Abstract
Rhizobia and legume plants establish symbiotic associations resulting in the formation of organs specialized in nitrogen fixation. In such organs, termed nodules, bacteria differentiate into bacteroids which convert atmospheric nitrogen and supply the plant with organic nitrogen. As a counterpart, bacteroids receive carbon substrates from the plant. This rather simple model of metabolite exchange underlies symbiosis but does not describe the complexity of bacteroids' central metabolism. A previous study using the tropical symbiotic model Aeschynomene indica/photosynthetic Bradyrhizobium sp. ORS278 suggested a role of the bacterial Calvin cycle during the symbiotic process. Herein we investigated the role of two RuBisCO gene clusters of Bradyrhizobium sp. ORS278 during symbiosis. Using gene reporter fusion strains, we showed that cbbL1 but not the paralogous cbbL2 is expressed during symbiosis. Congruently, CbbL1 was detected in bacteroids by proteome analysis. The importance of CbbL1 for symbiotic nitrogen fixation was proven by a reverse genetic approach. Interestingly, despite its symbiotic nitrogen fixation defect, the cbbL1 mutant was not affected in nitrogen fixation activity under free living state. This study demonstrates a critical role for bacterial RuBisCO during a rhizobia/legume symbiotic interaction.
Introduction
The Calvin Benson Bassham cycle (CBB cycle), also termed reductive pentose phosphate pathway, is the quantitatively most important mechanism of autotrophic CO2 fixation in nature [1]. It's key enzyme ribulose 1,5 bis-phosphate Carboxylase Oxygenase, RuBisCO catalyzes the formation of two molecules of 3 phosphoglycerate from ribulose bisphosphate and CO2 and has been subject to extensive studies since its discovery [2]. Besides fundamental interest in carbon assimilation, research in this field is also motivated by the objective to improve crop yield and by the capacity of CO2 fixation pathways to transform the greenhouse gas [3] [4].
Whereas in plants and other autotrophic organisms, the CBB cycle plays an essential role in biomass production, the presence of a functional CBB cycle in heterotrophic organisms is less obvious. The operation of the calvin cycle is costly and consumes both ATP and reducing power (3 ATP and 4 reducing equivalents per CO2 assimilated). It thus appears paradoxal that organic carbon compounds could be oxidized to CO2 at the same time that CO2 is assimilated by the cell. The phenomenon of substantial carbon fixation in the presence of multicarbon compounds was observed earlier and an alternative role of the CBB cycle in some microorganisms suggested that consists in redox cofactor balancing [5], [6], [7], [8], [9].
Sinorhizobium meliloti, Bradyrhizobium japonicum and the photosynthetic Bradyrhizobium strain BTAi1 are known to assimilate CO2 via the CBB cycle [10], [11], [12]. These microorganisms belong to the polyphyletic rhizobial group. Rhizobia are soil bacteria able to form symbiotic interactions with legume plants. During these symbiotic interactions, plants develop new organs on their root system. In these organs, called nodules, bacteria reside intracellularly and their metabolism is drastically modified as it is largely dedicated to the reduction of atmospheric nitrogen that is made available to the plant host. In return bacteria receive organic carbon in the form of dicarboxilic acids from the plant. This basic assimilated carbon versus nitrogen exchange is the basis of the symbiotic relationship between both partners [13] but the metabolites exchanges seems much more complex [14].
Photosynthetic bradyrhizobia induce and colonize nodules on semi-aquatic legume plants belonging to the Aeschynomene genus [15]. Beyond their photosynthetic trait, bacteria involved in these interactions present other atypical aspects compared to classical rhizobia. They can fix nitrogen in the free living state, an ability restricted among rhizobia to photosynthetic bradyrhizobia and to Azorhizobium caulinodans [16], [17]. Furthermore, photosynthetic bradyrhizobia lack the nodABC genes, indicating that these bacteria interact with their legume host according to a new molecular mechanism independent of the long-time considered universal Nod factors [18]. Finally, in accordance with the semi-aquatic nature of their Aeschynomene host, they distinguish from classical rhizobia by their adaptation to the aquatic environment beyond the classical rhizobial habitats (rhizo-, phyllo-sphere, soil and root nodules). As a consequence, photosynthetic bradyrhizobia nodulate not only the roots but also the stems of Aeschynomene plants when the latter are subjected to immersion. This capacity to colonize various environments (stem and root nodules, plant surfaces, soil, and flooded area) might rely on versatile metabolic skills. As an example, photosynthetic capacities of the model Bradyrhizobium sp. ORS278 were shown to be important for stem nodulation but not root nodulation [19].
