Abstract
The default lipid for the bulk of the crystallogenesis studies performed to date using the cubic mesophase method is monoolein. There is no good reason however, why this 18-carbon, cis-monounsaturated monoacylglycerol should be the preferred lipid for all target membrane proteins. The latter come from an array of biomembrane types with varying properties that include hydrophobic thickness, intrinsic curvature, lateral pressure profile, lipid and protein makeup, and compositional asymmetry. Thus, it seems reasonable that screening for crystallizability based on the identity of the lipid creating the hosting mesophase would be worthwhile. For this, monoacylglycerols with differing acyl chain characteristics, such as length and olefinic bond position, must be available. A lipid synthesis and purification program is in place in the author's laboratory to serve this need. In the current study with the outer membrane sugar transporter, OprB, we demonstrate the utility of host lipid screening as a means for generating diffraction-quality crystals. Host lipid screening is likely to prove a generally useful strategy for mesophase-based crystallization of membrane proteins.
Keywords: membrane protein structure, OprB, outer membrane proteins, porin, screen, sugar transporter, X-ray crystallography
1. Introduction
The in meso method for crystallizing membrane proteins for use in structure determination employs the lipidic mesophases, the cubic phase in particular.1, 2 The method works convincingly with a variety of membrane protein types. To date, the Membrane Protein Data Bank (www.mpdb.tcd.ie),3 includes 56 records for membrane protein (and peptide) structures covering six distinct types of integral membrane proteins that were solved using in meso-grown crystals. The method has received considerable notice of late having been used in the structure determination of a number of pharmaceutically important G protein-coupled receptors.4–8
A mechanism has been advanced for how in meso crystallization comes about at a molecular level (Figure 1).2 This involves an initial reconstitution of the target protein into the bilayer of a bicontinuous cubic mesophase. The latter consists of a continuous, highly curved and multiply branched membrane draped on either side by an aqueous channel that tracks the membrane along its 3-dimensional course. The surface defined by the bilayer mid-plane of this mesophase can be described topologically as a periodic minimal surface. Crystallization is initiated by bathing the bolus of mesophase in a precipitant solution. Solutes from the precipitant diffuse into its porous bulk and are proposed to stabilize locally lamellar domains that attract proteins from the surrounding mesophase. It is in these domains that the protein locally concentrates in a process that ultimately leads to nucleation and crystal growth. Crystal packing is of the layered or Type I type consistent with the mechanism just described.
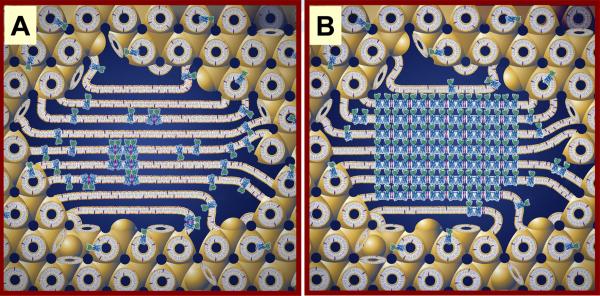
Figure 1.
Cartoon representation of the events proposed to take place during the crystallization of an integral membrane protein from the lipidic cubic mesophase. The process begins with the protein reconstituted into the curved bilayer of the `bicontinuous' cubic phase (tan). Added `precipitants' shift the equilibrium away from stability in the cubic membrane. This leads to phase separation wherein protein molecules diffuse from the bicontinuous bilayered reservoir of the cubic phase into a sheet-like or lamellar domain (A) and locally concentrate therein in a process that progresses to nucleation and crystal growth (B, from reference 9). Cocrystallization of the protein with native lipid (cholesterol) is shown in this illustration. As much as possible, the dimensions of the lipid (tan oval with tail), detergent (pink oval with tail), cholesterol (purple), protein (blue and green; β2-adrenergic receptor-T4 lysozyme fusion; PDB code 2RH1), bilayer and aqueous channels (dark blue) have been drawn to scale. The lipid bilayer is ~40 Å thick.
Lipids play an integral role in in meso crystallization. Importantly, they create the hosting mesophase into which the target membrane protein is reconstituted as a prelude to nucleation and crystal growth. Additionally, lipids can be added to the hosting mesophase to satisfy the protein's specific needs.9 A case in point is the set of G protein-coupled receptors that have yielded to the method but only when the mesophase is suitably doped with cholesterol.4–8 The goal of the current work is to describe how important the chemical constitution of the lipids that create the hosting mesophase is to membrane protein crystallization. The results highlight the need for the implementation of a crystallization screen that incorporates hosting lipids as a key variable.
