Abstract
Candidate gene-based studies have identified a handful of aberrant CpG DNA methylation events in prostate cancer. However, DNA methylation profiles have not been compared on a large scale between prostate tumor and normal prostate, and the mechanisms behind these alterations are unknown. In this study, we quantitatively profiled 95 primary prostate tumors and 86 benign adjacent prostate tissue samples for their DNA methylation levels at 26,333 CpGs representing 14,104 gene promoters by using the Illumina HumanMethylation27 platform. A 2-class Significance Analysis of this data set revealed 5912 CpG sites with increased DNA methylation and 2151 CpG sites with decreased DNA methylation in tumors (FDR < 0.8%). Prediction Analysis of this data set identified 87 CpGs that are the most predictive diagnostic methylation biomarkers of prostate cancer. By integrating available clinical follow-up data, we also identified 69 prognostic DNA methylation alterations that correlate with biochemical recurrence of the tumor. To identify the mechanisms responsible for these genome-wide DNA methylation alterations, we measured the gene expression levels of several DNA methyltransferases (DNMTs) and their interacting proteins by TaqMan qPCR and observed increased expression of DNMT3A2, DNMT3B, and EZH2 in tumors. Subsequent transient transfection assays in cultured primary prostate cells revealed that DNMT3B1 and DNMT3B2 overexpression resulted in increased methylation of a substantial subset of CpG sites that showed tumor-specific increased methylation.
Prostate cancer is the most commonly diagnosed malignancy for men in the United States, with an estimated 217,730 new cases projected for 2010 (Jemal et al. 2010). After more than two decades of widespread serum prostate-specific antigen (PSA) testing, clinical prostate cancer has shifted to a predominantly localized disease. However, two large-scale, randomized trials of PSA screening suggest that prostate cancer is overdiagnosed and overtreated, likely because many cancers that are detected are never destined to progress (Andriole et al. 2009; Schröder et al. 2009). However, prostate cancer can have an aggressive and lethal course, and an estimated 32,050 men were projected to die of prostate cancer in 2010 (Jemal et al. 2010). This broad range of clinical behavior is likely a reflection of the underlying genomic diversity of the tumors (Taylor et al. 2010). Previous studies of prostate tumors reported significant heterogeneity in the gene expression profiles and genomic structural alterations including DNA copy number changes and gene fusions often involving the ETS family of transcription factors detectable in approximately half of prostate tumors (Singh et al. 2002; Lapointe et al. 2004; Tomlins et al. 2005, 2008; King et al. 2009; Sboner et al. 2010; Taylor et al. 2010; Pflueger et al. 2011; Robbins et al. 2011). However, exon sequencing of known oncogenes and tumor suppressors has found few somatic mutations, and the calculated background mutation rate appears to be relatively low (Taylor et al. 2010). This suggests the presence of other forms of genomic aberrations that contribute to the observed gene expression variations, and, in turn, the diversity in tumor behavior.
DNA methylation has long been suspected to play a role in tumorigenesis and cancer progression in various tissue types (Lapeyre et al. 1981; Jones 1986; Laird and Jaenisch 1994, 1996; Ehrlich 2002; Esteller and Herman 2002; Patra et al. 2002; Das and Singal 2004). Early studies in cancer epigenetics revealed an overall reduction of 5-methylcytosine in various tumor genomes (Feinberg and Vogelstein 1983; Gama-Sosa et al. 1983). In contrast, more recent studies identified many hypermethylation events in CpG islands near known tumor-suppressor transcriptional start sites, which correlated with reduction in transcript levels (Lee et al. 1994; Brooks et al. 1998). Many of these candidate gene-based approaches have led to discovery of potentially prognostic DNA methylation events (Müller et al. 2003; Kim et al. 2008). However, recent advances in microarray and high-throughput massively parallel sequencing technologies have enabled investigators to study site-specific DNA methylation events on a much broader scale. Recent studies of the DNA methylome in colorectal cancer and glioblastomas have revealed valuable new insights into those diseases, including the discovery of hundreds of affected genes previously not identified (Cancer Genome Atlas Research Network 2008; Irizarry et al. 2009; Noushmehr et al. 2010).
In prostate cancer, hypermethylation of CpG islands within several tumor-suppressor promoters has been well documented (Lee et al. 1994; Brooks et al. 1998; Jerónimo et al. 2004). In addition, Kron et al. (2009) recently reported the DNA methylation profiles of 20 prostate tumors at CpG islands across the genome using a human CpG island microarray. However, this study did not determine the profiles of normal prostate tissues and was thus limited to comparisons between the prostate tumors and six cases of age-matched lymphocytes. While specific sites of methylation level heterogeneity among tumor samples were identified (Kron et al. 2010; Liu et al. 2011), the study design precluded the examination of changes in methylation between normal prostate and prostate tumors. In addition to these studies of prostate cancer methylation profiles, a few studies looking at DNA methyltransferases (DNMTs) and DNMT-interacting proteins have suggested that misregulation of these genes in prostate cancer is responsible for the improper DNA methylation events in primary tumors and cell lines (Hoffmann et al. 2007; Yaqinuddin et al. 2008; Ley et al. 2010).
Here, we quantitatively profiled 95 primary prostate tumors and 86 benign adjacent prostate tissues for their DNA methylation levels at 26,333 CpG sites in 14,104 gene promoters. Based on the results, we identified the differentially methylated CpGs and explored subsets of them that accurately distinguished tumor and benign adjacent prostate tissues. We then integrated available clinical data to discover novel prognostic markers of aggressive tumors. Finally, we investigated the DNMT protein family, as well as their interacting partners, for their role in the alteration of DNA methylation in prostate cancer.
Results
To explore the prostate DNA methylome, we profiled 95 primary prostate tumors and 86 benign adjacent prostate tissues, including 70 matched pairs, using the Illumina HumanMethylation27 microarrays. These tissue samples were harvested from men who underwent radical retropubic prostatectomy for clinically localized prostate cancer. Surgeries were performed between 1998 and 2007, and detailed clinical data, including follow-up and recurrence status, were available in 96 patients (88%). Mean patient age, pre-operative serum PSA levels, clinical stage, and pathological Gleason grade were compatible with the risk profiles of contemporary patients undergoing surgery for prostate cancer (Supplemental Table S1; Brooks et al. 2008).