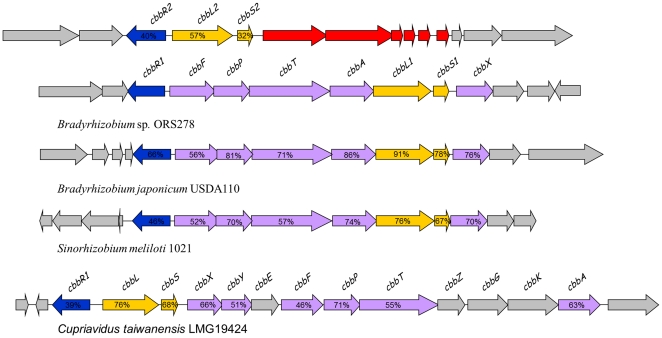
In a previous study aiming at identifying ORS278 mutants altered in the symbiotic process, four Tn5 insertional mutants potentially affected in the CBB cycle were described as altered in their symbiotic nitrogen fixation capacity (Fix- phenotype) [20]. ORS278 genome harbours two RuBisCO gene clusters [18] encoding for proteins which belong to the form I RuBisCO of heterodimeric proteins containing 8 large and 8 small subunits. Figure 1 describes the genetic organization of the RuBisCO genes clusters. The two enzymes, called above RubisCO 1 and 2, are encoded by cbbL1/cbbS1 and cbbL2/cbbS2 respectively. Genes corresponding to RuBisCO 1 are located in a locus containing most of the genes required for the reductive pentose phosphate pathway (Figure 1). RuBisCO 2 gene region harbours typical traits of a horizontally acquired gene cluster [18]. Downstream of cbbLS2, genes involved in carboxysome formation are found. Carboxysomes are proteic microcompartments found mainly in Cyanobacteria and some chemoautotrophs. These organelle-like structures are responsible for CO2 concentration around the RuBisCO enzyme [21], [22]. A gene coding for a CbbR regulator is located upstream of cbbLS2 and divergently transcribed. Such a regulator is also found upstream of RuBisCO 1 gene cluster (Figure 1). The existence of two RuBisCO genes, both associated with a regulator, suggests that ORS278 might differentially use one or the other enzyme depending on environmental conditions.
Figure 1. Genetic organization of RuBisCO 1 (encoded by BRADO1659 and BRADO1660) and RuBisCO 2 (encoded by BRADO2274 and BRADO2275) loci of Bradyrhizobium sp. ORS278 and corresponding synthenic regions of RuBisCO 1 in other rhizobial models.
LysR type regulator are represented in blue, RuBisCO genes in yellow, other CBB cycle genes in purple and carboxysome genes in red. Amino-acid identity level is represented as a percentage for the genes found in synteny.
Herein we investigated the importance of RuBisCO 1 and 2 during the symbiotic process and demonstrate the contribution of RuBisCO 1 to symbiosis.
Results
Bradyrhizobium spp. ORS278 genome houses two physically and phylogenetically distant RuBisCO gene clusters
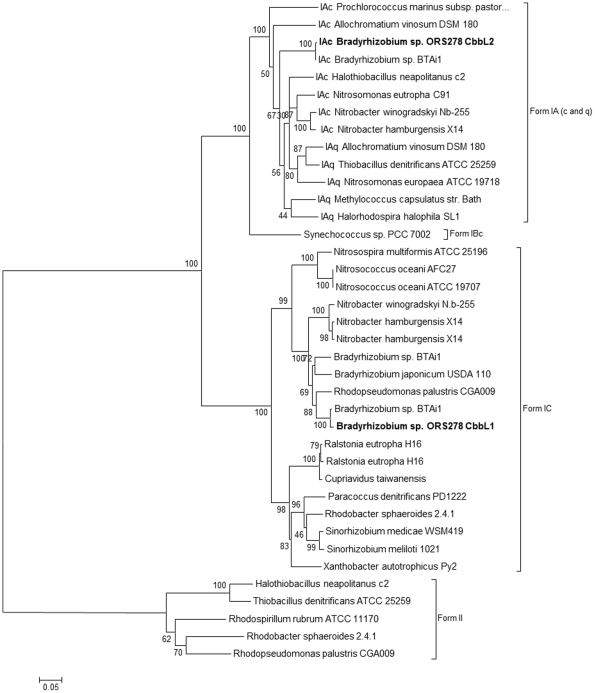
As mentioned above, the two predicted ORS278 CbbL proteins belong to the group of Type I RuBisCO enzymes, nevertheless they exhibit a sequence identity of only 57%. Neighbour joining analysis performed on various procaryotic RuBisCO large subunit sequences showed that the two ORS278 genes belong to two distinct clades (Figure 2). RuBisCO 1 belongs to the IC RuBisCO class, a group related to medium to high CO2 environments [23]. Beyond the CbbL1 closest ortholog in BTAi1 (99% of identity), predicted proteins are well conserved in some non-photosynthetic rhizobia such as Sinorhizobium meliloti with an identity level of 76% [24] or B. japonicum (91% identity). In these three strains, the genomic context is well conserved and the whole gene clusters exhibit a perfect microsyntheny (Figure 1). A CbbL1 homologue (76% of identity) is also present in the phyllogeneticaly distant beta-rhizobium Cupriavidus taiwanensis [25]. RuBisCO 2 belongs to the IAc RuBisCO class, which harbors orthologs from some other Alphaproteobacteria such as Nitrobacter strains, Beta- and Gammaproteobacteria as well as Cyanobacteria, this class is related to low CO2 environments [23]. The only known rhizobial ortholog in the IAc clade is Bradyrhizobium sp. BTAi1 (>99% of identity).
