Abstract
The relevance of serine 5 phosphorylation of RNA polymerase II carboxy-terminal domain during initiation has been difficult to determine in mammalian cells as no general in vivo Ser5 kinase has been identified. Here, we demonstrate that deletion of the TFIIH kinase subunit Mat1 in mouse fibroblasts leads to dramatically reduced Pol II Ser5 phosphorylation. This is associated with defective capping and reduced Ser2 phosphorylation, decreased Pol II progression into elongation and severely attenuated transcription detected through analysis of nascent mRNAs, establishing a general requirement for mammalian Mat1 in transcription. Surprisingly, the general defect in Pol II transcription in Mat1−/− fibroblasts is not reflected in the majority of steady-state mRNAs. This indicates widespread stabilization of mRNAs and points to the existence of a regulatory mechanism to stabilize mRNAs following transcriptional attenuation, thus revealing a potential caveat in similar studies limited to analysis of steady-state mRNAs.
INTRODUCTION
The general transcription factor IIH (TFIIH) is a conserved 10 subunit protein complex required in vitro for RNA Polymerase II (Pol II) transcription and for nucleotide excision repair (NER). In transcription, TFIIH consists of a core and a kinase submodule (Cdk-activating kinase, CAK). TFIIH core is required during initiation to melt promoter DNA through DNA-dependent helicase and ATPase activities (reviewed in 1), can support transcription in some reconstituted systems (2) and is functional in NER without CAK (3). CAK in mammalian cells contains the CDK/cyclin pair Cdk7 (4) and Cyclin H (5,6) and the regulatory subunit Mat1 (7) required for stability of the kinase submodule in cells (7,8) and in mouse tissues (9,10), and which links CAK to core TFIIH through association with XPD (2,11).
The role of mammalian TFIIH kinase in Pol II transcription remains controversial, but most proposed functions are linked to its ability to phosphorylate serine 5 of the Pol II C-terminal domain (4–6). The CTD consists of 52 heptapeptide repeats with the consensus sequence YS2PTS5PS7, where sequential phosphorylation of serines 5 and 2 leads to differential protein recruitment and regulation of specific co-transcriptional events (reviewed in 12). Ser5 phosphorylation occurs at initiation (13,14) and is critical for recruitment and activation of capping enzymes (reviewed in 15). Capping is coupled to Ser2 phosphorylation in yeast cells, as Ser2 kinases are recruited by capping enzymes (16–19). Ser5-P also promotes recruitment of several elongation factors and chromatin modifiers in yeast (20–23) and mammalian (24) cells. In addition to TFIIH kinase, Ser5 can be phosphorylated by at least Cdk9/P-TEFb (25,26) as well as the Mediator kinase Cdk8 (27), and the Mediator has been proposed to be involved in recruitment of P-TEFb in mammalian cells (14).
In reconstituted transcription systems, TFIIH kinase is required for both Ser5 phosphorylation and transcription (28,29), but studies in living cells provide a complicated picture. In budding yeast, temperature-sensitive (ts) alleles of the Cdk7 homolog KIN28 demonstrate dramatically reduced mRNA levels in the majority of genes (30), and strongly decreased Ser5 phosphorylation, capping and Pol II occupancy (31–33). Ts alleles of the Mat1 homolog RIG2/Tfb3 display either strong (34) or barely detectable (35) alterations in mRNA levels. Inhibition of analog-sensitive KIN28 in one study did not affect mRNA levels of the majority of genes (36), whereas in another study 58% of mRNAs were decreased (37); in both studies Ser5-P was decreased on specific genes. In fission yeast experiments analyzing ts alleles of the Cdk7 homolog Mcs6 and Mat1 homolog Pmh1, Ser5 phosphorylation was decreased and associated with a moderate (25–33%) general decrease in mRNA levels (38), yet inhibition of an analog-sensitive Mcs6 allele did not alter global mRNA levels although a general decrease in Ser5-P was reported (19). Partial loss of Cdk7 in Caenorhabditis elegans resulted in decreased global Ser5-P as well as defective transcription in mutant embryos (39), whereas temperature-sensitive mutants in Drosophila demonstrate no alterations (40), a general decrease in mRNA levels (41), or attenuated transcription and Ser5 phosphorylation on the Hsp70 locus (42).
In mammalian cells, genetic investigations have not demonstrated clear transcriptional defects following abrogation of TFIIH kinase activity. Analysis of mouse blastocysts with a targeted disruption of Mat1 did not reveal transcriptional defects despite moderately decreased Ser5 and Ser2 immunoreactivity (8). Similarly targeted cardiomyoctes only displayed specific alterations in mRNAs of mitochondrial metabolic enzymes (43). In Cdk7as HCT116 human cancer cells, Cdk7 inhibition did not detectably affect general transcription or result in Ser5-P changes at global levels or on the studied genes c-myc and GAPDH (13,44). siRNA-mediated knockdown of Cdk7 in human fibroblasts on the other hand did attenuate induction of 3 UV-induced mRNAs (3).
In an attempt to clarify the requirement of TFIIH kinase in Pol II CTD phosphorylation and transcription we have here analyzed effects of deleting the murine Mat1 subunit in a temporally controlled manner in cultured murine fibroblasts. Our results provide evidence indicating that TFIIH kinase represents the major CTD Ser5 kinase and is required for general transcription in mammalian cells.
MATERIALS AND METHODS
Affymetrix expression profiling
For Affymetrix expression profiling, mRNA was collected from three batches of exponentially growing Mat1−/flox cells 72 and 96 h following AdCre or AdLacZ infection. mRNA was purified using RNEasy Kit (QIAGEN). cDNA synthesis using oligo dT primers, target labeling, an hybridization were done using Affymetrix GeneChip Reagents according to the manufacturer’s protocol using Affymetrix high-density Mouse Genome 430 2.0 Arrays, and scanned with an Agilent microarray laser scanner. Data from all 12 arrays have been deposited at ArrayExpress (www.ebi.ac.uk/microarray-as/ae/). For data analysis, see Supplementary Data.
