Abstract
Background & Aims
Identification of a disease-specific H pylori virulence factors predictive of the outcome of infection remains unachieved.
Methods
We used the polymerase chain reaction and Southern blot to compare the presence of 14 vir homologue genes with clinical presentation of H pylori infection, mucosal histology, and mucosal interleukin (IL)-8 levels.
Results
We examined 500 H pylori strains from East Asia and South America, including 120 with gastritis, 140 with duodenal ulcer (DU), 110 with gastric ulcer (GU), and 130 with gastric cancer. Only 1 gene that encompassed both jhp0917 and jhp0918 called dupA (duodenal ulcer promoting gene) was associated with a specific clinical outcome. dupA was present in 42% of DU vs. 21% of gastritis (adjusted odds ratio [OR] = 3.1, 95% confidence interval [CI]: 1.7–5.7). Its presence was also associated with more intense antral neutrophil infiltration and IL-8 levels and was a marker for protection against gastric atrophy, intestinal metaplasia, and gastric cancer (OR for gastric cancer = 0.42, 95% CI: 0.2–0.9 compared with gastritis). In vitro studies in gastric epithelial cells using dupA-deleted and -complemented mutants showed that the dupA plays roles in IL-8 production, in activation of transcription factors responsible for IL-8 promoter activity, and in increased survivability at low pH.
Conclusions
dupA is a novel marker associated with an increased risk for DU and reduced risk for gastric atrophy and cancer. Its association with DU-promoting and -protective effects against atrophy/cancer was evident in both Asian and Western countries.
The outcome of a Helicobacter pylori infection is thought to reflect an interplay between the virulence of the infecting strain, host genetics, and environmental factors. Although a number of putative H pylori virulence genes such as the cag pathogenicity island (PAI), VacA, BabA, OipA, and HrgA have been associated with increased risks of a clinical outcome such as peptic ulcer or gastric cancer, none can be clearly linked to 1 specific H pylori-related disease (eg, duodenal ulcer).1–12 It is known that different patterns of gastritis are associated with different clinical outcomes.13,14 For example, antral-predominant gastritis is associated with hyperchlorhydria and duodenal ulcer (DU), whereas corpus gastritis leads to hypochlorhydria and gastric atrophy and an increased risk of distal gastric cancer.15,16
Comparison of the sequences of the whole genomes of 2 H pylori strains (26695 and J99) revealed several regions whose G + C content differed from that of H pylori, suggesting the presence of acquired DNA within the H pylori genome.17–19 One such region is the cag PAI. The other has been termed the “plasticity region,” based on the variability in gene content among different isolates (approximately 3% to 4% of the chromosome).17–19 Genome sequence comparisons indicated that nearly half of H pylori’s strain-specific genes are located in the plasticity region. There has been considerable recent interest in this plasticity region.20 –23 For example, Occhialini et al found that the jhp0940 and jhp0947 genes in the plasticity region were more likely to be found in H pylori from gastric cancer patients from a small group of 17 gastric cancer and 26 gastritis patients from Costa Rica.20 A follow-up to test the hypothetical relationship was done in 200 strains of H pylori from Brazil, including stains from DU, gastric cancer, and H pylori gastritis patients. The original hypothesis was not confirmed, but the new study suggested a relation between jhp0947 (not jhp0940) and both DU and gastric cancer.21 More recently, de Jonge et al reported that the presence of the jhp0947 and jhp0949 genes (which are linked) was significantly associated with DU disease when compared with gastritis from a small group of 26 duodenal ulcer and 19 gastritis patients from Dutch population.22 Kersulyte et al recently reported a novel 16.3-kb segment (tfs3) in the plasticity region, 7 of whose 16 open reading frames were homologues of the type IV secretion genes.23 Full-length and partial tfs3 elements were each found in about one fifth of H pylori strains. Overall, these data suggest that some genes or combination of genes in the plasticity region may play critical roles in the pathogenesis of H pylori-associated gastroduodenal diseases.
H pylori contains a number of vir genes homologues.17,18 For example, the cag PAI contains 7 vir genes homologues: virB4, virB7, virB8, virB9, virB10, virB11, and virD4 of the so-called VirB/D complex of type IV secretion systems known from diverse bacteria such as Agrobacterium tumefaciens and Bordetella pertussis.24 –28 Mutation studies suggest that VirB homologues in the cag PAI are involved in the induction of proinflammatory cytokine interleukin (IL)-8 and activation of transcription factors such as nuclear factor (NF)-κB in gastric epithelial cells.24,26,29,30 In this study, we examined the relationship between the presence of vir homologues and clinical outcomes and histologic changes in patients with different clinical presentations of H pylori infection. We show that the jhp0917 and jhp0918 genes (VirB4 homologue genes), which are present in the plasticity region, form 1 continuous gene. The gene is a marker for development of DU diseases and for protection against gastric atrophy, intestinal metaplasia, and gastric cancer. We have tentatively designated jhp0917-jhp0918 as the DU-promoting (dupA) gene of H pylori.
Materials and Methods
H Pylori Studied
For the study of the possible relation between vir homologues and clinical outcome in H pylori infection, we examined a large number of H pylori from different geographic regions to reduce the likelihood of spurious associations based on examining small numbers or restricted regional circulation of a particular genotype. We examined H pylori obtained from patients in East Asia (Kyoto Prefectural University of Medicine, Kyoto, Japan, and Guro Hospital, Korea University College of Medicine, Seoul, Korea) and from South America (Universidad Nacional de Colombia Bogotá, Colombia). We chose 4 different clinical presentations: simple H pylori gastritis, DU disease, gastric ulcer disease (GU), and noncardiac gastric adenocarcinoma. DU and GU were identified endoscopically; patients with ulcer scars were included. We excluded GU cases with DU. Gastritis was defined as histologic gastritis with no peptic ulcers or gastric cancer. No subjects had received treatment for H pylori infection.
H pylori isolates were chosen from those collected between 1997 and 2002. Thirty-seven percent (187 of 500) of H pylori had been used in our previous study for genotyping iceA, cagA, vacA, and/or hrgA genes.12,31 In addition, to assess the relation to gastric mucosal IL-8 levels, we used 98 strains from Japanese patients whose mucosal IL-8 levels in gastric biopsy specimens had been measured previously.32 None of these 98 patients were included among the 500 patients in the disease-association study. Informed consent had been obtained from all patients, and samples were taken under protocols approved by each local ethics committee.
Histology
Gastric mucosal biopsy specimens were fixed in 10% buffered formalin, embedded in paraffin, cut in sequential 4-μm sections, and stained with Genta stain33 or El-Zimaity triple stain34 (Korea and Colombia) or modified Giemsa stain (Japan). We examined sections unaware of the patient’s clinical diagnosis or the characteristics of the H pylori strain. Each specimen was scored for H pylori density, neutrophil infiltration, intestinal metaplasia, and atrophy. All the variables were graded using the visual analogue scale graded from 0 (absent/normal) to 5 (maximal intensity), as described previously.35
Analysis of vir Homologues
Chromosomal DNA was isolated from confluent plate cultures expanded from a single colony using the QIAamp Tissue kit (QIAGEN Inc. Santa Clarita, CA) according to the manufacturer’s instructions. We examined all 14 vir homologue genes from strain 26695 and/or J99: 7 genes in the cag PAI (jhp0473/hp0524: virD4 homologue, jhp0474/hp0525: virB11 homologue, jhp0476/hp0527: virB10 homologue, jhp0477/hp0528: virB9 homologue, jhp0479/hp0530: virB8 homologue, jhp0481/hp0532 [cagT]: virB7 homologue and jhp0492/hp0544 [cagE]: virB4 homologue), 4 in the plasticity region (hp0441, hp0459, jhp0917, and jhp0918: virB4 homologues), and 3 genes outside of the cag PAI and the plasticity region (jhp0015/hp0017: virB4 homologue [comB4], hp1006: virD4 homologue, and jhp1316/hp1421: virB11 homologue). Primers for jhp0473/hp0524, jhp0474/hp0525, jhp0476/hp0527, jhp0477/hp0528, cagT, and cagE had been reported previously.36,37 Primers for other genes are presented in Table 1. When the polymerase chain reaction (PCR) yielded negative results, Southern blot hybridization was performed to confirm the accuracy of PCR methods as previously described.36 In the case of a given region detected only by hybridization and not by PCR, the isolate was considered positive for this region.
Table 1.