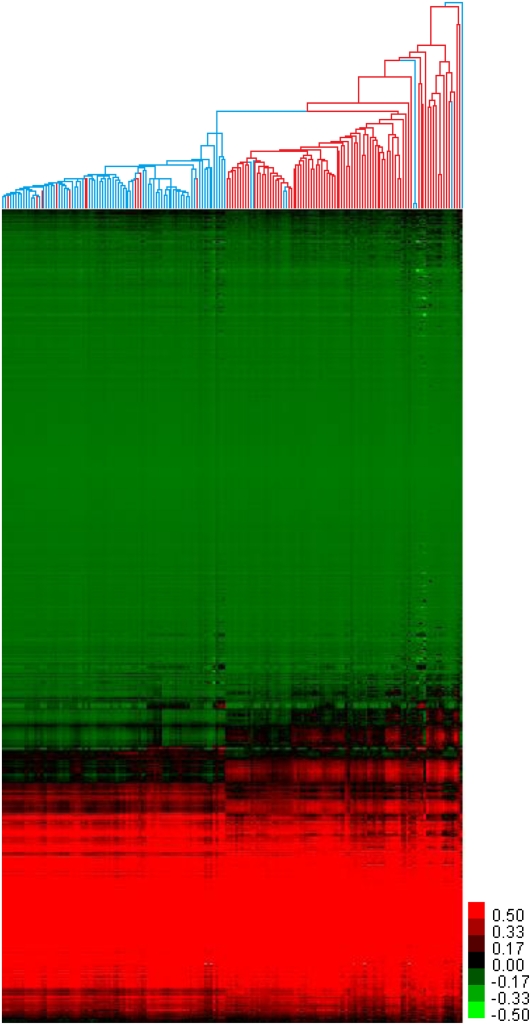
The Illumina HumanMethylation27 platform assays 27,578 CpG sites, almost all in the proximal promoter regions of 14,495 transcription start sites (Weisenberger et al. 2008; Hernandez et al. 2011). After batch-correcting and quality-filtering the data, we were able to determine quantitative methylation status (beta scores; range: 0 to 1) for 26,333 CpG sites in 14,104 promoters. To investigate the similarities and differences of the DNA methylation profiles of the benign adjacent samples and tumor samples, as well as their heterogeneity, we performed unsupervised hierarchical clustering on the entire data set (Fig. 1). When the data were clustered by sample, we observed two main clusters—one composed almost entirely of benign adjacent samples (77/88) and the other almost entirely of the tumor samples (67/71). The branch lengths in the benign adjacent sample cluster were generally shorter than the branch lengths in the tumor sample cluster, indicating more heterogeneity in methylation profiles among the tumor samples. Twenty-two of the samples did not fall into either of the two main clusters and formed long off-shooting branches or small clusters. Eighteen of these were tumor samples, further indicative of the heterogeneous nature of the tumor DNA methylome. By visual inspection, the majority of the samples showed relatively little methylation change between the tumor and benign adjacent clusters (Fig. 1), and most of these invariable CpG sites showed low levels of methylation in both benign adjacent and tumor samples. However, there were distinct CpG clusters with methylation patterns that distinguished the benign adjacent or tumor sample clusters, and, strikingly, a large number of CpG sites showed increased methylation in the tumor cluster compared to the benign adjacent cluster.
Figure 1.
Hierarchical clustering of prostate tissues by DNA methylation. Unsupervised hierarchical clustering of 181 prostate tissues and 26,333 CpGs, by sample and by CpG. (Red branches) Tumor samples; (blue branches) benign adjacent samples; (red pixels) high DNA methylation; (green pixels) low DNA methylation.
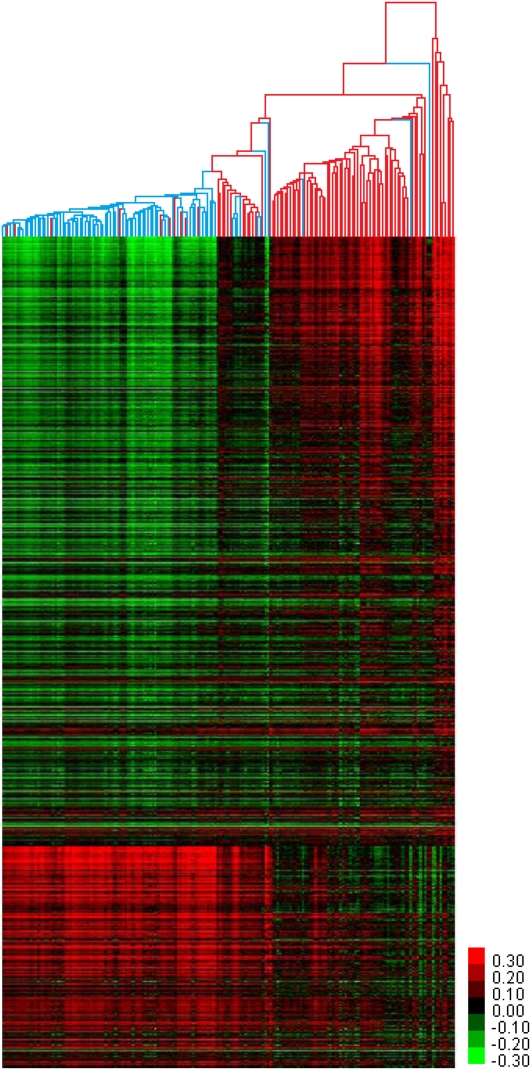
To identify the CpG sites with statistically different DNA methylation status between benign adjacent prostate tissues and tumors, we performed a two-class Significance Analysis of Microarrays (SAM) (Tusher et al. 2001). As we had matched benign adjacent tissues for only 70 of the 95 tumors used in this study, we conducted the SAM analysis as unpaired. The analysis identified 5912 CpG sites hypermethylated in tumors compared to benign adjacent tissues and 2151 CpG sites hypomethylated at FDR < 0.8% (Supplemental Fig. S1; Supplemental Table S2). We performed hierarchical clustering on all samples based on these 8063 differentially methylated CpG sites (Fig. 2). When the fold-change was examined for these sites, 1851 sites had a twofold or greater change and as high as a 141-fold increase in methylation for a CpG near the transcriptional start site of ZNF296 (average normal beta: 5.30×10−4, average tumor beta: 0.0756). CpG island hypermethylation of ZNF296 has been observed and implicated in its transcriptional silencing in oligodendroglioma (Hong et al. 2003; Noushmehr et al. 2010). In addition, ZNF296 has been reported to be drastically overexpressed in acute myeloid leukemia (Poland et al. 2009). This suggests that the aberrant gene expression of ZNF296, as a result of DNA methylation or otherwise, is a common event in tumorigenesis or tumor progression. All but 609 of the CpGs had a change of 5% or greater. While these 609 sites had a low level of fold-change, these were nonetheless identified as statistically significant changes that were detectable because of the large sample size (Supplemental Fig. S1).
Figure 2.
Differentially methylated CpGs of prostate tumors. Unsupervised hierarchical clustering of 181 prostate tissues based on the 5912 and 2151 CpG sites hypermethylated and hypomethylated in prostate tumors, respectively, as identified by 2-class SAM. (Red branches) Tumor samples; (blue branches) benign adjacent samples; (red pixels) high DNA methylation; (green pixels) low DNA methylation.
