5.5, with 170 milliunits of sialidase A from Arthrobacter ureafaciens (Glyko) for 24 h at 37 °C. Digestion with endo-β-galactosidase from Escherichia freundii (EC 3.2.1.103; Glyko) was carried out in 200 μl of 100 mm sodium acetate, pH 5.8, at 37 °C for 48 h, with aliquots of enzyme (20 milliunits) added at 0 and 24 h. All digested samples were lyophilized, permethylated, and purified on a C18 Sep-Pak (Waters). For HF treatment, dry glycans were incubated overnight at 4 °C with 50 μl of 48% HF, and the reaction was terminated by drying under a stream of nitrogen before permethylation.
For linkage analysis by gas chromatography-mass spectrometry, partially methylated alditol acetates were prepared, as described previously (25), and analyzed on a Clarus 500 instrument (PerkinElmer Life Sciences) fitted with an RTX-5 fused silica capillary column, 30 m × 0.32 mm (Restek Corp.). Each sample was dissolved in ∼50 μl of hexanes (Sigma) and injected manually (1–2 μl) into the injector set at 250 °C. The oven was initially heated at 65 °C for 1 min, following which the temperature was raised linearly to 290 °C at a rate of 8 °C per min, held at 290 °C for 5 min, and raised to 300 °C over 1 min.
Internalization of Glycoproteins by Cells
Proteins for fluorescent labeling, including neutrophil lactoferrin (1 mg/ml), BSA (5 mg/ml), ovalbumin (5 mg/ml, Sigma), and LNFPIII-BSA (0.2 mg/ml, Dextra) in 100 mm Bicine, pH 9.0, 150 mm NaCl, were reacted with fluorescein isothiocyanate (50 μg/ml). Fluorescein isothiocyanate was added as 5 aliquots of a stock solution at 1 mg/ml in dimethyl sulfoxide with vortexing between each addition. Reactions were incubated at 4 °C overnight in the dark. Excess dye was removed on a 10-ml column of Sepharose G-25 eluted with PBS, collecting 1-ml fractions. Dye-containing peak fractions were identified by eye, and samples of all fractions were analyzed by SDS-PAGE to confirm association of dye color with protein.
Transfected fibroblasts expressing SRCL grown on coverslips were incubated at 37 °C with fluorescein-labeled proteins diluted to 1 μg/ml in phenol red-free Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Coverslips were washed in Dulbecco's PBS, fixed for 15 min with 4% paraformaldehyde in PBS, washed, and blocked for 20 min with 10% normal goat serum in PBS. Cells were incubated for 60 min with rabbit anti-fluorescein (Abcam) at 5 μg/ml followed by 30 min with Alexa Fluor 488-labeled goat anti-rabbit IgG (Invitrogen) at 1 μg/ml in 2% normal goat serum in PBS. Coverslips were mounted with Vectashield aqueous mounting medium containing DAPI (Vector Laboratories) and sealed.
Immunofluorescence on Tissue Sections
Serial sets of tissue array slides of formalin-fixed, paraffin-embedded human normal organs were purchased from SuperBioChips. Slides were dried for 1 h in a 60 °C oven, deparaffinized in five changes of xylene, and rehydrated through a graded ethanol series before immersion in water. Heat-mediated antigen retrieval was performed by incubating slides at 95–99 °C for 20 min in antigen unmasking solution from Vector Laboratories. The TSA biotin kit (PerkinElmer Life Sciences) was used for tyramide signal amplification. A 15-min incubation with 3% hydrogen peroxide in PBS was included prior to blocking to quench endogenous peroxidase activity. Rabbit antibodies to SRCL (23) were used at 1 μg/ml for 90 min. Following removal of primary antibody and washing, cells were incubated for 30 min with horseradish peroxidase-conjugated goat anti-rabbit IgG (Vector Laboratories) at 1 μg/ml followed by 10 min of incubation with biotinyl tyramide and detection by 30 min of incubation with Alexa Fluor 488-labeled streptavidin (Invitrogen) at 2 μg/ml. Images were collected using a Nikon Eclipse E400 microscope with ×20 and ×40 objectives and a DXM1200 digital camera. Images from the color channels were acquired as 8-bit grayscale TIFF files with software package Lucia GF version 4.60 and converted to merged 24-bit RGB files using Corel PhotoPaint 12.
RESULTS
Granule Proteins as Dominant Neutrophil Ligands for SRCL
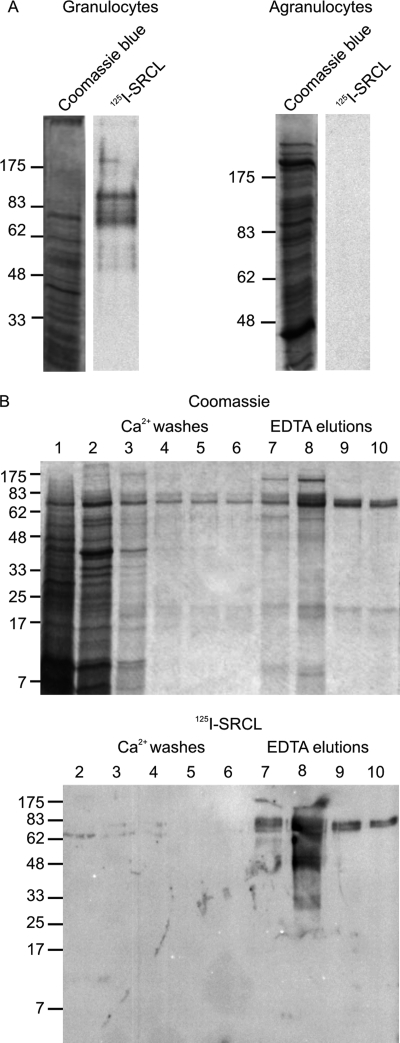
The reported expression of SRCL on endothelial cells suggested that the receptor might mediate leukocyte adhesion through binding to leukocyte surface glycoproteins. In an attempt to detect potential physiological ligands for SRCL on leukocytes, freshly isolated leukocytes were fractionated into a granulocyte population, consisting predominantly of neutrophils with some basophils and eosinophils, and an agranulocyte population, containing lymphocytes and monocytes. Cell extracts from these two populations were resolved by SDS-PAGE and blotted onto nitrocellulose. The blots were probed with 125I-labeled extracellular domain of SRCL, a soluble, trimeric fragment of the receptor that exhibits highly selective and high affinity binding to Lewisx (Fig. 1A) (6). Multiple species were detected in the blot of a granulocyte extract, indicating that several glycoproteins in granulocytes are recognized by SRCL. The presence of Lewisx epitopes on these proteins would be consistent with the high level of Lewisx expression in granulocytes (26). In contrast, agranulocytes do not contain significant quantities of protein recognized by SRCL, reflecting the low level of Lewisx expression in these cells (27).