Analysis of capped, FU-labeled and EU-labeled transcripts
Capped and methylated transcripts were isolated as previously described (36). Briefly, 5µg α-m7G-cap antibody (H20; K121 Calbiochem) was added to 20 µg of total RNA in cap binding buffer (150 nM NaCl, 0.1% NP-40, 10 mM Tris, pH 8.0) and kept on ice for 1 h. About 100 µl of ProtA Sepharose beads + 5 µl DTT + 1.25 µl RNAseOUT were added and the samples incubated overnight in +4°C. After five washes with ice-cold binding buffer containing 2.5 mM DTT, beads were resuspended in 200 µl of ProtK solution and nutated in +37°C for 30 min, after which the precipitated RNA was purified, measured and calculated as percent input RNA. For FU experiments, cells were pulsed with 1 mM FU in KH buffer (30 mM KCl, 10 mM HEPES, pH 7.4) for 10 min and pulse-chased for various times. IPs and washes were done as in capping analysis but with a-BrdU (DAKO). EU labeling experiments were done according to the protocol of Click-iT Nascent RNA Capture kit (Invitrogen). Briefly, cells were pulsed with 0.5 mM EU, total RNA was isolated and used in a copper catalyzed click reaction with azide-modified biotin. After this, the nascent transcripts were captured on streptavidin magnetic beads and cDNA synthesis performed with IP:d material directly on the beads using Superscript VILO cDNA synthesis kit (Invitrogen) followed by analysis with QRT-PCR. See Supplementary Data for primers.
Chromatin immunoprecipitation
Immunoprecipitation was performed using the following antibodies and amounts: N20 (Santa Cruz Biotechnology), 10 µg; 5131 (AbCam), 10 µg overnight. Binding was performed for 3 h at +4°C with 50% slurry protein-A sepharose followed by washes once with low salt buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton-X100, 0.1% SDS), three times in high salt buffer (20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% Triton-X100, 0.1% SDS), once in LiCl containing buffer (10 mM Tris–HCl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% DOC, 1% NP-40) and twice with TE, pH 8.0. Precipitated proteins were eluted twice from the beads using elution buffer (0.1 M NaHCO3, 1% SDS) containing 50 µg of proteinase K (Fermentas) in +42°C for 30 min. Reverse cross-linking of precipitated chromatin was performed at +67°C overnight with NaCl solution to 200 mM final concentration. Controls: antibodies: rabbit IgG and peroxisomal α–catalase (ab1877), negative genomic region: SBiosciences intergenic negative control primers. The ChIP samples were purified using QIAquick PCR purification kit spin columns (QIAGEN) and eluted twice in 10 mM Tris–HCl, pH 8.5, buffer. IP:d material was analyzed with qRT-PCR. Primers are provided in Supplementary Data.
RESULTS
Mat1 is required for Pol II CTD phosphorylation in mouse fibroblasts
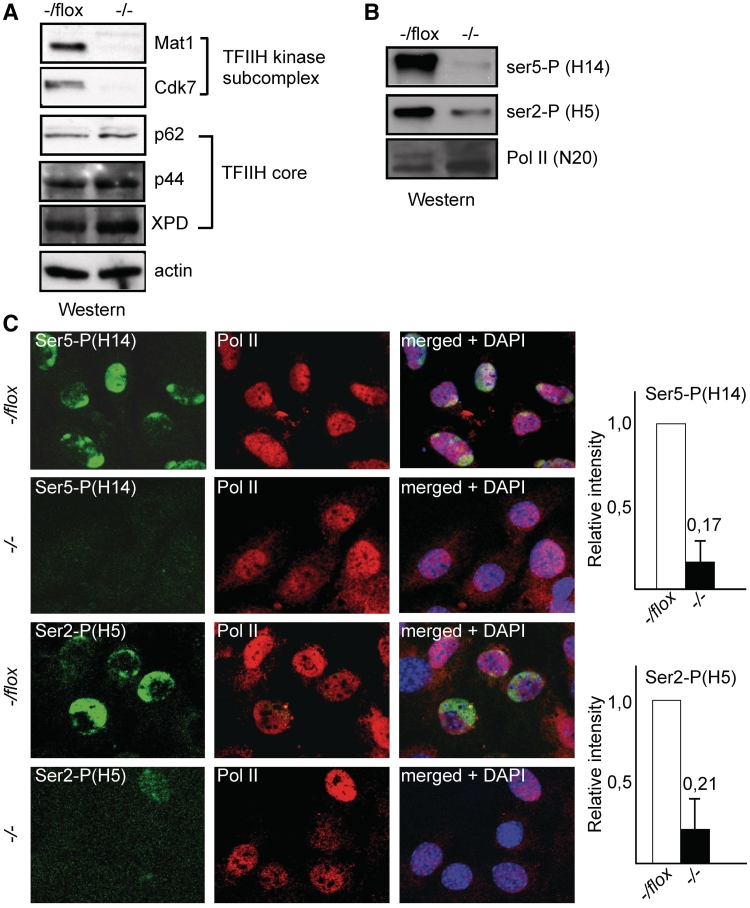
Deletion of the TFIIH subunit Mat1 from Mat1−/flox immortalized mouse embryonic fibroblasts (MEFs) resulted in the concomitant and expected (10,43) decrease in levels of the CAK subunit Cdk7 without affecting TFIIH core subunits XPD, p44 or p62 (Figure 1A) consistent with ability of core subunits to form functional complexes in the absence of the kinase submodule (2). Loss of Mat1 and Cdk7 was associated with a dramatic decrease in global phosphorylation of Pol II CTD serine 5 (Ser-P) and with a slightly smaller decrease in Ser2 phosphorylation noted in cell lysates (Figure 1B) as well as by immunostaining (Figure 1C) where quantitation of average signals indicated a 83% decrease in Ser5-P and 79% decrease in Ser2-P in Mat1−/− MEFs. Thus the TFIIH kinase component Mat1 is required for the majority of Ser5 and Ser2 phosphorylation in mammalian cells as detected in lysates and immunofluorescence.
