Abstract
Extracts of the sponge genus Candidaspongia showed selective cytotoxicity towards melanoma cells in the NCI 60-cell-line screen. Continued investigation of the Candidaspongia sp. extracts led to the isolation of three new tedanolide analogs, precandidaspongiolides A (1) and B (2), and candidaspongiolide B (4), as well as candidaspongiolide A (3) and tedanolide (5). Semi-synthetic derivatives were also generated to develop SAR. Candidaspongiolides A/B were the most potent and showed low nanomolar activity against several melanoma cell lines.
Melanoma is the most common life-threatening form of skin cancer, and lifetime incidence rates have been steadily rising over the past 30 years. Metastatic melanoma is notoriously resistant to a wide range of chemotherapeutic agents, and patient prognosis is generally poor.1 Consequently, the search for novel melanoma-specific agents continues.
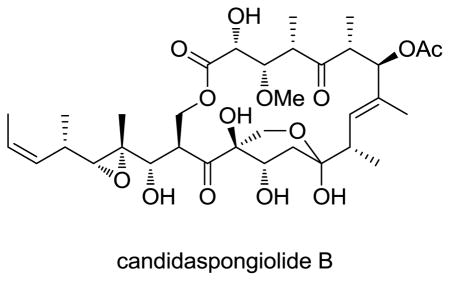
Several extracts of the sponge genus Candidaspongia were identified as potently cytotoxic and showed selectivity towards melanoma cells in the NCI 60-cell-line screen. Further examination of these Candidaspongia extracts yielded a complex mixture of acyl esters of a tedanolide-related macrolide; the candidaspongiolides.2 Lipase catalyzed hydrolysis of the complex mixture resulted in the isolation and identification of the macrolide core, named herein candidaspongiolide A (3). In the NCI 60-cell-line screen, the candidaspongiolide acyl ester mixture and candidaspongiolide A (3) exhibited GI50’s of ~14 nM and <4 nM, respectively, against seven melanoma cell lines (Tables S7 and S8). The candidaspongiolides belong to a small class of tedanolide macrolides, which includes tedanolide (5),3 13-deoxytedanolide4 and tedanolide C,5 all of which are cytotoxic in the subnanomolar to nanomolar range against various cancer cell lines.
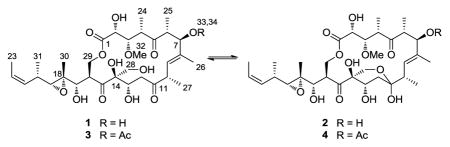
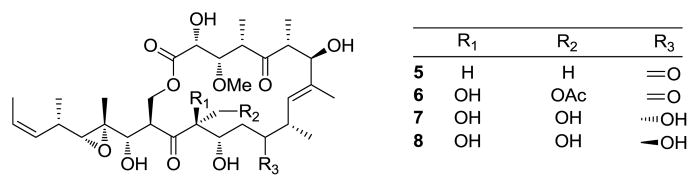
Continued investigation of the Papua New Guinea Candidaspongia sp. extracts for more melanoma-selective agents led to the isolation of three new compounds, precandidaspongiolides A (1) and B (2), and candidaspongiolide B (4), as well as candidaspongiolide A (3), (not previously isolated from natural sources), and (+)-tedanolide (5). Semi-synthetic derivatives (6–8) of precandidaspongiolide A (1) were also prepared to assess the importance of the hemiketal/hydroxy ketone for melanoma inhibitory activity (Figure 1).
Figure 1.
Tedanolide (5) and semi-synthetic precandidaspongiolides (6–8).
Mass-guided fractionation of the aqueous Candidaspongia extracts utilizing size exclusion chromatography, reversed-phase column chromatography and reversed-phase HPLC resulted in the isolation of 1–5.
