Abstract
Pfs230, surface protein of gametocyte/gamete of the human malaria parasite, Plasmodium falciparum, is a prime candidate of malaria transmission-blocking vaccine. P. vivax has an ortholog of Pfs230 (Pvs230), however, there has been no study in any aspects on Pvs230 to date. To investigate whether Pvs230 can be a vivax malaria transmission-blocking vaccine, we performed evolutionary and population genetic analysis of the Pvs230 gene (pvs230: PVX_003905). Our analysis of Pvs230 and its orthologs in seven Plasmodium species revealed two distinctive parts: an interspecies variable part (IVP) containing species-specific oligopeptide repeats at the N-terminus and a 7.5 kb interspecies conserved part (ICP) containing 14 cysteine-rich domains. Pvs230 was closely related to its orthologs, Pks230 and Pcys230, in monkey malaria parasites. Analysis of 113 pvs230 sequences obtained from worldwide, showed that nucleotide diversity is remarkably low in the non-repeat 8-kb region of pvs230 (θπ = 0.00118) with 77 polymorphic nucleotide sites, 40 of which resulting in amino acid replacements. A signature of purifying selection but not of balancing selection was seen on pvs230. Functional and/or structural constraints may limit the level of polymorphism in pvs230. The observed limited polymorphism in pvs230 should ground for utilization of Pvs230 as an effective transmission-blocking vaccine.
Keywords: malaria, Plasmodium vivax, Pvs230, gametocyte surface antigen, purifying selection, transmission-blocking vaccine
1. Introduction
Malaria is a major infectious disease caused by protozoa of the genus Plasmodium and transmitted by anopheline mosquito. There were an estimated 243 million clinical cases and 863,000 malaria-related deaths in 2008 [1]. Among four species of human malaria parasites, Plasmodium vivax was the most globally distributed. Although often malaria caused by P. vivax is regarded as a benign and self-limiting infection, there is increasing evidence that the overall burden, the economic impact, and the severity of disease associated with P. vivax have been underestimated. Particularly in Asia and Pacific, many endemic countries now wish to eliminate P. vivax malaria totally. The elimination strategies are however limited and confounded by emergence of multidrug resistant isolates and relapse from dormant hypnozoites stages at varying time intervals after the initial infection and there have not been available any optimal chemotherapeutic agents to combat against these situations [2,3]. Therefore, the development of vaccines against P. vivax is a necessary component towards malaria elimination [4]. Malaria vaccines are generally divided into three groups based on stages of the parasite life cycle that are targeted: pre-erythrocytic, asexual blood-stage, and transmission-blocking vaccines (TBVs). Pre-erythrocytic vaccines act against sporozoites and liver-stage parasites and are designed to prevent infection. Asexual blood-stage vaccines are aimed at reducing parasite multiplication and growth to protect against clinical symptoms, but not infection. TBVs are aimed at blocking malaria transmission by interrupting the parasite life cycle in the mosquito. TBVs as such do not directly protect vaccinated individuals from infection; however, they could contribute to elimination of the disease by lowering the parasite transmission efficiency.
TBVs elicit antibodies against surface antigens of sexual- and mosquito-stage parasites and, thus, arrest subsequent development of parasite in the mosquito midgut [5]. Target antigens for TBV development are sexual- and mosquito-stage specific surface molecules. Antigens specifically expressed by zygotes and ookinetes in the mosquito midgut (e.g., P25 and P28 in P. falciparum and P. vivax), referred to as post-fertilization target antigens, have been shown to be effective for inducing transmission-blocking immunity [6–8]. The ookinete surface antigens, Pfs25 in P. falciparum and Pvs25 in P. vivax, have been tested in Phase I clinical trials, and a positive correlation of the TBV efficacy with the antibody titer against each vaccine in the volunteers have been demonstrated [9–11]. However, P25 is not a boostable TBV candidate because it is not expressed by blood- stage parasites and hence it is not exposed to host immune response in natural infections [12]. In contrast to ookinete surface antigens, antigens that are involved in fertilization of male and female gametes, referred to as pre-fertilization target antigens, may be boostable TBV candidate antigens because they are also expressed in the gametocytes in the human blood and exposed to human immune response. Major proteins found on the surface of both male and female gametocytes/gametes such as Pfs48/45 and Pfs230 belong to a family defined by the presence of a unique arrangement of six cysteine-containing domains [13]. Pfs48/45, expressed on the surface of gametocytes/gametes, contains three cysteine rich domains (CRDs) [14,15]. Pfs230 is a 360-kDa surface protein, which is also expressed on the surface of gametocytes/gametes, contains 14 CRDs [16–19]. In P. falciparum the Pfs230 minus males have reduced ability to interact with erythrocytes and fewer oocysts are produced [20]. Monoclonal antibodies (mAbs) against the CRDs can potently be effective in blocking the infectivity of the parasites to mosquitoes [21–25]. Antibodies raised against a recombinant protein corresponding to a N-terminal 76-kDa part of the mature Pfs230 have been shown to reduce the ability of P. falciparum parasites to infect mosquitoes [26]. Importantly, Pfs230 elicits humoral immune responses in infected individuals that can mediate transmission-blocking immunity [27,28]. These accumulating evidence supports the priority of Pfs230 as candidate of falciparum TBV.
Most of the antigens expressed on the surface of asexual blood-stage parasites are highly polymorphic and under positive selection [29]. Polymorphic antigens are likely to induce immune responses in an allelic variant-dependent manner [30,31] and thus may limit the efficacy of vaccines based on such antigens. In contrast, antigens expressed in the sexual- and mosquito-stages show limited polymorphism; these include Pfs25 [9], Pfs28 [32], Pfs230 [17,33,34], Pvs25 and Pvs28 [35,36]. P. vivax has an ortholog of Pfs230 (Pvs230) [37]. Searching for immunodominant and conserved domains in this large molecule is of particular importance to design the vaccine antigen. There has been no study in any aspects on Pvs230 to date. We have therefore decided to investigate whether Pvs230 can be a promising TBV candidate. We performed evolutionary and population genetic analyses of the pvs230, specifically addressing (i) the evolutionary relatedness of pvs230 with its orthologs in other seven Plasmodium species and (ii) nucleotide polymorphism of pvs230 in P. vivax populations from diverse geographical areas. Comparative sequence analysis identified interspecies variable- and conserved-parts in pvs230, and provides evidence that Pvs230, particularly CRDs, has limited polymorphism. These results reinforce that Pvs230 can be a promising TBV candidate of P. vivax.