The test protein used in this study is OprB, a porin resident in the outer membrane of Pseudomonas aeruginosa. It is responsible for transporting sugars and related polyols, such as glycerol and mannitol.10, 11 OprB mediated transport of glucose has been demonstrated in vitro by liposome swelling 12 and black lipid membrane measurements13.
The long-term objective of our work on OprB is to solve its 3-dimensional X-ray structure at or close to atomic resolution with a view to deciphering its molecular mode of action and substrate specificity. For this, macromolecular crystallography is the only viable method. Since the in meso approach has proven successful in the crystallization and structure determination of other outer membrane proteins (BtuB,14 OpcA15), we set out to use it under default conditions to grow diffraction quality crystals of OprB. The latter conditions, which included using the monoacylglycerol (MAG), monoolein (9.9 MAG, Footnote 1), as the hosting lipid, did not work. On reflection, there is no reason why a single hosting lipid should serve all membrane proteins. They come from different organisms and source membranes and perhaps even distinct domains within membranes that have their own physicochemical and compositional characteristics. Thus, presenting the target protein with alternative hosting lipids made good sense and is the focus of the current work. In this paper, we describe the behaviour of OprB in the default lipid monoolein, our attempts at optimizing those default conditions, and our approach to choosing a new hosting lipid that produced diffraction-quality crystals. The results make a convincing case for performing in meso crystallization trials with different hosting MAGs and, by extension, for the routine implementation of host lipid screening.
2. Materials and Methods
2.1. Materials
Monoolein (9.9 MAG, Footnote 1, Lot M239-S16-S, 356 g/mol) was purchased from Nu-Chek prep (Elysian, MN). 6.8 MAG (Lot JPL52, 300 g/mol), 8.8 MAG (Lot JPL42, 328 g/mol), 6.9 MAG (Lot Con 5, 314 g/mol), 8.7 MAG (Lot JPL-3–71, 314 g/mol) and 7.8 MAG (Lots JPL-2–45, JPL-3–89, 314 g/mol) were synthesized and purified in-house following established procedures.16, 17 Sodium chloride (Cat. BP358-10, lot 060875) was purchased from Fisher Scientific (Loughborough, Leicestershire, UK). Agar (Cat. 1.01613.1000, lot VM135213 342) and ammonium sulphate (Cat. 1.01217.1000, lot A456917 420) were purchased from Merck Biosciences (Beeston, UK). The pET200-D/TOPO cloning kit (Cat. K200-1, lot 1296068) was obtained from Invitrogen (Carlsbad, CA). dATP (Cat. 1039401, lot 133198976), dCTP (Cat. 1039400, lot 133198284), dTTP (1039398, lot 133199118), dGTP (Cat. 1039399, lot 133199288) and Ni-NTA resin (Cat. 1018142, lot 136231239) were purchased from Qiagen (Hilden, Germany). PfuUltra II DNA polymerase (Cat. 600672-51, lot 0006051615) was sourced from Stratagene (Santa Clara, CA). Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Cat. MB1008, lot A21345), kanamycin (Cat. K0126, lot 18022) and glycerol (Cat. G1345, lot A21344) were purchased from Melford Labs (Ipswich, UK). Tris (Cat. T8,760-2, lot 1133AH-437), hydrochloric acid (Cat. 258148, lot. 1403487 23808281), imidazole (Cat. 56750 lot 1239193 21006157), tri-ethylene glycol (Cat. 95126, lot 1327740 20807065), glycine (G8898, lot 017K0031), sodium acetate (Cat. S2889, lot 079K0122), acetic acid (Cat. A6283, lot 522096HK), 2-(N-morpholino) ethanesulfonic acid (MES) (Cat. M3671, lot 068K5421), sodium hydroxide (Cat. S8045, lot 0001427117), tryptone (Cat. 95039, lot 1321423 20207068), yeast extract (Cat. 70161, lot 1180742 32505281), N,N-dimethyldodecylamine-N-oxide (LDAO) (Cat. 40236, lots 1242624 30207224, BCBB8394), agarose (Cat. A5304, lot 108K1012), guanidine hydrochloride (Cat. G4505, lot 119K5401), phenylmethanesulfonyl fluoride (PMSF, 78830, lot 1419691 31509037) and dialysis tube (D9402-100FT, lot. 3110) were from Sigma (St. Louis, MO). Crystallization screens: Salt RX HT (Cat. HR108, lot 213607-05), Index HT (Cat. HR20144, lot 214407), Crystal Screen HT (Cat. HR2-130, lots 211094 and 211251), MemFac HT (HR2-137, lot 213103-24-22) and Additive Screen HT (HR2-138, lot 213803) were purchaed from Hampton Research (Aliso Viejo, CA); JBScreen Classic HTS1S (Cat. CS201), JBScreen Classic HTS2S (Cat. CS202) and JBScreen Membrane HTSS (Cat. JBS 00011630, lots 2004/01 and 2003/02) were from Jena Bioscience GmbH (Jena, Germany); HT96 PACT Premier (Cat. MD1-36, lot BN002) and HT 96 Strcture Screen 1 and 2 (Cat. MD 1-30, BN002-1-30) were from Molecular Dimensions (Newmarket, Suffolk, UK); Wizard III (Cat. EBS-WIZ-3, lot EBS 0006152009299) was from Emerald Biosystems (Bainbridge Island, WA). Syringes (Cat. 81030) were obtained from Hamilton (Bonaduz, GR, Switzerland). Superdex 200 16/60 was purchased from GE Healthcare (Little Chalfont, Buckinghamshire, UK). Amicon Ultracel-50 membrane concentrators (Cat. UFC905008) were purchased from Millipore (Billerica, MA). All buffers were prepared using Milli-Q water (Catrige Cat. PROG000S2, lot F9HN95000, Filter Cat. MPGL04001, lot F5PN18060) (Millipore, Billerica, MA).
