Abstract
Nitration of tyrosine (Y) residues of proteins is a low abundant post-translational modification that modulates protein function or fate in animal systems. However, very little is known about the in vivo prevalence of this modification and its corresponding targets in plants. Immunoprecipitation, based on an anti-3-nitroY antibody, was performed to pull-down potential in vivo targets of Y nitration in the Arabidopsis thaliana proteome. Further shotgun liquid chromatography–mass spectrometry (LC-MS/MS) proteomic analysis of the immunoprecipitated proteins allowed the identification of 127 proteins. Around 35% of them corresponded to homologues of proteins that have been previously reported to be Y nitrated in other non-plant organisms. Some of the putative in vivo Y-nitrated proteins were further confirmed by western blot with specific antibodies. Furthermore, MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) analysis of protein spots, separated by two-dimensional electrophoresis from immunoprecipitated proteins, led to the identification of seven nitrated peptides corresponding to six different proteins. However, in vivo nitration sites among putative targets could not be identified by MS/MS. Nevertheless, an MS/MS spectrum with 3-aminoY318 instead of the expected 3-nitroY was found for cytosolic glyceraldehyde-3-phosphate dehydrogenase. Reduction of nitroY to aminoY during MS-based proteomic analysis together with the in vivo low abundance of these modifications made the identification of nitration sites difficult. In turn, in vitro nitration of methionine synthase, which was also found in the shotgun proteomic screening, allowed unequivocal identification of a nitration site at Y287.
Keywords: AminoY, Arabidopsis, nitric oxide, nitrotyrosine, nitroY, post-translational modification, protein nitration
Introduction
During the last 20 years, nitric oxide (NO) has been characterized as an essential regulator of many physiological processes in animals. More recently, NO has been characterized as a signal molecule regulating plant defence against pathogens (Romero-Puertas et al., 2004; Mur et al., 2006), resistance to abiotic stress (Zhang et al., 2006), and different developmental processes including seed dormancy and germination (Bethke et al., 2006; Liu et al., 2007), floral transition (He et al., 2004; Simpson, 2005), and leaf senescence (Mishina et al., 2007). NO acts as a regulator of gene expression at the transcriptional level by regulating disease resistance processes (Polverari et al., 2003) and the expression of stress-related transcription factors and signalling-related kinases (Parani et al. 2004), and by the interaction with other signalling molecules such as salicylic acid and jasmonic acid (Grün et al., 2006).
Some of the regulatory properties of NO are exerted through NO-mediated post-translational modifications including nitrosylation of thiol groups and nitration of tyrosine (Y) residues (Gow et al., 2004). This is thought to affect the activity, the stability, or the intracellular location of proteins, thus potentially altering their functions and eventually cell signalling. The regulation of protein function at the levels of NO-related post-translational modifications represents a new area of research in plant biology, and it will help to elucidate the mode of action of NO in regulating many processes in plants. Recent reports suggest that S-nitrosylation is specific and regulated (Lindermayr et al., 2005; Romero-Puertas et al., 2008), and it may play a regulatory role in central processes in plants such as ethylene biosynthesis (Lindermayr et al., 2006). The interaction between NO and superoxide leads to the formation of peroxynitrite, a reactive molecule with strong nitrating activity (Szabó et al., 2007). The production of peroxynitrite under physiological conditions in plants has been reported (Bechtold et al., 2009; Chaki et al., 2009). Some proteins are targets of peroxynitrite, and the nitration of Y residues to 3-nitrotyrosine represents a hallmark of post-translational protein modification associated with human pathologies and biological ageing (Hong et al., 2007). Although well characterized in mammals, scant information is available on nitration of Y residues of proteins in plants. Detection of nitrated proteins was first reported in tobacco plants with reduced nitrite reductase activity (Morot-Gaudry-Talarmain et al., 2002). Later, the detection of in vivo nitrated proteins in plants treated with exogenous nitrating reagents (Saito et al., 2006) as well as under physiological conditions in both unstressed conditions (Chaki et al., 2009) and upon pathogen challenge (Romero-Puertas et al., 2007; Cecconi et al., 2009) was reported. However, in all these recent reports there are no data about unequivocal identification of nitrated peptides or proteins (i.e. nitration sites). Here the identification of potential in vivo nitration sites of some Arabidopsis proteins is reported. Drawbacks in proteomic approaches to identify Y nitration post-translational modification under physiological conditions are also discussed. The analysis of the regulatory functions of Y nitration of proteins in any plant biological process will require, after initial identification of potential targets, a case-by-case analysis. Recent proteomic approaches based on the protection of the primary amino group by acetylation followed by the reduction of nitroY to aminoY residues, and further derivatization of the amino group from aminoY residues (Chiapetta et al., 2009; Tsumoto et al., 2010), will help to overcome some of the difficulties found due to the low abundance and limited stability of nitroY residues in proteins determined to be potentially nitrated in vivo in this work.
Materials and methods
Plant growth conditions
Seeds of the Col-0 wild-type accession of Arabidopsis thaliana were sown in moistened soil and grown under photoperiodic conditions (cycles of 8 h day and 16 h night for short days, at 22 °C and 20 °C, respectively) as mentioned in different experiments. Plants were illuminated with 150 μE m−2 s−1 cool white fluorescent lamps and grown under 60% relative humidity. Alternatively, surface-sterilized seeds were germinated and grown in sterile liquid or agar-supplemented Murashige and Skoog (MS) medium (Duchefa, Haarlem, The Netherlands) with 1% (w/v) sucrose.
Protein extraction and immunoprecipitation
Two-week-old seedlings were frozen and ground in liquid nitrogen. Proteins were extracted by adding extraction buffer [10 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 1% (v/v) protease inhibitor cocktail from Sigma, USA] and briefly vortexing. Protein extracts were obtained by centrifugation at 13 000 g at 4 °C. Protein extracts (4× 1 mg) were pre-cleared with 50 μl of protein A–agarose (EZView Sigma, USA) for 8 h at 4 °C. The unbound fractions were each incubated overnight with 0.1 μg of monoclonal anti-3-nitroY antibody (Cayman, USA) at 4 °C. To recover 3-nitroY-containing proteins, 60 μl of protein A–agarose were added and incubated for 8 h at 4 °C. After extensive washing with extraction buffer, proteins were eluted at 95 °C with elution buffer [1% SDS, 100 mM dithiothreitol (DTT), 50 mM TRIS-HCl pH 7.6] three times. After removing agarose beads with a 0.2 μm filter (Costar Corning, NY, USA), the proteins were precipitated, combined, and processed with a 2D-Clean Up Kit (GE, UK) for subsequent two-dimensional electrophoresis (2-DE) and liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
2-DE and image analysis
Protein samples (100 μg) were dissolved in DeStreak Rehydration solution (GE, UK) before electrophoresis. For the first dimension, 18 cm pH 3–10 NL (non-linear) strips were passively rehydrated overnight at room temperature. The set-up of the IPGphor3 (GE, UK) was 1 h at 50 V step-and-hold, 1 h at a 150 V gradient, 1 h 30 min at a 500 V gradient, 1 h 30 min at a 1000 V gradient, 2 h at a 4000 V gradient, 2 h at a 8000 V gradient, and 7 h at a 8000 V step-and-hold. The strips were then treated with 1 mg ml−1 DTT for 15 min and alkylated with 25 mg ml−1 iodoacetamide for 15 min in equilibration buffer (6 M urea, 75 mM TRIS-HCl pH 8.8, 29.3% glycerol, 2% SDS, and 0.002% bromophenol blue), and the focused proteins were then separated on 12.5% acrylamide gels in the EttanDalt six electrophoresis unit (GE, UK) as recommended by the manufacturers for an overnight run. The gels were stained with a DeepPurple (GE, UK) or PlusOne™ Silver Staining Kit (GE, UK), digitalized with Typhoon (GE, UK), and analysed by using Image Master Platinum 5.0 (GE, UK) software.
MS analysis
Samples were digested with sequencing grade trypsin (Promega, USA). Peptide separation by LC-MS/MS was performed using an Ultimate nano-LC system (LC Packings) and a QSTAR XL Q-TOF hybrid mass spectrometer (MDS Sciex-Applied Biosystems). Samples (5 μl) were delivered to the system using a FAMOS autosampler (LC Packings) at 40 μl min−1, and the peptides were trapped on a PepMap C18 pre-column (5 mm, i.d. 300 m; LC Packings). Peptides were then eluted from a PepMap C18 analytical column (15 cm, i.d. 75 m; LC Packings) at 200 nl min−1 and separated using a 55 min gradient of 15–50% acetonitrile (120 min for the mixtures). The QSTAR XL was operated in information-dependent acquisition mode, in which a 1 s time of flight (TOF) MS scan from 400 m/z to 2000 m/z, was performed, followed by 3 s product ion scans from 65 m/z to 2000 m/z on the three most intense doubly or triply charged ions. A database search on Swiss-Prot and NCBInr databases was performed using the MASCOT search engine (Matrix-Science). Searches were done with tryptic specificity allowing one missed cleavage and a tolerance on the mass measurement of 100 ppm in MS mode and 0.8 Da for MS/MS ions. Carbamidomethylation of C was used as a fixed modification, and oxidation of M, deamidation of D and E, and nitration or amination of Y as variable modifications.
Western blot
Protein extracts (10 μg) were separated by SDS–PAGE, blotted onto a nitrocellulose membrane, stained with Ponceau-S, and probed with antibodies at the followed dilutions: monoclonal anti-3-nitroY (Cayman Chemicals) 1:1000, anti-5×His (QIAGEN) 1:8000, polyclonal anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) 1:10 000, anti-CA (carbonic anhydrase) 1:3000, anti-PKL (PICKLE) 1:5000; anti-FBPase (fructose bisphosphatase) 1:2000, and anti-GRP (glycine-rich RNA-binding protein) 1:2500. Secondary antibody was anti-mouse or anti-rabbit, for monoclonal or polyclonal primary antibodies, respectively, coupled to horseradish peroxidase (GE, UK) at 1:10 000 dilution, and an ECL kit (GE, UK) was used for visualization of proteins.
GAPDH activity
Proteins were extracted in 50 mM TRIS-HCl pH 7.4 and quantified. GAPDH activity of the extracts was assayed according to Muñoz-Bertomeu et al. (2009) with minor modifications. Briefly, 50 μg of protein extracts from plants treated or not with 2 mM SIN-1 (3-morpholinosydnonimine) were incubated in reaction buffer (10 mM TRIS-HCl pH 7.4, 20 mM arsenate, 2 mM NAD, 0.5 mM DTT) and the reaction was initiated by the addition of 2 mM DL-GAPDH in a final volume of 1 ml. GAPDH activity was measured following the conversion of NAD to NADH at 340 nm during 4 min.
Synthesis, purification, and nitration of His-tagged methionine synthase AtMS1
A plasmid containing AtMS1 cDNA fused to a 6×His tag (Dixon et al., 2005) was used to transform BL21(DE3) competent cells (Sigma-Aldrich) for recombinant protein production. For protein induction, cell cultures with OD=0.7 were treated with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 5 h. Recombinant protein production was checked by SDS–PAGE and western blot analysis. Recombinant protein purification was carried out with the QIAexpress Ni-NTA Fast Start Kit (Qiagen) following the manufacturer's recommendations. Purified AtMS1 was treated or not with a nitrating buffer as described previously (Chen et al., 2008). Briefly, 10 μl of purified protein was incubated with 500 μM H2O2 and 500 μM NaNO2 in 0.1 M potassium phosphate buffer pH 7.2 for NO2 radical-mediated protein nitration at 37 °C in the dark for 2 h in a total volume of 500 μl. To clean nitrated protein, the nitrating reaction volume was filtered trough a 10 kDa cut-off filter (Microcon, Ambion). Proteins were then analysed by SDS–PAGE and western blot. Protein nitration was confirmed with anti-3nitroY antibody (Cayman) and the anti-5×His antibody supplied by the manufacturers (QIAGEN). A duplicate gel was run and stained with Coomassie blue, and the bands were excised, trypsin digested, and further analysed by LC-MS/MS as described above.
