SUMMARY
Sensory cilia are assembled and maintained by kinesin-2-dependent intraflagellar transport (IFT). We investigated if two C. elegans α- and β-tubulin isotypes, identified via mutants that lack their cilium distal segments, are delivered to their assembly sites by IFT. Mutations in conserved residues in both tubulins destabilize distal singlet microtubules (MTs). One isotype, TBB-4, assembles into MTs at the tips of the axoneme core and distal segments, where the MT tip-tracker, EB1, is found, and localizes all along the cilium, whereas the other, TBA-5, concentrates in distal singlets. IFT assays, FRAP analysis and modeling suggest that the continual transport of sub-stoichiometric numbers of these tubulin subunits by the IFT machinery can maintain sensory cilia at their steady state length.
INTRODUCTION
Sensory cilia detect and transmit signals that control gene expression, cell behavior and development1. They consist of a specialized ciliary membrane containing signaling molecules surrounding an axoneme that is differentiated longitudinally into a “middle segment” of nine MT doublets, and a “distal segment”, which defines a specialized signaling domain, consisting of nine singlet MTs that extend from the A-tubules of the doublets2–6.
Cilia are assembled by intraflagellar transport (IFT), a process discovered in Chlamydomonas and which involves the kinesin-2 driven movement of IFT particles from the base to the tip of the axoneme7–12. IFT-particles are multimeric protein complexes visible by EM as “trains”13,14 that are proposed to deliver assembly precursors, e.g. tubulin, to the tips of the axoneme15–17. Despite the progress in studying the transport of tubulin along axons18, the role of IFT in the delivery of tubulin subunits to their site of incorporation within axonemes remains poorly understood16.
In Caenorhabditis elegans, two members of the kinesin-2 family cooperate to drive IFT and assemble sensory cilia on chemosensory neurons. First, IFT particles, transported by the concerted action of heterotrimeric kinesin-II and homodimeric OSM-3, build the middle segment of the axoneme. Subsequently, kinesin-II dissociates from the IFT particles, which are then moved by OSM-3 alone to assemble the distal singlet MTs19–22. Significantly, the hypothesis that IFT moves tubulin subunits along these cilia has not been tested, but the use of two types of kinesin-2 motors to build specific parts of sensory cilia may be widespread. For example, in vertebrates, heterotrimeric kinesin-2 (KIF3) builds the axoneme core but homodimeric kinesin-2 (KIF17), which is targeted to cilia by the nuclear import machinery, builds distal singlets on zebrafish photoreceptors and targets signaling proteins to primary cilia2,23,24.
The C. elegans community has produced a valuable collection of mutants that affect IFT and ciliogenesis25. Previously, by screening such mutants for defects in the OSM-3/distal segment assembly pathway, we identified the IFT particle subcomplex B (IFT-B) associated protein, DYF-1/IFT7026, which we propose to be an OSM-3 activator19,27. Here we describe three IFT-B proteins and two tubulin isoforms that are also components of this pathway. Microscopy and modeling suggests that IFT transport delivers tubulins to the distal tips of axonemal MTs, where they become differentially localized.
RESULTS
Mutants lacking the distal segments of sensory cilia fall into two classes
Based on the morphology of cilia containing fluorescently-tagged IFT particle proteins19,27 the dyf mutants25 were organized into five groups (Supplementary Information, Fig. S1). Complementation tests revealed that many were allelic (e.g. ks69, qj42, qj16 and che-10) (Supplementary Information, Fig. S2, Tables S1 and S2). We focused on qj55, qj23, qj14 and dyf-12(sa127)27,28, the uncharacterized chemosensory mutants in class C that had intact middle segments but no distal segments (Fig. 1a–c, and see Supplementary Information, Fig. S1c, S3a,b, Table S3).
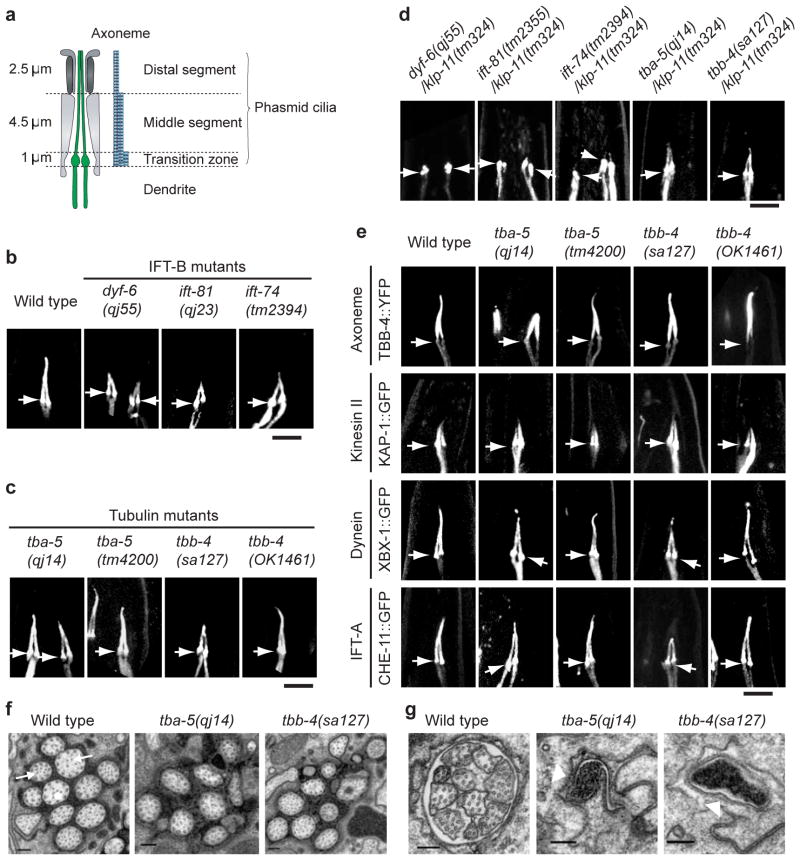
Figure 1.
Characterization of the dyf-6, ift-81, ift-74, tba-5 and tbb-4 mutants. (a) Cartoon illustrating the structure of the phasmid endings. (b–c) The DYF-1::GFP marker was used to visualize the phasmid ciliary morphology of wild type, three IFT-B mutants (b) and two tubulin mutants (c). (d) The DYF-1::GFP marker was used to visualize the phasmid ciliary morphology of the double mutants, dyf-6;klp-11, ift-81;klp-11 and ift-74;klp-11, tba-5/klp-11, tbb-4/klp-11, demonstrating that dyf-6, ift-81 and ift-74 are distinct from tba-5(qj14) and tbb-4(tm324). (e) Four different markers were used to visualize the phasmid ciliary morphology in the tubulin mutants. In b-e, bar=5 μm. Arrows point to transition zones with cilia oriented upward. See also Figures S3, which shows the ciliary morphology of these mutants in the phasmid and amphids using other markers. (f) Electron micrographs of amphid middle segments in wild-type (left), tba-5(qj14) (middle), and tbb-4(sa127) (right) adult animals. Arrows in the wild-type section point to singlet microtubules that occur less frequently in tbb-4(sa127) mutants. (g) Same as (f) except the amphid distal segment is shown. White arrow heads point to the empty distal channel. In (f) and (g), bar=200 nm.
qj55 and qj23, were shown to be new alleles of dyf-6 and ift-81, respectively (Supplementary Information, Table S1, and Fig. S2, S4a–d)29,30 whose products, DYF-6 and IFT-81, plus the IFT-81 binding partner, IFT-74, are IFT-B subunits13,28,29. Double mutants qj23;klp-11 and qj55;klp-11, which lack kinesin-II function, are missing the entire axoneme similar to the osm-3;klp-11 mutants (Fig 1d, and see Supplementary Information, Fig. S3c and Table S3). This, together with IFT assays (Supplementary Information, Table S4) suggests that IFT-74/81, which bound OSM-3 in yeast two hybrid assays (Supplementary Information, Fig. S4e), and DYF-6, which did not bind OSM-3, serve to activate OSM-3 motor activity, similar to the previously described IFT-B subunit, DYF-119 (Supplementary Information, Fig. S4f).
