p53 exhibits a single non-excitable pulse in response to UV radiation, which contrasts with the excitable series of undamped pulses induced by DNA double-strand breaks. The authors find that these behaviors are conferred by specific regulatory connections in the p53 network.
Keywords: DNA damage, dynamics, excitability, feedback, live-cell imaging
Abstract
Many biological networks respond to various inputs through a common signaling molecule that triggers distinct cellular outcomes. One potential mechanism for achieving specific input–output relationships is to trigger distinct dynamical patterns in response to different stimuli. Here we focused on the dynamics of p53, a tumor suppressor activated in response to cellular stress. We quantified the dynamics of p53 in individual cells in response to UV and observed a single pulse that increases in amplitude and duration in proportion to the UV dose. This graded response contrasts with the previously described series of fixed pulses in response to γ-radiation. We further found that while γ-triggered p53 pulses are excitable, the p53 response to UV is not excitable and depends on continuous signaling from the input-sensing kinases. Using mathematical modeling and experiments, we identified feedback loops that contribute to specific features of the stimulus-dependent dynamics of p53, including excitability and input-duration dependency. Our study shows that different stresses elicit different temporal profiles of p53, suggesting that modulation of p53 dynamics might be used to achieve specificity in this network.
Introduction
The tumor suppressor protein p53 is induced in response to many stress signals (Horn and Vousden, 2007) and activates various stress-response programs including cell-cycle arrest, senescence, and apoptosis. An important unanswered question is how can a single protein orchestrate such a complex response to different inputs? Much attention in this regard has been focused around the hypothesis that different post-translational modifications of p53 lead to activation of different downstream gene programs (Brooks and Gu, 2003; Bode and Dong, 2004). However, another potential level of regulation, which has not been explored as much in this system, is the temporal dynamics of p53.
The dynamical behavior of p53 has been extensively characterized in response to one particular form of DNA damage, double-strand breaks (DSBs) caused by γ-radiation or the radiomimetic drug neocarzinostatin (NCS) (Shiloh et al, 1983). DSBs trigger a series of p53 pulses with fixed amplitude and duration, independent of the damage dose, whereas the number of pulses increases with higher damage (Lahav et al, 2004; Geva-Zatorsky et al, 2006). It is not clear whether other types of DNA damage lead to similar dynamical behavior. In addition, the p53 response to DSBs was shown to be excitable—a full p53 pulse is triggered by either a sustained or a transient input (Batchelor et al, 2008; Loewer et al, 2010). The mechanism generating excitability in the p53 response to DSBs has not yet been determined.
Here we show that different stresses trigger different temporal profiles of p53. We found that the p53 dynamical response to UV significantly differs from the response to DSBs, and we provide new mechanistic insights about the role of feedback controlling specific features of p53 dynamics, such as excitability and input-strength dependency.