The 8063 differentially methylated sites corresponded to 4227 promoters with at least one hypermethylated CpG and 1795 promoters with at least one hypomethylated CpG. Of the 11,116 gene promoters represented by two or more CpG sites on the HumanMethylation27 platform, only 223 had opposite methylation effects (i.e., at least one hypermethylated CpG and at least one hypomethylated CpG) (Supplemental Table S3). When the distances from transcriptional start sites were compared in these 223 promoters with opposite methylation effects, we saw enrichment for hypermethylated CpGs in the −100-bp to +800-bp range, whereas we saw enrichment for the hypomethylated CpGs in the −700-bp to −200-bp range. Thus, overall, nearly one-third (8063/26,333) of assayed promoter CpGs had a statistically significant change in DNA methylation, with most of those showing an increase in methylation. Interestingly, 41% (5799/14,104) of all gene promoters assayed had at least one CpG with a tumor-specific methylation change. We repeated this analysis using two-class paired SAM on only the 70 matched sample pairs and observed similar results (Supplemental Text S1).
Diagnostic methylation markers
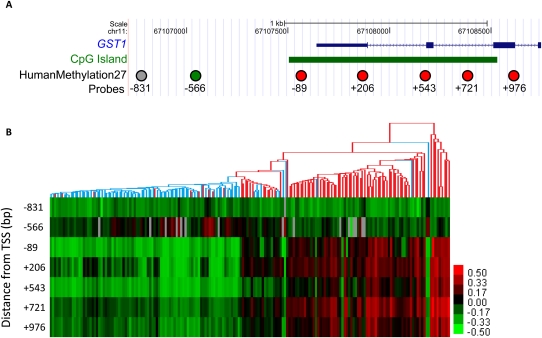
Among the CpG sites that we found to be differentially methylated in tumor versus benign adjacent prostate tissues by SAM, and shown clustered in Figure 2, were several sites that had been previously characterized in prostate tumors, most notably several CpG sites near or within the GSTP1 gene. Hypermethylation of the CpG island overlapping the transcriptional start site of the GSTP1 gene has been associated with transcriptional silencing and is described as the most common molecular alteration in prostate cancer identified to date (Lin et al. 2001; Woodson et al. 2008). Since GSTP1 promoter methylation is very common and specific for prostate cancer, many investigators have proposed using this methylation event as a diagnostic biomarker for prostate cancer (Cairns et al. 2001; Nakayama et al. 2004). The HumanMethylation27 arrays contain seven CpG sites in the GSTP1 promoter. Five of these sites showed significantly increased DNA methylation in tumors, four of which are located in the promoter CpG island that had been previously characterized as a site of hypermethylation in prostate cancer (Brooks et al. 1998), while the fifth lies 88 bp downstream from the annotated CpG island boundary (red circles in Fig. 3A). The two remaining CpGs showed either no differential methylation (gray circle in Fig. 3A) or slight but statistically significant hypomethylation (green circle in Fig. 3A); both lie further upstream of the transcriptional start site, outside of the promoter CpG island. Our data not only confirm the previously described hypermethylation of the GSTP1 promoter CpG island, but also show that CpG DNA methylation alteration is highly context-dependent even within a single promoter.
Figure 3.
GSTP1 CpG island hypermethylation in prostate tumors. (A) Diagram of the RefSeq annotation of the GSTP1 gene. (Green box) CpG island calculated by the UCSC Genome Browser. Circles are CpG sites assayed by HumanMethylation27. (Red circles) Probes that were identified to be hypermethylated in prostate tumors by 2-class SAM; (green circle) probe that was hypomethylated; (gray circle) probe that showed no significant change. The numbers below the circles indicate the relative distance in base pairs from the predicted TSS. (B) Heatmap depicts DNA methylation pattern of the seven probes near GSTP1. The dendrogram is based on the hierarchical clustering from Figure 2. (Red branches) Tumor samples; (blue branches) benign adjacent samples. Coordinates are based on the NCBI36/hg18 human genome assembly.
In addition to GSTP1, we also examined our data specifically for methylation changes in the promoters of APC and RASSF1, which have also been previously shown to have hypermethylation in prostate cancer (Jerónimo et al. 2004) and were represented by multiple probes on the HumanMethylation27 array. With APC, all six CpG sites represented on the array showed hypermethylation in tumors, located 122 bp upstream to 488 bp downstream of the TSS (Supplemental Fig. S3). With RASSF1, three CpGs sites were probed, located 58 bp upstream to 176 bp downstream of the TSS and within a CpG island boundary; all three were hypermethylated (Supplemental Fig. S4). However, five of the six probes located more than 2 kb downstream from the TSS in a second CpG island did not show differential methylation.
While hierarchical clustering of samples using the most differentially methylated CpG sites (the set shown in Fig. 2) was able to distinguish most tumors from benign adjacent tissues, the classification was not perfect, as indicated by the inclusion of benign adjacent tissue samples within the tumor cluster and vice versa. To identify CpG sites that could best predict either the tumor state or the benign adjacent state, we performed a Prediction Analysis of Microarrays (PAM) to perform sample classification (Tibshirani et al. 2002). This analysis generated a list of 87 predictive CpG sites, most of which had increased methylation in the tumor samples (83/87), and represented 82 gene promoters total (Supplemental Fig. S5; Supplemental Table S4). The CYBA, GSTP1, KLK10, PPT2, and CXCL1 promoters each had two CpGs represented in this list. Notably, in this ranked list of 87 predictive methylation alterations, the GSTP1 hypermethylation was ranked 57th (Supplemental Table S4). Thus, we have identified 56 molecular events, most of which had not been previously characterized, that are better identifiers of prostate cancer than is GSTP1. We validated several of these diagnostic methylation markers by PyroMark sequencing (Supplemental Text S2).
Prognostic methylation markers
To explore tumor heterogeneity, we compared the methylation profiles of the 86 tumors with respect to Gleason grade and time to biochemical recurrence (defined as serum PSA > 0.07 ng/mL after surgery) of the donors. Gleason grade is a powerful predictor of treatment failure, tumor progression, and death from prostate cancer; and biochemical recurrence has also been correlated with prostate cancer-specific mortality (Freedland et al. 2005). We conducted a multiclass SAM in an effort to identify methylation events that distinguished tumors of different Gleason grades but were unable to identify such events. Next, we conducted a SAM survival analysis with the time to biochemical recurrence as the survival variable. With a false discovery rate of 26.8%, we identified six CpGs that showed greater methylation in tumors from men who had shorter time to recurrence and 63 CpGs that showed lower methylation in patients with shorter time to recurrence (Supplemental Table S5). This strong bias toward lower methylation in aggressive tumors was striking as we observed a bias for CpG sites with increased methylation in the tumor/benign adjacent comparison. At a false discovery rate of 26.8%, we expect 18 of those calls to be false. At a lower false discovery rate cutoff of 1%, we only observed four CpGs that showed higher methylation in patients with shorter time to recurrence and none that showed lower methylation (Supplemental Table S5). Strikingly, we did not observe the differential methylation of the CpG island described by Liu et al. (2011), and this may be due to the different platforms used, different length of clinical follow-up, or differences in the distribution of samples of various Gleason scores. This discrepancy warrants further investigation. While we were only able to identify a small number of CpGs whose methylation state correlated with time to recurrence, we noted that several of these CpG sites are in the proximal promoter regions of known cancer-related genes, including three CpGs near MAGE gene family members that encode for strictly tumor-specific antigens (Chomez et al. 2001) and four CpGs near WT1, a transcription factor gene associated with Wilm's tumor.