Protein modelling and structural analysis
Three-dimensional (3-D) protein models were generated by homology modelling at the SWISS-MODEL workspace (Arnold et al., 2006) using the coordinates of GAPDH from rat (PDB code 2VYN), serine hydroxymethyltransferase from Mycobacterium tuberculosis (PDB code 3H7F), transketolase from maize (PDB code 1ITZ), Rubisco from spinach (PDB code 1IR1), and mannitol dehydrogenase from Cladosporium harbarum (PDB code 3GDF) as templates. For methionine synthase, the crystal structure from A. thaliana was used (PDB code 1U1J). Model qualities were evaluated by ANOLEA, Verify3D, and Procheck (Melo and Feytmans, 1998; Bowie et al., 1991; Laskowski et al., 1996, respectively). 3-D models were visualized and manipulated with Yasara (www.yasara.org) or PyMol (www.pymol.org). The distance between residues in Amstrongs (Å) and the presence of hydrogen bonds were analysed with both programs using default settings.
Results
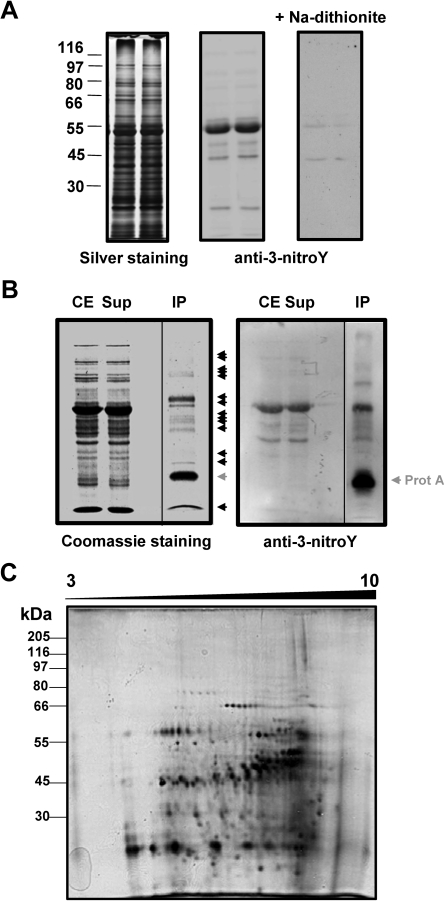
Crude protein extracts from A. thaliana plants contained a number of proteins spanning the whole range of molecular weights that cross-react with antibodies against 3-nitroY in western blot analysis (Fig. 1A). The specific cross-reaction of antibodies with 3-nitroY residues of those target proteins was checked by on-membrane protein reduction of 3-nitroY to 3-aminoY with sodium dithionite, as previously reported (Miyagi et al., 2002), thus resulting in no cross-reaction with the specific antibodies (Fig. 1A). Upon antibody validation, anti-3-nitroY antibodies were used as a specific immunoprecipitation reagent together with protein A–agarose to pull-down 3-nitroY-containing proteins from crude Arabidopsis seedling extracts. Figure 1B shows that a small number of proteins present in the crude extracts, <20 bands as detected by Coomassie staining, were recovered in the immunoprecipitated fraction. Those proteins were further checked for cross-reaction in western blots with anti-3-nitroY antibodies (Fig. 1B). A moderate enrichment in nitrated proteins was thus observed in the immunoprecipitated fraction (Fig. 1B). Considering the low resolution capacity of one-dimensional SDS–PAGE, the complexity of the immunopurified samples was further assessed by 2-DE and the more sensitive silver staining, resulting in the separation of ∼450 spots with isoelectric points in the 3–10 range (Fig. 1C).
Fig. 1.
Detection of 3-nitroY-containing proteins. (A) Crude protein extracts (10 μg per lane) were separated using 10% SDS–PAGE in duplicate. The left panel shows the silver-stained gel with the position of a molecular weight protein ladder. The central panel shows the corresponding western blot performed with anti-3-nitroY primary antibody, and the right panel the corresponding western blot after reduction of 3-nitroY to 3-aminoY with 100 mM sodium dithionite for 30 min. (B) In vivo immunoprecipitation of Arabidopsis 3-nitroY-containing proteins. Crude extracts (CE) were immunoprecipitated with antibody against 3-nitroY. The resulting supernatants (Sup) and immunoprecipitated proteins (IP) alongside the CE were separated by one-dimensional SDS–PAGE in duplicate and either Coomassie stained (left panel) or transferred to a nitrocellulose membrane and probed with anti-3-nitroY antibodies by western blot (right panel). Immunoprecipitated proteins detected by one-dimensional SDS–PAGE are marked with black arrowheads. The protein A which is released from the resin in the immunoprecipitates is marked with a grey arrowhead. (C) Immunoprecipitated proteins (0.1 mg) were separated by 2-DE with isoelectric focusing in the range of pH 3–10 NL and a second dimension 10% gel. The identification of spots corresponding to nitrated proteins was performed by comparing four independent sets of 2-DE gels corresponding to biologically independent replicates with similar spot patterns. Molecular mass marker positions are indicated in kDa on the left side. Proteins were silver stained.
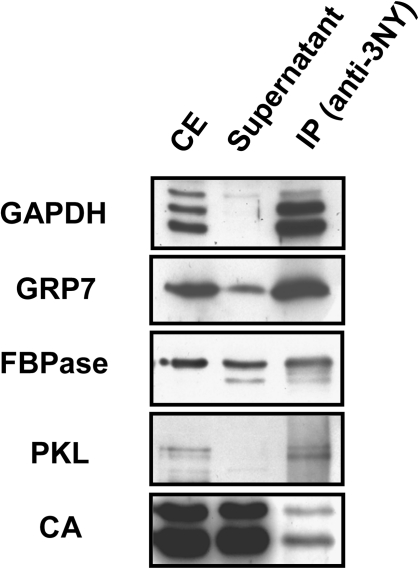
To identify potential in vivo targets of Y nitration in Arabidopsis, the immunopurified fraction was then analysed by MS following two different strategies. First, a shotgun analysis, based on LC-MS/MS of the immunoprecipitated proteins, was performed. Comparison of MS-generated data with the SwissProt database by specifying taxonomy for Arabidopsis allowed identification of 127 proteins with a statistically significant MASCOT score of at least 35 and more than two matched peptides (Table 1). Among identified proteins, 35% have homologue counterparts that have been previously reported as nitrated in non-plant organisms (Supplementary Table S1 available at JXB online), thus supporting the usefulness of the immunoprecipitation approach to enrich the purified fractions in Arabidopsis potential nitrated proteins. To validate the proteomic identification further, several of the identified proteins were detected by western blots with specific antibodies in the samples immunopurifed by precipitation with anti-3-nitroY antibodies. Some proteins identified with a MASCOT score >200, such as chloroplastic GAPDH, CA, or FBPase, and some others with a lower score such as GRP7 (score 66) and the CHD3-type chromatin remodelling factor PKL (score 58) were selected. All of them cross-reacted with proteins in the 3-nitroY-immunoprecipitated samples (Fig. 2), making the proteomic identification reliable. For proteins such as GAPDH or PKL showing no signal in the supernatant, most of the corresponding proteins were associated with IP resin and a significant proportion further recovered after washing in the IP. In contrast, the immunoprecipitation is far less efficient for others proteins such as FBPase or CA which show a similar amount of protein in the supernatant and crude extract, thus suggesting that the corresponding nitrated forms would not be abundant in the total protein population of crude extracts. Most of the proteins tested gave complex patterns of cross-reacting bands in both crude extracts and immunopurified samples (Fig. 2, Supplementary Fig. S5 at JXB online). This is probably due to different cross-reactive isoforms that are the result of potential post-translational modifications or to unespecific cross-reaction of the antibody.
Table 1.
Immunopurified Y-nitrated proteins identified in Arabidopsis thaliana seedlings by a shotgun LC-MS/MS approach Those proteins that have been previously reported as nitrated in other plant systems are been marked with a single (Chaki et al., 2009) or double asterisk (Cecconi et al., 2009).