2.2. Methods
2.2.1. Gene Cloning, OprB Refolding and Purification
The partial oprb gene encoding the mature protein from residues Ala32 to Phe454 was amplified by PCR from the PAO1 strain of P. aeruginosa. The PCR product was directly cloned into pET200D-TOPO using the TOPO reaction. The identity of the cloned gene was confirmed by sequencing (Eurofins MWG Operon, Ebersberg, Germany).
The production of OprB and its purification, primarily from inclusion bodies, were carried out as follows. BL21 (DE3) Star cells carrying the pET200D-TOPO-OprB plasmid and grown in Luria-Bertani broth were induced at an absorbance at 600 nm of 0.6 – 0.8 for 3 h at 37 °C with 1 mM IPTG. Biomass was harvested by centrifugation at 8,000 g for 15 min at 4 °C. The cell pellet from a 2 L culture was resuspended in 0.1 L Lysis Buffer (50 μg/mL DNAse I, 150 mM NaCl, 1 mM PMSF, 100 mM Tris/HCl pH 8.0), and broken by passage thrice through a French Press at 16,000 psi at room temperature (RT, 19–24 °C). After centrifugation at 9,000 g for 10 min at 4 °C, the pellet (containing inclusion bodies and unbroken cells) was washed twice with 0.1 L Buffer A (150 mM NaCl, 50 mM Tris/HCl, pH8.0) and once with 0.1 L 5 %(w/v) LDAO in Buffer A. The washing was performed at 4 °C for 1 h with vigorous stirring. The pellet was collected by centrifugation, as above, after each washing cycle. The pellet was resuspended at 20 °C by votexing in 25 mL 0.2 M Tris/HCl pH 8.0, and 75 mL 8 M GuHCl was added to solubilise the inclusion bodies. The solution was centrifuged at 9,000 g for 10 min at 4 °C and the supernatant, containing solubilised inclusion bodies, was collected. Refolding of OprB was effected by rapid dilution. To this end, the supernatant was diluted drop-wise into 1 L Refolding Buffer (10 %(v/v) glycerol, 5 %(w/v) LDAO, 300 mM NaCl, 50 mM Tris/HCl pH 8.0) at 20 °C. The resulting solution was dialysed (12 kDa cut-off) against 10 L Dialysis Buffer (0.1 %(w/v) LDAO, 150 mM NaCl, 20 mM Tris/HCl pH 8.0) overnight at 20 °C. The dialyzed solution (1.2 L), containing refolded protein, was centrifuged at 9,000 g for 10 min at 4 °C to remove precipitated protein. The clear, colorless supernatant was passed through a 0.22 μm membrane to remove large aggregates. Batch binding was performed by incubating the filtrate with 5 mL Ni-NTA resin for 1 h at 4 °C. After washing with 50 mL Washing Buffer (0.1 %(w/v) LDAO, 150 mM NaCl, 50 mM Tris/HCl pH 8.0) and 50 mL 50 mM imidazole in Washing Buffer, the protein was eluted with 300 mM imidazole in Washing Buffer. The protein was concentrated (Amicon Ultracel-50 membrane concentrator, Cat. UFC905008, Millipore, Billerica, MA) to 20 mg/mL (molar extinction coefficient at 280 nm, 115,405 M−1 cm−1; Gentle software. http://gentle.magnusmanske.de/) and subjected to gel filtration on a Superdex 200 26/60 column equilibrated with Gel Filtration Buffer (0.1 %(w/v) LDAO, 150 mM NaCl, 10 mM Tris/HCl pH 8.0) attached to an AKTA FPLC system (GE Healthcare). The protein was eluted at 240.7 mL as a symmetric peak. Peak fractions were pooled (8 mL total) and concentrated to 20 – 46 mg/mL using a spin concentrator, as above. The protein was aliquoted into PCR tubes at 10 μL/tube (0.2 mL PCR tube, Cat. 72.737.002, Sarstedt, Numbrecht, Germany), flash-frozen in liquid nitrogen and stored in −80 °C. The typical yield was 10 mg of purified OprB per liter of culture.