FIGURE 1.
Isolation of glycoprotein ligands for SRCL from granulocytes. A, extracts of granulocyte and agranulocyte populations analyzed on 10% SDS-polyacrylamide gels. Samples correspond to ∼5 × 105 cells. B, SDS-PAGE of granulocyte extract fractionated by affinity chromatography on immobilized CRD from SRCL. SRCL ligands were bound to the 2-ml column in the presence of Ca2+ and eluted with EDTA. Fractions of 1 ml were collected, and aliquots of 20 μl were analyzed on the gels. Total proteins were stained with Coomassie Blue, and SRCL ligands were visualized by incubating blots with 125I-SRCL.
Oligomerization of SRCL is required to generate a ligand-binding interaction of sufficiently high affinity for affinity purification of SRCL ligands. Because the naturally trimeric extracellular domain of SRCL is difficult to produce in sufficient quantity for preparation of an affinity column, artificial tetramers of the bacterially produced CRD were created by binding biotin-tagged CRDs to immobilized avidin. A subset of proteins from solubilized granulocytes was retained on the column and eluted with EDTA, indicating a specific interaction with the CRD of SRCL (Fig. 1B). Probing of a blot of the fractions with 125I-SRCL confirmed that the column efficiently retains SRCL-binding glycoproteins, with very little reactivity remaining in the wash fractions. The efficiency and specificity of the affinity chromatography procedure were demonstrated by the fact that glycoproteins in the bound and eluted fractions were almost completely retained when rerun on the affinity column (data not shown).
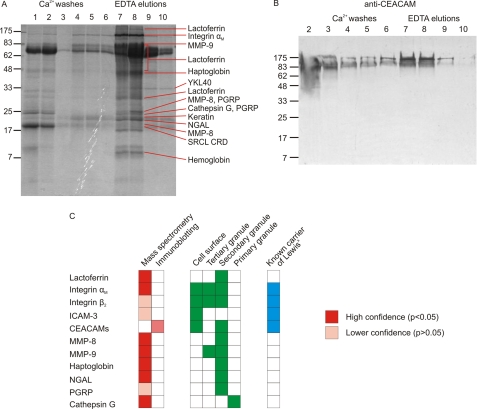
Proteins present in the repurified fractions were identified by proteomic analysis. Following gel electrophoresis, peptides released from individual bands by trypsin digestion were analyzed by mass spectrometry, and the resulting masses and partial sequences were screened against the SwissProt data base of human proteins. The most abundant glycoprotein identified was lactoferrin (Fig. 2A). Other glycoproteins detected at high levels were αM integrin, MMP-9, MMP-8, haptoglobin, neutrophil gelatinase-associated lipocalin, cathepsin G, and YKL40, although the slow release of YKL40 from the SRCL affinity column in the presence of EDTA suggests that it may be binding to the glycans on the immobilized avidin as a result of its intrinsic GlcNAc binding activity rather than interaction with SRCL (28, 29). The major nonglycosylated proteins identified were the common keratin contaminants as well as hemoglobin, which probably bound to haptoglobin during preparation of the granulocyte extract and co-purifies on the affinity column. Neutrophil integrin β2, ICAM-3, and CEACAM-8 are known to carry Lewisx epitopes (30–32). Although these proteins were not identified using stringent search parameters, peptides from these proteins would be hard to detect because of masking by the abundant peptides from lactoferrin, which covers a similar molecular weight range on the gel, and examination of the data for unassigned peptides in this region revealed peptides attributable to integrin β2 and ICAM-3 in the appropriate molecular weight bands. CEACAM would be particularly difficult to identify from the mass spectrometry data because the high degree of glycosylation of this protein prevents matching of many of the tryptic peptides. Therefore, a blot of fractions from the affinity column was also probed with anti-CEACAM antibodies, revealing that a proportion of CEACAM is also bound by the column and eluted with EDTA (Fig. 2B). The presence of CEACAM in the wash fractions indicates that only a subset of CEACAM proteins carry the appropriate glycosylation to be bound by SRCL.
FIGURE 2.
Identification of SRCL ligands isolated from neutrophils. A, SDS-polyacrylamide gel of fractions from separation of granulocyte extracts on the SRCL affinity column. The gel was stained with Coomassie Blue, and slices were subjected to proteomic analysis. Major proteins identified in each band are indicated. Details of protein identification are provided in supplemental Tables S1 and S2. B, confirmation that some forms of CEACAM are ligands for SRCL. Blot of an SDS-polyacrylamide gel was probed with anti-CEACAM antibody, which was visualized with phosphatase-conjugated protein A. C, summary of ligands identified by proteomics and blotting. The distribution of glycoproteins in granulocytes is shown for comparison.
The potential ligands for SRCL isolated by affinity chromatography can be categorized into two groups as follows: cell surface transmembrane glycoproteins, and soluble glycoproteins of secondary granules (Fig. 2C). The cell surface glycoproteins are also ligands for the Lewisx-binding C-type lectin DC-SIGN, which also recognizes Lewisx-containing glycans (30, 33, 34), suggesting that DC-SIGN and SRCL may interact with overlapping sets of neutrophil glycoproteins to mediate interaction of neutrophils with dendritic cells and endothelia, respectively. However, the most abundant ligand for SRCL is the soluble granule protein lactoferrin, and many of the additional SRCL ligands, including MMP-8, MMP-9, and haptoglobin, are also soluble proteins found in granules. These results suggest that a distinct group of soluble glycoproteins contained within secondary granules are major previously unidentified carriers of Lewisx in neutrophils, explaining early reports of the presence of CD15 epitopes on uncharacterized granule glycoproteins (35, 36).
The fact that the majority of the ligand molecules for SRCL in neutrophils are soluble granule glycoproteins rather than cell surface glycoproteins suggests that rather than functioning principally as an adhesion receptor, SRCL may have a role in clearing soluble secondary granule glycoproteins that are released by degranulation of neutrophils at sites of inflammation. Such a role would be consistent with the known endocytic capability of the receptor (6) and would complement that of the macrophage mannose receptor, which clears myeloperoxidase released from neutrophil primary granules (37). The Lewisx structure would therefore act as a recognition tag added to a range of soluble glycoproteins to facilitate their clearance following exocytosis.