While the qj14 and dyf-12(sa127) mutants also lack distal segments, they define a second class of distal segment mutants, since the qj14;klp-11 or dyf-12(sa127);klp-11 doubles, which lack kinesin-II activity, retain intact middle segments, plausibly assembled by OSM-3-driven IFT (Fig. 1c–g, and see Supplementary Information, S3b, S3c and Table S3). In qj14 or dyf-12(sa127) single mutants, IFT particles move along the residual middle segments at ~0.7 μm s−1, characteristic of OSM-3 and kinesin-II working together, but in qj14;klp-11 or dyf-12(sa127);klp-11 double mutants, they move at ~1.2 μm s−1, characteristic of OSM-3 alone (Table 1). Thus OSM-3 retains activity and drives IFT in the qj14 and dyf-12(sa127) mutants.
Table 1.
Anterograde IFT velocities in the middle segment measured by IFT assays in tba-5 and tbb-4
| Marker | Genetic background | IFT velocity (μm/s) | N |
|---|---|---|---|
| Wild type | 0.70 ± 0.09 | 121 | |
| OSM-3::GFP | tba-5 (qj14) | 0.78 ± 0.09 | 131 |
| tbb-4(sa127) | 0.76 ± 0.08 | 146 | |
| Wild type | 0.73 ± 0.08 | 145 | |
| tba-5 (qj14) | 0.74 ± 0.10 | 146 | |
| KAP-1::GFP | tba-5 (tm4200) | 0.74 ± 0.13 | 118 |
| tbb-4(sa127) | 0.75 ± 0.09 | 146 | |
| tbb-4(OK1461) | 0.76 ± 0.10 | 127 | |
| Wild type | 0.77 ± 0.08 | 105 | |
| tba-5 (qj14) | 0.79 ± 0.09 | 127 | |
| XBX-1::YFP | tba-5 (tm4200) | 0.75 ± 0.09 | 116 |
| tbb-4(sa127) | 0.79 ± 0.09 | 166 | |
| tbb-4(OK1461) | 0.79 ± 0.10 | 88 | |
| Wild type | 0.72 ± 0.09 | 113 | |
| tba-5 (qj14) | 0.78 ± 0.09 | 210 | |
| CHE-11::GFP | tba-5 (tm4200) | 0.74 ± 0.08 | 114 |
| tbb-4(sa127) | 0.70 ± 0.07 | 116 | |
| tbb-4(OK1461) | 0.78 ± 0.09 | 107 | |
| Wild type | 0.75 ± 0.07 | 155 | |
| tba-5 (qj14) | 0.77 ± 0.08 | 158 | |
| tba-5 (tm4200) | 0.73 ± 0.09 | 104 | |
| DYF-1::GFP | tbb-4(sa127) | 0.73 ± 0.10 | 196 |
| tbb-4(OK1461) | 0.79 ± 0.08 | 117 | |
| tba-5 (qj14)/klp-11 | 1.17 ± 0.20 | 146 | |
| tbb-4(sa127)/klp-11 | 1.20 ± 0.13 | 142 | |
| tba-5 (qj14)/bbs-8 | 1.15 ± 0.20 | 239 |
tbb-4 and their double mutants with klp-11(tm324) or bbs-8(nx77). N indicates the number of IFT particles. of IFT particles.
The second class of mutants, qj14 and dyf-12(sa127), occur in genes encoding the α-and β-tubulins, TBA-5 and TBB-4
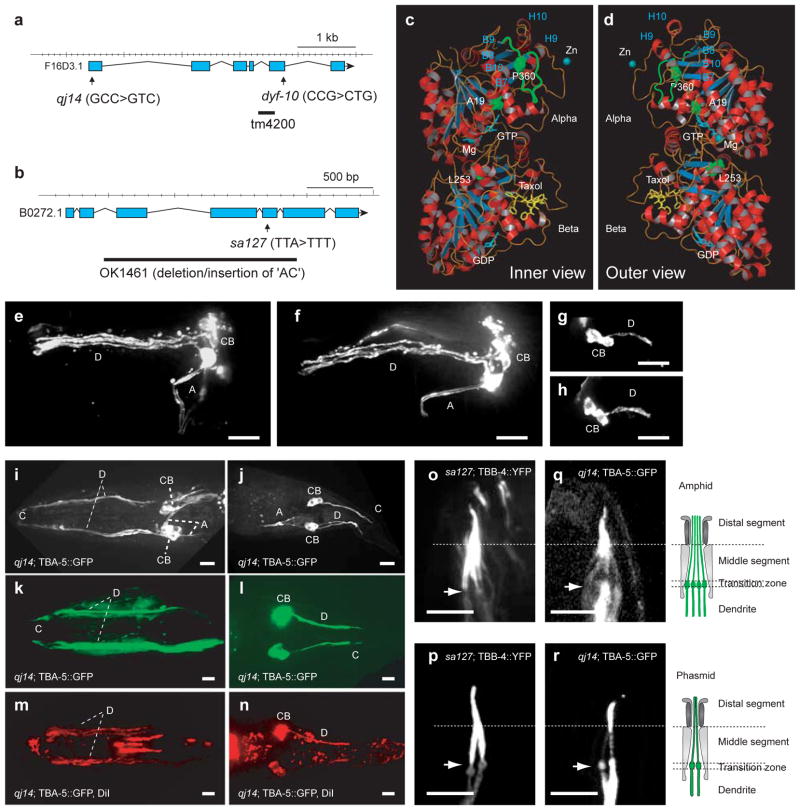
Complementation tests indicated that qj14 and dyf-10 are alleles of one another (Supplementary Information, Fig. S2 and Table S1). Mutant qj14 was SNP-mapped to the tba-5 gene locus (Fig. 2a), which encodes one of the nine C. elegans α-tubulins, TBA-5. Of these, tba-6, tba-9, but not tba-5, were proposed to be expressed in ciliated neurons based on genomic analysis31. SNP-mapping of the mutant dyf-12(sa127) showed that it encodes one of the 6 C. elegans β-tubulins, TBB-4, (Fig. 2b) which is expressed in cilia31.
Figure 2.
Expression and localization of two axonemal tubulins, TBA-5 and TBB-4 and characterization of their missense mutations. (a–b) Models of the tba-5 and tbb-4 gene. Two tba-5 missense mutations, qj14 and dyf-10, and the deletion mutation, tm4200 (a) and a tbb-4 missense mutation dyf-12(sa127) and a deletion mutation, tbb-4(OK1461) (b) are shown. (c–d) Inner (c) and outer (d) views of structure of predicted TBA-5 and TBB-4 heterodimer based on porcine brain tubulin dimer structure, 1JFF. The three point mutation sites (P360, A19 and L253) and the loop that contains P360 are shown in green. (e–h) Dyf assays of dpy-6 and dpy-6;tba-5(tm4200);tbb-4(OK1461) show no obvious defects in ciliary structure. Bar=10 μm. (i–j) A transgene tba-5p::tba-5::GFP is expressed in amphid neurons (i) and phasmid neurons (j) in tba-5(qj14). Bar=10 μm. (k–n) Cilium formation was rescued in amphids and phasmids of tba-5(qj14), by expression of the transgene, tba-5p::tba-5::GFP, demonstrated by the pseudocolor green in cilia and dendrites (k–l) and by the red color in Dyf assays (m–n). Bar=5 μm. (o–r) TBB-4::YFP restored the ciliary length of amphids (o) and phasmids (p) in tbb-4(sa127) and localized to the entire cilia nearly homogeneously. TBA-5::GFP restores the lengths of amphid (q) and phasmid (r) cilia in tba-5(qj14) and extended from the distal regions of middle segments to the distal tips of distal segments. The cartoons in the right depict the structure of the cilia and dendrites in amphids and phasmids. Bar=5 μm. Arrows point to transition zones with cilia oriented upward. A, axon; C, cilia; CB, cell body; D, dendrite.