PCR Primers for Amplification of the vir Homologue Genes
| Gene | Primer | Primer sequence (5′→3′) | Size (bp) of PCR product |
|---|---|---|---|
| jhp0479/hp0530 (virB8) | ORF16 (+) ORF16 (−) |
ATGTCAAACACTTTGAAGCC CCAATAGGACTTTCAAAAGGGC |
574 |
| hp0441 (virB4) | HP441 (+) HP441 (−) |
AGGCGCTCTTAAATTAGAGG TCATGCAAACGACCTCTAGC |
573 |
| hp0459 (virB4) | HP459 (+) HP459 (−) |
CTACGCAGAATACATCAATGGC CTCAAACGATTGGCTTGCAC |
363 |
| jhp0917 (virB4) | JHP917 (+) JHP917 (−) |
TGGTTTCTACTGACAGAGCGC AACACGCTGACAGGACAATCTCCC |
307 |
| jhp0918 (virB4) | JHP918 (+) JHP918 (−) |
CCTATATCGCTAACGCGCGCTC AAGCTGAAGCGTTTGTAACG |
276 |
| jhp0015/hp0017 (comB4, virB4) | JHP15 (+) JHP15 (−) |
AAACTTAGTGGGCATCCTCCG ACGCCATGCAAAGAGTGAGTG |
276 |
| hp1006 (virD4) | HP1006 (+) HP1006 (−) |
AAGTAGGAGCTATGGTGTGG GGTTTAGCCTTATGACCGCTTAC |
336 |
| jhp1316/hp1421 (virB11) | JHP1316 (+) JHP1316 (−) |
ACCCATAGAGTTTTACAAGCC ATTCGCGCTCACCCTATAGC |
267 |
Recently, Kersulyte et al reported a novel 16.3-kb segment (tfs3) in the plasticity region, 7 of whose 16 ORFs were vir homologues (virB4, virB7 to virB11, and virD4).23 Therefore, we also examined the presence of tfs3 by PCR as previously described.23 H pylori 26695 (ATCC700392: American Type Culture Collection, Rockville, MD) and J99 (ATCC700824) were used as reference H pylori strains.
Construction of Gene Deleted Strains and Complemented Strains of H pylori
For construction of the whole jhp0917-jhp0918-deleted mutants, we used gene replacement mutagenesis techniques. PCR fragments containing the most downstream fragment of the jhp0916 and the most upstream of the jhp0919 were cloned into the plasmid (pT7Blue; Novagen) resulting in pTJHP916 and pTJHP919, respectively. A chloramphenicol (cat) cassette (a gift from D. E. Taylor, University of Alberta, Edmonton, Canada) was inserted at the BamHI site of pTJHP916 resulting in pTJHP916::cat. Finally, the blunt fragment of jhp0916-cat (HincII/SmaI) was inserted at the HincII site of pTJHP919, resulting in pTJHP916/919::cat. The purified plasmids were used to inactivate chromosomal H pylori genes by natural transformation as previously described.38 Inactivation of the genes was confirmed by PCR amplification and/or Southern blot analyses (data not shown).
For construction of the jhp0917-jhp0918-complemented mutants, PCR fragments containing jhp0917-jhp0918 with the promoter of jhp0917 were cloned into the plasmid (pBlueScript; Stratagene, La Jolla, CA). The BamHI/HincII fragment was cut from the plasmid and cloned into the BamHI/NruI sites of H pylori/Escherichia coli shuttle vector (pHel3)39 (a gift from R. Haas, Max von Pettenkofer Institut, München, Germany) to generate the complementation construct pHel3-JHP917-918. The purified plasmid was used to introduce the gene into the jhp0917-jhp0918-deleted mutants with selection onto kanamycin and chloramphenicol. The plasmids were extracted from the complemented strains and subjected to restriction analyses to ensure their integrity. Transformants harboring intact plasmids were selected for further studies. The jhp0917-jhp0918-deleted mutants introduced by the empty shuttle vector (pHel3) were used as control.
IL-8 Levels From Gastric Cancer Cells Cocultured With H pylori
In vitro IL-8 secretion from gastric epithelial cells was examined as previously described.32 Briefly, gastric epithelial cell line, MKN45 cells (Japanese Cancer Research Resource Bank, Tsukuba, Japan) (approximately 5 × 105/mL) were plated into 24-well plates and cultured for 2 days (approximately 1 × 106/mL for each). Experiments were performed in duplicate for clinical samples and were performed at least 3 times for wild type and its isogenic mutants. H pylori (multiplicity of infection [MOI] of 100) or brain heart infusion (BHI) broth (control) were added to the cultured cells for 20 hours, and IL-8 in the supernatant was assayed by an enzyme-linked immunosorbent assay (ELISA) (R&D Systems, Minneapolis, MN) in duplicate.
Luciferase Reporter Gene Assay
The PathDetect cis-reporting plasmids pISREluci, pNF-κBluci, pAP-1luci, and pCREluci, which contain the luciferase reporter gene driven by the TATA box plus tandem repeats of the consensus-binding sequence for the corresponding transcription factor (interferon-stimulated responsive element [ISRE], NF-κB, activator protein [AP]-1, and cAMP responsive element [CRE]), respectively, were obtained from Stratagene. Luciferase reporter gene assay was performed as previously described40 in duplicate for clinical samples and was performed at least 4 times for wild type and its isogenic mutants. Luciferase activity was normalized to control renilla luciferase activity and expressed as firefly luciferase activity/renilla luciferase activity (normalized luciferase activity).
Protein/DNA Array for Different Components of the Signal Transduction System
Recently, a protein/DNA array that provides a profile to the DNA-binding activity of multiple transcription factors in a single array experiments has been introduced (TranSignal Arrays, Panomics, Inc., Redwood City, CA). The basic steps involved include the following: (1) A set of biotin-labeled DNA-binding oligonucleotides (TranSignal Probe Mix) are preincubated with a nuclear extract of interest to allow the formation of DNA/protein complexes; (2) the protein/DNA complexes are separated from the free probes; and (3) the probes in the complexes are then extracted and hybridized to the TranSignal Array. Each kit includes the reagents for HRP-based chemiluminescence detection. This array allows 54 transcription factors to be identified simultaneously. MKN45 cells (approximately 5 × 105/mL) were plated into 10-cm2-well plates and cultured for 2 days (approximately 1 × 106/mL for each). H pylori (MOI of 100) or BHI broth (control) was added to the cultured cells for 90 minutes, and nuclear extracts of gastric cells were prepared using hypotonic/nonionic detergent lysis. An equal amount of protein was used for protein/DNA array according to the manufacturer’s instruction. Assays were performed in triplicate. Quantitation of the activity of the transcription factors was determined using Scion Image beta 4.02 (Scion Corp., Frederick, MA).
Survival of H Pylori After Exposure to Low pH
Susceptibility of H pylori to low pH was measured as described previously in duplicate for clinical samples and at least 4 times for wild type and its isogenic mutants.41 Briefly, H pylori (approximately 108 CFU/mL) were grown in BHI broth adjusted to pH 6 with citrate-phosphate buffer (CPB) and supplemented with 5% horse serum on a rotating shaker for 24 hours. We preexposed H pylori to pH 6 based on previous studies showing that H pylori has an acid-tolerance response, and H pylori adapted at pH 6 survived exposure to pH 3 approximately 100-fold better than did unadapted cells (exposed to pH 7).42 H pylori were then centrifuged at 2000g, resuspended in CPB at low pH (3 to 5), and incubated at 37°C for 20 minutes. All buffer solutions were urea free to ensure that any effect was independent of urease enzymatic activity. Viable counts before and after acid stress was assessed by serial 10-fold dilution in BHI broth onto BHI agar plates for 3–5 days. Difference in viable counts before and after exposure to acid stress was presented as log10 kill. Intraassay variation in our laboratory was within 10%.
Data Analysis
For univariate analysis, Fisher exact test, χ2 test, and Mann-Whitney rank sum test were used, depending on the data set of concern. A multiple logistic regression analysis was performed to determine which vir homologue factor(s) was the most discriminating for clinical outcome, where vir homologues, age, and sex were explanatory variables. Multiple linear regression analyses were used for the histologic data because the data showed normal distributions. In the analyses, vir homologue factors, sex, age, and clinical outcomes were included as explanatory variables, and mutually adjusted associations with the criterion variables were calculated. Selections of variables were by backward stepwise deletion in the logistic regression and by F-out and F-in stepwise method in the linear regression, where F values were both 2.0. A P value of less than .05 was accepted as statistically significant. Calculations were carried out using statistical software “HALBAU” (Gendai-sugaku-sha, Kyoto, Japan).
Results
Prevalence of the vir Homologue Genes and Clinical Outcomes
We examined 500 H pylori isolates for the 14 known vir homologue genes previously detected within the H pylori genome. Specimens were obtained from 294 men and 206 women with a mean age of 52.4 years. There were 160 subjects from Japan (presentations included 50 gastritis, 30 DU, 50 GU, and 30 gastric cancer), 175 from Korea (presentations included 30 gastritis, 65 DU, 30 GU, and 50 gastric cancer), and 165 from Colombia (presentations included 40 gastritis, 45 DU, 30 GU, and 50 gastric cancer).
Tables 2 and 3 show the association among the prevalence of the 14 vir homologue genes in relation to clinical presentation. Independent univariate analysis showed that the prevalence of the jhp0917 and jhp0918 genes (virB4 homologue genes) in the plasticity region was significantly greater among strains isolated from patients with DU (42%) than from patients with non-DU H pylori gastritis (21%), with GU (27%) or with gastric cancer (9%) (P < .001 for DU vs. gastritis or cancer (P < .05 for GU vs. cancer). The prevalence of the jhp0917 and jhp0918 genes was closely linked, with only 10 of 500 (2%) strains being jhp0917 negative/jhp0918 positive (5 from DU, 3 from gastritis, and 2 from gastric cancer), and the status of the 2 genes in the remaining strains was identical (both positive or both negative). We therefore categorized strains as either genotype positive or negative based on the combination of the jhp0917 and jhp0918 genes (jhp0917-0918). Ten cases were jhp0917 negative and jhp0918 positive and were excluded for further analyses.