Fucose-dependent High Affinity Binding of Neutrophil Lactoferrin to SRCL
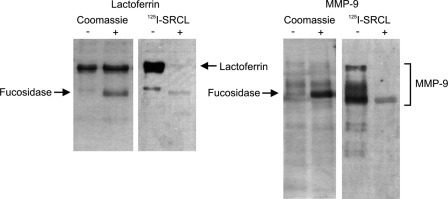
The idea that SRCL may serve to clear released granule proteins tagged with Lewisx-containing glycans was investigated further by examining the nature of the interaction of SRCL with several granule glycoproteins. Following digestion with bovine kidney α-l-fucosidase, blots of granule glycoproteins show almost complete loss of reactivity with SRCL as expected for fucose-dependent binding of SRCL to Lewisx groups on these glycoproteins (Fig. 3). Fucose-dependent binding of SRCL to MMP-8 and MMP-9 in addition to lactoferrin suggests that SRCL recognizes glycan structures common to a range of neutrophil granule proteins and that lactoferrin can be used as a model for this group of proteins.
FIGURE 3.
Fucose dependence of binding of SRCL to granule proteins. Samples of purified neutrophil lactoferrin and matrix metalloproteinase were incubated in the presence or absence of fucosidase and separated on SDS-polyacrylamide gels that were stained with Coomassie Blue or blotted onto nitrocellulose and probed with 125I-SRCL.
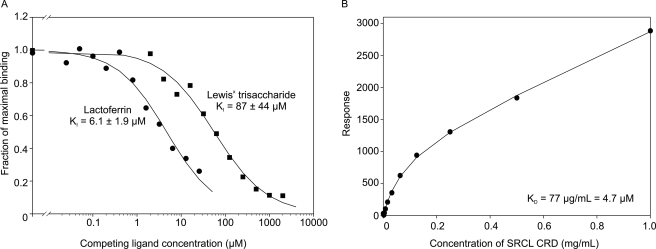
A competition assay for binding to immobilized CRD from SRCL, using LNFPIII-BSA as a reporter ligand, was employed to obtain quantitative information about lactoferrin binding to SRCL. The KI of 6.1 μm for lactoferrin is more than 10-fold higher than the KI of 87 μm for Lewisx (Fig. 4A). The affinity of SRCL for lactoferrin was also measured using surface plasmon resonance to monitor binding of the CRD from SRCL to lactoferrin immobilized on a sensor chip surface, which yielded a binding constant of 4.7 μm (Fig. 4B). Because the CRD of SRCL is monomeric, the binding detected represents the interaction of one protein molecule with one glycan. The similarity in the values in the competition and surface plasmon resonance experiments suggests that lactoferrin binds to immobilized CRDs through a single glycan at a time, possibly reflecting the disposition of the two N-glycans on lactoferrin, which project from diametrically opposite sides of the molecule. Although it is possible that the enhanced binding to lactoferrin compared with the oligosaccharide ligand could reflect some recognition of the lactoferrin polypeptide by SRCL, it has been observed that glycan-binding proteins often bind to glycoproteins with higher affinity than to free glycans, even where the enhancement cannot be due to multivalent effects or sequence-specific interactions with the polypeptide (38). The difference in binding affinity between lactoferrin and Lewisx may therefore be due to presentation of the Lewisx structure on a glycoprotein.
FIGURE 4.
Quantitative analysis of SRCL binding to neutrophil lactoferrin. A, competition binding assay with immobilized CRD from SRCL probed with 125I-LNFPIII-BSA in the presence of competing oligosaccharide or lactoferrin. Experimental data are shown as symbols and fitted curves for first order binding are shown as solid lines. Data shown are averages of duplicates, and KI values represent the average ± S.D. for two separate experiments. B, binding analyzed by surface plasmon resonance. CRD from SRCL at varying concentrations was flowed over a sensor surface coated with neutrophil lactoferrin.
Modulation of SRCL Binding by Tissue-specific Glycosylation of Lactoferrin
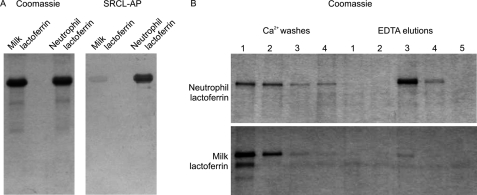
In addition to being found in neutrophil granules, lactoferrin secreted from mammary epithelial cells is present in milk (39), making it possible to investigate cell type-specific differences in lactoferrin glycosylation. In a blotting experiment, alkaline phosphatase-conjugated SRCL was far more reactive toward neutrophil-derived lactoferrin than milk lactoferrin (Fig. 5A), suggesting that the neutrophil protein carries more Lewisx structures that are recognized by SRCL. Correspondingly, a KI value for milk lactoferrin in the solid phase competition binding assay could not be measured because sufficiently high protein concentrations could not be achieved (data not shown).
FIGURE 5.
Differential binding of SRCL to milk and neutrophil lactoferrin. A, SDS-polyacrylamide gels of lactoferrin were stained with Coomassie Blue or blotted onto nitrocellulose and probed with SRCL conjugated to alkaline phosphatase (SRCL-AP). B, comparison of fractionation of neutrophil and milk lactoferrin on immobilized SRCL. Fractions from the affinity column were analyzed on SDS-polyacrylamide gels that were stained with Coomassie Blue.
The basis for differential interaction of SRCL with neutrophil and milk lactoferrin was initially investigated by fractionation of the lactoferrin samples on an SRCL affinity column (Fig. 5B). Some neutrophil lactoferrin was washed off the column with Ca2+-containing buffer, but most was only released using EDTA, indicating that the majority of neutrophil lactoferrin molecules bind with high affinity to SRCL. In contrast, almost all of the milk-derived protein was washed off the column with Ca2+-containing buffer, with only a very minor proportion being eluted with EDTA, indicating that the majority of molecules do not bind to SRCL or bind with low affinity. Although SRCL does not bind to sialyl Lewisx structures (38), the possibility that poor interaction with milk lactoferrin is due to capping with sialic acid was dismissed because neuraminidase digestion does not affect the reactivity of SRCL with either the milk or the neutrophil lactoferrin samples, indicating that few sialyl-Lewisx groups are present in either sample (data not shown).
Analysis of N-Glycans from Lactoferrin
Each of the two lobes of human lactoferrin is glycosylated at a single asparagine residue (40). It was previously reported that human neutrophil lactoferrin glycans are homogeneous biantennary structures with terminal sialic acid and the complete absence of fucose, even from the core (41). The reported absence of fucose is surprising, given the documented abundance of the Lewisx modification in neutrophils (42). The fucose dependence of SRCL binding demonstrated here is in direct contradiction with this report and indicates that the structures of neutrophil lactoferrin glycans need to be re-examined.