Correlation of tumor hypermethylation with DNA methyltransferase expression
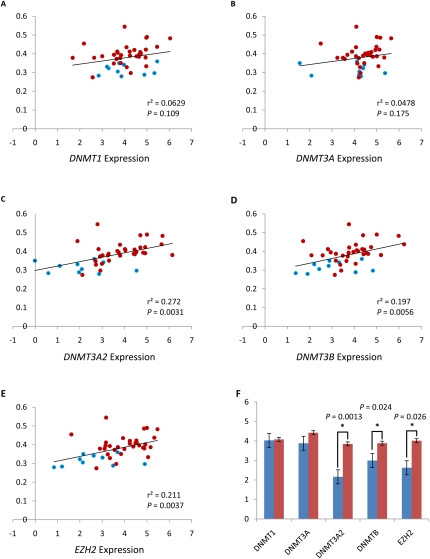
With nearly one-third of assayed CpGs showing changes in DNA methylation between tumor and benign adjacent samples, we hypothesized that one or more of the DNA methyltransferases (DNMTs), or a protein that interacts with a DNMT, had altered activity, possibly due to changes in transcript abundance, in prostate tumors. Such alterations in activity could, in turn, lead to global DNA methylation changes. To test this hypothesis, we selected RNA from 10 of the benign adjacent and 36 of the tumor samples, and measured the transcript abundance of DNMT1, DNMT3A, DNMT3A2, DNMT3B, DNMT3L, and EZH2 using the TaqMan Gene Expression assay. These genes comprise the known maintenance methyltransferase (DNMT1) (Chuang et al. 1997), all known methyltransferases with de novo capability (DNMT1 [Estève et al. 2005], DNMT3A [Okano et al. 1999], DNMT3B [Okano et al. 1999]), and two interacting proteins thought to target methyltransferases to specific genomic regions (DNMT3L [El-Maarri et al. 2009] and EZH2 [Okano et al. 1999]). In addition, we uniquely assayed DNMT3A and its alternative promoter variant DNMT3A2 by using transcript-specific primers and probes. While several splice variants of DNMT3B have been characterized, we were unable to design variant-specific primers and probes for them, so instead we designed primers and probes to the common region of all DNMT3B variants. We did not observe detectable levels of DNMT3L transcript abundance from either tumor or benign adjacent samples (data not shown). When the transcript levels of the remaining genes were compared between benign adjacent and tumor samples with a two-tailed t-test, three showed significant changes: DNMT3A2 (P = 0.0013), DNMT3B (P = 0.024), and EZH2 (P = 0.026), while DNMT1 and DNMT3A did not (Fig. 4F).
Figure 4.
Expression of DNMTs and EZH2 correlates with global hypermethylation in prostate tumors. Comparison of transcript levels of DNMTs and EZH2 measured by TaqMan qPCR with the average DNA methylation levels of CpG sites that are hypermethylated in prostate tumors. (Blue circles) Benign adjacent samples; (red circles) tumor samples. The P-value was calculated by linear regression analysis. y-axis: average DNA methylation levels (beta score); x-axis: relative gene expression levels [log2(RQ)]; (black line) linear regression. (A) DNMT1 expression. (B) DNMT3A expression. (C) DNMT3A2 expression. (D) DNMT3B expression. (E) EZH2 expression. (F) Comparison of DNMTs and EZH2 transcript levels between benign adjacent tissues (blue) and tumors (red). Significant differences are indicated by asterisks; P-values were calculated by t-test. Standard errors are depicted by error bars. For part F, the y-axis is relative gene expression levels [log2(RQ)].
We compared the expression values for these five genes to global DNA methylation levels. Specifically, we plotted the mean percent methylation of all 5912 hypermethylated CpG sites against relative expression of each methyltransferase or interacting protein, and calculated regression and the goodness-of-fit of the regression for each sample. Again, DNMT3A2 (r2 = 0.272, P = 0.0031), DNMT3B (r2 = 0.197, P = 0.0056), and EZH2 (r2 = 0.211, P = 0.0037) all showed significant correlation between expression and global hypermethylation, while DNMT1 and DNMT3A did not (Fig. 4A–E). The correlation between DNMT3A2, DNMT3B, and EZH2 expression and global hypermethylation, in conjunction with the observed overexpression of the same genes in tumors, suggests a possible causal role in the global methylation changes seen in prostate tumor.
DNMT overexpression recapitulates hypermethylation events seen in prostate tumors
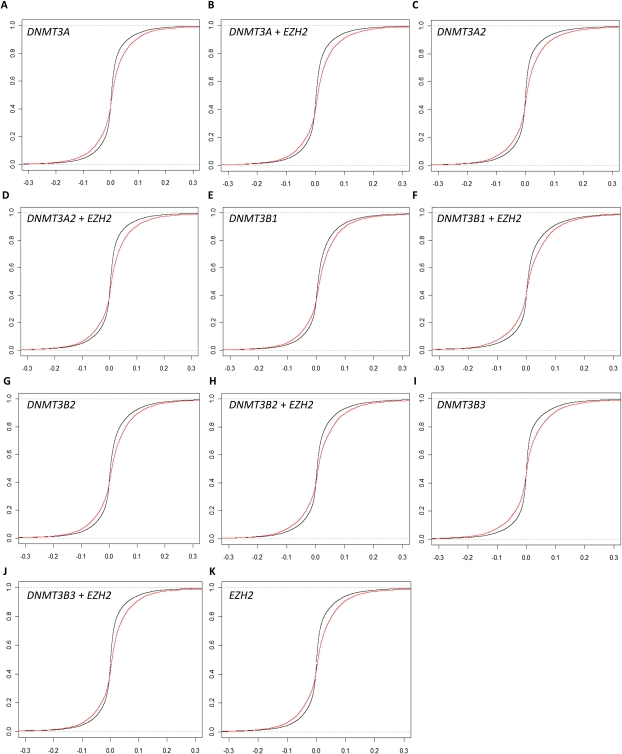
To determine whether the increased transcript abundance of DNMT3A2, DNMT3B, and EZH2 in tumor cells has a causal role in the hypermethylation of a large number of promoter CpGs, we expressed these genes from the CMV promoter in transient transfection assays in primary cultures of normal prostatic epithelial cells. We used plasmids expressing DNMT3A, DNMT3A2, DNMT3B1, DNMT3B2, and DNMT3B3, an EZH2-cDNA plasmid, and a no-insert plasmid. We co-transfected each cDNA plasmid with the no-insert plasmid, and independently with the EZH2 plasmid, and also included a mock “no-insert plasmid only” transfection. We calculated the change in DNA methylation for each CpG between each cDNA transfection and the mock transfection after 48 h. We then plotted the ideal cumulative distribution function of the DNA methylation level change at all 26,333 CpG sites along with the empirical cumulative distribution function of just the changes at the 5912 CpG sites hypermethylated in tumors (Fig. 5A–K), and tested the difference between the two distribution functions using the Kolmogorov-Smirnov (K-S) test. In all 11 experimental transfections, the distribution of the 5912 CpG sites was significantly enriched compared to the null: DNMT3A (P = 6.0×10−45), DNMT3A2 (P = 3.5×10−62), DNMT3B1 (P = 1.2×10−31), DNMT3B2 (P = 5.2×10−39), DNMT3B3 (P = 4.6×10−44), EZH2 (P = 1.1×10−59), DNMT3A+EZH2 (P = 7.8×10−64), DNMT3A2+EZH2 (P = 9.8×10−65), DNMT3B1+EZH2 (P = 2.1×10−29), DNMT3B2+EZH2 (P = 6.7×10−42), and DNMT3B3+EZH2 (P = 2.5×10−67). Consistent with our hypothesis, when the plots of the empirical cumulative distribution functions were visually inspected, we observed that the low P-value of the K-S test appeared to be driven more by the CpGs of increased methylation rather than CpGs of decreased methylation in all 11 conditions.