2. Materials and methods
2.1. Parasite isolates and DNA extraction
P. vivax isolates were collected from patients in seven endemic countries; Brazil, Turkey, Madagascar, China, Thailand, Papua New Guinea (PNG) and Solomon Islands. In Brazil, 22 isolates were collected from Acre state, northwestern Brazil, between 1999 and 2006 [38,39]; in Turkey, 20 isolates were collected from Siverek and Harran in Sanliurfa province, southeastern Turkey in June to November 2007–2008 [40]; in Madagascar, 16 isolates were collected from various regions (Sainte Marie in eastern coast, Taolagnaro in southern coast, Antananarivo and Ankazobe in central highland, Saharevo in eastern foothill, and Ampasimpotsy in western foothill) in 2000 to 2005; in China, 22 isolates were collected from Hubei, southeastern China in July to October 2000; in Thailand, 37 isolates were collected from Mae Kasa and Mae Sod in Tak Province, western Thailand in January 1999 to October 2000; in PNG, 29 isolates were collected from Kiniambu and Jawia villages, Wewak, East Sepik Province in northeast coast in August to September 2001 [41]; in Solomon Islands, 18 isolates were collected from area B of northern Guadalcanal island in February 2007–2008 [42]. In all cases, ethical clearance for sampling was approved from relevant ethical committees, and informed consent was obtained from patients or their guardians. In Turkey, PNG and Solomon Islands, finger-prick blood was collected on Whatman® 31ETCHR filter paper. Parasite genomic DNA was extracted from filter blots using the EZ1 DNA Investigator kit on the EZ1 BioRobot™ (Qiagen, Germany). In other countries, parasite DNA was extracted from venous blood, using QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). Three Aotus monkey-adapted P. vivax isolates were additionally obtained from ATCC (American Type Culture Collection): Chesson strain (PNG) (ATCC #30060), Nicaragua strain (ATCC # 30073) and Panama strain (ATCC # 30138). Genomic DNA from P. cynomolgi (B strain) (ATCC # 30129), a P. vivax-related monkey malaria parasite [43] was also used. Genomic DNA from P. yoelii (17XNL strain) was extracted from infected mouse blood, using QIAamp DNA Blood Mini Kit (QIAGEN).
2.2. DNA sequencing
The Pvs230 gene (pvs230) was amplified by PCR. PCR primers were designed from the P. vivax Sal-1 pvs230 sequence (PlasmoDB, Gene ID PVX_003905; http://plasmodb.org/plasmo/) (Suppl. Table 1). Amplification was carried out in a 20 μl reaction mixture containing 0.4 μM each of forward and reverse primers, 400 μM each of dNTP, 1 unit of LA-Taq (Takara, Otsu, Japan), 2 μl of 10×PCR buffer, 2.5 mM of MgCl2, and 1 μl of genomic DNA. PCR conditions were as follows: initial denaturation at 93 °C for 1 min, and amplification for 40 cycles at 93 °C for 20 s and 62 °C for 4 min, followed by a final extension at 72 °C for 10 min in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). The PCR products were subjected to DNA sequencing using the Terminator Cycle Sequencing Kit v1.1 (Applied Biosystems) in an Applied Biosystems 3130xl Genetic Analyzer. Sequencing primers were designed to cover target regions from both directions (Suppl. Table 1). Sequences from fifty-four samples showed superimposed electropherogram peaks, indicative of mixed genotype infections, and thus they were excluded from further analysis (Suppl. Table 2). Sequences were verified by at least two independent amplifications from the same DNA. We confirmed the Sal-1 pvs230 sequence and used as a reference sequence. For those samples from PNG and Solomon Islands, in which PCR products were not sufficient for direct sequencing, the nested PCR was performed using internal primers (Suppl. Table 1). P230 sequence from a P. vivax-related monkey malaria parasite, P. cynomolgi (B strain) [43], pcys230, was also obtained by direct sequencing as described above, using specific primers (Suppl. Table 1). P230 gene sequence from a rodent malaria parasite, P. yoelii (17XNL strain), pys230, was also obtained as described above.
Sequences obtained in this study have been deposited to DDBJ/EMBL/GenBank under accession numbers AB574508-AB574621.
2.3. Sequence alignments and analyses
We obtained a total of 112 pvs230 sequences from P. vivax isolates in seven countries (Suppl. Table 2). P230 sequences were also determined from P. cynomolgi (pcys230) and P. yoelii (pys230). P230 sequences from other malaria parasite species retrieved from PlasmoDB were P. vivax (pvs230; Gene ID PVX_003905), P. falciparum (pfs230; PFB0405w), P. knowlesi (pks230; PKH_041100) and P. chabaudi (pchs230; PCAS_030830). Additionally, P. berghei P230 (pbs230) sequence was obtained from Sanger Institute database. P. gallinaceum P230 partial sequence (Pgs230: aa 1-1527) was obtained by NCBI Blast search in P. gallinaceum genome, Pg_2265551.c000320551.Contig1. Sequences were aligned using CLUSTAL W [44] implemented in MEGA version 4 software [45] with manual corrections. A phylogenetic tree was constructed using the Neighbor-Joining method [46] with the Jukes and Cantor correction implemented in MEGA. Bootstrap values were obtained by 1,000 heuristic replications.
Sequence polymorphism was estimated by the following parameters: (1) S, the number of polymorphic nucleotide sites, (2) the number of haplotypes (h), (3) haplotype diversity (Hd), and (4) the observed average number of pairwise nucleotide difference per site (θπ). These estimates were calculated using the DnaSP v4.10.9 software [47]. To estimate the proportion of genetic variance due to population subdivision, the Wright’s fixation index [48] of inter-population variance in allele frequencies, termed Fst, was calculated using the Arlequin software [49]. Overall Fst for all six parasite populations, in which more than seven sequences were available, and pairwise Fst were estimated.
2.4. Tests for departure from neutrality
In this study, we used the Nei and Gojobori method [50] with the Jukes and Cantor correction as implemented in the MEGA [45] to examine departure from neutrality. This method estimates the difference between the numbers of synonymous substitutions per synonymous site (dS) and of nonsynonymous substitutions per nonsynonymous site (dN). Standard error was determined by 1,000 bootstrap replications, and dN and dS were compared with a Z-test using MEGA. If dN is significantly higher than dS, positive selection (diversifying selection) appears to be acting, while if dS is higher than dN, purifying selection is predicted.
We also used Tajima’s D test, and McDonald-Kreitman (MK) test to examine departure from neutrality. Tajima’s D test measures allele frequency spectrum by comparing θπ and θS, the latter of which is the standardized number of polymorphic sites per site [51]. Under neutrality a value of Tajima’s D is expected to be 0. Significantly positive values of Tajima’s D suggest recent population bottleneck or balancing selection, whereas negative values suggest population growth or directional selection. Fu and Li’s D* and F* tests were also used to test for excess or lack of singleton nucleotides by comparing estimates of θS based on the number of singletons vs. that derived from S (the D* index) or θπ (the F* index) [52]. All of these estimates were calculated using DnaSP. The MK test [53] was used to assess a signature for selection, in which the ratio of nonsynonymous to synonymous substitutions was compared between polymorphic difference (within species) and fixed difference (between closely related species) using DnaSP. Under neutrality, these ratios will be similar, whereas an excess of intraspecific nonsynonymous polymorphisms is suggestive of balancing selection. It should be mentioned that the MK test is greatly affected by the presence of rare alleles or singleton alleles, and positive values could be produced even for a gene under purifying selection [54,55]. Pcys230 sequence was used as an outgroup in this test. Fisher’s exact test was used to test statistical significance.