Figure 1.
Mat1 is required for Ser5 and Ser2 phosphorylation. (A) Western blotting analysis Mat1−/flox MEFs with antibodies against indicated TFIIH subunits or actin as a loading control 72 h after infection with AdCre (−/−) or control virus (−/flox). (B) Western blotting analysis as in (A) using antibodies against Pol II Ser5-P (H14), Ser2-P (H5) or total Pol II (N20). (C) Immunostaining of Mat1−/flox and Mat1−/− MEFs with antibodies against Pol II Ser5-P (H14), Ser2-P (H5) or total Pol II (N20). DAPI staining is shown to identify nuclei in merged panels. Columns on right indicate the average intensity of signal from Ser5-P and Ser2-P in Mat1−/− MEFs (black bars with ratios shown as number on top) relative to Mat1−/flox MEFs (white bars) in three biological replicates with error bars showing standard between replicates.
Deficient c-Fos and Hsp70 mRNA induction in Mat1−/− cells
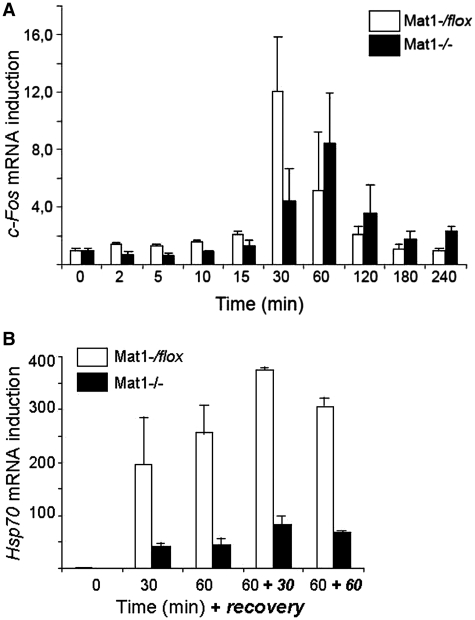
To investigate whether the decreased CTD phosphorylation correlates with defective transcription two induced transcription models were utilized. Analysis of c-Fos mRNA induction during serum stimulation was studied in serum-starved Mat1−/flox and Mat1−/− MEFs treated with 10% serum for indicated times (Figure 2A). Notably the mRNA levels of c-Fos prior to induction (0 min) as well as Gapdh (data not shown) used for normalization were comparable between the genotypes. c-Fos mRNA levels were strongly induced in control cells reaching a 12-fold induction at 30 min followed by a rapid decline (Figure 2A). In Mat1−/− MEFs induction was delayed and attenuated with a maximally 8-fold induction at 60 min. Interestingly, the decline of c-Fos levels was also slower, and did not reach baseline levels during 240 min suggesting mRNA stabilization in Mat1−/− MEFs. Induction of Hsp70 mRNA following a heat shock was also severely defective, where the 200–375-fold induction noted in control cells was reduced to a 40–80-fold induction in Mat1−/− MEFs (Figure 2B) remarkably comparable to the 3.6-fold reduction of hsp70 mRNA induction in Cdk7ts flies (42).
Figure 2.
Deficient c-Fos and Hsp70 mRNA induction in Mat1−/− MEFs. (A) c-Fos mRNA induction relative to 0 min following serum stimulation at indicated time points in Mat1−/flox (white bars) and Mat1−/− (black bars) MEFs. (B) Hsp70 mRNA induction relative to 0 min following heat shocks at 42°C for 30 or 60 min, or after a recovery of 30 or 60 min as indicated in Mat1−/flox (white bars) and Mat1−/− (black bars) MEFs. Error bars indicate standard deviation in three independent experiments. All mRNAs were normalized to Gapdh mRNA levels.
Genome-wide alterations in mRNA levels following Mat1 deletion
Genome-wide alterations in steady-state mRNA levels in Mat1−/− MEFs was investigated using Affymetrix MG430 2.0 microarrays from three biological replicates of Mat1−/− or Mat1−/flox MEFs 72 or 96 h following infection. Gene ontology (GO) analysis of differentially expressed mRNAs in the 96 h samples suggested a G2/M cell cycle arrest (Supplementary Figure S1A) supported by subsequent cell cycle analyses (Supplementary Figure S1B and C). To focus on Mat1 transcriptional regulation, subsequent analyses were limited to the 72 h samples, where no evidence for cell cycle effects were noted in Affymetrix analyses as expected (10).
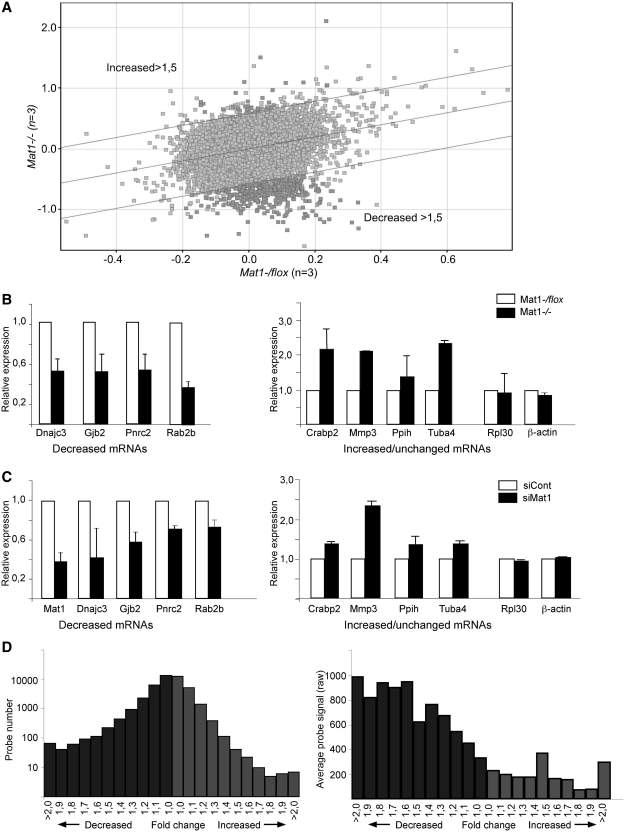
Identification of differential mRNAs by conventional criteria indicated that <1% of mRNA levels were altered in Mat1−/− cells (>1.5-fold change, P < 0.05) mostly representing decreases (365 decreased probe sets; 35 elevated; Figure 3A). These changes were reproducible as indicated by qRT-PCR analysis of four decreased, four elevated and two unchanged mRNAs (Figure 3B) and also could be detected following depletion of Mat1 through transfection of siRNAs targeting Mat1 mRNA (Figure 3C). GO analysis identified several mRNA processing GO categories (Supplementary Figure S2) as enriched in the decreased mRNAs with 52 probe sets.