Sequencing revealed that tba-5(qj14), tba-5(dyf-10) and tbb-4(sa127) contain missense mutations in highly conserved residues, A19V, P360L, and L253F, respectively (Supplementary Information, Fig. S5). In contrast to these mutants, the deletion mutants, tba-5(tm4200) and tbb-4(OK1461) (Fig. 2a, b) displayed negligible cilium defects based on normal: (i) dye-filling assays (Fig. 2e–h, 3a); (ii) cilium morphology using TBB-4::YFP and IFT markers (Fig. 1c and see Supplementary Information, Fig. S3b); and (iii) rates of IFT along the middle segments (Table 1). Thus the deletion of TBA-5 and TBB-4 has minor effects on cilia compared to the presence of tubulins containing the aforementioned missense mutations. Since the missense mutations do not cause dominant phenotypes in genetic tests, we conclude that they are “recessive, gain-of-function” mutations, similar to tubulin mutations causing defects in MT dynamics in C. elegans embryos32.
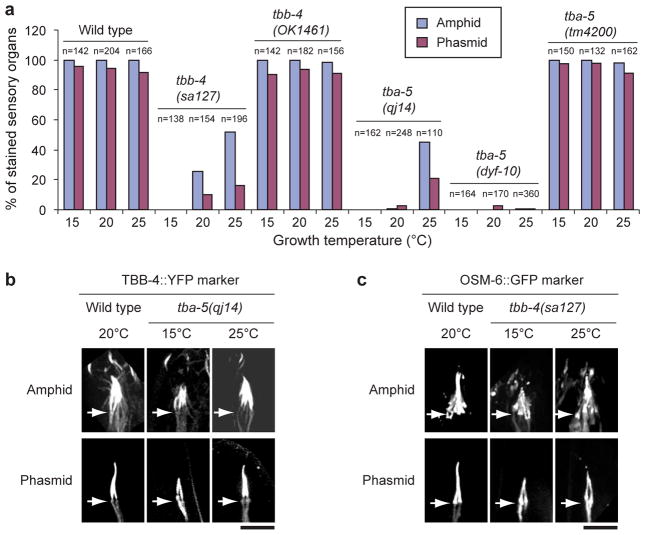
Figure 3.
Tubulin point mutants are temperature sensitive. (a) Wild type and the two deletion mutants, tbb-4(OK1461) and tba-5(tm4200), were nearly 100% stained in the amphid and phasmid neurons at 15°C, 20°C and 25°C. tbb-4(sa127) and tba-5(qj14) worms were not stained when grown at 15°C. But at 25°C, around 50% of the amphids and 20% phasmids are stained while tba-5(dyf-10) has very little temperature effect. n indicates the number of amphids or phasmids. (b) Visualized with a TBB-4::YFP tubulin marker, tba-5(qj14) possessed only the middle segment of the amphid and phasmid cilia at 15°C, but full-length cilia could be seen in tba-5(qj14) at 25°C. Bar=5 μm. Arrows point to transition zones with cilia oriented upward. (c) Visualized with an OSM-6::GFP marker, tbb-4(sa127) possessed only the middle segment of the amphid and phasmid cilia at 15°C, but full-length cilia could be seen in tbb-4(sa127) at 25°C. Bar=5 μm. Arrows point to transition zones with cilia oriented upward. The wild type image of the TBB-4::YFP marker in b and c was the same as used in Fig. 1e and Fig. S3b.
To assess the impact of these distal singlet-destabilizing missense mutations, we examined their localization on TBA-5 and TBB-4 polypeptides “docked” onto the 0.35 nm αβ-tubulin structure33 (Fig. 2c, d). This suggests that: (i) none of the mutated residues lie in helix H12, a major site for tubulin interaction with kinesin motors34, concordant with them not interfering with OSM-3-driven IFT; (ii) α-tubulin mutations A19V and P360L in TBA-5, lie within helix H1 and between loops S9 and S10, respectively, which are important for proper lateral interactions between adjacent MT protofilaments (pfs), so these mutations may destabilize distal singlet MTs by interfering with pf-pf interactions; and (iii) β-tubulin mutation L253F in TBB-4 lies at the junction between loop T7 and helix H8 which contribute to longitudinal tubulin-tubulin contacts and adjacent to a conserved lysine that may be critical for E-site GTP hydrolysis33, so this mutation may destabilize distal singlet MTs by interfering with GTP hydrolysis and with interactions between α- and β-tubulin polypeptides.
The tba-5 and tbb-4 missense mutants destabilize singlet MTs in cilia
To test whether the missense mutations in tba-5 and tbb-4 destabilize distal singlet MTs, we used dye-filling assays to monitor cilium integrity in strains cultured at 15°C, 20°C and 25°C. MTs are destabilized at low temperatures, so we predicted that lower temperatures would cause disassembly of axoneme distal segments and defective dye-filling, whereas high temperatures would stabilize them allowing normal dye-filling. DiI uptake was constant in the amphids and phasmids of wild-types, both deletion strains, and tba-5(dyf-10) at all three temperatures (Fig. 3a). However, in the missense mutants, tbb-4(sa127) and tba-5(qj14), DiI uptake was not observed at 15°C consistent with loss of cilium integrity, but dye-filling increased with increasing temperature (50% amphids and 20% phasmids were stained at 25°C). Accordingly, tba-5(qj14) and tbb-4(sa127) expressing fluorescent ciliary markers assembled only middle segments at 15°C and full-length cilia at 25°C (Fig. 3b, c). These results support the hypothesis that these missense mutations destabilize the singlet MT polymer lattice. The specific destabilization of ciliary singlet MTs is underscored by TEM which revealed loss of all distal singlet MTs and, in the case of tbb-4(sa127), loss of many central singlets in the middle segments (Fig. 1f, g).
TBA-5 and TBB-4 localize differentially within sensory cilia
The β-tubulin, tbb-4 gene is expressed in sensory cilia in a DAF-19 transcription factor-dependent fashion31,35. We observed that the TBB-4::YFP protein restores the length of the cilia present on amphid (Fig. 2o) and phasmid (Fig. 2p) sensory cilia in tbb-4(dyf12) mutants, suggesting that it is functional. TBB-4::YFP localized along the full length of the cilium but was not observed at the transition zone or in dendrites.
Examination of TBA-5::GFP introduced into tba-5(qj14) mutant worms revealed that this α-tubulin is also expressed in amphid and phasmid sensory neurons (Fig. 2i, j). TBA-5::GFP protein expression rescued the dye-filling (Fig. 2k–n) and short cilia phenotypes of tba-5(qj14) mutants (Fig. 2q–r), suggesting that the expressed tubulin isotype is functional. Also the tagged TBA-5 protein localized along dendrites and around the basal bodies as well as within sensory cilia. Unlike TBB-4, it was more concentrated in the distal than in the middle segment within cilia, consistent with distal-singlet MT associated functions.