Figure 2. Evolutionary relationships of RuBisCO large subunit sequences.
Neighbour joining method was used. Bootstrap support values (10000 replicates with Mega4) are provided as percentage at the corresponding nodes. CbbL1 and CbbL2 proteins from ORS278 strain are highlighted in bold characters. For RuBisCO IA members, the RuBisCO subclasses IAc and IAq is indicated before bacterial name.
Only cbbL1 is expressed in planta
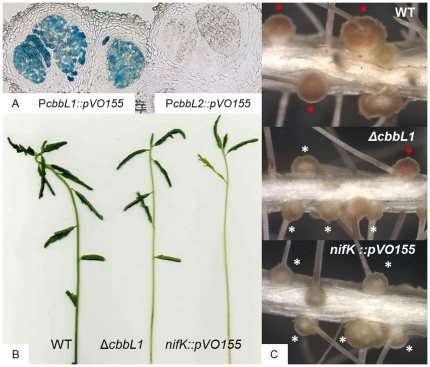
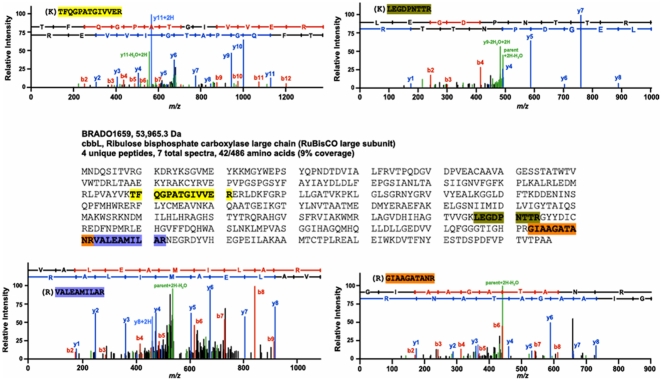
In order to determine any potential contribution of RuBisCO 1 or 2 during the symbiotic process, reporter strains carrying gus fusion downstream of the predicted promoter region of the RuBisCO gene clusters were constructed. A. indica nodules of different age induced by these strains (5, 7, 10, 14, 21 days post-inoculation) were sliced into thin layers, stained with X-Gluc and reporter gene expression was monitored by standard light microscopy. No detectable activity of gus reporter fusion was revealed for the cbbL2 promoter region. In contrast, the 5′ region of the cbbL1 gene cluster displayed a transcriptional activity detectable at every stages of nodule development examined even at five dpi when first young nodules appeared. This expression did not seem to diminish in mature nodules (Figure 3A corresponds to 14 dpi nodule). In order to determine if CbbL1 protein is produced in nodules, a proteomics analysis was performed on bacteroids isolated from A. indica 14 dpi nodules. Protein extract was separated by SDS-PAGE and a gel section corresponding to CbbL1 and 2 predicted molecular weight was analysed by trypsin digestion followed by LC-MS/MS. Fragmentation profiles allowed the identification of four peptides from CbbL1 (Figure 4) but none of CbbL2.
Figure 3. cbbL1 and cbbL2 expression and symbiotic phenotypes of the cbbL1 mutant compared with the WT-strain and the nifK mutant on A. indica.
A. Transcriptional activity of the cbbL1 and cbbL2 putative promoter regions fused to gusA gene revealed on 40 µm nodule sections stained with X-gluc. B. Aerial part and C. root systems of plants inoculated with Bradyrhizobium sp. ORS278 and cbbL1 and nifK derivative strains, white and red asterisk indicate fix minus like and WT-like nodules respectively.
