Abstract
The pluripotency control regions (PluCRs) are defined as genomic regions that are bound by POU5F1, SOX2, and NANOG in vivo. We utilized a high-throughput binding assay to record more than 270,000 different DNA/protein binding measurements along incrementally tiled windows of DNA within these PluCRs. This high-resolution binding map is then used to systematically define the context of POU factor binding, and reveals patterns of cooperativity and competition in the pluripotency network. The most prominent pattern is a pervasive binding competition between POU5F1 and the forkhead transcription factors. Like many transcription factors, POU5F1 is co-expressed with a paralog, POU2F1, that shares an apparently identical binding specificity. By analyzing thousands of binding measurements, we discover context effects that discriminate POU2F1 from POU5F1 binding. Proximal NANOG binding promotes POU5F1 binding, whereas nearby SOX2 binding favors POU2F1. We demonstrate by cross-species comparison and by chromatin immunoprecipitation (ChIP) that the contextual sequence determinants learned in vitro are sufficient to predict POU2F1 binding in vivo.
POU5F1 (formerly Oct4), SOX2, and NANOG are generally regarded as the core set of transcription factors necessary for maintaining the pluripotent state in stem cells (Boyer et al. 2005). The loss of POU5F1 or NANOG causes embryonic stem (ES) cells to lose pluripotency and differentiate inappropriately (Hough et al. 2006). Precise POU5F1 levels are also critical to maintaining pluripotency and are at least partially controlled by SOX2, a factor that can heterodimerize to potentially change POU5F1 binding specificity (Niwa et al. 2000; Remenyi et al. 2003). POU5F1 can be regarded as a central pillar in that SOX2 and NANOG interact with POU5F1 but are not known to interact with each other; POU5F1 can be regarded as the hub of this pluripotency network (Zhang et al. 2007).
POU5F1 is part of a larger family of POU factors (formerly known as Oct factors) that share DNA binding specificity and overlapping domains of expression (Phillips and Luisi 2000). The entire family of POU factors was identified on the basis of their ability to bind the octamer sequence ATGCAAAT (Rosales et al. 1987). In addition to POU5F1, the paralog POU2F1 (formerly Oct1) is expressed in ES cells and is capable of heterodimerizing with POU5F1 (Okamoto et al. 1990; Tomilin et al. 2000). This additional complexity raises the question of how distinct biological functions emerge from DNA binding proteins that have similar specificity.
POU binding can be influenced by POU factor–specific interactions with other DNA binding proteins. The human protein reference database reports 36 DNA binding proteins that interact with POU2F1 and 10 that interact with POU5F1, suggesting a large repertoire of complexes that may lend distinct specificities to POU paralogs (Peri et al. 2003). The presence of cofactors can influence binding and distinguish one POU factor from another. For example, POU2F1 and POU2F2, but not other POU factors, interact with the DNA binding tissue-specific cofactor POU2AF1 (also known as OCA-B and OBF-1) (Gstaiger et al. 1996). POU5F1 is known to dimerize with SOX2 and can be immunoprecipitated with NANOG (Zhang et al. 2007).
Many transcription factors bind DNA via alternate motifs (Badis et al. 2009). Distinct binding specificities between POU factors may lie in their ability to use noncanonical modes of binding octamer-derived motifs on DNA. The octamer motif is a bipartite sequence (i.e., ATGC, AAAT) recognized by two subdomains that together compose the POU domain. As dimers, POU factors are capable of binding a variety of palindromic arrangements of halfsites such as the palindromic octamer recognition element (PORE), ATTTGAAATGCAAAT, or the MORE (more PORE), ATGCATATGCAT (Phillips and Luisi 2000; Tantin et al. 2008). The distinct conformations that POU factors form on different classes of binding sites could modulate the access of co-activators (Gstaiger et al. 1996; Kang et al. 2009) and give rise to distinct biological activities. However, there is evidence that the current three classes of binding sites do not describe all the modes through which POU5F1 binds DNA (Tantin et al. 2008).
Here, we present the application of a high-throughput binding assay to map protein/DNA complexes on ∼400 kb of transcriptional control regions that regulate pluripotency in ES cells (i.e., regions bound simultaneously by POU5F1, SOX2, and NANOG in vivo). We mapped protein occupancy on these DNA targets at a resolution of 10 nucleotides (nt). We discovered that while POU2F1 and POU5F1 share specificity for the octamer, they differ in their ability to bind complex sites. Both POU factors appear to bind their targets through clusters of sites that sometimes stretch for hundreds of nucleotides. Numerous forkhead factor-binding sites co-occur with POU binding sites. We demonstrate that POU5F1 competes with FOXO1 and synergizes with NANOG, while POU2F1 binds synergistically with SOX2. In the process of determining this network of biochemical interactions, we also created a rich and detailed map of cis-elements on 316 transcription control regions that are important for early development.
Results
POU5F1/SOX2/NANOG modules define 316 pluripotency control regions
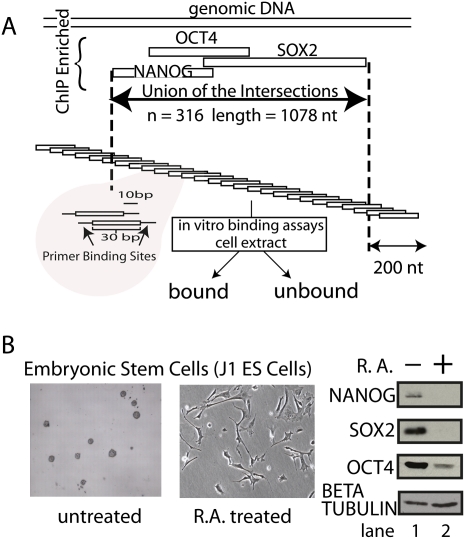
We define pluripotency control regions (PluCRs)as genomic regions that bind POU5F1, SOX2 and NANOG in vivo. Based on previous chromatin immunoprecipitation (ChIP) data, we identified 316 PluCRs by demanding physically overlapping regions of ChIP enrichment (Fig. 1A; Boyer et al. 2005). ChIP enrichment identified regions that ranged in size from 180–3108 nt (average, 1078 nt). The goal of this study was to map precisely which windows in these regions were bound by the POU factors present in ES cells (POU2F1 and POU5F1) and SOX2 and NANOG. This high-resolution map was then used to discover higher-order patterns and contexts of transcription factor binding in PluCRs. An oligonucleotide pool was constructed by tiling across all 316 PluCRs. (Fig. 1A). Cell extract was added to the oligonucleotide pool, and bound proteins were recovered by immunoprecipitation. The enrichment of each oligonucleotide in the bound fraction relative to the starting pool was quantified by microarray (Tantin et al. 2008; Watkins et al. 2009).
Figure 1.
Resynthesizing 316 pluripotency control regions (PluCRs). (A) Regions identified by chromatin immunoprecipitation (ChIP) as binding sites for POU5F1 (Oct4), SOX2, and NANOG were resynthesized as an oligonucleotide pool. The pool tiles through the regions, resulting in a population of 45,793 overlapping oligonucleotides that can be amplified by a universal primer pair. The P32 radiolabeled oligonucleotide pool was incubated in untreated J1 embryonic cell extract and in extract from RA-treated J1 cells and separated into a bound and unbound fraction by two methods. Ligands for specific factors were separated by coimmunoprecipitation. Ligands for unknown cellular DNA binding proteins were identified by their retention in nitrocellulose filter (see below). (B) Morphological changes associated with RA differentiation are recorded in the left panels. Pluripotency markers POU5F1, SOX2, and NANOG were assayed by Western before (lane 1) and after (lane 2) RA treatment.