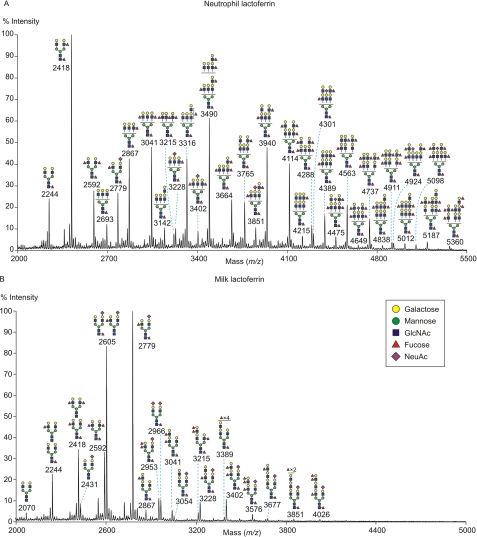
The glycomic profile of the permethylated N-glycans on neutrophil lactoferrin was obtained from MALDI-MS analysis (Fig. 6A). The deduced monosaccharide compositions for the major peaks indicate that all the glycans have been processed to complex type structures rather than remaining as high mannose or hybrid structures (supplemental Table S3). Based on evidence described in detail below, although a portion of the glycans have a minimal bi-antennary HexNAc4Hex5 skeleton, many also have additional N-acetyllactosamine units that either extend the antennae or add additional branches up to tetra-antennary N-glycans, and virtually all of the glycans are decorated with Lewisx epitopes, either on different antennae as terminal groups or as multiple Lewisx groups on the same antenna. Glycans with more than one fucose residue carry at least a proportion of outer arm fucose as part of a Lewisx epitope that could mediate binding to SRCL.
FIGURE 6.
Analysis of permethylated N-linked glycans from lactoferrin by MALDI-TOF mass spectrometry. A, glycans released from neutrophil lactoferrin. B, glycans released from milk lactoferrin. Spectra for the 50% acetonitrile fraction following C18 Sep-Pak fractionation are shown. Putative structures of the most abundant species present in each fraction, deduced from composition, MS/MS analysis, endoglycosidase digestion, linkage analysis, and information of biosynthetic pathways, are shown schematically. Structures that show sugars outside a bracket have not been unequivocally defined. All molecular ions are [M + Na]+.
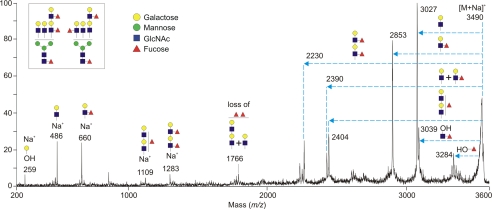
An example of MS/MS analysis of the second most abundant glycan, HexNAc6Hex7Fuc3 (m/z 3490), is shown in Fig. 7. This molecular ion corresponds mainly to a mixed population of core-fucosylated tri-antennary structures with different arrangements of Lewisx epitopes as shown by loss of both terminal and extended Lewisx groups (m/z 2853, 2230, 2390, and 2404). The presence of the Lewisx epitope was demonstrated by the elimination of fucose from the molecular ion to generate the m/z 3284 fragment, indicating that the fucose residue is linked at the 3-position of GlcNAc. This result is consistent with previous evidence that the antennae on neutrophil glycans contain fucose in the α1,3-linkage of the Lewisx epitope but not the α1,4-linkage of the Lewisa structure (43).
FIGURE 7.
Fragmentation of an abundant neutrophil glycan. The [M + Na]+ molecular ion at m/z 3490 from Fig. 6A was subjected to MS/MS analysis to demonstrate the presence of multiple fucose residues on outer arms of a tri-antennary oligosaccharide. The horizontal arrows on the spectrum indicate losses from the molecular ion [M + Na]+ of the designated sequences in the inset. The signal at m/z 3039 corresponds to loss of HexNAcFuc from the reducing side of the glycans, indicative of core fucosylation, and the ion at m/z 3284 results from elimination of a fucose residue in a Lewisx epitope. The ions at m/z 486 and 660 establish the presence of a terminal N-acetyllactosamine unit and a terminal Lewisx terminal epitope, whereas the ions at m/z 1109 and 1283 correspond to single elongated and double Lewisx epitopes.
Consistent with many of the N-glycans being tri- and tetra-antennary structures, gas chromatography-mass spectrometry analysis indicated that of the total α-mannose residues in the pool of neutrophil lactoferrin glycans, 47% are 2-linked α-mannose, 20% are 2,4-linked α-mannose, and 32% are 2,6-linked α-mannose (supplemental Table S4). MS/MS fragmentation spectra (supplemental Fig. S2) indicated that tri-antennary forms predominate over the tetra-antennary structures and that additional N-acetyllactosamine units are distributed across the antennae to give branches of similar length, rather than all being added to a single antenna. MS/MS analysis of glycans following endo-β-galactosidase digestion (supplemental Fig. S3A) or HF treatment (supplemental Fig. S4) confirms that both tri- and tetra-antennary glycans are present.
MS analysis after endo-β-galactosidase digestion (supplemental Fig. S3A and Table S5) revealed structures that were not affected by the enzyme, supporting the conclusion that neutrophil lactoferrin N-glycans frequently carry multiple Lewisx groups attached on the same antenna, in which the internal fucose residues block the action of the enzyme. Analysis of fragments released by endo-β-galactosidase digestion (supplemental Fig. S3B) also showed the presence of large amounts of Lewisx-containing tetrasaccharide (m/z 896), indicating that terminal Lewisx groups are abundant. The absence of GlcNAcβ1–3Gal oligosaccharides at m/z 518 also shows that there are no long N-acetyllactosamine extensions that do not bear fucose residues. Finally, release of fucose by HF treatment indicated that around 80% of glycans are core-fucosylated (supplemental Fig. S4 and Table S6).
Sialylated structures were detected only as a minor fraction of neutrophil lactoferrin oligosaccharides (m/z 3402, 4924, and 5098; Fig. 6A). Gas chromatography-mass spectrometry linkage analysis on untreated and desialylated neutrophil lactoferrin N-glycans revealed that both α2,3- and α2,6-linked sialic acids are present, with 58% of sialic acid in the α2,6-linkage (supplemental Table S7). In sialylated glycans, the sialic acid and fucose residues are always found on separate antennae, so that the sialyl Lewisx epitope is not observed (supplemental Fig. S2, D and H).