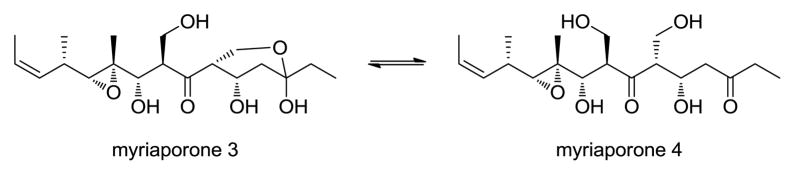
Precandidaspongiolides A (1) and B (2) were isolated as an inseparable mixture of two isomers in equilibrium, clearly related to the tedanolides, with the major isomer containing a primary alcohol (1) and the minor isomer containing a hemiketal (2) (ratio 1:2 = 4.5:1). Interestingly, the myriaporones, isolated from a bryozoan and structurally related to the southern hemisphere of the tedanolides, have also been isolated as an equilibrium mixture (Figure 2).6 In subsequent synthetic studies of the myriaporones, equilibrium mixtures were also reported.7,8 Therefore, structure determination of the two isomers was carried out on the mixture of 1 and 2. HRESIMS data (m/z 665.3146 [M + Na]+, Δ = 0.3 ppm) indicated that the molecular formula for both 1 and 2, was C32H50O13; two oxygens more than the molecular formula for tedanolide (5), and 42 Da less than the molecular weight of candidaspongiolide A (3). The aforementioned data suggested that 1 and 2 lacked the large fatty acid esters, as well as the C-7 acetoxy moiety present in the original candidaspongiolide mixtures. Analysis of the combined NMR data confirmed this supposition (Table 1).
Figure 2.
Myriaporones 3 and 4
Table 1.
NMR Data for Precandidaspongiolides A (1) and B (2) and Candidaspongiolides A (3) and B (4) (600 MHz, CD3OD)
| precandidaspongiolide A (1) | precandidaspongiolide B (2) | candidaspongiolide A (3) | candidaspongiolide B (4) | |||||
|---|---|---|---|---|---|---|---|---|
| no. | δC | δH, mult (J, Hz) | δC | δH, mult (J, Hz) | δC | δH, mult (J, Hz) | δC | δH, mult (J, Hz) |
| 1 | 173.4 | — | 173.3 | — | 173.2 | — | 173.1 | — |
| 2 | 73.1 | 3.76a (2.0) | 72.7 | 3.60, d (1.9) | 73.0 | 3.77, d (1.9) | 72.7 | 3.57, d (1.9) |
| 3 | 84.8 | 3.81, dd (9.5, 2.0) | 84.9 | 3.66, dd (9.5, 1.9) | 85.4 | 3.80, dd (9.4, 1.9) | 85.1 | 3.68, dd (9.8, 1.9) |
| 4 | 49.6 | 3.15,a (9.5, 7.1) | 48.5 | 3.34, dq (9.5, 7.2) | 48.9 | 3.21, dq (9.4, 7.0) | 48.2 | 3.31,d (9.8, 6.9) |
| 5 | 217.8 | — | 217.2 | — | 216.1 | — | 215.4 | — |
| 6 | 51.3 | 3.15,a (10.0, 7.0) | 51.6 | 3.15, dq (10.2, 6.9) | 50.0 | 3.41, dq (10.2, 7.1) | 49.7 | 3.35,c (6.7) |
| 7 | 80.0 | 4.04, d (10.0) | 79.2 | 4.15, d (10.2) | 81.4 | 5.39, d (10.2) | 81.3 | 5.48,c |
| 8 | 139.1 | — | 135.8 | — | 134.7 | — | 131.4 | — |
| 9 | 129.9 | 5.33, br d (9.5)b | 132.5 | 5.40, br d (10.0) | 133.3 | 5.45, br d (9.8) | 135.9 | 5.55 br d (10.0) |
| 10 | 46.7 | 3.38, dq (9.5, 6.8) | 44.3 | 2.35, dq (10.0, 6.9) | 46.6 | 3.35, dq (9.8, 6.8) | 44.6 | 2.32, dq (10.0, 6.8) |
| 11 | 212.9 | — | 100.5 | — | 212.2 | — | 100.4 | — |