Table 2.
Association Between the vir Homologue Genes Outside of the cag PAI and Disease Outcomes
| No. | hp0441 (virB4) % | hp0459 (virB4) % | jhp0917 (virB4) % | jhp0918 (virB4) % | jhp0917-0918 (dupA) % | jhp0015/hp0017 (virB4) % | hp1006 (virD4) % | jhp1316/hp1421 (virB11) % | |
|---|---|---|---|---|---|---|---|---|---|
| Total | |||||||||
| Gastritis | n = 120 | 3 | 35 | 20a | 23a | 21a | 82 | 38 | 100 |
| DU | n = 140 | 1 | 38 | 41b,c,d | 44b,c,d | 42b,c,d | 86 | 44 | 100 |
| GU | n = 110 | 2 | 37 | 27c | 27c | 27c | 83 | 41 | 100 |
| Gastric cancer | n = 130 | 2 | 37 | 9 | 11 | 9 | 82 | 37 | 100 |
| Japan | |||||||||
| Gastritis | n = 50 | 2 | 40 | 14 | 16 | 14 | 82 | 34 | 100 |
| DU | n = 30 | 0 | 47 | 37a,e | 37 | 37a,e | 83 | 43 | 100 |
| GU | n = 50 | 2 | 40 | 26 | 26 | 26 | 78 | 40 | 100 |
| Gastric cancer | n = 30 | 7 | 43 | 10 | 13 | 10 | 83 | 37 | 100 |
| Korea | |||||||||
| Gastritis | n = 30 | 3 | 30 | 7 | 7 | 7 | 67 | 33 | 100 |
| DU | n = 65 | 2 | 37 | 37b,c | 37b,c | 37b,c | 80 | 51 | 100 |
| GU | n = 30 | 3 | 37 | 17 | 17 | 17 | 83 | 43 | 100 |
| Gastric cancer | n = 50 | 0 | 32 | 6 | 8 | 6 | 78 | 36 | 100 |
| Colombia | |||||||||
| Gastritis | n = 40 | 5 | 33 | 38c | 43c | 39c | 93 | 45 | 100 |
| DU | n = 45 | 2 | 33 | 49c | 60c | 55c | 96 | 36 | 100 |
| GU | n = 30 | 0 | 33 | 40a | 40a | 40a | 90 | 40 | 100 |
| Gastric cancer | n = 50 | 2 | 38 | 12 | 12 | 12 | 84 | 38 | 100 |
NOTE. P value was determined by Fisher exact test or χ2 test.
For jhp0917–0918, 10 samples with jhp0917-negative/jhp0918-positive samples were excluded.
P < .05 compared with gastric cancer.
P < .01 compared with gastritis.
P < .01 compared with gastric cancer.
P < .05 compared with gastric ulcer.
P < .05 compared with gastritis.
Table 3.
Association Between the vir Homologue Genes in the cag PAI and Disease Outcomes
| No. | jhp0473/hp0524 (virD4) % | jhp0474/hp0525 (virB11) % | jhp0476/hp0527 (virB10) % | jhp0477/hp0528 (virB9) % | jhp0478/hp0529 (virB8) % | cagT (virB7) % | cagE (virB4) % | cag PAI (intact) % | |
|---|---|---|---|---|---|---|---|---|---|
| Total | |||||||||
| Gastritis | n = 120 | 93 | 93 | 93 | 93 | 93 | 93 | 94 | 95 |
| DU | n = 140 | 94 | 94 | 95 | 95 | 95 | 95 | 96 | 96 |
| GU | n = 110 | 96 | 96 | 96 | 96 | 96 | 95 | 95 | 97 |
| Gastric cancer | n = 130 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 |
| Japan | |||||||||
| Gastritis | n = 50 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| DU | n = 30 | 97 | 97 | 97 | 97 | 97 | 97 | 100 | 100 |
| GU | n = 50 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Gastric cancer | n = 30 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Korea | |||||||||
| Gastritis | n = 30 | 97 | 97 | 97 | 97 | 97 | 97 | 97 | 100 |
| DU | n = 65 | 97 | 97 | 98 | 98 | 98 | 98 | 98 | 98 |
| GU | n = 30 | 100 | 100 | 100 | 100 | 100 | 97 | 97 | 100 |
| Gastric cancer | n = 50 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Colombia | |||||||||
| Gastritis | n = 40 | 80 | 80 | 80 | 80 | 80 | 83 | 85 | 84 |
| DU | n = 45 | 89 | 89 | 89 | 89 | 89 | 89 | 89 | 89 |
| GU | n = 30 | 87 | 87 | 87 | 87 | 87 | 87 | 87 | 87 |
| Gastric cancer | n = 50 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 |
NOTE. For the cag PAI, 7 samples with partial deleted cag PAI were excluded from analyses.
None of the other genes examined were related to clinical outcome (Table 3). However, there was a trend for the prevalence of an intact cag PAI with all 7 vir homologue genes in the island being positive to be greater among strains from gastric cancer patients (94%) compared with those from gastritis patients (80%) (P = .06). Only 7 of 500 strains (1%) possessed a partially deleted cag PAI, and they were excluded for further analyses. The cag PAI status was then categorized into 2 types: positive for all 7 cag PAI genes (intact cag PAI) or cag PAI negative. The hp0441 gene was very rarely present (3%). All strains studied possessed the jhp1316/hp1421 gene.
Multiple logistic regression analysis was performed for 14 vir homologue genes, adjusting for age, sex, and country of origin. As shown in Table 4, only the presence of the jhp0917-0918 genes was related to the clinical outcomes. The presence of the jhp0917-0918 genes was an independent determinant predictor of DU and was protective against gastric cancer (DU vs. gastric cancer; OR = 8.3; 95% CI: 4.0 –17.4). Importantly, this pattern was similar in all 3 geographic regions examined, confirming that the observation was not a reflection of restricted regional circulation of a particular genotype. In Colombia, the presence of the jhp0917-0918 genes was also an independent determinant protective (or linked to another protective factor) against gastric cancer vs. gastritis and GU (Table 4).
Table 4.
Multiple Logistic Regression Analysis: H pylori Factors Associated With Clinical Presentation
| Factors (positive vs. negative) | P value | Adjusted OR | 95% CI | |
|---|---|---|---|---|
| Duodenal ulcer vs. gastric cancer | ||||
| Total | jhp0917–jhp0918 | <.001 | 8.3 | 4.0–17.4 |
| Japan | jhp0917–jhp0918 | .023 | 5.4 | 1.3–23.0 |
| Korea | jhp0917–jhp0918 | <.001 | 10.9 | 1.8–66.9 |
| Colombia | jhp0917–jhp0918 | <.001 | 9.0 | 3.0–26.6 |
| Duodenal ulcer vs. gastritis | ||||
| Total | jhp0917–jhp0918 | <.001 | 3.1 | 1.7–5.7 |
| Japan | jhp0917–jhp0918 | .027 | 3.7 | 1.2–11.5 |
| Korea | jhp0917–jhp0918 | .005 | 9.2 | 2.0–43.0 |
| Gastric cancer vs. gastritis | ||||
| Total | jhp0917–jhp0918 | .042 | 0.42 | 0.2–0.9 |
| Colombia | jhp0917–jhp0918 | .012 | 0.22 | 0.1–0.7 |
| Gastric cancer vs. gastric ulcer | ||||
| Colombia | jhp0917–jhp0918 | .034 | 0.29 | 0.1–0.8 |
NOTE. The data with statistically significant are presented. When we analyzed data in combinations of 3 counties (total), countries were also included in explanatory variables.
We also examine the presence of tfs3 in 24 strains (12 jhp0917-0918 positive and 12 jhp0917 0918-negative) for each country. Complete tfs3 was detected in 8 (33%) of Colombian strains, 2 (8%) of Japanese strains, and 3 (13%) of Korean strains. The presence of complete tfs3 was not linked to other vir homologue genes (eg, 7 [19%] of the jhp0917-0918-positive strains and 6 [17%] of the jhp0917-0918-negative strains possessed complete tfs3). The presence of partial tfs3 was also not linked to other vir homologue genes, except for that of hp0459 (hp0459 is a member of tfs3) (data not shown).
Prevalence of the vir Homologue Genes and Gastric Mucosal Histology
Gastric mucosal biopsy specimens were analyzed histologically and scored for H pylori density, neutrophil infiltration, intestinal metaplasia, and atrophy. All the variables were graded using the visual analogue scale graded from 0 (absent/normal) to 5 (maximal intensity), as described previously.35 Histologic analyses were made using biopsy specimens from normal-appearing gastric mucosa of Japanese and Colombian patients. Twenty-one Colombian patients (11 from gastric cancer, 6 from GU, 2 from DU, and 2 from gastritis) were excluded from these analyses because we had only 1 biopsy sample in either antrum or corpus because gastric cancer was widely spread in the antrum or corpus.