Sensory cilium MT plus ends exchange tubulin subunits relatively slowly at the middle and distal segment tips
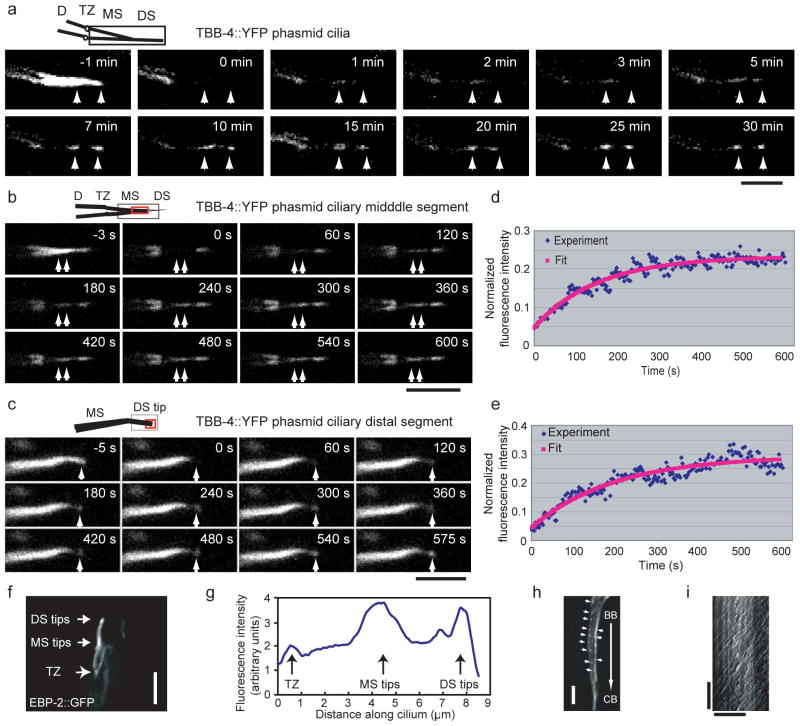
We investigated the dynamics of TBB-4, which is present in both middle and distal segments of the axoneme, using fluorescence recovery after photobleaching (FRAP). Photobleaching of full-length cilia within wild type phasmids expressing TBB-4::YFP, resulted in a striking pattern of fluorescence recovery at two regions corresponding to the middle and distal segment tips and revealed that the plus ends of both the A- and B-tubules of these axonemal MTs are dynamic (Fig. 4a). Accordingly, EBP-2::GFP, an EB1-related MT end binding protein that binds polymerizing MT plus ends, displayed relatively high concentrations at the middle and distal segment tips, although, as in vertebrate cells, it localized all along the cilium36 (Fig. 4f, g). Photobleaching of a TBB-4::YFP area covering only the middle (Fig. 4b) or distal (Fig. 4c) segment, surprisingly revealed similar rates of recovery (t1/2=77.2±23.7 s, middle; 90.3±20.1 s, distal). These rates were slower than in dendrites (t1/2=4.2±2.1 s) and unlike diffusible GFP (t1/2 ~ 1–5 s) and persistently moving IFT proteins (t1/2 ~ 5–10 s) which recover all along the cilium, not only at the MT tips suggesting that the middle and distal segment MTs share similar dynamic properties (Fig. 4d, e). However, the extent of recovery was higher in the distal (42.8±18.1%) than in the middle (26.7±5.6%) segments. Because axonemal A and B tubules emanate from the transition zone with their plus ends lying at the distal (A-tubule) and middle (B-tubule) segment tips, respectively (Fig. 1A), this is consistent with turnover of A and B tubules being due to dynamic instability of their plus ends.
Figure 4.
Dynamics of axonemal MTs at the middle segment and distal segment tips. (a–c) Cilia expressing TBB-4::YFP were photobleached in different regions and recovery was recorded for entire cilia (a), tips of middle segments (b) and distal segments (c) in phasmids. In each case, images are shown before, at (0 s) and after photobleaching. The arrows point to the recovery regions. The cartoon at the upper left for each set represents the region of cilia that was analyzed; the photobleached region is shown by a black square and the region used for recovery analysis is shown by a red square. Bar=5 μm. (d–e) The kinetics of FRAP recovery at the tips of middle segments (d) and the distal segments (e) were fit with a single exponential equation (red line). The fluorescence intensity is normalized to the prebleach. (f) EBP-2::GFP proteins are more concentrated at the tips of middle segments and distal segments. Bar=5 μm. (g) A line scan along the cilia in (f) is shown. (h) Dynamics of EBP-2::GFP in dendrites where the EB1 homolog tracks the tips of the MTs. The arrows point to the comets. Bar=5 μm. (i) A kymograph of EBP-2::GFP comets from (h) is shown. Horizontal bar=10 μm and vertical bar=10 s. The labels are: BB, basal body (equivalent to TZ); CB, cell Body; D, dendrite; DS, distal segment; MS, middle segment; TZ, transition zone.
No EBP-2::GFP movement was detected in cilia, suggesting a stable association with slowly turning over MT plus ends at the middle and distal segment tips, whereas robust movement of EBP-2 tracking the tips of the more dynamic dendritic MTs was observed (see comets in Fig. 4h). In dendrites, EBP-2::GFP comets moved in both directions, consistent with an antiparallel organization of dendritic MTs, but the majority (94% out of 321 MTs) moved from the basal body toward the cell body at ~0.25±0.05 μm s−1 (Fig. 4i) suggesting that the minus ends of most of these dendritic MTs face the cilium. Thus we can picture the IFT machinery being moved along dynamic dendritic MTs by minus end-directed motors to the basal body, where the IFT proteins are unloaded, enter cilia and move along very stable axonemal MT tracks.
Delivery of tubulin to the tips of the middle and distal segments: IFT versus diffusion
How do tubulin subunits translocate from the transition zone to their site of incorporation at the middle and distal segment tips? Possible mechanisms include passive diffusion and active transport by IFT. However, diffusion is inconsistent with experimental observations. GFP alone can only diffuse part of the way along the cilium and substantial amounts never reach the distal tip (37 and our unpublished observation). Using FRAP we estimate that GFP has a diffusion coefficient ~1–5 μm2 s−1 in these cilia (Methods), and since GFP-tubulin is significantly larger, its diffusion coefficient must be considerably lower, suggesting that tubulin could never reach the distal segment tips by diffusion alone. In contrast, tubulin could diffuse to the tip of the MS and a simple diffusion model would be consistent with the FRAP recovery observed at the middle segment tip (after bleach of almost the entire cilium e.g. Fig.4a), provided that a enough free tubulin is available at the TZ to maintain a concentration gradient steep enough for effective diffusion. However, an intense fluorescent tubulin signal at the TZ was not observed. Moreover, this “passive diffusion to middle segment tip” model predicts a rapid and extensive recovery all along the MS not just at the tips, which is not observed experimentally either (Fig. 4a).