Figure 5.
Overexpression of DNMTs and EZH2 results in increased methylation at a subset of prostate tumor hypermethylation sites. Ideal (black) and empirical (red) cumulative distribution functions of change in DNA methylation after DNMT or EZH2 transfection into cultured normal prostate cells. The empirical distribution functions are based on the 5912 CpGs that were hypermethylated in prostate tumors, while the ideal distribution functions are based on all 26,333 CpGs assayed on the array. Overexpression of (A) DNMT3A, (B) DNMT3A2, (C) DNMT3B1, (D) DNMT3B2, (E) DNMT3B3, (F) EZH2, (G) DNMT3A and EZH2, (H) DNMT3A2 and EZH2, (I) DNMT3B1 and EZH2, (J) DNMT3B2 and EZH2, and (K) DNMT3B3 and EZH2.
To test specifically whether the list of 5912 CpG sites was statistically enriched for CpGs with substantially increased DNA methylation, we performed a series of chi-square tests. Based on the distribution of CpG methylation levels in tumor and benign adjacent tissues at these CpG sites, we set a cutoff value of 0.05. In other words, CpG sites where the methylation increased by 5% or greater in the experimental transfection compared to the mock transfection were considered to have substantially increased DNA methylation. We calculated expected values based on the distribution of these CpGs with substantially increased DNA methylation in the entire set of 26,333 CpGs. When chi-square tests were performed, all 11 experimental conditions had very low P-values: DNMT3A (P = 1.1×10−45), DNMT3A2 (P = 1.7×10−66), DNMT3B1 (P = 8.9×10−127), DNMT3B2 (P = 1.8×10−157), DNMT3B3 (P = 6.6×10−10), EZH2 (P = 9.4×10−31), DNMT3A+EZH2 (P = 1.5×10−13), DNMT3A2+EZH2 (P = 1.1×10−11), DNMT3B1+EZH2 (P = 1.9×10−185), DNMT3B2+EZH2 (P = 9.4×10−107), and DNMT3B3+EZH2 (P = 2.3×10−68). DNMT3B1 and DNMT3B2, which are alternative splicing isoforms differing by the presence of one exon, both in the presence and absence of EZH2 co-transfection, showed the lowest P-values, all <1×10−100. From these data, we conclude that our list of 5912 CpGs is, indeed, enriched for CpGs with substantially increased methylation when DNMTs or EZH2 were overexpressed, with DNMT3B1 and DNMT3B2 appearing to have the strongest impact on the DNA methylation levels at these sites.
Based on these data, we further investigated the altered DNA methylation in the DNMT3B1 and DNMT3B2 overexpression experiments. Because these splice isoforms differ by only one exon coding for 21 amino acids in a linker region (Sakai et al. 2004), we suspected that they would share many targets. To identify the CpGs targeted by DNMT3B1 and DNMT3B2 in prostate tumors, we examined the list of CpGs that were hypermethylated in prostate tumors and in the overexpression experiments. Specifically, we looked for overlaps in the list of CpGs with 5% or greater increase in methylation compared to the mock in the DNMT3B1 (1267 CpGs), DNMT3B1+EZH2 (1322 CpGs), DNMT3B2 (1261 CpGs), and DNMT3B2+EZH2 (1235 CpGs) overexpression experiments. Four hundred and thirty-eight CpGs were represented in all four lists, and an additional 425 CpGs were represented in three of the four lists. We performed two permutation tests to determine the likelihood of our results. In the first permutation test, we generated four lists of CpGs (1267, 1322, 1261, and 1235 CpGs, respectively) drawn randomly from the whole list of 26,333 CpGs and counted the number of incidences where there was an overlap of 438 CpGs in all four lists. This level of overlap (438 CpGs) was never observed in the 10,000 permutations. In our second permutation test, we repeated the first permutation test but changed the criteria to observing at least 863 CpGs overlapping in three of the four lists. This, too, was never observed in 10,000 permutations. This provided further evidence that the differentially methylated CpGs in the DNMT3B1 and DNMT3B2 overexpression experiments, indeed, significantly deviated from random sampling and are likely to be those that are specifically, directly or indirectly, targeted by these methyltransferases.
Discussion
Alterations in DNA methylation have been shown to play a role in tumorigenesis and cancer progression in many malignancies, including prostate cancer. Until recently, technical limitations have restricted these findings to either characterization of a handful of candidate loci or of overall abundance of 5-methylcytosine in the genome. Although a previously published study reported the DNA methylation profiles of prostate tumors at CpG islands across the genome (Kron et al. 2009), no study has examined the methylation profiles of normal prostate tissue necessary to determine the methylation changes that occur during or as a result of tumorigenesis. Here, we present quantitative DNA methylation levels at more than 26,000 loci across 14,000 gene promoters. Because we assayed 95 cancers and 86 benign adjacent prostate tissues in parallel at CpGs specifically enriched at gene promoters, we were able to show that 41% of gene promoters represented in our assay had a tumor-specific methylation change. In addition to confirming methylation changes seen in previously published candidate loci studies, we also identified thousands of novel changes, including a set of hypermethylated loci more strongly predictive of prostate cancer than GSTP1. Our data show that DNA methylation changes in prostate cancer occur on a broad scale, at many loci throughout the genome.
DNA methylation alteration has been observed in early cancers and precursor lesions, suggesting that methylation changes drive malignant initiation rather than tumor progression (Belinsky et al. 1998; Brooks et al. 1998; Baylin et al. 2001; Guerrero-Preston et al. 2009). Our observations are largely consistent with this hypothesis. If the acquisition of DNA methylation alterations continues throughout tumor progression, variation in methylation profiles should be observed in tumors of different histological grades and clinical outcomes. Although we detected more heterogeneity among tumors than among benign adjacent tissues, the vast majority of tumors fell in a single cluster and we did not observe obvious subclassifications, although some tumor samples did cluster with benign adjacent samples. We compared clinical outcomes of the donors of the tumors that clustered with benign adjacent tissues against the donors of the other tumors but did not observe any differences in Gleason grades or time-to-recurrence (data not show). However, from the little inter-tumor heterogeneity that did exist, we identified several dozen DNA methylation changes that correlated with patients' time-to-recurrence. While the sites we identified were different from those identified by Liu et al. (2011), both our study and theirs were consistent in the overall absence of many differentially methylated CpG islands across tumors.