Results and discussion
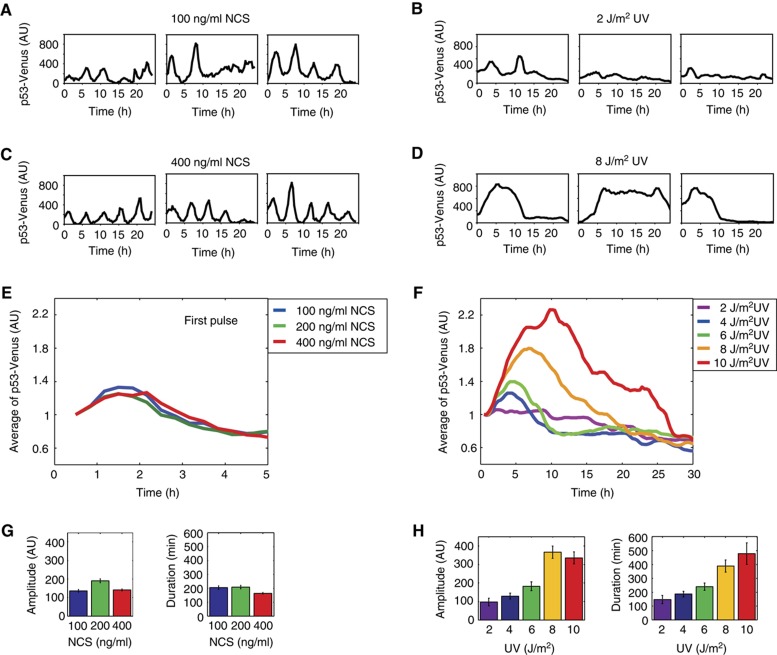
We first set out to determine whether a distinct form of DNA damage leads to a series of undamped pulses as was previously shown in response to DSBs (Lahav et al, 2004; Geva-Zatorsky et al, 2006; Batchelor et al, 2008). We chose UV light, which is known to cross-link consecutive pyrimidine bases leading to the exposure of single-stranded DNA (ssDNA). Previous studies showed that population-averaged analysis can mask true dynamical responses (Lahav et al, 2004; Tay et al, 2010); we therefore analyzed p53 dynamics in single cells using a p53-Venus fusion (Batchelor et al, 2008). We found that in response to a single short burst of 2 J/m2 UV, p53 showed pulses that were similar in amplitude and duration to the pulses observed in response to DSBs caused by NCS (Figure 1A and B). However, the frequency of the pulses in response to UV was lower and did not appear to be as regular as the pulses in response to DSBs.
Figure 1.
Stimulus-dependent dynamics of p53 in response to NCS (DSBs) and to UV. (A–D) Representative traces of MCF7 cells expressing p53-Venus exposed to 100 (A) or 400 ng/ml (C) of NCS or to 2 (B) or 8 J/m2 (D) UV at the indicated time following treatment. (E, F) p53-Venus levels averaged over the traces of cells in response to various levels of NCS and UV. Note that for NCS, the averaged response is shown only for the first p53 pulse, as synchrony between cells is lost in subsequent pulses. Each experiment includes between 60 and 110 cells. The mean trace was normalized to the mean p53-Venus of the first time point. (G, H) Quantification of the relative amplitude and full-width half-maximum duration of all p53 pulses in response to NCS (G) or the first pulse in response to UV (H). Error bars represent s.e.m.
To determine whether there was a principal frequency of the UV-induced pulses, we performed pitch detection as was previously described (Geva-Zatorsky et al, 2006). In contrast to the DSB response that results in a principal period of 4–7 h (Geva-Zatorsky et al, 2006), a broad distribution of pitches was determined for the UV response (Supplementary Figure S1), indicating that there was no dominant frequency of p53 pulses. The irregularity of the pulse frequency suggests that later pulses in response to UV might be independent of the extrinsic UV damage, and likely represent pulses resulting from intrinsic damage during normal proliferation as was recently shown (Loewer et al, 2010).
The amplitude, duration, and frequency of individual p53 pulses in response to DSBs are fixed and do not depend on the damage dose (Lahav et al, 2004; Geva-Zatorsky et al, 2006; Figure 1A, C, E, and G). To determine whether this is also the case in response to UV, we irradiated cells with a broad range of UV doses, each delivered in a single burst. We found that higher UV doses increased both the amplitude and the duration of the p53 response (Figure 1B, D, F, and H). No principal frequency was identified by pitch detection for any of the UV doses (Supplementary Figure S1), indicating that there is no periodicity in the p53 response to UV. This dose-dependent, single p53 pulse in response to UV is in stark contrast to the repeated fixed pulses in response to DSBs (Figure 1; Supplementary Movies 1 and 2).