High-throughput binding assay produces a comprehensive map of binding elements on 316 PluCRs
We used this assay to compare DNA/protein complexes in ES cell extract before and after differentiation. The extract was prepared from the pluripotent J1 ES cells (Li et al. 1992) before and after retinoic acid (RA) differentiation (Bain et al. 1995). Differentiated ES cells adopt a neuronal morphology, stop dividing, and repress POU5F1, SOX2, and NANOG expression (Fig. 1B). We also detected a loss of the repressor of neuronal differentiation REST, which is expressed in ES cells and other non-neural cell types (like HeLa cells) but not in neurons (data not shown). We concluded that our differentiation protocol successfully shifted J1 cells from an ES state to a neuronal-like state.
The oligonucleotide pool was incubated in extract derived from undifferentiated ES cells. POU5F1, POU2F1, SOX2, and NANOG immunoprecipitations were used to separate the oligonucleotide pool into a bound and unbound fraction. The degree of oligonucleotide enrichment in the immunoprecipitate was measured by hybridizing differentially Cy-labeled bound pool and starting pool to a custom oligonucleotide microarray (for detailed methods and comparisons between replicates, see Supplemental Data). A single round of immunoprecipitation achieved a maximal enrichment of 100-fold in our assays. Throughout this article, we refer to the log10 normalized red/green ratios as “enrichment,” and this value is used throughout the article to rank the oligonucleotides by their binding. As these oligonucleotides represent tiled windows of genomic sequence, the enrichment from all microarray probes that overlap each nucleotide position was averaged and rendered on a genomic coordinate system (Fig. 2A). To estimate the selective pressure on these binding elements, the top 1% of oligonucleotides enriched in the immunoprecipitated fractions in these four experiments were examined across 17 vertebrate species. Binding elements of these specific transcription factors were between three- and fourfold more conserved than background levels, suggesting that these elements are functional (Supplemental Table S1).
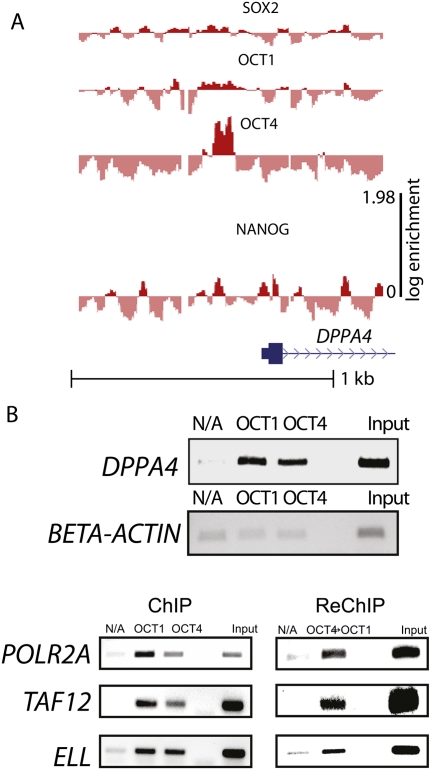
Figure 2.
A map of transcriptional complexes on DPPA4, a known target of POU5F1 (Oct4). (A) The oligonucleotide pool was subjected to binding assays to identify specific protein DNA complexes by coimmunoprecipitation with SOX2, POU2F1 (Oct1), POU5F1 (Oct4), and NANOG antibodies. Specific oligonucleotide enrichment in the bound fraction was analyzed by two color microarray (bound vs. starting pool). The oligonucleotides were mapped back to genomic coordinates (x-axis), and their average log10 red/green ratio (enrichment) is plotted on the y-axis. UCSC known gene annotation of the DPPA4 indicates transcription and translation start sites and a portion of the first intron of the DPPA4 gene. Dotted vertical lines mark the boundaries of complexes identified in the J1 extracted. (B) The enrichment of DPPA4 and a control region (beta-ACTIN) by chromatin immunoprecipitation with anti-POU2F1 (anti-Oct1), anti-POU5F1 (anti-Oct4), and irrelevant (p53) antibodies measure binding in vivo in J1 ES cells. No antibody and input controls are labeled on the figure. ChIP and sequential ChIP (Re-ChIP) performed on previously identified POU2F1 (Oct1) targets are labeled similarly.
A well-known target of POU5F1, DPPA4, is enriched in the POU5F1 bound fraction
To consider a specific functional element within the more than 316 kb analyzed in this study, we considered a well-characterized target of POU5F1 regulation, the DPPA4 gene. The DPPA4 promoter was one of the most enriched signals in the original POU5F1 ChIP experiment in human ES cells, is highly expressed in ES cells, and is regulated by mutationally characterized POU5F1 sites (Boyer et al. 2005; Babaie et al. 2007; Chakravarthy et al. 2008). This example of the DPPA4 gene illustrates two generalizations about POU5F1 binding that can be made across the 316 PluCRs: (1) Binding of POU5F1 can occur across large regions that can extend hundreds of nucleotides; and (2) the ubiquitously expressed paralog POU2F1 can also bind POU5F1 sites. As POU5F1 possess a nonredundant function in maintaining stemness in ES cells, a competition among POU paralogs could modulate this stemness function within the pluripotency control network.
To verify the results of the array and confirm that POU2F1 was binding in vivo, we performed ChIP on the characterized POU5F1 binding site in DPPA4 (Fig. 2B). While the relative contribution of each POU factor cannot be determined by this assay, both POU2F1 and POU5F1 are enriched at this locus in vivo relative to the beta-ACTIN control (Fig. 2B). Sequential ChIP (re-ChIP) experiments with a variety of previously identified POU2F1 ligands suggest that POU2F1 and POU5F1 bind together in vivo, perhaps by forming heterogeneous complexes similar to those shown in vitro (Tomilin et al. 2000). While ChIP is not capable of resolving events at the level of single complexes, the re-ChIP clearly demonstrates that the POU2F1/POU5F1 binding ratio varies with local sequence context.
POU5F1 binding is more octamer-based and clustered than POU2F1 binding
To discover signals that influence which paralog joins a POU factor complex, we compared sequence motifs that were discovered de novo in the bound fraction of the POU2F1 immunoprecipitate against those from the POU5F1 immunoprecipitate. As expected, the canonical octamer binding site ATGCAAAT was the dominant motif returned in both the POU2F1 and POU5F1 binding assays (Fig. 3A). The ATGC submotif appeared more prominently in the POU5F1 immunoprecipitate. POU2F1 and POU5F1 also differ in their preference for the previously characterized noncanonical MORE and PORE binding elements. Each of these elements represents palindromic combinations of halfsites (sequences in Fig. 3A) that support POU dimer formation. Although oligonucleotides that contained any octamer-based motif were more enriched in POU5F1 than POU2F1, both POU factors appeared to bind a simple octamer and the PORE element. However, oligonucleotides containing a MORE sequence were differentially enriched for POU5F1 (98th percentile) but not POU2F1 (Fig. 3A; Supplemental Table S5). This result suggests that POU5F1 binds octamer-based oligonucleotides with higher affinity, and MOREs are more “tuned” to POU5F1 than POU2F1. Consistent with this observation, POU dimers bound in a MORE confirmation are structurally incompatible with the POU2F1 cofactor POU2AF1. However, when bound in the PORE conformation, these contacts are permitted (Tomilin et al. 2000).