The MALDI-MS glycomic profile of milk lactoferrin N-glycans is considerably simpler than that of the neutrophil protein (Fig. 6B). The glycans are also of generally smaller size than those from neutrophil lactoferrin, reflecting the fact that milk lactoferrin glycans are predominantly without additional N-acetyllactosamine units added to the basic core (supplemental Table S8). Unlike the N-glycans of neutrophil lactoferrin, the most abundant glycans from milk lactoferrin have one antenna capped with sialic acid, which has previously been found to be exclusively in α2–6-linkage (44). MS/MS analysis indicated that the low abundance milk lactoferrin glycans that have additional N-acetyllactosamine units are biantennary rather than tri- or tetra-antennary and that sialic acid is present only on the unextended antenna (supplemental Fig. S5, G–J), in agreement with previous studies (44, 45). In addition, the milk lactoferrin glycans contain outer arm fucose (supplemental Fig. S5), which has previously been reported to be α1,3-linked to form Lewisx (44). The MS/MS indicates that sialic acid and fucose residues are always found on separate antennae, so that the sialyl-Lewisx epitope is not observed (supplemental Fig. S5). Unlike neutrophil lactoferrin, milk lactoferrin contains Lewisy structures (supplemental Fig. S5, F and J), consistent with the expression of α1,2-fucosyltransferase in epithelial cells but not neutrophils (46).
The much reduced abundance of terminal Lewisx groups in milk lactoferrin and the absence of clusters of this epitope compared with neutrophil lactoferrin is consistent with the reduced affinity of the milk form of the protein for SRCL. High affinity binding leading to retention of proteins on the SRCL column probably requires multivalent interaction of multiple Lewisx groups, either on separate glycans or on different branches of a single glycan. Almost every neutrophil lactoferrin molecule carries multiple Lewisx groups per molecule and is thus tightly retained, whereas milk lactoferrin molecules carry on average at most a single Lewisx group, which is insufficient to cause retention on the SRCL column.
Abundance of High Affinity Ligands for SRCL in Neutrophil Lactoferrin
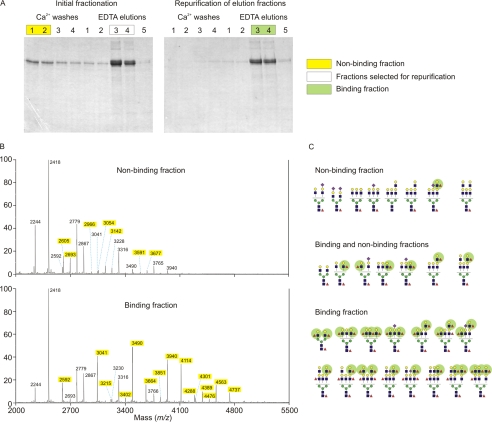
To establish a specific correlation between the abundance of Lewisx epitopes on neutrophil lactoferrin and the interaction with SRCL, lactoferrin was fractionated based on its affinity for the SRCL affinity column and glycans associated with each fraction were analyzed (Fig. 8A). The small fraction of lactoferrin molecules that did not bind to the column, or bound weakly, were washed off with buffer containing Ca2+, whereas lactoferrin molecules that bound strongly were eluted with EDTA. The molecules were re-applied to the column in the presence of Ca2+ and again eluted with EDTA. This strategy ensures that at least one of the glycans on the bound population of lactoferrin binds tightly to SRCL, although these molecules may bear a second glycan that is not a high affinity ligand, so the pool of glycans released from the binding fraction is enriched for high affinity ligands but does not consist exclusively of high affinity ligands.
FIGURE 8.
Comparison of neutrophil lactoferrin samples with high and low affinity for SRCL. A, SDS-polyacrylamide gel analysis of neutrophil lactoferrin fractionated by two rounds of affinity chromatography on immobilized SRCL. Gels were stained with Coomassie Blue, and the indicated fractions were pooled for glycan analysis. B, mass spectrometry of glycans in binding and nonbinding fractions. Species unique to each pool are highlighted in yellow. C, summary of major glycans present in binding and nonbinding fractions, based on data in B and additional MS/MS results (data not shown), with Lewisx groups highlighted in green.
The molecular ion profiles obtained for the binding and nonbinding pools of glycans are strikingly different, particularly in the region of the spectrum above m/z 3400, which includes several major peaks for the binding sample but only a very minor fraction of the glycans in the nonbinding sample (Fig. 8B). Comparing the glycan compositions found in the two samples shows that the extent to which a glycan is retained on the SRCL column depends strongly on the number of fucose residues in the structure (Fig. 8C and supplemental Tables S9 and S10). Glycans with only one fucose residue are for the most part found in the nonbinding fraction, because this single fucose is usually attached to the core GlcNAc residue and is not part of a Lewisx group. In contrast, glycans with three or more fucose residues, generally corresponding to two or more outer arm fucose residues, were found almost exclusively in the binding fraction. Glycans with two fucose residues, one of which is usually on an outer arm, were found in both fractions. Thus, the presence of more than one Lewisx group on a glycan significantly enhances binding to SRCL and may be sufficient to cause retention of the protein molecule even if the glycan at the other position does not carry any Lewisx. The isolation of a lactoferrin fraction that does not bind to SRCL served to enrich rare glycans that were not discernable in the molecular ion profile of total neutrophil lactoferrin glycans (supplemental Table S9), including a biantennary glycan in which both antennae are capped with sialic acid, which is common in serum glycoproteins but very rare in neutrophil lactoferrin. The results provide strong evidence that the efficient binding of neutrophil lactoferrin to SRCL results from the enrichment in glycans bearing multiple terminal Lewisx epitopes.
Internalization of Neutrophil Lactoferrin Mediated by SRCL
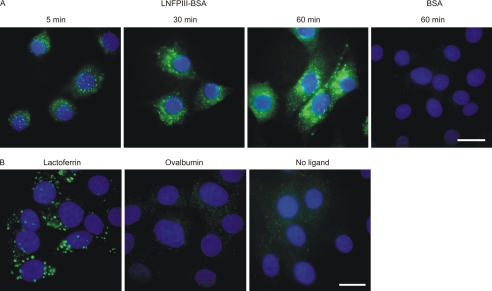
Uptake of Lewisx-bearing ligands mediated by SRCL was visualized directly by incubating fluorescein-labeled glycoprotein with SRCL-transfected fibroblasts and detected with anti-fluorescein antibody followed by fluorescently labeled secondary antibody. The assay was validated using the neoglycoprotein LNFPIII-BSA, which associates with SRCL-transfected fibroblasts within 5 min and accumulates in intracellular compartments over an hour (Fig. 9A). Fluorescein-labeled BSA that does not carry Lewisx groups is not internalized. These results are consistent with earlier indirect studies based on detection of released degradation products in the medium following uptake of LNFPIII-BSA by SRCL-transfected fibroblasts (6). When similar ligand uptake experiments were performed with neutrophil lactoferrin, internalization of fluorescein-tagged lactoferrin was readily observed in the SRCL-transfected fibroblasts (Fig. 9B). The specificity of the uptake observed was confirmed by the finding that fluorescein-tagged ovalbumin, which carries high mannose and hybrid N-glycans without Lewisx groups, was not bound and internalized. The demonstration that SRCL can mediate internalization of neutrophil granule glycoproteins is consistent with the proposal that the receptor clears such glycoproteins at sites of inflammation.
