Abstract
Telomeres play a central role in human cancer, cardiovascular aging and possibly longevity. However, present methods to measure telomere length are fraught with shortcomings that limit their use. Here, we describe a novel method to measure the relative telomere DNA content by dot blot analysis. In each dot, the DNA content is measured by a DNA stain (Dx) and the telomeric DNA content is measured with a telomeric probe (T). The T normalized for Dx (T/Dx) of each dot is a measure of telomere content. The method requires ∼20 ng of DNA per assay. Moreover, the T/Dx data are highly correlated linearly with mean telomere lengths derived from Southern blots of the terminal restriction fragments (r > 0.96, P < 0.0001). The method is also simple to use, has a relatively low interassay coefficient of variation (<6%), retains its precision in moderately degraded DNA and can be forged for high throughput analysis. The method might help researchers and clinicians alike in understanding risks for and extent of human diseases.
INTRODUCTION
Telomeres, the TTAGGG repeats at the ends of chromosomes, undergo progressive shortening in replicating cells that lack active telomerase—the reverse transcriptase that adds telomere repeats to the end of the chromosomes (1,2). In these cells, telomere length is not only a record of cell replication but it is also an index of replicative potential. Cellular replication figures in mammalian development, tissue maintenance and in aging; it is also a key player in the runaway cellular replication that is the hallmark of cancer (3,4). Accordingly, telomere dynamics (telomere length and its shortening) have been the focus of intense interest in both clinical and epidemiological studies, which have primarily measured leukocyte telomere length (LTL). These studies have established that LTL is inversely related to age and is relatively short in individuals who suffer from aging-related diseases, principally atherosclerosis (5–7). Relatively short LTL has also been found to be associated with diminished survival of elderly persons (8–10). Moreover, shortened LTL is often expressed in various forms of aplastic anemia (11).
Methods to measure telomere length/content include Southern blot analysis of the terminal restriction fragment (TRF) lengths (1), quantitative PCR (qPCR) (12,13), fluorescent in situ hybridization (FISH) (14,15), slot blot or dot blot analysis (16–18) and single telomere length analysis (STELA) (19). Although these techniques have been successfully employed in broadening understanding the roles of telomeres in human health and disease, they are also fraught with shortcomings (20,21) that have contributed to a growing debate about their research applications and usefulness in clinical settings. For epidemiological research, the TRF length analysis and qPCR method are typically preferred over the other methods, but they too display considerable problems that limit their practical use.
Clearly, there is a growing need for a simple, specific and sufficiently precise method to measure telomere length for research and clinical purposes. This need will only escalate with the anticipated expansions of research and clinical applications that will require telomere length measurements. We have developed such a method. Based on dot blot analysis, the method requires little amount of DNA (∼20 ng/assay), and moreover is inexpensive and easy to perform.
MATERIALS AND METHODS
General procedures
In the first set of experiments, we used leukocyte DNA donated by 28 young and middle-age adults. As LTLs in this group did not encompass the overall spectrum of LTLs seen throughout the entire human lifespan, in a second set of experiments we determined LTL in leukocyte samples from seven newborns and seven elderly persons (aged 90–96 years).
We isolated DNA by Gentra Puregene Blood kit (Qiagen). The telomere probe we used for both the TRF length analysis and dot blot analysis consisted of three CCCTAA oligonucleotide repeats, which are complementary to the canonical sequence of TTAGGG repeats, and it is labeled at the 3′-end with digoxigenin (DIG).
Telomere repeats/DNA ratio by dot blot analysis
The method proceeds as follows: (i) the analysis is carried out using DNA dot blots; (ii) the DNA content of the sample on the blot is measured by SYBR Dx DNA Blot Stain (S-7550, Invitrogen); (iii) the blot is hybridized with a DIG labeled telomeric probe; and (iv) the ratio of telomere repeats/DNA (T/Dx) is the amount of the repeats per unit DNA.