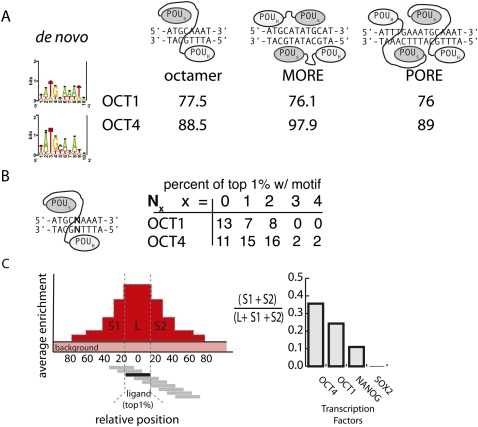
Figure 3.
Identifying the specificity and clustering tendency of transcription factors. (A) De novo motif finding was used to identify sequence motifs enriched in the top 1% of oligonucleotides ranked by enrichment in the POU2F1 (Oct1) and POU5F1 (Oct4) immunoprecipitate. The average percentile rank of oligonucleotides matching the three existing POU factor binding models was recorded. Reported conformations of POU factor binding are shown. (B) The distribution of the top 1% of POU2F1 (Oct1) and POU5F1 (Oct4) ligands within gapped octamers is depicted. (C) The tendency of binding events to cluster was analyzed by measuring the average enrichment in the neighborhood of the top 1% of ligands. The percentage of area of the peak that fell outside of the ligand was used to measure a transcription factor's clustering tendency.
As the flexible linker connecting the two POU subdomains allows some spacing between the ATGC and the AAAT halfsites, the tolerance for intervening nucleotides within the octamer was compared for POU2F1 and POU5F1. Annotating the top 1% of POU2F1 and POU5F1 ligands with various gapped binding models suggests both POU factors tolerate up to 2 nt of spacer (Fig. 3B). However, a greater fraction of high-affinity POU5F1 binding events are found on gapped octamers than perfect octamers.
After determining the primary sequence motifs for POU2F1 and POU5F1, we characterized the transcription factors' affinity for higher-order motif patterns. In the DPPA4 gene, POU binding stretches over multiple oligonucleotides encompassing an enriched region of 92 nt (Fig. 2A). An examination of POU binding across all 316 PluCR reveals 42 regions where POU binding seems to span more than a hundred consecutive positions, suggesting a clustering of sites. In order to measure the tendency of sites to cluster, we considered the top 1% of oligonucleotides enriched in the immunoprecipitate to be bona fide POU5F1 “ligands.” We reasoned that an architecture of clustered sites would be evident from examining the oligonucleotides neighboring these ligands. If clustered binding was a prevalent mode of POU binding, the oligonucleotides adjacent to the strong ligands would also be enriched above background in the POU bound fraction. We used this residual enrichment observed in the areas of the peak that lie beyond the dominant “ligand” oligonucleotide to measure a transcription factor's tendency to bind in cluster (Fig. 3C). For POU5F1 ligands in the PluCRs, we observed a sloping shoulder of enrichment that, on average, accounted for 34% enrichment of the peak and reached background enrichment levels 70 nt away from the ligand. The degree to which binding is clustered is slightly less for POU2F1 and considerably less for NANOG and SOX2 (Fig. 3C; Supplemental Table S2). These findings indicate that POU factor binding extends well beyond the footprint of a traditionally defined transcription factor binding site.
PluCRs contain shared POU2F1/POU5F1 sites, a subpopulation of POU2F1-specific sites and patterns of local associations between POU and other transcription factors
To explore the differences in binding specificity between POU5F1 and POU2F1 at higher resolution, we returned to the output of the high-throughput binding assay. This assay shows that the affinity of all 45,793 oligonucleotides for POU2F1 or POU5F1 can be inferred from their enrichment in the POU2F1 and POU5F1 coimmunoprecipitate. We report a clear positive relationship between the affinity of an oligonucleotide for POU2F1 and its affinity for POU5F1 (Fig. 4A, diagonal trend) and a significant population of oligonucleotides (102 oligos observed, five expected) that are ligands for both factors (Fig. 4A, region 2). This high-resolution comparison of POU factor specificity suggests that the overlapping specificity observed represents shared sites and not two similar classes of sites that are occupied exclusively by POU2F1 or POU5F1. Motif sampling performed on subregions of this distribution indicate that these shared sites contain stronger octamer motifs than found in either comparison alone (Fig. 4A vs. Fig. 3A). While few oligonucleotides recognize only POU5F1 at this threshold, there is a prominent subpopulation of oligonucleotides that bound POU2F1 but not POU5F1 (Fig. 4A, region 3 vs. region 1). In contrast to the populations of POU5F1 binding oligonucleotides (region 1 and 2), distinct nonoctamer motifs were found to be enriched in the POU2F1 subpopulation, implying alternate modes of POU2F1 binding or indirect recruitment via factors with alternate DNA binding specificity (Fig. 4B; Supplemental Fig. S3).
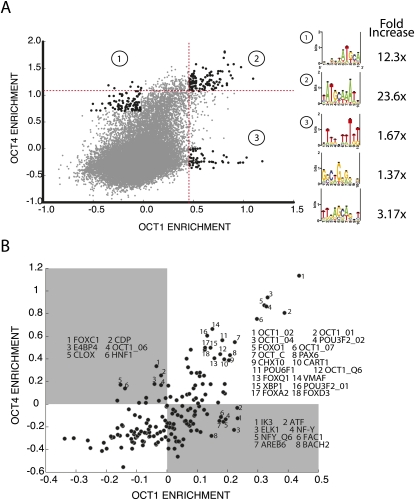
Figure 4.
Defining the relative specificity of PluCR oligonucleotides for POU2F1 (Oct1) and POU5F1 (Oct4). (A) The enrichment of all 40,000 oligonucleotides in the bound fraction of POU5F1 (Oct4) and POU2F1 (Oct1) was compared by scatterplot. The top 1% of oligonucleotides defined in each experiment is indicated by vertical (POU2F1) and horizontal (POU5F1) dashed lines. Motifs derived from the doubly enriched and POU specific fraction were identified as in Figure 3. Their enrichment in the bolded data points is printed to the right of the motifs. (B) The association between TRANSFAC/JASPER binding models and POU2F1/POU5F1 binding tendencies. The entire set of vertebrate TRANSFAC/JASPER binding models was used to annotate the oligonucleotide pool. The average POU2F1 (Oct1; x-axis) and POU5F1 (Oct4; y-axis) binding enrichment of the set of oligonucleotides that contain a significant match to a TRANSFAC motif was plotted for each TRANSFAC motif that contained multiple matches.
To systematically assess the contribution of other transcription factors, we annotated all oligonucleotides with a data set of transcription factor binding motifs and calculated the average POU2F1 and POU5F1 enrichments for the set of oligonucleotides that contain a particular motif. These average enrichments (POU2F1, POU5F1) were plotted as transcription factor binding site “centroids” (Fig. 4B) and summarized the relationship between the TRANSFAC or JASPAR binding models and POU binding. Most centroids clustered at zero enrichment in the immunoprecipitated fraction for both factors, indicating no relationship with POU binding. However, numerous centroids fell along the upper right diagonal, indicating an association with POU factor binding. Similar to the comparison of POU2F1 to POU5F1 oligonucleotide binding enrichment observed in the pool, we observed an overrepresentation of TRANSFAC centroids in the region that represents POU2F1-specific sequences (Fig. 4B, gray areas).