Figure 3.
Genome-wide alterations in mRNA levels following Mat1 deletion. (A) Scatter plot of Affymetrix MG430 2.0 average probe set signals plotted according to genotype as indicated, with lines delineating probe sets >1.5-fold increased or decreased in Mat1−/− samples as indicated. Probe sets differing significantly (P < 0.05) according to genotype are highlighted in dark gray. (B) mRNA levels of indicated genes in Mat1−/− samples (black bars) relative to control (white bars) assessed by qRT-PCR grouped to genes decreased, increased or unchanged (Rpl30 and Actb) in Affymetrix analysis. Error bars indicate standard deviation between at least five experiments. (C) Analysis of indicated mRNA levels as in (B) from MEFs transfected with control (siCont) or Mat1 targeting siRNAs (siMat1). Error bars indicate standard deviation between four experiments. (D) Distribution of Affymetrix probe sets (see A) according to fold change (0.1-fold change increments) in Mat1−/− samples with decreased samples in dark gray and increased in light gray. Note that probe set numbers are plotted on a logarithmic scale. (E) Distribution of average probe signal (raw) in categories described in (D) demonstrating that probe sets with stronger signal are significantly more likely to demonstrate decreased signal in Mat1−/− samples.
To investigate minor general alterations and to avoid potential skews by normalization algorithms in standard microarray data analyses (37) raw signal (triplicate average) ratios (Mat1−/−/Mat1flox/−) were compared after normalizing only with raw signals from relevant external control probes (n = 45; probes with CRE sequences excluded). Interestingly, the cumulative signal of all mouse probe sets (45 049 probe sets) was significantly decreased (0.964, P < 0.001) in Mat1−/−samples. Also a pairwise comparison of probe sets indicated a significant increase in ratios below 1 (expected 22 525; observed 24 471; 8.6% increase, P < 0.0001), which was more prominent if low signal (raw value <50 in control samples) probe sets were excluded (n = 20311; expected 10156; observed 12134; 19.5% increase, P < 0.0001). Plotting the distribution of the probe sets according to fold change (Figure 3D) revealed significantly (P < 0.0001) more decreased (dark gray) probe sets in all fold-change categories with the difference increasing with fold change. The result indicates a wide-spread small decrease in steady-state mRNA levels in Mat1−/− cells partly masked by the noise inherent in this approach especially with low signals. In support of this, it was noted that probes sets with robust signal were significantly (P < 0.0001) more likely to demonstrate decreased signal in Mat1−/− cells in all fold change categories (Figure 3E).
Decreased RNA Pol II occupancy in gene bodies in Mat1−/− cells
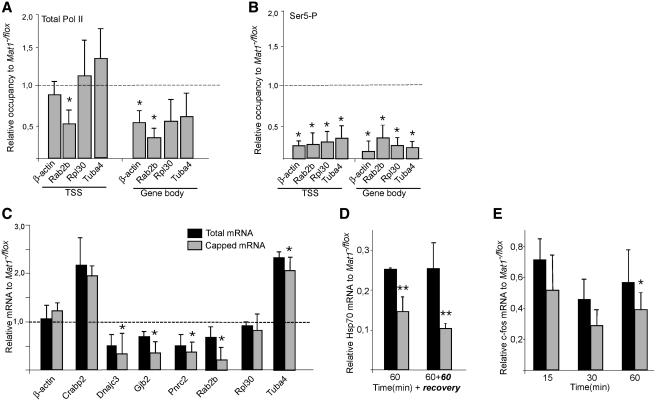
To investigate whether the altered levels of specific mRNAs in Mat1−/− cells reflected Pol II occupancy, genes with unchanged (b-actin, Rpl30), decreased (Rab2b) or elevated (Tuba4) mRNA levels were analyzed from total Pol II (N20) chromatin immunoprecipitates. Mat1 deletion did not affect Pol II occupancy in the promoter-proximal region (Figure 4A, TSS) except on Rab2b, where Pol II levels were decreased in correlation with decreased mRNA levels. By contrast, levels of elongating Pol II in gene bodies was decreased in all analyzed genes in Mat1−/− cells indicating deficient progression into elongation (Figure 4A). Ser5 phosphorylation of Pol II as expected was strongly reduced both at the promoters and in elongating polymerase (Figure 4B) consistent with the global Ser5-P reduction (Figure 1).
Figure 4.
Reduced levels of elongating Pol II and deficient capping following Mat1 deletion (A and B) Chromatin immunoprecipitation (ChIP) analysis from Mat1−/flox and Mat1−/− MEF lysates with antibodies recognizing total Pol II (N20) and Ser5-P Pol II (ab5131). ChIP-enriched DNA was quantified by qPCR with primers amplifying transcription start site (TSS) and gene bodies of the indicated genes. Columns indicate relative levels of Pol II in Mat1−/− samples comparedwith Mat1−/flox levels (indicated as dotted line). Error bars indicate standard deviation within three biological replicates and asterisks indicate statistically significant alterations (P < 0.05). (C) Relative levels of m7G-capped (gray columns) and total mRNA of indicated genes in Mat1−/− MEFs compared to Mat1−/flox levels (indicated as dotted line). Levels of capped mRNAs were determined by qRT-PCR from RNA immunoprecipitated with an α-m7Gcap antibody (H2O) and levels of total mRNAs were quantitated with qRT-PCR from the same experiments. (D) Total and capped Hsp70 mRNA levels following a 1 h heat shock and 1 h recovery showing total and capped mRNA levels in Mat1−/− MEFs in relation to Mat1−/flox MEFs. (E) Total and capped c-Fos mRNA levels from samples collected at indicated time points following a serum stimulation analyzed as in (C). All bars in (A–C) represent averages from three independent experiments, with standard deviations as error bars.