We performed the assays as follows: DNA concentration in all samples, including those used for generation of standards, was initially measured in solution by UV absorbance at 260 nm. DNA for the standard was from a leukocyte sample with a mean TRF length of 7.29 kb. DNA (3.3 µl; 20 ng/µl for samples and 5, 15, 25 and 35 ng/µl for the standard) was diluted into 16.5 µl of denaturing solution (0.5 M NaOH, 1.5 M NaCl) and incubated at 55°C for 30 min. Neutralizing solution (495 µl; 0.5 M Tris–HCl, 1.5 M NaCl) was added. A positively charged nylon membrane (Roche) was soaked in distilled water for 10 min and Bio-Dot Microfiltration Apparatus was assembled according to the manufacturer's instructions. Each well was washed once with 200 µl water. The 156 µl of neutralized sample or standard was loaded into each well (in triplicate) and subjected to gentle vacuum. Thereafter, each well was washed once with 200 µl 2× saline–sodium citrate buffer (SSC), the membrane was removed, rinsed in 2× SSC, dried and UV-cross-linked (UV dose = 120 mJ/cm2) on both sides of the membrane.
For DNA blot staining, the membrane was washed in distilled water for 10 min, rinsed with 0.5 × Tris–borate–EDTA buffer (TBE) and stained with 5 ml SYBR Dx stain (1000× dilution in 0.5× TBE) for 30 min. The fluorescence signal was measured by Typhoon 9400 (GE Healthcare). The DNA amount of each sample was calculated from the standard.
The membrane was pre-hybridized in 5× SSC, 0.1% Sarkosyl, 0.02% sodium dodecyl sulfate (SDS) and 1% blocking reagent (Roche) at 65°C for 2 h and hybridized at 65°C with the telomeric probe overnight in the same solution. The membrane was washed three times at room temperature in 2× SSC and 0.1% SDS each for 15 min and once in 2× SSC for 15 min. The DIG-labeled probe was detected by the DIG luminescent detection procedure using CDP-Star (Roche) and exposed on X-ray film (21). The amount of telomere repeats from each sample was calculated from the standard.
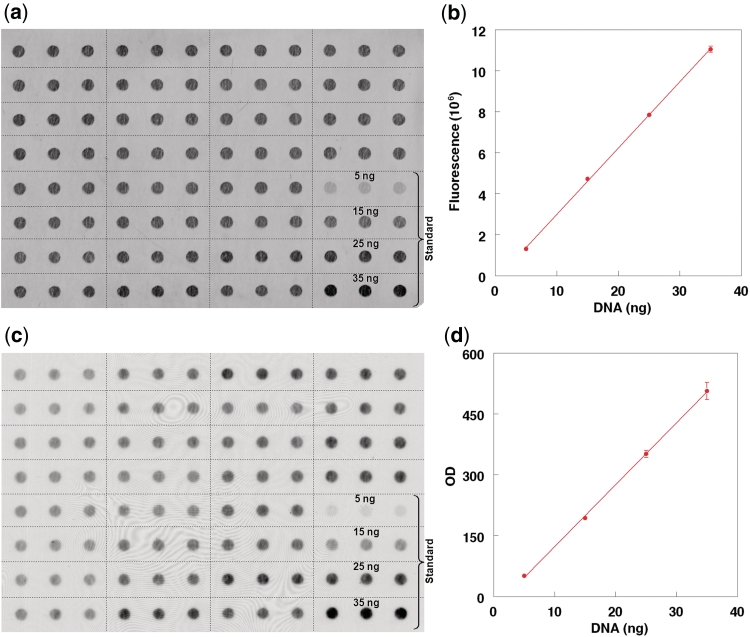
Using ImageQuant, the total intensity above local background surrounding each dot was independently determined for each of the three dots in each rectangle shown in Figure 1a and c. The mean value of the three dots was used for the standard curves displayed in Figure 1b and d. For each sample, the T/Dx for each of the three dots was independently obtained and the mean of the three T/Dx values was derived.
Figure 1.
Dot blot staining of SYBR Dx and the telomere (TTAGGG) signal. (a) The SYBR Dx dot blot staining of different samples in triplicate. Samples used for the DNA standard are at the right lower corner. (b) The linearity of the staining between 5 and 35 ng of standard DNA. Each data point is the mean of triplicate measurements. (c) The telomere signal, while (d) shows the linearity of the telomere signal within the 5–35 ng of standard DNA. Each data point is the mean of triplicate measurements. Vertical bars denote SD. No vertical bar indicates that the SD is within the space of the data point.