Unsurprisingly, the strongest associations with POU5F1 and POU2F1 binding are found in the numerous highly similar (i.e., overlapping) POU factor binding motifs in the database. However in addition to the many POU factor binding models, certain other transcription factor binding sites were associated with oligonucleotides that bound both POU factors (Fig. 4B, upper right). These factors could be synergistic cobinders or competitors or coincident with POU factors. Many of these transcription factors (FOXO1, PAX6, VSX2, FOXA2, FOXQ1, and FOXD3) have central roles in development. The strongest nonoctamer nonoverlapping motif associated with POU binding is the binding site for FOXO1. FOXO1 is an important regulator of cellular stress associated with aging and has overlapping specificity with POU5F1 and POU2F1 in the pluripotency network. Like POU5F1, FOXO1 is expressed in ES cells, but unlike POU5F1, it persists throughout differentiation (Fig. 5D). While mostly associated with aging and the response to oxidative stress, FOXO1-null mice are characterized by an expanded population of neuronal stem cells, indicating that FOXO1 is capable of restricting multipotency (Brunet et al. 2004; Salih and Brunet 2008; Paik et al. 2009). These genetic data suggest that FOXO1 may function as a POU factor antagonist.
Figure 5.
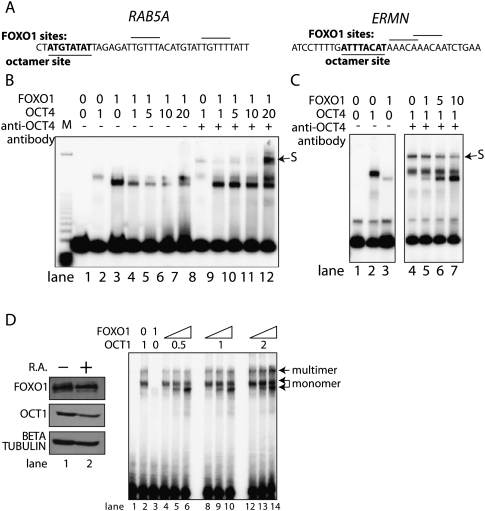
Defining a competition between FOXO1 and POU5F1 (Oct4) for DNA binding at the RAB5A and ERMN loci. (A) Oligonucleotides originating from the RAB5A and ERMN upstream control regions that contain annotated octamer and FOXO1 sites were analyzed for FOXO1 or POU5F1 (Oct4) binding. (B) A radiolabeled oligonucleotide was incubated with recombinant POU5F1 (Oct4) or FOXO1 and fractionated by native PAGE. Antibody added prior to loading was used to supershift POU5F1 (Oct4) containing species (marked S). A panel of binding assays containing increasing amounts of POU5F1 (Oct4) was used to assay the ability of POU5F1 (Oct4) to displace FOXO1. (C) An analogous binding assay performed with increasing the amounts of FOXO1 was used to test binding competition at the ERMN loci. (D) POU2F1 (Oct1) and FOXO1 protein levels were measured in differentiated and undifferentiated extract. Binding competition between POU2F1 (Oct1) and FOXO1 was tested by gel shift on the RAB5A probe. Increasing concentration (1, 5, 10 μL) of FOXO1 was incubated with a fixed concentration of POU2F1 (Oct1). The ratio of multiply bound to monomeric probe was quantified by phosphoimager.
Binding competition between POU5F1 and FOXO1 forms a molecular switch in PluCRs
In order to mechanistically classify the relationship between FOXO1 and POU5F1 as cooperative or competitive, we used recombinant POU5F1 and probes from the RAB5A and ERMN gene in an in vitro binding assay. Both genomic regions contain equally strong matches to the octamer site (ATGCA[A/T]AT, 4C→T in both regions) and two FOXO1 binding sites (TTGTTT) (Fig. 5A). These oligonucleotides map to in vivo regions of POU5F1 binding and are located upstream of ERMN and RAB5A genes. We performed a series of gel mobility shift assays with varying concentrations of recombinant POU5F1 and FOXO1 in order to determine the nature of the binding events. Although each binding site was tuned to bind either FOXO1 (e.g., the RAB5A probe) (Fig. 5B, lane 3 vs. lane 2) or POU5F1 (e.g., the ERMN probe) (Fig. 5B, lane 2 vs. lane 3), both probes were capable of binding both proteins; however, there was no evidence of an additional species corresponding to both factors binding simultaneously. FOXO1 binding immediately decreased with increasing POU5F1 (Fig. 5B, lanes 4–7). Incubation with antibodies against POU5F1 had a supershifting effect (“←S”; Fig. 5B, lane 8). This antibody also had a blocking effect, thereby reducing the binding of POU5F1 to the probe, which allowed FOXO1 to remain bound (Fig. 5B, lanes 9–12). POU5F1 binds ERMN with higher affinity than RAB5A (Fig. 5C); however, the quantity of POU5F1-containing species is reduced by increased concentration of FOXO1 (Fig. 5C, decreasing supershift, lanes 4–7). We conclude that POU5F1 and FOXO1 bind competitively to these sequences. As FOXO1 and POU5F1 are expressed together in ES cells, this competition could also be occurring in vivo (Fig. 5D). To test whether POU2F1 also competes with FOXO1, we purified recombinant POU2F1 and assayed binding on the RAB5A probe. POU2F1 exhibited the same binding preference as POU5F1 on the two substrates tested binding with equal or perhaps slightly lower affinity and exhibiting a greater tendency for dimerization that increased with POU2F1 concentration (Fig. 5D, lane 2, upper band; data not shown). Unlike POU5F1, there was no detectable decrease in monomeric POU2F1 binding with increasing FOXO1 over several different concentrations (Fig. 5D, lanes 4–14) but rather an increase in multiply bound probe (lane 14).
Proximal binding of NANOG or SOX2 influences POU2F1 versus POU5F1 binding
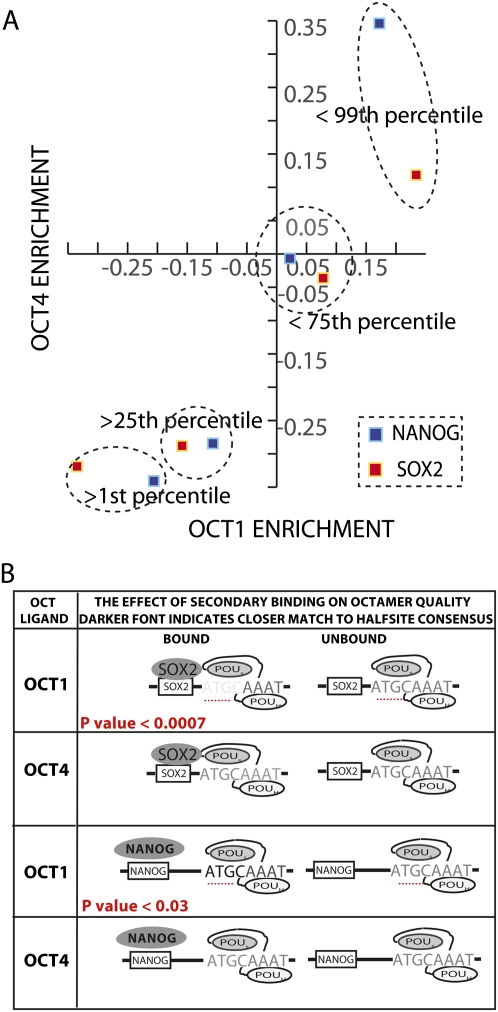
While there are many more potential cooperative and competitive relationships suggested by this analysis, the most obvious hypothesis for secondary influences on POU5F1 binding is the nearby binding of NANOG or SOX2. To understand the effect of SOX2 and/or NANOG binding on POU specificity, the oligonucleotide pool was ranked by NANOG binding. The nested sets from the top and bottom of this ranked list were then analyzed for POU2F1 and POU5F1 binding. One striking observation is that oligonucleotides enriched in the bound fraction of both NANOG and SOX2 are also enriched in the bound fraction of POU5F1 and POU2F1, respectively (Fig. 6, diagonal trend toward the upper right quadrant with increasing enrichment in SOX2 and NANOG). While the association between these binding patterns validates previous reports of interactions, the resolution and scale of this binding study afforded the opportunity to search hundreds of biologically relevant binding sites to determine the relative binding locations of POU5F1 to NANOG and SOX2.