| SwissProt locus | AGI code | Description | MASCOT score | Peptides matched (no.) | Best two peptides (ion score) |
| ATPB_ARATH | AtCg00480 | ATP synthase subunit beta | 1150 | 23 | R.FVQAGSEVSALLGR.M (85) |
| K.IGLFGGAGVGK.T (80) | |||||
| METE_ARATH | At5g17920 | Methionine synthase 1 | 1014 | 27 | K.DEALFSANAAALASR.R (97) |
| K.MLAVLEQNILWVNPDCGLK.T(91) | |||||
| G3PB_ARATH | At1g42970 | GAPDH B, chloroplast | 884 | 22 | K.IVDNETISVDGK.L (85) |
| R.KDSPLEVVVLNDSGGVK.N (75) | |||||
| G3PA_ARATH | At3g26650 | GAPDH A, chloroplast | 831 | 18 | R.VPTPNVSVVDLVVQVSK.K (68) |
| K.KVIITAPGK.G (60) | |||||
| RCA_ARATH | At2g39730 | Rubisco activase, chloroplast** | 761 | 20 | R.GLAYDTSDDQQDITR.G (81) |
| R.VQLAETYLSQAALGDANADAIGR.G (72) | |||||
| GOX1_ARATH | At3g14415 | Probable peroxisomal glycolate oxidase1 | 670 | 16 | R.AASAAGTIMTLSSWATSSVEEVASTGPGIR.F (101) |
| K.DIQWLQTITNMPILVK.G (58) | |||||
| GOX2_ARATH | At3g14420 | Probable peroxisomal glycolate oxidase2 | 651 | 16 | R.AASAAGTIMTLSSWATSSVEEVASTGPGIR.F (101) |
| R.IPVFLDGGVR.R (52) | |||||
| SAHH1_ARATH | At4g13940 | Adenosyl homocysteinase 1* | 581 | 18 | K.VALLHLGK.L (55) |
| R.DSAAVFAWK.G (54) | |||||
| PGKH_ARATH | At1g56190 | Phosphoglycerate kinase, chloroplast | 542 | 14 | K.LASLADLYVNDAFGTAHR.A (77) |
| K.FAAGTEAIANK.L (75) | |||||
| ATPA_ARATH | AtCg00120 | ATP synthase subunit alpha** | 504 | 12 | R.EAYPGDVFYLHSR.L (64) |
| R.EQHTLIIYDDLSK.Q (62) | |||||
| EFTU_ARATH | At4g20360 | Elongation factor Tu, chloroplast | 491 | 13 | K.KYDEIDAAPEER.A (72) |
| R.SYTVTGVEMFQK.I (54) | |||||
| G3PC_ARATH | At3g04120 | GAPDH C, cytosolic | 479 | 13 | R.VPTVDVSVVDLTVR.L (71) |
| K.KVVISAPSK.D (52) | |||||
| CAHC_ARATH | At3g01500 | Carbonic anhydrase 1, chloroplast | 475 | 13 | K.YGGVGAAIEYAVLHLK.V (64) |
| R.EAVNVSLANLLTYPFVR.E (60) | |||||
| EF1A_ARATH | At1g07940 | Elongation factor 1-alpha | 450 | 11 | R.EHALLAFTLGVK.Q (103) |
| K.FHINIVVIGHVDSGK.S (82) | |||||
| ACT7_ARATH | At5g09810 | Actin-7 | 448 | 12 | K.SEYDESGPSIVHR.K (75) |
| K.NYELPDGQVITIGAER.F (57) | |||||
| ACT2_ARATH | At3g18780 | Actin-2 | 430 | 12 | K.NYELPDGQVITIGAER.F (57) |
| K.AGFAGDDAPR.A (52) | |||||
| KPPR_ARATH | At1g32060 | Phosphoribulokinase, chloroplast | 418 | 13 | R.LDELIYVESHLSNLSTK.F (55) |
| K.ILVIEGLHPMFDER.V (52) | |||||
| RUBB_ARATH | At1g55490 | Rubisco large subunit beta | 389 | 13 | R.GYISPYFVTDSEK.M (71) |
| K.YEDLMAAGIIDPTK.V (52) | |||||
| CAH2_ARATH | At5g14740 | Carbonic anhydrase 2 | 379 | 11 | R.EAVNVSLANLLTYPFVR.E (60) |
| K.VENIVVIGHSACGGIK.G (59) | |||||
| TBA6_ARATH | At4g14960 | Tubulin alpha-6 chain | 358 | 11 | R.AVFVDLEPTVIDEVR.T (67) |
| R.LVSQVISSLTASLR.F (50) | |||||
| METK1_ARATH | At1g02500 | S-Adenosyl methionine synthetase 1 | 334 | 11 | R.FVIGGPHGDAGLTGR.K (73) |
| K.IIIDTYGGWGAHGGGAFSGK.D (64) | |||||
| RUBA_ARATH | At2g28000 | Rubisco large subunit alpha, chloroplast | 331 | 11 | K.VVNDGVTIAR.A (60) |
| K.TNDSAGDGTTTASILAR.E (56) | |||||
| METK2_ARATH | At4g01850 | S-Adenosyl methionine synthetase 2 | 326 | 11 | R.FVIGGPHGDAGLTGR.K (73) |
| K.IIIDTYGGWGAHGGGAFSGK.D (64) | |||||
| GLNA2_ARATH | At5g35630 | Glutamine synthetase, chloroplast/mitochondrial** | 314 | 10 | K.VSGEVPWFGIEQEYTLLQQNVK.W (76) |
| K.HETASIDQFSWGVANR.G (42) | |||||
| SGAT_ARATH | At2g13360 | Serine-glyoxylate aminotransferase | 306 | 10 | R.AALDLIFEEGLENIIAR.H (61) |
| K.VFFDWNDYLK.F (42) | |||||
| RBS1A_ARATH | At1g67090 | Rubisco small subunit 1A, chloroplast | 299 | 9 | K.LPLFGCTDSAQVLK.E (71) |
| K.EVDYLIR.N (46) | |||||
| TBA3_ARATH | At5g19770 | Tubulin alpha-3/alpha-5 chain | 284 | 8 | R.AVFVDLEPTVIDEVR.T (67) |
| R.LISQIISSLTTSLR.F (65) | |||||
| PORB_ARATH | At4g27440 | Protochlorophyllide reductase B | 263 | 12 | R.LLLDDLKK.S (53) |
| K.GYVSETESGKR.L (46) | |||||
| RBS1B_ARATH | At5g38430 | Rubisco small subunit 1B, chloroplast | 254 | 7 | K.LPLFGCTDSAQVLK.E (71) |
| K.EVDYLLR.N (46) | |||||
| ILV5_ARATH | At3g58610 | Ketol-acid reductoisomerase, chloroplast | 240 | 9 | K.VSLAGYEEYIVR.G (44) |
| K.APVSLDFETSVFK.K (43) | |||||
| TBB4_ARATH | At5g44340 | Tubulin beta-4 chain | 226 | 8 | K.LAVNLIPFPR.L (54) |
| R.YLTASAVFR.G (35) | |||||
| HSP71_ARATH | At5g02500 | Heat shock cognate 70 kDa protein 1* | 217 | 10 | R.MVNHFVQEFK.R (40) |
| K.ATAGDTHLGGEDFDNR.M (35) | |||||
| F16P1_ARATH | At3g54050 | Fructose-1,6-bisphosphatase | 214 | 10 | R.TLLYGGIYGYPR.D (58) |
| R.VLDIQPTEIHQR.V (42) | |||||
| TBB2_ARATH | At5g62690 | Tubulin beta-2/beta-3 chain | 203 | 9 | K.LAVNLIPFPR.L (54) |
| R.AVLMDLEPGTMDSLR.S (35) | |||||
| TBB1_ARATH | At1g75780 | Tubulin beta-1 chain | 193 | 8 | K.LAVNLIPFPR.L (54) |
| R.AVLMDLEPGTMDSIR.S (35) | |||||
| PGMP_ARATH | At5g51820 | Phosphoglucomutase, chloroplast | 173 | 9 | K.SLPTKPIEGQK.T (30) |
| K.LPFFEVPTGWK.F (26) | |||||
| P2SAF_ARATH | At5g23120 | Photosystem II stability/assembly factor HCF136 | 172 | 8 | R.ADGGLWLLVR.G (40) |
| K.GTGITEEFEEVPVQSR.G (34) | |||||
| HSP73_ARATH | At3g09440 | Heat shock cognate 70 kDa protein 3* | 172 | 7 | R.MVNHFVQEFK.R (40) |
| K.ATAGDTHLGGEDFDNR.M (35) | |||||
| APX1_ARATH | At1g07890 | L-Ascorbate peroxidase 1, cytosolic | 161 | 5 | K.EGLLQLVSDK.A (44) |
| K.QMGLSDKDIVALSGAHTLGR.C (35) | |||||
| MTDH_ARATH | At4g39330 | Probable mannitol dehydrogenase | 139 | 5 | K.NYGGYSENIVVDQR.F (47) |
| K.NYGGYSENIVVDQR.F (34) | |||||
| CD48A_ARATH | At3g09840 | Cell division control protein 48 A | 120 | 6 | R.KGDLFLVR.G (29) |
| R.IVSQLLTLMDGLK.S (29) | |||||
| GME_ARATH | At5g28840 | GDP-mannose 3,5-epimerase | 112 | 5 | R.SFTFIDECVEGVLR.L (43) |
| K.KLPIHHIPGPEGVR.G (31) | |||||
| GBLP_ARATH | At1g18080 | Guanine nucleotide-binding protein subunit beta | 103 | 4 | R.LWDLAAGVSTR.R (42) |
| K.DGVVLLWDLAEGK.K (27) | |||||
| CLPP_ARATH | AtCg00670 | ATP-dependent Clp protease | 99 | 2 | R.SPGEGDTSWVDIYNR.L (70) |
| R.TGKPIWVISEDMER.D (30) | |||||
| GCST_ARATH | At1g11860 | Aminomethyltransferase, mitochondrial | 99 | 5 | K.GGDVSWHIHDER.S (25) |
| R.AEGGFLGADVILQQLK.D (24) | |||||
| AAT5_ARATH | At4g31990 | Aspartate aminotransferase, chloroplast | 98 | 5 | K.ATAELLFGAGHPVIK.E (27) |
| R.VATIQGLSGTGSLR.L (24) | |||||
| ACA9_ARATH | At3g21180 | Ca-transporting ATPase 9, plasma membrane | 98 | 7 | R.VAIDSMAK.N (28) |
| R.QAALVLNASRR.F (21) | |||||
| RH56_ARATH | At5g11200 | DEAD-box ATP-dependent RNA helicase 56 | 97 | 5 | K.LSEMEKNR.K (30) |
| K.VSVFYGGVNIK.I (25) | |||||
| ENO_ARATH | At2g36530 | Enolase | 96 | 6 | K.AGAVVSGIPLYK.H (30) |
| K.LAMQEFMILPVGAASFK.E (30) | |||||
| MRP7_ARATH | At3g13100 | Multidrug resistance-associated protein 7 | 86 | 7 | R.YGPHLPMVLRGLTCTFR.G (20) |
| R.GIEAGWLK.K (17) | |||||
| AFB3_ARATH | At1g12820 | AUXIN SIGNALLING F-BOX 3 | 84 | 6 | R.LWILDSIGDK.G (23) |
| R.LMSCAPQLVDLGVGSYENEPDPESFAK.L (17) | |||||
| PDX13_ARATH | At5g01410 | Pyridoxal biosynthesis protein | 79 | 4 | K.VGLAQMLR.G (43) |
| R.NMDDDEVFTFAK.K (14) | |||||
| PDX11_ARATH | At2g38230 | Pyridoxal biosynthesis protein | 75 | 3 | K.VGLAQMLR.G (43) |
| K.IAAPYDLVVQTK.E (20) | |||||
| EFTM_ARATH | At4g02930 | Elongation factor Tu, mitochondrial | 75 | 2 | R.GSALSALQGTNDEIGR.Q (49) |
| K.LMDAVDEYIPDPVR.V (26) | |||||
| MDR11_ARATH | At3g28860 | Multidrug resistance protein 11 (P-glycoprotein 19) | 73 | 6 | K.SSVIAMIER.F (24) |
| R.AVLKNPTVLLLDEATSALDAESECVLQEALERLMR.G (22) | |||||
| MDHP_ARATH | At3g47520 | Malate dehydrogenase, chloroplast | 70 | 3 | K.DVNVVVIPAGVPR.K (35) |
| K.LFGVTTLDVVR.A (22) | |||||
| SR54C_ARATH | At5g03940 | Signal recognition particle 54 kDa protein, chloroplast | 70 | 5 | R.GVKPDQQLVK.I (16) |
| R.QEDAEDLQKK.I (16) | |||||
| MDHG1_ARATH | At5g09660 | Malate dehydrogenase, glyoxysomal | 70 | 3 | R.TGAEEVYQLGPLNEYER.I (31) |
| K.LLGVTTLDVAR.A (30) | |||||
| TAF1B_ARATH | At3g19040 | Transcription initiation factor TFIID subunit 1-B | 69 | 7 | R.ENLKQLNSDARGR.L (20) |
| K.EIGTPICQMKKILK.E (17) | |||||
| TYW23_ARATH | At4g04670 | tRNA wybutosine-synthesizing protein | 69 | 5 | R.ADPLNILNDVWR.L (24) |
| K.RVIIAIRCSIR.M (15) | |||||
| CATA3_ARATH | At1g20620 | Catalase-3 | 69 | 3 | R.LGPNYLQLPVNAPK.C (32) |
| K.GFFEVTHDISNLTCADFLR.A (28) | |||||
| KASC1_ARATH | At5g46290 | 3-Oxoacyl-[acyl-carrier-protein] synthase I, chloroplast | 68 | 3 | K.LLSGESGISLIDR.F (53) |
| R.ADGLGVSSCIER.C (9) | |||||
| ATPG1_ARATH | At4g04640 | ATP synthase gamma chain 1, chloroplast | 68 | 2 | R.ALQESLASELAAR.M (52) |
| R.ASSVSPLQASLRELR.D (16) | |||||
| GRP7_ARATH | At2g21660 | Glycine-rich RNA-binding protein 7 | 66 | 1 | R.ALETAFAQYGDVIDSK.I (66) |
| FDH_ARATH | At5g14780 | Formate dehydrogenase, mitochondrial | 66 | 5 | R.QAVVDAVESGHIGGYSGDVWDPQPAPK.D (18) |