Telomere Length/DNA content measurements by Southern blot analysis of the TRFs and by qPCR
We obtained the mean TRF length using either HinfI/RsaI or HphI/MnlI, as previously described (21). HinfI/RsaI, which typically is used in TRF length analysis in most laboratories, cuts the DNA within the non-canonical subtelomeric region. However, HphI/MnlI cut the DNA at telomere repeat variants that are in more proximal regions of the telomeres (22). Therefore, digestion with HinfI/RsaI usually results in a mean TRF length that is longer by ∼1 kb than that resulting from HphI and MnlI digestion (21).
Measurements of the mean TRF length were performed as previously described (21). Briefly, we first evaluated the DNA integrity by SYBR Green I, after resolving each sample (10 ng) on 1% agarose gel at 200 V for 60 min. Thereafter, samples were digested with restriction enzymes Hinf I (10 U) and Rsa I (10 U; Roche) or HphI (3.1 U)/MnlI (3.1 U) (New England Biolabs, Ipswich, MA, USA). DNA samples (3 µg each), and DNA ladders (1 kb DNA ladder plus λ DNA/Hind III fragments; Invitrogen, Carlsbad, CA, USA) were resolved on 0.5% agarose gels for most subjects and on a 0.6% agarose gels for subjects 90–96 years (20 cm × 20 cm) at 50 V (GNA-200 Pharmacia Biotech). After 16 h, the DNA was depurinated for 15 min in 0.25 N HCl, denatured 30 min in 0.5 M NaOH/1.5 mol/l NaCl and neutralized for 30 min in 0.5 M Tris, pH 8/1.5 M NaCl. The DNA was transferred for 1 h to a positively charged nylon membrane (Roche) using a vacuum blotter (Boeckel Scientific, Feasterville, PA, USA). Thereafter, membranes were hybridized at 65°C with the DIG-labelled telomeric probe overnight in 5 x SSC, 0.1% Sarkosyl, 0.02% SDS and 1% blocking reagent (Roche). The membranes were washed three times at room temperature in 2× SSC, 0.1% SDS each for 15 min and once in 2× SSC for 15 min. The DIG-labeled probe was detected by the DIG luminescent detection procedure (Roche) and exposed on X-ray film. All autoradiographs were scanned, and the TRF signal was digitized. The optical density values versus DNA migration distances were converted to optical density (adjusted for background)/molecular weight versus molecular weight (21).
The measurement of telomere repeats by qPCR provides the ratio of the telomeric product (T) normalized for a single-copy gene product (S) (12,13). This measurement was performed as previously described (23), using minor modifications of the original method (12) and β-globin as S.
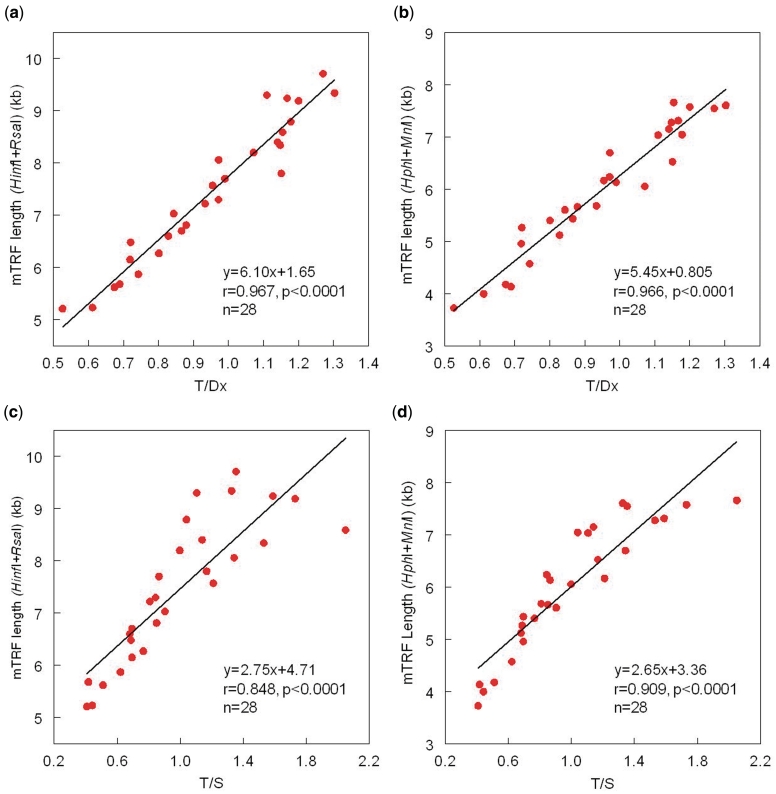
Each of the data points described in Figures 2 and 3 represents (i) the mean of two different runs for the mean TRF length (one measure/run); (ii) the mean of two T/Dx runs (three measures/run); and (iii) the mean of three T/S runs (three measures/run). The coefficient of variations for the intraassay and interassay measurements were computed as the ratios of the standard deviation (SD) to the mean of measurements performed on the same run and the mean of measurements performed on different runs, respectively.