Independent univariate analysis showed that the presence of the jhp0917-0918 genes was positively related to neutrophil infiltration and inversely related to intestinal metaplasia and atrophy (Table 5). When we examined each disease separately, similar tendency was observed in each disease in both countries (Figure 1). The effect of the cag PAI was restricted to biopsy specimens from Colombia because cag PAI-negative strains are rarely found in East Asia. Scores for gastric mucosal atrophy were significantly higher in patients with infection with intact cag PAI compared with PAI-negative cases (Table 5). No other genes examined were related to changes in gastric histology (data not shown).
Table 5.
Univariate Analysis of Relationship Between the jhp0917–0918 and cag PAI Status and Histology
| No. | Antrum
|
Corpus
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| H pylori density | Neutrophil infiltration | Intestinal metaplasia | Atrophy | H pylori density | Neutrophil infiltration | Intestinal metaplasia | Atrophy | ||
| Japan | |||||||||
| jhp0917–0918 | |||||||||
| Positive | 34 | 3.0 (3) | 3.5 (3) | 0.4 (0) | 0.6 (0) | 2.6 (3) | 2.3 (3) | 0.3 (0) | 0.3 (0) |
| Negative | 124 | 2.7 (3) | 2.1 (2) | 1.1 (1) | 2.1 (2) | 2.1 (2) | 1.8 (2) | 1.0 (1) | 1.8 (1) |
| P value | .07 | <.001 | .006 | <.001 | NS | <.001 | .043 | <.001 | |
| Colombia | |||||||||
| jhp0917–0918 | |||||||||
| Positive | 50 | 2.5 (2) | 2.8 (3) | 0.2 (0) | 0.6 (0) | 2.4 (2) | 2.3 (2) | 0.1 (0) | 0.3 (0) |
| Negative | 87 | 2.2 (2) | 1.7 (2) | 1.1 (0) | 1.7 (1) | 2.2 (2) | 1.7 (2) | 1.0 (0) | 1.1 (0.5) |
| P value | NS | .002 | .004 | <.001 | NS | .023 | .003 | <.001 | |
| cag PAI | |||||||||
| Positive | 125 | 2.4 (2) | 2.5 (2) | 0.9 (0) | 1.4 (1) | 2.3 (2) | 2.0 (2) | 0.7 (0) | 0.9 (0) |
| Negative | 17 | 2.3 (2.5) | 1.6 (2) | 0.4 (0) | 0.5 (0) | 2.2 (2) | 1.3 (1) | 0.1 (0) | 0 (0) |
| P value | NS | .057 | NS | .037 | NS | .063 | NS | .009 | |
NOTE. For histologic scores (minimum 0 to maximum 5), mean (median) scores are presented. The cag PAI data in Japan did not show because there were no cag PAI negative strains in Japan. Other vir factors were not related to changes in gastric histology and were not shown.
NS, not significant; P > .10.
Figure 1.
Univariate analysis showing the relationship between the jhp0917-0918 gene (dupA) and the gastric mucosal histology. The scores (0: absent/normal to 5: maximal intensity) for neutrophil infiltration, intestinal metaplasia, and atrophy scores are presented separately in relation to clinical presentation in 1 East Asian and 1 Western country (Japan and Colombia, respectively). The strains from Japanese patients included 34 dupA-positive strains (7 from gastritis, 11 from DU, 13 from GU, and 3 from gastric cancer) and 124 dupA-negative strains (42 from gastritis, 19 from DU, 37 from GU, and 26 from gastric cancer). Strains from Colombia included 50 dupA-positive strains (14 from gastritis, 21 from DU, 9 from GU, and 6 from gastric cancer) and 87 dupA-negative strains (22 from gastritis, 17 from DU, 15 from GU, and 33 from gastric cancer). The data are expressed as mean ± standard error.
A backward stepwise multiple linear regression analysis showed the similar results for determining the vir factors related to the severity of histology (Table 6). For these analyses, we used the vir factors as well as age, sex, and clinical outcomes as explanatory variables. The presence of the jhp0917-0918 genes was significantly associated with severe neutrophil infiltration and with reduced intestinal metaplasia and atrophy, confirming that the jhp0917-0918 genes were markers for DU, which typically is associated with severe antral inflammation and high acid secretion. Importantly, these patterns were confirmed in both Japan and Colombia (Table 6). For example, patients infected with the jhp0917-0918-positive strains would be expected to have 1.21 ± 0.26 (= partial regression coefficient ± SE) higher scores for the neutrophil infiltration than those with the jhp0917-0918-negative strains. In contrast to the jhp0917-0918 status, the cag PAI-positive status was associated with intestinal metaplasia and atrophy in agreement with previous studies.10 In fact, no intestinal metaplasia and atrophy were observed in patients infected with the jhp0917-0918-positive/cag PAI-negative strains (n = 10).
Table 6.
Final Model Using Multiple Linear Regression Analysis: H pylori vir Homologue Genes Associated with Histology
| Histology | Site | Factors (positive vs. negative) | Partial regression coefficient ± SE | P value |
|---|---|---|---|---|
| Japan | ||||
| Neutrophil infiltration | Antrum | jhp0917–jhp0918 | 1.21 ± 0.26 | <.001 |
| Corpus | jhp0917–jhp0918 | 0.49 ± 0.25 | .021 | |
| Intestinal metaplasia | Antrum | jhp0917–jhp0918 | −0.52 ± 0.21 | .023 |
| Corpus | jhp0917–jhp0918 | −0.47 ± 0.23 | .034 | |
| Atrophy | Antrum | jhp0917–jhp0918 | −1.31 ± 0.25 | <.001 |
| Corpus | jhp0917–jhp0918 | −1.03 ± 0.24 | <.001 | |
| Colombia | ||||
| Neutrophil infiltration | Antrum | jhp0917–jhp0918 | 0.84 ± 0.26 | .002 |
| cag PAI | 0.74 ± 0.36 | .068 | ||
| Corpus | cag PAI | 0.70 ± 0.32 | .049 | |
| jhp0917–jhp0918 | 0.41 ± 0.24 | .076 | ||
| Intestinal metaplasia | Antrum | jhp0917–jhp0918 | −0.62 ± 0.23 | .009 |
| Corpus | jhp0917–jhp0918 | −0.53 ± 0.23 | .018 | |
| cag PAI | 0.54 ± 0.32 | .079 | ||
| Atrophy | Antrum | jhp0917–jhp0918 | −0.81 ± 0.27 | .004 |
| cag PAI | 0.74 ± 0.39 | .077 | ||
| Corpus | cag PAI | 0.96 ± 0.35 | .004 | |
| jhp0917–jhp0918 | −0.63 ± 0.25 | .009 | ||
NOTE. Factors that remained in final model are presented. The partial regression coefficient provides an estimate of the expected changes of the histologic scores (minimum 0 to maximum 5) between the different diseases.
In Vivo Mucosal IL-8 Production and the jhp0917-0918 Status
In a previous study, we measured in vivo gastric mucosal IL-8 levels and H pylori density in gastric mucosal biopsy specimens from 98 Japanese patients infected with the cag PAI-positive strains (50 from DU and 48 from gastritis patients).32 Antral IL-8 levels were measured from all patients, and corpus IL-8 levels were measured in 35 patients for each disease. We therefore used those data to examine the relationship between mucosal IL-8 production and the jhp0917-0918 status determined using PCR. The results confirmed that the presence of the jhp0917-0918 genes was strongly related to the presence of DU because 23 of 50 (46%) strains from DU were jhp0917-0918-positive compared with 15% (7 of 48) with simple gastritis (P < .001). The presence of the jhp0917-0918 genes was also strongly related to the cellular inflammation and negatively related to intestinal metaplasia and atrophy (data not shown).
Antral mucosal IL-8 levels were significantly higher in biopsy specimens obtained from DU patients infected with jhp0917-0918-positive strains compared with those with jhp0917-0918-negative strains (160.9 ± 19 vs. 101.2 ± 14 pg/mg protein, respectively) (P < .05) (Figure 2). Adjustment of antral IL-8 levels for H pylori density (IL-8 per H pylori) also showed a similar tendency (347 ± 10 vs. 258 ± 7 pg/CFU × 103, respectively) (P < .05). Antral IL-8 levels were also higher in gastritis patients with jhp0917-0918-positive infections (IL-8 per H pylori = 302 ± 7 vs. 150 ± 19 pg/CFU × 103, respectively) (P < .01). In contrast, we did not find a relation between IL-8 levels in the corpus and the presence of the jhp0917-0918 genes in either DU or gastritis patients (Figure 2). These close relationships between the presences of the jhp0917-0918 genes and high antral IL-8 production are consistent with prior studies showing that DU is typically associated with severe antral inflammation with high antral IL-8 production.32
Figure 2.