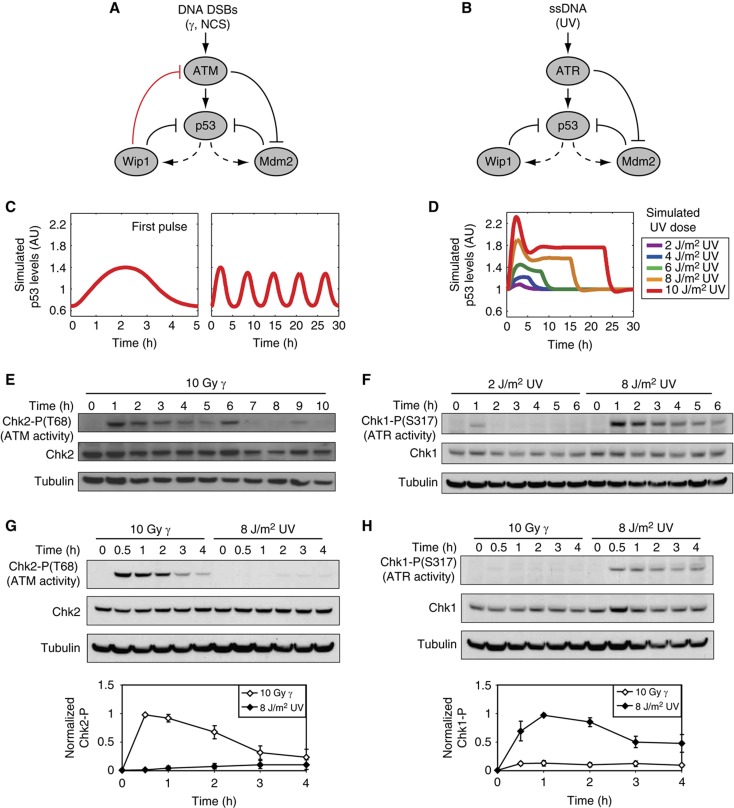
How can a single protein respond to different inputs with such markedly different dynamics? To address this question, we searched for differences in the architecture of the networks responding to DSBs or to UV. In both networks, PI3 kinase-related kinases (ATM or ATR) relay the damage signal to p53, activating two core negative-feedback loops (Figure 2A and B). One feedback loop is between p53 and the E3 ubiquitin ligase Mdm2, in which p53 activates Mdm2 expression and Mdm2 targets p53 for degradation (Barak et al, 1993; Wu et al, 1993; Haupt et al, 1997; Kubbutat et al, 1997). The other feedback is between p53 and the phosphatase Wip1, in which p53 activates Wip1 expression and Wip1 dephosphorylates p53, thus reducing its stability (Fiscella et al, 1997; Lu et al, 2005). However, we noted one main difference: the network responding to DSBs includes a negative feedback between p53 and ATM mediated by Wip1 (Shreeram et al, 2006; Figure 2A, red inhibitory arrow). In contrast, as Wip1 acts through dephosphorylation, and ATR does not require phosphorylation for its activity (Cimprich and Cortez, 2008), we assumed no direct inhibition of ATR by Wip1 (Figure 2B). To determine the contribution of the Wip1-ATM feedback to p53 dynamics, we used our mathematical model of p53 pulses in response to DSBs (Batchelor et al, 2008; Figure 2C) and removed the negative feedback from Wip1 to ATM (Supplementary information). This perturbation was sufficient for switching p53 dynamics from repeated pulses into a single pulse (Supplementary Figure S2). In addition, our model predicts that, in contrast to the pulsatile dynamics of ATM in response to DSBs (Batchelor et al, 2008; Figure 2E), active ATR will show a single continuous pulse with a duration that depends on the level of UV (Supplementary Figure S2). Western blot analysis of ATR activity in response to low and high UV was consistent with this prediction (Figure 2F). To computationally recapitulate the dose-dependent amplitude of p53 pulses observed experimentally in response to UV (Figure 1F), we were required to vary the production rate of active ATR in a dose-dependent manner (Supplementary information; Figure 2D). Indeed, we found experimentally that the levels of ATR substrate, Chk1-P, depended on the UV dose (Figure 2F). Although these data cannot rule out the possibility that Chk1-P levels might be regulated by an ATR-independent phosphatase, our results are consistent with a dose-dependent ATR activity. In agreement, in our previous work we showed that modulation of the DSB network by Wip1 knockdown resembled the UV response, generating a dose-dependent p53 response to DSBs (Batchelor et al, 2008).