Figure 4. Identification of CbbL1 (BRADO1659) peptides in bacteroids protein extract by tandem mass spectrometry and database search.
cbbL1 is required for efficient symbiotic nitrogen fixation
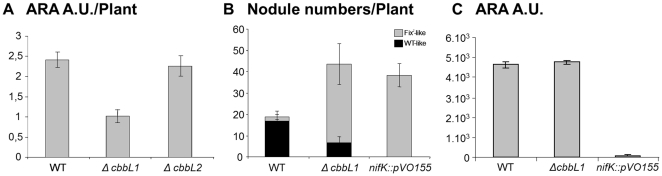
In order to evaluate the importance of RuBisCO 1 or 2 during the symbiotic process, mutant strains harbouring a deletion in one of the two genes coding for the large RuBisCO subunit CbbL were constructed and inoculated on A. indica. To prevent any effect of bacterial photosynthesis and mimic natural conditions, plants were root inoculated and roots systems were maintained in the darkness. Two weeks after inoculation, acetylene reduction assays were conducted to evaluate nitrogenase activity of plants housing the different strains. Whereas no significant defect was observed for plants inoculated with the cbbL2 mutant, plants inoculated with the cbbL1 mutants exhibited a nitrogenase activity defect ranging from 30 to 50% to that of the wild type (WT) strain (Figure 5A). This result was consistent with phenotypic observations of three week old plants which revealed that plants inoculated with a cbbL1 mutant but not those inoculated with a cbbL2 mutant exhibited typical nitrogen starvation symptoms such as foliage chlorosis or stem thinning (Figure 3B). Symptom intensity did not reach those observed on plants inoculated with a nifK mutant which was altered in the nitrogenase enzyme itself and therefore displayed a strict Fix- phenotype.
Figure 5. RuBisCO 1 mutant displays typical traits of Fix- mutant on A. indica but is able to reduce acetylene in vitro.
A. Symbiotic acetylene reduction assays: A. indica plants were inoculated with the indicated Bradyrhizobium sp. ORS278 strain one week after germination. Acetylene reduction activities were measured on individual plants two weeks after inoculation. B. Nodule numbers induced by the indicated strain on A. indica were determined two weeks after infection. C. Acetylene reduction activities were measured during growth on BNM medium supplemented with oxo-glutarate. Error bars represent standard deviations.
The cbbL1 mutant induces more nodules which are heterogenic in phenotype
Root systems of plants inoculated with the cbbL1 mutant harboured two times more nodules than those inoculated with the WT strain (Figure 5B) and was thus in the same range as observed upon inoculation of a nifK mutant. On every root system, cbbL1 induced more than 80% of nodules with a yellow colour typically indicative of nitrogen fixation deficiency, the remaining being WT-like reddish nodules (Figure 3C, 5B). In order to determine if a secondary mutation occurred in cbbL1 mutants present in WT-like nodules, bacteria were rescued from these nodules. After cultivation, mutation in cbbL1 was confirmed by PCR and these cbbL1 mutants derived from WT-like nodules were inoculated on new plants. The resulting root systems displayed also a mixture of WT-like and yellowish nodules. This result demonstrated the heritable nature of the heterogenic phenotype and excluded the possibility of secondary mutations in cbbL1 mutants forming WT-like nodules.
cbbL1 is not required for in vitro nitrogen fixation
In order to determine whether the symbiotic defect observed with the cbbL1 mutant was directly related to nitrogen fixation capacity, we investigated the bacterial capacity to fix nitrogen under free-living conditions. Using succinate, malate or 2-oxoglutarate as a sole source of carbon and energy, no significant nitrogenase activity defect was revealed by acetylene reduction assays (Figure 5C and not shown) demonstrating the absence of direct correlation between the presence of RuBisCO 1 and the capacity to fix nitrogen.
Discussion
The presence of an active RuBisCO has been reported in several rhizobial strains including photosynthetic and non-photosynthetic rhizobium; however, to our knowledge, so far no role of this enzyme during the symbiotic process has been reported. In this study, we demonstrate that one of the two RuBisCO enzymes identified in the photosynthetic Brayrhizobium strain (ORS278) is required for an efficient symbiosis with A indica plants. First, we showed expression of the cbbL1 but not cbbL2 gene during the symbiotic process and second, we observed a reduced symbiotic nitrogenase activity level of plants inoculated with cbbL1 mutant leading to typical nitrogen starvation symptoms such as foliage chlorosis and stem thinning. cbbL1 interacting plants display nitrogen starvation symptoms weaker than plants housing nitrogenase minus mutants. This intermediary phenotype can be explained by the heterogeneity of the induced nodules. Indeed, cbbL1 induced a mixture of WT-like, reddish, and fix minus-like, yellowish, nodules on the same plants. Yellowish nodules are likely nitrogen fixation deficient whereas the WT-like nodules probably fix and transfer nitrogen to the host, partially compensating nitrogen deprivation. This heterogeneous phenotype and cbbL1 mutant capacity to fix nitrogen in vitro indicate that cbbL1 is not essential for nitrogen fixation. Thus, the importance of RuBisCO 1 during the symbiotic process is probably not related to mature bacteroid metabolism but to an early step of the symbiosis.