The fact that we observed changes at a very specific subset of CpG sites across most tumors, rather than a global DNA methylation deregulation or instability, suggests a common mechanism of tumorigenesis among prostate cancers. This specificity in target sites was particularly apparent in gene promoters assayed by multiple probes and by the PyroMark assay (Supplemental Text S2). The case of GSTP1 illustrates this point well, where the methylation changes were highly context-dependent: Only the CpG island overlapping the transcriptional start site was hypermethylated. Based on these findings, we suspect that cellular processes involved with targeted CpG methylation regulation are themselves misregulated or altered in early tumor initiation. The most likely candidates are DNMTs and DNMT-interacting proteins. In support of this hypothesis, we observed significant correlations between the gene expression levels and levels of global hypermethylation for several of these candidates. In vitro experiments in normal prostatic epithelial cells confirmed that overexpression of DNMT3B1 and DNMT3B2 leads to the hypermethylation of a subset of the prostate tumor-specific changes. These data, together with previous observations, strongly suggests that dysregulation of DNMTs and possibly DNMT- interacting proteins are among the earliest events in tumorigenesis.
While we did not address the mechanism for the observed decreased methylation of some CpGs in tumors, there are three likely possibilities. First, there may be aberrations in the maintenance DNA methyltransferase gene DNMT1. Although we did not observe a decrease in the DNMT1 transcript level, there may be a post-transcriptional dysregulation of this gene or mutations that lead to decreased activity. Decrease in DNMT1 activity may lead to improper maintenance and gradual loss of methylation with every DNA replication. However, this would likely lead to a global loss rather than a targeted loss at particular CpGs and, therefore, is the least likely scenario. A second possibility is the dysregulation of a direct or indirect DNA demethylase. While there have been a few reports of such enzymes in mammalian cells, none has been conclusive, and their existence is still speculative (Iyer et al. 2009; Bhutani et al. 2010; Okada et al. 2010). Finally, the targeted hypomethylation may be the result of dysregulation of an interacting protein of DNMT1 or the hypothetical DNA demethylase. With more than 20 DNMT1-interacting proteins already identified, it is conceivable that one or more of them are involved in DNMT1 targeting. However, a better understanding of the biology behind DNA demethylation is needed to answer this question.
By approaching DNA methylation in cancer from a genomic perspective, we were able to gain new insights into the underlying biology of prostate cancer, as well as discover novel markers for more accurate diagnosis of the disease. However, our study was limited in scale by technology and practicality: With only 26,333 assayed CpGs, mostly biased toward gene promoters, it is likely that these results are not representative of the 28 million CpGs found in the human genome. Even among nearby CpGs in the same promoter, the PyroMark assay revealed variability in methylation levels (Supplemental Text S2; Supplemental Fig. S7). Thus, using Reduced Representation Bisulfite Sequencing (RRBS) (Meissner et al. 2005) and/or the new Illumina HumanMethylation450 array will likely uncover additional sites of interest, probably including ones with better diagnostic and prognostic value. In addition, this is the first study comparing methylation in prostate cancer to benign adjacent tissue; while our cohort was clinically representative of patients presenting with the disease, it is paramount that the findings be now verified in an independent replication set of samples. Furthermore, recent success in integrative analysis of copy-number variation (CNV) and gene expression data highlights the great value in studying prostate cancer from multiple perspectives (Taylor et al. 2010). Expanding such an integrative analysis to include DNA methylation data along with gene expression and CNV data is likely to lead to a better understanding of prostate cancer biology, and if robust diagnostic and prognostic markers can be identified, they will need to be developed into biomarkers for use in a clinical setting. Finally, as methylation profile data of more tumor types become available, researchers will be able to identify common and type-specific alterations. While we were largely unable to do this among existing data sets due to drastically different study designs, investigations of these similarities and distinctions are likely to lead to a deeper understanding of cancer biology as a whole.
Methods
Sample collection and preparation
All prostate samples used for this study were collected at the Stanford University Medical Center between 1999 and 2007 with patient's informed consent under an IRB-approved protocol. Multiple tissue samples were harvested from each prostate, flash-frozen, and stored at −80°C. Sections of each prostate tissue sample were evaluated by a genitourinary pathologist. The tumor and non-tumor areas were marked, and contaminating tissues were trimmed away from the block as described previously (Lapointe et al. 2004). Tumor samples in which at least 90% of the epithelial cells were cancerous and non-tumor samples having no observable tumor epithelium were selected for extraction of DNA and RNA. Clinical information associated with prostate samples included in the analysis is summarized in Supplemental Table S1.
Primary prostate cell culture and transfection assays
A primary culture of human prostatic epithelial cells (E-PZ-231) was established from benign tissue of the peripheral zone of the prostate of a 56-yr-old man who underwent radical prostatectomy to treat prostate cancer. Using previously described methods (Peehl 2002), primary cultures were serially passaged. When tertiary passage cells were ∼50% confluent, they were fed Complete PFMR- 4A medium (Peehl 2002) without gentamycin until they reached ∼85% confluency. Cells in each 60-mm, collagen-coated dish were then transfected with 10 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 48 h, cells from three 60-mm dishes per condition were dissociated with TrypLE Express (Invitrogen), centrifuged, and snap-frozen in liquid nitrogen. These cell pellets were then used for DNA isolation.
Nucleic acid isolation
DNA and RNA were isolated from tissue samples or cell cultures using QIAGEN AllPrep DNA/RNA mini kit (QIAGEN) following the manufacturer's protocol, with the exception of the RNA from primary prostate cell cultures. This RNA was isolated with TRIzol Reagent (Invitrogen) according to the manufacturer's instructions.
Sodium bisulfite conversion
Sodium bisulfite conversion of genomic DNA was performed using the EZ-96 DNA Methylation Kit (Deep-Well format; ZymoResearch). The conversion was completed using the alternative incubation protocol for the Illumina Infinium Methylation Assay, as described by the manufacturer.
Methylation analysis by Illumina Infinium HumanMethylation27
Five hundred nanograms of sodium bisulfite–converted genomic DNA from patient samples or cultured cells were assayed by Infinium HumanMethylaton27, RevB Beadchip Kits (Illumina). The assay was performed using the protocol as described by the manufacturer.
Beta score calculations, quality filtering, and batch normalization
HumanMethylation27 array results were initially extracted and analyzed using Illumina BeadStudio software with the Methylation Module v3.2. Beta scores were calculated manually using values exported from BeadStudio. For each probe intensity value, we subtracted the median negative background control probe value based on the color channel. The beta score was calculated using the background subtracted intensity values as: β = IntensityMethylated/(IntensityMethylated + IntensityUnmethylated). Any negative beta scores were converted to a zero. Any beta scores with an associated detection P-value of >0.01 were converted to “missing values.” To correct for any array-by-array variation, we imputed all missing values using KNN Impute, then performed normalization using the ComBat R-package (Johnson et al. 2007). All previously imputed values were converted back to “missing values” for subsequent analyses.