Figure 2.
Relationship between mean telomere length, measured by Southern blots of the TRFs, versus the ratio of T (telomere amount)/Dx (DNA amount), measured by dot blots, and versus the ratio of T (telomere product)/S(single gene product), measured by qPCR. (a and b) The TRF products generated by HinfI/RsaI and by HphI/MnlI versus the T/Dx. (c and d) The TRF product generated by HinfI/RsaI and by HphI/MnlI versus T/S.
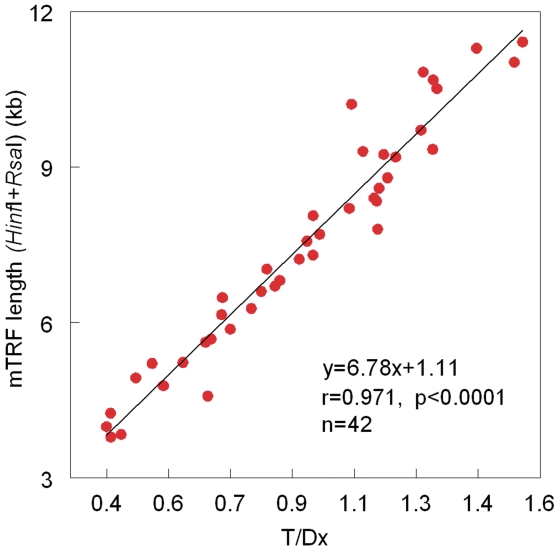
Figure 3.
Relationship between mean telomere length, measured by Southern blots of the TRFs, generated by HinfI/RsaI, versus the ratio of T (telomere amount)/Dx (DNA amount), measured by dot blots. Data displayed in the figure are a composite of data displayed in Figure 2 plus an additional set of measurements in leukocytes from newborns and exceptionally old persons.
Informed consent
DNA donors or their parents/guardians provided written consent. The study was approved by the Institutional Review Board of University of Medicine and Dentistry of NJ, NJ Medical School, USA.
RESULTS
Figure 1 displays dot blot staining of SYBR Dx (a) and the telomere signal (c) of triplicate samples. Clearly, at the range of 5–35 ng the standard curves of both the SYBR Dx dye (b) and the telomere signals (d) are highly linear. The intraassay coefficient of variations (%) of triplicate samples (analyzed at the same time) were: T = 4.4, Dx = 2.6, T/Dx = 5.4 (N = 56). The interassay coefficient of variation of T/Dx for samples analyzed on different days was 5.7 (N = 28).
Figure 2 shows the relations of T/Dx and T/S with mean TRF length generated either by HinfI/RsaI or HphI/MnlI. For the T/Dx (a and b), strong correlations are observed with mean TRF length regardless of the restriction enzymes used to generate the TRFs. Strong correlations are also observed for the relation of T/S (c and d), with the mean TRF length, although the correlations are not as robust as those between T/Dx and the mean TRF length, particularly for the HinfI/RsaI product.