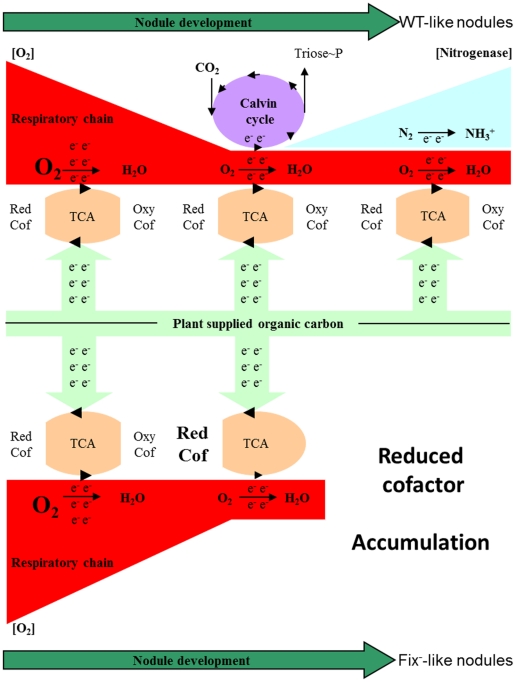
Results presented here were obtained with plant root systems maintained in the darkness. Under this condition bacteria produce energy by oxidation of reduced carbon compounds supplied by the plant. In this context, the operation of the CBB cycle as a mean to produce biomass seems futile since reduced carbon is provided by the plant. In some photosynthetic bacteria, the CBB cycle plays an essential role during growth on reduced carbon substrates in the absence of oxygen [7], [26]. In these bacteria, the CBB cycle acts as an electron sink driving the cell electron excess to CO2 and thus oxidizing reduced cofactor and contributing in a significant way to equilibrate the redox state of the cell [6]. Interestingly, in Rhodospirillum rubrum and Rhodobacter sphaeroides, a RuBisCO mutant which is unable to grow photoheterotrophically, can be rescued by acquiring secondary mutation de-repressing nitrogen fixation genes [27]. Joshi and Tabita (1996) and more recently McKinley and Harwood (2010) explained these observations by the high reductant requirement of nitrogenase which, like the Calvin cycle, can play an electron sink role. We propose ORS278 strain uses the Calvin cycle for this purpose at an early stage of the symbiotic process: when oxygen concentration in the nodule has already significantly decreased and before the establishment of functional nitrogenase. At such a step, reduced cofactors pool produced primarily by the Krebs cycle might not be balanced and the CBB cycle might fulfil this function which, in developed bacteroids, is fulfilled by nitrogenase activity (Figure 6). The dynamical dimension of this hypothetical model offers an explanation to the observed WT-like nodules induced by cbbL1 mutant. Indeed, oxygen concentration decreasing rate in the nodule and nitrogenase production might be influenced by diverse parameters and we may consider the possibility that in some nodules oxygen concentration decreases more slowly and nitrogenase production starts more vigorously than on the average nodule which would not allowed the accumulation of reductant making useless the Calvin cycle.
Figure 6. Hypothetical model of ORS278 central metabolism during the symbiotic process.
Upper part of the figure represents the WT and WT-like situation: Plant supplied organic carbon to bacteria which use it via the tricarboxylic acid cycle (TCA) resulting in the production of reduced cofactors (Red Cof). Reduced cofactors are totally oxidized by the respiratory chain at the early stage of nodule formation, then oxygen concentration drops and Calvin cycle drives the electron excess to CO2. Once nitrogenase synthesis has started, electron excess is consumed by the nitrogen fixation process. Lower part of the figure represents the situation with a cbbL1 mutant leading to a Fix–like nodule: electron excess cannot be drive to CO2 therefore, reduced cofactors accumulate and disturbed bacterial metabolism preventing normal nodule development.
More work will be required to identify a role for RuBisCO 2 as the cbbL2 mutant did not display any phenotype during the symbiotic process or nitrogen fixation in vitro. Transcriptional activity from cbbL2 promoter was not detected during growth on rich or minimal medium using various carbon compounds. It is possible that ORS278 strain uses this enzyme under yet unidentified environmental conditions or that this cluster is not yet functional in this host strain due to its recent acquisition.