Figure 6.
POU2F1(Oct1)/POU5F1(Oct4) binding is influenced by proximal SOX2 and NANOG binding. (A) Subsets of the oligonucleotide pool were created by ranking the pool by SOX2 (red squares) and NANOG (blue squares) enrichment. For each subset, a data point was plotted comparing the average POU2F1 (x-axis) and POU5F1 (y-axis) binding enrichment. (B) The top 1% of POU2F1 (Oct1) and POU5F1 (Oct4) enrichments are denoted as POU2F1 and POU5F1 “ligands” and annotated with an octamer binding model that allowed up to three mismatches. The number (n) and quality of octamer sites were compared for the POU ligands that did or did not bind SOX2. The darker font indicates closer agreement of a halfsite to the octamer consensus in the subset of POU ligands bound by SOX2. The lighter font indicates less agreement. The same analysis was repeated for NANOG. Statistically significant differences in comparisons of half-site strength are listed in red.
Previous studies have demonstrated that SOX2 positioned upstream of the octamer ATGCAAAT can be separated by up to 3 nt and retain protein–protein complexes; however, no structural information has been reported for NANOG (Remenyi et al. 2003; Williams et al. 2004). Comparing the distribution of NANOG sites relative to the octamer motif revealed that high-affinity NANOG sites were significantly more likely (P-value = 0.016) to contain the NANOG recognition element CATT in the forward orientation 3 or 4 nt upstream of the octamer to make the combined motif CATTNNN(N)ATGCAAAT (Supplemental Fig. S2; Supplemental Table S3). In comparing NANOG (as a cofactor) versus SOX2, proximal NANOG binding is more strongly associated with POU5F1 binding than POU2F1 binding. Conversely, SOX2 is more highly associated with POU2F1 binding than POU5F1 binding in this assay. This is consistent with reports that while NANOG is known to interact with POU5F1 alone, SOX2 can also interact with POU2F1 (Williams et al. 2004). In addition, the structure of POU5F1/SOX2 contact was determined by homology modeling a complex containing POU2F1's and not POU5F1's POU domain (Remenyi et al. 2003).
It is interesting to note that in the tripartite structure (DNA/POU domain/SOX2), the SOX2 protein contacts DNA and the POUs domain of the POU factor. The POU factor's POUs subdomain recognizes the ATGC of the octamer through contacts that are presumably stabilized by the interacting SOX2. As an POU factor in complex with SOX2 has the added stability of protein–protein complexes, we hypothesized a lesser requirement for the DNA contacts in the ATGC motif than would be observed in POU2F1 or POU5F1 binding in the absence of SOX2. To test this idea that strong SOX2 binding rescues suboptimal binding of the POUs domain to DNA, we compared the tolerance of mismatched octamers in POU2F1 ligands that bind SOX2 (top quartile) versus ligands that do not (bottom three quartiles). Within the set of octamer containing POU2F1 ligands, the subset of regions that also bind SOX2 has significantly weaker octamer sequences than regions that do not bind SOX2 (P-value = 0.0007). This result suggests that SOX2 can rescue a deficiency in octamer binding sites in POU2F1 but not POU5F1 binding. The region most tolerant of mismatches is the ATGC halfsite that is recognized by the POUs domain. As the SOX2 interaction domain is contained within the POUs subdomain, this result suggests that the interaction with SOX2 can rescue a loss in canonical protein–DNA contacts with the ATGC halfsite. This phenomenon of protein–protein interactions rescuing suboptimal protein–DNA interactions is only observed in POU2F1/SOX2 complexes and not in POU5F1/SOX2 or POU2F1/NANOG complexes. On the contrary, stronger octamer sites are required for POU2F1 binding in the presence of NANOG binding, suggesting that POU2F1 needs a stronger octamer perhaps to compete with POU5F1 that can be stabilized by NANOG. Consistent with this, octamers in POU5F1 ligands that also bind NANOG are slightly weaker, although this difference did not reach the threshold of statistical significance. This analysis suggests that SOX2 more effectively stabilizes POU2F1 than POU5F1 on DNA and that NANOG is most associated with POU5F1.
Sequence motifs enriched in the POU2F1 subclass of POU ligands are predictive of POU2F1 binding in vivo
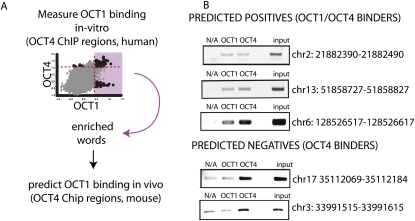
The analysis of the POU2F1-specific ligands suggests that there are sequence determinants that allow for the prediction of POU2F1 binding within POU5F1 regulated genes. These sequence determinants are complex and integrate the contribution of a multitude of unknown secondary binding events (perhaps similar to SOX2 and NANOG) with the intrinsic difference in POU binding specificities to types of sites (such as MORE, POREs, gapped octamers). We developed a prediction tool that captures these sequence determinants for the purpose of detecting POU2F1 binding in vivo within POU5F1 ChIP enriched regions, such as the PluCRs (Fig. 7A, prediction scheme). We identified a subset of 5-mers (Supplemental Table S6) enriched in POU2F1 binding sites that were drawn from human POU5F1 ChIP enriched regions. We scored PluCRs for agreement to this word set to return a score that predicted whether an entire PluCR would be predominantly bound by POU2F1 or POU5F1.
Figure 7.
In vivo validation of predicted POU2F1 (Oct1) binding. The prediction algorithm is depicted in A. Significantly enriched words in the top 1% of POU2F1 (Oct1) binding oligonucleotides were added to the predictor with their enrichment score (χ2 statistic). This score calculated over a window of 100 nt was used to the predict POU5F1 ChIP regions in mouse and orthologous regions in human, cow, and dog. (B) High-scoring regions predicting POU2F1 binding were tested by ChIP in mouse. Low-scoring regions predicting background levels of POU2F1 binding were also tested.