Figure 2.
A single feedback switches repeated, fixed p53 pulses into a single dose-dependent pulse. (A, B) Diagrams showing key species of the p53 signaling networks in response to DSBs (A) and UV (B). Dashed lines indicate transcription; solid lines indicate protein–protein interactions. (C, D) Simulated p53 levels in response to DSBs (C) and UV (D). See Supplementary information for a description of the model. (E–H) Western blot analysis of ATM activity (measured by Chk2-P) and ATR activity (measured by Chk1-P) in cells irradiated with γ or UV. Quantification of Chk1-P and Chk2-P levels were normalized to tubulin levels. Error bars represent s.e.m. of triplicate experiments.
Our model and network diagrams (Figure 2A and B) display a complete separation between the DSBs/ATM and the UV/ATR pathways. However, previous studies have shown cross-talk between the two pathways (Jazayeri et al, 2006; Yajima et al, 2009). We therefore sought to determine the extent of cross-talk in our system and its potential impact on p53 dynamics. We measured the levels of Chk2-P (ATM target) and the levels of Chk1-P (ATR target) in response to γ and UV. Our results show relatively low levels of cross-talk between the pathways (Figure 2G and H; Supplementary information). We simulated the experimentally measured cross-talk and found minimal effect on p53 dynamics compared with simulations in which cross-talk was absent (Supplementary Figure S3; Supplementary information). We concluded that the low level of cross-talk between the DSB and UV pathways does not alter the dynamical behavior of p53.
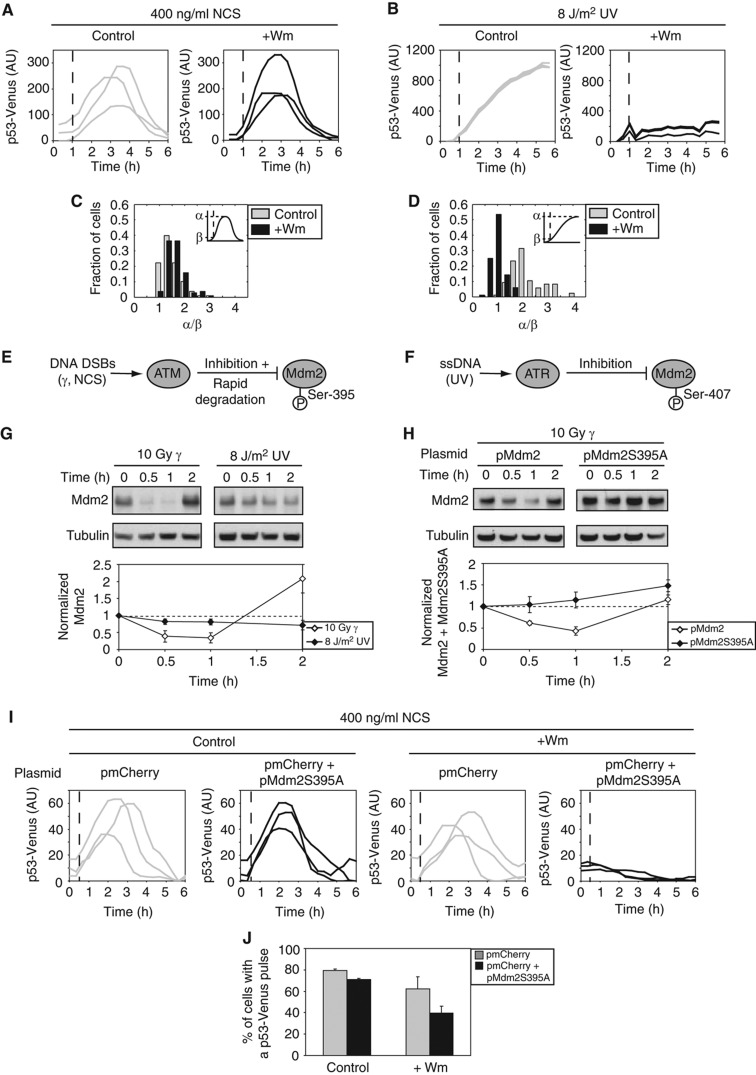
We next asked whether the p53 response to UV exhibits excitable behavior as was shown in response to DSBs (Batchelor et al, 2008; Loewer et al, 2010; Figure 3A and C). A hallmark of excitability is that a transient input is sufficient for triggering a full response. We generated a transient input signal by challenging cells with UV and inhibiting ATR 1 h post-damage, using the PI3-kinase-like kinase inhibitor wortmannin (Sarkaria et al, 1998; Supplementary Figure S4). We found that in response to UV, p53 immediately stopped accumulating when the upstream damage signal was inhibited (Figure 3B and D; Supplementary Figure S5). These results show that the p53 response to UV, as opposed to the response to DSBs, is not excitable. Wortmannin is a broad-range inhibitor and we therefore cannot exclude the possibility that the p53 pulse is halted because of the inhibition of other PI3-kinase-like kinases. However, as ATR is the kinase dominating the response to UV, and we show a complete inhibition of ATR activity in the presence of wortmannin (Supplementary Figure S4), lack of excitability in response to UV supports the suggestion that the p53 response depends on continuous activation of ATR.