| 12a | 44.4 | 2.75, dd (17.6, 9.6) | 41.1 | 2.01, dd (13.6, 4.3) | 44.5 | 2.70, dd (17.6, 9.6) | 41.2 | 1.99, dd (13.8, 4.2) |
| 12b | 2.24, dd (17.6, 1.7) | 1.03,c (13.6, 12.7) | 2.24, dd (17.6, 1.9) | 1.00,c (13.8, 12.6) | ||||
| 13 | 69.5 | 4.44, dd (9.6, 1.7) | 71.9 | 4.22, dd (12.7, 4.3) | 69.4 | 4.46, dd (9.6, 1.9) | 71.9 | 4.21, dd (12.6, 4.2) |
| 14 | 85.4 | — | 79.1 | — | 85.5 | — | 79.1 | — |
| 15 | 216.6 | — | 212.1 | — | 216.5 | — | 212.1 | — |
| 16 | 48.9 | 4.08, ddd (11.2, 10.8, 4.1) | 51.9 | 4.36,c (11.7, 10.4, 3.8) | 49.0 | 4.04, ddd (11.0, 10.7, 3.9) | 52.0 | 4.35,c (11.9, 10.4, 3.9) |
| 17 | 78.5 | 3.21, d (10.8) | 78.9 | 3.02, d (10.4) | 78.4 | 3.23, d (10.7) | 78.9 | 3.00, d (10.4) |
| 18 | 64.1 | — | 63.9 | — | 63.7 | — | 63.8 | — |
| 19 | 67.5 | 2.64, d (9.4) | 67.3 | 2.66, d (9.2)b | 67.5 | 2.64, d (9.2) | 67.3 | 2.62, d (9.2) |
| 20 | 32.6 | 2.48, ddq (10.7, 9.4, 6.6) | 32.6 | 2.64,c (9.2, 7.2) | 32.5 | 2.48, ddq (10.7, 9.2, 6.5) | 32.5 | 2.50,c |
| 21 | 131.8 | 5.33, ddq (10.9, 10.7, 1.5)b | 132.0 | 5.32,c (1.6) | 131.8 | 5.35, ddq (10.9, 10.7, 1.5) | 132.0 | 5.32, ddq (10.9, 10.6, 1.4) |
| 22 | 126.3 | 5.51, dq (10.9, 6.8) | 126.1 | 5.50,c (6.9) | 126.3 | 5.51, dq (10.9, 6.7) | 126.1 | 5.50,c (6.8) |
| 23 | 13.7 | 1.63, dd (6.8, 1.5) | 13.7 | 1.60, dd (6.9, 1.6) | 13.7 | 1.63, dd (6.7, 1.5) | 13.7 | 1.60, dd (6.8, 1.5) |
| 24 | 15.2 | 1.23, d (7.1) | 15.5 | 1.25, d (7.2)b | 15.0 | 1.23, d (7.0) | 15.6 | 1.15, d (6.9) |
| 25 | 15.7 | 1.26, d (7.0) | 15.2 | 1.14, d (6.9) | 14.8 | 1.18, d (7.1) | 14.5 | 1.13, d (6.7) |
| 26 | 10.5 | 1.66, br s | 10.4 | 1.51, br s | 11.0 | 1.66, br s | 10.6 | 1.50, br s |
| 27 | 15.8 | 1.04, d (6.8) | 13.0 | 0.95, d (6.9) | 15.6 | 1.00, d (6.8) | 12.8 | 0.91, d (6.8) |
| 28a | 65.8 | 3.75,a s | 66.8 | 3.63, d (11.2) | 65.8 | 3.75, s | 66.8 | 3.62, d (11.2) |
| 28b | — | 3.53, d (11.2) | — | 3.53, d (11.2) | ||||
| 29a | 65.3 | 4.34, dd (10.8, 4.1) | 65.4 | 4.31, dd (11.7, 10.0) | 65.0 | 4.33, dd (10.8, 3.9) | 65.4 | 4.31, dd (11.9, 10.0) |
| 29b | 3.93, dd (11.2, 10.8) | 3.98, dd (10.0, 3.8) | 3.95, dd (11.0, 10.8) | 3.98, dd (10.0, 3.9) | ||||
| 30 | 11.6 | 1.35, s | 11.5 | 1.38, s | 11.6 | 1.35, s | 11.5 | 1.35, s |
| 31 | 18.8 | 1.11, d (6.6) | 18.9 | 1.12, d (7.2)b | 18.9 | 1.10, d (6.5) | 18.9 | 1.10, d (6.5) |
| 32 | 61.5 | 3.39, s | 61.2 | 3.31, s | 61.5 | 3.41, s | 61.3 | 3.32, s |
| 33 | 171.9 | — | 172.0 | — | ||||
| 34 | 21.0 | 2.03, s | 21.0 | 2.03, s | ||||
Signals overlapped.
Measured in d6-acetone.
Signal obscurred by major/minor isomer.
Buried under CD3OD signal.