In vivo gastric mucosal IL-8 levels and the jhp0917-0918 status. Mucosal IL-8 levels from biopsy specimens measured by ELISA are presented on the left panel, and adjusted IL-8 levels for H pylori density (IL-8 per H pylori) are presented on the right panel. IL-8 in the supernatant was assayed in duplicate. Data are expressed as mean ± standard error.
Structural Analyses of the jhp0917-0918 Genes
Sequence analyses (BLAST: http://www.ddbj.nig.ac.jp/search/blast-e.html, GTOP: http://spock.genes.nig.ac.jp/%7Egenome/gtop.html and Pfam: http://www.sanger.ac.uk/Software/Pfam/) shows that neither jhp0917 nor jhp0918 genes possess a complete functional virB4 homologue gene in genome sequence strain J99. The jhp0917 gene consists of 475 amino acids and lacks most of the 3′ end of the virB4 gene. The jhp0918 gene consists of 140 amino acids and contains a fragment of the virB4 gene from the 3′ end. We therefore sequenced the jhp0917-0918 genes in 24 jhp0917-0918-positive strains (8 randomly selected from each country). The total size from the 5′ end of the jhp0917 gene to 3′ end of the jhp0918 gene in all clinical isolates was the same as that of strain J99 by PCR; however, all the clinical isolates contains 1 base pair insertion (C or T) in the 3′ region of the jhp0917 gene (after position 1385), resulting in a frameshift leading to a continuous open reading frame (Figure 3). In contrast, J99 strain in our stock showed exactly the same sequences as those deposited in GenBank, forming separate 2 genes: jhp0917 and jhp0918 genes. Therefore, we conclude that jhp0917-0918 is 1 gene homologous to virB4 gene, with strain J99 being an exception.
Figure 3.
Schematic structure of the jhp0917 gene and jhp0918 gene in strain J99 and that of the dupA gene in the clinical isolates. All 24 clinical isolates examined contains “C or T” insertion after position 1385 to produce 1 continuous open-reading frame homologue to the virB4 gene. We denote the gene containing the sequences of the jhp0917 gene and jhp0918 as the duodenal ulcer-promoting (dupA) gene.
The above in vivo data clearly showed that the presence of the jhp0917-0918 genes were strongly related to the existence of DU and that the prevalence of the jhp0917 gene and jhp0918 gene was closely linked to each other. We therefore designated the jhp0917-0918 as the DU-promoting (dupA) gene of H pylori. Full sequences of the dupA gene has been deposited in GenBank (accession number: AB196363).
In Vitro IL-8 Production From the Epithelial Cells and dupA Status
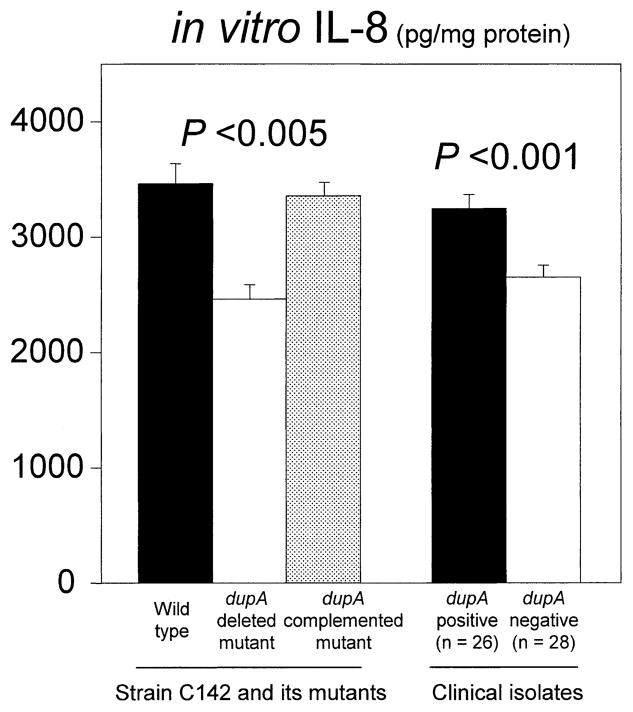
We examined the relationship between in vitro IL-8 induction from the epithelial cells and dupA status. We used a wild-type dupA-positive strain, its dupA-deleted mutant, and its complemented dupA mutant. We selected strain C142 as the dupA-positive parental strain, because it contains 13 of the 14 vir homologue genes (only jhp0441 gene is absent), for use to investigate the specific effect of the dupA gene among the vir homologue genes. Strain C142 was originally isolated from Colombian DU patients, and sequence analyses showed that this strain contained intact dupA gene (continuous from the 5′ end of the jhp0917 to 3′ end of the jhp0918) (GenBank accession number: AB196363). IL-8 levels were significantly higher in supernatants cocultured with C142 wild type than in those with its dupA-deleted mutant (mean ± SE = 3421 ± 175 vs. 2480 ± 124 pg/mL, respectively) (P < .005). IL-8 induction was restored using the complemented dupA mutant (3354 ± 216 pg/mL), confirming that the dupA gene is involved in IL-8 induction (Figure 4). The dupA-deleted mutants introduced by empty shuttle vector (control) failed to restore IL-8 production (data not shown).
Figure 4.
In vitro IL-8 production from MKN45 cells cocultured with wild-type strain C142 and its dupA-deleted mutant and dupA-complemented mutant and clinical isolates is shown. H pylori (MOI of 100) was cocultured with MKN45 cells for 20 hours. Experiments were performed at least 4 times for the wild type and its isogenic mutants and were performed in duplicate for clinical samples. IL-8 in the supernatant was assayed by an ELISA in duplicate. Data are expressed as mean ± standard error.
To confirm the relationship between in vitro IL-8 induction and the dupA status, we also examined clinical isolates. Because both the cag PAI and OipA affect IL-8 induction in vitro,38,40 we limited the analysis to the cag PAI-positive/oipA “on” strains. We selected 26 dupA-positive strains and 28 dupA-negative strains from among the 500 strains (6 strains with each disease from each country). IL-8 levels from MKN45 cells were significantly higher in supernatants cocultured with dupA-positive strains than in those with dupA-negative strains (mean ± SE = 3207 ± 123 vs. 2681 ± 118 pg/mL, respectivley) (P < .001) (Figure 4).
Relationship Between Gene Promoter Activation and dupA Status
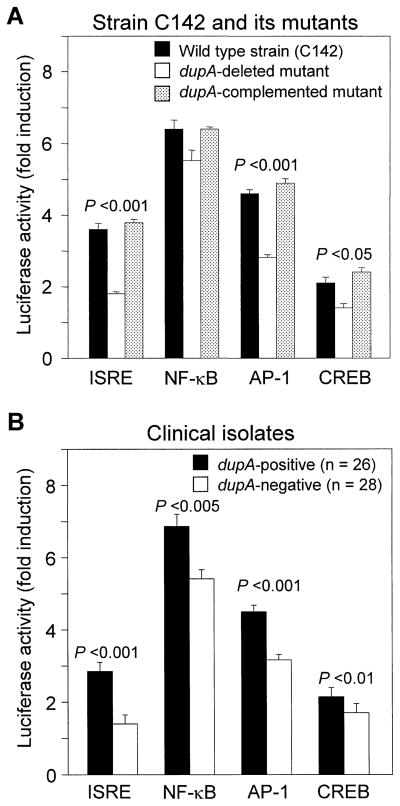
Transcription of genes is regulated by transcription factors that bind to their promoter. For example, we previously reported that full activation of the IL-8 promoter in H pylori infection required binding sites for NF-κB, AP-1, and ISRE-like elements.40 Full activation of IL-6 and RANTES promoters, which are also important inflammatory cytokine and chemokine, in H pylori infection required binding sites for CRE-binding protein (CREB) (unpublished observation). The luciferase reporter gene assay was performed to investigate the relationship between the dupA status and gene promoter activation by the ISRE, NF-κB, AP-1, and CREB. When we used the wild-type strain (C142) and its dupA-deleted mutants, luciferase activity for ISRE, AP-1, and CRE was significantly higher in cases cocultured with the wild-type H pylori compared with those with the dupA-deleted mutants (fold induction: 3.6 ± 0.2 vs. 1.8 ± 0.1, respectively [P < .001], for ISRE; 4.6 ± 0.2 vs. 2.8 ± 0.2, respectively [P < .001], for AP-1; and 2.1 ± 0.1 vs. 1.4 ± 0.1, respectively [P < .05] for CREB) (Figure 5A). The dupA-complemented mutants restored the activity for these 3 factors (fold induction: 3.8 ± 0.2 for ISRE, 4.9 ± 0.2 for AP, and 2.4 ± 0.1 for CREB). The dupA-deleted mutants introduced by empty shuttle vector (control) did not show restoration (data not shown). The dupA-positive wild-type H pylori also induced higher luciferase activity for NF-κB compared with the dupA-deleted mutants, although the differences were not statistically significant (fold induction: 6.4 ± 0.3 vs. 5.5 ± 0.2, respectively). Luciferase activity for all 4 factors was similar between strain J99 and its jhp0917-0918-deleted mutant (data not shown), indicating that jhp0917 region of the dupA gene is not involved in activation of these transcription factors.
