Abstract
The synthesis of ribosomal subunits in the nucleolus is a conserved, essential process that results in cytoplasmic ribosomes with precisely processed and folded rRNAs assembled with ribosomal proteins. It has been proposed, but never directly demonstrated, that the U3 small nucleolar RNA (snoRNA), a nucleolar component required for ribosome biogenesis, is a chaperone for pre-18S rRNA folding. To test this, we used in vivo chemical probing with dimethyl sulfate to detect changes in pre-rRNA structure upon genetic manipulation of the yeast, Saccharomyces cerevisiae. Based on changes in nucleotide reactivity, we found that the U3 snoRNA is indeed required for folding of the pre-18S rRNA. Furthermore, we detected a new essential base pairing interaction that is likely the initial anchor that recruits the U3 snoRNA to the pre-rRNA, is a prerequisite for the subsequent interactions, and is required for the small subunit processome formation. Substitution of the 5′-ETS nucleotides of the pre-rRNA involved in this initial base pairing interaction is lethal, but growth is restored when a complementary U3 snoRNA is expressed. The U3 snoRNP, via base pairing, and its associated proteins, are part of the required machinery that orchestrates the folding of pre-rRNA that results in the assembly of the small ribosomal subunit.
INTRODUCTION
The biogenesis of ribosomes is a highly complex process that requires the coordinated activity of a vast number of factors (1–3) and a large proportion of the energy available in a growing cell (4). In all eukaryotes the primary ribosomal RNA (rRNA) transcript, which is synthesized in the nucleolus, is processed through a series of structural re-arrangements, nucleotide modifications and cleavage events that ultimately lead to formation of mature ribosomes. Biochemical and genetic approaches have revealed the many factors involved in this process (1,3,5). Yet the function of many of these factors, the details of their interaction in the pre-ribosome and the structural rearrangements that take place in the pre-rRNA are still undefined.
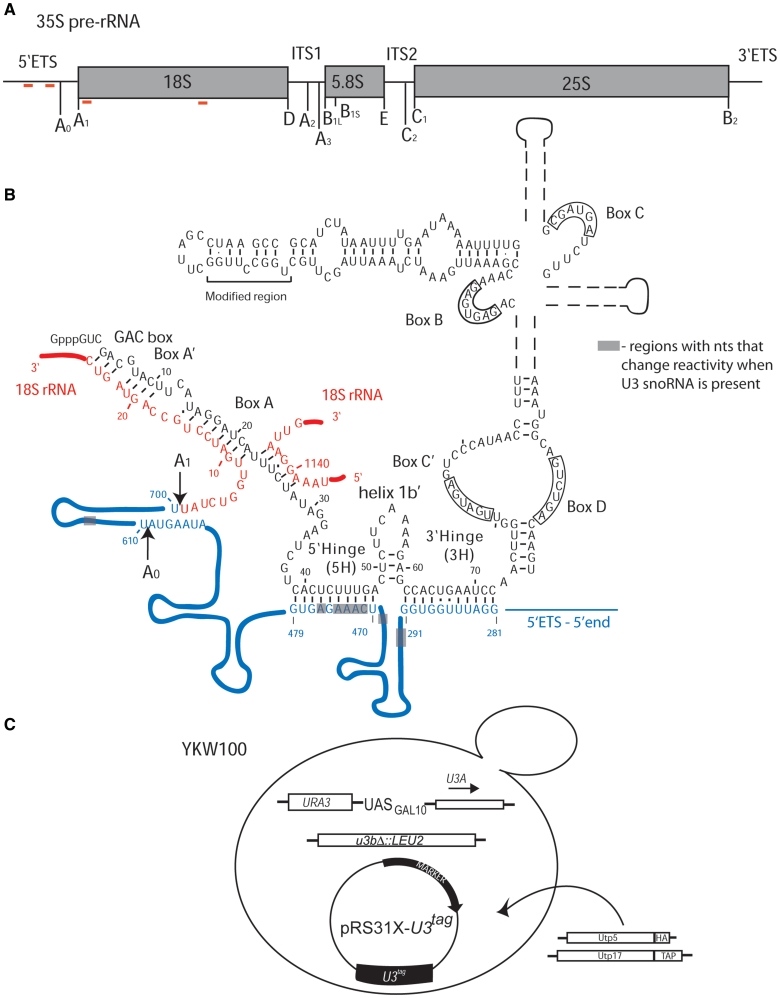
Pre-rRNA processing has been most extensively studied in the yeast Saccharomyces cerevisiae, as a result of the available genetic tools and the ease of manipulation (1–3). In yeast, the pre-rRNA contains three of the four rRNAs separated by spacers, and is processed to form the mature 18S, 5.8S and 25S rRNAs (Figure 1A) (1). Co-transcriptional 2′-O-ribose methylations and pseudouridylations along with cleavage of the internal and external transcribed spacers (ITS and ETS) involve a series of small nucleolar ribonucleoproteins (snoRNPs). The small nucleolar RNAs (snoRNAs) present in the snoRNPs base pair to pre-rRNA sequences and thus guide the RNPs to the site of modification. The U3 snoRNP is unusual for a snoRNA in that it is essential for the cleavage of the 35S primary transcript at sites A0, A1 and A2 (6) and has also been proposed to be a chaperone for 18S rRNA folding (1,7).
Figure 1.
(A) Schematic of the 35S primary pre-rRNA transcript in S. cerevisiae with the processing sites indicated. The 35S pre-rRNA is composed of the 18S, 5.8 and 25S rRNAs, separated by the internal transcribed spacers (ITS1 and ITS2) and the two external transcribed spacers (5′-ETS and 3′-ETS). The red lines indicate the base pairing sites for the U3 snoRNA. (B) The secondary structure of the U3 snoRNA base paired to the 35S pre-rRNA adapted from ‘The Yeast snoRNA database’ (43) and Mereau et al. (14). The U3 snoRNA is shown in black and its different structural and functional elements are indicated, including the modified region used for detection by northern blotting. The 5′-ETS region of the 35S pre-rRNA is shown in blue, the 18S rRNA is shown in red and the cleavage sites are indicated. The gray boxes indicate regions where nucleotides change reactivity to DMS when U3 snoRNA is present. The U3 snoRNA, 5′-ETS and the shaded boxes are not drawn to scale. (C) The YKW100 strain used to analyze the role of the U3 snoRNA in pre-rRNA folding. In the haploid S. cerevisiae strain YKW100 the U3B (SNR17b) gene is disrupted, while the U3A (SNR17a) is under the control of a galactose-inducible/dextrose repressible promoter (GAL10). Tagged U3 snoRNAs, wild-type or mutant, were expressed from plasmids carrying either TRP1 (pRS314) or HIS3 (pRS313) auxotrophic markers. For the co-immunoprecipitation experiments, Utp5 was HA-tagged (KANR) and Utp17 was TAP-tagged (TRP1).
The U3 snoRNA performs its function as a component of a large ribonucleoprotein complex also containing the pre-rRNA, ribosomal proteins (r-proteins) and non-ribosomal proteins, termed the small subunit (SSU) processome (8), and also called the 90S pre-ribosome (9). The SSU processome likely assembles from preformed independent sub-complexes on the nascent pre-rRNA transcript (3,10–13). Several sub-complexes of the SSU processome have been characterized and an order of assembly is emerging. For example, the UtpA/t-Utp sub-complex links transcription and pre-rRNA processing and may be the module that binds first to the pre-rRNA (11,12). Association of the other sub-complexes, including the U3 snoRNP, is dependent on this event (12).
In S. cerevisiae the U3 snoRNA is considered to be composed of two secondary structural domains: a short 5′-domain (nucleotides 1–39) linked to the larger 3′-domain (nucleotides 73 to the 3′-end) through a hinge region (Figure 1B) (14,15). There are two conserved regions of the U3 snoRNA that base pair with regions in the pre-18S rRNA: (i) the 5′-end of the U3 snoRNA (the GAC box and boxes A′ and A) base pairs to the 18S rRNA region of the pre-rRNA, downstream of the A1 cleavage site and with an internal site in the 18S rRNA and (ii) the 5′-part of the hinge (5′-hinge) base pairs with the 5′-ETS of the pre-rRNA (Figure 1B) (7,16–20). The 3′-part of the hinge (3′-hinge) of the U3 snoRNA has also been predicted to base pair with the 5′-ETS, based on results obtained in other eukaryotes (21–24), but this base pairing has not yet been tested in yeast. In addition to its role in the processing of the pre-18S rRNA, the U3 snoRNA also participates in the formation of the conserved 5′-end pseudoknot in the 18S rRNA, leading to the proposal that it acts as a chaperone in the folding of the rRNA (1,7). There is, as yet, no direct evidence that has confirmed the role of the U3 snoRNA as a pre-18S rRNA chaperone. This remains one of the outstanding questions in the field of ribosome biogenesis.