Figure 3.
Differential p53 excitability between DSBs and UV results from differential regulation of Mdm2. (A–D) Cells expressing p53-Venus were treated with NCS (A) or UV (B). One hour after damage, medium containing DMSO (control) or wortmannin (+Wm) (Sarkaria et al, 1998) was added (dashed line). Representative single-cell traces of average p53-Venus intensity are shown for each condition. (C) Histogram of the ratio of peak p53-Venus intensity, α, to p53-Venus intensity at time of DMSO or Wm addition, β, for the experiment shown in (A) (>100 cells/condition). (D) Histogram of the ratio of p53-Venus intensity at 6 h after UV, α, to p53-Venus intensity at time of DMSO or Wm addition, β, for the experiment shown in (B) (>80 cells/condition). (E, F) Diagrams showing the differences in the negative regulation of Mdm2 by ATM (E) and ATR (F). (G) Western blot analysis and quantification of Mdm2 levels in cells irradiated with γ or UV. Error bars represent s.e.m. of triplicate experiments. (H) Western blot analysis and quantification of WT and mutant Mdm2 levels following γ treatment. Note that bands include both endogenous and exogenous Mdm2 or Mdm2S395A. Error bars represent s.e.m. of triplicate experiments. (I) Representative traces of average p53-Venus intensity for cells transfected with control or mutant Mdm2 plasmid in response to NCS. DMSO (control) or wortmannin (+Wm) was added 30 min after NCS treatment (dashed line). (J) The percentage of cells that show a p53-Venus pulse for the experiment in (I) (60–100 cells/condition). Error bars represent s.e.m.
We next sought to identify possible mechanisms that could generate excitability in response to DSBs but not in response to UV. One mechanism for generating excitability is to have a fast positive feedback (Hodgkin and Huxley, 1952; Suel et al, 2006). The currently known positive feedbacks in the p53 response to DSBs function on a transcriptional timescale (Harris and Levine, 2005), and therefore are not likely to act quickly enough to generate the observed behavior. Another possible mechanism for generating excitable behavior is fast removal of an inhibitor, (Suel et al, 2006; Howlett et al, 2008), such as Mdm2. Indeed, Stommel and Wahl (2004) showed that Mdm2 is rapidly degraded in response to NCS (Stommel and Wahl, 2004). In our previous work, we showed computationally that ATM-mediated degradation of Mdm2 contributes to the accumulation of p53 in response to DSBs (Batchelor et al, 2008). It is possible that Mdm2 is not rapidly degraded in the early response to UV, preventing p53 excitability. Indeed, we found that Mdm2 levels slowly decrease in response to UV compared with a rapid, relatively large degradation in response to DSBs (Figure 3G).