Both the present dot blot analysis and the qPCR-based methods measure only the canonical part of the telomeres, which consist of strictly TTAGGG repeats. In contrast, the TRF length generated by the Southern blots includes both the canonical and non-canonical region extending to the nearest restriction site. Estimates of the extrapolated non-canonical regions can be obtained from the regressions displayed in Figure 2 when T/Dx = 0 or T/S = 0, with the assumption that linearity of the regressions is maintained beyond the empirical data. Accordingly, for TRFs generated by HinfI/RsaI, when T/Dx = 0, mean TRF = 1.65 kb; for TRFs generated by HphI/MnlI, when T/Dx = 0, mean TRF = 0.805 kb. However, for TRFs generated by HinfI/RsaI, when T/S = 0, mean TRF = 4.71 kb; for TRFs generated by HphI/MnlI, when T/S = 0, mean TRF = 3.36 kb. These differences in the extrapolated lengths of the non-canonical segment of the TRFs probably stem from deviation from linearity of relation between T/S versus mean TRF length (Figure 2c and d), which is already observed for the empirical data of the regressions displayed in Figure 2c and d. This finding has been shown by others (12,13,24). The underlying causes are not certain but we presume that they relate to problems with the PCR of S, T or both.
The data displayed in Figure 2 do not cover the entire spectrum of LTL seen throughout the entire human lifespan. For this reason, in a second set of experiments we measured LTL (by dot blot analysis and Southern blots of the TRFs generated by HinfI/RsaI) in leukocytes from newborns and exceptionally old persons. Figure 3 consists of data derived from results generated in the first set of experiments (Figure 2a) and the second set of experiments. Clearly, the linear relation between the mean TRF and the T/Dx is maintained in leukocytes from donors whose age range essentially covers the entire human lifespan.
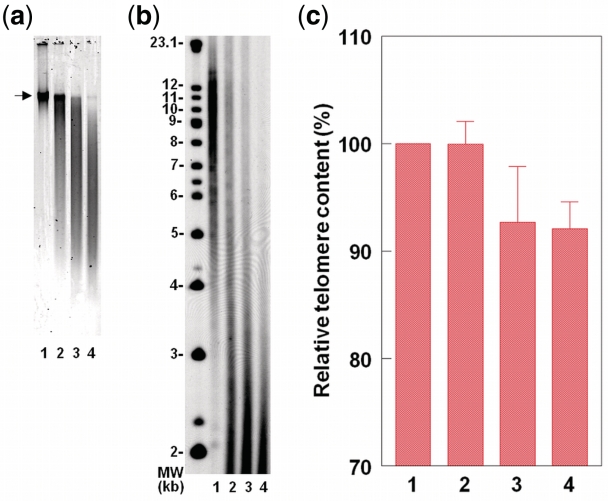
The effect of DNA degradation on the TRF length and the T/Dx measurements is shown in Figure 4. DNA was degraded by sonication (Ultrasonic Processor XL2020, microtip probe) and integrity determined (using SYBR Green I) by resolving 10 ng DNA on 1% (wt/vol) agarose gel at 200 V for 60 min (a). Moderate DNA degradation, where the DNA crown is still compact but a long tail of DNA smear is observed, already compromises any meaningful analysis of the TRFs. That is because the TRFs are considerably shortened, so that the TRF smear extends to the edge of the gel (b), outside the routine scanning region of the TRFs, which rarely extends lower than 1.2 kb (6). More degradation further exacerbates this effect. In contrast, The T/Dx results are not modified by moderate DNA degradation (c). However, more severe DNA degradation caused 7–8% decline in the T/Dx values, probably because the stretches of telomere repeats become too short for an effective annealing of the telomere probe.
Figure 4.
The effect of DNA sonication on the TRF and T/Dx measurements. For all panels, lane 1 = no sonication; lane 2 = 0.2 s × 5 pulse (Position 1.5); Lane 3 = 0.2 s × 5 pulse (Position 2); lane 4 = 0.2 s × 10 pulse (position 2). (a) Illustrates DNA integrity (arrow indicates the DNA crown). (b) Illustrates TRF length distribution. (c) Shows T/Dx results based on three DNA samples.