Figure 5.
Shows the luciferase reporter activity of ISRE, NF-κB, AP-1, and CRE associated with H pylori infection. Luciferase reporter gene assay was performed as previously described35 at least 4 times for (A) wild type and its isogenic mutant and was performed in duplicate for (B) clinical samples. Luciferase activity was normalized by renilla luciferase vector DNA (Promega), and normalized luciferase activity (fold induction) is presented. Data are expressed as mean ± standard error of normalized luciferase activity.
We next used the clinical isolates that are the same strains as used for in vitro IL-8 levels as shown above. The dupA-positive H pylori induced luciferase activity for all 4 factors significantly higher compared with the dupA-negative strains (Figure 5B). These data confirmed that the dupA genes are involved in the activation of transcription factors responsible for the IL-8 promoter.
Protein/DNA Array for Different Components of the Signal Transduction System
Using the novel protein/DNA array, we assessed relationship between levels of activated transcription factors and dupA status. Wild-type H pylori C142 produced more than a 2-fold induction of 34 of 54 transcription factors evaluated compared with uninfected control (Table 7). Twelve transcription factors were activated in both wild-type strains and dupA-deleted mutants (eg, YY1, Ets-1/PEA3), whereas 22 transcription factors were activated only by wild-type strains (eg, AP-1, CREB, p53). Results for ISRE, AP-1, and CREB were in agreement with the above results using luciferase reporter gene assay. The accuracy of the protein/DNA array was further confirmed by using electrophoretic mobility shift assay for NF-κB, AP-1, CREB, and ISRE (unpublished observation).
Table 7.
Differential Activation of Transcription Factors Induced by H pylori: Comparison Between Wild Type Strain (C142) and its dupA-Deleted Mutant
| Name | Wild type (C142) | dupA-deleted mutants | Ratio for wild type/dupA deleted |
|---|---|---|---|
| Transcription factors activated in both wild type strain C142 and dupA deleted mutant | |||
| YY1 | 2.1 | 2.9 | 0.8 |
| Ets | 2.5 | 2.2 | 1.1 |
| NFκB | 2.5 | 2.0 | 1.3 |
| c-Myb | 2.6 | 2.2 | 1.2 |
| GATA | 2.7 | 2.1 | 1.5 |
| Brn-3 | 2.8 | 2.2 | 1.3 |
| Ets-1/PEA3 | 4.5 | 6.6 | 0.8 |
| Interferon regulatory factor 1 (IRF-1) | 4.6 | 4.5 | 1.2 |
| Pre-B-cell leukemia transcription factor 1 (Pbx1) | 4.7 | 2.3 | 2.2 |
| Signal transducer and activator of transcription 1 (Stat1) | 5.2 | 2.2 | 2.7 |
| Estrogen receptor (ERE) | 5.9 | 2.4 | 2.6 |
| Hepatocyte nuclear factor 4 (HNF-4) | 9.4 | 2.2 | 4.6 |
| Transcription factors activated only in wild type strain C142 | |||
| E2F transcription factor 1 | 2.1 | 1.4 | 1.6 |
| Retinoic acid receptor (RAR) | 2.2 | 1.1 | 1.9 |
| Heat shock element (HSE) | 2.2 | 1.6 | 1.4 |
| Upstream transcription factor 1 (USF-1) | 2.2 | 1.7 | 1.4 |
| AP-1 | 2.2 | 1.2 | 2.4 |
| Sp1 | 2.4 | 1.4 | 1.7 |
| Stat4 | 2.4 | 1.9 | 1.3 |
| Nuclear factor of activated T-cells, cytoplasmic (NFATc) | 2.5 | 1.5 | 1.7 |
| Nuclear factor (erythroid-derived 2) (NF-E2) | 2.7 | 1.9 | 1.5 |
| CREB | 2.7 | 1.3 | 2.1 |
| Stat5/Stat6 | 2.9 | 1.5 | 1.9 |
| p53 | 3.0 | 1.3 | 2.4 |
| Glucocorticoid receptor element (GRE) | 3.1 | 1.9 | 1.8 |
| Thyroid hormone receptor (TR) | 3.2 | 1.3 | 2.4 |
| CAAT box General (CEF) | 4.0 | 1.9 | 2.3 |
| Nuclear factor I (NF-1) | 4.0 | 1.7 | 2.3 |
| Stat3 | 4.4 | 1.8 | 3.0 |
| Progesterone receptor element (PRE) | 4.9 | 1.6 | 3.5 |
| Serum response element (SRE) | 7.4 | 0.9 | 9.0 |
| Myocyte enhancing factor 2 (MEF2) | 8.9 | 1.0 | 9.3 |
| FAST-1 | 9.9 | ND | |
| ISRE | 17.4 | ND | |
NOTE. Fold induction compared with control is presented for wild type strains and the dupA-deleted mutants. Assays were performed in triplicate, and mean value are presented.
ND, Not detected.
Susceptibility of H pylori to Low pH
The above in vivo experiments clearly show that the dupA status was associated with intestinal metaplasia and atrophy. Because intestinal metaplasia and atrophy were related to acid secretion, we evaluated the acid tolerance. We used wild-type strain (C142) and its dupA-deleted mutant and dupA-complemented mutant. Viable counts of H pylori grown at pH 6 for 24 hours were approximately 1010 CFU/mL, irrespective of the strains. We exposed H pylori to pH 3 BHI broth for 20 minutes after preexposure of pH 6 BHI broth for 24 hours. The dupA deleted mutant was more susceptible to pH 3 BHI broth than wild-type strains in 5 independent experiments (log10 killing = 6.6 ± 0.1 vs. 5.8 ± 0.1, respectively) (P < .001), and the dupA complemented mutant had susceptibility similar to the wild type (6.8 ± 0.1) (P < .001 compared with dupA-deleted mutant) (Figure 6). When H pylori was exposed to pH 4 BHI broth for 20 minutes, similar results were observed (Figure 6). In contrast, most strains survived exposure of pH 5 BHI broth, irrespective of the strain used (data not shown). The cag PAI-deleted knockout mutants were not related to acid tolerance (data not shown).
Figure 6.
Susceptibility of H pylori to exposure to low pH. Log10 kill represents the difference in viable H pylori before and after exposure to low pH. Assays were performed at least 4 times. Data are expressed as mean ± standard error.
We next examined 54 clinical isolates that were also used for in vitro IL-8 induction. We exposed H pylori to pH 3 BHI broth for 20 minutes after pre-exposure of pH 6 BHI broth for 24 hours. The dupA-negative strains were significantly more susceptible to pH 3 BHI broth than the dupA-positive strains (log10 killing = 6.4 ± 0.9 vs. 6.0 ± 0.9, respectively; P < .005). These findings suggest that the dupA-positive strains were resistant to high acid conditions (eg, DU) and were in agreement with the above in vivo data.
Discussion
Although a number of putative virulence factors of H pylori had been reported (eg, the cag PAI, vacA, babA, and oipA), their presence has typically been associated with an increased risk of both gastric cancer and peptic ulcers.1–10 We report an H pylori gene (jhp0917-0918: dupA) whose presence was related to increased risk of DU and with neutrophil infiltration as well as protection against atrophy, intestinal metaplasia, and gastric cancer. This pattern mirrors the clinical observation that the presence of a DU is known to be protective against gastric cancer and may eventually prove to solve that conundrum.43,44 To reduce known causes of bias associated with nonrandom selections of disease or by restricted circulation of a particular strain or strains that may produce spurious associations, we used both a large number of isolates as well as isolates from different geographic regions and clinical presentations.45 With the exception of strain J99, the jhp0917-jhp0918 gene forms 1 continuous reading frame and has been designated as the duodenal ulcer promoting (dupA) gene of H pylori. The results of the current study were consistent with the dupA gene being a marker for duodenal DU disease and also for a protective effect for atrophy, intestinal metaplasia, and gastric cancer. It is unknown whether either or both of these effects are directly related to the gene or whether the gene is a marker for other disease-modifying virulence factors. However, we believe that the results from complementation experiments strengthen our hypothesis that the dupA gene plays important roles in development of DU. The effects were consistent in both Asian and Western countries. We propose that, basically, the presence of the dupA gene is related to development of a pattern of nonatrophic gastritis. The presence of the dupA gene was also associated with increased IL-8 production from the antral gastric mucosa in vivo as well as from the gastric epithelial cells in vitro. We further confirmed that the dupA gene is involved in the activation of transcription factors responsible for IL-8 promoter. These results are in agreement with in vivo results showing higher neutrophil infiltration and IL-8 production in the antral gastric mucosa of DU patients compared with other diseases.32,46
However, more than half of all isolates from DU patients did not possess the dupA gene. In addition, the prevalence of the dupA gene was higher in Colombia compared with East Asia, irrespective of the disease outcomes, consistent with the notion that outcome results from a combination of factors including H pylori virulence factors, host genetics (eg, genetic polymorphisms affecting the immune response), and environmental factors (eg, diet or smoking) as well as combinations and interactions among various factors.