2.2.2. Reconstitution In Meso
OprB was reconstituted into the bilayer of the cubic phase following a standard protocol.9 The stock protein solution, usually at 20 mg/mL, was homogenized with monoolein (9.9 MAG) in a coupled syringe device18 at RT using two volumes of protein solution for every three volumes of lipid. For 6.9 MAG, 7.8 MAG, 8.7 MAG and 8.8 MAG, which have slightly different phase behaviours compared to monoolein, the volume ratio used was 1/1.
2.2.3. Crystallization In Meso
In meso crystallization trials were set up by transferring 50 nL of the OprB/lipid cubic mesophase onto a 96-well glass crystallization plate which was subsequently covered with 0.8 or 1 μL precipitant solution using an in meso robot.9, 19 Wells were sealed with a glass coverslide. The glass sandwich plates were stored in incubators/imagers (RockImager RI54 at 4–6 °C, RockImager RI1500 at 20 °C, Formulatrix, Inc., Waltham, MA, USA) for crystal growth. Crystallization progress was monitored automatically in the two imagers and manually using normal and polarized light microscopy (Eclipse E 400 Pol, Nikon, Melville, NY, USA). For the more successful hosting lipid, 10 μm-long rod-shaped crystals usually appeared after 16 hours in precipitants containing 19–26 %(v/v) tri-ethyleneglycol, 0.05–0.15 M glycine, 0.05–0.15 M ammonium sulphate, as detailed under Results.
All trials were performed in `duplicate' where the inverted commas highlight the need for the following qualification. Setting up an in meso crystallization plate with an 8-tip robot takes about 7 minutes. Thus, wells filled first, on the left hand side of the plate, remain open and exposed on the deck of the robot for considerably longer than wells on the right side of the plate, set up last, and are more likely to succumb to changes (mostly in solute concentration) due to equilibration with the ambient atmosphere. Since precipitant blocks are aligned usually with well A1 in the upper left corner, such an arrangement could introduce bias if only one such arrangement was used with each precipitant block. In an attempt to remove bias the entire process is repeated by filling a second, so-called `duplicate' plate but with the precipitant block rotated by 180° such that well H12 is now at the upper left corner. Very often we find hits that appear in conditions with the precipitant block oriented in the 0° or forward direction also appear in the same conditions on the `duplicate' plate with the block oriented in the 180° or reverse direction.
2.2.4. Harvesting
Harvesting was done as described.9, 20 Briefly, plates with crystals were removed from the incubator/imager and transferred to a 20 °C room. The cover glass of the glass sandwich plates was scored manually using a tungsten carbide glass cutter (Model: 633657, TCT Scriber & Glass Cutter, Silverline, Yeovil, UK) and gently removed from the well. To minimize changes in composition of well contents during harvesting, the mesophase bolus in the well was immediately overlain with 1 μL precipitant solution. Cryo-loops (Micro Mounts, MiTeGen, Ithaca, NY, USA), ranging in size from 30 to 100 μm, were used to harvest crystals. Crystals were directly cryo-cooled and stored in liquid nitrogen.9
2.2.5. Diffraction Measurements
Diffraction data were collected on GM/CA CAT beamline 23ID-B, the Advanced Photon Source, with a MAR 300 CCD detector using 1.033 Å X-rays. Data were collected using 1° oscillation and 1 s exposure per image, a collimated beam size of 10 μm21 and a sample-to-detector distance of 400 mm. Best diffraction to 2.8 Å was observed with crystals grown in the lipidic cubic phase prepared with 7.8 MAG.