To remove CpG probes with potentially problematic hybridization, we performed BLAT on all 27,578 probe sequences against the GRCh27/hg19 build of the human genome. One thousand and twenty-eight probes showed questionable mapping and therefore were removed from analysis. We also identified 217 probes that included a SNP of >3% minor allele frequency within 15 bp of the assayed CpG. These probes were also rejected with consideration to potential variation in probe hybridization due to the common SNP.
Clustering
Prior to each hierarchical clustering, the beta scores were mean-centered. Hierarchical clustering of the arrays was done using the software Cluster 3.0 with Average Linkage. Because these data sets were too large to cluster the genes by Cluster 3.0, gene clustering was done using XCluster, available through the Stanford Microarray Database, using non-centered Pearson correlation to perform the hierarchical clustering.
Significance Analysis of Microarray (SAM)
Each SAM was performed as described in the software manual. The data were analyzed using the latest version of SAM available at the time of this manuscript preparation, which was version 3.09c. SAM was implemented using R version 2.10.0.
Prediction Analysis of Microarray (PAM)
Prior to PAM, the CpGs were sorted by standard deviation across all tumors and benign adjacent tissue. To improve statistical power, only CpGs that had a standard deviation of 0.04 or greater were analyzed. PAM was performed as described in the software manual. The data were analyzed using the latest version of PAM available at the time of this manuscript preparation, which was version 2.11. PAM was implemented using R version 2.10.0. Based on visual examination of the training errors and the cross-validation results, we set the shrinkage threshold to 10.5.
PyroMark assays
PyroMark assays were performed at the Stanford Protein and Nucleic Acid Facility using the manufacturer's recommended protocol (QIAGEN). For each target region, three primers were used: a forward and reverse PCR primer and a sequencing primer. Primer sequences are listed in Supplemental Table S6.
TaqMan gene expression assay
Expression levels of genes encoding several DNMT and DNMT-interacting proteins, as well as beta-2-microglobulin as an endogenous control, were measured in 10 benign adjacent and 36 tumor samples by the TaqMan Gene Expression Assay. We used the following Applied Biosystems inventoried assays with FAM/MGD labeled probes (Assay ID in parentheses): DNMT1 (Hs00945900_g1), DNMT3A (Hs00173377_m1), DNMT3A2 (Hs00601097_m1), DNMT3B (Hs01003405_m1), DNMT3L (Hs01081364_m1), EZH2 (Hs01016789_m1), and the Human B2M (beta-2-microglobulin) Endogenous Control. Twenty-five nanograms of cDNA were assayed in triplicate for each target, using the protocol as described by the manufacturer, on the ABI PRISM 7900HT instrument. The results were analyzed using the ABI SDS 2.4 and ABI RQ Manager 1.2.1 software. Briefly, the average CT and delta-CT were calculated for each DNMT and EZH2. By integrating the average CT value from the B2M CT, we calculated the delta-delta-CT. All sample delta-delta-CT values were normalized to that of a tumor sample PC625T to generate an RQ value. To present the RQ value as a positive value, we added 5 to each RQ value.
Expression vectors
The pcDNA3/Myc-EZH2 construct was a generous gift from A. Chinnaiyan (University of Michigan) (Okano et al. 1999). The pcDNA3/Myc-DNMT3A, pcDNA3/Myc-DNMT3A2, pcDNA3/Myc-DNMT3B1, pcDNA3/Myc-DNMT3B2, and pcDNA3/Myc-DNMT3B3 constructs were a generous gift from A. Riggs (City of Hope) (Chen et al. 2005).
Acknowledgments
We thank Kenneth Day, Kevin Roberts, and Krista Stanton for help with the HumanMethylation27 and TaqMan assays. We thank Preti Jain for assistance with HumanMethylation27 quality filtering. We thank the Stanford Protein and Nucleic Acid Facility for generating the PyroMark data. We thank Kevin Bowling, Marie Cross, Chris Gunter, Preti Jain, Brittany Lasseigne, Jun Li, Jonathan Pollack, Rob Tibshirani, Katherine Varley, and Daniela Witten for stimulating discussions. We also thank Marie Cross, Barbara Dunn, Dan Kvitek, and Jared Wenger for critical reading of this manuscript. This project was funded by NIH (CA111782 to J.D.B.), an Institutional Training Grant in Genome Science from the NIH/NHGRI (5 T32 HG0000044) (to Y.K.), and HudsonAlpha funds (to R.M.M. and D.M.A.).
Authors' contributions: Y.K., D.M.A., Z.G.G., S.R.Y., J.K.M., D.M.P., J.D.B., R.M.M., and G.S. agree with the manuscript's results and conclusions. Y.K., D.M.A., S.R.Y., D.M.P., J.D.B., R.M.M., and G.S. designed the experiments. Y.K., D.M.A., J.D.B., R.M.M., and G.S. analyzed the data. Y.K., D.M.A., Z.G.G., and S.R.Y. collected data and performed experiments for the study. J.K.M. and J.D.B. prepared tissue samples. Y.K. wrote the first draft of the paper. Y.K., D.M.A., D.M.P., J.D.B., R.M.M., and G.S. contributed to the writing of the paper.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE26126.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.119487.110.