Chemical probing can be used to investigate conformational changes of RNA in living cells because it determines changes in reactivity with nucleotide resolution (14,25,26). The variations in reactivity can be attributed to differences in the accessibility of nucleotides in the probed molecule under different conditions. Dimethyl sulfate (DMS) is the most widely used probe for in vivo chemical probing as it penetrates yeast cells rapidly without prior permeabilization, allowing for a quick snapshot of the RNA structure in vivo. The reactivity towards DMS is reduced if the base is involved in hydrogen bonding or if it is not solvent accessible. As DMS is a base-specific chemical probe, methylation of A and C residues can be detected by primer extension without further manipulation. The modified nucleotides are detected as a pause one nt before the site of methylation. Recently, in vivo DMS probing was used to analyze the structure of the pre-rRNA from a region encompassing the D cleavage site (27). Previous attempts at chemical probing of rRNA in vivo in the yeast S. cerevisiae have been limited to small regions of the large subunit of the ribosome (28–30), and were successful. In vivo chemical probing with DMS is thus a powerful tool for the analysis of RNA structure and conformational changes in the native environment.
To analyze the role of the U3 snoRNA in ribosome biogenesis we used an approach combining the power of yeast genetics with in vivo chemical probing. We tested whether the U3 snoRNA is required for folding of the pre-18S rRNA by in vivo chemical probing with DMS when the U3 snoRNA is conditionally expressed, and determined that there were indeed changes in the structure of the pre-rRNA dependent on the presence of the U3 snoRNA (Figure 1B). We go on to show that a specific chemical modification pattern is a result of a base pairing interaction, previously undescribed in yeast, between the 3′-hinge of the U3 snoRNA and the 5′-ETS sequence of the pre-rRNA. Both mutational analysis and genetic ‘rescue’ experiments support this initial interaction as a pre-requisite for the subsequent base pairing interactions and show that it is necessary for the assembly of the SSU processome.
MATERIALS AND METHODS
Strains and media
The strain YKW100 (a ura3-52 his3-Δleu2 lys2-801amber trp1-Δ63 u3aΔ UASGAL:U3A::URA3 u3bΔ::LEU2) (10) was used in all the chemical probing and co-immunoprecipitation experiments. The NOY504 (a rrn4::LEU2 ade2-101 ura3-1 trp1-1 leu2-3, 112 his3-11 can1-100) (31) strain was used for the experiments involving mutations in the 5′-ETS of the pre-rRNA. In general, yeast were grown in YPD (1% yeast extract, 2% peptone and 2% dextrose), YPG/R (1% yeast extract, 2% peptone, 2% galactose and 2% raffinose), or yeast selective media (SC-Trp, SC-His or SC-Ura-Trp; Clontech) supplemented with either 2% dextrose or 2% galactose and 2% raffinose. The solid media contained 2% Bactoagar. A standard lithium acetate protocol (32) was used for the yeast transformations. For depletion experiments the YKW100 strain with the appropriate plasmid was grown in selective media with galactose and raffinose to an optical density at 600 nm of 0.4-0.8 and shifted to YPD for the indicated time. The NOY504 strain was grown at 25°C and then shifted to 37°C.
Plasmids
The plasmid pRS314 U3 WT contains a copy of the U3 snoRNA gene without its intron that carries a unique sequence that enables detection by northern blotting in the yeast expression vector pRS314 (AMPR, TRP, CEN/ARS) (10). The plasmid pRS313 U3 WT is identical but carries a different auxotrophic marker (AMPR, HIS, CEN/ARS). pRS314 U3 Box A, pRS314 U3-63, pRS314 U3-72 were obtained by excising the XhoI-EcoRI U3 fragments from pRS313 Box A, pRS313 U3-63 and pRS313 U3-72, described as pRU3*[Box A : subst], pR*[-63], pR*[trunc/-72] in previous studies (15,33). The plasmids pRS314 U3 5H, pRS314 U3 3H7 and pRS314 U3 3H11 were created by site directed mutagenesis with the Quick Change kit from Stratagene, and subsequently subcloned in the pRS313 plasmid, as described above. All the mutations were confirmed by DNA sequencing. The pRS31× U3 snoRNA containing plasmids were transformed in YKW100 and maintained on the corresponding selective media SC-His/Trp.
To generate the mutations in the 5′-ETS region of the pre-rRNA, a BamHI-SacII fragment (1560 nt) of the pNOY102 plasmid (31) was PCR amplified and subcloned into the PCR4-TOPO vector (TOPO® TA Cloning® Kit; Invitrogen). After site directed mutagenesis (Quick Change kit; Stratagene) the fragments containing the altered sequences were used to replace the same fragment in pNOY102. All plasmid constructs were verified by DNA sequencing at the DNA Analysis Facility at Yale University. The mutagenized plasmids were transformed into NOY504 alone or with pRS314 U3 snoRNA containing plasmids and maintained on SC-Ura or SC-Ura-Trp, respectively.
Chemical probing in vivo
The chemical probing protocol was adapted from ref. (25). Briefly, cells grown to an optical density at 600 nm of 0.5 in 15 ml of media were treated with DMS diluted 1:10 in ethanol to a final concentration of 20 mM for the analysis of the 5′-ETS and 10 mM for the analysis of the 18S region of the pre-rRNA. The reaction was stopped after 4 min at 30°C with 10 ml ice-cold 1 M 2-mercaptoethanol and the cells pelleted by centrifugation. A second addition of 10 ml of 0.6 M ice-cold 2-mercaptoethanol ensured that all the DMS was neutralized.
RNA manipulations: primer extension and northern blots
Total RNA used for primer extension and northern blotting was obtained by hot phenol extraction (34). The quality of the RNA was assessed on 1.25% agarose gels in 1× TAE. Northern blots were performed as described earlier (15,33). Briefly, 7 µg of RNA were separated on 8% denaturing polyacrylamide gels, transferred to a Hybond N+ membrane (GE Healthcare) and a series of oligonucleotides were used to detect the desired snoRNAs. The oligonucleotides were: SD13 to detect U3 snoRNA expressed from the plasmid that carries a unique sequence to facilitate detection, SD74 which hybridizes to all forms of U3 snoRNA (15,33) and an oligonucleotide specific for the U14 snoRNA.
The primer extension protocol was adapted from (25,35). The primers used for primer extension are listed in Supplementary Table S2. The primer is named for the first nucleotide incorporated by the reverse transcriptase and the part of the pre-rRNA where it anneals. For analyzing the 5′-ETS region, 5 µg of total RNA were used while 2.5 µg of total RNA were necessary for the analysis of the 18S rRNA region. Briefly, 0.3 pmoles of a 5′-end 32P labeled oligonucleotide were hybridized to the target RNA in a final volume of 4.5 µl (50 mM K+ HEPES pH 7.0, 100 mM KCl) at 92°C for 2 min and afterwards cooled slowly to 42°C. Extension buffer and dNTP mix were added to a final concentration of 0.13 M Tris–Cl pH 8.5, 10 mM MgCl2, 10 mM DTT and 0.125 mM dNTP mix with 2 U of AMV-RT (20 U/µl from Northstar BioProducts). The final volume was 6.5 µl and the extension was performed at 42°C for 30 min. Sequencing ladders were generated by adding 1 µl of the corresponding 1 mM ddNTP, in the same conditions. The reaction was stopped by adding 11 µl stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol from USB). The samples were heated for 2 min at 95°C, cooled on ice and loaded (2.5 µl) on a 6% acrylamide denaturing gel in 1× TBE. Some films were scanned and quantitation performed with Image J.
Co-immunoprecipitations
The co-immunoprecipitations were performed as described (36) with the following modification: 10% of the input was used for RNA extraction and northern blotting as described in the RNA manipulation section.
RESULTS
In vivo chemical probing to determine the role of the U3 snoRNA in pre-rRNA folding—the system
To acquire information on the role(s) of the U3 snoRNA in the cleavage and folding of pre-rRNA during ribosome biogenesis we analyzed the modification patterns by chemical probing and primer extension of pre-rRNA in the absence and presence of the U3 snoRNA in vivo. For this purpose we used a system previously described (7,10,15,33). In the yeast strain YKW100 one of the U3 snoRNA genes is disrupted and the second one has been placed under a galactose-inducible/dextrose-repressible promoter (7,10,15,33) (Figure 1C). Wild-type U3 snoRNA or U3 snoRNA containing mutations can be constitutively expressed from a plasmid (Figure 1C, Supplementary Table S1). The plasmid-encoded U3 snoRNA carries a unique sequence that facilitates its detection (Figure 1B and C). To establish the most suitable experimental conditions for chemical probing and the best time after carbon source shift at which to probe, a series of experiments were performed. When the yeast were shifted from galactose to dextrose containing medium, the U3 snoRNA levels were reduced as soon as 3 h (Figure 2A), while the 18S rRNA levels began to decrease only after 10 h (Figure 2B). We chose to perform in vivo chemical probing ∼16 h after the transfer to dextrose containing medium, when both the U3 snoRNA and the 18S rRNA were greatly depleted. Otherwise, the presence of high quantities of mature 18S rRNA would mask the pre-rRNA, while the high levels of U3 snoRNA wild-type would obscure the effects of mutations. The optimal conditions for in vivo chemical probing (concentration, temperature and time) and primer extension were established by determining when full length cDNAs transcribed from the unmodified and modified RNA were comparable in levels and when a good modification pattern was obtained (25,26) (data not shown). To ensure that the modification reaction was stopped, controls were also performed by adding 2-mercaptoethanol (the quenching reagent) before adding the DMS (25) (data not shown). A series of primers were designed to efficiently cover the length of the pre-18S rRNA (Supplementary Table S2), and also to provide overlapping sequence as necessary. The quantities of RNA and primer used for primer extension were also determined experimentally to obtain a good signal to noise ratio (data not shown).