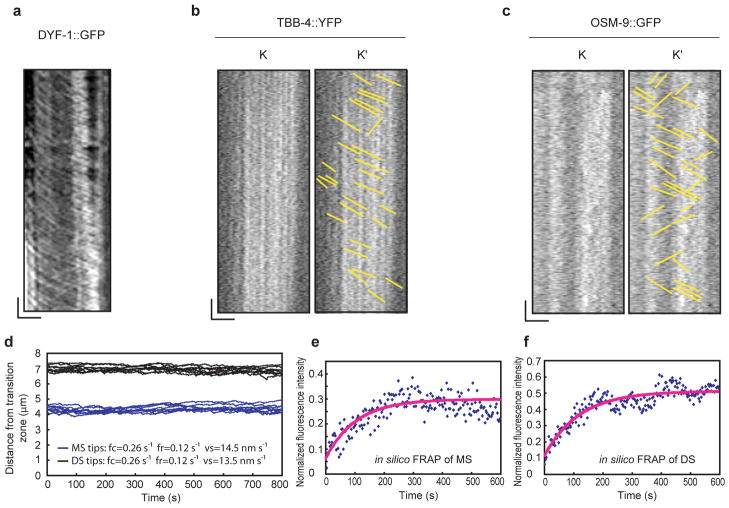
To test the role of IFT in the delivery of tubulin, we compared the transport of TBB-4::YFP with that of the IFT particle component, DYF-1::GFP using IFT assays. Although robust tracks of moving IFT particles were observed (Fig. 5a), tubulin movement could not be detected above the high levels of fluorescence arising from tubulin assembled into axonemes (e.g. Fig. 4a). Therefore we performed photo-bleaching and looked for diagonal tracks of TBB-4::YFP moving across the bleach zone using kymography. This revealed faint diagonal tracks (Fig. 5b), similar to those seen with another presumptive IFT cargo, OSM-938(Fig. 5c). The average rate of TBB-4::YFP transport was 0.8±0.1 μm s−1 but, as with OSM-938, it was not possible to discriminate characteristic middle and distal segments rates21. Identical results were obtained for TBA-5::GFP (data not shown). One plausible interpretation is that these transported cargoes are sub-stoichiometric to the IFT particles, which presumably bind multiple distinct molecules, so that the number of tubulin molecules per particle is much smaller than the number of IFT components per particle, but testing this requires measurement of the relevant molar ratios.
Figure 5.
Analysis of TBB-4::YFP transport rate in cilia. Kymographs of DYF-1::GFP and TBB-4::YFP in IFT assays under exactly same conditions except that TBB-4::YFP was photobleached with a mercury lamp before recording to reduce the background. (a) DYF-1::GFP represents the IFT transport in cilia, and the IFT tracks are clear and thick in the kymograph. (b) The tracks of TBB-4::YFP in cilia are faint and thin compared to IFT tracks, e.g. in (a). (c) For comparison, OSM-9::GFP, which is proposed to be transported by IFT was used as control. All the recorded movies were processed using the basic filters (Sharpen (High) and Low pass) before creating kymographs. K is the kymograph that was created along the cilia and K′ is a drawing of the kymograph lines in K. In (a–c), Horizontal bar=2.5 μm and vertical bar=5 s. (d–f) Modeling of MT dynamics in a cilium. Dynamic instability and the delivery of tubulin subunits via IFT can constrain the length fluctuations of MTs in both the middle (blue) and the distal (black) segments to a narrow range (d). In silico FRAP of the cilium shown in (d) for both the middle (e) and distal (f) segments indicates similar recovery curves to the experimental results (Figure 4d, e). The fluorescence intensity is normalized to the prebleach. The labels are: DS, distal segment; MS, middle segment.
Quantitative modeling of tubulin dynamics and transport
While consistent with a role for IFT in tubulin (and OSM-9) transport, the faint tracks seen in IFT assays are not definitive. Thus, we used modeling to determine if active transport of tubulin by IFT could account for our tubulin FRAP data. We developed a stochastic model which describes the evolution of the doublet and singlet MT tips undergoing dynamic instability (i.e. stochastic switches between polymerization/depolymerization), concurrent with the vectorial transport of tubulin subunits along the axoneme by IFT (Supplemental Material). Our model is derived from the deterministic balance point model, in which the steady-state length of the axoneme is established by a balance between IFT and the turnover of axonemal tubulin subunits16. We asked if the delivery of tubulin subunits by IFT is compatible with data on: (i) the dynamic tips being constrained to the observed narrow recovery region of less than a micron; (ii) the lack of fluorescence recovery everywhere along the axoneme except for the tips of the MS and DS; and (iii) the observed rate and extent of recovery of MT doublet and singlet tips undergoing dynamic instability (Fig. 4).
We varied the parameters of MT dynamic instability and the quantity of tubulin subunits transported by the IFT machinery to identify conditions that could maintain the tips within a micrometer region while also reproducing the FRAP results (half time and percent recovery). We used the fewest possible parameters, only essential restrictions and the simplest assumptions in order to identify the minimal system compatible with the experimental data. We found parameters yielding solutions that fit the data very well in the framework of this stochastic model, so long as the variance in the number of IFT particles moving along each MT remains small; i.e. if approximately equivalent numbers of IFT motors plus their cargo are loaded onto each MT at the base of the cilium, then the distal tips of these MTs are maintained within the experimentally observed range of one micrometer, and In Silico FRAP of this model cilium yields recovery curves similar to those observed experimentally (Fig. 5d–f). Moreover, this good fit is obtained using rates that are characteristic of the functional cooperation between kinesin-II and OSM-3 as seen in wild type cilia, but not using rates characteristic of either motor acting alone (Supplementary Information). We conclude that IFT driven by kinesin-II and OSM-3 in these cilia provides an efficient mechanism for maintaining MTs at their steady state lengths by controlling the supply of tubulin subunits such that the growth velocity is regulated and cilium length is tightly maintained in the face of MT dynamic instability, concordant with the “balance point model’ 16,17.
DISCUSSION
This screen revealed that mutants lacking distal singlets of sensory cilia fall into two classes; (i) those disrupting OSM-3-driven IFT, including the IFT-B subunits, DYF-119, DYF-6, and IFT-81/74; and (ii) those affecting MT tracks but not OSM-3 activity, namely missense mutations in two sensory cilium-associated α- and β-tubulin isoforms, TBA-5 and TBB-4.
Our work combined with genomics31 identifies the α- and β –tubulins, TBA-5, TBA-6, TBA-9 and TBB-4 in sensory cilia. Of these, deletion of TBA-5 and TBB-4 yielded negligible ciliary phenotypes suggesting that they can be substituted by other tubulins. Interestingly, TBB-4 functions in all ciliated sensory neurons and distributes all along the cilium whereas TBA-5 functions only in a subset of these cells and is more concentrated in the distal segments, suggesting a functional differentiation of these tubulin isotypes within sensory cilia.
Missense mutations in TBA-5 and TBB-4 cause more severe phenotypes than the deletion mutants, similar to missense mutations in certain vertebrate tubulin isotypes that yield stronger neurological phenotypes than RNAi knockdown39,40, and they resemble “recessive, gain of function” missense mutations found in C. elegans embryos32. We propose that these mutations, which occur at conserved residues, may directly destabilize polymerized distal singlet and middle segment central singlet MTs, although indirect mechanisms e.g. sequestration of a chaperone required for singlet assembly are also possible. Axoneme-specific tubulin residues like A19 and P360 in TBA-5 and L253 in TBB-4 are proposed to be evolutionarily conserved because they are required to build specific parts of axonemes41, in this instance because of their importance in maintaining singlet MTs that form specific parts of sensory cilia.
How do TBA-5 and TBB-4 assemble into axonemes? FRAP reveals that axonemal MTs turn over with t1/2 around 1–2 min, an order of magnitude slower than dendritic MTs in the same neurons, and incorporate tubulin subunits at the plus ends of both the A-tubule (at distal segment tip) and the B-tubule (at middle segment tip) where the growing MT plus-end tip tracker, EBP-2 is concentrated.
To enter the cilium, IFT proteins and their tubulin cargo must move along dendritic MTs from the cell body to the transition zone. Since the majority of MTs appear to be oriented with their minus ends facing the cilium, minus-end-directed MT motors may mediate this dendritic transport, although counter-arguments include the possibility that plus-end motors could select the minor population of opposite polarity MTs.