Defective capping and splicing in Mat1−/− cells
The role of the TFIIH subunit Mat1 in capping was investigated by isolating capped and methylated RNAs (36) by immunoprecipitation followed by RT-PCR analysis. Decreased levels of capped mRNA levels were noted in Mat1−/− cells for all analyzed genes with largest decreases in genes with reduced steady-state mRNAs (Figure 4C). Decreased capped mRNAs were also evident in the induced and rapidly transcribed Hsp70 and c-Fos mRNAs in Mat1−/− cells (Figure 4D and E). The results indicate a requirement of intact TFIIH kinase for efficient capping, which was however not reflected in steady-state levels of all mRNAs.
Deficient Ser5-P Ser2-P and capping in other models would be expected to affect splicing. U2- and U12-type splicing was investigated for Dock1, Pex16, Tcea2 and Pcyt2 genes previously identified to display differential amounts of U2- and U12-type introns (45). Whereas no major alterations were noted in relative amounts of U2- and U12- splicing (data not shown), levels of both U2- and U12-type intron-containing RNAs were slightly increased in Mat1−/− cells for Dock1, Pex16 and Tcea2 (Supplementary Figure S3), and a significant increase in unspliced transcripts was also noted using pooled data of all genes, demonstrating a small deficiency in splicing likely affecting the majority of mRNAs.
General requirement of mammalian Mat1 for transcription
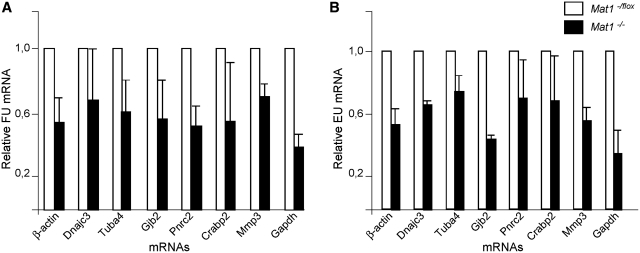
Given the decreased Ser5 phosphorylation and elongating Pol II in Mat1−/− MEFs, the alterations in steady-state mRNA levels identified in Affymetrix analyses were unexpectedly small. To investigate transcription more directly Mat1−/flox and Mat1−/− MEFs were pulse-labeled for 10 min with 5′-fluorouridine (FU) (46) followed by a 1-h chase and purification of FU-labeled nascent RNA using antibodies recognizing halogenated uridine. Analysis of levels of FU-labeled specific mRNAs indicated a general decrease (on average 57%; from 38% to 70%) noted in all analyzed genes (Figure 5A), independent of their steady-state mRNA levels (Figure 3). mRNA purified from wild-type MEFs treated with α-amanitin served as a control and showed barely quantifiable amounts of labeled mRNA (Figure 6A). Rpl30 and Rab2b mRNAs were not detectable following a 1 h pulse probably reflecting their slow transcription rate (47). A second approach for analyzing nascent RNA using 5-ethynyluridine (EU) labeling followed by purification of labeled RNA using click chemistry (48) validated the decrease (on average 56%; from 36% to 74%) in all analyzed mRNAs (Figure 5B), confirming the general defect in RNA Pol II transcription in Mat1−/− MEFs.
Figure 5.
Nascent mRNA analysis reveals requirement of Mat1 for Pol II transcription. (A) Analysis of levels of FU-labeled mRNAs from Mat1−/− cells (black bars) relative to Mat1−/flox samples (white bar). Mat1−/flox and Mat1−/− cells exposed to a 10-min FU pulse followed by a 1-h chase were used for RNA isolation and subsequent immunoprecipitation with an antibody recognizing halogenated uridine. Columns indicate relative amounts of mRNAs in immunoprecipitates quantified by qRT-PCR compared with Mat1−/flox. Error bars indicate standard deviation from biological triplicates. (B) Analysis of levels of 5-ethynyluridine (EU) labeled mRNAs from Mat1−/− cells chased for 1 h prior to lysis and purification of EU-labeled RNA using click-chemistry based biotin cross-linking. Values are shown relative to Mat1−/flox samples (white bar). Error bars indicate standard deviation in three independent experiments.
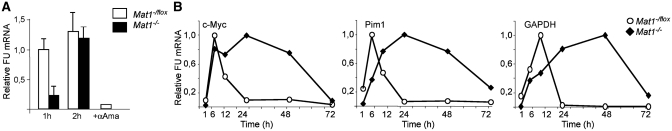
Figure 6.
Stabilization of mRNAs in Mat1−/− MEFs. (A) Mat1−/flox and Mat1−/− cells exposed to a 10-min FU pulse followed by a 1 or 2 h chase analyzed for nascent FU-labeled RNA as in Figure 5 demonstrate almost comparable amounts of labeled RNA after a 2-h chase in Mat1−/flox and Mat1−/− cells. α-Amanitin treated wild-type MEFs chased for 2 h are used as control. Error bars indicate standard deviation in three independent experiments. (B) Levels of three FU-labeled mRNAs (c-Myc, Pim1 and Gapdh) from Mat1−/flox (white circles) and Mat1−/− (black diamonds) MEFs quantified by qRT-PCR at various time points after 10 min FU pulse. FU mRNA values are relative to the highest value within experiment.