3. Results and Discussion
3.1. Characteristics and Purification of the Protein
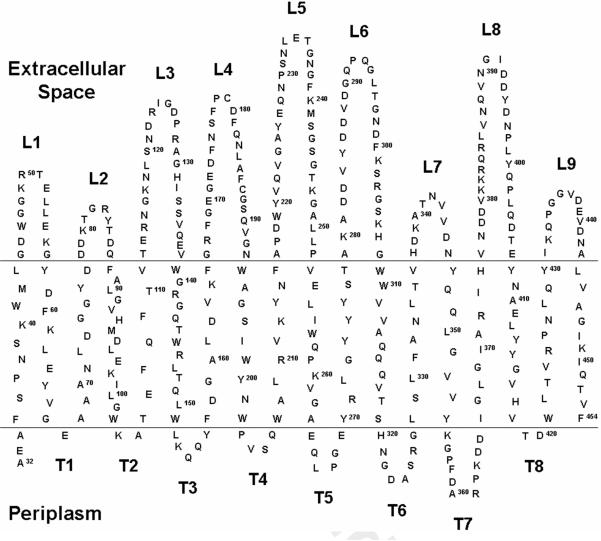
OprB is a porin from the outer membrane of P. aeruginosa. It is synthesized in vivo as a 454 residue protein with an N-terminal signal peptide.13 The mature protein includes residues 32 to 454. A membrane topology map for OprB (Figure 2) predicts it to exist as an 18-stranded β-barrel with short periplasmic turns (T) and extracellular loops (L) of varying sizes. Certain loops, such as L2 and L7, include just 10 residues whilst others, L5 for example, is predicted to be 37 amino acids long. The transmembrane strands range in size from 7 to 15 residues with an average strand length of 10 residues.
Figure 2.
Predicted topology map for OprB. The prediction was performed using the PRED-TMBB web server.22 Residues 1 to 31 correspond to the signal peptide and are not shown. Extracellular loops (L) and periplasmic turns (T) are numbered sequentially from the N- to the C-terminus. Predicted transmembrane β-strands are shown between horizontal lines representing the aqueous/apolar interfaces of the membrane. The topology suggests that the protein crosses the membrane as an 18-stranded β-barrel. The composition of the mature protein is as follows: 27 Ala, 17 Arg, 25 Asn, 36 Asp, 2 Cys, 28 Gln, 19 Glu, 47 Gly, 7 His, 12 Ile, 34 Leu, 24 Lys, 3 Met, 18 Phe, 16 Pro, 18 Ser, 19 Thr, 15 Trp, 22 Tyr, 34 Val.
An N-terminally hexa-histidine-tagged variant of the mature protein was produced recombinantly in E. coli. Lacking a signal sequences forces the protein into inclusion bodies which can be conveniently isolated for subsequent solubilisation in a strong denaturant. The protein, thus produced, was refolded by rapidly diluting the solubilized inclusion bodies into a detergent (LDAO) solution and was purified by sequential nickel affinity and size exclusion chromatography. The protein eluted from the size exclusion column as a symmetric Gaussian-shaped peak with an elution volume corresponding to a molecular weight of 152 kDa (Figure 2A). This suggests that the protein in solution exists as a dimer or a trimer, given that the molecular weight of the mature protein (plus its His-tag) is 51.7 kDa and that of an LDAO micelle ranges in size from 17 to 22 kDa23, 24. The purity of the protein was judged by SDS-PAGE (Figure 2B). At a low protein loading, a single band with an apparent molecular weight of 53 kDa was seen. Higher loading revealed a number of faster running contaminants. Based on these data the protein was estimated to have a purity in excess of 90 % and thus, considered suitable for crystallization trials. The protein could be concentrated as an optically clear solution to at least 46 mg/mL.
3.2. In Meso Crystallization
3.2.1. Default Hosting Lipid, Monoolein (9.9 MAG)
To date, the structures of two outer membrane proteins crystallized by the in meso method, have been reported.14, 15 In both cases, monoolein was used as the lipid creating the hosting mesophase from which crystals grew. Accordingly, monoolein was the lipid of first choice with which to perform crystallization trials on OprB. Initial trials were carried out using ten 96-condition screens and were performed `in duplicate' as described under Methods. All ten screen kits were commercially sourced. Initial trials were conducted under standard conditions at 20 °C with 50 nL OprB-loaded mesophase and 0.8 – 1 μL precipitant solution. A hit was registered in one of the JBS Membrane HT screen conditions that included 25 %(v/v) tri-ethylene glycol, 0.1 M ammonium sulphate and 0.1 M glycine. Small birefringent crystals appeared within a couple of days (Figure 4) that grew to a maximum size of 5 μm in two weeks. In the absence of a UV microscope or other means to ascertain if the crystals were made of protein, control plates were set up using protein-free buffer. These did not produce crystals suggesting that the micro-crystals observed were indeed composed of OprB.
Figure 4.