IFT assays were consistent with the hypothesis that TBA-5 and TBB-4 are transported from the base to the tip of the axoneme by IFT in amounts that are sub-stoichiometric to subunits of IFT particles and motors, which yield more robust tracks in kymographs21,27. Quantitative modeling supports this hypothesis by providing an excellent fit to experimental FRAP data. In the model, tubulin subunits transported along a specific MT can only incorporate onto the tips of the same MT, and to maintain all MT tips within the sub-micron region observed during FRAP recovery, we had to keep the number of IFT particles per MT approximately equal. An elaborate “gated import” machinery is thought to regulate entry of IFT particles at the basal body24,42 and could target equivalent numbers of IFT particles to each MT. Alternative, untested explanations for the tight MT length regulation observed are; (a) tubulin subunits are unequally loaded onto MTs at the base but following unloading at the tips, they diffuse to dissipate any tubulin accumulation and then incorporate equivalently into all MTs as needed or; (b) additional factors regulate MT dynamics to maintain their length.
The delivery of tubulin subunits to the tips of axonemal MTs by IFT is a cornerstone of the ‘balance point’ model for cilium length control16,17. Our finding that a refined, stochastic version of this model can explain new experimental data on tubulin dynamics using reasonable parameters supports this hypothesis. Thus we suggest that, reminiscent of the delivery of tubulin by axonal transport motors for axonal MT assembly18, IFT motors transport tubulin subunits along axonemes to maintain sensory cilia.
METHODS
Constructs, nematode culture and worm genetics
Worms were cultured using standard methods43. The fluorescently labeled markers were introduced into single mutants by genetic crossing. The double mutants comprising dyf-6, ift-81, ift-74, tba-5 and tbb-4 with klp-11(tm324) or bbs-8(nx77) were produced using genetic crosses and monitoring of the mutant background, Dyf phenotype or deletion sequence (by PCR). The double deletion mutant, tba-5(tm4200) and tbb-4(OK1461) was facilitated using a dpy-6 marker linked to tbb-4. Rescue of tba-5(qj14) was performed by injection of a TBA-5 construct, which was made by cloning its cDNA and upstream 7.4 kb promoter region into pPD95.75. For observation of EBP-2 in dendrites and cilia, a construct of ebp-2::GFP driven by an osm-6 promoter was introduced into wild type worms.
Cloning of qj55, qj23, qj14 and dyf-12
Complementation tests between two dyf mutants were done by crossing N2 male worms with one dyf mutant to generate heterozygous males carrying the mutated gene (heterozygous males were used because most dyf worms have low mating efficiency). These males were then mated with hermaphrodites of the second dyf mutant. The crossed progeny were analyzed by dye-filling assays to determine whether the two mutants are alleles or not. The SNP mappings were done based on documented single nucleotide polymorphisms between the N2 and the Hawaiian strains (CB4856)44. In brief, a double mutant of qj23 with its linkage marker gene dpy-8 (the worms are dumpy) was made and allowed to mate with CB4856 to obtain the heterozygote worms. From their progeny, eleven Dpy non Dyf and five Dyf non Dpy recombinants were selected and analyzed by SNP markers. qj23 was narrowed down to a region containing nine genes. These genes were analyzed by sequencing to determine the mutation. The same SNP cloning strategy was applied to qj14 and dyf-12, and dpy-5 and dpy-6 were used as their linkage markers.
Dye-filling assay
Worms were washed off the culturing plates with M9 buffer and collected in a 15 ml tube by centrifugation at 3000 rpm for 3 min. The DiI (Molecular Probes, Invitrogen, Carlsbad, California, USA) solution was added to a final concentration of 10 μg ml−1. After incubation for 2–4 h, the stained worms were spun down and washed three times with M9 buffer. The worms were then transferred to 2% agarose pads with a drop of 10 mM NaN3 and observed under a compound microscope with a 60× objective. The staining ratio is the number of stained amphids or phasmids divided by the total number of amphids or phasmids.
Electron microscopy
Animals were prepared and sectioned for electron microscopy using standard methods45. Imaging was performed with an FEI Tecnai G2 Spirit BioTwin transmission electron microscope equipped with a Gatan 4K × 4K digital camera.
IFT assay and cilium length measurement
IFT and cilium morphology were assayed as described previously19,21,46. The worms were immobilized on a 2% agarose pad by anesthetizing them in 10 mM levamisole. The images were collected with an Olympus microscope equipped with a 100×, 1.35NA objective and an Ultraview spinning disc confocal head. The IFT was recorded at 300 ms/frame at 21°C for 3 min using a CCD camera (ORCA-ER; Hamamatsu, Bridgewater, New Jersey, USA). Acquired images were analyzed in MetaMorph (Molecular Devices, Sunnyvale, California, United States) to create kymographs and calculate the transport rate. For the transport assay of TBB-4::YFP, OSM-9::GFP and DYF-1::GFP, the recorded movies were processed using the basic filters (Sharpen High and Low pass) before creating kymographs. Cilia lengths were measured on projection images, created in MetaMorph from recorded z-stacks of the cilia. Shown are projection images edited in Adobe Photoshop 7.0 and assembled in Adobe Illustrator 10. During editing, the brightness and contrast of projection images were slightly adjusted in Photoshop.
Y2H
The yeast strain used in this study was PJ69-4A (genotype: MATa; trp1-901; leu2-3,112; ura3-52; his3-200; gal4D; gal80D; GAL2-2ADE;. LYS2::GAL1-HIS3; met2::GAL7-lacZ). The yeast two hybrid plasmids were pGAD-C1, pGBD-C1 containing GAL4 AD (activation domain) and GAL4 BD (DNA binding domain) respectively47. The ift-81 gene was cloned from cDNA of C. elegans, and the ift-74, osm-3, and dyf-6 genes were cloned from their EST clones. To eliminate self activation of the expression of His reporter, the genes cloned in pGAD-C1 were co-transformed with empty pGBD-C1 and genes cloned in pGBD-C1 were co-transformed with empty pGAD-C1. Combinations of pGAD-C1 and pGBD-C1 plasmids each carrying one gene to be tested were co-transformed into yeast strain PJ69-4A. Six transformant colonies from each selective plate were streaked onto Leu-, Trp- and His-lacking selective plate to detect the activation of His reporter. In each set of experiments both positive and negative controls were included.
Bioinformatics analysis
The domain analyses of DYF-6 and IFT-81 were performed using SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) and Coils (e.g.48). TBA-5 or TBB-4 and their orthologs were aligned with Clustal X249. The heterodimeric structure of TBA-5 and TBB-4 was predicted with Modller9v6 using the porcine brain tubulin heterodimer structure 1JFF33 as a template. The predicted structure was visualized with PyMOL 0.99 (http://www.pymol.org/).
FRAP
FRAP experiments were performed on a laser-scanning Olympus confocal microscope (FV1000) with a 60× 1.40 NA objective at 23°C, and images were acquired using the Fluoview software (version 1.5; Olympus). A 405 nm laser at 40% power was used for photo-bleaching and images were acquired with a 514 nm laser every 3 or 5 s. The data were normalized to the fluorescence before the bleach. The recovery curve was fit with an exponential equation F(t)=F0+(Finf−F0)(1−e−kt), where F(t) is the total fluorescence at time t after the bleach, and k is a constant describing the rate of recovery. F0 is the fluorescence immediately after the bleach and Finf is the maximum recovered fluorescence. The recovery half time was calculated by t1/2=ln2/k and the percentage of fluorescence recovery was given by (Finf−F0)/(Fpre−F0)50, where Fpre is the fluorescence intensity before the bleach. It is difficult to determine the exact area of recovery directly so we used linescans along the cilia to determine the maximum recovered intensity i.e. Finf in the equation above.