The major isomer, precandidaspongiolide A (1) clearly lacked the acetyl group at C-7, as indicated by the upfield shift of H-7 (δH 4.04) compared to 3 (δH 5.39). Additionally, only one ester was present in 1; the macrolide lactone (δc 173.4, C-1). The presence of a primary alcohol was also apparent in 1, as evidenced by the degenerate chemical shifts of the C-28 methylene (δH 3.75, s). The NMR data (Table 1) supported the remaining structure of precandidaspongiolide A (1), (or 7-deacetyl-candidaspongiolide A), as drawn.
The minor isomer, precandidaspongiolide B (2) also lacked the acetoxy group at C-7 (Table 1), and had a large number of chemical shift deviations from 1, particularly in the C-10 to C-14 portion of the molecule. Compound 2 contained only two ketones (δc 217.2, 212.1), that were identified as C-5 and C-15, respectively, and suggested a modification to the C-11 ketone. Additionally, the diastereotopicity of the C-28 methylene (δH 3.63, 3.53) and a downfield quaternary carbon (δC 100.5) supported the presence of a hemiketal ring in 2. The hemiketal identity was confirmed based on HMBC correlations from H-28a/b, H-10, and H-12a/b to C-11. The NMR data (Table 1) corroborated the remaining portion of 2, the tedanolide core, as drawn.
Candidaspongiolides A (3) and B (4) were also isolated as an equilibrium mixture (ratio 3:4 = 1.7:1). HRESIMS data (m/z 707.3235 [M + Na]+, Δ = 2.1 ppm) indicated that the molecular formula for 3 and 4, was C34H52O14, matching the molecular formula for candidaspongiolide A, originally isolated from lipase catalyzed hydrolysis of the candidaspongiolide acyl esters.2 The molecular formula and NMR data (Table 1) suggested that 3 was the acetylated version of 1. Comparison of the spectroscopic data for the macrolide core (candidaspongiolide A)2 and the major isomer 3 (in acetone-d6, Table S19) confirmed the identity and structure, as drawn.10
The minor isomer, candidaspongiolide B (4) closely resembled precandidaspongiolide B (2), with the major differences appearing around C-7. Analysis of the NMR data (Table 1) confirmed the identity of candidaspongiolide B (4) as the C-7 acetyl analog of 2.
HRESIMS data (m/z 633.3232 [M + Na]+, Δ = 2.2 ppm) supported a molecular formula of C32H50O11 for 5, identical to that of tedanolide. Compound 5 was confirmed as (+)-tedanolide upon comparison of spectroscopic data with those reported in the literature.3,11
Analysis of the coupling constants and ROESY data for 1–4 suggested that their relative configurations were identical to tedanolide,3 13-deoxytedanolide,4 and the original candidaspongiolides.2 Biosynthetic principles support 1–4 having the same absolute configuration as (+)-tedanolide (5), given that 1–4 were concurrently isolated with 5.
Within the small tedanolide class, there are a number of structural features that are unique to the candidaspongiolides, particularly the hemiketal. Three semi-synthetic analogs were generated from the equilibrium mixture of 1/2 to develop SARs among the candidaspongiolides, and assess the importance of the hemiketal for melanoma cytotoxicity. The primary alcohol of 3 was selectively acetylated,12 using AcCl and 2,4,6-trimethylpyridine, to give 28-acetyl-precandidaspongiolide (6), and the 11-keto group was preferentially reduced13 using NaBH4 in MeOH to yield a pair of diastereomers, 11R- and 11S- dihydroprecandidaspongiolide A (7, 8), respectively. The configurations of C-11 in 7 and 8 were assigned based on interpretation of ROESY data, coupling constants, and comparison of 1H NMR chemical shifts with 11R- and 11S-dihydro-13-dexoytedanolide.12 ROESY cross peaks between H-10/H-11 and H-11/H-13 suggested an 11R stereochemistry for 7, while cross peaks between H-9/H-11 and a number of upfield 1H NMR shifts in 8 compared to 7 implied a 11S stereochemistry for 8. In addition, the 11R diastereomer was reported as the major product from the reduction of 13-deoxytedanolide (confirmed by Mosher’s analysis),12 which is in agreement with the major product (7–11R) from the reduction of 1.