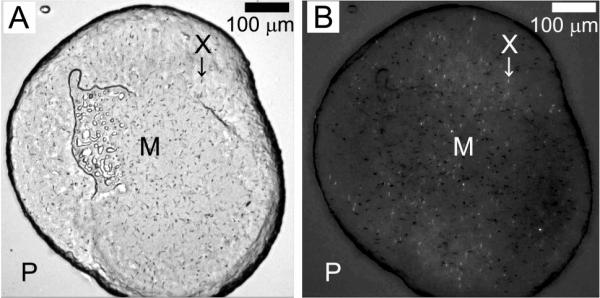
Micro-crystals of OprB growing at 20 °C in the lipidic cubic mesophase formed using monoolein. Small crystal are seen as dark flecks when viewed with normal light (A) and as bright flecks when viewed with polarized light (B). The contrast and brightness of the image in panel B have been adjusted to make the small crystals a little more obvious. The precipitant solution contained 25 %(v/v) tri-ethylene glycol, 0.1 M ammonium sulphate and 0.1 M glycine. Other conditions are as described in Section 2.2.3. The precipitant (P), mesophase bolus (M) and micro-crystals (X) are appropriately labelled.
Whilst the initial screen was successful in that it yielded crystals, the micro-crystals produced were not big enough to be useful in diffraction measurements. Optimization was called for that included varying hit condition ingredients in a systematic way. Thus, for example, glycine, tri-ethylene glycol and ammonium sulphate concentrations were varied in the ranges 0 to 0.2 M, 12 to 38 %(v/v), and 0 to 0.2 M, respectively. pH values from 4.6 to 7.6 in steps of 0.1 pH units were screened, and OprB concentrations from 8 to 46 mg protein/mL were tried. Trials were carried out at 4, 16 and 20 °C, and bolus volumes of 37, 50, 100 and 150 nL with 800 nL precipitant solution were tested. Well depth was adjusted by using 1, 2 and 3 spacers to change the geometry of the bolus and the precipitant/bolus interface. The objective of these assorted `optimization' trials, which involved a total of 8,256 conditions tested, was to grow bigger crystals. This proved successful to a degree in that best conditions produced rod shaped crystals 30 μm long (Figure 5). They appeared to be well formed and were clearly birefringent (Figure 5B) with obvious 3-dimensional bulk when viewed by microscope manually. Unfortunately, they were not considered of suitable size for use in diffraction.
Figure 5.
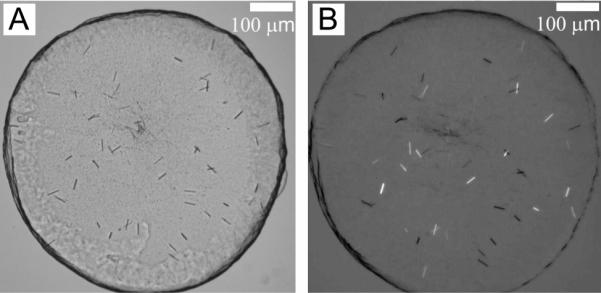
Crystals of OprB obtained in the lipidic mesophase formed by monoolein following extensive optimization. The precipitant used in this case contained 22 %(v/v) tri-ethylene glycol, 0.1 M glycine, 0.05 M ammonium sulphate, and 0.1 M sodium acetate, pH 5.0, and the trial was conducted using 50 nL mesophase and 800 nL precipitant solution at 20 °C for 14 days. Images recorded with normal and with polarized light are shown in panels A and B, respectively. Other conditions are as described in Section 2.2.3 and in the legend to Figure 4.
3.2.2. Rationally Designing a Hosting Lipid
3.2.2.1. Hydrophobic Matching
Despite extensive optimization, crystallization trials with monoolein did not produce crystals deemed suitable for diffraction. An alternative strategy for generating bigger crystals of diffraction quality was therefore considered that involved adjusting the characteristics of the hosting mesophase from which crystals grew. Crystal growth in meso has been described as involving a partitioning between two mesophase domains; one cubic and of high interfacial curvature, the other lamellar composed of stacked planar bilayers (Figure 1). The objective therefore is to favour partitioning of the target protein into the latter wherein it locally concentrates in a process that leads to nucleation and crystal growth. The relevant partition coefficient will depend on the characteristics of the mesophase microstructure which, in turn, will reflect the lipid used to create the mesophase. The latter is referred to as the hosting lipid. Another way to conveniently adjust mesophase character is to employ hosting lipids in combination, or to dope the hosting system with additive lipids. In this study, we chose to examine the effect that hosting lipids, used singly, have on crystallogenesis.
Cis-monounsaturated MAGs are the most common lipids used to create the cubic mesophase for in meso crystallogenesis, and to date, monoolein (9.9 MAG, Footnote 1) is the most tested (and successful) of the MAGs (www.mpdb.tcd.ie). Chain homologs of monoolein are available where the length of the acyl chain and position of the cis-olefinic bond on the chain vary. These, in turn, give rise to mesophases whose microstructure changes with the identity of the acyl chain. Some are commercially available; others have been synthesized and purified in-house.16, 17 For any to be suitable for in meso crystallization trials it is necessary that they form the cubic mesophase under conditions of full hydration at or around 20 °C.