3. Results
3.1. Sequence divergence of P230 among Plasmodium species
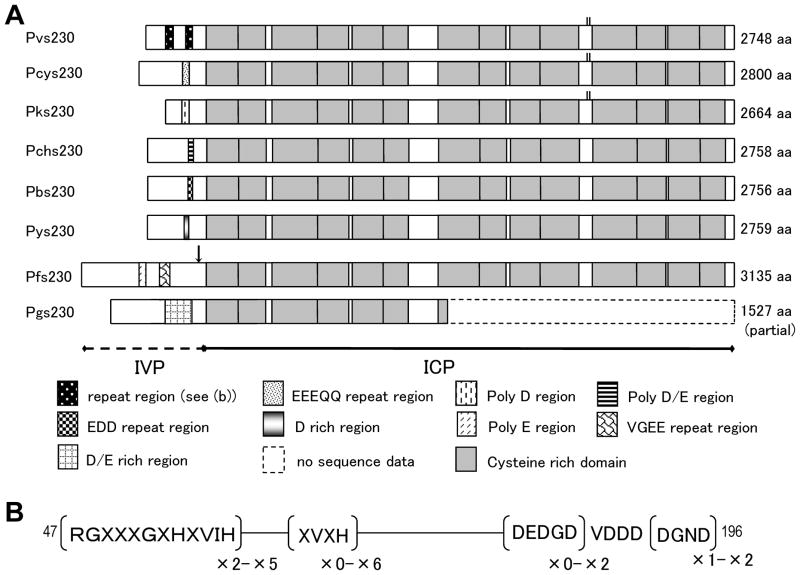
An amino acid sequence alignment of P230 from P. vivax (Pvs230), P. cynomolgi (Pcys230), P. knowlesi (Pks230), P. berghei (Pbs230), P. chabaudi (Pchs230), P. yoelii (Pys230), P. falciparum (Pfs230) and partial sequence of P. gallinaceum (Pgs230: aa 1-1527) revealed two distinctive sequence parts: one is an interspecies variable part (IVP) at the N-terminus, in which sequences are highly variable with tandem arrays of oligopeptide repeats, and the other is an interspecies conserved part (ICP), which contains14 cysteine-rich domains (CRDs) in a 7.5 kb region [16,17] (Fig. 1A and Suppl. Fig. 1). The boundary of IVP and ICP does not coincide with the putative cleavage sites proposed for Pfs230 [56], which reside in a C-terminal part of IVP. The cleavage sites are apparently not conserved among other species (Suppl. Fig. 1). In IVP, oligopeptide repeat sequences are species specific: thus, in Pfs230, poly-E and E(E/G)(V/E)E repeats occur as reported earlier [18], poly-(D/E) in Pks230, and degenerative EEEQQ repeat and poly-E in Pcys230 (Fig. 1A and Suppl. Fig. 1). In rodent parasite species, poly-D/E and repeats of EDD are arrayed in Pchs230 and Pbs230, respectively. Pys230 and Pgs230 also contain a D/E-rich sequence region. In Pvs230, there are two repeat sequence regions identified: degenerative repeats of RGXXXGXHXVIH, XVXH, DEDGD and DGND (Fig. 1B). Thus, no primary sequence motifs are shared by Plasmodium species examined in the repeat region of P230. It is noteworthy that repeats are rich in acidic residues, E, D and Q. Non-repeat sequences in IVP are also highly divergent, making an alignment not reliable (Suppl. Fig. 1). Despite this, three rodent species show relatively high sequence similarities, and sequences from P. vivax, P. cynomolgi and P. knowlesi show moderate similarities.
Fig. 1.
Primary structure of Pvs230 and its orthologs. In (a), deduced amino acid sequences obtained from P. vivax (Pvs230), P. cynomolgi (Pcys230), P. knowlesi (Pks230), P. chabaudi (Pchs230), P. berghei (Pbs230), P. yoelii (Pys230), P. falciparum (Pfs230), and partial sequence of P. gallinaceum (Pgs230: aa 1-1527) are aligned. In interspecies variable part (IVP), tandem repeat regions are represented as variously marked boxes, and 14 cysteine-rich domains in interspecies conserved part (ICP) are represented as half-tone boxes. Predicted cleavage sites of Pfs230 are marked by an arrow. Two cysteine residues between CRD10 and CRD11 in Pvs230, Pcys230 and Pks230 are shown in small bars. Predicted amino acid sizes are shown in right of respective amino acid sequences. In (b), repeat motifs and their repeat number in Pvs230 are shown, in which X denotes any amino acid residues. Major repeat units in RGXXGXHVIH are RGSYEGIHQVIH, RGRCEGIHQVIH, and RGRCDGGHHVIH, those in XVXH are RVVH, CVVH, RVAH, and RVIH (Suppl. Fig. 2). Amino acid positions are numbered after the Pvs230 sequence of Sal-1 strain (GenBank accession #XM_001612970).
ICP is primarily composed of 14 CRDs, in which sequences are somewhat conserved among the seven Plasmodium species (Fig. 1A and Suppl. Fig. 1). Each CRD contains 2 to 6 cysteine residues, all of which are perfectly conserved. In Pvs230, Pcys230 and Pks230, there are two additional cysteine residues identified between CRD10 and CRD11 (Suppl. Fig. 1), which are absent in Pfs230 and P230 of rodent parasites. Amino acid sequence divergence in ICP is 9% to 59% among the seven species, and 21% to 25% between P. vivax, P. cynomolgi and P. knowlesi (Suppl. Table 3A). A phylogenetic tree constructed using aligned sequences (7230 bp) revealed three major monophyletic groups: a group of Pvs230, Pcys230 and Pks230, a group of three rodent P230s, and Pfs230 (Fig. 2A). When the partial Pgs230 was included in this analysis, Pgs230 was in the same group of Pfs230 (Fig. 2B). The sequence that is most closely related to Pvs230 is Pcys230, and that most distantly related to Pvs230 is Pfs230.
Fig. 2.
A phylogenetic tree of Plasmodium P230. This tree was constructed by the Neighbor-Joining method. Shown along nodes are bootstrap values with >50%. Nucleotide sequences in ICP among the seven full-length olthologs (A), or among eight olthologs calculated using orthologous sequences to the partial Pgs230 ICP (B).
3.2. Sequence polymorphism in pvs230
We sequenced a total of 112 full-length pvs230 sequences of isolates collected from Brazil (n=20), Turkey (n=20), Madagascar (n=7), China (n=20), Thailand (n=20), Papua New Guinea (n=21), Solomon Islands (n=2), Nicaragua (n=1) and Panama (n=1) (Suppl. Table 2). The rate of mixed infections, as inferred from superimposed electropherogram peaks, varied greatly, depending on countries, from 0% (Turkey) to 89% (Solomon Islands) (Suppl. Table 2).