DISCUSSION
Among the methods that measure telomere length or DNA content, the qPCR (12,13) and to a lesser extent the Southern blots of the TRFs (1,21) have been the main techniques used in epidemiological research. The FISH method to measure telomere content (14,15) requires intact nuclei and samples must be processed within a short amount of time. Although STELA (19) is an ingenious approach to measure telomere length, it is a complex method with limited application for large-scale epidemiological studies and in clinical settings.
Telomere DNA content has also been measured in slot/dot blot analysis by normalizing for alphoid centromere repeats (16,17). However, the length of alphoid centromeric repeats is highly variable among individuals (26). Moreover, in those few published works in which this method has been employed, the measurements of telomeric content and centromeric content were performed in duplicate blots (one for telomeric content and the other for centromeric content) (16) or in the same blot in which the hybridization with the telomeric probe was followed by stripping the probe and re-hybridizing with the centromeric probe (17). These approaches increase the measurement error because the DNA input in the different blots and different slots/dots might not be precisely the same. Also stripping the membranes and re-probing may raise the background signal, resulting in an increase in the measurement error.
One report described dot blot analysis in which telomere DNA content on the blots was normalized for total DNA measured in solution by PicoGreen of samples applied to the blots (18). The following are three intrinsic errors that take place when the DNA content is measured in solution while the telomere DNA is quantified on the blot. The first error is in the measurement of DNA itself. No matter how accurate the measurement in solution might be, it has its own intrinsic error, including a potential shift in the DNA standard on different runs, which would increase the interassay variation. The second error level is the fact that small amounts of DNA might adhere to the walls of tubes, pipette tips and the blot apparatus. Thus, the amount of DNA actually in the dot is not the amount that is presumed to be pipetted onto it. The third error level is in the pipetting into the dot; again, no matter how accurate this pipetting might be, the DNA input still varies for different dots. Together, these three tiers of error introduce considerable variation to the results.
Our approach avoids these errors altogether by taking advantage of the unique feature of the SYBR Dx DNA Blot Stain to measure the total DNA content in the dot without interference with the hybridization of the telomere probe. Such a feature enables measuring the relative amount of telomere DNA content to total DNA in the dot itself regardless of the extent of the error in the input DNA. This applies to the standard and the samples. For instance, suppose that the true DNA content in a given dot is <20% what one thinks it is, the telomere DNA signal will also be proportionally lower by 20%. Therefore, the ratio between the telomere DNA signal and the total DNA signal, which is the measure of telomere content in the sample, would not be affected.
Normalizing the telomeric content for the DNA content of a given sample is based on the premise that the amount of DNA in nuclei of somatic cells is constant. Thus, T/Dx provides a measure of mean telomere per DNA content in a sample. Such a conclusion is reasonable provided that the amount of telomeric DNA is within the discernible range of the method and the DNA does not contain large amounts of heterogeneous telomere-like sequences within its bulk. This is apparently the case for human DNA and other mammals. The DNA content doubles in cells undergoing mitosis, but so does the amount of telomere repeats. Therefore, the T/Dx ratio is still constant in mitotic cells. In addition, females have two X chromosomes, while males have only one X chromosome and the much smaller Y chromosome. In principle, the nuclei of females should have more DNA, but evidently this effect is small, amounting to a difference of ∼1.6% (25). If future large-scale studies show that the sex difference in DNA content impacts the T/Dx ratio in relation to indices of interest, a sex adjustment factor can be introduced or standard curves can be developed separately for females and males.
We underscore the following features of the T/Dx approach:
The SYBR Dx DNA stain is used to quantify single-stranded DNA directly on the dot itself.
The SYBR Dx DNA Blot Stain does not interfere with the telomeric probe hybridization, as indicated in the ‘Product Information’ of the SYBR Dx DNA Blot Stain (Invitrogen).
The detections of both DNA and telomeric signals are performed within their linear ranges.
The measurement of DNA content is completed within 1 h.
Small quantities of DNA (∼20 ng and as little as 5 ng) are used per dot, yet the analysis does not rely on PCR.
The method only measures the canonical component of telomeres.
Moderate DNA degradation has no effect on the T/Dx ratio.