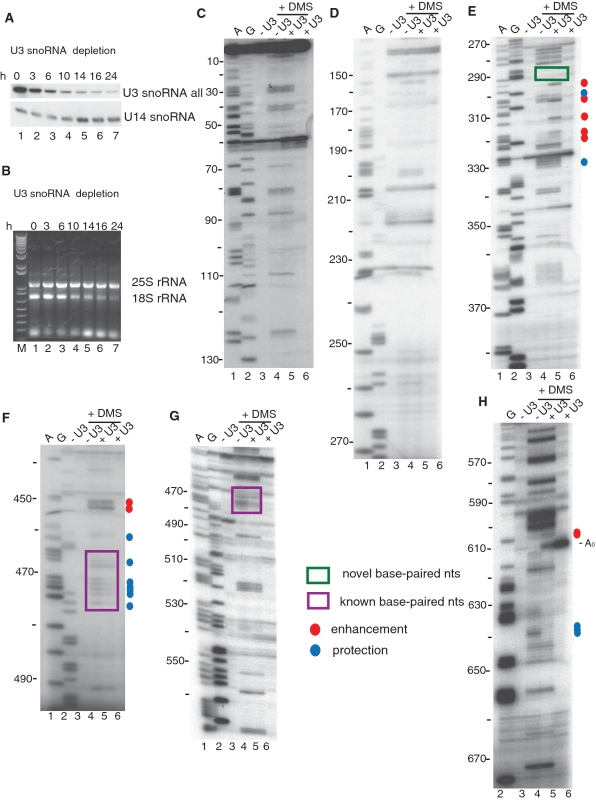
Figure 2.
Depletion of the U3 snoRNA results in changes in 5′-ETS nucleotide reactivity to DMS as determined by in vivo chemical probing. (A) Total RNA was extracted from yeast cells shifted from galactose containing media (0 h) to dextrose containing media for the indicated time and analyzed. Time course for the depletion of the endogenous U3 snoRNA in the YKW100 strain detected by northern blotting. The U3 snoRNA was probed with an oligonucleotide (SD74) (15) that recognizes all forms of the U3 snoRNA, while for the U14 snoRNA a specific oligonucleotide was used. (B) Depletion of the U3 snoRNA leads to reduced 18S, but not 25S rRNA levels. Mature 18S and 25S rRNA levels were followed on an agarose gel with ethidium bromide staining for the samples analyzed by northern blotting in (A). The M lane is a marker lane (1 Kb plus DNA ladder; Invitrogen). (C–H) In vivo DMS modification of the 5′-ETS of the pre-rRNA. DMS was added at 16 h after shifting the yeast to dextrose to deplete the endogenous U3 snoRNA. Yeast either contained a plasmid that expressed the tagged U3 snoRNA (+U3; U3 WT) or an empty vector (−U3; EV). Total RNA was extracted from unmodified and modified yeast and analyzed by primer extension with the following primers: (C) 157-5′-ETS; (D) 277-5′-ETS; (E) 400-5′-ETS; (F) 500-5′-ETS; (G) 611-5′-ETS and (H) 693-5′-ETS. Lanes A and G are dideoxy sequencing lanes, and the nucleotide numbers from the start site of transcription are indicated on the left side of the gels, and the A0 cleavage site is indicated. Blue circles denote nucleotides with lower reactivity to DMS, while red circles denote nucleotides with higher reactivity. The size of the dot represents the intensity of the change. The purple box highlights the nucleotides of the 5′-ETS that were previously known to base pair the U3 snoRNA, while the green boxes highlight the nucleotides involved in the novel base pairing.
The presence of U3 snoRNA causes changes in the structure of the 5′-ETS of the pre-rRNA
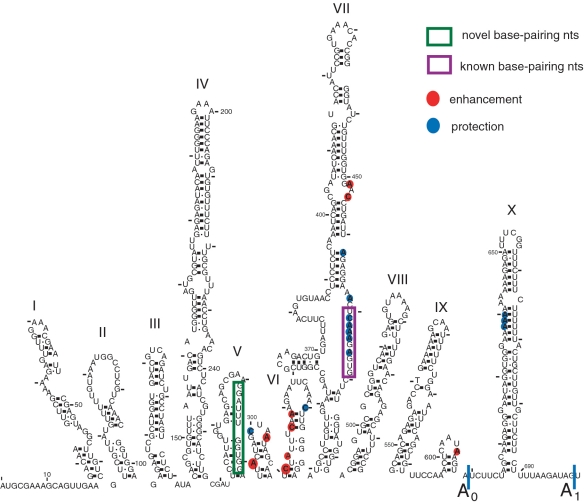
Genetic and biochemical experiments have shown that the yeast U3 snoRNA base pairs with the pre-rRNA in the 5′-ETS (5′-hinge with nucleotides 470–479; Figure 1B) (16–18) and with the 18S rRNA (Box A with nucleotides 9–16; Figure 1B) (7,19,20). Genetic experiments also suggest that Boxes GAC and A′ base pair to nucleotides 19–25 and 1139–1143 of the 18S rRNA (Figure 1B) (7,19,20). Consequently, we surveyed the DMS modification patterns in the 5′-ETS when the U3 snoRNA is present compared to when it is not (Figure 2C–H, compare lanes 4 and 5). No differences in the reactivity of nucleotides 5–290 of the 5′-ETS were observed under these two conditions (Figure 2C and D compare lanes 4 and 5, summarized in Figures 1B and 3, Supplementary Table S3), implying that the presence of the U3 snoRNA is not required to maintain the structure in this region. However, differences in the reactivity of the nucleotides of the pre-18S rRNA were detected in the nucleotides 290–330 region (Figure 2E compare lanes 4 and 5, summarized in Figures 1B and 3, Supplementary Table S3). In addition, some nucleotides in the nucleotide 470 region (471–474, 476), where the U3 snoRNA base pairs with the pre-18S rRNA, became less reactive in the presence of U3 snoRNA (Figure 2F–G compare lanes 4 and 5, summarized in Figures 1B and 3, Supplementary Table S3). This protection in the presence of the U3 snoRNA suggests that base pairing of the U3 snoRNA to the 5′-ETS is detectable by this chemical probing technique. Several changes were also observable in the region close to the A0 and A1 cleavage sites, regions 605 and 635 (Figure 2H compare lanes 4 and 5, summarized in Figures 1B and 3, Supplementary Table S3).
Figure 3.
Summary of the results from in vivo DMS modification of the 5′-ETS with and without the U3 snoRNA (data shown in Figure 2). Nucleotides with altered reactivity as a result of the presence of the U3 snoRNA, compared to absence of the U3 snoRNA as a reference point, are shown on the secondary structure of the 5′-ETS adapted from (44). Blue circles denote nucleotides with lower reactivity to DMS, while red circles denote nucleotides with higher reactivity. The size of the dot represents the intensity of the change. The purple box highlights the nucleotides of the 5′-ETS that were previously known to base pair the U3 snoRNA, the green box highlights the nucleotides involved in the novel base pairing and the cleavage sites are indicated.
The U3 snoRNA causes changes in the structure of the pre-rRNA near the 5′-end pseudoknot of the 18S rRNA
The conserved pseudoknot located at the 5′-end of the mature 18S rRNA involves nucleotides from the 5′-end of the 18S rRNA (nucleotides 4–20) in which nucleotides 12–14 base pair with nucleotides 1140–1142 of the 18S rRNA to form the tertiary interaction. Surveying changes in reactivity around the nucleotides involved in the 5′-pseudoknot, we found that nucleotides 12–14, directly involved in the 5′-pseudoknot, were not reactive to DMS under our conditions (Supplementary Figure S1A compare lanes 4 and 5 dark and light exposure, Supplementary Figure S2 and Table S4). In contrast, many nucleotides located in the 5′-domain of the 18S rRNA, starting around nucleotide 40, were much more reactive when U3 snoRNA was repressed (Supplementary Figure S1A compare lanes 4 and 5, S1B and S1C compare lanes 5 and 6, Supplementary Figure S2 and Table S4). At increased distance from the 5′-end, the difference in reactivity of nucleotides induced by the presence of the U3 snoRNA decreased (Supplementary Figure S1A compare lanes 4 and 5, S1B and S1C compare lanes 5 and 6, Supplementary Figure S2 and Table S4). Few differences in reactivity to DMS with and without the U3 snoRNA were observed in the region close to nucleotide 1100 (Supplementary Figure S1D–E compare lanes 5 and 6, Supplementary Figure S2 and Table S4) including at nucleotides 1140–1142, which are also directly involved in the 5′-pseudoknot. Lack of RNA reactivity to DMS can generally be attributed to base pairing, inaccessible structure or protection by protein components. In contrast, nucleotide 1139, the nucleotide immediately adjacent to the pseudoknot nucleotides, became less reactive when U3 snoRNA was expressed (Supplementary Figure S1E compare lanes 5 and 6, Supplementary Figure S2 and Table S4), indicating formation of the 5′-end pseudoknot. In summary, we were unable to obtain information on the nucleotides directly involved in formation of the 5′-end pseudoknot by chemical probing in vivo with DMS; however, we do detect decreased accessibility to DMS in the region surrounding the 5′-end pseudoknot nucleotides when U3 snoRNA is present. The higher reactivity of 18S rRNA sequences in general in the absence of the U3 snoRNA is evidence of a less structured state of the pre-rRNA.