Initially, we sought to evaluate the quality of the POU2F1/POU5F1 predictions by particular cross-species comparisons. POU2F1 is found in all vertebrates, whereas POU5F1 is restricted to vertebrates that undergo implantation during embryogenesis. Therefore, we reasoned that if computational prediction correctly distinguishes POU5F1 from POU2F1 binding, then the PluCR targets predicted to only bind POU5F1 should be less conserved than the POU2F1 targets in egg-laying vertebrates. It has recently been demonstrated that sites of POU5F1 binding are rapidly evolving (Kunarso et al. 2010). Therefore this comparison was performed on the subset of PluCRs that were retained across three of the four mammalian genomes used in this study. Comparing the conservation of this subset of human PluCR in chicken reveals that orthologs of the predicted POU2F1-bound PluCRs can be found at a higher rate (56%) than orthologs of predicted POU5F1 PluCRs (37%) for cases where the gene is conserved (P-value = 0.18). This observation that POU5F1 predicted targets are under reduced selection in a species lacking POU5F1 suggests that the POU2F1/POU5F1 predictor is accurate; however, the statistical confidence of this claim is modest because of the low numbers of successful double alignments (22 PluCR + gene alignments) and the fact that POU5F1 is only one of many trans-acting factors that bind PluCRs.
To directly test the validity of the POU factor predictions, we performed ChIP on predicted POU2F1 and POU5F1 ligands. Positive and negative predictions for POU2F1 binding were selected on the basis of consistently high or low scores in three vertebrate orthologous regions. We found windows that contain words enriched in POU2F1-specific ligands bind POU2F1 in vivo (Fig. 7B, predicted positives), and that the opposite is also true; regions that lack these words are not enriched in POU2F1 ChIP'ed regions and appear to bind POU5F1 exclusively (Fig. 7B, predicted negatives). By incorporating the local context of binding, these findings demonstrate how data derived from in vitro binding results performed in extract can be predictive of transcription factor binding in vivo.
Discussion
We have developed a high-throughput implementation of a classic biochemical binding assay to map 400 Mb of genomic sequence for the precise sites of complex formation in ES cell extract. Mapping DNA binding at the resolution of 10 nt over thousands of individual sites facilitates the detection of nuanced differences in binding between highly similar paralogs and discovering cooperative and competitive binding events on a genomic or semi-genomic scale. This approach will help solve a common generic problem: There are many families of nucleic acid binding proteins that share similar biological specificity but possess nonredundant functions.
Here we compared binding between POU2F1 and POU5F1, two factors that are expressed in ES cells and were originally discovered (and named) by their affinity for the same octamer sequence, ATGCAAAT. As POU factors are well-studied proteins, many early biochemical characterizations of POU binding performed on synthetic sequences can be revisited with endogenous sequences on a genomic scale. The ability of POU factors to bind palindromic combinations of halfsites (MORE and PORE motifs) is a point of difference between POU5F1 and POU2F1; POU5F1 has a higher affinity for MORE elements (Fig. 3A). Structural studies demonstrated that the flexible linker connecting the two POU subdomains enables POU binding to tolerate a space of up to 2 nt between halfsites (Tomilin et al. 2000). This analysis finds that the thousands of binding events observed on real sites conform almost perfectly to this limit, but again, there are differences between paralogs. While the single largest class POU2F1 ligand is the perfect octamer, POU5F1 ligands appear to be almost three times more likely to contain a space (i.e., a 1- or 2-nt gap) than to be perfect octamers (Fig. 3B).
Here we report that POU2F1 and POU5F1 both bind the octamer sequence with some important distinctions. POU5F1's predominant mode of interacting with DNA is dominated by full or combinations of half octamer sites. Relative to POU2F1, the more prominent ATGC halfsite is found in POU5F1 ligands, and binding is spread over broader genomic regions. While POU5F1 mostly binds DNA through some combination of octamer halfsites, motif analysis suggests POU2F1 can bind or be recruited through alternate mechanisms (note nonoctamer motifs in Fig. 4A, region 3). The human protein reference database reports 36 interactions between POU2F1 and other DNA binding proteins compared with 10 for POU5F1 (Peri et al. 2003). While there are many potential biases in the interactome data, this study does suggest that POU5F1's primary mode of binding may be more direct than that of POU2F1 in the PluCRs. It is possible that POU2F1 is serving as an adapter and interacting with the transcriptional machinery as heterogeneous complexes with distinct functions. The need to spatially separate these distinct signals may explain the lower tendency of POU2F1 sites to cluster. Indeed, a side population of oligonucleotides seems specific for POU2F1 and not for POU5F1. Scoring the ligands in the oligonucleotide pool for matches to the TRANSFAC motifs suggested patterns of a second transcription factor binding with POU2F1 or POU5F1. Some of these second factors have been implicated in cooperative binding events with a specific POU factor. For example, NF-Y binding motifs are associated with POU2F1 binding across the PluCRs. This trend is supported by reports of NF-Y forming complexes with POU2F1 on the GADD45 promoter (Fan et al. 2002).
In addition to this interesting class of elements, there are sites that are strongly associated with the binding profile of both POU factors. Not surprisingly, half of the most enriched binding models presented in Figure 4B belong to the multiple TRANSFAC POU position weight matrices. More than a fifth of the remaining transcription factor binding sites belong to forkhead (FOX) transcription factor motifs. We show at multiple loci that POU5F1 and FOXO1 are engaged in a mutually exclusive binding pattern. A similar competition pattern was reported with FOXD3 and POU5F1 on the osteopontin promoter (Guo et al. 2002). However, here we present an analysis that suggests that this relationship exists between the entire family of forkhead and POU factors at numerous regulatory regions within the PluCRs.
Evidence suggests that this antagonistic binding relationship between POU and forkhead transcription factors is mirrored by counteracting roles in maintaining stemness. At an organismal level, studies on null FOXO allele mice reveal defects in tissue homeostasis of various adult stem cell populations. The defects are characterized by a small stem cell compartment in the adult and, in some cases, a transient bloom in differentiation at younger stages. FOXO3-deficient animals undergo premature follicle hyperactivation, which depletes oocytes and causes infertility (Castrillon et al. 2003). FOXO-deficient animals develop increased brain size and proliferation of neural stem cells in early life, followed by a greatly reduced stem cell compartment and neurodegeneration in adult life (Paik et al. 2009). This transient increase in a stem cell compartment following FOXO depletion argues for an antagonistic function between forkhead factors and factors that maintain a pluripotent state. In the FOXO-deficient background, the hemotopoetic stem cell compartment is reduced and marked by elevated levels of oxidative stress (Tothova et al. 2007). This loss of homeostatic control is reversed by antioxidants. FOXO1's role in controlling oxidative stress and in restricting the population of stem cells antagonizes POU factor function, as POU factors have recently been found to increase their binding activity in response to cellular stress (Kang et al. 2009). Taken together, the forkhead and POU factors represent two large transcription factor families that are important in development, have roles in cellular stress response, and bind regulatory regions competitively with overlapping specificities. Furthermore, FOX proteins appear to restrict adult stem cell populations in early development and, in this regard, antagonize the pluripotent function of POU factors.
In addition to competition, the proximal binding of SOX2 and NANOG appears to play a large role in the modulation of POU specificity in the PluCR regions. The association between strong NANOG and POU5F1 binding is compelling and is supported by the finding that NANOG and POU5F1 can be coimmunoprecipitated in vivo (Zhang et al. 2007). The optimal binding location of NANOG lies 3–4 nt apart from the octamer motif, raising the possibility that, at least on these substrates, NANOG may not directly contact POU5F1. SOX2, on the other hand, is associated with both binding events, but strong SOX2 sites are tilted toward POU2F1 but not POU5F1 binding.