The method can be forged for high-throughput analysis.
It is useful to compare these features with those of the Southern blot analysis of the TRFs and the qPCR method to measure telomere DNA content.
Considered the gold standard of telomere length measurement, the Southern blot analysis of TRF lengths provides in absolute values (kb) the entire distribution of telomere lengths in the DNA sample. The TRF analysis requires a large amount of DNA (∼2.5 µg per assay) and is labor intensive and costly; it also requires a considerable degree of expertise (21). Since the TRFs include the non-canonical portion of telomeres up to the nearest restriction site, in theory, polymorphisms in these sites might impact the findings. Of importance, DNA integrity is essential for obtaining reliable results.
The qPCR method normalizes T for S and it measures the mean content of the canonical component of telomeres. The method is high throughput, relatively inexpensive and like the dot blot method, it requires little DNA (∼20 ng per assay). However, confounders that affect the telomere and reference signals are poorly understood. For instance, regardless of which gene is used as a reference S, copy number variations might affect the findings. Moreover, the PCR might amplify any measurement error.
Like the qPCR and FISH techniques, the dot blot analysis measures telomere DNA content in relative units rather than in units of telomere length (kb). The relation between the mean TRF length and the qPCR results (T/S) is nonlinear. Therefore, expressing the T/S to telomere length (in kb) based on a linear model would generate a considerable distortion in the results. In principle, the dot blot results (T/Dx) can be expressed in unit telomere length based on linear models. This entails generating for each study cohort a formula that expresses the correlation between the mean TRF length and the T/Dx based on a reference panel of DNA samples with known mean TRFs across the expected range of telomere lengths for that cohort. However, little is known about the impact of potential polymorphism in the restriction sites on the mean TRF length. It is possible, therefore, that DNA panels from different individuals might show slightly different association between the mean TRF length and T/Dx, but we anticipate that this effect will be small.
A major shortcoming of the T/Dx method is that it cannot be applied to various forms of cancer. Hematopoietic malignancies and solid cancers display an array of DNA changes, including deletions and amplifications of genes and chromosomal segments, which would alter the DNA content. This might also apply to the qPCR method.
Finally, the lack of an optimal method to measure telomere length has frustrated efforts to translate the knowledge generated by basic and epidemiological telomere research to answer practical and pressing questions in the clinical domain. The measurement of the true telomere length or content, like the measurement of any parameter, is a standard that can only be approached asymptotically. That said, in the future telomere length measurement methods, tailored to meet specific needs, will be central to screening programs to detect persons with short (or long) telomeres and enable the biomedical community to gain vital information above and beyond conventional biomarkers about persons at risk for cardiovascular disease and premature death. This knowledge might ultimately guide preventive and therapeutic interventions defined by the new era of personalized medicine.
FUNDING
Funding for open access charge: The National Institutes of Health (grant numbers AG030678 and AG20132).
Conflict of interest statement. None declared.