Precandidaspongiolides A (1) and B (2) showed excellent selectivity against melanoma cell lines in the NCI 60-cell line screen (Figures S3 and S4). The LC50 values for 1/2 against melanoma cell lines were significantly lower than other tumor cell lines; seven of the nine melanoma cell lines in the panel had nanomolar LC50 values (19–174 nM), while a majority of the other tumor cell lines had LC50’s greater than 100 M (Figure S4).
To develop SARs, compounds 1–8 were tested against three NCI-60 melanoma cell lines (UACC-257, LOX-IMVI, and M14), as well as a breast (MCF7) and lung cancer (NCI-H460) cell line (Table 2). Compounds 1–8 showed nanomolar activity among the various cell lines. Identical patterns were observed between melanoma, breast and lung tumor cell lines, which are still consistent with the original NCI 60-cell line data for 1/2 (Figures S3 and S4). In the interest of preserving the limited amount of material available for 3–8, the compounds were tested in the IC50 range (= NCI 60-cell line GI50, see Supporting Information S7) and not the LC50 range. It is likely that the LC50 values and melanoma selectivity of 3–8 would mimic those of 1/2. Candidaspongiolides A/B (3/4) were the most active against all cell lines, while 8 was the least active. Comparing the activities for 1/2 and the semi-synthetic derivatives (6–8), it appears that the hemiketal is not essential for activity, as 7 retains the same level of potency as 1/2. However, potency is affected when the primary alcohol is substituted, as in 6, which is ~ 5–15× less potent than 1/2. This is consistent with the NCI 60-cell line data for the candidaspongiolide acyl esters and candidaspongiolide A (3) (Tables S7 and S8); compound 3 is over 3× more potent than the candidaspongiolide fatty acid esters. Interestingly, compound 7 is significantly more potent than 8. Fusetani et al. reported that the 11S diastereomer of 13-deoxytedanolide was more potent than the 11R in p388 murine leukemia cells.12 This apparent discrepancy may be due to different tumor cell type specificity, and would be an interesting area for further investigation. Additionally, when comparing 1/2 and 3/4, it appears that C-7 acetylation increases potency. However, differences in the equilibrium mixture ratios may also contribute to the potency differences. The somewhat simpler tedanolide (5) retains low nanomolar activity without the oxygenation at C-14 and C-28, the hemiketal moiety, and acetylation at C-7.
Table 2.
Biological activity of 1–8.
| IC 50 (nM)a | |||||
|---|---|---|---|---|---|
| compound | UACC257 | LOX-IMVI | M14 | MCF7 | NCI-H460 |
| melanoma | breast | lung | |||
| precandidaspongiolide A/B (1/2) | 14.2 ± 0.2 | 6.9 ± 1.0 | 17.9 ± 4.3 | 8.3 ± 0.5 | 12.3 ± 1.0 |
| candidaspongiolide A/B (3/4) | 1.6 ± 0.5 | 2.0 ± | 7.5 ± 1.5 | 2.0 ± | 3.4 ± 0.2 |
| tedanolide (5) | 5.9 ± 0.1 | 2.5 ± 0.4 | 8.6 ± 2.4 | 3.6 ± 0.3 | 7.0 ± 3.9 |
| 28-acetyl-precandidaspongiolide A (6) | 103.8 ± 3.4 | 34.2 ± 9.4 | 261.0 ± 63.9 | 98.6 ± 12.5 | 96.8 ± 15.7 |
| 11R-dihydro-precandidaspongiolide A (7) | 16.7 ± 2.2 | 10.3 ± 0.3 | 25.1 ± 0.5 | 12.4 ± 1.2 | 43.7 ± 16.6 |
| 11S-dihydro-precandidaspongiolide A (8) | 423.1 ± 57.4 | 275.3 ± 20.0 | 505.6 ± 94.0 | 368.8 ± 19.1 | 433.4 ± 17.5 |
IC50 cytotoxicity values were determined as the drug concentration that reduced cell growth to 50% of the untreated control.