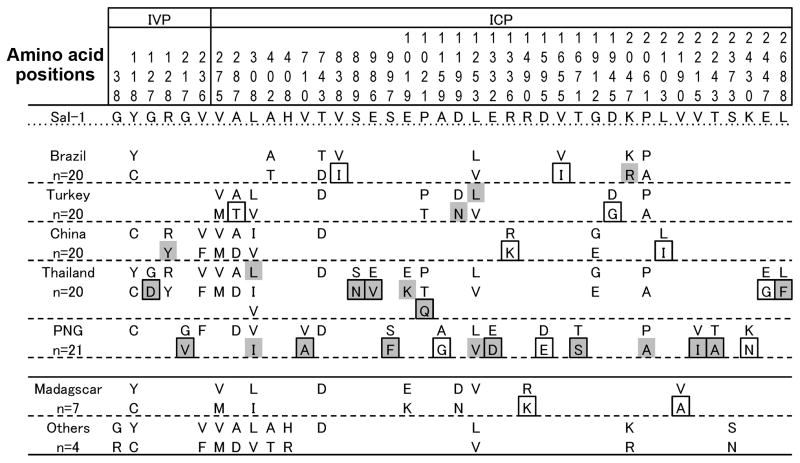
In IVP, there are two repeat sequence regions: repeats of 12-mer (RGXXXGXHXVIH) and 4-mer (XVXH) and degenerative repeats of DEDGD and DGND, which are separated by 69 non-repeat amino acids (Fig. 1B). The number of repeats varied among isolates: 2 to 5 times of 12-mer RGXXXGXHXVIH repeat, and 0 to 6 times of 4-mer XVXH repeat. In the second repeat, the main repeat motif is DEDGD-VDDD-DGND. Variation in the number of this repeat was limited and geographically restricted: for example, one DEDGD in all countries, except PNG and Solomon Islands, where the repeat number was 0 to 2 (Suppl. Fig. 2).
In non-repeat sequence regions (8001 bp), there are 77 polymorphic nucleotide sites (Table 1), 40 of which resulting in amino acid replacements (6 sites in IVP and 34 sites in ICP) (Fig. 3). Of the 40 sites, 35 were found in 5 countries (Brazil, Turkey, China, Thailand and PNG), where ≥20 sequences were available in each country for geographical comparisons. Of the 35 sites, 22 sites (60%) were country-specific. A half of these substitutions (12/22) are those with minor allele frequency of ≤5% in the five countries. The number of haplotypes was 72 for worldwide samples with Hd of 0.983 (Table 1). Hd varied geographically, low in Brazil and Turkey and high in Madagascar, China, Thailand, and PNG. In Brazil and Turkey, identical haplotypes were frequently obtained from multiple isolates (Suppl. Fig. 3), contributing to the reduction in Hd. No haplotype was shared by two or more countries, suggesting a strong geographical clustering of pvs230 alleles.
Table 1.
Polymorphism in Pvs230 and tests for neutrality
| Area | Region | No. of polymorphic sites | No. of singletons | No. of haplotypes | Haplotype diversity Hd ± SD | Nucleotide diversity θπ ± SD | dN ± SE | dS ± SE | P value |
|---|---|---|---|---|---|---|---|---|---|
| Brazil (N=20) | Entire gene | 15 | 4 | 6 | 0.747 ± 0.076 | 0.00060 ± 0.00007 | 0.00059 ± 0.00024 | 0.00065 ± 0.00030 | 0.8752 |
| IVP | 1 | 0 | 2 | 0.395 ± 0.101 | 0.00070 ± 0.00018 | 0.00092 ± 0.00092 | 0.00000 ± 0.00000 | 0.3105 | |
| ICP | 14 | 4 | 6 | 0.747 ± 0.076 | 0.00059 ± 0.00006 | 0.00056 ± 0.00027 | 0.00070 ± 0.00030 | 0.7405 | |
| Turkey (N=20) | Entire gene | 13 | 2 | 6 | 0.763 ± 0.066 | 0.00054 ± 0.00009 | 0.00030 ± 0.00011 | 0.00140 ± 0.00054 | 0.0496 |
| IVP | 0 | 0 | 1 | 0.000 ± 0.000 | 0.00000 ± 0.00000 | 0.00000 ± 0.00000 | 0.00000 ± 0.00000 | 1.0000 | |
| ICP | 13 | 2 | 6 | 0.763 ± 0.066 | 0.00059 ± 0.00010 | 0.00032 ± 0.00012 | 0.00152 ± 0.00062 | 0.0617 | |
| Madagascar (N=7) | Entire gene | 14 | 9 | 7 | 1.000 ± 0.076 | 0.00063 ± 0.00012 | 0.00040 ± 0.00016 | 0.00144 ± 0.00063 | 0.1171 |
| IVP | 1 | 0 | 2 | 0.571 ± 0.119 | 0.00101 ± 0.00021 | 0.00133 ± 0.00133 | 0.00000 ± 0.00000 | 0.3105 | |
| ICP | 13 | 9 | 7 | 1.000 ± 0.076 | 0.00060 ± 0.00012 | 0.00033 ± 0.00014 | 0.00156 ± 0.00067 | 0.0719 | |
| China(N=20) | Entire gene | 13 | 4 | 14 | 0.958 ± 0.028 | 0.00051 ± 0.00005 | 0.00050 ± 0.00018 | 0.00056 ± 0.00035 | 0.8789 |
| IVP | 4 | 3 | 4 | 0.574 ± 0.090 | 0.00143 ± 0.00038 | 0.00164 ± 0.00121 | 0.00075 ± 0.00073 | 0.5396 | |
| ICP | 9 | 1 | 14 | 0.958 ± 0.028 | 0.00044 ± 0.00003 | 0.00041 ± 0.00018 | 0.00054 ± 0.00037 | 0.7456 | |
| Thailand (N=20) | Entire gene | 32 | 14 | 18 | 0.989 ± 0.019 | 0.00084 ± 0.00010 | 0.00065 ± 0.00016 | 0.00153 ± 0.00047 | 0.0723 |
| IVP | 5 | 1 | 6 | 0.726 ± 0.090 | 0.00230 ± 0.00045 | 0.00303 ± 0.00172 | 0.00000 ± 0.00000 | 0.0728 | |
| ICP | 27 | 13 | 18 | 0.989 ± 0.019 | 0.00073 ± 0.00009 | 0.00047 ± 0.00015 | 0.00165 ± 0.00047 | 0.0241 | |
| PNG (N=21) | Entire gene | 28 | 17 | 16 | 0.976 ± 0.020 | 0.00062 ± 0.00010 | 0.00030 ± 0.00011 | 0.00175 ± 0.00056 | 0.0130 |
| IVP | 1 | 1 | 2 | 0.095 ± 0.084 | 0.00017 ± 0.00015 | 0.00022 ± 0.00021 | 0.00000 ± 0.00000 | 0.2976 | |
| ICP | 27 | 16 | 16 | 0.976 ± 0.020 | 0.00065 ± 0.00011 | 0.00030 ± 0.00012 | 0.00189 ± 0.00060 | 0.0123 | |
| Worldwide (N=113) | Entire gene | 77 | 32 | 72 | 0.983 ± 0.005 | 0.00118 ± 0.00003 | 0.00089 ± 0.00022 | 0.00221 ± 0.00064 | 0.0494 |
| IVP | 8 | 4 | 9 | 0.731 ± 0.017 | 0.00203 ± 0.00013 | 0.00263 ± 0.00154 | 0.00013 ± 0.00013 | 0.1128 | |
| ICP | 69 | 28 | 71 | 0.983 ± 0.005 | 0.00111 ± 0.00003 | 0.00076 ± 0.00020 | 0.00238 ± 0.00067 | 0.0218 |
Sequence lengths are 564 bp and 7437 by for interspecies variable part and interspecies conserved part, respectively. IVP, interspecies variable part; ICP, interspecies conserved part.