FIGURE 9.
Internalization of glycoproteins by transfected fibroblasts expressing SRCL. A, neoglycoprotein LNFPIII-BSA and control BSA ligands. B, neutrophil lactoferrin and ovalbumin ligands. Cells were incubated for the indicated times at 37 °C with fluorescein-labeled glycoproteins. After fixation, bound and internalized glycoproteins were visualized with antibodies to fluorescein followed by Alexa 488-labeled secondary antibodies. Nuclei were visualized with DAPI stain. Bar, 20 μm.
Broad Distribution of SRCL in Endothelial Cells
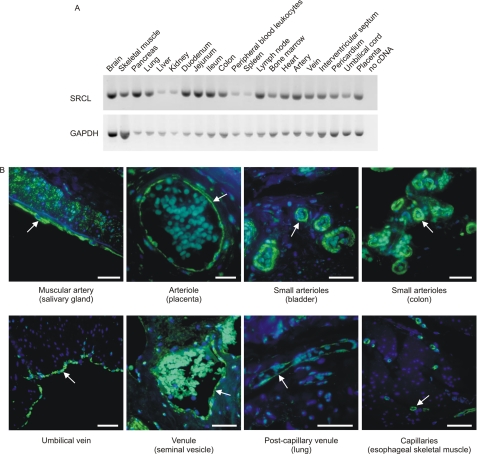
Expression of SRCL has previously been characterized in human umbilical vein endothelial cells and in a subset of endothelia, but efficient local clearance would require a broad distribution of SRCL near potential sites of glycoprotein release. A survey of the distribution of SRCL in a panel of cDNA libraries confirmed that the receptor is expressed, at varying levels, in all tissues examined, including a range of cardiovascular tissues not previously examined (Fig. 10A). A follow-up study using immunohistochemistry to examine SRCL localization revealed that SRCL is expressed throughout the vasculature in arteries, arterioles, veins, venules, and continuous capillaries (Fig. 10B). In most tissues the expression is confined to endothelial cells. Combined with earlier reports of SRCL expression patterns, these results are consistent with the idea that SRCL is widely expressed in endothelia.
FIGURE 10.
Tissue distribution of SRCL expression. A, PCR analysis of the tissue distribution of SRCL. A fragment of 492 bases, covering the CRD-encoding region, was amplified for 35 cycles. Control amplification of the cDNA for glyceraldehyde-3-hydrogenase (GAPDH) for 25 cycles was performed in parallel. B, immunohistochemical localization of SRCL in tissues using rabbit polyclonal antibodies raised against the CRD of SRCL, counterstained with peroxidase-conjugated goat anti rabbit antibody, followed by tyramide treatment and incubation with Alexa 488-labeled streptavidin. Nuclei were visualized with DAPI stain. A summary of all tissues examined is provided in supplemental Table S11. Arrows highlight examples of endothelia, and bars indicate 50 μm.
DISCUSSION
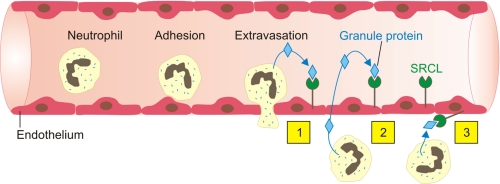
The results presented here suggest a role for SRCL in scavenging for glycoproteins released by degranulation of neutrophils in inflamed tissues. SRCL expressed on endothelial cells could clear neutrophil granule glycoproteins through multiple routes (Fig. 11). Because of the localization of SRCL throughout the vasculature, released granule glycoproteins could be cleared locally from the tissue surrounding the blood vessel by SRCL molecules expressed on the surface of the endothelial cell. In addition, glycoprotein could be cleared systemically following either direct release into the bloodstream or following diffusion into the bloodstream after release in subendothelial tissues. In either case, clearance of potentially destructive enzymes and immunomodulatory factors such as the matrix metalloproteinases and lactoferrin would have a protective effect.
FIGURE 11.
Summary diagram showing potential modes of neutrophil granule glycoprotein clearance by SRCL on endothelial cells. Possible routes of uptake include the following: 1) removal from the luminal surface of the cell following release of granule proteins from circulating neutrophils; 2) removal from the luminal surface following release in tissue and leaking back in the lumen; or 3) uptake from the tissue following local release.
Analysis of the N-glycans of neutrophil lactoferrin by mass spectrometry revealed a range of large and highly fucosylated structures. Despite considerable heterogeneity in the number and positioning of additional N-acetyllactosamine units and of fucose residues, the vast majority of the glycans carry Lewisx groups, often on multiple antennae. The glycan skeleton thus serves as a scaffold for the display of Lewisx groups, which are recognized by SRCL. The display of Lewisx structures on multiple similarly sized antennae appears to represent the optimum disposition of carbohydrate groups for binding to SRCL, because such glycans bind to the receptor with higher affinity than small glycans with only one Lewisx group as found on milk lactoferrin. Tagging of a glycoprotein with multiple copies of a receptor-targeting epitope is essential in the case of clearance receptors, because this allows them to achieve multivalent ligand binding to a soluble glycoprotein ligand. This arrangement contrasts with that found in PSGL-1, the ligand for the C-type lectin P-selectin, which displays a single sialyl-Lewisx group on one extended O-glycan antenna. This alternative arrangement of glycan ligands is effective for cell adhesion, because multiple copies of the receptor and ligand can interact when two cell surfaces come into contact (47).
The abundance of Lewisx groups in neutrophil glycoproteins is likely to be due to expression of the α1,3-fucosyltransferase FUT9, which is particularly selective for synthesis of this structure and is responsible for addition of Lewisx epitopes to neutrophil cell surface proteins (33, 48, 49). A recent study showed that SRCL recognizes CEACAM-1 expressed in transfected cells only when cells are also transfected with FUT9, as well as confirming the recognition by SRCL of proteins in a blot of neutrophil extract (50). Expression of FUT9 is limited to a small number of cell types, including granulocytes, neurons, and an unidentified cell type in the kidney (48, 51), indicating that the synthesis of Lewisx may be regulated to enable its use as a specific recognition tag. Because neutrophil secondary granule proteins are all synthesized during the same phase of neutrophil development (52, 53), they are all exposed to the same repertoire of glycosylation enzymes and could be expected to receive similar glycosylation to lactoferrin. The distinct carbohydrate recognition events mediated by the selectins and SRCL are made possible by the different pathways of synthesis of the sialyl-Lewisx and Lewisx epitopes from terminal type II N-acetyllactosamine units as follows: sialyl Lewisx is formed by the sequential actions of sialyltransferase ST3GalIV and fucosyltransferase FUT7 (47), and Lewisx is formed by the action of FUT9 and is far more abundant (42). The substrate selectivity of the enzymes involved in the synthesis of these structures is matched by the selectivity of glycan recognition in the cognate receptors (6, 54).