Materials and Methods
Phyllogenetic analysis
RuBisCO large sub-unit sequences from various bacteria YP_001735042.1 (Synechococcus sp. PCC 7002); NP_892668.1 (Prochlorococcus marinus subsp. pastoris str. CCMP1986); YP_001416820.1 (Xanthobacter autotrophicus Py2); YP_001203774.1 and YP_001204346.1 (Bradyrhizobium sp. ORS278); YP_002008347.1 (Cupriavidus taiwanensis); NP_769225.1 (Bradyrhizobium japonicum USDA 110); NP_841943.1 (Nitrosomonas europaea ATCC 19718); YP_115143.1 (Methylococcus capsulatus str. Bath); NP_946905.1 and NP_949975.1(Rhodopseudomonas palustris CGA009); NP_436731.1 (Sinorhizobium meliloti 1021); YP_427487.1 (Rhodospirillum rubrum ATCC 11170); YP_354363.1; YP_354780.1 (Rhodobacter sphaeroides 2.4.1); YP_001236643.1; YP_001238690.1 and YP_001242213.1 (Bradyrhizobium sp. BTAi1); YP_316382.1 and YP_316396.1 (Thiobacillus denitrificans ATCC 25259); YP_915492.1 (Paracoccus denitrificans PD1222); YP_571545.1, YP_571759.1 and YP_578935.1 (Nitrobacter hamburgensis X14); YP_318598.1; YP_319531.1 (Nitrobacter winogradskyi Nb-255); YP_342389.1 (Nitrosococcus oceani ATCC 19707); YP_411385.1 (Nitrosospira multiformis ATCC 25196); YP_747036.1 (Nitrosomonas eutropha C91); YP_001002624.1 (Halorhodospira halophila SL1); YP_001312667.1 (Sinorhizobium medicae WSM419); NP_943062.1 and YP_840914.1 (Ralstonia eutropha H16); ZP_05049432.1 (Nitrosococcus oceani AFC27); YP_003262812.1 and YP_003262978.1 (Halothiobacillus neapolitanus c2); YP_003443332.1; YP_003444691.1 (Allochromatium vinosum DSM 180)) were aligned using clustalW multiple alignment [28] with a bootstrap of 10000 replicates. Phyllogenetic tree was produced using Mega 4 [29].
Bacterial Strains and Growth Conditions
Bradyrhizobium sp. strain ORS278 [30], and derivatives ΔcbbL1, ΔcbbL2, PcbbL1::pVO155 and PcbbL2::pVO155 constructed in this work were grown in a modified YM medium (pH 6.8) as described in [19]. Strains were grown aerobically in a gyratory shaker (170 rpm) at 37°C. Standard methods were used for Escherichia coli growth in Luria–Bertani (LB) medium supplemented with the appropriate antibiotics.
Strains constructions
In order to construct mutant strains deleted in Brado1659 and Brado2274, DNA fragments were amplify from ORS278 genomic DNA using primer pairs Brado1659F (5′- TGCAGGACATGTTCAATCAGTTC-3′) /Brado1659R (5′-GGCCGGTTCACGATGAAGGACAG-3′) and Brado2274F (5′- GAGTTGTTTGCACTGCATCGAAAC-3′) /Brado2274R (5′-CACAGGAGCCGGTGAAGCAATC-3′). Resulting fragments were cloned in pGEMT easy and transformed in thermocompetent XL2-blue. Brado2274 and Brado1659 DNA fragments were excised from pGEMT using ApaI/SpeI and NotI respectively and cloned into pJQ200uc1 [31] (Brado 1659) and pJQ200sk [31](Brado 2274) opened with NotI and ApaI/SpeI respectively resulted in pBG1659 and pBG2274. Ligation products were transformed into XL2-Blue cells. Once check by restriction profiles, resulting plasmids were digested with XhoI/SalI (pBGBrado2274) and SalI (pBGBrado1659) and kanamycin resistance cassette excised from pKOK5 [32] using SalI was cloned into linearized vectors. Plasmids were transferred in E. coli S17-1 [33] by electroporation prior to mating with ORS278. Transconjugants were selected using nalidixic acid and kanamycin. Then transconjugants were replicated on sucrose medium containing kanamycin. Resulting colonies were check by PCR amplification of cassette flanking regions.
nifK mutant: Brado5438R/Brado5438F (5′-TCTAGACGTGTCGAACACATTCGAGTTGTC-3′/5′GTGTCGACCTGCATGGCGGAAGTC-3′) were used to amplify the central part of nifK open reading frame. DNA fragment was cloned into pGEMT easy and ligation product was transformed into XL2-Blue. After verification, insert was excise using SalI/XbaI and cloned into SalI/XbaI linearised pVO155 [34]. Resulting plasmid was successively transferred into XL1-Blue and S17-1. Resulting S17-1 strain was used to introduce the construction in ORS278 by mating.
Reporter strains: putative cbbL1 and cbbL2 promoter regions were PCR amplified using Brado1655PromF/Brado1655PromR (5′-CGATGATGATGTGCGGATGCTTG-3′ and 5′-GTGTCTAGAGATGATGATCCATCGGGTGATTG-3′) and Brado2274PromF/Brado2274PromR (5′- GAGGTCGACCTGATTGTCGGCCAAG-3′ and 5′-CCTTCTAGACGATCTTGAAGACCGCGAGAAG-3′) primer pairs.
DNA fragments were cloned in pGEMT easy resulting in PcbbL1_pGEMt and PcbbL2_pGEMt. SalI/XbaI insert were then excised, purified and ligated in pVO155 suicide vector [34] linearised with SalI/XbaI. Ligation products were introduced in XL2-Blue and S17-1 successively and finally transferred to ORS278 by mating.