Fig. 3.
Amino acid substitutions in Pvs230 of P. vivax populations used in this study. IVP and ICP are interspecies variable part and interspecies conserved part, respectively. Amino acid positions are numbered after the Sal-1 sequences. Country-specific amino acid changes are boxed. The category “Others” include Nicaragua, and Panama strains and two isolates from Solomon Islands. Amino acid substitutions with minor allele frequency of ≤0.5% are highlighted in grey.
Nucleotide diversity (θπ) was 0.00118 for worldwide samples and 0.00051 to 0.00084 in six countries (Table 1). θπ was about 1.8-fold higher in non-repeat region of IVP (θπ 0.00203) than in ICP (θπ 0.00111) (Table 1). θπ in each country was lower than that in worldwide samples, suggestive of country-specific substitutions. θπ was relatively high in Thailand and low in Turkey and China. Sliding window plot of θπ revealed a peak (θπ 0.01577) at nucleotide positions 844 – 943 (positions after the Sal-1 sequence) occurring in CRD1 (Suppl. Fig. 4), in which 5 of a total of 40 amino acid changes in Pvs230 occur. In contrast, in CRD4, where 8 of a total of 27 amino acid changes are clustered in Pfs230 [17], only two amino acid changes occurred in Pvs230. The θπ (0.00118) of pvs230 in worldwide samples is at least one order lower than that of known blood stage antigen genes such as pvmsp1, pvmsp3a and pvdbp with a few exceptions (Suppl. Table 4).
3.3. Inter-population differentiation of pvs230 among geographic areas
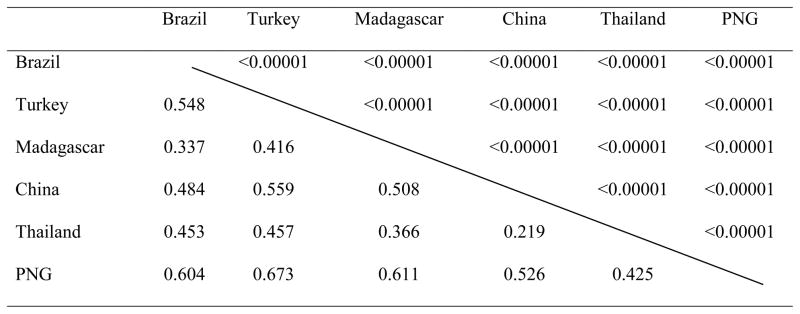
Overall Fst estimate for worldwide populations was 0.51 (95% CI = 0.42 – 0.56), indicating about half of variation was apportioned within parasite populations (Table 2). This high Fst value suggests a high degree of differentiation of pvs230 among geographic parasite populations examined. Pairwise comparisons of Fst revealed a significant difference in all pairs, with relatively low values between China and Thailand and high values between PNG and any of other five countries.
Table 2.
Inter-population differentiation (Fst) of pvs230 between six geographic areas.
|
Overall Fst is 0.505 (95% CI, 0.423 - 0.564) with variance of 5.104. Fst values are shown in the bottom left and P values are shown in the upper right.
3.4. Departure from neutrality
The entire coding sequence (8001 bp), except the N-terminal repeat regions, did show a significant excess of dS over dN in worldwide samples (Table 1). When the sequence was separated into IVP and ICP, a significantly higher dS than dN was noted for ICP (7437 bp) in worldwide samples, suggesting purifying selection. An excess of dS over dN was noted in some (but not in all) countries. In IVP (564 bp), dN was higher than dS in worldwide samples, but this difference was not significant. Tajima’s D test and Fu and Li’s D* and F* tests did not detect significant values in all countries and worldwide samples (data not shown).
The MK test for the ICP region detected an excess of fixed synonymous substitutions over nonsynonymous substitutions in worldwide samples (P = 0.003) (Suppl. Table 5), again suggesting purifying selection. This excess was not significant in six individual parasite populations, probably due to limited numbers of substitutions. A slightly higher number of intraspecific nonsynonymous substitutions over synonymous substitutions can be seen. However, caution is required for evaluating this difference because the sequences contain a number of singleton mutations (32 singletons in 77 polymorphic sites) (Table 1), suggesting that the balancing selection (positive selection) is unlikely in pvs230. The abundance of singleton mutations, most of which are geographic area-specific and thus contributing to high geographic structure (Fst > 0.5 among some areas), limits the validity of the MK test for testing neutrality of pvs230.
4. Discussion
The present sequence analysis of P230, a Plasmodium-specific gametocyte/gamete surface antigen, identified 2 distinctive parts in the protein: IVP at the N-terminus and ICP, a 7.5-kb component containing 14 CRDs. IVP consists of tandem arrays of species-specific repeats with varying lengths and non-repeat unique sequences, both of which are highly divergent among species, with relatively less divergence in three rodent parasite species and P. vivax and related monkey parasites. Despite high divergence, species-specific repeats of P230 are rich in acidic residues, such as D and E. The role of these acidic residue-rich repeats remains unknown. Since the N-terminal repeat region of Pfs230, which induce human antibody and T cell response, is shed into the plasma after processing to form the mature protein during gametogenesis [56], a mechanism for immune evasion by the repeat region has been postulated [57].
In contrast to a highly divergent IVP, ICP is relatively conserved with complete conservation of all cysteine residues in 14 CRDs, which have been predicted to form disulfide bonds in Pfs230 [16,17]. These strongly suggest conformational conservation of the CRDs among all Plasmodium species. It remains to be elucidated whether two additional cysteine residues found between CRD10 and CRD11 in P230 from P. vivax and related monkey parasites form an additional disulfide bond. Although no strong evidence, suggesting disulfide bonding of the two cysteines, was obtained with available disulfide bond-prediction algorithms (data not shown), a loop structure formed by these two cysteines cannot be excluded. P230 is a member of 6-cys protein family [19], in which Pf12, Pf38 and Pf41 play a role in recognition and invasion of erythrocyte entry by the merozoite [58], and Pbs36 and Pbs36p do so in sporozoite’s invasion into liver cells [59]. Pfs230 has been suggested to be associated with male gamete’s binding to human erythrocyte during the formation of exflagellation center [20]. Thus, P230 is likely to be involved in interacting with host cells and probably gamete recognition. Such important interactions may impose constraints of sequence variations in Pvs230.
The phylogenetic tree of P230s among seven Plasmodium species examined is in good consistency with that of the mitochondrial genomes and 18S rRNA genes [60–62]. In a group of P. vivax and P. vivax-related monkey malaria species, P. cynomolgi P230 is the closest relative to P. vivax P230. Genes for gametocytes/gamete surface proteins of rodent malaria parasites are rapidly evolving [19]. Consistent with this, branch lengths of the P230 phylogenetic tree are relatively long, suggesting fast evolution in the P230 gene. A phylogenetic tree of msp1, a major immune target merozoite surface protein 1 gene, has recently been shown to significantly differ from that of the mitochondrial genome, with a striking displacement of P. vivax from a position close to P. cynomolgi in the mitochondrial genome tree to an outlier of Asian monkey parasites, suggesting positive selection in pvmsp1 [63]. We therefore consider that pvs230 has not been subjected to such strong positive selection. Positive selection with the ratio of dN/dS over 1 on CRD4 of P230 has recently been detected among three rodent species [19]. In P. vivax, P. cynomolgi and P. knowlesi, dN/dS over 1 was not detected (data not shown), suggesting that positive selection in the P230 gene is lineage specific.