Uncoupling of transcription and steady-state mRNA levels in Mat1−/− fibroblasts
Severely reduced transcription together with only minor changes in mRNA levels suggested stabilization of mRNAs in Mat1−/− MEFs. This was supported by the observation that extending the in vivo labeling time decreased the difference between control and Mat1−/− MEFs (Figure 6A). The stabilization was also noted in a follow up of FU-labeled mRNAs after a short FU pulse of three mRNAs with variable half-lives (49): c-Myc (22 min), Pim1 (52 min) and Gapdh (313 min), none of which demonstrated significant alterations in steady-state mRNA levels. Decreased accumulation of FU-labeled mRNAs at early time points was followed by a delayed decrease (Figure 6B) with all genes, where the kinetics correlated with the reported half-lives. These results indicate that the decreased transcription in Mat1−/− MEFs is associated with decreased mRNA decay.
DISCUSSION
This study reveals that the TFIIH kinase subunit Mat1 is required for Ser5 phosphorylation and general transcription in cultured mouse fibroblasts. Loss of Mat1 is associated with loss of TFIIH kinase activity (10,43), and destabilization of Cdk7 (Figure 1) (8–10,43) and cyclin H (43). Depletion of the kinase submodule is not associated with alterations in TFIIH core components (Figure 1) (43) as expected based on the observed dynamic dissociation/association of kinase and core submodules in fibroblasts (3). The depletion of TFIIH kinase did not generally affect preinitiation complex formation (Figure 4) unlike when using a temperature-sensitive kinase allele (36). Although in an attenuated fashion, Pol II in this system was also capable of initiation, which requires a functional TFIIH core in vitro (2,28) and in vivo (50,51). Together these observations provide evidence indicating that the notable defects in Mat1−/− cells are due to lack of TFIIH kinase-mediated effects on Pol II CTD and possible other substrates although contributions from modified TFIIH core or as of yet unidentified TFIIH kinase-independent functions of Mat1 have not been ruled out.
Inactivating TFIIH kinase by depletion distinguishes this study from those using temperature-sensitive alleles (30,34,35,38) where larger structural alterations in TFIIH and the preinitiation complex have been proposed to underlie general transcriptional defects (19,36). TFIIH kinase depletion in mammalian cells through deletion of the Mat1 locus leads to a gradual complete loss of function as opposed to siRNA-mediated knockdown (3) or to chemical inhibition of ATP analog-sensitive Cdk7 mutants (Cdk7as) (13,44). Although the use of analog-sensitive mutants potentially enables rapid analysis of transcriptional effects following inhibition, Cdk7as transcriptional effects have been analyzed 8 (13) or 14 h (44) after inhibition. In yeast cells, alterations in mRNA levels have been noted 15 min (budding yeast) (37) or 30 min (fission yeast) (19) following inhibitor addition.
TFIIH kinase is required for global CTD Ser5 and Ser2 phosphorylation
The observation that depletion of murine Mat1 resulted in decreased Pol II Ser5-P both globally and on all analyzed genes extends the identified role of TFIIH kinase in Ser5 phosphorylation from other model systems (19,36–39,42) to mammalian cells. In human cells, inhibition of Cdk7as did not detectable decrease Ser5-P globally (44) or on c-myc and GAPDH (13). This could be due to the presence of an inactive but structurally intact Cdk7 in that approach compared to Mat1−/− cells, enabling recruitment of e.g. alternative Ser5 kinases. We consider this unlikely since both Kin28as (36) and Mcs6as (19) demonstrate reduced Ser5-P levels similar to ours. It is also possible that the difference is due to low residual activity of Cdk7as sufficient for Ser5-P. A third possibility is differential activity of putative alternative Ser5 kinases (13,14) in Cdk7as HCT116 colon carcinoma cells and Mat1−/− mouse fibroblasts. Loss of Mat1 also led to a very significant decrease in Ser2–P as noted previously in fission yeast (19), attributed to defective Cdk9 recruitment. As Cdk7 does not phosphorylate Ser2 in vitro (5), defective recruitment of Cdk9 via capping enzymes (16–19) may also be the mechanism in Mat1−/− MEFs, consistent with the observed decrease in capped transcripts.
Mammalian Mat1 is required for efficient global transcription
Global Pol II transcription and the transcription of all studied individual mRNAs is severely defective following Mat1 deletion. The finding that Mat1 deficient MEFs contain less capped transcripts suggests that Ser5-P CTD is needed for proper recruitment of the mammalian capping machinery and that this is dependent on TFIIH kinase activity, supported by studies in vitro (52,53) and in vivo (31,32). In this model, the capping machinery then recruits P-TEFb and enables Pol II progression into elongation (17). Although a recent report suggested a link between Cdk7 activity and Pol II pausing (13,14), in this study we observed decreased labeling of both paused and non-paused genes, demonstrating a more general role for TFIIH kinase in transcription.
In addition to this capping-dependent mechanism, at least two alternative or additive models for the transcriptional defect can be envisioned. First, it may be mediated directly through a Cdk7 substrate different from Ser5. Pol II CTD Ser7 is phosphorylated by Cdk7 in vitro (33) and chemical inhibition of Cdk7 resulted in decreased Ser7-P on the studied genes both in yeast (54) and human HCT116 cells (13). However, decreased Ser7-P is unlikely to be the main cause of the general transcriptional defect observed in our system, as Ser7-P has been shown to be specifically required for transcription of snRNA but not protein-encoding mRNAs (55). Additionally, Cdk7 has been shown to phosphorylate elongation factors Spt5 and Cdk11 (56), which are involved in progression into elongation and stimulation of elongation rate. The observed general transcription defect in Mat1-deficient cells may therefore be partly mediated through direct decreased activation of elongation factors by Cdk7.