Plant cultivations and inoculation
A. indica seeds were surface sterilized by immersion in sulphuric acid under shaking during 45 minutes. Seeds were abundantly washed with sterile distilled water and incubated overnight in sterile water. Seeds were then transferred for two days at 37°C in the darkness on 0.8% agar plate for germination. Plantlets were then transferred on the top of test tubes covered by aluminium paper for hydroponic culture in buffered nodulation medium (BNM) [35]. Plants were grown in closed mini green house in a 28°C growth chamber with a 16-h light and 8-h dark regime and 70% humidity. Seven days after transfer, each seedling was inoculated with one milliliter of cell suspension resulting from a 5 day-old bacterial culture washed in BNM and adjusted to reach an optical density of one at 600 nm.
Histochemical analysis and microscopy
30-to 40-µm-thick vibratome (VT1000S; Leica, Nanterre, France) sections from fresh nodule samples were observed under bright-field illumination with an optical microscope (PROVIS; Olympus, Rungis, France). To follow glucuronidase activity, nodule thick sections were incubated at 37°C in the darkness in a GUS assay buffer for 4 hours. This reaction buffer was described in [36] and contained 0.075% (wt/vol) of X-Gluc (5-bromo-4-chloro-3-indolyl-â-D-glucuronide) in 0.1 M K/Na phosphate buffer (pH 7.0), 20 mM NaEDTA, 0.02% Triton X-100, 1 mM K-ferricyanide, and 1 mM K-ferrocyanide.
Proteomics analysis
21 days post inoculation nodules were harvested and immediately flash frozen in liquid nitrogen. Bacteroids were isolated on a sucrose density gradient using a procedure derived from [37] using modifications described in [38] and [39]. Proteins were separated and analysed as described in [40].
Acetylene reduction assays
In planta acetylene reduction assays were conducted as described in [20] with minor modifications. Briefly: one plant was placed into 125 mL glass vials sealed with rubber septa. To avoid overpressure, 12.5 ml of air were removed before injecting the same volume of acetylene. After 3 hours of incubation at room temperature, the ethylene produced was measured by GC-FID using one ml of sampled gas. At least ten plants were used per condition. Plants inoculated with ORS278 strain were used as a reference. In vitro acetylene reduction assays were performed as described in [16].
Acknowledgments
We are grateful to Claudia Knief (ETH Zurich) for helpful advices on bacteroids isolation.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the French national research agency (ANR-NEWNOD-2006-BLAN-0095 and ANR-SESAM-2010-BLAN-170801); BG was supported by a grant from the European Molecular Biology Organization. This work has benefited from the facilities and expertise of the Functional Genomic Center Zurich. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Berg IA. Ecological aspects of distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson I, Backlund A. Structure and function of RuBisCO. Plant Physiol Biochem. 2008;46:275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Andersson I. Catalysis and regulation in RuBisCO. J Exp Bot. 2008;59:1555–1568. doi: 10.1093/jxb/ern091. [DOI] [PubMed] [Google Scholar]
- 4.Gutteridge S, Gatenby AA. RuBisCO Synthesis, Assembly, Mechanism, and Regulation. Plant Cell. 1995;7:809–819. doi: 10.1105/tpc.7.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillmer P, Gest H. H2 metabolism in the photosynthetic bacterium Rhodopseudomonas capsulata: H2 production by growing cultures. J Bacteriol. 1977;129:724–731. doi: 10.1128/jb.129.2.724-731.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKinlay JB, Harwood CS. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc Natl Acad Sci U S A. 2010;107:11669–11675. doi: 10.1073/pnas.1006175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson DJ, King GF, Kelly DJ, McEwan AG, Ferguson SJ, et al. The role of auxiliary oxidants in maintaining redox balance during phototrophic growth of Rhodobacter capsulatus. Archive of Microbiology. 1988;150:131–137. [Google Scholar]
- 8.Falcone DL, Tabita FR. Expression of endogenous and foreign ribulose 1,5-bisphosphate carboxylase-oxygenase (RuBisCO) genes in a RubisCO deletion mutant of Rhodobacter sphaeroides. J Bacteriol. 1991;173:2099–2108. doi: 10.1128/jb.173.6.2099-2108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallenbeck PL, Lerchen R, Hessler P, Kaplan S. Roles of CfxA, CfxB, and external electron acceptors in regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase expression in Rhodobacter sphaeroides. J Bacteriol. 1990;172:1736–1748. doi: 10.1128/jb.172.4.1736-1748.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickering BS, Oresnik IJ. Formate-dependent autotrophic growth in Sinorhizobium meliloti. J Bacteriol. 2008;190:6409–6418. doi: 10.1128/JB.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier RJ. Rhizobium japonicum mutant strains unable to grow chemoautotrophically with H2. J Bacteriol. 1981;145:533–540. doi: 10.1128/jb.145.1.533-540.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hungria M, Ellis JM, Hardy RWF, Eaglesham ARJ. Light-stimulated 14CO2 uptake and acetylene reduction by bacteriochlorophyll containing stem nodule isolate BTAi 1. Biology and Fertility of Soils. 1993;15:208–214. [Google Scholar]