The observed polymorphism level of pvs230 (θπ = 0.00118) is much lower than that of most blood stage antigen genes (Suppl. Table 4). Since blood stage antigens are targets of host antibody responses, it is likely that high genetic diversity in these antigens is a mechanism for parasite’s immune evasion. Balancing selection has been inferred to maintain high levels of polymorphism in pvmsp1 [43,63], pvmsp3a [64], pvmsp3b [65], pvmsp5 [66,67], and pvdbp [68]. In pvs230, however, no evidence for balancing selection was obtained in this study but data suggest purifying selection in this gene. Functional and/or structural constrains of Pvs230 would probably limit the accumulation of point mutations, resulting in low level of polymorphism. A higher nucleotide diversity in IVP than in ICP also supports this notion because in Pfs230, most parts of IVP is presumed to be cleaved off from the mature protein containing CRDs. Selective sweep, as often seen in drug resistant genes, potentially contributes to reduction in polymorphism. In this case, only variant haplotypes become predominant in a population. Selective sweep is unlikely for pvs230 because haplotype diversity was high in most parasite populations. Recent population growth may also reduce genetic diversity; however, significantly negative values were not obtained by Tajima’s D test and Fu and Li’s D* and F* tests, making population expansion unlikely for pvs230. A comparison of polymorphism between pvs230 and pfs230, although the number of pfs230 sequences analyzed was small [17], reveals a difference in the distribution of polymorphism across the genes: major amino acid substitutions is clustered within CRD4 in Pfs230, whereas it is within CRD1 in Pvs230. Different intragenic distribution of polymorphisms between P. falciparum and P. vivax may suggest different constraints in these distantly related Plasmodium species.
This study showed a high genetic differentiation (Fst = 0.51) of pvs230 among global parasite populations. This indicates very limited gene flow between populations. Some proteins on the surface of gametes are involved in gamete recognition and fertilization, and genes for these proteins tend to be highly divergent among populations [69,70]. This has been suggested to be true for Pfs48/45, in which Fst is as high as 0.69 among parasite populations from Africa, Asia and South America [71], given that Fst of two housekeeping genes of P. falciparum populations is 0.20 for worldwide parasite populations [41]. Fst of pfs230 is not known. Previous estimates of Fst among geographic P. vivax populations, which have been derived from microsatellite data (0.13–0.26 between Southeast Asia and Colombia [72]) and predominantly silent single-nucleotide polymorphisms (0.228 between Brazil and Asia [73]), are remarkably lower than the Fst of pvs230 obtained in this study. Comparisons of Fst between P. vivax housekeeping genes and pvs230 as well as other gamete surface protein genes, such as pvs47 and pvs48/45 would be required to assess divergent (directional) selection on P. vivax gamete surface protein genes.
In conclusion, the limited polymorphism of pvs230 observed in this study would provide a strong ground for developing effective TBV based on Pvs230, and help to identify polypeptide regions suitable for designing vaccines. Since it would not be practical to include all 14 CRDs in a TBV, several CRDs that can effectively induce transmission blocking immunity (TBI) should be targeted. The present study identified short sequence regions with relatively high polymorphism, particularly in the N-terminal part of CRD1. In our parallel studies, we have found that a recombinant DNA vaccine, encompassing CRD1 of Pvs230, induces TBI (Tachibana et al., unpublished). Whether polymorphism in small polymorphic regions in CRD1 is involved in strain-specific TBI awaits further studies. Additionally, this study revealed a substantial endemic area-specific SNPs in pvs230. If a vaccine includes a region having such polymorphism, caution is required, and the reactivity of the serum antibodies to variants and hence efficiency of TBI in individuals living in endemic areas should be monitored.
Supplementary Material
Acknowledgments
We thank all those who participated in the epidemiological studies for their kind cooperation, particularly T. Tsukahara, F. Hombhanje, Fehmi Yuksel, Nebiye Doni, and B. Bakote’e. We also thank Thangavelu U. Arumugam for the critical reading of the manuscript. This research was supported by the Ministry of Education, Culture, Sports, Science and Technology (18073013, 18GS03140013, 20390120, 19406009, 21022034, 22406012), and by the Ministry of Health, Labour, and Welfare, Japan (H20-Shinkou-ippan-013, H21-Chikyukibo-ippan-005). Field work in Brazil was funded by the National Institutes of Health of USA (RO1 AI 075416-01), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 470570/2006-7), and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 05/51988-0 and 07/51199-0). This work was also supported by grant from the National Natural Science Foundation of China (30972774).
Footnotes
Disclosure: The view of the author, J. Sattabongkot, does not purport to reflect the position of the US Department of the Army or Department of Defense.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO. World Malaria Report 2009. Geneva, Switzerland: WHO Press; 2009. [Google Scholar]
- 2.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009;9:555–66. doi: 10.1016/S1473-3099(09)70177-X. [DOI] [PubMed] [Google Scholar]
- 4.Arevalo-Herrera M, Chitnis C, Herrera S. Current status of Plasmodium vivax vaccine. Hum Vaccine. 2010;6:124–32. doi: 10.4161/hv.6.1.9931. [DOI] [PubMed] [Google Scholar]
- 5.Carter R, Mendis KN, Miller LH, Molineaux L, Saul A. Malaria transmission-blocking vaccines--how can their development be supported? Nat Med. 2000;6:241–4. doi: 10.1038/73062. [DOI] [PubMed] [Google Scholar]
- 6.Kaslow DC, Quakyi IA, Syin C, Raum MG, Keister DB, Coligan JE, et al. A vaccine candidate from the sexual stage of human malaria that contains EGF-like domains. Nature. 1988;333:74–6. doi: 10.1038/333074a0. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y, Przysiecki C, Flanagan E, Bello-Irizarry SN, Ionescu R, Muratova O, et al. Sustained high-titer antibody responses induced by conjugating a malarial vaccine candidate to outer-membrane protein complex. Proc Natl Acad Sci USA. 2006;103:18243–8. doi: 10.1073/pnas.0608545103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hisaeda H, Stowers AW, Tsuboi T, Collins WE, Sattabongkot J, Suwanabun N, et al. Antibodies to malaria vaccine candidates Pvs25 and Pvs28 completely block the ability of Plasmodium vivax to infect mosquitoes. Infect Immun. 2000;68:6618–23. doi: 10.1128/iai.68.12.6618-6623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaslow DC. Transmission-blocking vaccines. Chem Immunol. 2002;80:287–307. doi: 10.1159/000058850. [DOI] [PubMed] [Google Scholar]