References
- Andriole GL, Grubb RL, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, et al. 2009. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med 360: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 10: 687–692 [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG 1998. Aberrant methylation of p16INK4a is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci 95: 11891–11896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, Epstein JI, Isaacs WB, Nelson WG 1998. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev 7: 531–536 [PubMed] [Google Scholar]
- Brooks JD, Tibshirani R, Ferrari M, Joseph PC, Harcharan G, King CR 2008. The impact of tumor volume on outcomes after radical prostatectomy: Implications for prostate cancer screening. Open Prost Cancer J 1: 1–8 [Google Scholar]
- Cairns P, Esteller M, Herman JG, Schoenberg M, Jeronimo C, Sanchez-Cespedes M, Chow N, Grasso M, Wu L, Westra WB, et al. 2001. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res 7: 2727–2730 [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Mann JR, Hsieh C, Riggs AD, Chédin F 2005. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem 95: 902–917 [DOI] [PubMed] [Google Scholar]
- Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S 2001. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res 61: 5544–5551 [PubMed] [Google Scholar]
- Chuang LS, Ian H, Koh T, Ng H, Xu G, Li BF 1997. Human DNA-(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science 277: 1996–2000 [DOI] [PubMed] [Google Scholar]
- Das PM, Singal R 2004. DNA methylation and cancer. J Clin Oncol 22: 4632–4642 [DOI] [PubMed] [Google Scholar]
- Ehrlich M 2002. DNA methylation in cancer: too much, but also too little. Oncogene 21: 5400–5413 [DOI] [PubMed] [Google Scholar]
- El-Maarri O, Kareta MS, Mikeska T, Becker T, Diaz-Lacava A, Junen J, Nusgen N, Behne F, Wienker T, Waha A, et al. 2009. A systematic search for DNA methyltransferase polymorphisms reveals a rare DNMT3L variant associated with subtelomeric hypomethylation. Hum Mol Genet 18: 1755–1768 [DOI] [PubMed] [Google Scholar]
- Esteller M, Herman JG 2002. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol 196: 1–7 [DOI] [PubMed] [Google Scholar]
- Estève P, Chin HG, Pradhan S 2005. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci 102: 1000–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89–92 [DOI] [PubMed] [Google Scholar]
- Freedland SJ, Humphreys EB, Mangold LA, Eisenberger M, Dorey FJ, Walsh PC, Partin AW 2005. Risk of prostate cancer–specific mortality following biochemical recurrence after radical prostatectomy. JAMA 294: 433–439 [DOI] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M 1983. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 11: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Preston R, Báez A, Blanco A, Berdasco M, Fraga M, Esteller M 2009. Global DNA methylation: a common early event in oral cancer cases with exposure to environmental carcinogens or viral agents. P R Health Sci J 28: 24–29 [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, et al. 2011. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet 20: 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann MJ, Engers R, Florl AR, Otte AP, Muller M, Schulz WA 2007. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer. Cancer Biol Ther 6: 1403–1412 [DOI] [PubMed] [Google Scholar]
- Hong C, Bollen AW, Costello JF 2003. The contribution of genetic and epigenetic mechanisms to gene silencing in oligodendrogliomas. Cancer Res 63: 7600–7605 [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L 2009. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 8: 1698–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E 2010. Cancer Statistics, 2010. CA Cancer J Clin 60: 277–300 [DOI] [PubMed] [Google Scholar]
- Jerónimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, Oliveira J, Teixeira MR, Lopes C, Sidransky D 2004. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res 10: 8472–8478 [DOI] [PubMed] [Google Scholar]
- Johnson WE, Rabinovic A, Li C 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127 [DOI] [PubMed] [Google Scholar]
- Jones PA 1986. DNA methylation and cancer. Cancer Res 46: 461–466 [PubMed] [Google Scholar]
- Kim E, Kim Y, Jeong P, Ha Y, Bae S, Kim W 2008. Methylation of the RUNX3 promoter as a potential prognostic marker for bladder tumor. J Urol 180: 1141–1145 [DOI] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. 2009. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet 41: 524–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron K, Pethe V, Briollais L, Sadikovic B, Ozcelik H, Sunderji A, Venkateswaran V, Pinthus J, Fleshner N, van der Kwast T, et al. 2009. Discovery of novel hypermethylated genes in prostate cancer using genomic CpG island microarrays. PLoS ONE 4: e4830 doi: 10.1371/journal.pone.0004830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron KJ, Liu L, Pethe VV, Demetrashvili N, Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L, van der Kwast TH, et al. 2010. DNA methylation of HOXD3 as a marker of prostate cancer progression. Lab Invest 90: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Laird PW, Jaenisch R 1994. DNA methylation and cancer. Hum Mol Genet 3: 1487–1495 [DOI] [PubMed] [Google Scholar]
- Laird PW, Jaenisch R 1996. The role of DNA methylation in cancer genetic and epigenetics. Annu Rev Genet 30: 441–464 [DOI] [PubMed] [Google Scholar]
- Lapeyre JN, Walker MS, Becker FF 1981. DNA methylation and methylase levels in normal and malignant mouse hepatic tissues. Carcinogenesis 2: 873–878 [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. 2004. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci 101: 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS, Hsieh WS, Isaacs WB, Nelson WG 1994. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci 91: 11733–11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Tascilar M, Lee W, Vles W, Lee B, Veeraswamy R, Asgari K, Freije D, Van Rees B, Gage W, et al. 2001. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. Am J Pathol 159: 1815–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Kron KJ, Pethe VV, Demetrashvili N, Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L, van der Kwast TH, et al. 2011. Association of tissue promoter methylation levels of APC, TGFβ2, HOXD3, and RASSF1A with prostate cancer progression. Int J Cancer doi: 10.1002/ijc.25908 [DOI] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R 2005. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 33: 5868–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, Perkmann E, Marth C, Widschwendter M 2003. DNA methylation in serum of breast cancer patients. Cancer Res 63: 7641–7645 [PubMed] [Google Scholar]
- Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG 2004. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem 91: 540–552 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y 2010. A role for the elongator complex in zygotic paternal genome demethylation. Nature 463: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Patra SK, Patra A, Zhao H, Dahiya R 2002. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog 33: 163–171 [DOI] [PubMed] [Google Scholar]
- Peehl DM 2002. Human prostatic epithelial cells. In Culture of epithelial cells, 2nd ed (ed. RI Freshney, MG Freshney), Chapter 6 Wiley, New York [Google Scholar]
- Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin P, Svensson MA, Kitabayashi N, Moss BJ, MacDonald TY, et al. 2011. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res 21: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland KS, Shardy DL, Azim M, Naeem R, Krance RA, Dreyer ZE, Neeley ES, Zhang N, Qiu YH, Kornblau SM, et al. 2009. Overexpression of ZNF342 by juxtaposition with MPO promoter/enhancer in the novel translocation t(17;19)(q23;q13.32) in pediatric acute myeloid leukemia and analysis of ZNF342 expression in leukemia. Genes Chromosomes Cancer 48: 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, Beckstrom-Sternberg J, Barrett M, Long J, Chinnaiyan A, et al. 2011. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res 21: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Suetake I, Shinozaki F, Yamashina S, Tajima S 2004. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr Patterns 5: 231–237 [DOI] [PubMed] [Google Scholar]
- Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur S, Hoshida Y, Perner S, Adami H, Fall K, Mucci L, et al. 2010. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics 3: 8 doi: 10.1186/1755-8794-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. 2009. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 360: 1320–1328 [DOI] [PubMed] [Google Scholar]
- Singh D, Febbo P, Ross K, Jackson D, Manola J, Ladd C, Tamayo P, Renshaw A, D'Amico A, Richie J, et al. 2002. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1: 203–209 [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G 2002. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci 99: 6567–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, Demichelis F, Helgeson BE, Laxman B, Morris DS, et al. 2008. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell 13: 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D, Van Den Berg D, Pan F, Berman B, and Laird PW 2008. Comprehensive DNA methylation analysis on the Illumina Infinium assay platform. http://www.illumina.com/documents/products/appnotes/appnote_dna_methylation_analysis_infinium.pdf
- Woodson K, O'Reilly KJ, Hanson JC, Nelson D, Walk EL, Tangrea JA 2008. The usefulness of the detection of GSTP1 methylation in urine as a biomarker in the diagnosis of prostate cancer. J Urol 179: 508–512 [DOI] [PubMed] [Google Scholar]
- Yaqinuddin A, Qureshi S, Qazi R, Abbas F 2008. Down-regulation of DNMT3b in PC3 cells effects locus-specific DNA methylation, and represses cellular growth and migration. Cancer Cell Int 8: 13 doi: 10.1186/1475-2867-8-13 [DOI] [PMC free article] [PubMed] [Google Scholar]