| R.LQMAPELEK.E (17) | |||||
| HSP83_ARATH | At5g56010 | Heat shock protein 81-3* | 62 | 5 | K.GIEVLYMVDAIDEYAIGQLK.E (21) |
| K.EGQNDIFYITGESK.K (16) | |||||
| TGA2_ARATH | At5g06950 | Transcription factor TGA2 | 61 | 4 | K.LTQLEQELQR.A (19) |
| R.LQTLQQMIR.V (15) | |||||
| TCPA_ARATH | At3g20050 | T-complex protein 1 subunit alpha | 61 | 6 | R.NKIHPTSIISGYR.L (19) |
| R.GANDYMLDEMER.A (15) | |||||
| CAPP3_ARATH | At3g14940 | Phosphoenolpyruvate carboxylase 3 | 60 | 4 | K.LLVSEDLWAFGEKLR.A (22) |
| K.RLVSDLGK.S (15) | |||||
| WRK19_ARATH | At4g12020 | WRKY transcription factor 19 | 60 | 6 | K.CTYLGCPSKRK.V (19) |
| K.LCQVEGCQKGAR.D (16) | |||||
| THI4_ARATH | At5g54770 | Thiazole biosynthetic enzyme, chloroplast | 59 | 2 | K.HAALFTSTIMSK.L (33) |
| K.ALDMNTAEDAIVR.L (26) | |||||
| OMT1_ARATH | At5g54160 | Quercetin 3-O-methyltransferase 1 | 59 | 2 | K.NPEAPVMLDR.I (34) |
| K.VLMESWYHLK.D (25) | |||||
| IF5A2_ARATH | At1g26630 | Eukaryotic translation initiation factor 5A-2 (eIF-5A) | 59 | 2 | K.LPTDDGLTAQMR.L (33) |
| K.CHFVAIDIFTAK.K (26) | |||||
| PKL_ARATH | At2g25170 | PICKLE chromatin-remodelling factor | 58 | 6 | K.GLLHPYQLEGLNFLR.F (19) |
| K.AYKSNHRLK.T (14) | |||||
| Y1934_ARATH | At1g09340 | Uncharacterized protein chloroplast | 57 | 3 | K.SSLSAEGFDVVYDINGR.E (26) |
| R.FIGLFLSR.I (16) | |||||
| VIN3_ARATH | At5g57380 | VERNALIZATION-INSENSITIVE 3 | 56 | 5 | R.GIVNRLSSGVHVQKLCSQAMEALDK.V (27) |
| R.NEIMKIICAEMGKER.K (14) | |||||
| PME4_ARATH | At2g47030 | Pectinesterase-4 (VANGUARD1-like protein 1) | 54 | 6 | K.AVQGICQSTSDKASCVK.T (16) |
| K.NTAGPMGHQAAAIRVNGDRAVIFNCR.F (12) | |||||
| APT1_ARATH | At1g27450 | Adenine phosphoribosyltransferase 1 (APRT 1) | 54 | 3 | R.AIIIDDLIATGGTLAAAIR.L (35) |
| K.DTIALFVDR.Y (15) | |||||
| DRL19_ARATH | At1g63350 | Putative disease resistance protein At1g63350 | 54 | 4 | R.NAELQRLCLCGFCSKSLTTSYR.Y (17) |
| K.MCLLYCALFPEDAK.I (16) | |||||
| FABG_ARATH | At1g24360 | 3-Oxoacyl-[acyl-carrier-protein] reductase, chloroplast | 54 | 3 | K.WGTIDVVVNNAGITR.D (25) |
| K.ILGTIPLGR.Y (19) | |||||
| BSL1_ARATH | At4g03080 | Serine/threonine-protein phosphatase BSL1 | 53 | 4 | K.IICMHGGIGR.S (16) |
| R.HGAASVGIRIYVHGGLR.G (16) | |||||
| PER9_ARATH | At1g44970 | Peroxidase 9 | 52 | 3 | K.AYAEDERLFFQQFAK.S (26) |
| K.EPRMAASLLR.L (13) | |||||
| UPL1_ARATH | At1g55860 | E3 ubiquitin-protein ligase UPL1 | 52 | 5 | K.LLSDIVLMYSHGTSVILR.R (20) |
| R.LIDFDNKKAYFR.S (16) | |||||
| HDA5_ARATH | At5g61060 | Histone deacetylase 5 | 51 | 3 | R.KVGLIYDETMCK.H (24) |
| K.LQLAGVSQR.C (18) | |||||
| HAC12_ARATH | At1g16710 | HAC12 histone acetyltransferase | 51 | 5 | K.LTTHPSLADQNAQNK.E (14) |
| K.ASGQSDFSGNASK.D (13) | |||||
| MRP14_ARATH | At3g59140 | Multidrug resistance-associated protein 14 | 50 | 7 | R.IATFLEAPELQGGERRR.K (16) |
| R.VVAVENPTKPVK.E (11) | |||||
| ASHH2_ARATH | At1g77300 | Histone-lysine N-methyltransferase ASHH2 | 50 | 6 | K.ILPRPRPR.M (13) |
| K.SPSENGSHLIPNAKKAK.H (13) | |||||
| ATM_ARATH | At3g48190 | Serine/threonine-protein kinase ATM (PI3Kc_related) | 47 | 8 | R.RVLLQILGCEKCTMQHLLQSASLLR.K (14) |
| K.QIPMAQLHENEGRK.S (11) | |||||
| FBX10_ARATH | At1g51290 | Putative F-box only protein 10 | 47 | 4 | R.LVICCYDETQQVYIYIVRR.N (16) |
| K.YVIGYDNKK.R (14) | |||||
| PSBP1_ARATH | At1g06680 | Oxygen-evolving enhancer protein 2-1, chloroplast | 45 | 3 | K.TNTDFLPYNGDGFK.V (25) |
| K.EIEYPGQVLR.F (12) | |||||
| CHLD_ARATH | At4g18480 | Magnesium-chelatase subunit chlD, chloroplast | 45 | 3 | K.IYKAGMSLLVIDTENK.F (26) |
| R.VAAVGIATQFQERCNEVFR.M (22) | |||||
| FBK38_ARATH | At2g29800 | Putative F-box/Kelch-repeat | 44 | 3 | K.MANFGGKLVILGCYR.S (20) |
| R.HLRNMKR.D (16) | |||||
| GLYM_ARATH | At4g37930 | Serine hydroxymethyltransferase mitochondrial | 44 | 4 | R.GFVEEDFAK.V (22) |
| K.VLEAVHIASNK.N (11) | |||||
| SCP37_ARATH | At3g52010 | Serine carboxypeptidase-like 37 | 44 | 3 | K.AIHANTTK.L (19) |
| K.KLPGQPSGVSFR.Q (18) | |||||
| COL14_ARATH | At2g33500 | CONSTANS-LIKE 14 | 44 | 3 | K.LCLPCDQHVHSANLLSR.K (20) |
| K.SNNIPAAIHSHK.S (14) | |||||
| SYV_ARATH | At1g14610 | Valyl-tRNA synthetase | 43 | 7 | K.SDLFKADAK.S (16) |
| K.INLDILRVVGYR.Q (13) | |||||
| DRP1D_ARATH | At2g44590 | Dynamin-related protein 1D | 43 | 3 | R.MQCAKRLELYK.K (22) |
| R.MGSEYLAK.L (14) | |||||
| VATB_ARATH | At1g76030 | Vacuolar ATP synthase subunit B | 43 | 3 | R.NIFQSLDLAWTLLR.I (16) |
| R.KFVMQGAYDTR.N (15) | |||||
| SIZ1_ARATH | At5g60410 | E3 SUMO-protein ligase SIZ1 | 42 | 5 | K.WQCPICLK.N (15) |
| R.HRSLNKICIILCAGK.N (12) | |||||
| HAC2_ARATH | At1g67220 | HAC2 histone acetyltransferase | 42 | 4 | R.ACTGCYTKNRTLR.H (16) |
| K.LGTVVDIIEPMKCDER.S (11) | |||||
| TMK1_ARATH | At1g66150 | Putative receptor protein kinase TMK1 precursor | 42 | 4 | K.GNDPCTNWIGIACSNGNITVISLEK.M (18) |
| K.VVNLTNNHLQGPVPVFK.S (12) | |||||
| SYM_ARATH | At4g13780 | Probable methionyl-tRNA synthetase | 42 | 3 | R.LVEGSCPFEGCNYDSAR.G (26) |
| K.CKVCQNTPR.I (12) | |||||
| WEE1_ARATH | At1g02970 | Wee1-like protein kinase | 41 | 3 | R.AMPPPCLK.N (19) |
| K.LPLLPGHSLQLQQLLK.T (15) | |||||
| ARR12_ARATH | At2g25180 | Two-component response regulator | 41 | 5 | –.MTVEQNLEALDQFPVGMR.V (17) |
| R.HCQYHVTTTNQAQK.A (9) | |||||
| CESA4_ARATH | At5g44030 | Cellulose synthase A catalytic subunit 4 | 41 | 4 | K.KAGAMNAMVR.V (22) |
| K.SSLMSQKNFEKR.F (12) | |||||
| AUR2_ARATH | At2g25880 | Serine/threonine-protein kinase Aurora-2 | 41 | 3 | R.LYGYFYDQKRVYLILEYAVR.G (18) |
| M.LYQAASEAAQK.R (14) | |||||
| Y1838_ARATH | At1g18380 | Uncharacterized protein At1g18380 | 41 | 3 | R.YIMEDKACR.R (32) |
| R.SSDSDEGCMKYAEIPMLR.S (8) | |||||
| 2AAA_ARATH | At1g25490 | Serine/threonine-protein phosphatase 2A regulatory subunit A alpha | 41 | 4 | R.LAGGEWFAAR.V (17) |
| R.RAAASNLGK.F (11) | |||||
| FBK84_ARATH | At4g19865 | F-box/Kelch-repeat protein At4g19865 | 40 | 3 | K.IEFGNVNEMCAYDTKLCK.W (20) |
| K.IYVMGGCQGLKDEPWAEVFNTK.T (10) | |||||
| MSH3_ARATH | At4g25540 | DNA mismatch repair protein MSH3 | 40 | 4 | R.LVNAGYKIGVVK.Q (17) |
| R.LVNAGYK.I (13) | |||||
| DCDA1_ARATH | At3g14390 | Diaminopimelate decarboxylase 1, chloroplast | 39 | 1 | R.DAAVLMIEYIDEIR.R (39) |
| GL25_ARATH | At5g26700 | Probable germin-like protein subfamily 2–5 | 39 | 3 | R.IDYAPNGLNPPHVHPR.A (17) |
| K.LPGLNTLGLSMSR.I (14) | |||||
| CYSK1_ARATH | At4g14880 | Cysteine synthase (OAS-TL A) | 39 | 3 | K.IDGFVSGIGTGGTITGAGK.Y (21) |
| R.IGFSMISDAEK.K (15) | |||||
| MRP13_ARATH | At1g30410 | Multidrug resistance-associated protein 13 | 39 | 4 | R.KKYYNCVLGLLACYCVVEPVLR.L (22) |
| R.SVLIKQEER.E (14) | |||||
| ERG11_ARATH | At5g24150 | Squalene monooxygenase 1,1 | 39 | 3 | R.RLLQPLSNLGNAQK.I (18) |
| R.LFGLAMKMLVPHLK.A (13) | |||||
| DPOLA_ARATH | At5g67100 | DNA polymerase alpha catalytic subunit | 38 | 4 | K.NGCNVLSIENSERALLNRLFLELNK.L (14) |
| R.KRSGILSHFTVVR.N (13) | |||||
| CWP17_ARATH | At2g06850 | 23 kDa cell wall protein | 38 | 3 | –.IPCRKAIDVPFGTR.Y (19) |
| R.KAIDVPFGPR.Y (13) | |||||
| MOCOS_ARATH | At1g16540 | Molybdenum cofactor sulphurase (ABA3) | 38 | 7 | K.LLKSLTPSAIWMHTTSLSIYVK.K (12) |
| R.YEIDEKR.Q (10) | |||||
| ALA11_ARATH | At1g13210 | Phospholipid-transporting ATPase 11 | 38 | 5 | K.SLTYALEDDFKK.K (18) |
| R.SMAMRSNGSSLVGDDLDVVVDQSGPK.I (10) | |||||
| TAP1_ARATH | At1g70610 | Antigen peptide transporter-like 1, chloroplast | 38 | 3 | R.GCFFGIANMILVKRMR.E (16) |
| R.QRIGYVGQEPK.L (12) | |||||
| AGO1_ARATH | At1g48410 | Protein argonaute | 37 | 2 | R.INLLDEEVGAGGQR.R (36) |
| R.GYGQPPQQQQQYGGPQEYQGRGR.G (4) | |||||
| FBK19_ARATH | At1g32430 | Putative F-box/Kelch-repeat protein At1g32430 | 37 | 2 | K.VEVRELTLNNPGLK.A (22) |
| R.CIKLEVNEPSLDFLGIGYDNNK.R (14) | |||||
| LUMI_ARATH | At4g02560 | LUMINIDEPENDENS | 37 | 2 | K.KHMLGSNPSYNK.E (21) |
| K.HDSSTHPYWNQNK.R (18) | |||||
| CAPP1_ARATH | At1g53310 | Phosphoenolpyruvate carboxylase 1 | 36 | 2 | K.LEELGSVLTSLDPGDSIVIAK.A (23) |
| K.GIAAGLQNTG.– (14) | |||||
| WBC16_ARATH | At3g55090 | Probable white–brown complex homologue protein 16 | 36 | 2 | K.TIIGDEGHR.G (29) |
| R.ILFYLCLLLGSKNK.R (8) | |||||
| CNGC4_ARATH | At5g54250 | Cyclic nucleotide-gated ion channel 4 | 36 | 3 | R.IGLTCGGR.R (36) |
| R.GVDECEMVQNLPEGLR.R (5) | |||||
| U496I_ARATH | At2g18630 | UPF0496 protein At2g18630 | 36 | 2 | K.INSEYTEHLSSYER.A (21) |
| K.YEKVVRGQK.E (13) | |||||
| ARFM_ARATH | At1g34170 | Auxin response factor 13 | 36 | 2 | K.FVDAMNNNYIVGSR.F (20) |
| K.FVDAMNNNYIVGSRFR.M (16) | |||||
| CYSKM_ARATH | At3g59760 | Cysteine synthase, mitochondrial (OAS-TL C) | 35 | 3 | K.IQGIGAGFIPK.N (15) |
| R.IGYSMVTDAEQKGFISPGK.S (15) |
Fig. 2.