The POU/SOX2 interaction surface that forms along the POUS domain of POU2F1 ligands appears to have relaxed the stringency of the ATGC halfsite that the POUS domain recognizes. Recently, molecular simulation between the POUS domain of POU2F1 and SOX2 have kinetically modeled the binding of the POUS domain of POU2F1 to its ATGC substrate with and without SOX2 (Lian et al. 2010). Extensive hydrophobic protein/protein interface between POUS and HMG (e.g., SOX2) is largely responsible for ensuring a stable ternary complex formation. Using mutations that perturb the protein/protein interface and the POUS/DNA interface, we conclude that the protein/protein interface effectively prevents the dissociation of POUS from the DNA. This result explains the loss of the requirement of ATGC for POU2F1 when SOX2 is bound proximately. Given this, it is interesting to note that POU5F1 binding motifs observed in this study feature more prominent ATGC motifs in the octamer than POU2F1 (Figs. 3A, 4A).
While the idea that POU2F1 and not POU5F1 binding synergizes more strongly with SOX2 may seem surprising, there is additional evidence that supports this claim. There are examples that these factors interact to coregulate genes outside ES cells. POU2F1 and SOX2 have been shown to function synergistically in the regulation of the PAX6 and nestin genes later in neural development (Donner et al. 2007; Jin et al. 2009). SOX2 is widely expressed and therefore present with POU2F1 in many more tissues than POU5F1, including cell types and cell lines (e.g., HeLa) where POU2F1 is the only POU factor detectable (M Gemberling and L Ferraris, unpubl.). All structural evidence of POU domain/HMG interactions comes from studies on POU2F1. The solution structure of the HMG/POU domain interaction on DNA was performed with POU2F1, not POU5F1 (Williams et al. 2004). Both structures of POU5F1-SOX2 on the UTF1 and FGF4 promoter elements were crystallized with POU2F1 and converted to POU5F1 coordinates by homology modeling (Remenyi et al. 2003).
Finally, using this high-throughput binding assay as a means of interrogating the context in which DNA/protein interactions occur, we effectively model DNA/protein interactions in vivo. While chromatin features and epigenetic marks are unlikely to be recapitulated in this assay, we are performing these assays on ChIP'ed regions that are both accessible to and bound by POU, SOX2, and NANOG in vivo. The resolution of ChIPs is on a different scale than the 10-nt resolution achieved in vitro because of the technical limitations of the shearing step during the ChIP protocol. Despite this difficulty, averaging the in vitro–generated prediction over longer windows distinguishes genomic regions that bind POU2F1 from those that bind only POU5F1. This tool can be applied to PluCRs to reevaluate the network and provisionally identify genes that are more likely to be regulated by POU5F1 than POU2F1. High-POU2F1 scoring genes included HDGF, NFIB, HES1, and TGFA. The most significant GO term associated with POU2F1-predicted genes was “positive regulation of cell proliferation.” The most strongly associated motif was FOXO1, which suggests the possibility that POU2F1 and FOXO1 interact to regulate cell fate (Subramanian et al. 2005). Given the overlapping specificity of POU factors and their distinct binding relationship with FOXO1, it is possible that the exchange of POU factors on these targets during differentiation acts as a switch that triggers FOXO1 binding. Understanding which targets are important to reprogramming a pluripotent state in somatic cells will be of major interest as the therapeutic potential of induced pluripotent stem cells are better evaluated. Likely POU5F1-only genes included POU5F1 itself, GAL, FOXH1, FGFR1, and RAB2B. As POU5F1 is a central player in this triad of factors and POU2F1 and SOX2 are already present in many somatic cells, we intend to focus on predicted POU5F1-specific targets for refining the pluripotency control network in ES cells.
Methods
Cell culture and cell extracts
ES cells were cultured in DMEM + HEPES supplemented with 1 mM glutamine, 1 mM sodium pyruvate, and 1 mM MEM nonessential amino acids (Invitrogen) plus 15% ES cell-qualified heat-inactivated fetal bovine serum (HyClone), 50 mM 2-mercaptoethanol (Sigma), and leukemia inhibitory factor (LIF/ESGRO; Chemicon). Differentiation of J1 ES cells occurred in the presence of 10-7 M (or 100 nM) RA over a 16-d period. Whole-cell extracts were obtained from J1 ES (male) undifferentiated and RA differentiated cells. Cells were pelleted, resuspended, and incubated in extraction buffer (200 mM KCl, 100 mM Tris at pH 8.0, 0.2 mM EDTA, 0.1% Igepal, 10% glycerol, and 1 mM PMSF) for 50 min on ice. Cell debris was pelleted, and extracts were frozen using liquid N2 and stored at −80°C.
Library design, oligonucleotide synthesis, cloning, and sequencing
A complex pool of 60-mer oligonucleotides was synthesized to contain the union of the intersections of ChIP-ChIP fragments of POU5F1, NANOG, and SOX2, as found in core transcriptional regulatory circuitry in human ES cells (Boyer et al. 2005). The previously described MEGAshift protocol was used to map binding to the PluCRs (Reid et al. 2009). This pool was synthesized as a custom oligonucleotide microarray and liberated from the slide by boiling for 1 h at 99°C. Low-cycle PCR was used to amplify the pool. Electrophoretic analysis was used to ensure log linear amplification as described previously (Reid et al. 2009). Each oligonucleotide was designed as a tiled genomic 30-mer flanked by the common sequences CCAGTAGATCTGCCA and ATGGAGTCCAGGTTG, which were used as the universal primer binding pair. This oligonucleotide pool was used in coimmunoprecipitation binding studies with antibodies identified below. Nonuniform representations of oligonucleotides are normalized by the two color array approach described in the next section.
Microarrays
We used 8×15k and 2×104k Custom Agilent oligonucleotide microarrays designed complementary to oligonucleotides in each oligonucleotide pool. DNA oligos isolated from four different coimmunoprecipitation reactions (POU5F1, POU2F1, SOX2, NANOG) were amplified with T7 tailed primers and used as templates to produce RNA targets for array analysis. The starting pool was subjected to equal PCR cycles/treatments. RNA targets were produced containing amino-allyl UTP using MEGAshortscript High-Yield Transcription kit (Ambion) after appending a T7 promoter to the oligonucleotides. Amino-Allyl UTPs were then coupled to Cy3 and Cy5 dyes, through a labeling procedure. RNA was pelleted and resuspended in 1 M NA2CO3; monoreactive dyes were added; and the reaction was allowed to continue for 1 h at room temperature. We used 4 M hydroxylamine to quench the reaction followed by phenol:chloroform and ethanol precipitation to remove remaining free nucleotides. Microarrays were hybridized for 3 h at 50°C using an optimized Agilent gene expression hybridization kit and protocol. Microarrays were scanned at 5 μm using a GenePix 4000B scanner and were analyzed using Feature Extraction Software from Agilent. Raw data and UCSC Genome Browser tracks can be downloaded at http:/fairbrother.biomed.brown.edu/data/pluripotent-2010 or through GEO at http://www.ncbi.nlm.nih.gov/geo/ using accession no. GSE27535. Replicate measurements were averaged, and oligonucleotides were ranked according to enrichment. Oligonucleotides were separated into categories according to enrichment, and a variety of sequence analyses and comparisons were performed on these sets as described in the text. Data analysis is described in detail in the Supplement.