The differential regulation of Mdm2 in response to DSBs and UV can be explained by the differential phosphorylation of Mdm2 in response to these stresses. In response to DSBs, Mdm2 is phosphorylated by ATM on Ser-395 (Khosravi et al, 1999; Maya et al, 2001; Michael and Oren, 2003). This phosphorylation destabilizes Mdm2 by increasing its autoubiquitination and subsequent proteasomal degradation (Stommel and Wahl, 2004) (Figure 3E). In contrast, UV radiation leads to phosphorylation on residue Ser-407 (Shinozaki et al, 2003), which inhibits Mdm2 activity (Figure 3F). UV therefore reduced p53 access by Mdm2 through a post-translational modification, which is rapidly reversible; in contrast, DSBs cause degradation of Mdm2, which takes longer to reverse as mRNA and protein synthesis operate on a slower timescale than dephosphorylation (hours compared with minutes).
To determine whether the rapid degradation of Mdm2 in response to DSBs contributes to the excitability of p53, we mutated Ser-395 of Mdm2 to alanine and monitored Mdm2 and p53 dynamics. The alanine substitution prevented the degradation of Mdm2 in the initial response to DNA DSBs (Figure 3H). Transient transfection of the mutated Mdm2 plasmid revealed that this form of Mdm2, even in the presence of endogenous Mdm2, reduced the percentage of cells that showed an excitable p53 pulse (Figure 3I and J) in response to DSBs. These results suggest that the rapid degradation of Mdm2, mediated by ATM in response to DSBs, contributes to the excitable behavior of p53. In contrast, lack of rapid Mdm2 degradation in the initial UV response results in non-excitable activation of p53.
In summary, we showed how p53, a single transcription factor responding to multiple stresses in human cells, can transmit distinct signals by eliciting different dynamical patterns; DSBs lead to a series of excitable pulses, whereas UV leads to a graded, non-excitable single pulse. Comparison of the networks responding to these types of damage allowed us to identify and validate, both experimentally and computationally, the molecular mechanisms responsible for specific features of p53 dynamics; activation of the core p53–Mdm2-feedback loop by ATR generates a graded single pulse, the feedback from Wip1 to ATM is required for repeated fixed pulses, and the rapid degradation of Mdm2 contributes to the excitable behavior of p53.
It is intriguing to ask why excitability evolved in response to DSBs but not UV. One potential explanation is that DSBs are more harmful to cells than UV-induced DNA lesions, as they can give rise to gross chromosomal aberrations. It makes sense that the system would be poised to respond to DSBs by triggering a p53 pulse as a precaution (Loewer et al, 2010). In addition, the frequency of ATR-activating stress (e.g. ssDNA) during normal growth is much greater than the frequency of DSBs. It may be detrimental to excite a full p53 response every time ssDNA is detected, unnecessarily activating cell-cycle arrest or apoptosis.
Our study also raises the question of whether the dynamical response of p53 to other types of stress (such as hypoxia, ribosomal stress, and oncogenic activation) correspond to one of the identified classes of dynamics (excitable pulses or a graded response), or whether new dynamical behaviors can be identified. Moreover, as DSBs and UV are known to activate distinct targets of p53 (Zhao et al, 2000), it is tempting to speculate that the stimulus-dependent dynamics of p53 contribute to this differential regulation. Such a connection has already been shown in other organisms including bacteria (Suel et al, 2006) and yeast (Hao et al, 2008), and in other systems in mammalian cells. For example, stimulation of PC-12 cells with epidermal growth factor was found to generate a transient Erk kinase activity, resulting in cellular proliferation. In contrast, stimulation with neuronal growth factor led to sustained Erk activity, resulting in differentiation (Marshall, 1995; Santos et al, 2007). Further quantitative investigations of p53 dynamics and cellular outcomes in response to various sources of stress are required to determine whether distinct p53 dynamics are decoded differently by various downstream programs, which will provide new opportunities for controlling individual cell fate decisions.