Confirmation of the presence of proteins identified through shotgun proteomic analysis in the immnunopurified nitroproteome. Crude protein extracts (CE) were immunoprecipitated with anti-3-nitroY (anti-3-NY) antibodies. The CE, supernatant, and immunoprecipitate (IP) were separated by 12% SDS–PAGE, transferred to a nitrocellulose membrane, and probed with specific antibodies raised against chloroplastic glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glycine-rich protein 7 (GRP7), fructose bisphosphatase (FBPase), PICKEL (PKL), or carbonic anhydrase (CA). The procedure started from 1mg of total protein in the crude extract that was immunoprecipitated as described in the Materials and methods, and then the whole IP was loaded on the gel along with 1% of the CE input and the corresponding supernatant.
Despite the success in identifying a large number of potentially nitrated proteins, no MS/MS spectrum with a good enough MASCOT score was obtained for nitrated peptides, thus preventing the identification of unequivocal nitration sites. To overcome this, and because the amount of protein required for the identification of nitrated peptides is often a limitation in the method, the most abundant proteins in 2-DE gels from 3-nitroY-immunoprecipitated proteins were excised from the gels, digested with trypsin, and further analysed by MALDI-TOF (matrix-assisted laser desorption ionization-time of flight). Supplementary Table S2 at JXB online summarizes the identified proteins, their MASCOT scores, the number of non-redundant peptides, and the corresponding sequence coverage. Twenty-two proteins were identified with a MASCOT score >59, considered as significant in the proteomic analysis. Unfortunately, no MS/MS spectra with a high enough score corresponding to a bona fide Y-nitrated peptide could be obtained. However, six out of 22 identified proteins showed MALDI-TOF spectra for potentially nitrated peptides with a signal/noise ratio >25, considered as significant in the analysis. The simultaneous identification of nitrated peptides and their unmodified forms in addition to the length of the nitrated peptides identified (≥7 amino acid residues) makes the identification more reliable (Stevens et al., 2008). Table 2 shows the identity of those proteins and the corresponding nitrated peptides with the signal/noise ratio, the molecular mass of the unmodified and modified peptides, and the corresponding +45 shift to the modification of Y to nitroY. Three out of those six proteins (Rubisco, Rubisco activase, and transketolase) showed nitrated peptides containing a single Y residue and a +45 mass shift, thus allowing the assignment of a putative nitration site for those proteins. For serine hydroxymethyltransferase, the nitrated peptide contained two Y residues and showed a mass shift of +90, compatible with two Y nitration sites. Finally, for the other two proteins, a cytosolic GAPDH and a putative mannitol dehydrogenase, the nitrated peptides contained two Y residues and showed a mass shift of +45, corresponding to a single nitration event, so no nitration site could be proposed for these proteins (Table 2).
Table 2.
Putative Y-nitrated peptides identified by MALDI-TOF from 2D gel-excised spots
| Description | AGI identifier | Peptide sequence | Error | Signal-to-noise | Mr (observed) (unmodified) | Mr (observed) | Shift | Modification |
| Rubisco activase, chloroplast precursor | At2g39730 | 351R.VYDDEVR.K359 | 0.01 | 110 | 895.34 | 940.41 | +45.07 | NitroY (+45) |
| 72R.GLAYDTSDDQQDITR.G88 | –0.05 | 25 | 1697.66 | 1744.66 | +46.97 | 2 Deamination (+2) NitroY (+45) | ||
| Serine hydroxymethyl transferase | At4g13930 | 160K.VNFTTGYIDYDKLEEK.A177 | 0.03 | 60 | 1934.83 | 2025.92 | +91.09 | Deamination (+1)2 NitroY (+90) |
| Transketolase, putative* | At3g60750 | 333K.ANSYSVHGAALGEKEVEATR.N354 | 0.15 | 57 | (2090.15) | 2135.15 | (+45) | NitroY (+45) |
| Glyceraldehyde-3-phosphate dehydrogenase, cytosolic | At3g04120 | 313K.LVSWYDNEWGYSSR.V328 | –0.06 | 50 | 1761.72 | 1806.72 | +45 | NitroY (+45) |
| Probable mannitol dehydrogenase | At4g39330 | 133K.NYGGYSENIVVDQR.F148 | –0.04 | 27 | 1613.63 | 1658.70 | +45.07 | NitroY (+45) |
| Rubisco large chain precursor** | AtCg00490 | 236K.GHYLNATAGTCEEMIK.R253 | 0.04 | 25 | (1794.84) | 1839.84 | (+45) | NitroY (+45) |
Samples containing 3-nitroY immunopurified proteins were separated by 2-DE and identified by MALDI-TOF as described in the Materials and methods. The AGI identifiers for each identified protein are included along with the corresponding Y-nitrated peptide sequence (the residues susceptible to Y nitration are underlined and unequivocal nitration of Y is indicated in bold). Error (difference between the experimental and calculated masses); signal-to-noise ratio, relative molecular mass (Mr) observed for the modified and the corresponding unmodified peptide that appeared in the same MASCOT search. Values in parentheses indicate the absence of the unmodified peptide. The mass shift (Shift) and the modifications of the corresponding peptide with their respective mass increases are also shown. Those proteins that have been previously reported as nitrated in other plant systems have been marked with a single (Chaki et al., 2009) or double asterisk (Cecconi et al.. 2009).
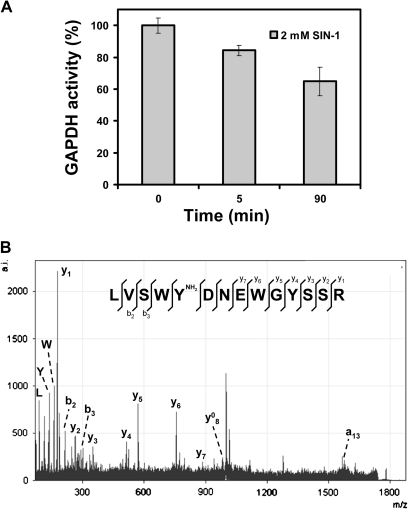
Y residues contained in the nitrated peptides were checked to see if they fulfilled the previously characterized factors determining the selectivity of Y nitration in proteins. These factors include the proximity of a basic amino acid within the primary sequence, the exposure of the aromatic ring to the surface of the protein, the location of the Y residue on a loop structure, its association with a neighbouring negative charge, and the proximity of the proteins to the site of generation of nitrating agents (Souza et al., 1999; Ischiropoulos, 2003; Chaki et al., 2009). With the exception of Rubisco activase, for which no structural model is available, the structures of the proteins were modelled as indicated in the Materials and methods. All putative nitrated Y residues had acidic residues close enough (<10 Å from the Y target) and all of them have basic amino acids in the primary sequence flanking the Y residue (Table 3). However, only Y337 and Y135 from transketolase and putative mannitol dehydrogenase, respectively, were located in loops, and most of them showed accessibile solvent area (ASA) indexes <70 (Table 3), thus having a low probability of being efficiently exposed to the solvent. Regarding the proximity of the proteins to the site of generation of nitrating agents, all the proteins identified are located in subcellular compartments previously characterized as sites of NO and superoxide production in plants, such as apoplasts, mitochondria, and chloroplasts (Corpas et al., 2001; Bethke et al., 2004; Gupta et al., 2005; Jasid et al., 2006; Flores-Pérez et al., 2008; Igamberdiev and Hill, 2009). In addition, the fact that some of the Y residues found to be potentially nitrated are highly conserved Y residues in proteins functionally homologous in other organisms (Supplementary Fig. S1 at JXB online) confers potential functional relevance to this post-translational modification as a regulatory mechanism of their activity/function. Regarding this, it has been confirmed that treatment of seedlings with a peroxynitrite donor, such as SIN-1, led to inhibition of GAPDH activity (Fig. 3A).
Table 3.
Structural features of potential Y targets of nitration in MALDI-TOF-identified proteins Protein annotation and AGI code along with the Putative nitrated Y are indicated. Parameters were calculated as described in the Materials and methods. Accessible solvent area (ASA) was calculated by NetSurfP software (Petersen et al. 2009).
| Protein/AGI | Putative nitrated Y | Distance to D/E | Proximal basic amino acids in primary sequence | Location in loop | ASA |
| Rubisco activase, chloroplast precursor_ At2g39730 | Y353 | (No model) | R351, R358, K359 | (No model) | 5.45 |
| Y76 | R72 | 79.13 | |||
| Serine hydroxymethyl transferase_ At4g13930 | Y167 | 5.99 Å to E342 | K160, K172, K176 | No | 62.38 |
| Y170 | 5.04 Å to D197 | K160, K172, K176 | No | 23.35 | |
| Transketolase, putative_ At3g60750 | Y337 | 9.01 Å to D268 | K333, H340, K347 | Yes | 66.16 |
| Glyceraldehyde-3-phosphate dehydrogenase, cytosolic_ At3g04120 | Y318 | 6.08 Å to D319 | K313, R327 | No | 7.35 |
| Y324 | 6.61 Å to E321 | K313, R327 | No | 19.17 | |
| Probable mannitol dehydrogenase_ At4g39330 | Y135 | 4.31 Å to E8 | K133, R147 | Yes | 34.66 |
| Y138 | 3.75 Å to D53 | K133, R147 | No | 13.55 | |
| Rubisco large subunit precursor_ AtCg00490 | Y239 | 6.33 Å to E158 | K236, H238, K252, R253 | No | 6.37 |
Fig. 3.