Mutations in the U3 snoRNA that are predicted to disrupt base pairing with the pre-18S rRNA also lead to changes in pre-rRNA structure
As noted in the Introduction, the U3 snoRNA base pairs with the pre-rRNA at at least three different sites. U3 snoRNAs with extensive substitutions in the sequences that base pair to the pre-rRNA do not support growth (data not shown) (33). However, we do not know whether disrupting these base pairing interactions leads to changes in pre-rRNA folding or structure.
We tested whether expression of U3 snoRNAs bearing substitutions of these essential sequences leads to changes in pre-rRNA structure by constructing yeast strains expressing only the mutant U3 snoRNAs, and by analyzing the reactivity of the 5′-ETS following in vivo DMS modification. Plasmids containing the unmutated U3 (U3 WT), the U3 snoRNA with 6 nt of the 5′-hinge region mutated (U3 5H) and the U3 snoRNA with mutations in the Box A sequence (U3 Box A) (Supplementary Table S1) were transformed into the yeast strain YKW100, where the endogenous U3 snoRNA can be depleted by growth in dextrose. As expected, when the strain was grown in dextrose, the endogenous U3 snoRNA was depleted (Figure 4A, lane 5). Expression of the WT and the two mutant U3 snoRNAs from the plasmid was readily detectable via a unique sequence tag when the endogenous U3 snoRNA expression was repressed (Figure 4A, lanes 6 to 8), though the levels of the two mutant U3 snoRNAs were somewhat lower than U3 WT. Consistent with the results that showed that these mutant U3 snoRNAs do not support yeast growth (data not shown) (33), the levels of 18S rRNA were also reduced in yeast when only these mutant U3 snoRNAs were expressed (Figure 4B, lanes 7 and 8).
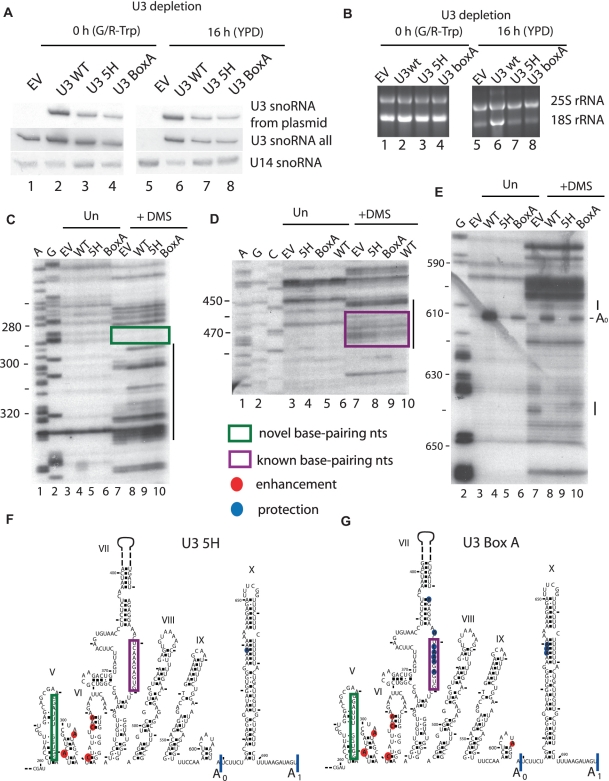
Figure 4.
Expression of U3 snoRNAs mutated in nucleotides that base pair changes the reactivity of 5′-ETS nucleotides to DMS in vivo. Total RNA was extracted from yeast cells depleted of the endogenous U3 snoRNA as described in the legend to Figure 2. Yeast contained either a plasmid expressing no U3 snoRNA (empty vector; EV), U3 snoRNA wild-type (U3 WT), a U3 snoRNA with the 5′-hinge mutated (U3 5H) (17) or a U3 snoRNA with the Box A mutated (U3 Box A) (33). (A) Expression of U3 snoRNA mutants from plasmids was detected via their unique sequence tag by northern blotting (oligo SD13) (15), while another oligonucleotide was used to visualize all forms of U3 snoRNA (SD74). The U14 snoRNA was detected with a specific oligonucleotide. (B) rRNA levels analyzed by ethidium bromide staining on agarose gels. (C–E) In vivo DMS modification of the RNA at 16 h after U3 snoRNA depletion was carried out in cells containing the plasmids described in (A). Total RNA was extracted from unmodified and modified cells and analyzed by primer extension with the following oligonucleotides: (C) 400-5′-ETS; (D) 611-5′-ETS; (E) 693-5′-ETS. Lanes A and G are dideoxy sequencing lanes. The nucleotide numbers from the start site of transcription are indicated on the left side of the gels. The lines on the right side of the gels highlight regions were nucleotides with changed reactivity to DMS are present. Nucleotides with altered reactivity as a result of the expression of the mutated U3 snoRNAs depicted on a portion (nucleotides 260–700) of the secondary structure of the 5′-ETS: (F) U3 5H and (G) U3 Box A. Blue circles denote nucleotides with lower reactivity, while red circles denote nucleotides with higher reactivity when the U3 snoRNA mutants are expressed, using nucleotide reactivity when no U3 snoRNA is expressed (EV) as a reference point. The size of the circles represents the intensity of the change. The purple boxes highlight the nucleotides of the 5′-ETS that were previously known to base pair the U3 snoRNA, the green boxes highlight the nucleotides involved in the novel base pairing and the cleavage sites are indicated.
We surveyed the changes in reactivity in the 5′-ETS in the presence of the two mutant U3 snoRNAs following in vivo DMS modification only in the areas where we observed differences in reactivity when U3 snoRNA was expressed (Figure 2C–H, summarized in Figures 1B and 3). Interestingly, we observed that the enhanced reactivity in the nucleotides 290–330 region occurred even in the presence of the mutations in the 5′-hinge and Box A sequences of the U3 snoRNA (Figure 4C, compare lanes 8–10, 4F, 4G, Supplementary Table S3), suggesting that these reactivity changes are independent of mutation in these essential elements. However, the few protections we previously observed in this region were lost when any substitution in the U3 snoRNA was made.
As shown in Figure 2, we were able to detect the base pairing interaction of the 5′-hinge nucleotides (nucleotides 39–48) to the 5′-ETS nucleotides (470–479) by changes in reactivity of these nucleotides in the presence and absence of U3 snoRNA. When we tested a U3 snoRNA with a mutation in the 5′-hinge sequence (U3 5H) that is also predicted to disrupt the interaction, we again found that the reactivity of these nucleotides in the 5′-ETS was increased similar to the levels obtained with the empty vector (Figure 4D compare lanes 7, 8 and 10, 4F, Supplementary Table S3). This indicates, as others have previously found (18), that mutations in the 5′-hinge sequence of the U3 snoRNA disrupt its base pairing with the 5′-ETS.
In contrast, mutation in the Box A sequence of the U3 snoRNA, which is not predicted to disrupt the base pairing interaction with nucleotides 470–479, conferred DMS reactivity similar to that of the WT U3 snoRNA for these nucleotides (Figure 4D compare lanes 9 and 10, 4G, Supplementary Table S3). Therefore, mutations in the Box A sequence permit the base pairing interaction to occur between the 5′-hinge sequence of the U3 snoRNA and nucleotides 470–479 of the 5′-ETS.
We also detected several differences in the chemical modification pattern for the different mutations in the U3 snoRNA in the area close to the A0 cleavage site and in the region situated between the A0 and A1 cleavage sites (nucleotide 605–700; Figure 4E–G, Supplementary Table S3). The enhancement observed for the U3 WT at nucleotide 605 and the protection at nucleotides 638 and 640, compared to the empty vector, were not present when the 5′-hinge region was mutated (U3 5H), while a weak protection was still present at 639 (Figure 4E, compare lanes 8 and 9, 4F, Supplementary Table S3). In contrast, when Box A was substituted (U3 BoxA) a weaker enhancement compared to U3 WT was observed, and the protections at nucleotides 638–640 were still present (Figure 4E, compare lanes 8 and 10, 4G, Supplementary Table S3). Consequently, both mutations in the 5′-hinge region and in Box A of the U3 snoRNA impede, to varying extents, the changes in reactivity attributable to the presence of U3 WT.