Of interest, preliminary in vitro experiments showed that the absence of the dupA gene was associated with increased susceptibility to low pH, and this observation was confirmed using the dupA-deleted and -complemented mutants. Changes in acid secretion, whether caused by drugs or by surgery, are associated with a change in the ability of H pylori to interact with the gastric mucosa.47,48 For example, in humans, it is known that antral predominant gastritis in DU patients spreads into the corpus after surgery to reduce acid (eg, selective vagotomy),47,49,50 suggesting that the ability of the organism to interact with acid may be an important outcome determinant. Subsequent experiments will be done to identify and characterize the pH-regulated genes that are responsible for the difference between the dupA-positive and the dupA-deleted mutants and whether there is a requirement for coinducers required in addition to pH stress.51
The dupA gene encodes homologues of VirB4 ATPase. Pfam search shows that jhp0917 region (position 3–201 of dupA) contains CagE_TrbE_VirB domain (Pfam accession number: PF03135) and FtsK/SpoIIIE family (PF01580). The region from the 3′ region jhp0917 to jhp0918 region (position 203–610) is homologue to TraG/TraD family (PF02534). The FtsK/SpoIIIE domain contains a putative ATP-binding P-loop motif and is involved in cell division and peptidoglycan synthesis or modification and is implicated in intercellular chromosomal DNA transfer.52,53 Members of the TraG/TraD family are potential NTP hydrolases that are essential for DNA transfer in bacterial conjugation and are thought to mediate interactions between the DNA processing and the mating pair formation systems.54,55 Many H pylori contain at least 2 functional, independent type IV transport systems: the secretion system for effector proteins such as CagA (cag PAI system) and the DNA import system via natural transformation (ComB system).28,56 –58 VirB4-homologous ATPases are contained within both systems (jhp0492/hp0544 [cagE] and jhp0015/hp0017 [comB4], respectively). Such ATPases typically provide energy for the correct assembly of the secretion apparatus or the delivery of the effector molecules. However, in A tumefaciens, it has been suggested that the oligomeric structure of VirB4, but not its capacity to bind ATP, is important for the assembly of VirB proteins as a DNA uptake system.59 It remains to be shown whether ATPase activity is necessary for the function of the dupA VirB4-homologues in type IV secretion system. The details of the relation between the dupA and type IV secretion remain to be elucidated, particularly because a search for genes in the plasticity region in strain J99 did not find partner genes with the dupA genes as components of a type IV secretion system. Kersulyte et al recently reported a new 16.3-kb segment (tfs3) located in the plasticity region in which 7 of the 16 ORFs were homologues of type IV secretion genes (virB4, virB7 to virB11, and virD4), the third putative type IV secretion system found in H pylori.23 However, we did not find any linkage between the presence of the dupA gene and full-length or partial tfs3 elements, which is in agreement with their report that the presence of the tfs3 elements was independent of clinical diseases.23 The function of the dupA gene is unknown; however, it might be possible that the dupA gene in combination with other novel vir homologue genes in the plasticity region might form a type IV secretion system similar to the cag PAI, which is also related to IL-8 secretion.
Preliminary data examining the effect of the presence of the dupA gene on transcription factors showed that activation of transcription factors for tumor suppressor gene p53 was decreased and activation of the YY1 was increased using the dupA-deleted mutants (Table 7). This pattern, shown in the dupA-deleted mutants, has been reported to be related to cancer development, with YY1 inhibiting activation of the p53 via binding competitively to the p53-binding site that contains the ACAT sequence.60 – 62 The activation of the Ets-1/PEA3 was induced by the absence of the dupA gene (Table 7). Ets-1/PEA3 is an oncogene and has been used as a marker of malignant potential in gastric cancer63 and also plays an important role in angiogenesis in the early stage of ulcer healing.64 Additional studies in vitro and confirmatory studies in vivo are needed to put these in vitro observations into a clinical context.
Acknowledgments
Supported in part by the Office of Research and Development Medical Research Service Department of Veterans Affairs (to D.Y.G.), by National Institutes of Health grants R01 DK62813 (to Y.Y.), and by Public Health Service grant DK56338, which funds the Texas Gulf Coast Digestive Diseases Center.
The authors thank Dr. Oscar Gutierrez (Universidad Nacional de Colombia, Bogota, Colombia), the late Dr. Jong G. Kim (Guru Hospital, Korea University College of Medicine, Seoul, Korea), Dr. Tadashi Kodama, Shoji Mitsufuji, and Dr. Takeshi Okanoue (Kyoto Prefectural University of Medicine, Kyoto, Japan) for providing clinical samples; Dr. Hala M. El-Zimaity (Baylor College of Medicine, Houston, Texas) for histologic analyses; an anonymous reviewer for giving us the clue to ask whether jhp0917 and jhp0918 form 1 continuous gene; Dr. Rainer Haas for providing H pylori/E coli shuttle vectors (pHel3); and Dr. Diane E. Taylor for providing chloramphenicol resistance gene cassette.
Abbreviations used in this paper
- AP-1
activator protein-1
- BHI
brain heart infusion
- CRE
cAMP responsive element
- CREB
CRE binding protein
- CPB
citrate-phosphate buffer
- DU
duodenal ulcer
- ELISA
enzyme-linked immunosorbent assay
- GU
gastric ulcer
- IL
interleukin
- ISRE
interferon-stimulated responsive element
- MOI
multiplicity of infection
- NF
nuclear factor
- PAI
pathogenicity island
- PCR
polymerase chain reaction
References
- 1.Graham DY, Yamaoka Y. H pylori and cagA: relationships with gastric cancer, duodenal ulcer, and reflux esophagitis and its complications. Helicobacter. 1998;3:145–151. doi: 10.1046/j.1523-5378.1998.08031.x. [DOI] [PubMed] [Google Scholar]
- 2.van Doorn LJ, Figueriedo C, Sanna R, Plaisier A, Schneeberger P, Boer WD, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 3.Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- 4.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atherton JC, Peek RM, Tham KT, Cover TL, Blaser MJ. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 6.Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter. 1998;4:241–253. doi: 10.1046/j.1523-5378.1998.08056.x. [DOI] [PubMed] [Google Scholar]
- 7.Prinz C, Schoniger M, Rad R, Becker I, Keiditsch E, Wagenpfeil S, Classen M, Rosch T, Schepp W, Gerhard M. Key importance of the Helicobacter pylori adherence factor blood group antigen binding adhesin during chronic gastric inflammation. Cancer Res. 2001;61:1903–1909. [PubMed] [Google Scholar]
- 8.Gerhard M, Lehn N, Neumayer N, Boren T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci U S A. 1999;96:12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaoka Y, Souchek J, Odenbreit S, Haas R, Arnqvist A, Borén T, Kodama T, Osato MS, Gutierrez O, Kim JG, Graham DY. Discrimination between cases of duodenal ulcer and gastritis on the basis of putative virulence factors of Helicobacter pylori. J Clin Microbiol. 2002;40:2244–2246. doi: 10.1128/JCM.40.6.2244-2246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaoka Y, Kikuchi S, El-Zimaity HMT, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori OipA in clinical presentation, gastric inflammation, and mucosal interleukin-8 production. Gastroenterology. 2002;123:414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 11.Ando T, Wassenaar TM, Peek RM, Jr, Aras RA, Tschumi AI, van Doorn LJ, Kusugami K, Blaser MJ. A Helicobacter pylori restriction endonuclease-replacing gene, hrgA, is associated with gastric cancer in Asian strains. Cancer Res. 2002;62:2385–2389. [PubMed] [Google Scholar]
- 12.Lu H, Graham DY, Yamaoka Y. Helicobacter pylori restriction endonuclease-replacing gene, hrgA, and clinical outcome in East Asia and Western countries. Dig Dis Sci. 2004;49:1551–1555. doi: 10.1023/b:ddas.0000042263.18541.ec. [DOI] [PubMed] [Google Scholar]
- 13.Graham DY. Campylobacter pylori and peptic ulcer disease. Gastroenterology. 1989;96(Suppl):615–625. doi: 10.1016/s0016-5085(89)80057-5. [DOI] [PubMed] [Google Scholar]
- 14.McColl KE, El-Omar E, Gillen D. Helicobacter pylori gastritis and gastric physiology. Gastroenterol Clin North Am. 2000;29:687–703. doi: 10.1016/s0889-8553(05)70138-2. [DOI] [PubMed] [Google Scholar]
- 15.Sipponen P, Stolte M. Clinical impact of routine biopsies of the gastric antrum and body. Endoscopy. 1997;29:671–678. doi: 10.1055/s-2007-1004278. [DOI] [PubMed] [Google Scholar]