Effect of nitration on GAPDH. (A) Arabidopsis seedlings were treated with SIN-1. After the indicated times, the GAPDH activity levels were measured in crude protein extracts from whole seedlings as described in the Materials and methods. Measurements for activity were performed in triplicate and the average values ±SD are shown. (B) MS/MS spectrum of aminated LVSWYDNEWGYSSR peptide from Arabidopsis glyceraldehyde-3-phosphate dehydrogenase. Detected peaks of y and b series as well as immonium ions of L, Y, and W are indicated.
Despite efforts made to identify sites of in vivo Y nitration among proteins immunoprecipitated with anti-3-nitroY, not a single MS/MS spectrum corresponding to a nitrated peptide was identified. There might be two explanations for this lack of success. First, the nitrated form of the identified proteins could be naturally very low abundant in the analysed samples, thus making MS/MS-based identification extremely difficult. Secondly, the lack of detection of nitrated peptides may be the result of the unstable nature of nitroY under the conditions used to process samples by MS. Regarding the latter, it has been proposed that the nitro group linked to Y residues of proteins can be reduced to an amino group (Sarver et al., 2001; Tsumoto et al., 2010). When crude proteomic data from spots excised from gels after 2-DE were searched for aminoY instead of nitroY post-translational modification, an MS/MS fragmentation spectrum corresponding to LVSWYDNEWGYSSR peptide (monoisotopic mass of neutral peptide of 1776.7631; ion score 43; expected 0.00088) was found for cytosolic GADPH (G3PC). This spectrum included a +15 shift compatible with an amino modification of Y318 (Fig. 3B). These data suggest that from two Y residues found as potential nitration targets in G3PC (Table 2), part of the 3-nitroY318 residues in the protein population might undergo reduction to 3-aminoY318 under the conditions used for MS analysis. Since reduction may occur with any nitroY, the crude data from LC-MS/MS shotgun analysis were searched for aminoY modification, and 51 putative aminoY-containing peptides with ion scores >15 were found that corresponded to 47 different proteins (Supplementary Table S3 at JXB online). Comparison of nitroY and aminoY searches led to only five peptides that were detected to be Y nitrated or aminated in the same residue, but all of them had ion scores <10, thus suggesting that the partial reduction of nitroY may lower the abundance of both modifications, making MS identification even more difficult.
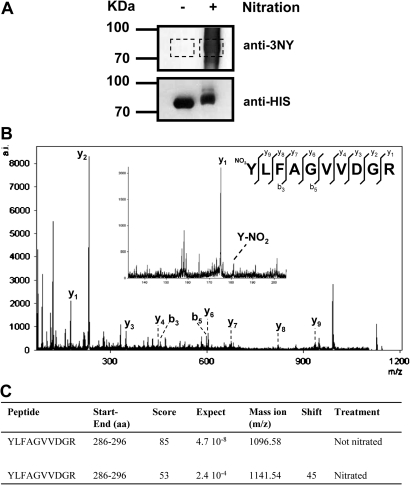
To overcome the low abundance of nitrated forms of proteins found in vivo, one of the proteins identified in the screen as potentially nitrated, 5-methyl tetrahydropteroyltriglutamate-homocysteine S-methyltransferase or methionine synthase 1 (AtMS1) was expressed as a 6×His-tagged version in bacteria. The tagged recombinant protein was expressed to moderately high levels by 5 h after induction with IPTG (data not shown). Crude recombinant extracts were checked for AtMS1 protein content by western blot with anti-5×His tag antibodies and subsequently purified with Ni-resin. The purified protein was then split into two equivalent samples, one of them being nitrated in vitro whereas the other was used as control of no exogenous nitration. The efficiency of nitration was then checked by western blot with anti-3-nitroY. No cross-reacting band was detected in the control protein but a strong signal was observed in the nitrated recombinant AtMS1 protein (Fig. 4A). Both samples had comparable levels of recombinant protein as confirmed by western blot with anti-5×His antibodies (Fig. 4A). A duplicate one-dimensional SDS–polyacrylamide gel was stained with Coomassie blue and the bands corresponding to nitrated and non-nitrated proteins were excised from the gel, digested in gel with trypsin, and further analysed by LC-MS/MS. The same YLFAGVVDGR peptide was found from control non-nitrated protein (m/z 1096.58, score 85) and nitrated protein (m/z 1141.54, score 53), showing a shift of 44.96 equivalent to the typical shift of a single nitration (Fig. 4C). The MS/MS spectrum of nitrated peptide showed most of the peaks corresponding to the y and b series and also the immonium ion of a nitrated Y287 residue (Fig. 4B). These data allowed identification of an unequivocal site of nitration in AtMS1 at Y287. Whether this post-translational modification of AtMS1 may alter its activity, stability, subcellular location, or other post-translational modifications will require further study. Nevertheless, Y287 is conserved in plant methionine synthases but not in the enzymes from yeast (Supplementary Fig. S2A at JXB online), and it is located in a loop on the external surface of the protein far from the 5-methyl tetrahydropteroyltriglutamate (THG)- and homocysteine (HC)-binding sites inside the catalytic pocket (Supplementary Fig. S2B). Y287 forms hydrogen bonds with two proximal residues, T262 and F264, which may be important to maintain suitable folding of the protein but which do not interfere directly with substrate binding or cofactor function. However, it has been described that methionine synthase activity is regulated by NO. NO treatment impairs methionine synthase activity in different models both in vitro (Brouwer et al., 1996, Nicolaou et al., 1996, 1997) and in vivo (Danishpajooh et al., 2001), suggesting that tyrosine nitration might be responsible for the NO-dependent reduction of methionine synthase activity.
Fig. 4.
Identification of the nitration site in recombinant tagged methionine synthase 1 from Arabidopsis. (A) Equal amounts (5 μg) of recombinant AtMS1 protein were nitrated (+) or not (–), separated by one-dimensional SDS–PAGE, and blotted onto nitrocellulose to be probed by western blot with anti-3-nitroY (anti-3NY) antibodies. After stripping, membranes were further probed with anti-5×His antibodies. Molecular size markers are shown on the left side of the panels. (B) MS/MS spectrum of nitrated YLFAGVVDGR peptide from AtMS1. The insert shows the detected y and b series as well as a detail of the spectrum showing the immonium ion corresponding to nitrated Y (C).
Discussion
Although several reports regarding proteomic approaches for the identification of nitrated proteins in mammals have been published recently (Suzuki et al., 2005; Sultana et al., 2006; Hong et al., 2007; Zhang et al., 2007) and the detection of nitrated proteins in pathogen-challenged plants was also reported (Romero-Puertas et al., 2007), the first two reports focusing on general proteomic approaches to nitrated plant protein identification were not published until very recently (Cecconi et al., 2009; Chaki et al., 2009). Both groups described the use of anti-3-nitroY antibodies for the detection of plant putatively nitrated proteins in western blot and the subsequent identification of the immunoreactive proteins by MALDI-TOF/TOF. A total of 8 and 21 proteins were identified in these reports (Cecconi et al., 2009; Chaki et al., 2009), respectively. However, no nitrated peptides and consequently no nitration sites were identified in either of those reports, probably due to the low level of nitration under non-stressed conditions (Chaki et al., 2009) and technical limitations (Cecconi et al., 2009), as described by the authors. In this work, a proteomic methodology has been used to purify and identify proteins nitrated in vivo at Y residues in A. thaliana. The method is based on the purification of nitrated proteins by immunoprecipitation with well characterized anti-3-nitroY antibodies (Schmidt et al., 2003; Gokulrangan et al., 2007), and further identification by LC-MS/MS. This method has been previously reported to be useful in identifying nitrated proteins in mammals (Turko et al., 2003; Liu et al., 2009; Zhan and Desiderio, 2009). The procedure was sensitive enough to identify 127 potentially nitrated proteins from Arabidopsis seedlings. These results are in the range of the best proteomic methods reported in animal systems (Suzuki et al., 2005; Sultana et al., 2006; Hong et al., 2007; Zhang et al., 2007), and they represent the description of the widest potential in vivo nitroproteome of a plant to date. A literature search showed that ∼35% of the identified Arabidopsis Y-nitrated proteins were previously described as Y nitrated in other organisms (Supplementary Table S1, and references therein), which supports the reliability of the method in identifying potentially Y-nitrated proteins. Moreover, a large proportion of the proteins reported to be potential targets of nitration in the two previous reports on plants (Cecconi et al., 2009; Chaki et al., 2009) were also identified as putatively nitrated in the present work. Moreover, some of the MS-based protein identifications used have been technically validated by detection of the corresponding proteins in the immunopurified samples by western blot with specific antibodies (Fig. 2). Although the methodology presented in this work seems to be reliable and robust enough to be considered a good starting point for the characterization of Y-nitrated plant proteins, no unequivocal nitration sites were found by MS/MS. Due to the low abundance of Y residues in proteins and because the nitration sites were usually restricted to one or two Y residues per protein (Abello et al., 2009), a low level of occurrence of Y nitration is expected.
The most abundant protein spots in 2-DE gels from anti-3-nitroY-immunoprecipitated proteins were analysed and searched for Y nitration modification. Nitrated peptides for GAPDH, ribulose bisphosphate carboxylase large subunit, Rubisco activase, mannitol dehydrogenase, and transketolase were identified (Table 2). The identifications are based on peptide mass fingerprinting data obtained by MALDI-TOF because no good fragmentation MS/MS spectra were obtained. Only molecular ions with a signal-to-noise ratio >25 and a difference between the experimental and calculated masses of <0.15 were selected. Furthermore, in silico analysis of potentially nitrated peptides showed that most of them fulfilled most of the criteria to be nitration targets: Y residues were located in loops with a large solvent-accessible area and had a basic amino acid in the vicinity and a proximal negative charge (Table 3). Gene Ontology tools for the analysis of the potentially Y-nitrated identified proteins showed a significant over-representation of proteins located in the chloroplast, peroxisome, mitochondria, and apoplast, subcellular compartments that have been proposed as a source of NO and superoxide anions in plants (Corpas et al., 2001; Bethke et al., 2004; Gupta et al., 2005; Jasid et al., 2006; Flores-Pérez et al., 2008; Igamberdiev and Hill, 2009), thus representing cellular domains where the nitrating reagent peroxynitrite is produced (Szabó et al., 2007). These data support the previously proposed idea that the proximity of proteins to the site of generation of nitrating agents is a main factor in directing protein nitration (Ischiropoulos, 2003).