At this juncture in our attempts to produce usable crystals of OprB it was necessary to consider which of the many available MAGs would be suitable for testing. A decision was arrived at by examining the hydrophobic match, or mismatch, between the target protein and the bilayer of the hosting mesophase. The logic behind this approach was that a good match would introduce less strain on the protein thereby creating a more native-like environment for the reconstituted target. A native conformation would, in turn, more likely produce a crystal of high quality. In the absence of a 3-dimensional structure for OprB, a best estimate of its membrane exposed hydrophobic dimensions was arrived at by using the corresponding values for homologs of known structure. OprB belongs to the maltoporin sub-group of porins of which crystal structures are available for two of its members.26–29 These include the maltose porin, LamB, from E. coli and Salmonella typhimurium, and the sucrose porin, ScrY, from S. typhimurium. An established algorithm (http://opm.phar.umich.edu/) was used to calculate hydrophobic thickness values based on crystal structures. These ranged from 23.4 Å for ScrY to 24.0 Å and 25.4 Å for LamB. Accordingly, a hydrophobic thickness for OprB of 23 to 25 Å was considered reasonable.
The thickness of the bilayer in the cubic phase of monoolein has been calculated at 32.3 Å (Footnote 2).30 It may be therefore that the mismatch between the hydrophobic thickness of OprB (23 – 25 Å) and the membrane of the cubic phase formed by monoolein was too great. As such, it was less than ideal as a hosting mesophase from which to grow crystals. Thus, a shorter chained MAG, with a cubic phase bilayer that better matched the hydrophobic thickness of OprB was deemed appropriate for the next round of crystallization trials. 7.7 MAG has a bilayer thickness of 25.8 Å (Footnote 2)30 which better matches the hydrophobic thickness of OprB. In separate studies, this MAG, with an acyl chain just 14 carbon atoms long, was shown to support in meso crystallization of a 22-stranded β-barrel outer membrane protein, the vitamin B12 transporter, BtuB.30 It was therefore considered a reasonable alternative to monoolein with which to continue crystallization trials on OprB.
3.2.2.2. In Meso Trials with Shorter Chain Hosting MAGs
Following the rationale outlined in the previous section, 7.7 MAG was tested in extensive crystallization trials with OprB. It failed to produce crystals.
Continuing this exploration of MAGs having acyl chains 14 carbon atoms long, the 6.8 MAG homolog was examined. Unfortunately, as with 7.7 MAG, no hits were observed with it either.
Thus far in the investigation, an 18-carbon MAG, monoolein or 9.9 MAG, produced small crystals while two 14-carbon MAG homologs did not support detectable crystallization of any type. As noted, there was good agreement between the predicted hydrophobic thickness of OprB and the bilayer thickness of the only representative of the 14-carbon MAGs for which we have reliable mensuration, 7.7 MAG. Nonetheless, crystals did not form in either of the 14-carbon MAGs tested. It is possible that the hydrophobic fit in this case was simply `too good' in the sense that the protein opted to remain in the hosting mesophase created by these 14-carbon MAGs and not partition into the ordered lattice of the crystal. It seemed reasonable therefore to investigate next MAGs with chain lengths longer than 14-carbons. The 16-carbon lipid, 8.8 MAG, was an obvious choice. After extensive in meso trials, while hits were observed with this lipid, the crystals formed were, at most, only 2 μm in maximum dimension (Figure 6).
Figure 6.
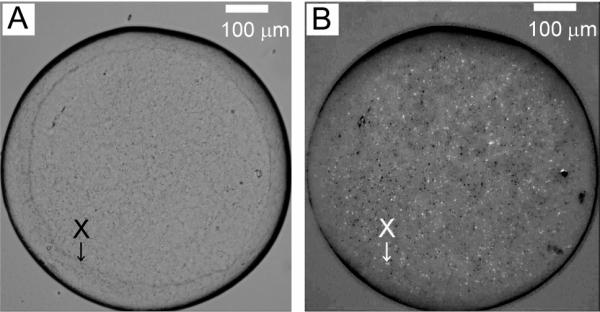
Crystals observed growing in the lipidic cubic phase formed by 8.8 MAG. Precipitant conditions include 21 %(v/v) tri-ethylene glycol, 0.1 M ammonium sulphate, 0.1 M glycine. Micro-crystals appeared in 3 days at 20 °C. Images recorded with normal and with polarized light are shown in panels A and B, respectively. Other conditions are as described in Section 2.2.3 and in the legend to Figure 4.
Fortunately, we had several MAGs to choose from within the 15-carbon homolog series and these were, in turn, tested. They included 6.9 MAG, 7.8 MAG and 8.7 MAG. The latter produced reasonably promising crystalline rods 50 μm long (Figure 7). Crystals appeared after 3 days and reached maximum size in a week. By contrast, 6.9 MAG grew large birefringent crystals that formed rapidly and were visible within 16 hours post set up.