The functions of the Lewisx structure in neutrophils and other cells have remained unclear even though the epitope has been known for many years. The finding that the Lewisx epitope is used as a recognition tag may reflect the fact that it is an optimal structure for recognition by glycan-binding proteins because it is an accessible terminal structure that adopts a single fixed conformation whether it is free in solution or bound to a protein (55), so there is little entropy penalty for binding. This arrangement is advantageous because glycan-binding proteins typically make only limited contacts with a bound glycan (56). A role for Lewisx in mediating communication between neutrophils and dendritic cells through binding to the glycan-binding receptor DC-SIGN has been proposed (57). The work presented here is consistent with a function for Lewisx in mediating protein clearance through interaction with SRCL, suggesting that the Lewisx tag may have multiple functions resulting from binding to different receptors in different cell types.
It is proposed that SRCL functions as a distributed clearance system, found throughout the vasculature, capable of picking up glycoproteins locally as well as from the circulation. This arrangement contrasts with the asialoglycoprotein receptor, which represents a centralized system for clearance of circulating glycoproteins into the liver. The SRCL arrangement is somewhat more reminiscent of the macrophage mannose receptor, which is able to scavenge locally released glycoproteins as well as glycoproteins in serum. However, for the mannose receptor this diversity in function is a result of expression on both tissue macrophages and sinusoidal endothelial cells in the liver, although SRCL could function in both roles through expression on a single cell type. Future studies with mice lacking the murine homolog of SRCL will provide a useful in vivo complement to the biochemical and cell biological characterization presented here.
Supplementary Material
Acknowledgment
We thank Alex Powlesland for advice on the glycomic and proteomic analysis.
This work was supported by Grant 075565 from the Wellcome Trust (to M. E. T. and K. D.) and Grants B19088, SF19107, BBF0083091, and BBC5196701 from the Biotechnology and Biological Sciences Research Council (to A. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S11 and Figs. S1–S10.
- SRCL
- scavenger receptor C-type lectin
- CRD
- carbohydrate-recognition domain
- LNFP
- lacto-N-fucopentaose
- CEACAM
- carcinoembryonic antigen cell adhesion molecule
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1. Nakamura K., Funakoshi H., Miyamoto K., Tokunaga F., Nakamura T. (2001) Biochem. Biophys. Res. Commun. 280, 1028–1035 [DOI] [PubMed] [Google Scholar]
- 2. Ohtani K., Suzuki Y., Eda S., Kawai T., Kase T., Keshi H., Sakai Y., Fukuoh A., Sakamoto T., Itabe H., Suzutani T., Ogasawara M., Yoshida I., Wakamiya N. (2001) J. Biol. Chem. 276, 44222–44228 [DOI] [PubMed] [Google Scholar]
- 3. Jang S., Ohtani K., Fukuoh A., Yoshizaki T., Fukuda M., Motomura W., Mori K., Fukuzawa J., Kitamoto N., Yoshida I., Suzuki Y., Wakamiya N. (2009) J. Biol. Chem. 284, 3956–3965 [DOI] [PubMed] [Google Scholar]
- 4. Resnick D., Chatterton J. E., Schwartz K., Slayter H., Krieger M. (1996) J. Biol. Chem. 271, 26924–26930 [DOI] [PubMed] [Google Scholar]
- 5. Peiser L., Mukhopadhyay S., Gordon S. (2002) Curr. Opin. Immunol. 14, 123–128 [DOI] [PubMed] [Google Scholar]
- 6. Coombs P. J., Graham S. A., Drickamer K., Taylor M. E. (2005) J. Biol. Chem. 280, 22993–22999 [DOI] [PubMed] [Google Scholar]
- 7. McEver R. P., Zhu C. (2010) Annu. Rev. Cell Dev. Biol. 26, 363–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kerr M. A., Stocks S. C. (1992) Histochem. J. 24, 811–826 [DOI] [PubMed] [Google Scholar]
- 9. Elola M. T., Capurro M. I., Barrio M. M., Coombs P. J., Taylor M. E., Drickamer K., Mordoh J. (2007) Breast Cancer Res. Treat. 101, 161–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor M. E., Drickamer K. (2007) Curr. Opin. Cell Biol. 19, 572–577 [DOI] [PubMed] [Google Scholar]
- 11. Fiete D. J., Beranek M. C., Baenziger J. U. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2089–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee S. J., Evers S., Roeder D., Parlow A. F., Risteli J., Risteli L., Lee Y. C., Feizi T., Langen H., Nussenzweig M. C. (2002) Science 295, 1898–1901 [DOI] [PubMed] [Google Scholar]
- 13. Park E. I., Mi Y., Unverzagt C., Gabius H. J., Baenziger J. U. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17125–17129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steirer L. M., Park E. I., Townsend R. R., Baenziger J. U. (2009) J. Biol. Chem. 284, 3777–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schatz P. J. (1993) Bio/Technology 11, 1138–1143 [DOI] [PubMed] [Google Scholar]
- 16. Eisenberg S. P., Evans R. J., Arend W. P., Verderber E., Brewer M. T., Hannum C. H., Thompson R. C. (1990) Nature 343, 341–346 [DOI] [PubMed] [Google Scholar]
- 17. Smith P. A., Tripp B. C., DiBlasio-Smith E. A., Lu Z., LaVallie E. R., McCoy J. M. (1998) Nucleic Acids Res. 26, 1414–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pipirou Z., Powlesland A. S., Steffen I., Pöhlmann S., Taylor M. E., Drickamer K. (2011) Glycobiology 21, 806–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burnette W. N. (1981) Anal. Biochem. 112, 195–203 [DOI] [PubMed] [Google Scholar]