Materials and methods
Plasmids and cells
We maintained human breast cancer epithelial MCF7 cells at 37°C in RPMI supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml fungizone (Gemini Bio-Products). MCF7-p53-Venus (Batchelor et al, 2008) was grown in medium supplemented with 400 μg/ml of G418. For UV-irradiated cells, we used RPMI lacking riboflavin and phenol red (transparent medium). For experiments involving wortmannin, we replaced the medium with fresh medium containing 100 μM wortmannin every hour because of the instability of this compound in solution.
To generate the plasmid pMdm2, we used MultiSite-Gateway recombination (Invitrogen). We amplified an ∼3.6-kb portion of the Mdm2 promoter, the Mdm2 cDNA, and a polyA tail individually by PCR with primers containing attB sites, and cloned each into pDONR plasmids by recombination (BP clonase, Invitrogen). We then generated a fusion construct by three-fragment recombination (LR Clonase Plus, Invitrogen) using the pDONR plasmids and a modified pDESTR4R3 vector containing a puromycin selection marker. To generate pMdm2S395A, we first used site-directed mutagenesis (Stratagene) to convert the TCT codon 395 to a GCT codon in the Mdm2 cDNA donor vector. We then used three-fragment recombination with the donor vectors for the Mdm2 promoter and the polyA tail. We transfected plasmid pMdm2S395A into MCF7 cells expressing p53-Venus using FuGene6 transfection reagent (Roche). Westerns were performed 48 h after transfection. In microscopy experiments, we identified cells that received the plasmid by co-transfecting an additional plasmid that constitutively expresses mCherry from the CMV promoter (pmCherry). pmCherry was constructed by inserting the sequence encoding the red fluorescent protein mCherry into the C1 expression vector (Clontech). Cells were imaged 3 days after transfection as described below in time-lapse microscopy. Only cells expressing mCherry were analyzed.
DNA damage was induced in cells using a cobalt-60 γ source, NCS (Sigma), or a UV-C 254 nm light source (Ushio). γ-Irradiation was given in a single burst lasting <4 min. NCS has been shown to act solely within 5 min following addition of the drug to the cell culture medium (Shiloh et al, 1983). The timescale of NCS treatment was therefore comparable to that of γ-irradiation. UV was delivered to cells using a UV lamp with a rate of 1.5 J/m2/s. All UV treatments, therefore, were performed in a single burst lasting <7 s.
Immunoblots
We harvested cells and obtained protein samples by lysis, quantifying the total protein concentration by Bradford assay. We separated equal amounts of total protein by electrophoresis on 4–12% Bis-Tris gradient gels (Invitrogen). We detected p53 using DO-1 monoclonal antibody (mAb; Santa Cruz Biotechnology), Mdm2 using SMP14 mAb (Santa Cruz Biotechnology), phospho-Chk1(S317) using a rabbit polyclonal antibody (Cell Signaling Technology), phospho-ATM(S1981) using a mAb (Rockland), β-tubulin using E7 mAb (Developmental Studies Hybridoma Bank), and actin using AC-74 mAb (Sigma). We quantified blot images using ImageJ (NIH).