Furthermore, decreased Ser5-P may result in inefficient recruitment of chromatin modifiers/elongation factors (20,21,23,24). The Set1 Histone 3 Lysine 4 (H3K4) methylase family members interact directly with Ser5-P Pol II (24,57) and H3K4 trimethylation strongly correlates with Ser5-P CTD at promoter regions (reviewed in 12). Indeed, knockout MEFs of MLL, one of the mammalian H3K4 methylases, are defective in elongation and have decreased Pol II occupancy on transcribed genes (57). Although a recent genome-wide screen of MLL−/− MEFs showed that only 5% of genes require MLL for efficient transcription and Pol II occupancy (58), it should be noted that mammals contain at least 6 H3K4 methylases. Therefore, a global decrease in H3K4 methylation resulting from decreased Ser5-P could lead to a general defect in transcription.
Global stabilization of mRNAs in response to defective transcription
The global strong decrease in Pol II transcription following Mat1 deletion was only partly reflected in steady-state mRNA levels. Together with the observed decrease in mRNA decay of all analyzed genes, this indicates a general stabilization of mRNAs in Mat1−/− MEFs. This suggests that the specific changes noted in steady-state mRNA levels may reflect specificity in transcription/mRNA processing or specificity in stabilization. The former is supported indirectly by the enrichment of mRNA processing genes in Mat1-sensitive genes and by the correlation of decreased mRNA levels to decreased capped transcripts (Figures 3 and 4). On the other hand, the specificity in capped mRNA amounts could be due to selective stabilization of mRNAs as the analysis in this study was performed from steady-state RNAs here as well as previously when TFIIH was proposed to selectively modulate capping (19). An obvious mechanism providing specificity in mRNA stabilization is mRNA half-life, but no correlation was noted between Mat1-sensitive mRNAs and mRNA half-life (47,49) even after a probe set level comparison (using data from ArrayExpress E-GEOD-10011; data not shown).
Widespread stabilization of mRNAs has been described in budding yeast in response to oxidative stress, where 36.2% of mRNAs show altered stability and 14% of transcripts display a pattern similar to the majority of mRNAs following Mat1 loss observed here; no significant change in steady-state mRNAs despite decreased transcription (59). Interestingly, the Upf1 RNA helicase previously linked to nonsense-mediated mRNA decay (NMD) has been identified as a mediator of the transcriptional response to oxidative stress in fission yeast (60), and Ddx5/p68 involved in mRNA degradation with Upf1 (61) was strongly downregulated in Mat1−/− MEFs.
In mammalian cells, stabilization of mRNAs represents a significant part of the transcriptional responses following heat shock (62) and ER stress (63) at least partly through inhibition of translation. mRNA stabilization can also occur through inhibition of nuclear export leading to mRNA accumulation at sites of transcription (64). Interestingly, Pol II CTD has been noted to be required for release of spliced and polyadenylated transcripts from the site of transcription (65), suggesting another mechanism by which compromised activity of the hypophosphorylated CTD in Mat1−/− MEFs regulates mRNA turnover.
While generalized stabilization of mRNAs following inhibition of transcription has not been widely recognized, it has been noted in studies utilizing kinetic RNA measurements in conjunction with transcriptional inhibitors. mRNA half-lives measured following inhibition of transcription by actinomycin D were longer than when measured using kinetic labeling with 4-thiouridine (47,49), and even longer when the transcription inhibition time was extended (49) in NIH3T3 mouse fibroblasts. Specific inhibition of Pol II transcription using α-amanitin led to efficient CTD-dependent inhibition of nascent transcription without leading to noticeable changes in steady-state mRNAs in human B-cells (66). Considering these data, it is plausible that stabilization of mRNAs in Mat1−/− MEFs results from inhibition of transcription, the mechanism of which will be interesting to investigate in future studies. The present findings highlight the importance of utilizing kinetic mRNA measurements in addressing transcriptional effects in living cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biomedicum Helsinki Foundation; Nylands Nation Foundation; Finnish Cultural Foundation; Research and Science Foundation of Farmos; Academy of Finland; EU FP6 Program (ENFIN); Finnish Cancer Organizations and Sigrid Juselius Foundation. Funding for open access charge: University of Helsinki.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Yuan Zhu for AdCre construct. We thank Jenny Bärlund, Saana Laine and Outi Kokkonen for technical assistance. Biomedicum Helsinki Molecular Imaging Unit, Biomedicum Biochip Center and Biomedicum Virus Core are acknowledged for services. Damien Hermand, Pekka Katajisto, Emilia Kuuluvainen, Saara Ollila and Tea Vallenius are thanked for critical comments on the article. Pekka Katajisto is also acknowledged and thanked for technical critique. K.H. is a graduate student in Helsinki Biomedical Graduate School.