When the Gene Ontology tools were used for the analysis of the Y-nitrated identified proteins, it was found that >60% were involved in primary metabolism. Post-translational nitration of key enzymes and the subsequent alteration of their catalytic properties may represent a new level of regulation of primary metabolism. It is noteworthy that one of the proteins identified as putatively nitrated in this work (S-adenosyl homocysteine hydrolase, Table 1) has also been reported to be nitrated in sunflower hypocotyls (Chaki et al., 2009). The activity of the enzyme was inhibited upon nitration (Chaki et al., 2009), thus suggesting that the activity of the Arabidopsis counterpart may also be regulated through nitration. Moreover, Rubisco activase, ATP synthase subunit α, and glutamine synthetase 2 have also been identified as putative nitrated proteins in pathogen-challenged Arabidopsis (Cecconi et al., 2009). It has been discussed that nitration of these proteins may be a way to modulate defence-related responses including the hypersensitive response (Cecconi et al., 2009). Alternatively, nitration of abundant proteins such as those involved in photosynthesis and carbon metabolism may represent just a non-selective scavenging system for reactive nitrogen and oxygen species produced under standard or stress-related conditions. Moreover, the functional relevance of this post-translational modification on these targets is further supported by the fact that most of the identified nitrated Y residues are strictly conserved in the amino acid sequence of homologous proteins from other organisms (Supplementary Fig. S1 at JXB online), thus supporting a potential functional effect of this post-translational modification.
In the case of GADPH, the two Y residues identified as nitrated in peptide LVSWY*DNEWGY*SSR were not only conserved in the rabbit GAPDH but were actually also identified as nitrated LISWY*DNEFGY*SNR, resulting in complete loss of catalytic activity (Palamalai and Miyagi, 2010). GAPDH models for rat and Arabidopsis overlapped greatly throughout the molecule and particularly on nitrated Y residues (Supplementary Fig. S3). In addition, as reported for yeast and mammals (Buchczyk et al., 2000; Palamalai and Miyagi, 2010), Arabidopsis GAPDH activity was also inhibited by peroxynitrite (Fig. 3A). Notwithstanding, several proteins participating with GAPDH in the gluconeogenesis conversion of malate to sucrose were also identified as nitrated forms in Arabidopsis (Table 1 and Supplementary Fig. S4), thus suggesting a potential for Y nitration as a significant regulatory level on this principal metabolic pathway. Interestingly, among potential targets of Y nitration in Arabidopsis were also three enzymes involved in the biosynthesis of methionine, the 5-methyl tetrahydropteroyltriglutamate-homocysteine methyltransferase, also called methionine synthase, the S-adenosylmethionine synthetases 1 and 2, and S-adenosylhomocysteinase 1 (Supplementary Fig. S4). It has been previously reported that NO probably inhibits mammalian methionine synthase activity by reaction with cobalt-containing cobalamin cofactor (Brouwer et al., 1996; Nicolaou et al., 1997; Danishpajooh et al., 2001). Nevertheless, in the light of the results obtained here, this mode of action for NO is compatible with the mechanism of control of methionine synthase activity through nitration of key Y residues of the protein. Moreover, the fact that not only a key regulatory step but most of the enzymes involved in methionine biosynthesis are potentially nitrated in Arabidopsis suggests that Y nitration may represent an important regulatory level to control the biosynthesis of this amino acid in plants. Furthermore, nitration of S-adenosylmethionine synthetases could also represent an interesting regulatory point in ethylene production. Regarding this, the S-nitrosylation of S-adenosylmethionine synthetase 1 resulting in reduced activity and decreased ethylene production in Arabidopsis has recently been reported (Lindermayr et al. 2006).
The fact that neither in this work nor in the two previous reports on protein nitration in plants (Cecconi et al., 2009; Chaki et al., 2009) were any nitrated peptide and the corresponding nitration site unequivocally identified needs further discussion. It is well known that Y nitration is a very low abundant post-translational modification as compared with other protein modifications such as phosphorylation (Abello et al., 2009). In fact, only 0.033–0.43 mmol of nitroY per mol of Y has been detected in plant proteins, depending on the tissue or species studied (Bechtold et al., 2009; Chaki et al., 2009). Moreover, it is also likely that under non-stressed conditions, when only basal levels of NO and superoxide and thus low amounts of peroxynitrite are generated by cells, even lower abundance is expected. Nevertheless, because the presented methodology enriched samples in potentially Y-nitrated-containing proteins by immunoprecipitation with a specific anti-3-nitroY antibody, the identification of some nitrated peptides by MS/MS should be expected. A survey of the literature on identification of nitrated proteins in different organisms points to a very low number of nitrated sites identified, thus suggesting the existence of technical difficulties intrinsically associated with MS-based analysis of this kind of protein modification. A possible explanation for the lack of nitroY signatures could be related to alterations produced by the treatments performed before mass spectrometry analysis or during the ionization of the protein samples. It has been reported that the treatment of nitrated proteins with DTT and elevated temperature, as used for trypsin digestion, can reduce the nitroY to aminoY or other related species (Söderling et al., 2007). Moreover, the ionization energy for MALDI or electrospray ionization (ESI) technologies is too aggressive for the nitrated Y residues, and the laser-induced photochemical decomposition of nitroY to aminoY during MALDI-MS analysis has been reported (Sarver et al., 2001). Therefore, a conversion of nitroY to aminoY in the samples during sample processing before MS analysis may explain the lack of detection of nitrated peptides in the present studies. To validate this hypothesis, the proteomic experiments were searched for aminoY modification instead of nitroY modification. By selecting aminoY as a variable modification in the MASCOT data analysis in MALDI-TOF/TOF experiments, a fragmentation MS/MS spectrum corresponding to a peptide containing a 3-aminoY residue was found in the protein spot corresponding to GADPH (Fig. 3). More precisely a peptide containing aminoY318 was found, suggesting that from the two Y residues found as potential targets to be nitrated in G3PC (Table 2), part of the 3-nitroY318 residues in the protein population might undergo reduction to 3-aminoY318 under the conditions used for MS analysis. Moreover, although no further MS/MS spectra corresponding to aminoY-containing peptides were obtained, ∼50 additional putative aminoY-containing peptides with an ion score >15 were found (Supplementary Table S3 at JXB online). This confirms the hypothesis that the lack of identification of nitrated peptides in this work, and probably in others, may be due to the conversion of the nitroY to aminoY. Such a conclusion leads to the proposal that future analysis of Y nitration of proteins should be based on a simultaneous search for both nitroY and aminoY variable modifications. Eventually, the chemical reduction of all nitroY to aminoY by means of a strong reducing reagent such as sodium dithionite may represent an advantage in further proteomic analysis either searching directly for aminoY or after derivatization of aminoY (Ghesquière et al., 2009; Abello et al., 2010).
The proteomic method described in this work represents a tool to identify proteins undergoing in vivo Y nitration in plants. The application of this methodology, with the improvements discussed above, to the analysis of different biological processes in plants will allow the identification of Y nitration protein targets. Because of the low abundance and limited stability of this post-translational modification, the obtained data suggest that after identification of in vivo targets, the confirmation of the modification sites and the functional consequences have to be addressed through in vitro assays with larger amounts of modified protein. These Y-nitrated proteins may represent nodes for a new unexplored level of regulation of proteins exerted by NO through post-translational modification. Further characterization of the identified Y-nitrated proteins will provide key information about new regulatory features of NO in many aspects of plant growth, development, and defence.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Potential Y nitration targets in glyceraldehyde-3-phopsphate dehydrogenase, serine hydroxymethyltransferase, transketolase, Rubisco large subunit, and Rubisco activase are conserved in different plants and other organisms.
Figure S2. Conservation and structural modelling analysis of plant methionine synthases.
Figure S3. Alignment of 3D structure models of rat and Arabidopsis glyceraldehyde-3-phosphate dehydrogenases.
Figure S4. Scheme displaying primary carbon and sulphur metabolism enzymes highlighting those that have been identified in this work as potentially nitrated in Arabidopsis.
Figure S5. Confirmation of the presence of proteins identified through shotgun proteomic analysis in the immnunopurified nitroproteome. The entire gels for western blots performed in Fig. 2 are shown to account for the specificity of the antibodies.
Figure S6. ROS and NO detection in roots of wild-type plants grown under standard conditions. Nitroblue tetrazolium (NBT) staining of roots in different zones (A, B). Roots were pre-incubated with 10 U ml−1 superoxide dismutase (SOD) prior to NBT staining (C, D). DAF-FM DA staining of roots pre-treated (G, H) or not (E, F) with the NO scavenger cPTIO under UV illumination (E, G) or bright field (F, H).
Table S1. Putative Y-nitrated proteins identified from Arabidopsis and the corresponding functional Y-nitrated counterparts in other organisms.
Table S2. Identification of potentially Y-nitrated proteins by MALDI-TOF peptide fingerprinting of the most abundant 2D gel-excised spots from anti-3-nitroY-immunoprecipitated Arabidopsis proteins.
Table S3. Identification of potential targets of 3-aminoY modification by shotgun LC-MS/MS analysis.
Acknowledgments
We thank Rafael Ruiz-Partida (CIPF) for his valuable help with protein modelling. We also thank Renate Scheibe (Universität Osnabrück, Germany), Dorothee Staiger (University of Bielefeld, Germany), Mariam Sahrawy (EEZ-CSIC, Granada, Spain), Joe Ogas (Purdue University, USA), and Dominique Rumeau (Université de la Méditerranée, France) for their kind donation of antibodies against GAPDH, GRP7, fructose bisphosphatase, PICKEL, and carbonic anhydrase, respectively. The AtMS1 cDNA fused to the 6×His tag was kindly donated by David Dixon (University of Durham, UK). MS-based protein identification was performed by the Proteomic Service of CIPF-PROTEORED (Valencia, Spain). This work was supported by Ministerio de Educación y Ciencia from Spain and FEDER funds from EU grants GEN2003-20477-C02-02, BIO2005-00222, BIO2008-00839, and CONSOLIDER TRANSPLANTA CSD2007-00057 (to JL), a fellowship from the Bancaja-CSIC Programme (to JLJ), and a contract of the I3P Programme of CSIC (co-financed with FEDER funds of the EU, to RC-M).