Time-lapse microscopy
Two days before microscopy, we grew cells in poly-D-lysine-coated glass-bottom plates (MatTek Corporation) in transparent medium supplemented with 5% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml fungizone (Gemini Bio-Products). We imaged cells with a Nikon Eclipse Ti-inverted fluorescence microscope (Nikon) on which the stage was surrounded by an enclosure to maintain constant temperature, CO2 concentration, and humidity. Images of cells in all experiments were taken every 20 min. The Venus filter set was 500/20 nm excitation, 515 nm dichroic beam splitter, and 520 nm emission (Chroma). The mCherry filter set was 545/20–25 nm excitation, 570 nm dichroic beam splitter, and 600/50–25 nm emission (Chroma). We analyzed images using MetaMorph software (Molecular Devices) and custom-written Matlab software (Mathworks). We used a semi-automated method in which cells are segmented manually using brightfield images, followed by automated analysis of the fluorescence images using custom Matlab software (Batchelor et al, 2008; Loewer et al, 2010). Background-subtracted fluorescence levels were used when reporting traces of p53-Venus intensity. Fluorescence levels without background subtraction were used when reporting histograms of the ratios of fluorescence levels.
Supplementary Material
Supplementary Tables S1–2, Supplementary Figures 1–5
Eight compressed Matlab m-files for the four models described in the text
Acknowledgments
We thank U Alon, R Kishony, M Springer, J Toettcher, J Purvis, A Puszynska, all members of our laboratory for helpful discussions; J Waters and the Nikon Imaging Center at Harvard Medical School for advice on live-cell imaging. This research was supported by National Institutes of Health grant GM083303. EB was supported by the American Cancer Society, Pamela and Edward Taft Postdoctoral Fellowship. AL was supported by fellowships from the German Research Foundation and the Charles A King Trust.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barak Y, Juven T, Haffner R, Oren M (1993) mdm2 expression is induced by wild type p53 activity. EMBO J 12: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G (2008) Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell 30: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode AM, Dong Z (2004) Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4: 793–805 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2003) Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol 15: 164–171 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O'Connor PM, Appella E (1997) Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA 94: 6048–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, Yarnitzky T, Liron Y, Polak P, Lahav G, Alon U (2006) Oscillations and variability in the p53 system. Mol Syst Biol 2: 2006.003310.1038/msb4100068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG (2008) Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell 30: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Levine AJ (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899–2908 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF (1952) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117: 500–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Vousden KH (2007) Coping with stress: multiple ways to activate p53. Oncogene 26: 1306–1316 [DOI] [PubMed] [Google Scholar]
- Howlett E, Lin CC, Lavery W, Stern M (2008) A PI3-kinase-mediated negative feedback regulates neuronal excitability. PLoS Genet 4: e1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D (1999) Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA 96: 14973–14977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U (2004) Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet 36: 147–150 [DOI] [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G (2010) Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell 142: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Nannenga B, Donehower LA (2005) PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev 19: 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185 [DOI] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, Kastan MB, Katzir E, Oren M (2001) ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 15: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael D, Oren M (2003) The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol 13: 49–58 [DOI] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, Bastiaens PI (2007) Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol 9: 324–330 [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT (1998) Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res 58: 4375–4382 [PubMed] [Google Scholar]
- Shiloh Y, van der Schans GP, Lohman PH, Becker Y (1983) Induction and repair of DNA damage in normal and ataxia-telangiectasia skin fibroblasts treated with neocarzinostatin. Carcinogenesis 4: 917–921 [DOI] [PubMed] [Google Scholar]
- Shinozaki T, Nota A, Taya Y, Okamoto K (2003) Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene 22: 8870–8880 [DOI] [PubMed] [Google Scholar]
- Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV (2006) Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell 23: 757–764 [DOI] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM (2004) Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 23: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB (2006) An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440: 545–550 [DOI] [PubMed] [Google Scholar]
- Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW (2010) Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature 466: 267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ (1993) The p53-mdm-2 autoregulatory feedback loop. Genes Dev 7: 1126–1132 [DOI] [PubMed] [Google Scholar]
- Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP (2009) DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J Mol Biol 385: 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ (2000) Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev 14: 981–993 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1–2, Supplementary Figures 1–5
Eight compressed Matlab m-files for the four models described in the text