The 60-cell line data for 1/2 revealed a number of candidaspongiolide insensitive cell lines (NCI/ADR-RES; HCT-15) that are known to express P-glycoprotein (P-gp), a multidrug resistance transporter. To test whether 1/2 were P-gp substrates, KB-3-1 (adenocarcinoma, P-gp-deficient) and KB-V1 (P-gp-overexpressing) cells were treated with 1/2 (Figure S10). KB-3-1 cells were sensitive to 1/2, while KB-V1 cells were resistant. Resistance is defined as the ratio of cytotoxicity against P-gp-expressing cell pairs versus parental cells (resistance ratio (RR) = 26.7). Both cell lines were co-incubated with 1/2 and tariquidar (TQR), a specific P-gp inhibitor. TQR did not affect the toxicity of 1/2 against the P-gp deficient KB-3-1 cells but completely reversed resistance in the KB-V1 cells, confirming that 1/2 are in fact P-gp substrates. P-gp is, however, not thought to play a role in drug-resistant melanoma.14
The similarities between the myriaporones and the new candidaspongiolides offers further evidence in support of microorganisms as producers of these compounds. 13-deoxytedanolide15 and candidaspongiolide A (3)16 both inhibit protein synthesis. However, the underlying reasons for the candidaspongiolides’ melanoma selectivity have yet to be determined and are currently being investigated.
Supplementary Material
Acknowledgments
The authors would like to thank P. Colin (CRRF) for sustained collecting efforts; P. Bergquist (U. of Auckland, deceased) and S. Sorokin (SARDI) for taxonomic analysis of the sponge samples; D. Newman (NPB) for contract collection; T. McCloud (NPSG) for extraction; and S. Tarasov and M. Dyba (Biophysics Resource Core, SBL, CCR) for assistance with high-resolution mass spectrometry. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supporting Information Available: General experimental details, organism collection, isolation details, reaction conditions, bioassay methods, spectral data, and NCI-60 cell line data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Soengas MS, Lowe SW. Oncogene. 2003;22:3138. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 2.Meragelman TL, Willis RH, Woldemichael GM, Heaton A, Murphy PT, Snader KM, Newman DJ, van Soest R, Boyd MR, Cardellina JH, II, McKee TC. J Nat Prod. 2007;70:1133. doi: 10.1021/np0700974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz FJ, Gunasekera SP, Yalamanchili G, Hossain MB, van der Helm D. J Am Chem Soc. 1984;106:7251. [Google Scholar]
- 4.Fusetani N, Sugawara T, Matsunaga S, Hirota H. J Org Chem. 1991;56:4971. [Google Scholar]
- 5.Chevallier C, Bugni TS, Feng X, Harper MK, Orendt AM, Ireland CM. J Org Chem. 2006;71:2510. doi: 10.1021/jo052285+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng JF, Lee JS, Sakai R, Jares-Erijman EA, Silva MV, Rinehart KL. J Nat Prod. 2007;70:332. doi: 10.1021/np060308p. [DOI] [PubMed] [Google Scholar]
- 7.Fleming KN, Taylor RE. Angew Chem Int Ed. 2004;43:1728. doi: 10.1002/anie.200353348. [DOI] [PubMed] [Google Scholar]
- 8.Perez M, del Pozo C, Reyes F, Rodriguez A, Francesch A, Echavarren AM, Cuevas C. Angew Chem Int Ed. 2004;43:1724. doi: 10.1002/anie.200353313. [DOI] [PubMed] [Google Scholar]
- 9.Minor revisions have been made to the NMR assignments of candidaspongiolide A (3) in acetone-d6.
- 10.There was no evidence of equilibrium isomers in the NMR data for the lipase derived macrolide core reported previously.2
- 11.See Supporting Information for a complete assignment of (+)-tedanolide (5) in CDCl3 and CD3OD.
- 12.Ishihara K, Kurihara H, Yamamoto H. J Org Chem. 1993;58:3791. [Google Scholar]
- 13.Nishimura S, Matsunaga S, Yoshida S, Nakao Y, Hirota H, Fusetani N. Bioorg Med Chem. 2005;13:455. doi: 10.1016/j.bmc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Chen KG, Valencia JC, Gillet JP, Hearing VJ, Gottesman MM. Pigment Cell Melanoma Res. 2009;22:740. doi: 10.1111/j.1755-148X.2009.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura S, Matsunaga S, Yoshida M, Hirota H, Yokoyama S, Fusetani N. Bioorg Med Chem. 2005;13:449. doi: 10.1016/j.bmc.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Trisciuoglio D, Uranchimeg B, Cardellina JH, Meragelman TL, Matsunaga S, Fustetani N, Del Bufalo D, Shoemaker RH, Melillo G. J Natl Cancer Inst. 2008;100:1233. doi: 10.1093/jnci/djn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.