Using U3 snoRNAs that are mutated in sequences known to base pair with the pre-18S rRNA, we were thus able to detect the known base pairing interaction between the 5′-hinge sequence of the U3 snoRNA and the 5′-ETS. Not unexpectedly, mutations in the Box A sequence of the U3 snoRNA do not affect this base pairing interaction. However, the chemical modification pattern observed between nucleotides 290 and 330 (nucleotide 300 region of the pre-rRNA) was quite unexpected as it was independent of substitutions in the known functionally important regions of the U3 snoRNA (Figure 4C). It raises the question of whether there is another base pairing interaction between the U3 snoRNA and the 5′-ETS that is responsible for this chemical modification pattern. To answer this question we analyzed the chemical modification patterns present when truncated U3 snoRNAs are expressed.
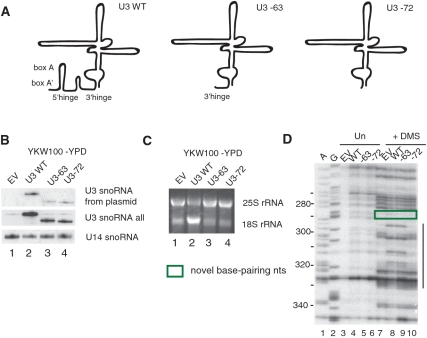
The 3′-hinge sequence of the U3 snoRNA influences the structure of the nucleotides 290–330 region of the 5′-ETS via an initial base pairing interaction
To dissect the potential role of additional base pairing interactions between the U3 snoRNA and the pre-rRNA, we analyzed the DMS reactivity patterns when truncated forms of U3 snoRNAs were expressed. We tested two previously described 5′-end truncated U3 snoRNAs (Supplementary Table S1), a form missing the first 63 nts and one missing the first 72 nts from the 5′-end (Figure 5A–C) (15,33). Consistent with the observation that these mutant U3 snoRNAs do not support growth (15,33), the levels of 18S rRNA were lower when either of the truncated forms were expressed compared to the U3 WT (Figure 5C, lanes 2–4). When the U3 snoRNA was truncated by 72 nts from its 5′-end, the enhancements in the nucleotides 290–330 region were no longer observed (Figure 5D, compare lanes 8 and 10). However, when a longer form in which only 63 nts were removed from the 5′-end was expressed, the enhancements were still present (Figure 5D, compare lanes 8 and 9). This is surprising as neither form contains the functionally important 5′-hinge or Box A sequences that base pair with the pre-rRNA. Therefore nucleotides 63–72 of the U3 snoRNA, encompassing the 3′-hinge, are essential for the observed changes in chemical reactivity among nucleotides 290–330 of the 5′-ETS. The 3′-hinge sequence of the U3 snoRNA would be predicted to base pair with the 5′-ETS at nucleotides 281–291 (Figure 1B) (22), immediately 5′-to the observed chemical modification pattern.
Figure 5.
The presence of the 3′-hinge nucleotides (62–72) of the U3 snoRNA affects the in vivo chemical modification pattern present in the 290–330 region of the 5′-ETS. (A) Schematic drawing of U3 snoRNA WT, U3 snoRNA-63 and U3 snoRNA-72 with the functionally important base pairing sequences indicated. (B–D) Total RNA was extracted from cells expressing the empty vector (EV), U3 snoRNA wild-type (U3 WT), U3 snoRNA–63 (U3-63) and U3 snoRNA-72 (U3-72) and analyzed by different methods. (B) Expression of U3 snoRNA truncations detected by northern blotting. U3 snoRNA expressed from plasmid was detected with an oligonucleotide (SD13) that recognizes the unique sequence, while another oligonucleotide (SD74) was used to detect all forms of the U3 snoRNA. The U14 snoRNA was visualized with a specific oligonucleotide. (C) rRNA levels in the presence of the indicated U3 snoRNAs analyzed by ethidium bromide staining on agarose gels. (D) In vivo DMS modification of RNA, at 16 h after U3 snoRNA depletion, was performed simultaneously in yeast where U3 snoRNAs were expressed from a plasmid (WT, -63, -72) and in yeast where U3 snoRNA was absent (EV) for comparison. Total RNA was extracted from unmodified and modified cells and analyzed by primer extension with the primer 400-5′-ETS. Lanes A and G are dideoxy sequencing lanes, and the nucleotides are indicated on the left side of the gel. The line on the right side of the gel highlights the region where nucleotides with changed reactivity to DMS are present. The green box highlights the nucleotides of the 5′-ETS involved in the novel base pairing.
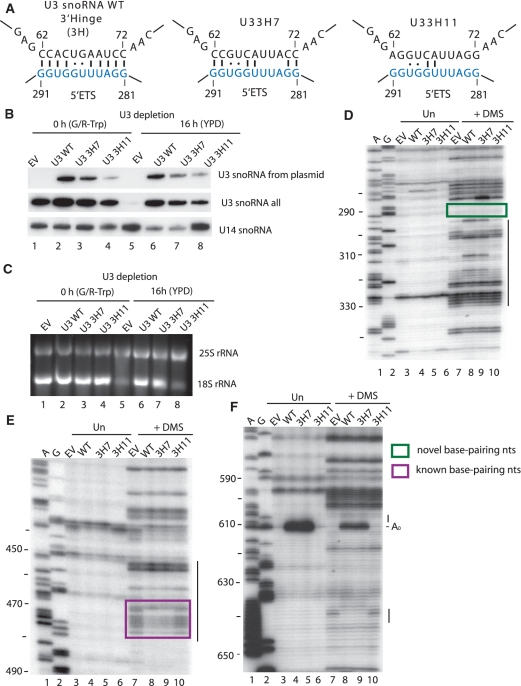
To further analyze the role of the 3′-hinge region of the U3 snoRNA, we substituted the nucleotides from 62 to 72 en bloc. Two different 3′-hinge mutants of U3 snoRNA were generated, the first, U3 3H7, which maintains 8/11 predicted base pairing interactions and U3 3H11, which maintains only 4/11 predicted base pairing interactions (Supplementary Table S1, Figure 6A). Both mutant U3 snoRNAs were expressed, though not to the same levels as WT (Figure 6B, lanes 6–8). When the U3 3H7 snoRNA was expressed the yeast grew as well as yeast expressing the WT U3 snoRNA (data not shown) and 18S rRNA maturation was not affected (Figure 6C, compare lanes 6 and 7), demonstrating that this extent of mutation is not enough to disrupt this base pairing interaction. In contrast, the U3 3H11 snoRNA did not support growth (data not shown) and the levels of 18S rRNA were drastically reduced, comparable to the levels observed in the absence of U3 snoRNA (Figure 6C, compare lanes 5 and 8). Thus, the U3 3H7 mutation has no effect on the function of U3 snoRNA while the U3 3H11 mutation is detrimental. Since the latter disrupts the predicted base pairing more extensively, it is likely that the 3′-hinge sequence of the U3 snoRNA base pairs with nucleotides 281–291 of the 5′-ETS, and is required for U3 snoRNA function.
Figure 6.
Disruption of the U3 snoRNA 3′-hinge:5′-ETS interaction results in reduced levels of 18S rRNA and the disappearance of the in vivo chemical modification pattern observed at nucleotides 290–330. (A) base pairing between the 5′-ETS of the pre-rRNA and the U3 snoRNA WT, the U3 snoRNA with 7 nt of the 3′-hinge mutated (U3 3H7) and the U3 snoRNA with 11 nt mutated (U3 3H11). (B–C) Total RNA was extracted from cells expressing the empty vector (EV), U3 snoRNA wild-type (U3 WT), U3 3H7 and U3 3H11 and analyzed by different methods. (B) Expression of U3 snoRNA 3′-hinge mutants analyzed by northern blotting. U3 snoRNA expressed from a plasmid was detected with an oligonucleotide (SD13) that recognizes the unique sequence, while another oligonucleotide (SD74) was used to detect all forms of U3 snoRNA. The U14 snoRNA was visualized with a specific oligonucleotide. (C) rRNA levels analyzed by ethidium bromide staining on agarose gels. (D–F) In vivo DMS modification of the RNA was performed, at 16 h after endogenous U3 snoRNA was depleted, in yeast expressing the indicated U3 snoRNAs and was compared to yeast where the U3 snoRNA was absent (EV). Total RNA was extracted from unmodified (Un) and modified (+DMS) cells and analyzed by primer extension with the following primers: (D) 400-5′-ETS (E) 611-5′-ETS; (F) 693-5′-ETS. The A0 cleavage site is indicated. Lanes A and G are dideoxy sequencing lanes, and the nucleotide numbers are indicated on the left side of the gels. The lines on the right side of the gels highlight regions were nucleotides with changed reactivity to DMS are present. The purple box highlights the nucleotides of the 5′-ETS that were previously known to base pair the U3 snoRNA, the green box highlights the nucleotides involved in the novel base pairing and the A0 cleavage site is indicated.