Estimating the diffusion coefficient of GFP in the cilium
Worms expressing free GFP were photobleached and fluorescence profiles along the ciliary length were obtained before and after the bleach. The postbleach fluorescence profiles were subtracted from the prebleach profile. The difference profiles obtained were then fitted to a Gaussian curve. Diffusion coefficients were obtained from these plots, by fitting the normalized bleach depth over time as described in 51. We estimate a value of ~ 1–5 μm2 s−1 for GFP.
Modeling
See “Supplemental Material: Modeling”. The modeling codes will be made available upon request.
Primers used for cloning the genes and identifying the mutants
For tba-5: pKP1056F, CCTCGGAGGAATTTCAAACG; pKP1056R, AGCTCCGTAAAGCAGCTTC; pKP1057F, ATCATTCTCCAGGCCACGTTAC; pKP1057R, CTGAACTAGTCGAACAAACCCC; pKP1082F, AATGAGATGCAAGACCGGGACC; pKP1082R, CTTTCCCACGACCTTTCTTGC; pKP1114F, AGATTGAGGCTGAAATATGGTG; pKP1114R, GTCGAGCAGCACCAGTTATTG; pKP1058F, CCAGTGTCCCGATAGAAAAC; pKP1058R, GAATCACCGCCAACATGAGA; pKP1059F, CATCTGGGACGTTCTTTCAC; pKP1059R, TTCAGGCTCCACTTTATGCC; pKP1117F, CGAATCCATATCGATGCGAC; pKP1117R, ACATCTCTGCGTGGCTCTTC; pKP1119F, TCAAATTTGGCACGTCATCAG; pKP1119R, CTCCATTTTGGAACTCCCAG; CE1-153F, CCGTGAAGCAAGTTCAAATGC; CE1-153R, CTTAACAAGAATTGGTGACCAAC; CE1-170F, CATGTCCGGCGAATGGATTC; CE1-170R, AGCCATGGAATCAGCTGTGG; F10D11.2F, CGCAGATTTGATGACTCCAC; F10D11.2R, TGGGAACTGGATAAACTGGC; uCE1-969F, ATACAGTCTAGTGGGGATTGC; uCE1-969R, CTCAGTGTTACTTGCAGCGG; F02E9F, AGAGAAGCTTATGCGGTTCG; F02E9R, AGTGCCGATTTACGATCTCG; F16D3.1-1, CTATGAGTACCTTCAAACCTG; F16D3.1-2, AAACTTGGCACTCCGTGTAC; F16D3.1-3, AAACTTGGCACTCCGTGTAC; F16D3.1-4, GTTGGACTTCTGACACCTAG; F16D3.1-5, CAGCAATGGTAGAGCCATAC; F16D3.1-6, TGTTCTAAGCCTATCTTGACC; F16D3.1-7, GCAATTTCGCTTGTTCTAACG; F16D3.1-8, TCCAAATGAACCCTTGTGCC; F16D3.1-9F(PstI), AAAActgcagGAGCATGAAGTAGTGTCCTTG; F16D3.1-10R(BamHI), CACGGGATCCCATTTTTCCATTTGGAGCCATGG; F16D3.1-11F(SalI), AAAAgtcgacggatccATGCGTGAAATAGTTTCGATTC; F16D3.1-12R(BamHI), CACGggatccTTTTCCATTTGGAGCCATGG; F16D3.1-12R(XmaI), CCTACCCGGGGATATTCTTCATCATTTGGATCGA; F16D3.1-13F(SphI), GAAAGACCTCGCATGCAAATTTA; F16D3.1-14R(BglII), CCTAagatctCTAATATTCTTCATCATTTGGATCGA; F16D3.1-14R(SphI), TAAATTTGCATGCGAGGTCTTTC; F16D3.1-15F, CGGAAATGtCTGTTGGGAACTG; F16D3.1-16R, CAGTTCCCAACAGACATTTCCG; F16D3.1-17F, TATCAACCACtGACTGTTGTT; F16D3.1-18R, AACAACAGTCAGTGGTTGATA. For tbb-4, pKP6127F, TTGGATCTCCGTAGACGTCAC; pKP6127R, GTCTTCATTGCATGATGTGGC; pKP6112F, GCGTGAGGGCAACTTTTTTG; pKP6112R, TAGGGATTCTCGCGTCATTG; pKP6135F, TCTTTGCTTGTGAGCCAATTGG; pKP6135R, CGGCACGTGTTTTCAAAATAAC; uCE6-1111F, TCTACATGACCTACATGTCTG; uCE6-1111R, TGGACATTTACACAGAACCTG; pas23221F, GTCGAGAAGTTATGTGTGCAG; pas23221R, AAGATGTCCATCTATGGACCG; uCE6-1120F, CAACCACATCGGATATGGTAG; uCE6-1120R, CGTTGGCTTTGACGTACGTTC; tbb4-1F, GCCATTTAAGGACACACCTCC; tbb4-2R, CACGCGTAAGGCGTTGAACC; tbb4-3F, CGGAAATTGAGCGACATCTCC; tbb4-4R, GCCATCATATTCTTGGCGTCG; tbb4-5F, TCCAGAGGAAGCCAGCAATAC; tbb4-6R, CTATGCATTTGTAGTAATGTATTACTG. For ift-81: pKP6103F, TGTCTAGTTCAAAAGCCCGG; pKP6103R, TTGTAGCAGATCCTACCCTACC; pKP6104F, TCCAATGTTACGCTACCAGC; pKP6104R, TGACAAGGCAACCACCATTG; pKP6120F, GATTCAGATCAAACAGAGGTGG; pKP6120R, TCGTGGCACCATAAAAGTG; pKP6151F, AGCAATTATAGTGTCATTGCCG; pKP6151R, TTAAAAGCTGGCTCTAGTGTTG; pKP6150F, AATCGTCCTAGTTATCCACGG; pKP6150R, TGGGGGTGAAAGAGATATGTC; uCE6-907F, GCAGACATGGGAAGAAGATG; uCE6-907R, GTGACGCATGAATGGCTGG; uCE6-929F, CGATGGAATTGAGTACTTCGATG; uCE6-929R, GTACATTTACTTACCTCCCACAC; pas16937F, AACGTGGTGAGAACGTGATG; pas16937R, GTACTGAACTCATCTCTGCC; Y34B4A-F,CTCAGATTCAGCTGTACCTC; Y34B4A-R, TCATTCCATTCTGCCGAAGG; pas16936F, ATCTAATTGTCTCGAGTGCG; pas16936R, GTCTCGCTCATTGAAATCTG; IFT-81-1F, ATAGCAAAGAGCCCAGCAAC; IFT-81-2R, CGCACATTGTAACTTTGTGCC; IFT-81-3F, TATCAGCAGGTCCACTTGGG; IFT-81-4R, CTAACACGATGAATTCAGATAGC; IFT-81-5F, AAGTAAGGGAGTTCTTTAGCG; IFT-81-6R, CTGTCGGCTGCACATTTATC; IFT-81-7F, AATGGCTTCAGACGTCAGAG; IFT-81-8R, ACGCAGATTGTGTCTCTTAGC; IFT-81-9F, AAGCAAAACCAGGTGATGAAC; IFT-81-10R, GTTAGCAGAGGTATCTGATAC; IFT-81-11F, TGCGTTCCCGATTTTGCAAG; IFT-81-12R, TGAAATGTCACTCTGCAACTG. For dyf-6: Dyf-6-1F, CTCAATGACCTAATATGCTC; Dyf-6-2R, AGAATGTCAGAAACGTCTGC; Dyf-6-3F, TTTGAATCCGTTTCTTCGGG; Dyf-6-4R, GTCActgcagCAGGTGACTCTATTCATTGAAGC; Dyf-6-5F, CTAGcccgggAAGTTCCAATCTGTCCATTGTTTC; Dyf-6-6R, CAGTCCCGGGCTCGCATGCGAGCTCCATTGGATTTTCCAATGCCTG; Dyf-6-7F, TTAAgagctcATGGCGGCAAACGGAGAGT; Dyf-6-8R(XmaI), CTAGCCCGGGAAAGTTCCAATCTGTCCATTGTTTC; Dyf-6-9F, CGTTGAATCCGACAGATACC.