- 10.Malkin EM, Durbin AP, Diemert DJ, Sattabongkot J, Wu Y, Miura K, et al. Phase 1 vaccine trial of Pvs25H: a transmission blocking vaccine for Plasmodium vivax malaria. Vaccine. 2005;23:3131–8. doi: 10.1016/j.vaccine.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Ellis RD, Shaffer D, Fontes E, Malkin EM, Mahanty S, et al. Phase 1 trial of malaria transmission blocking vaccine candidates Pfs25 and Pvs25 formulated with montanide ISA 51. PLoS One. 2008;3:e2636. doi: 10.1371/journal.pone.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuboi T, Tachibana M, Kaneko O, Torii M. Transmission-blocking vaccine of vivax malaria. Parasitol Int. 2003;52:1–11. doi: 10.1016/s1383-5769(02)00037-5. [DOI] [PubMed] [Google Scholar]
- 13.Templeton TJ, Kaslow DC. Identification of additional members define a Plasmodium falciparum gene superfamily which includes Pfs48/45 and Pfs230. Mol Biochem Parasitol. 1999;101:223–7. doi: 10.1016/s0166-6851(99)00066-3. [DOI] [PubMed] [Google Scholar]
- 14.Kocken CH, Jansen J, Kaan AM, Beckers PJ, Ponnudurai T, Kaslow DC, et al. Cloning and expression of the gene coding for the transmission blocking target antigen Pfs48/45 of Plasmodium falciparum. Mol Biochem Parasitol. 1993;61:59–68. doi: 10.1016/0166-6851(93)90158-t. [DOI] [PubMed] [Google Scholar]
- 15.van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JA, Dodemont HJ, et al. A central role for P48/45 in malaria parasite male gamete fertility. Cell. 2001;104:153–64. doi: 10.1016/s0092-8674(01)00199-4. [DOI] [PubMed] [Google Scholar]
- 16.Carter R, Coulson A, Bhatti S, Taylor BJ, Elliott JF. Predicted disulfide-bonded structures for three uniquely related proteins of Plasmodium falciparum, Pfs230, Pfs48/45 and Pf12. Mol Biochem Parasitol. 1995;71:203–10. doi: 10.1016/0166-6851(94)00054-q. [DOI] [PubMed] [Google Scholar]
- 17.Gerloff DL, Creasey A, Maslau S, Carter R. Structural models for the protein family characterized by gamete surface protein Pfs230 of Plasmodium falciparum. Proc Natl Acad Sci USA. 2005;102:13598–603. doi: 10.1073/pnas.0502378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson KC, Criscio MD, Kaslow DC. Cloning and expression of the gene for Plasmodium falciparum transmission-blocking target antigen, Pfs230. Mol Biochem Parasitol. 1993;58:355–8. doi: 10.1016/0166-6851(93)90058-6. [DOI] [PubMed] [Google Scholar]
- 19.van Dijk MR, van Schaijk BC, Khan SM, van Dooren MW, Ramesar J, Kaczanowski S, et al. Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog. 2010;6:e1000853. doi: 10.1371/journal.ppat.1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eksi S, Czesny B, van Gemert GJ, Sauerwein RW, Eling W, Williamson KC. Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol Microbiol. 2006;61:991–8. doi: 10.1111/j.1365-2958.2006.05284.x. [DOI] [PubMed] [Google Scholar]
- 21.Rener J, Graves PM, Carter R, Williams JL, Burkot TR. Target antigens of transmission-blocking immunity on gametes of plasmodium falciparum. J Exp Med. 1983;158:976–81. doi: 10.1084/jem.158.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermeulen AN, Ponnudurai T, Beckers PJ, Verhave JP, Smits MA, Meuwissen JH. Sequential expression of antigens on sexual stages of Plasmodium falciparum accessible to transmission-blocking antibodies in the mosquito. J Exp Med. 1985;162:1460–76. doi: 10.1084/jem.162.5.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter R, Graves PM, Keister DB, Quakyi IA. Properties of epitopes of Pfs48/45, a target of transmission blocking monoclonal antibodies, on gametes of different isolates of Plasmodium falciparum. Parasite Immunol. 1990;12:587–603. doi: 10.1111/j.1365-3024.1990.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 24.Read D, Lensen AH, Begarnie S, Haley S, Raza A, Carter R. Transmission-blocking antibodies against multiple, non-variant target epitopes of the Plasmodium falciparum gamete surface antigen Pfs230 are all complement-fixing. Parasite Immunol. 1994;16:511–9. doi: 10.1111/j.1365-3024.1994.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 25.Quakyi IA, Carter R, Rener J, Kumar N, Good MF, Miller LH. The 230-kDa gamete surface protein of Plasmodium falciparum is also a target for transmission-blocking antibodies. J Immunol. 1987;139:4213–7. [PubMed] [Google Scholar]
- 26.Williamson KC, Keister DB, Muratova O, Kaslow DC. Recombinant Pfs230, a Plasmodium falciparum gametocyte protein, induces antisera that reduce the infectivity of Plasmodium falciparum to mosquitoes. Mol Biochem Parasitol. 1995;75:33–42. doi: 10.1016/0166-6851(95)02507-3. [DOI] [PubMed] [Google Scholar]
- 27.Healer J, McGuinness D, Carter R, Riley E. Transmission-blocking immunity to Plasmodium falciparum in malaria-immune individuals is associated with antibodies to the gamete surface protein Pfs230. Parasitology. 1999;119:425–33. doi: 10.1017/s0031182099005041. [DOI] [PubMed] [Google Scholar]
- 28.Graves PM, Carter R, Burkot TR, Quakyi IA, Kumar N. Antibodies to Plasmodium falciparum gamete surface antigens in Papua New Guinea sera. Parasite Immunol. 1988;10:209–18. doi: 10.1111/j.1365-3024.1988.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 29.Hughes MK, Hughes AL. Natural selection on Plasmodium surface proteins. Mol Biochem Parasitol. 1995;71:99–113. doi: 10.1016/0166-6851(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 30.Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, et al. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol. 2004;52:159–68. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 31.Martinelli A, Cheesman S, Hunt P, Culleton R, Raza A, Mackinnon M, et al. A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proc Natl Acad Sci USA. 2005;102:814–9. doi: 10.1073/pnas.0405097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richards JS, MacDonald NJ, Eisen DP. Limited polymorphism in Plasmodium falciparum ookinete surface antigen, von Willebrand factor A domain-related protein from clinical isolates. Malar J. 2006;5:55. doi: 10.1186/1475-2875-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niederwieser I, Felger I, Beck HP. Limited polymorphism in Plasmodium falciparum sexual-stage antigens. Am J Trop Med Hyg. 2001:649–11. doi: 10.4269/ajtmh.2001.64.9. [DOI] [PubMed] [Google Scholar]
- 34.Williamson KC, Kaslow DC. Strain polymorphism of Plasmodium falciparum transmission-blocking target antigen Pfs230. Mol Biochem Parasitol. 1993;62:125–7. doi: 10.1016/0166-6851(93)90186-2. [DOI] [PubMed] [Google Scholar]
- 35.Tsuboi T, Kaslow DC, Gozar MM, Tachibana M, Cao YM, Torii M. Sequence polymorphism in two novel Plasmodium vivax ookinete surface proteins, Pvs25 and Pvs28, that are malaria transmission-blocking vaccine candidates. Mol Med. 1998;4:772–82. [PMC free article] [PubMed] [Google Scholar]