We asked whether substitution of the nucleotides in the 3′-hinge interfered with the characteristic chemical modification pattern observed in the 290–330 region upon in vivo chemical probing. When the fully functional U3 3H7 mutant was expressed, chemical probing revealed very few changes compared to the presence of U3 WT (Figure 6D, compare lanes 8 and 9, Supplementary Table S3). However, the more extensive disruption of base pairing with the U3 3H11 snoRNA resulted in the absence of the modification pattern in the nucleotides 290–330 region (Figure 6D, compare lanes 8 and 10, Supplementary Table S3). Our results indicate that mutation of nucleotides in the 3′-hinge that would prevent its base pairing to nucleotides 281–291 in the 5′-ETS causes loss of the chemical modification pattern in the nucleotides 290–330 region. Thus, these changes in pre-rRNA structure can be attributed to base pairing of the 3′-hinge sequence of the U3 snoRNA with the 5′-ETS at nucleotides 281–291.
Similarly, our results indicate that base pairing between the 3′-hinge sequence and the 5′-ETS is a prerequisite for the other U3 snoRNA:5′-ETS base pairing interaction. When the 3′-hinge base pairing was extensively disrupted with the 3H11 mutation, we found increased reactivity of the 5′-ETS nucleotides (nucleotides 470–479) that base pair with the 5′-hinge (Figure 6E, compare lanes 8 and 10, Supplementary Table S3). The levels of increased reactivity were similar to those observed when no U3 snoRNA was present (empty vector; Figure 6E, compare lanes 7 and 10, Supplementary Table S3). However, no changes in reactivity in nucleotides 470–479 were observed when a U3 snoRNA that does not disrupt the base pairing interaction (3H7) was expressed (Figure 6E, compare lanes 8 and 9, Supplementary Table S3) compared to the reactivity detected when U3 snoRNA is expressed. The modification pattern detected at nucleotides 470–479 with the disrupted 3′-hinge interaction (3H11) is indicative of lack of 5′-hinge base pairing. Thus, base pairing of the U3 snoRNA with its 3′-hinge sequence is required for subsequent 5′-hinge base pairing.
Likewise, when the 3′-hinge base pairing was disrupted extensively with the 3H11 U3 snoRNA mutant, the modification pattern in the 5′-ETS nucleotides between the A0 and A1 cleavage sites was similar to that found when no U3 snoRNA was expressed (empty vector; Figure 6F, compare lanes 7, 8 and 10). When the U3 snoRNA mutant that does not disrupt base pairing was expressed (3H7; Figure 6F, compare lanes 8 and 9, Supplementary Table S3) the modification pattern was the same as for the U3 WT. Thus, the 3′-hinge base pairing is required for the structural changes in the 5′-ETS attributed to the presence of the U3 snoRNA.
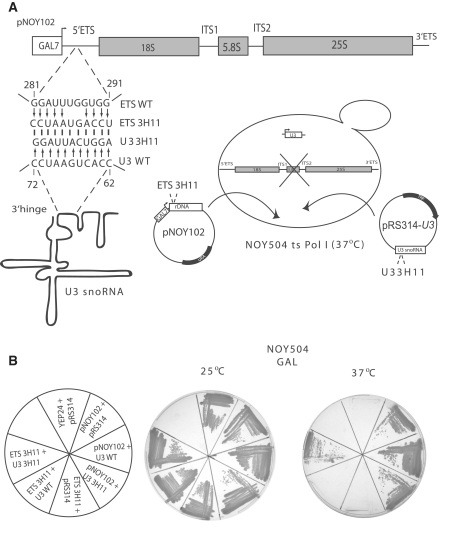
Substitution of the 5′-ETS nucleotides that base pair to the 3′-hinge of the U3 snoRNA is lethal, but growth can be restored by expressing a U3 snoRNA with a complementary 3′-hinge sequence
The strain NOY504 (31) carries a temperature sensitive mutation in RNA polymerase I, and consequently pre-rRNA is not synthesized at 37°C (Figure 7A). The plasmid pNOY102 (31) harbors the 35S rDNA gene under the control of a galactose inducible promoter, and can be used as a source for transcription of the pre-rRNA at 37°C in the presence of galactose (Figure 7A). The endogenous U3 snoRNA is expressed constitutively in this strain. As expected, the pNOY102 plasmid supports growth in the presence of empty vector (pNOY102 + pRS314), the WT U3 snoRNA expressed from a plasmid (pNOY102 + U3 WT), or in the presence of the 3′-hinge mutated U3 snoRNA (pNOY102 + U3 3H11) at 37°C (Figure 7B). To analyze the role of the 5′-ETS nucleotides involved in the base pairing interaction with the 3′-hinge of the U3 snoRNA we mutated them as shown in Figure 7A. When the resulting rDNA mutant construct (ETS 3H11) was transformed into NOY504, the yeast did not grow on galactose at 37°C (Figure 7B), either in the presence of an empty vector (ETS 3H11 + pRS314) or in the presence of the WT U3 snoRNA expressed from a plasmid (ETS 3H11 + U3 WT). Since the yeast are completely dependent on the plasmid derived pre-rRNA for survival at 37°C, the absence of growth indicates that functional ribosomes are not produced in the absence of the 5′-ETS:3′-hinge of U3 snoRNA base pairing. To prove that indeed the absence of growth was due to the reduced capacity of base pairing between the 5′-ETS and the 3′-hinge of the U3 snoRNA, we co-expressed the mutated 5′-ETS plasmid and the U3 3H11 mutant that can base pair to the mutated 5′-ETS construct. Growth at 37°C was restored to the yeast cells that contained both plasmids (ETS 3H11+U3 3H11; Figure 7B). Thus, substitution of the 5′-ETS nucleotides of the pre-rRNA that base pair to the 3′-hinge of the U3 snoRNA inhibits growth; however, expression of a U3 snoRNA with a compensatory mutation suppresses the lethal effect. Therefore, base pairing of the U3 snoRNA 3′-hinge nucleotides to the 5′-ETS of the pre-rRNA is essential for growth and ribosome biogenesis.
Figure 7.
The growth defect conferred by substitution of the 5′-ETS nucleotides that base pair with the 3′-hinge of the U3 snoRNA is suppressed by co-expression of a U3 snoRNA with compensatory mutations. (A) The rDNA (wild-type or mutant) in the pNOY102 plasmid is under a galactose-inducible/dextrose repressible promoter (GAL7), and the pNOY102 plasmid has a URA auxotrophic marker. U3 snoRNA (wild-type or mutant) is expressed from the plasmid pRS314 that carries a TRP auxotrophic marker. The nucleotides substituted in the 5′-ETS and the U3 snoRNA to generate ETS 3H11 and U3 3H11 are indicated. Schematic representation of the NOY504 strain that has a temperature sensitive mutation in RNA polymerase I. The pre-rRNA and the U3 snoRNA are co-expressed from the plasmids described above at 37°C, but the endogenous U3 snoRNA is also expressed constitutively. (B) Growth of the NOY504 strain co-expressing the indicated plasmids on galactose containing medium at 25 and 37°C. YEP24 is a plasmid used as a negative control.