- 20. Miroli A. A., James B. M., Spitz M. (1986) J. Immunol. Methods 88, 91–96 [DOI] [PubMed] [Google Scholar]
- 21. Powlesland A. S., Hitchen P. G., Parry S., Graham S. A., Barrio M. M., Elola M. T., Mordoh J., Dell A., Drickamer K., Taylor M. E. (2009) Glycobiology 19, 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 23. Jang-Lee J., North S. J., Sutton-Smith M., Goldberg D., Panico M., Morris H., Haslam S., Dell A. (2006) Methods Enzymol. 415, 59–86 [DOI] [PubMed] [Google Scholar]
- 24. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., Haslam S. M. (2008) J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 25. Dell A., Khoo K. H., Panico M., McDowell R. A., Etienne A. T., Reason A. J., Morris H. R. (1993) in Glycobiology: A Practical Approach (Fukuda M., Kabata A. eds) pp. 187–222, Oxford University Press, Oxford [Google Scholar]
- 26. McCarthy N. C., Albrechtsen M. T., Kerr M. A. (1985) Biosci. Rep. 5, 933–941 [DOI] [PubMed] [Google Scholar]
- 27. Terstappen L. W., Hollander Z., Meiners H., Loken M. R. (1990) J. Leukocyte Biol. 48, 138–148 [DOI] [PubMed] [Google Scholar]
- 28. Houston D. R., Recklies A. D., Krupa J. C., van Aalten D. M. (2003) J. Biol. Chem. 278, 30206–30212 [DOI] [PubMed] [Google Scholar]
- 29. Volck B., Price P. A., Johansen J. S., Sørensen O., Benfield T. L., Nielsen H. J., Calafat J., Borregaard N. (1998) Proc. Assoc. Am. Physicians 110, 351–360 [PubMed] [Google Scholar]
- 30. Bogoevska V., Nollau P., Lucka L., Grunow D., Klampe B., Uotila L. M., Samsen A., Gahmberg C. G., Wagener C. (2007) Glycobiology 17, 324–333 [DOI] [PubMed] [Google Scholar]
- 31. Lucka L., Fernando M., Grunow D., Kannicht C., Horst A. K., Nollau P., Wagener C. (2005) Glycobiology 25, 87–100 [DOI] [PubMed] [Google Scholar]
- 32. Skubitz K. M., Snook R. W., 2nd (1987) J. Immunol. 139, 1631–1639 [PubMed] [Google Scholar]
- 33. Bogoevska V., Horst A., Klampe B., Lucka L., Wagener C., Nollau P. (2006) Glycobiology 16, 197–209 [DOI] [PubMed] [Google Scholar]
- 34. van Gisbergen K. P., Ludwig I. S., Geijtenbeek T. B., van Kooyk Y. (2005) FEBS Lett. 579, 6159–6168 [DOI] [PubMed] [Google Scholar]
- 35. Albrechtsen M., Kerr M. A. (1989) Br. J. Haematol. 72, 312–320 [DOI] [PubMed] [Google Scholar]
- 36. Buescher E. S., Livesey S. A., Linner J. G., Skubitz K. M., McIlheran S. M. (1990) Anat. Rec. 228, 306–314 [DOI] [PubMed] [Google Scholar]
- 37. Shepherd V. L., Hoidal J. R. (1990) Am. J. Respir. Cell Mol. Biol. 2, 335–340 [DOI] [PubMed] [Google Scholar]
- 38. Coombs P. J., Harrison R., Pemberton S., Quintero-Martinez A., Parry S., Haslam S. M., Dell A., Taylor M. E., Drickamer K. (2010) J. Mol. Biol. 396, 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Legrand D., Pierce A., Elass E., Carpentier M., Mariller C., Mazurier J. (2008) Adv. Exp. Med. Biol. 606, 163–194 [DOI] [PubMed] [Google Scholar]
- 40. van Berkel P. H., van Veen H. A., Geerts M. E., de Boer H. A., Nuijens J. H. (1996) Biochem. J. 319, 117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Derisbourg P., Wieruszeski J. M., Montreuil J., Spik G. (1990) Biochem. J. 269, 821–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Babu P., North S. J., Jang-Lee J., Chalabi S., Mackerness K., Stowell S. R., Cummings R. D., Rankin S., Dell A., Haslam S. M. (2009) Glycoconj. J. 26, 975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fukuda M., Spooncer E., Oates J. E., Dell A., Klock J. C. (1984) J. Biol. Chem. 259, 10925–10935 [PubMed] [Google Scholar]
- 44. Spik G., Strecker G., Fournet B., Bouquelet S., Montreuil J., Dorland L., van Halbeek H., Vliegenthart J. F. (1982) Eur. J. Biochem. 121, 413–419 [DOI] [PubMed] [Google Scholar]
- 45. Matsumoto A., Yoshima H., Takasaki S., Kobata A. (1982) J. Biochem. 91, 143–155 [DOI] [PubMed] [Google Scholar]
- 46. Mollicone R., Cailleau A., Oriol R. (1995) Transfus. Clin. Biol. 2, 235–242 [DOI] [PubMed] [Google Scholar]
- 47. Sperandio M., Gleissner C. A., Ley K. (2009) Immunol. Rev. 230, 97–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakayama F., Nishihara S., Iwasaki H., Kudo T., Okubo R., Kaneko M., Nakamura M., Karube M., Sasaki K., Narimatsu H. (2001) J. Biol. Chem. 276, 16100–16106 [DOI] [PubMed] [Google Scholar]
- 49. Nishihara S., Iwasaki H., Kaneko M., Tawada A., Ito M., Narimatsu H. (1999) FEBS Lett. 462, 289–294 [DOI] [PubMed] [Google Scholar]
- 50. Samsen A., Bogoevska V., Klampe B., Bamberger A. M., Lucka L., Horst A. K., Nollau P., Wagener C. (2010) Eur. J. Cell Biol. 89, 87–94 [DOI] [PubMed] [Google Scholar]
- 51. Comelli E. M., Head S. R., Gilmartin T., Whisenant T., Haslam S. M., North S. J., Wong N. K., Kudo T., Narimatsu H., Esko J. D., Drickamer K., Dell A., Paulson J. C. (2006) Glycobiology 16, 117–131 [DOI] [PubMed] [Google Scholar]
- 52. Nauseef W. M. (1999) J. Leukocyte Biol. 66, 867–868 [DOI] [PubMed] [Google Scholar]
- 53. Faurschou M., Borregaard N. (2003) Microbes Infect. 5, 1317–1327 [DOI] [PubMed] [Google Scholar]
- 54. Revelle B. M., Scott D., Kogan T. P., Zheng J., Beck P. J. (1996) J. Biol. Chem. 271, 4289–4297 [DOI] [PubMed] [Google Scholar]
- 55. Imberty A. (1997) Curr. Opin. Struct. Biol. 7, 617–623 [DOI] [PubMed] [Google Scholar]
- 56. Weis W. I., Drickamer K. (1996) Annu. Rev. Biochem. 65, 441–473 [DOI] [PubMed] [Google Scholar]
- 57. van Gisbergen K. P., Geijtenbeek T. B., van Kooyk Y. (2005) Trends Immunol. 26, 626–631 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.