References
- Abello N, Barroso B, Kerstjens HA, Postma DS, Bischoff R. Chemical labeling and enrichment of nitrotyrosine-containing peptides. Talanta. 2010;80:1503–1512. doi: 10.1016/j.talanta.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Abello N, Kerstjens HA, Postma DS, Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. Journal of Proteome Research. 2009;8:3222–3328. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Bechtold U, Rabbani N, Mullineaux PM, Thornalley PJ. Quantitative measurement of specific biomarkers for protein oxidation, nitration and glycation in Arabidopsis leaves. The Plant Journal. 2009;59:661–671. doi: 10.1111/j.1365-313X.2009.03898.x. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Badger MR, Jones RL. Apoplastic synthesis of nitric oxide by plant tissues. The Plant Cell. 2004;16:332–41. doi: 10.1105/tpc.017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Jones RL. Nitric oxide reduces seed dormancy in Arabidopsis. Journal of Experimental Botany. 2006;57:517–526. doi: 10.1093/jxb/erj060. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Lüthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Brouwer M, Chamulitrat W, Ferruzzi G, Sauls DL, Weinberg JB. Nitric oxide interactions with cobalamins: biochemical and functional consequences. Blood. 1996;88:1857–1864. [PubMed] [Google Scholar]
- Buchczyk DP, Briviba K, Hartl FU, Sies H. Responses to peroxynitrite in yeast: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a sensitive intracellular target for nitration and enhancement of chaperone expression and ubiquitination. Biological Chemistry. 2000;381:121–126. doi: 10.1515/BC.2000.017. [DOI] [PubMed] [Google Scholar]
- Cecconi D, Orzetti S, Vandelle E, Rinalducci S, Zolla L, Delledonne M. Protein nitration during defense response in. Arabidopsis thaliana. Electrophoresis. 2009;30:2460–2468. doi: 10.1002/elps.200800826. [DOI] [PubMed] [Google Scholar]
- Chaki M, Valderrama R, Fernández-Ocaña AM, et al. Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. Journal of Experimental Botany. 2009;60:4221–4234. doi: 10.1093/jxb/erp263. [DOI] [PubMed] [Google Scholar]
- Chang GG, Huang TM. Involvement of tyrosyl residues in the substrate binding of pigeon liver malic enzyme. Biochimica et Biophysica Acta. 1980;611:217–226. doi: 10.1016/0005-2744(80)90058-3. [DOI] [PubMed] [Google Scholar]
- Chen H-JC, Chang C-M, Lin W-P, Cheng D-L, Leong M-I. H2O2/nitrite-induced post-translational modifications of human hemoglobin determined by mass spectometry: redox regulation of tyrosine nitration and 3-nitrotyrosine reductions by antioxidants. Chembiochem. 2008;9:312–323. doi: 10.1002/cbic.200700541. [DOI] [PubMed] [Google Scholar]
- Chiappetta G, Corbo C, Palmese A, Galli F, Piroddi M, Marino G, Amoresano A. Quantitative identification of protein nitration sites. Proteomics. 2009;9:1524–1537. doi: 10.1002/pmic.200800493. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends in Plant Science. 2001;6:145–150. doi: 10.1016/s1360-1385(01)01898-2. [DOI] [PubMed] [Google Scholar]
- Danishpajooh IO, Gudi T, Chen Y, Kharitonov VG, Sharma VS, Boss GR. Nitric oxide inhibits methionine synthase activity in vivo and disrupts carbon flow through the folate pathway. Journal of Biological Chemistry. 2001;20:27296–27303. doi: 10.1074/jbc.M104043200. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Grundy NM, Edwards R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiology. 2005;138:2233–2244. doi: 10.1104/pp.104.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez U, Sauret-Güeto S, Gas E, Jarvis P, Rodríguez-Concepción M. A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. The Plant Cell. 2008;20:1303–1315. doi: 10.1105/tpc.108.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghesquière B, Colaert N, Helsens K, et al. In vitro and in vivo protein-bound tyrosine nitration characterized by diagonal chromatography. Molecular and Cellular Proteomics. 2009;8:2642–2652. doi: 10.1074/mcp.M900259-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokulrangan G, Zaidi A, Michaelis ML, Schöneich C. Proteomic analysis of protein nitration in rat cerebellum: effect of biological aging. Journal of Neurochemistry. 2007;100:1494–1504. doi: 10.1111/j.1471-4159.2006.04334.x. [DOI] [PubMed] [Google Scholar]
- Gow AJ, Farkouh CR, Munson DA, Posencheg MA, Ischiropoulos H. Biological significance of nitric oxide-mediated protein modifications. American Journal of Physiology: Lung Cellular and Molecular Physiology. 2004;287:L262–L268. doi: 10.1152/ajplung.00295.2003. [DOI] [PubMed] [Google Scholar]
- Grün S, Lindermayr C, Sell S, Durner J. Nitric oxide and gene regulation in plants. Journal of Experimental Botany. 2006;57:507–516. doi: 10.1093/jxb/erj053. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Stoimenova M, Kaiser WM. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO. in vitro and in situ. Journal of Experimental Botany. 2005;56:2601–2609. doi: 10.1093/jxb/eri252. [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, et al. Nitric oxide represses the Arabidopsis floral transition. Science. 2004;305:1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- Hong SJ, Gokulrangan G, Schöneich C. Proteomic analysis of age dependent nitration of rat cardiac proteins by solution isoelectric focusing coupled to nanoHPLC tandem mass spectrometry. Experimental Gerontology. 2007;42:639–651. doi: 10.1016/j.exger.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev AU, Hill RD. Plant mitochondrial function during anaerobiosis. Annals of Botany. 2009;103:259–268. doi: 10.1093/aob/mcn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochemical and Biophysical Research Communication. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- Jasid S, Simontacchi M, Bartoli CG, Puntarulo S. Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiology. 2006;142:1246–1255. doi: 10.1104/pp.106.086918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Bahnweg G, Durner J. Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. Journal of Biological Chemistry. 2006;281:4285–4291. doi: 10.1074/jbc.M511635200. [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiology. 2005;137:921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Tewari AK, Zhang L, Green-Church KB, Zweier JL, Chen YR, He G. Proteomic analysis of protein tyrosine nitration after ischemia reperfusion injury: mitochondria as the major target. Biochimica et Biophysica Acta. 2009;1794:476–485. doi: 10.1016/j.bbapap.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Yu X, Cui DY, Sun MH, Sun WN, Tang ZC, Kwak SS, Su WA. The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Research. 2007;17:638–649. doi: 10.1038/cr.2007.34. [DOI] [PubMed] [Google Scholar]
- Melo F, Feytmans E. Assessing protein structures with a non-local atomic interaction energy. Journal of Molecular Biology. 1998;277:1141–1152. doi: 10.1006/jmbi.1998.1665. [DOI] [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J. Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant, Cell and Environment. 2007;30:39–52. doi: 10.1111/j.1365-3040.2006.01604.x. [DOI] [PubMed] [Google Scholar]
- Miyagi M, Sakaguchi H, Darrow RM, Yan L, West KA, Aulak KS, Stuehr DJ, Hollyfield JG, Organisciak DT, Crabb JW. Evidence that light modulates protein nitration in rat retina. Molecular and Cellular Proteomics. 2002;1:293–303. doi: 10.1074/mcp.m100034-mcp200. [DOI] [PubMed] [Google Scholar]
- Morot-Gaudry-Talarmain Y, Rockel P, Moureaux T, Quilleré I, Leydecker MT, Kaiser WM, Morot-Gaudry JF. Nitrite accumulation and nitric oxide emission in relation to cellular signaling in nitrite reductase antisense tobacco. Planta. 2002;215:708–715. doi: 10.1007/s00425-002-0816-3. [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernández E, Pozueta-Romero J, Kuhn JM, Segura J, Ros R. Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiology. 2009;151:541–558. doi: 10.1104/pp.109.143701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Carver TL, Prats E. NO way to live; the various roles of nitric oxide in plant–pathogen interactions. Journal of Experimental Botany. 2006;57:489–505. doi: 10.1093/jxb/erj052. [DOI] [PubMed] [Google Scholar]
- Nicolaou A, Kenyon SH, Gibbons JM, Ast T, Gibbons WA. In vitro inactivation of mammalian methionine synthase by nitric oxide. European Journal of Clinical Investigation. 1996;26:167–170. doi: 10.1046/j.1365-2362.1996.122254.x. [DOI] [PubMed] [Google Scholar]
- Nicolaou A, Warefield CJ, Kenyon SH, Gibbons WA. The inactivation of methionine synthase in isolated rat hepatocytes by sodium nitroprusside. European Journal of Biochemisyry. 1997;244:876–882. doi: 10.1111/j.1432-1033.1997.00876.x. [DOI] [PubMed] [Google Scholar]
- Palamalai V, Miyagi M. Mechanism of glyceraldehyde-3-phosphate dehydrogenase inactivation by tyrosine nitration. Protein Science. 2010;19:255–262. doi: 10.1002/pro.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parani M, Rudrabhatla S, Myers R, Weirich H, Smith B, Leaman DW, Goldman SL. Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnology Journal. 2004;2:359–366. doi: 10.1111/j.1467-7652.2004.00085.x. [DOI] [PubMed] [Google Scholar]
- Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Structural Biology. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polverari A, Molesini B, Pezzotti M, Buonaurio R, Marte M, Delledonne M. Nitric oxide-mediated transcriptional changes in. Arabidopsis thaliana. Molecular Plant-Microbe Interactions. 2003;16:1094–1105. doi: 10.1094/MPMI.2003.16.12.1094. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Campostrini N, Mattè A, Righetti PG, Perazzolli M, Zolla L, Roepstorff P, Delledonne M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics. 2008;8:1459–1469. doi: 10.1002/pmic.200700536. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Laxa M, Mattè A, Zaninotto F, Finkemeier I, Jones AM, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M. S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. The Plant Cell. 2007;19:4120–30. doi: 10.1105/tpc.107.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M. Nitric oxide signalling functions in plant–pathogen interactions. Cell Microbiology. 2004;6:795–803. doi: 10.1111/j.1462-5822.2004.00428.x. [DOI] [PubMed] [Google Scholar]
- Saito S, Yamamoto-Katou A, Yoshioka H, Doke N, Kawakita K. Peroxynitrite generation and tyrosine nitration in defense responses in tobacco BY-2 cells. Plant and Cell Physiology. 2006;47:689–697. doi: 10.1093/pcp/pcj038. [DOI] [PubMed] [Google Scholar]
- Sarver A, Scheffler NK, Shetlar MD, Gibson BW. Analysis of peptides and proteins containing nitrotyrosine by matrix-assisted laser desorption/ionization mass spectrometry. Journal of the American Society of Mass Spectrometry. 2001;12:439–448. doi: 10.1016/S1044-0305(01)00213-6. [DOI] [PubMed] [Google Scholar]
- Schmidt P, Youhnovski N, Daiber A, Balan A, Arsic M, Bachschmid M, Przybylski M, Ullrich V. Specific nitration at tyrosine 430 revealed by high resolution mass spectrometry as basis for redox regulation of bovine prostacyclin synthase. Journal of Biological Chemistry. 2003;278:12813–12819. doi: 10.1074/jbc.M208080200. [DOI] [PubMed] [Google Scholar]
- Simpson GG. NO flowering. Bioessays. 2005;27:239–241. doi: 10.1002/bies.20201. [DOI] [PubMed] [Google Scholar]
- Söderling AS, Hultman L, Delbro D, Højrup P, Caidahl K. Reduction of the nitro group during sample preparation may cause underestimation of the nitration level in 3-nitrotyrosine immunoblotting. Journal of Chromatography B. 2007;851:277–286. doi: 10.1016/j.jchromb.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. Factors determining the selectivity of protein tyrosine nitration. Archives of Biochemistry and Biophysics. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- Stevens SM, Jr., Prokai-Tatrai K, Prokai L. Factors that contribute to the misidentification of tyrosine nitration by shotgun proteomics. Molecular and Cellular Proteomics. 2008;7:2442–2451. doi: 10.1074/mcp.M800065-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, Markesbery WR, Butterfield DA. Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiology of Disease. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Tanaka M, Sohmiya M, Ichinose S, Omori A, Okamoto K. Identification of nitrated proteins in the normal rat brain using a proteomics approach. Neurological Research. 2005;27:630–633. doi: 10.1179/016164105X22039. [DOI] [PubMed] [Google Scholar]
- Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nature Reviews on Drug Discovery. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- Tsumoto H, Taguchi R, Kohda K. Efficient identification and quantification of peptides containing nitrotyrosine by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry after derivatization. Chemical and Pharmaceutical Bulletin. 2010;58:488–494. doi: 10.1248/cpb.58.488. [DOI] [PubMed] [Google Scholar]
- Turko IV, Li L, Aulak KS, Stuehr DJ, Chang JY, Murad F. Protein tyrosine nitration in the mitochondria from diabetic mouse heart. Implications to dysfunctional mitochondria in diabetes. Journal of Biological Chemistry. 2003;278:33972–33977. doi: 10.1074/jbc.M303734200. [DOI] [PubMed] [Google Scholar]
- Zhan X, Desiderio DM. Mass spectrometric identification of in vivo nitrotyrosine sites in the human pituitary tumor proteome. Methods in Molecular Biology. 2009;566:137–163. doi: 10.1007/978-1-59745-562-6_10. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Qian WJ, Knyushko TV, et al. A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. Journal of Proteome Research. 2007;6:2257–2268. doi: 10.1021/pr0606934. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Liu Y, Zhang Q, Wei Q, Zhang W. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta. 2006;224:545–555. doi: 10.1007/s00425-006-0242-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.