- 36.Zakeri S, Razavi S, Djadid ND. Genetic diversity of transmission blocking vaccine candidate (Pvs25 and Pvs28) antigen in Plasmodium vivax clinical isolates from Iran. Acta Trop. 2009;109:176–80. doi: 10.1016/j.actatropica.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–63. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira MU, Karunaweera ND, da Silva-Nunes M, da Silva NS, Wirth DF, Hartl DL. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J Infect Dis. 2007;195:1218–26. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- 39.Orjuela-Sanchez P, da Silva NS, da Silva-Nunes M, Ferreira MU. Recurrent parasitemias and population dynamics of Plasmodium vivax polymorphisms in rural Amazonia. Am J Trop Med Hyg. 2009;81:961–8. doi: 10.4269/ajtmh.2009.09-0337. [DOI] [PubMed] [Google Scholar]
- 40.Zeyrek FY, Babaoglu A, Demirel S, Erdogan DD, Ak M, Korkmaz M, et al. Analysis of naturally acquired antibody responses to the 19-kd C-terminal region of merozoite surface protein-1 of Plasmodium vivax from individuals in Sanliurfa, Turkey. Am J Trop Med Hyg. 2008;78:729–32. [PubMed] [Google Scholar]
- 41.Tanabe K, Mita T, Jombart T, Eriksson A, Horibe S, Palacpac N, et al. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 2010;20:1283–9. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- 42.Yamauchi T, Nakazawa M, Ohmae H, Kamei K, Sato K, Bakote’e B. Impact of ethnic conflict on the nutritional status and quality of life of suburban villagers in the Solomon Islands. J Nutri Sci Vitaminol. 2010;56:227–34. doi: 10.3177/jnsv.56.227. [DOI] [PubMed] [Google Scholar]
- 43.Tanabe K, Escalante A, Sakihama N, Honda M, Arisue N, Horii T, et al. Recent independent evolution of msp1 polymorphism in Plasmodium vivax and related simian malaria parasites. Mol Biochem Parasitol. 2007;156:74–9. doi: 10.1016/j.molbiopara.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 46.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 47.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–7. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 48.Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]
- 49.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 51.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–4. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 54.Nachman MW, Brown WM, Stoneking M, Aquadro CF. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics. 1996;142:953–63. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tetteh KK, Stewart LB, Ochola LI, Amambua-Ngwa A, Thomas AW, Marsh K, et al. Prospective identification of malaria parasite genes under balancing selection. PLoS One. 2009;4:e5568. doi: 10.1371/journal.pone.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks SR, Williamson KC. Proteolysis of Plasmodium falciparum surface antigen, Pfs230, during gametogenesis. Mol Biochem Parasitol. 2000;106:77–82. doi: 10.1016/s0166-6851(99)00201-7. [DOI] [PubMed] [Google Scholar]
- 57.Riley EM, Williamson KC, Greenwood BM, Kaslow DC. Human immune recognition of recombinant proteins representing discrete domains of the Plasmodium falciparum gamete surface protein, Pfs230. Parasite Immunol. 1995;17:11–9. doi: 10.1111/j.1365-3024.1995.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 58.Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, et al. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005;280:40169–76. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- 59.Ishino T, Chinzei Y, Yuda M. Two proteins with 6-cys motifs are required for malarial parasites to commit to infection of the hepatocyte. Mol Microbiol. 2005;58:1264–75. doi: 10.1111/j.1365-2958.2005.04801.x. [DOI] [PubMed] [Google Scholar]
- 60.Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95:8124–9. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayakawa T, Culleton R, Otani H, Horii T, Tanabe K. Big bang in the evolution of extant malaria parasites. Mol Biol Evol. 2008;25:2233–9. doi: 10.1093/molbev/msn171. [DOI] [PubMed] [Google Scholar]
- 62.Mitsui H, Arisue N, Sakihama N, Inagaki Y, Horii T, Hasegawa M, et al. Phylogeny of Asian primate malaria parasites inferred from apicoplast genome-encoded genes with special emphasis on the positions of Plasmodium vivax and P. fragile. Gene. 2010;450:32–8. doi: 10.1016/j.gene.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Sawai H, Otani H, Arisue N, Palacpac N, de Oliveira Martins L, Pathirana S, et al. Lineage-specific positive selection at the merozoite surface protein 1 (msp1) locus of Plasmodium vivax and related simian malaria parasites. BMC Evol Biol. 2010;10:52. doi: 10.1186/1471-2148-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ord R, Polley S, Tami A, Sutherland CJ. High sequence diversity and evidence of balancing selection in the Pvmsp3alpha gene of Plasmodium vivax in the Venezuelan Amazon. Mol Biochem Parasitol. 2005;144:86–93. doi: 10.1016/j.molbiopara.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Rayner JC, Huber CS, Feldman D, Ingravallo P, Galinski MR, Barnwell JW. Plasmodium vivax merozoite surface protein PvMSP-3 beta is radically polymorphic through mutation and large insertions and deletions. Infect Genet Evol. 2004;4:309–19. doi: 10.1016/j.meegid.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 66.Gomez A, Suarez CF, Martinez P, Saravia C, Patarroyo MA. High polymorphism in Plasmodium vivax merozoite surface protein-5 (MSP5) Parasitology. 2006;133:661–72. doi: 10.1017/S0031182006001168. [DOI] [PubMed] [Google Scholar]
- 67.Putaporntip C, Udomsangpetch R, Pattanawong U, Cui L, Jongwutiwes S. Genetic diversity of the Plasmodium vivax merozoite surface protein-5 locus from diverse geographic origins. Gene. 2010;456:24–35. doi: 10.1016/j.gene.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez P, Suarez CF, Cardenas PP, Patarroyo MA. Plasmodium vivax Duffy binding protein: a modular evolutionary proposal. Parasitology. 2004;128:353–66. doi: 10.1017/s0031182003004773. [DOI] [PubMed] [Google Scholar]
- 69.Palumbi SR. Speciation and the evolution of gamete recognition genes: pattern and process. Heredity. 2009;102:66–76. doi: 10.1038/hdy.2008.104. [DOI] [PubMed] [Google Scholar]
- 70.Tsaur SC, Ting CT, Wu CI. Sex in Drosophila mauritiana: a very high level of amino acid polymorphism in a male reproductive protein gene, Acp26Aa. Mol Biol Evol. 2001;18:22–6. doi: 10.1093/oxfordjournals.molbev.a003716. [DOI] [PubMed] [Google Scholar]
- 71.Conway DJ, Machado RL, Singh B, Dessert P, Mikes ZS, Povoa MM, et al. Extreme geographical fixation of variation in the Plasmodium falciparum gamete surface protein gene Pfs48/45 compared with microsatellite loci. Mol Biochem Parasitol. 2001;115:145–56. doi: 10.1016/s0166-6851(01)00278-x. [DOI] [PubMed] [Google Scholar]
- 72.Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, et al. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol. 2007;37:1013–22. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 73.Orjuela-Sanchez P, Karunaweera ND, da Silva-Nunes M, da Silva NS, Scopel KK, Goncalves RM, et al. Single-nucleotide polymorphism, linkage disequilibrium and geographic structure in the malaria parasite Plasmodium vivax: prospects for genome-wide association studies. BMC Genet. 2010;11:65. doi: 10.1186/1471-2156-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.