EMSA
Oligonucleotides were prepared for EMSA by end labeling PCR products with g-32P-ATP. Samples were prepared in 20 μL (0.6× Buffer D, 50 ng/μL Poly dI•dC, 1 μg/μL BSA, 1 mM DTT, and 20 ng of probe). Samples were incubated at room temperature for 30 min. Native 4% polyacrylamide gels (29:1 acrylamide:bisacrylamide, 1% glycerol, 0.5× TBE) were prerun for 1 h at 80 V; samples were loaded and run for 1.75 h at 80 V. A POU5F1 antibody (Santa Cruz Biotechnology) was used for the supershift.
Predictor of POU2F1 binding
Briefly, this tool was trained on the human in vitro binding data described above, and the occurrence of frequency of motifs associated with POU2F1 binding was used to score a test set derived from a mouse POU5F1 ChIP study. To detect words preferentially enriched in the POU2F1 binding subpopulation, we fragmented each 30mer oligo into its constituent k-mer and then compared the frequency of each of these k-mers in the POU2F1 enriched set against the background of the total pool. A power calculation indicates 90% of twofold enrichment events can be detected with 5-mers.
Annotation of POU5F1 ChIP regions for potential POU2F1 binding
We proceeded to use the chi-square scores of 5-mers from our POU2F1 data to determine regions that could be reasonably predicted to interact with POU2F1. The cross-species comparative analysis of POU2F1 word enrichment in known POU5F1-binding regions began by mapping POU5F1 ChIP regions in Homo sapiens to Rattus norvegicus, Bos taurus, and Pan troglodytes with the UCSC tool, liftover. We scanned through all possible 100-mer windows in these four species and annotated regions in which three of the four species had average chi-square scores that were above or below threshold (average chi-square >100). Regions for which at least three of four species had data and in which the average of the window exceeded threshold were used for validation (Fig. 7) and for comparison of conservation in chicken, a species that lacks POU5F1. This final comparison recorded the success rate of identifying the ortholog of the regulatory region (i.e., the PluCR) and its associated gene in chicken.
Western blotting and immunodetection
Following SDS-PAGE separation, proteins were transferred electrophoretically onto a nitrocellulose membrane for 1 h at 30 V following Western blotting. The antibodies used to blot the membrane were anti-POU5F1, anti-SOX2, anti-POU2F1, anti-FOXO1 (Santa Cruz Biotechnology), anti-NANOG (Cosmo Bio), anti-mouse, and goat anti-rabbit HRP-linked (Cell Signaling Technology). The results were visualized by a chemoluminescent reaction using Pierce ECL Substrate Western blot detection (Pierce). For blot development, the membranes were exposed to Kodax Bio Max Light Film.
Acknowledgments
We thank Dr. Lisa Cirillo (Medical College of Wisconsin, Department of Cell Biology, Neurobiology and Anatomy) for the extremely generous gift of FOXO1 proteins and expression constructs, and Richard Freiman and Alex Brodsky for discussions and reagents. The work in the laboratory is partially supported by Award Number R21HG004524 from the National Human Genome Research Institute and Brown University. We also thank the NSF UBM-Group for summer support to A.P.S. (DUE-0734234), a CCMB Scholarship Award (to W.G.F), the Brown University Research Seed Fund Program (to W.G.F), and grants from the American Cancer Society (GMC-115196) and National Cancer Institute (1R21CA141009) to D.T. Finally, we thank the anonymous reviewers for specific suggestions that were incorporated into the manuscript.
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE27535.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.115824.110.
References
- Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J 2007. Analysis of POU5F1-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells 25: 500–510 [DOI] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI 1995. Embryonic stem cells express neuronal properties in vitro. Dev Biol 168: 342–357 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA 2003. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science 301: 215–218 [DOI] [PubMed] [Google Scholar]
- Chakravarthy H, Boer B, Desler M, Mallanna SK, McKeithan TW, Rizzino A 2008. Identification of DPPA4 and other genes as putative SOX2:POU-3/4 target genes using a combination of in silico analysis and transcription-based assays. J Cell Physiol 216: 651–662 [DOI] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL 2007. SOX2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol 303: 784–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Jin S, Tong T, Zhao H, Fan F, Antinore MJ, Rajasekaran B, Wu M, Zhan Q 2002. BRCA1 regulates GADD45 through its interactions with the POU-1 and CAAT motifs. J Biol Chem 277: 8061–8067 [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W 1996. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with POU-1/POU-2 factors, leading to differential promoter activation. EMBO J 15: 2781–2790 [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R 2002. The embryonic stem cell transcription factors POU-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci 99: 3663–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough SR, Clements I, Welch PJ, Wiederholt KA 2006. Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of POU5F1 and NANOG. Stem Cells 24: 1467–1475 [DOI] [PubMed] [Google Scholar]
- Jin Z, Liu L, Bian W, Chen Y, Xu G, Cheng L, Jing N 2009. Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. J Biol Chem 284: 8160–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D 2009. A general mechanism for transcription regulation by POU2F1 and POU5F1 in response to genotoxic and oxidative stress. Genes Dev 23: 208–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Lian P, Angela Liu L, Shi Y, Bu Y, Wei D 2010. Tethered-hopping model for protein-DNA binding and unbinding based on SOX2-POU2F1-Hoxb1 ternary complex simulations. Biophys J 98: 1285–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG 2000. Quantitative expression of POU-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H 1990. A novel Octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60: 461–472 [DOI] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, et al. 2009. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5: 540–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. 2003. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 13: 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K, Luisi B 2000. The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol 302: 1023–1039 [DOI] [PubMed] [Google Scholar]
- Reid DC, Chang BL, Gunderson SI, Alpert L, Thompson WA, Fairbrother WG 2009. Next-generation SELEX identifies sequence and structural determinants of splicing factor binding in human pre-mRNA sequence. RNA 15: 2385–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of POU5F1 and SOX2 on two enhancers. Genes Dev 17: 2048–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales R, Vigneron M, Macchi M, Davidson I, Xiao JH, Chambon P 1987. In vitro binding of cell-specific and ubiquitous nuclear proteins to the octamer motif of the SV40 enhancer and related motifs present in other promoters and enhancers. EMBO J 6: 3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Brunet A 2008. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 20: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D, Gemberling M, Callister C, Fairbrother W 2008. High-throughput biochemical analysis of in vivo location data reveals novel distinct classes of POU5F1(POU5F1)/DNA complexes. Genome Res 18: 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilin A, Remenyi A, Lins K, Bak H, Leidel S, Vriend G, Wilmanns M, Scholer HR 2000. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell 103: 853–864 [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339 [DOI] [PubMed] [Google Scholar]
- Watkins KH, Stewart A, and Fairbrother W 2009. A rapid high-throughput method for mapping ribonucleoproteins (RNPs) on human pre-mRNA. J Vis Exp 34: 1622 doi: 10.3791/1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DC Jr, Cai M, Clore GM 2004. Molecular basis for synergistic transcriptional activation by POU2F1 and SOX2 revealed from the solution structure of the 42-kDa POU2F1.SOX2.Hoxb1-DNA ternary transcription factor complex. J Biol Chem 279: 1449–1457 [DOI] [PubMed] [Google Scholar]
- Zhang L, Rayner S, Katoku-Kikyo N, Romanova L, Kikyo N 2007. Successful co-immunoprecipitation of POU5F1 and NANOG using cross-linking. Biochem Biophys Res Commun 361: 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]