- 13.White J, Prell J, James EK, Poole P. Nutrient sharing between symbionts. Plant Physiol. 2007;144:604–614. doi: 10.1104/pp.107.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prell J, Bourdes A, Kumar S, Lodwig E, Hosie A, et al. Role of symbiotic auxotrophy in the Rhizobium-legume symbioses. PLoS One. 2010;5:e13933. doi: 10.1371/journal.pone.0013933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud E, Fleischman D. Nitrogen-fixing symbiosis between photosynthetic bacteria and legumes. Photosynth Res. 2004;82:115–130. doi: 10.1007/s11120-004-1768-1. [DOI] [PubMed] [Google Scholar]
- 16.Alazard D. Nitrogen fixation in pure culture by rhizobia isolated from stem nodules of tropical Aeschynomene species. FEMS Microbiology Letters. 1990;68:177–182. [Google Scholar]
- 17.Dreyfus BL, Elmerich C, Dommergues YR. Free-living Rhizobium strain able to grow on N2 as the sole nitrogen source. Appl Environ Microbiol. 1983;45:711–713. doi: 10.1128/aem.45.2.711-713.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, et al. Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science. 2007;316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- 19.Giraud E, Hannibal L, Fardoux J, Vermeglio A, Dreyfus B. Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc Natl Acad Sci U S A. 2000;97:14795–14800. doi: 10.1073/pnas.250484097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonaldi K, Gourion B, Fardoux J, Hannibal L, Cartieaux F, et al. Large-scale transposon mutagenesis of photosynthetic Bradyrhizobium sp. strain ORS278 reveals new genetic loci putatively important for nod-independent symbiosis with Aeschynomene indica. Mol Plant Microbe Interact. 2010;23:760–770. doi: 10.1094/MPMI-23-6-0760. [DOI] [PubMed] [Google Scholar]
- 21.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 22.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 23.Badger MR, Bek EJ. Multiple RuBisCO forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot. 2008;59:1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 24.Galibert F, Finan TM, Long SR, Puhler A, Abola P, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 25.Amadou C, Pascal G, Mangenot S, Glew M, Bontemps C, et al. Genome sequence of the beta-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res. 2008;18:1472–1483. doi: 10.1101/gr.076448.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller F. On the metabolism of the purple sulfur bacteria in organic media. Arch Microbiol. 1933;4:131–166. [Google Scholar]
- 27.Joshi HM, Tabita FR. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci U S A. 1996;93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 30.Lorquin J, Molouba F, Dreyfus BL. Identification of the carotenoid pigment canthaxanthin from photosynthetic Bradyrhizobium strains. Appl Environ Microbiol. 1997;63:1151–1154. doi: 10.1128/aem.63.3.1151-1154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 32.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 33.Simon R, Priefer U, Puhler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat Biotech. 1983;1:784–791. [Google Scholar]
- 34.Oke V, Long SR. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol Microbiol. 1999;32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 35.Ehrhardt DW, Atkinson EM, Long SR. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 36.Bonaldi K, Gherbi H, Franche C, Bastien G, Fardoux J, et al. The Nod Factor-Independent Symbiotic Signaling Pathway: Development of Agrobacterium rhizogenes-Mediated Transformation for the Legume Aeschynomene indica. Mol Plant Microbe Interact. 2010;23:1537–1544. doi: 10.1094/MPMI-06-10-0137. [DOI] [PubMed] [Google Scholar]
- 37.Sarma AD, Emerich DW. Global protein expression pattern of Bradyrhizobium japonicum bacteroids: a prelude to functional proteomics. Proteomics. 2005;5:4170–4184. doi: 10.1002/pmic.200401296. [DOI] [PubMed] [Google Scholar]
- 38.Delmotte N, Ahrens CH, Knief C, Qeli E, Koch M, et al. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics. 2010;10:1391–1400. doi: 10.1002/pmic.200900710. [DOI] [PubMed] [Google Scholar]
- 39.Koch M, Delmotte N, Rehrauer H, Vorholt JA, Pessi G, et al. Rhizobial adaptation to hosts, a new facet in the legume root-nodule symbiosis. Mol Plant Microbe Interact. 2010;23:784–790. doi: 10.1094/MPMI-23-6-0784. [DOI] [PubMed] [Google Scholar]
- 40.Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, et al. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci U S A. 2009;106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]