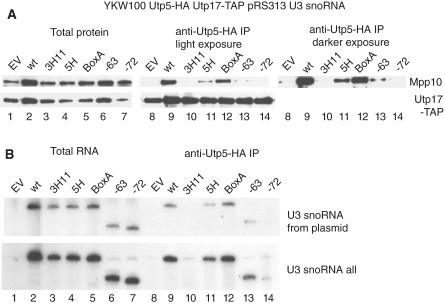
Interaction of the 3′-hinge sequence of the U3 snoRNA with the 5′-ETS is required for SSU processome formation
We performed immunoprecipitations between two SSU processome proteins that are members of different sub-complexes to test whether the 3′-hinge U3 snoRNA:5′-ETS interaction is required for the formation of the SSU processome. The YKW100 strain was engineered to express tagged versions of two UtpA/t-Utp proteins: Utp5 (HA-tagged) and Utp17 (TAP-tagged) (Figure 1C). Immunoprecipitations of Utp5-HA with an anti-HA antibody were performed, and detection of the SSU processome was carried out by western blotting with anti-Mpp10 antibodies. The immunoprecipitations were performed in the presence of the wild-type U3 snoRNA, in the presence of the mutant U3 snoRNAs, and in the absence of U3 snoRNA (empty vector). As expected, in the presence of the U3 snoRNA, Utp-5HA co-immunoprecipitated Mpp10, indicating formation of the SSU processome (Figure 8A, lane 9, top panel). Similarly, in the absence of the U3 snoRNA, Utp-5HA did not co-immunoprecipitate Mpp10 (Figure 8A, compare lanes 8 and 9, top panel), indicating that the SSU processome does not form. When we tested the U3 snoRNA 3′-hinge mutant that disrupts base pairing (U3 3H11) and the U3-72 truncation, Utp5-HA did not co-immunoprecipitate Mpp10 (Figure 8A, lanes 10 and 14, top panel), indicating that these two mutations prevent formation of the SSU processome. In the presence of the U3 snoRNA 5′-hinge (U3 5H) mutant and the U3-63 truncation, anti-HA immunoprecipitation of Utp5-HA resulted in small amounts of co-immunoprecipitating Mpp10 (Figure 8A, lanes 11 and 13, top panel), indicating that some SSU processome is formed. Anti-HA immunoprecipitation of Utp5-HA from yeast expressing U3 snoRNA mutated in Box A resulted in almost as much co-immunoprecipitating Mpp10 as that observed with the wild-type U3 snoRNA (Figure 8A compare lanes 9 and 12, top panel). We have previously shown that the UtpA/t-Utp subcomplex is maintained even in the absence of the U3 snoRNA (11). Likewise, we found that Utp5-HA co-immunoprecipitated Utp17-TAP independent of the presence of the mutated U3 snoRNAs (Figure 8A, lanes 8–14, bottom panel), indicating that the UtpA/t-Utp subcomplex is maintained. Thus, disruption of base pairing of the 3′-hinge of the U3 snoRNA with the 5′-ETS results in loss of SSU processome formation. U3 snoRNAs that can make the base pairing interaction with the 3′-hinge but not the base pairing interaction with the 5′-hinge result in some SSU processome formation, while the U3 snoRNAs that can make both the 5′- and 3′-hinge interactions but not the Box A interaction with the 5′-ETS result in almost normal levels of the SSU processome. Therefore, SSU processome assembly requires base pairing of the 3′-hinge sequence of the U3 snoRNA with the 5′-ETS, but does not depend as much on base pairing of the 5′-hinge or of Box A.
Figure 8.
Interaction of the 3′-hinge sequence of the U3 snoRNA with the 5′-ETS is required for SSU processome formation. Utp5-HA was immunoprecipitated with an anti-HA antibody from total cellular extract from yeast cells expressing the indicated U3 snoRNA mutants, at 16 h after the endogenous U3 snoRNA was depleted. (A) Total (5%) and immunoprecipitated proteins were separated by SDS–PAGE and transferred to Immobilon PVDF membranes. SSU processome assembly was assayed by western blotting for Mpp10 with an anti-Mpp10 antibody. Two different exposures are presented for the Mpp10 detection, a lighter (30 s) and a darker one (5 min). Formation of the UtpA/t-Utp subcomplex and its association with other components of the SSU processome was assayed by western blotting for Utp17-TAP with peroxidase-antiperoxidase antibodies (PAP), another UtpA/t-Utp subcomplex member. (B) The RNA extracted from either the total cellular extract (10%) or the immunoprecipitates was separated on denaturing polyacrylamide gels, transferred to Hybond N+ and the presence of U3 snoRNA was detected by northern blotting. U3 snoRNA expressed from a plasmid was detected with an oligonucleotide (SD13) that recognizes the unique sequence, while another oligonucleotide (SD74) was used to detect all forms of U3 snoRNA.
In the same way, we tested co-immunoprecipitation of the different mutant forms of the U3 snoRNA by HA tagged-Utp5 (Figure 8B) as an indication of SSU processome formation. On northern blots, wild-type U3 snoRNA was immunoprecipitable by HA-tagged Utp5 (Figure 8B, lane 9) but as expected, when no U3 snoRNA was expressed from the plasmid, no co-immunoprecipitating U3 was detectable (Figure 8B, lane 8). U3 snoRNAs mutated or truncated in the 3′-hinge sequence (U3 3H11 and U3-72; Figure 8B, lanes 10 and 14) similarly were not immunoprecipitable by HA-tagged Utp5, indicating that they are not incorporated into the SSU processome. In contrast, U3 snoRNAs mutated in the 5′-hinge sequence, in Box A, or that maintain the 3′-hinge nucleotides (U3-63) were immunoprecipitated to varying extents by anti-HA antibodies (Figure 8B, lanes 11–13). Therefore, the 3′-hinge interaction between the U3 snoRNA and the 5′-ETS is required for SSU processome formation while interruption of the two other base pairing interactions has much less of an effect.
DISCUSSION
The U3 snoRNA is an exceptional snoRNA in that its base pairing with the pre-18S rRNA leads to pre-rRNA cleavage events. We have asked whether the U3 snoRNA plays a role in the pre-rRNA structural changes in the 5′-ETS and in the pre-18S rRNA sequences that participate in the 5′-end pseudoknot (nucleotides 1–220; 980-1150). Using in vivo chemical probing, we demonstrate that the structure of these pre-ribosomal sequences depends on the presence of the U3 snoRNA, and thus provide direct evidence for a function in pre-18S folding for the U3 snoRNP and its associated proteins. Furthermore, in vivo chemical probing, mutational analysis and genetic ‘rescue’ revealed a third previously unexplored (in yeast) U3 snoRNA:pre-rRNA base pairing interaction where the 3′-hinge at nucleotides 62–72 in the U3 snoRNA is base paired to nucleotides 281–291 in the 5′-ETS. This U3 snoRNA:5′-ETS interaction is a prerequisite for the earlier reported interaction between the 5′-hinge of the U3 snoRNA and nucleotides 470–479. We show that the 3′-hinge interaction of the U3 snoRNA with the 5′-ETS is essential for growth and required for the subsequent binding of other SSU processome proteins.
These results suggest that, temporally, the 3′-hinge base pairing with the 5′-ETS is likely the first RNA:RNA interaction to occur between the U3 snoRNP and the pre-rRNA. The 5′-ETS sequence that base pairs to the 3′-hinge is the first among the base pairing sequences to be transcribed by RNA polymerase I. Furthermore, while the 3′-hinge interaction is required for the 5′-hinge interaction to occur, the reverse is not true.
Our results are in agreement with previous chemical probing studies of the yeast U3 snoRNA in an RNP in vivo and in vitro (14). Analysis by chemical probing of U3 snoRNA sequences confirmed the base pairing of the 5′-hinge and of Box A, Box A′ and the GAC Box to the pre-rRNA. Interestingly, because of the lack of nucleotide reactivity to DMS in vivo, Mereau et al. (14) also proposed that nucleotides 60–64 of the 3′-hinge are base paired to the pre-rRNA. Additionally, they observed a decrease in reactivity of nucleotides 68–72 of the 3′-hinge in vivo versus in vitro, further supporting the hypothesis that these sequences are involved in base pairing. These studies were limited to probing the U3 snoRNA, so they did not query the base paired nucleotides in the pre-rRNA.
We have found that base pairing of the 3′-hinge sequence of the U3 snoRNA with the 5′-ETS and its essential function is conserved to budding yeast. This base pairing interaction has not previously been reported in S. cerevisiae, though it had been previously described in both Xenopus laevis and in Trypanosoma brucei as essential for SSU biogenesis in these organisms (22,23,37,38). Interestingly, the A′ cleavage site, which is U3-dependent, is upstream of the 3′-hinge base pairing site in both X. laevis and T. brucei. Since we have shown here that the 3′-hinge base pairs in yeast as well, it is also plausible that there is an A′ cleavage site associated with it. The presence of an A′ cleavage site in yeast would confirm that indeed the pre-rRNA cleavage events dependent on the U3 snoRNA are conserved among eukaryotes, since the U3 dependent cleavage sites (A′, A0-A2) have also been observed in mice (39) and humans (40). Furthermore, results from mutational analysis of the corresponding 3′-hinge of the human U3 snoRNA are also similar to those presented here (41). Among the predicted three human U3 snoRNA base pairing sites, disruption of the 3′-hinge sequence reduced human Mpp10 co-immunoprecipitation to the greatest extent. Moreover, an intact 3′-hinge sequence was required for subnucleolar localization of the U3 snoRNA.
In vivo chemical probing is a powerful tool to monitor changes in RNA structure. The particular nucleotides examined by in vivo chemical probing are determined by the characteristic reactivity of bases to DMS and by the capacity of reverse transcriptase to stall at the modified base. However, not all nucleotides are equally reactive or accessible to DMS since they may be base paired or protected by proteins. For example, we can directly detect the 5′-hinge base pairing to the 5′-ETS (nucleotides 470–479) because many of the nucleotides involved are A’s and C’s whose modification is easily detected by primer extension without further manipulation. Nevertheless, it is not possible to directly ascertain the base pairing of the 3′-hinge of U3 snoRNA to nucleotides 281–291 of the 5′-ETS because these nucleotides are mainly G’s and U’s, and their modification is not detectable in this assay. Instead, we were able to detect a specific chemical modification pattern associated with the 3′-hinge base pairing interaction nearby between nucleotides 290–330. Thus, it is the combination of mutational analysis with in vivo DMS probing that has enabled us to delineate a new U3 snoRNA:5′-ETS base pairing interaction.