REFERENCES
- 1.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 2.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 3.Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell. 2008;31:9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JH, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription- DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 5.Serizawa H, Makela TP, Conaway JW, Conaway RC, Weinberg RA, Young RA. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 6.Shiekhattar R, Mermelstein F, Fisher RP, Drapkin R, Dynlacht B, Wessling HC, Morgan DO, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 7.Adamczewski JP, Rossignol M, Tassan JP, Nigg EA, Moncollin V, Egly JM. MAT1, cdk7 and cyclin H form a kinase complex which is UV light- sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 8.Rossi DJ, Londesborough A, Korsisaari N, Pihlak A, Lehtonen E, Henkemeyer M, Makela TP. Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. EMBO J. 2001;20:2844–2856. doi: 10.1093/emboj/20.11.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korsisaari N, Rossi DJ, Paetau A, Charnay P, Henkemeyer M, Makela TP. Conditional ablation of the Mat1 subunit of TFIIH in Schwann cells provides evidence that Mat1 is not required for general transcription. J. Cell Sci. 2002;115:4275–4284. doi: 10.1242/jcs.00121. [DOI] [PubMed] [Google Scholar]
- 10.Helenius K, Yang Y, Alasaari J, Makela TP. Mat1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipocyte differentiation. Mol. Cell. Biol. 2009;29:315–323. doi: 10.1128/MCB.00347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang WH, Kornberg RD. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell. 2000;102:609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 12.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei Y, Schwer B, Shuman S. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 2003;278:7180–7188. doi: 10.1074/jbc.M211713200. [DOI] [PubMed] [Google Scholar]
- 17.Guiguen A, Soutourina J, Dewez M, Tafforeau L, Dieu M, Raes M, Vandenhaute J, Werner M, Hermand D. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 2007;26:1552–1559. doi: 10.1038/sj.emboj.7601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viladevall L, St Amour CV, Rosebrock A, Schneider S, Zhang C, Allen JJ, Shokat KM, Schwer B, Leatherwood JK, Fisher RP. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell. 2009;33:738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 21.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol. Cell. Biol. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol. Cell. Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 2000;20:6958–6969. doi: 10.1128/mcb.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- 28.Mäkelä TP, Parvin JD, Kim J, Huber LJ, Sharp PA, Weinberg RA. A kinase-deficient transcription factor IIH is functional in basal and activated transcription. Proc. Natl Acad. Sci. USA. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akoulitchev S, Mäkelä TP, Weinberg RA, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 30.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 31.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 2009;284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faye G, Simon M, Valay JG, Fesquet D, Facca C. Rig2, a RING finger protein that interacts with the Kin28/Ccl1 CTD kinase in yeast. Mol. Gen. Genet. 1997;255:460–466. doi: 10.1007/s004380050518. [DOI] [PubMed] [Google Scholar]
- 35.Jona G, Livi LL, Gileadi O. Mutations in the RING domain of TFB3, a subunit of yeast transcription factor IIH, reveal a role in cell cycle progression. J. Biol. Chem. 2002;277:39409–39416. doi: 10.1074/jbc.M202733200. [DOI] [PubMed] [Google Scholar]
- 36.Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl Acad. Sci. USA. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong SW, Hong SM, Yoo JW, Lee YC, Kim S, Lis JT, Lee DK. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. USA. 2009;106:14276–14280. doi: 10.1073/pnas.0903642106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee KM, Miklos I, Du H, Watt S, Szilagyi Z, Saiz JE, Madabhushi R, Penkett CJ, Sipiczki M, Bahler J, et al. Impairment of the TFIIH-associated CDK-activating kinase selectively affects cell cycle-regulated gene expression in fission yeast. Mol. Biol. Cell. 2005;16:2734–2745. doi: 10.1091/mbc.E04-11-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallenfang MR, Seydoux G. cdk-7 is required for mRNA transcription and cell cycle progression in Caenorhabditis elegans embryos. Proc. Natl Acad. Sci. USA. 2002;99:5527–5532. doi: 10.1073/pnas.082618399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larochelle S, Pandur J, Fisher RP, Salz HK, Suter B. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 1998;12:370–381. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leclerc V, Raisin S, Leopold P. Dominant-negative mutants reveal a role for the Cdk7 kinase at the mid-blastula transition in Drosophila embryos. EMBO J. 2000;19:1567–1575. doi: 10.1093/emboj/19.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz BE, Larochelle S, Suter B, Lis JT. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol. Cell. Biol. 2003;23:6876–6886. doi: 10.1128/MCB.23.19.6876-6886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sano M, Izumi Y, Helenius K, Asakura M, Rossi DJ, Xie M, Taffet G, Hu L, Pautler RG, Wilson CR, et al. Menage-a-trois 1 is critical for the transcriptional function of PPARgamma coactivator 1. Cell Metab. 2007;5:129–142. doi: 10.1016/j.cmet.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Larochelle S, Merrick K, Terret M, Wohbold L, Barboza NM, Zhang C, Shokat KM, Jallepalli PV, Fisher RP. Requirements for Cdk7 in the Assembly of Cdk1/Cyclin B and Activation of Cdk2 Revealed by Chemical Genetics in Human Cells. Mol. Cell. 2007;25:839–850. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pessa HK, Ruokolainen A, Frilander MJ. The abundance of the spliceosomal snRNPs is not limiting the splicing of U12-type introns. RNA. 2006;12:1883–1892. doi: 10.1261/rna.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guzder SN, Qiu H, Sommers CH, Sung P, Prakash L, Prakash S. DNA repair gene RAD3 of S. cerevisiae is essential for transcription by RNA polymerase II. Nature. 1994;367:91–94. doi: 10.1038/367091a0. [DOI] [PubMed] [Google Scholar]
- 51.Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- 52.Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 53.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larochelle S, Batliner J, Gamble MJ, Barboza NM, Kraybill BC, Blethrow JD, Shokat KM, Fisher RP. Dichotomous but stringent substrate selection by the dual-function Cdk7 complex revealed by chemical genetics. Nat. Struct. Mol. Biol. 2006;13:55–62. doi: 10.1038/nsmb1028. [DOI] [PubMed] [Google Scholar]
- 57.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl Acad. Sci. USA. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molina-Navarro MM, Castells-Roca L, Belli G, Garcia-Martinez J, Marin-Navarro J, Moreno J, Perez-Ortin JE, Herrero E. Comprehensive transcriptional analysis of the oxidative response in yeast. J. Biol. Chem. 2008;283:17908–17918. doi: 10.1074/jbc.M800295200. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Gabriel MA, Watt S, Bahler J, Russell P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol. Cell. Biol. 2006;26:6347–6356. doi: 10.1128/MCB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bond AT, Mangus DA, He F, Jacobson A. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 2001;21:7366–7379. doi: 10.1128/MCB.21.21.7366-7379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan J, Yang X, Wang W, Wood WH, III, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawai T, Fan J, Mazan-Mamczarz K, Gorospe M. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell. Biol. 2004;24:6773–6787. doi: 10.1128/MCB.24.15.6773-6787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- 65.Custodio N, Vivo M, Antoniou M, Carmo-Fonseca M. Splicing- and cleavage-independent requirement of RNA polymerase II CTD for mRNA release from the transcription site. J. Cell Biol. 2007;179:199–207. doi: 10.1083/jcb.200612109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meininghaus M, Chapman RD, Horndasch M, Eick D. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxy-terminal domain of the large subunit affects early steps in transcription. J. Biol. Chem. 2000;275:24375–24382. doi: 10.1074/jbc.M001883200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.