Supplementary Material
Acknowledgments
We thank Xiaoping Zhang (Heyer Laboratory), Arshad Desai and Guangshuo Ou for valuable discussion; Mitani Shohei for the deletion mutants, tm2355, tm2394 and tm4200; the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources) for strains; and Yuji Kohara, Center for Genetic Resource Information, National Institute of Genetics, Japan for EST clones. This work was supported by NIH grants # GM50718 to J.M.S. and R01NS064273 to S.S.
Footnotes
AUTHOR CONTRIBUTIONS
J.M.S is PI of the grant and laboratory that support this IFT project. L.H. and J.M.S. designed the experiments and drafted the manuscript. J.M.S. wrote the manuscript. L.H. performed most of the experiments. M.T. characterized the qj14 mutant by crossing all the used IFT markers into it, made the TBB-4::YFP transgenic worms and performed the EBP-2 experiments. I.B.M. implemented the stochastic tubulin transport and dynamics model and the in silico FRAP model and analyzed the results, and helped with the FRAP experiment and transport assay of TBB-4::YFP and analysis of the results. G.C.S. designed and wrote the stochastic tubulin transport and dynamics model and the in silico FRAP model scripts. Y.L. and S.S. performed the EM studies and analyzed the results. S.A. and B.P. performed the Y2H assays. All the authors read the manuscript.
References
- 1.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesland DA, Hoffman JL, Caligor E, Goodenough UW. Flagellar tip activation stimulated by membrane adhesions in Chlamydomonas gametes. J Cell Biol. 1980;84:599–617. doi: 10.1083/jcb.84.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran DT, Rowley JC, 3rd, Jafek BW, Lovell MA. The fine structure of the olfactory mucosa in man. J Neurocytol. 1982;11:721–746. doi: 10.1007/BF01153516. [DOI] [PubMed] [Google Scholar]
- 5.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Pan J, Snell WJ. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell. 2006;125:549–562. doi: 10.1016/j.cell.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 7.Cole DG, et al. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 8.Cole DG, et al. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci U S A. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shakir MA, Fukushige T, Yasuda H, Miwa J, Siddiqui SS. C. elegans osm-3 gene mediating osmotic avoidance behaviour encodes a kinesin-like protein. Neuroreport. 1993;4:891–894. doi: 10.1097/00001756-199307000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Pedersen LB, Rosenbaum JL. Intraflagellar Transport (IFT) Role in Ciliary Assembly, Resorption and Signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 12.Hao L, Scholey JM. Intraflagellar transport at a glance. J Cell Sci. 2009;122:889–892. doi: 10.1242/jcs.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucker BF, Miller MS, Dziedzic SA, Blackmarr PT, Cole DG. Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46. J Biol Chem. 2010;285:21508–21518. doi: 10.1074/jbc.M110.106997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pigino G, et al. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J Cell Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel BD, Ludington WB, Marshall WF. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J Cell Biol. 2009;187:81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah JV, Cleveland DW. Slow axonal transport: fast motors in the slow lane. Curr Opin Cell Biol. 2002;14:58–62. doi: 10.1016/s0955-0674(01)00294-0. [DOI] [PubMed] [Google Scholar]
- 19.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 20.Pan X, et al. Mechanism of transport of IFT particles in C. elegans cilia by the concerted action of kinesin-II and OSM-3 motors. J Cell Biol. 2006;174:1035–1045. doi: 10.1083/jcb.200606003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snow JJ, et al. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 22.Burghoorn J, et al. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:7157–7162. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins PM, et al. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Dishinger JF, et al. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inglis PN, Ou G, Leroux MR, Scholey JM. The sensory cilia of Caenorhabditis elegans. WormBook. 2007:1–22. doi: 10.1895/wormbook.1.126.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan ZC, et al. Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex B and is required for flagellar assembly. Mol Biol Cell. 2010;21:2696–2706. doi: 10.1091/mbc.E10-03-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou G, et al. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol Biol Cell. 2007;18:1554–1569. doi: 10.1091/mbc.E06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, Gengyo-Ando K, Ishihara T, Katsura I, Mitani S. IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells. 2007;12:593–602. doi: 10.1111/j.1365-2443.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- 29.Bell LR, Stone S, Yochem J, Shaw JE, Herman RK. The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics. 2006;173:1275–1286. doi: 10.1534/genetics.106.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou Y, et al. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurd DD, Miller RM, Nunez L, Portman DS. Specific {alpha}- and {beta}-Tubulin Isotypes Optimize the Functions of Sensory Cilia in Caenorhabditis elegans. Genetics. 2010;185:883–896. doi: 10.1534/genetics.110.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright AJ, Hunter CP. Mutations in a beta-tubulin disrupt spindle orientation and microtubule dynamics in the early Caenorhabditis elegans embryo. Mol Biol Cell. 2003;14:4512–4525. doi: 10.1091/mbc.E03-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 34.Uchimura S, Oguchi Y, Hachikubo Y, Ishiwata S, Muto E. Key residues on microtubule responsible for activation of kinesin ATPase. EMBO J. 2010;29:1167–1175. doi: 10.1038/emboj.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efimenko E, et al. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- 36.Schroder JM, Schneider L, Christensen ST, Pedersen LB. EB1 is required for primary cilia assembly in fibroblasts. Curr Biol. 2007;17:1134–1139. doi: 10.1016/j.cub.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 37.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci U S A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin H, et al. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 39.Jaglin XH, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tischfield MA, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen MG, Turner FR, Hutchens JA, Raff EC. Axoneme-specific beta-tubulin specialization: a conserved C-terminal motif specifies the central pair. Curr Biol. 2001;11:529–533. doi: 10.1016/s0960-9822(01)00150-6. [DOI] [PubMed] [Google Scholar]
- 42.Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- 43.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 45.Yoshimura S, Murray JI, Lu Y, Waterston RH, Shaham S. mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development. 2008;135:2263–2275. doi: 10.1242/dev.019547. [DOI] [PubMed] [Google Scholar]
- 46.Hao L, Acar S, Evans J, Ou G, Scholey JM. Analysis of intraflagellar transport in C. elegans sensory cilia. Methods Cell Biol. 2009;93:235–266. doi: 10.1016/S0091-679X(08)93013-2. [DOI] [PubMed] [Google Scholar]
- 47.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Signor D, Wedaman KP, Rose LS, Scholey JM. Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol Biol Cell. 1999;10:345–360. doi: 10.1091/mbc.10.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Cheerambathur DK, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM. Dynamic partitioning of mitotic kinesin-5 cross-linkers between microtubule-bound and freely diffusing states. J Cell Biol. 2008;182:429–436. doi: 10.1083/jcb.200804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullineaux CW, Nenninger A, Ray N, Robinson C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J Bacteriol. 2006;188:3442–3448. doi: 10.1128/JB.188.10.3442-3448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.