In contrast, the presence or absence of the U3 snoRNA does not influence the reactivity of the first 280 nucleotides of the 5′-ETS of the pre-rRNA. This suggests that the first 280 nucleotides of the 5′-ETS likely interact with other components of the SSU processome, before the first U3 snoRNA base pairing occurs at nucleotides 281–291. One strong candidate is the SSU processome subcomplex, UtpA/t-Utp, which is required for both optimal transcription of the rDNA and pre-rRNA processing (11). The UtpA/t-Utp sub-complex interacts with the pre-rRNA even when U3 snoRNA is not expressed (11). Furthermore, its assembly onto the pre-rRNA is independent of the presence of other SSU subcomplexes (12) and prior binding of the UtpA/t-Utp subcomplex to the pre-rRNA is required for the subsequent assembly of other SSU processome subcomplexes (12).
Using the 3′-hinge, 5′-hinge and Box A mutant U3 snoRNAs, we asked whether any or all of these base pairing interactions are necessary for SSU processome formation. To assay for SSU processome formation, we turned to co-immunoprecipitation of two proteins from different subcomplexes (Utp5 and Mpp10) and co-immunoprecipitation of the U3 snoRNA with Utp5. The Mpp10 protein was originally used as one of the ‘hooks’ to purify the SSU processome (8), and its presence is required for SSU processome formation as detected in Miller spreads (42). The UtpA/t-Utp subcomplex, of which Utp5 is a member, does not co-immunoprecipitate the U3 snoRNA except in the presence of the pre-rRNA (11). Co-assembly of Utp5 with Mpp10, and co-immunoprecipitation of the U3 snoRNA with Utp5, are therefore both good indications of macromolecular assembly of the SSU processome. When we assay the effect of disruption of the U3 snoRNA base pairing interactions, we find that the 3′-hinge base pairing interaction with the pre-rRNA is most important for SSU processome formation, followed by the 5′-hinge interaction and then by the Box A interaction. Indeed, the 3′-hinge interaction is sufficient for some degree of SSU processome formation as a 5′-truncated U3 snoRNA that cannot base pair with either the 5′-hinge or Box A complementary sequences but can base pair with the 3′-hinge complementary sequence (-63) becomes incorporated into the SSU processome while a truncated form missing all three interactions (-72) does not.
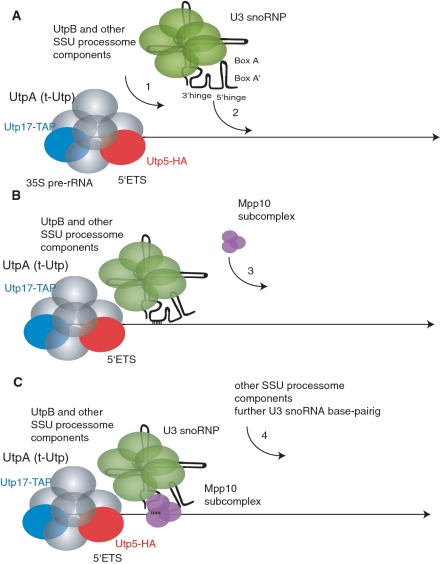
Together with previously published findings, the results presented here provide a more detailed picture of the initial steps in SSU processome assembly. Assembly begins by protein subcomplex interactions with the nascent pre-rRNA, with the UtpA/t-Utp subcomplex likely the first to bind (Figure 9A). Because the UtpA/t-Utp and UtpB subcomplexes co-immunoprecipitate in the absence of the U3 snoRNA (data not shown), the UtpB subcomplex is likely to assemble on the pre-rRNA following UtpA/t-Utp (13) and prior to the U3 snoRNP. Other, yet to be identified, SSU processome proteins may also assemble at this point. Next, the U3 snoRNP assembles into the nascent SSU processome via base pairing to the pre-rRNA using its 3′-hinge sequence (Figure 9B). This RNA:RNA interaction recruits the Mpp10 subcomplex (Figure 9C). The two subsequent U3 snoRNA: pre-rRNA interactions occur, and other components likely assemble. The sequential binding at each U3 snoRNA base pairing site from 5′ to 3′ on the pre-rRNA is likely to be a signal for the assembly of other components of the SSU processome. Therefore the U3 snoRNP and its associated proteins coordinate, via U3 snoRNA base pairing, the pre-18S rRNA folding that leads to the cleavage events that release the mature 18S rRNA. These results thus support the proposal that the U3 snoRNP and its associated proteins play an important role in the folding of pre-18S rRNA.
Figure 9.
Model for the initial assembly steps of the SSU processome. (A) The UtpA/t-Utp subcomplex is assembled first onto the nascent pre-rRNA, followed by the UtpB subcomplex and, probably, some other components of the SSU processome. (B) The U3 snoRNP anchors to the pre-rRNA through the base pairing of the 3′-hinge. (C) The base pairing of the 3′-hinge of the U3 snoRNA permits assembly of the Mpp10 subcomplex. The pre-rRNA is represented by the black straight line, the UtpA/t-Utp subcomplex is pictured in gray, with two of its components, Utp17 and Utp5, shown in blue and red respectively. The U3 snoRNA is drawn schematically with its base pairing regions indicated, and the protein components of the U3 snoRNP are shown in green, while the Mpp10 subcomplex is shown in purple.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (GM52581 to S.J.B.); Anna Fuller Fund Fellowship in Molecular Oncology (to L.M.D.); National Institutes of Health, pre-doctoral fellowship (GM20905 to J.E.G.G.). Funding for open access charge: National Institutes of Health (GM52581).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Gloria Culver, Joan Steitz and Sandra Wolin and the members of the Baserga laboratory for critical reading of the manuscript.
REFERENCES
- 1.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Phipps KR, Charette JM, Baserga SJ. The SSU Processome in Ribosome Biogenesis – Progress and Prospects. WIREs RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 5.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 6.Hughes JM, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes JM. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 8.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 10.Wehner KA, Gallagher JE, Baserga SJ. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 2002;22:7258–7267. doi: 10.1128/MCB.22.20.7258-7267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 14.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 15.Samarsky DA, Fournier MJ. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma K, Venema J, Tollervey D. The 5′ end of the 18S rRNA can be positioned from within the mature rRNA. RNA. 1999;5:678–686. doi: 10.1017/s1355838299990052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borovjagin AV, Gerbi SA. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 1999;286:1347–1363. doi: 10.1006/jmbi.1999.2527. [DOI] [PubMed] [Google Scholar]
- 22.Borovjagin AV, Gerbi SA. The spacing between functional Cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 2000;300:57–74. doi: 10.1006/jmbi.2000.3798. [DOI] [PubMed] [Google Scholar]
- 23.Hartshorne T, Toyofuku W. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27:3300–3309. doi: 10.1093/nar/27.16.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartshorne T, Toyofuku W, Hollenbaugh J. Trypanosoma brucei 5′ETS A'-cleavage is directed by 3′-adjacent sequences, but not two U3 snoRNA-binding elements, which are all required for subsequent pre-small subunit rRNA processing events. J. Mol. Biol. 2001;313:733–749. doi: 10.1006/jmbi.2001.5078. [DOI] [PubMed] [Google Scholar]
- 25.Wells SE, Hughes JM, Igel AH, Ares M., Jr Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- 26.Liebeg A, Waldsich C. Probing RNA structure within living cells. Methods Enzymol. 2009;468:219–238. doi: 10.1016/S0076-6879(09)68011-3. [DOI] [PubMed] [Google Scholar]
- 27.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 29.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Muldoon-Jacobs KL, Dinman JD. Specific effects of ribosome-tethered molecular chaperones on programmed -1 ribosomal frameshifting. Eukaryot. Cell. 2006;5:762–770. doi: 10.1128/EC.5.4.762-770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl Acad. Sci. USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 33.Wormsley S, Samarsky DA, Fournier MJ, Baserga SJ. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA. 2001;7:904–919. doi: 10.1017/s1355838201010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Simth JA, Struhl K, editors. Short Protocols in Molecular Biology. 3rd edn. New York, NY: John Wiley & Sons Inc.; 1995. [Google Scholar]
- 35.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 36.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc. Natl Acad. Sci. USA. 2006;103:9464–9469. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borovjagin AV, Gerbi SA. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005;33:4995–5005. doi: 10.1093/nar/gki815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartshorne T. Distinct regions of U3 snoRNA interact at two sites within the 5′ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 1998;26:2541–2553. doi: 10.1093/nar/26.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent T, Lapik YR, Pestov DG. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA. 2009;15:14–20. doi: 10.1261/rna.1384709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granneman S, Vogelzangs J, Luhrmann R, van Venrooij WJ, Pruijn GJ, Watkins NJ. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell. Biol. 2004;24:8600–8610. doi: 10.1128/MCB.24.19.8600-8610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 43.Piekna-Przybylska D, Decatur WA, Fournier MJ. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA. 2007;13:305–312. doi: 10.1261/rna.373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh LC, Lee JC. Structure analysis of the 5′ external transcribed spacer of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 1992;228:827–839. doi: 10.1016/0022-2836(92)90867-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.