- 16.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 17.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quacken-bush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenny K, Fitzegerald LM, Lee NM, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hays WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 18.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 19.Alm RA, Trust TJ. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J Mol Med. 1999;77:834–846. doi: 10.1007/s001099900067. [DOI] [PubMed] [Google Scholar]
- 20.Occhialini A, Marais A, Alm R, Garcia F, Sierra R, Megraud F. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2000;68:6240–6249. doi: 10.1128/iai.68.11.6240-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos A, Queiroz DM, Menard A, Marais A, Rocha GA, Oliveira CA, Nogueira AM, Uzeda M, Megraud F. New pathogenicity marker found in the plasticity region of the Helicobacter pylori genome. J Clin Microbiol. 2003;41:1651–1655. doi: 10.1128/JCM.41.4.1651-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jonge R, Kuipers EJ, Langeveld SC, Loffeld RJ, Stoof J, van Vliet AH, Kusters JG. The Helicobacter pylori plasticity region locus jhp0947-jhp0949 is associated with duodenal ulcer disease and interleukin-12 production in monocyte cells. FEMS Immunol Med Microbiol. 2004;41:161–167. doi: 10.1016/j.femsim.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Kersulyte D, Velapatino B, Mukhopadhyay AK, Cahuayme L, Bussalleu A, Combe J, Gilman RH, Berg DE. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J Bacteriol. 2003;185:3764 –3772. doi: 10.1128/JB.185.13.3764-3772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 26.Selbach M, Moese S, Meyer TF, Backert S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. Infect Immun. 2002;70:665–671. doi: 10.1128/iai.70.2.665-671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buhrdorf R, Forster C, Haas R, Fischer W. Topological analysis of a putative virB8 homologue essential for the cag type IV secretion system in Helicobacter pylori. Int J Med Microbiol. 2003;293:213–217. doi: 10.1078/1438-4221-00260. [DOI] [PubMed] [Google Scholar]
- 28.Carroll IM, Khan AA, Ahmed N. Revisiting the pestilence of Helicobacter pylori: insights into geographical genomics and pathogen evolution. Infect Genet Evol. 2004;4:81–90. doi: 10.1016/j.meegid.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 30.Glocker E, Lange C, Covacci A, Bereswill S, Kist M, Pahl HL. Proteins encoded by the cag pathogenicity island of Helicobacter pylori are required for NF-κB activation. Infect Immun. 1998;66:2346–2348. doi: 10.1128/iai.66.5.2346-2348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relation between clinical presentation, Helicobacter pylori density, interleukin-1β and -8 production and cagA status. Gut. 1999;45:804–811. doi: 10.1136/gut.45.6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genta RM, Robason GO, Graham DY. Simultaneous visualization of Helicobacter pylori and gastric morphology: a new stain. Hum Pathol. 1994;25:221–226. doi: 10.1016/0046-8177(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 34.El-Zimaity HMT, Ota H, Scott S, Killen DE, Graham DY. A new stain for Helicobacter pylori suitable for the autostainer. Arch Pathol Lab Med. 1998;122:732–736. [PubMed] [Google Scholar]
- 35.El-Zimaity HMT, Graham DY, Al-Assi MT, Malaty H, Karttunen TJ, Graham DP, Huberman RM, Genta RM. Interobserver variation in the histopathological assessment of Helicobacter pylori gastritis. Hum Pathol. 1996;27:35–41. doi: 10.1016/s0046-8177(96)90135-5. [DOI] [PubMed] [Google Scholar]
- 36.Hsu PI, Hwang IR, Cittelly D, Lai KH, El-Zimaity HMT, Gutierrez O, Kim JG, Osato MS, Graham DY, Yamaoka Y. Clinical presentation in relation to diversity within the Helicobacter pylori cag pathogenicity island. Am J Gastroenterol. 2002;97:2231–2238. doi: 10.1111/j.1572-0241.2002.05977.x. [DOI] [PubMed] [Google Scholar]
- 37.Occhialini A, Marais A, Urdaci M, Sierra R, Munoz N, Covacci A, Megraud F. Composition and gene expression of the cag pathogenicity island in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2001;69:1902–1908. doi: 10.1128/IAI.69.3.1902-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaoka Y, Kwon DH, Graham DY. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci U S A. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heuermann D, Haas R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol Gen Genet. 1998;257:519–528. doi: 10.1007/s004380050677. [DOI] [PubMed] [Google Scholar]
- 40.Yamaoka Y, Kudo T, Lu H, Casola A, Brasier A, Graham DY. Role of interferon stimulated responsive element-like element in interleukin-8 promoter in Helicobacter pylori infection. Gastroenterology. 2004;126:1030–1043. doi: 10.1053/j.gastro.2003.12.048. [DOI] [PubMed] [Google Scholar]
- 41.Yamaoka Y, El-Zimaity HMT, Gutierrez O, Figura N, Kim JG, Kodama T, Kashima K, Graham DY. Relationship between subtypes of the cagA 3′ repeat region, gastric histology, and susceptibility to low pH. Gastroenterology. 1999;117:342–349. doi: 10.1053/gast.1999.0029900342. [DOI] [PubMed] [Google Scholar]
- 42.Karita M, Blaser MJ. Acid-tolerance response in Helicobacter pylori and differences between cagA+ and cagA− strains. J Infect Dis. 1998;178:213–219. doi: 10.1086/515606. [DOI] [PubMed] [Google Scholar]
- 43.Sipponen P, Stolte M. Clinical impact of routine biopsies of the gastric antrum and body. Endoscopy. 1997;29:671–678. doi: 10.1055/s-2007-1004278. [DOI] [PubMed] [Google Scholar]
- 44.Molloy RM, Sonnenberg A. Relation between gastric cancer and previous peptic ulcer disease. Gut. 1997;40:247–252. doi: 10.1136/gut.40.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graham DY, Yamaoka Y. Disease-specific Helicobacter pylori virulence factors: the unfulfilled promise. Helicobacter. 2000;5(Suppl 1):S3–S9. doi: 10.1046/j.1523-5378.2000.0050s1003.x. [DOI] [PubMed] [Google Scholar]
- 46.Yamaoka Y, Kita M, Kodama T, Sawai N, Tanahashi T, Kashima K, Imanishi J. Chemokines in the gastric mucosa in Helicobacter pylori infection. Gut. 1998;42:609–617. doi: 10.1136/gut.42.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graham DY. Campylobacter pylori and peptic ulcer disease. Gastroenterology. 1989;96:615–625. doi: 10.1016/s0016-5085(89)80057-5. [DOI] [PubMed] [Google Scholar]
- 48.Graham DY, Opekun AR, Yamaoka Y, Osato MS, El Zimaity HM. Early events in proton pump inhibitor-associated exacerbation of corpus gastritis. Aliment Pharmacol Ther. 2003;17:193–200. doi: 10.1046/j.1365-2036.2003.01400.x. [DOI] [PubMed] [Google Scholar]
- 49.Roland M, Berstad A, Liavag I. A histological study of gastric mucosa before and after proximal gastric vagotomy in duodenal ulcer patients. Scand J Gastroenterol. 1975;10:181–186. [PubMed] [Google Scholar]
- 50.Peetsalu A, Maaroos HI, Sipponen P, Peetsalu M. Long-term effect of vagotomy on gastric mucosa and Helicobacter pylori in duodenal ulcer patients. Scand J Gastroenterol. 1991;186:77–83. [PubMed] [Google Scholar]
- 51.Hall HK, Karem KL, Foster JW. Molecular responses of microbes to environmental pH stress. Adv Microb Physiol. 1995;37:229–272. doi: 10.1016/s0065-2911(08)60147-2. [DOI] [PubMed] [Google Scholar]
- 52.Wu LJ, Errington J. Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 53.Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dougherty BA, Hill C, Weidman JF, Richardson DR, Venter JC, Ross RP. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol Microbiol. 1998;29:1029–1038. doi: 10.1046/j.1365-2958.1998.00988.x. [DOI] [PubMed] [Google Scholar]
- 55.Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofreuter D, Odenbreit S, Henke G, Haas R. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol Microbiol. 1998;28:1027–1038. doi: 10.1046/j.1365-2958.1998.00879.x. [DOI] [PubMed] [Google Scholar]
- 57.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 58.Smeets LC, Kusters JG. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends Microbiol. 2002;10:159–162. doi: 10.1016/s0966-842x(02)02314-4. [DOI] [PubMed] [Google Scholar]
- 59.Berger BR, Christie PJ. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci U S A. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Yakovleva T, Kolesnikova L, Vukojevic V, Gileva I, Tan-No K, Austen M, Luscher B, Ekstrom TJ, Terenius L, Bakalkin G. YY1 binding to a subset of p53 DNA-target sites regulates p53-dependent transcription. Biochem Biophys Res Commun. 2004;318:615–624. doi: 10.1016/j.bbrc.2004.04.065. [DOI] [PubMed] [Google Scholar]
- 63.Yu Y, Zhang YC, Zhang WZ, Shen LS, Hertzog P, Wilson TJ, Xu DK. Ets1 as a marker of malignant potential in gastric carcinoma. World J Gastroenterol. 2003;9:2154–2159. doi: 10.3748/wjg.v9.i10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ito M, Nakayama T, Naito S, Matsuu M, Shichijo K, Sekine I. Expression of Ets-1 transcription factor in relation to angiogenesis in the healing process of gastric ulcer. Biochem Biophys Res Commun. 1998;246:123–127. doi: 10.1006/bbrc.1998.8585. [DOI] [PubMed] [Google Scholar]