REFERENCES
- 1.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 3.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 4.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willeit P, Willeit J, Brandstätter A, Ehrlenbach S, Mayr A, Gasperi A, Weger S, Oberhollenzer F, Reindl M, Kronenberg F. Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler. Thromb. Vasc. Biol. 2010;30:1649–1656. doi: 10.1161/ATVBAHA.110.205492. [DOI] [PubMed] [Google Scholar]
- 6.Brouilette SW, Moore JS, McMahon AD, Thompson JR, Ford I, Shepherd J, Packard CJ, Samani NJ West of Scotland Coronary Prevention Study Group. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet. 2007;369:107–114. doi: 10.1016/S0140-6736(07)60071-3. [DOI] [PubMed] [Google Scholar]
- 7.O'Donnell CJ, Demissie S, Kimura M, Levy D, Gardner JP, White C, D'Agostino RB, Wolf PA, Polak J, Cupples LA. Leukocyte telomere length and carotid artery intimal medial thickness: The Framingham Heart Study. Arterioscler. Thromb. Vasc. Biol. 2008;28:1165–1171. doi: 10.1161/ATVBAHA.107.154849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura M, Hjelmborg JV, Gardner JP, Bathum L, Brimacombe M, Lu X, Christiansen L, Vaupel JW, Aviv A, Christensen K. Short leukocyte telomeres forecast mortality: a study in elderly Danish twins. Am. J. Epidemiol. 2008;167:799–806. doi: 10.1093/aje/kwm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakaysa SL, Mucci LA, Slagboom PE, Boomsma DI, McClearn GE, Johansson B, Pedersen NL. Telomere length predicts survival independent of genetic influences. Aging Cell. 2007;6:769–774. doi: 10.1111/j.1474-9726.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- 10.Fitzpatrick AL, Kronmal RA, Kimura M, Gardner JP, Psaty BM, Jenny NS, Tracy RP, Hardikar S, Aviv A. Leukocyte Telomere Length and Mortality in the Cardiovascular Health Study. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66A:421–429. doi: 10.1093/gerona/glq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calado RT, Young NS. Telomere diseases. N. Engl. J. Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat. Protoc. 2006;1:2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 15.Canela A, Vera E, Klatt P, Blasco MA. High-throughput telomere length quantification by FISH and its application to human population studies. Proc. Natl Acad. Sci. USA. 2007;104:5300–5305. doi: 10.1073/pnas.0609367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryant JE, Hutchings KG, Moyzis RK, Griffith JK. Measurement of telomeric DNA content in human tissues. Biotechniques. 1997;23:476–478. doi: 10.2144/97233st05. [DOI] [PubMed] [Google Scholar]
- 17.Ogami M, Ikura Y, Ohsawa M, Matsuo T, Kayo S, Yoshimi N, Hai E, Shirai N, Ehara S, Komatsu R, et al. Telomere shortening in human coronary artery diseases. Arterioscler. Thromb. Vasc. Biol. 2004;24:546–550. doi: 10.1161/01.ATV.0000117200.46938.e7. [DOI] [PubMed] [Google Scholar]
- 18.Fordyce CA, Heaphy CM, Griffith JK. Chemiluminescent measurement of telomere DNA content in biopsies. Biotechniques. 2002;33:144–146, 148. doi: 10.2144/02331md02. [DOI] [PubMed] [Google Scholar]
- 19.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 20.Baird DM. New developments in telomere length analysis. Exp. Gerontol. 2005;40:363–368. doi: 10.1016/j.exger.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M, Stone RC, Hunt SC, Skurnick J, Lu X, Cao X, Harley CB, Aviv A. Measurement of telomere length by the southern blot analysis of the terminal restriction fragment lengths. Nat. Protoc. 2010;5:1596–1607. doi: 10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
- 22.Baird DM, Britt-Compton B, Rowson J, Amso NN, Gregory L, Kipling D. Telomere instability in the male germline. Hum. Mol. Genet. 2006;15:45–51. doi: 10.1093/hmg/ddi424. [DOI] [PubMed] [Google Scholar]
- 23.Gardner JP, Kimura M, Chai W, Durrani JF, Tchakmakjian L, Cao X, Lu X, Li G, Peppas AP, Skurnick J, et al. Telomere dynamics in macaques and humans. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:367–374. doi: 10.1093/gerona/62.4.367. [DOI] [PubMed] [Google Scholar]
- 24.Ehrlenbach S, Willeit P, Kiechl S, Willeit J, Reindl M, Schanda K, Kronenberg F, Brandstätter A. Influences on the reduction of relative telomere length over 10 years in the population-based Bruneck Study: introduction of a well-controlled high-throughput assay. Int. J. Epidemiol. 2009;38:1725–1734. doi: 10.1093/ije/dyp273. [DOI] [PubMed] [Google Scholar]
- 25.Repin MV, Golubev PI, Repina LA. New sequence-based data on the relative DNA contents of chromosomes in the normal male and female human diploid genomes for radiation molecular cytogenetics. Mol. Cytogenet. 2009;2:13. doi: 10.1186/1755-8166-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jabs EW, Goble CA, Cutting GR. Macromolecular organization of human centromeric regions reveals high-frequency, polymorphic macro DNA repeats. Proc. Natl Acad. Sci. USA. 1989;86:202–206. doi: 10.1073/pnas.86.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]