Figure 7.
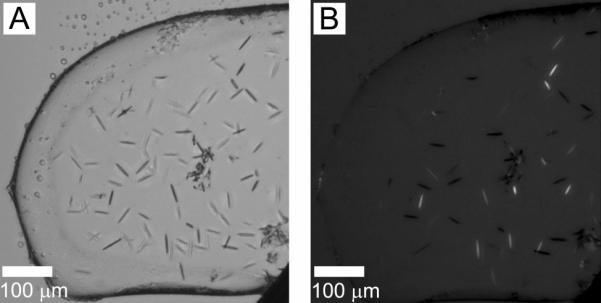
OprB crystals growing in the cubic mesophase formed by 8.7 MAG. Precipitant conditions include 20 %(v/v) tri-ethylene glycol, 0.08 M ammonium sulphate, and 0.15 M glycine. Crystals appeared on day 3 and reached maximum size of 50 μm on day 7. Images recorded with normal and with polarized light are shown in panels A and B, respectively. Other conditions are as described in Section 2.2.3 and in the legend to Figure 4.
They continued to grow and reached a maximum size of 40 μm × 100 μm in 7 days (Figure 8). These lath-shaped crystals were now of a size suitable for harvesting and for diffraction measurement. Alas, the crystals from 6.9 MAG did not diffract well, with measurable reflections out to only 12 Å.
Figure 8.
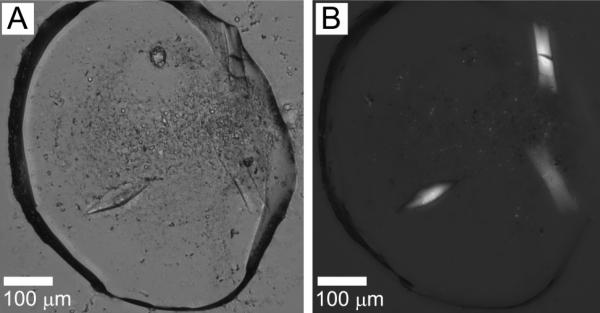
OprB crystals growing in the cubic mesophase formed by 6.9 MAG. Precipitant conditions include 21 %(v/v) tri-ethylene glycol, 0.05 M ammonium sulphate, and 0.15 M glycine. Crystals appeared after 16 h and grew to a maximum size of 40 μm × 100 μm in 7 days. Images recorded with normal and with polarized light are shown in panels A and B, respectively. Other conditions are as described in Section 2.2.3 and in the legend to Figure 4.
The biggest crystals of OprB grew in 7.8 MAG (Figure 9). These appeared as well formed, highly birefringent rods 100 to 300 μm long and 20 μm in diameter. Each well contained just a few crystals that were distributed in a way that made for facile harvesting. These crystals diffracted to 2.8 Å.
Figure 9.

OprB crystals growing in the cubic mesophase formed by 7.8 MAG. Precipitant conditions include 26 %(v/v) tri-ethylene glycol, 0.1 M ammonium sulphate, and 0.15 M glycine. Crystals appeared after 16 h and grew to a maximum size of 20 μm × 100 – 300 μm in 7 days. Images recorded with normal and with polarized light are shown in panels A and B, respectively. Other conditions are as described in Section 2.2.3 and in the legend to Figure 4.
3.2.3. A Recapitulation
The above results show clearly that the acyl chain characteristics of the MAG used to create the hosting mesophase has a profound effect on the in meso crystallogenesis of OprB. It is likely that these results are of relevance to other porins and, indeed, to other membrane proteins. It is accepted that a crystal does not equate to a solved structure. However, in the absence of a crystal there is no structure by the method of macromolecular crystallography. Nonetheless, the direction taken in the current investigation was dictated primarily by the size and visual appearance of the crystals that grew in meso with diffraction used only when crystals of a suitable size became available. In 7.8 MAG, crystals hundreds of micrometers in size that diffracted to 2.8 Å were produced. These are being used for structure determination, as will be reported on separately.
The thrust of the current study was to evaluate different MAGs and to establish how they influence the crystallogenesis of a membrane protein, in this instance an outer membrane porin. What emerges from the work is a very clear message that the acyl chain identity of the hosting lipid plays a significant role in the generation of diffraction-quality crystals. Specifically, we found that the 18-carbon MAG, monoolein, produced small crystals while the two 14-carbon homologs examined proved singularly ineffective crystallogenically. Like monoolein, the 16-carbon lipid also produced small crystals. However, MAGs with chains 15-carbon atoms long proved most interesting from a crystallogenesis perspective. Within that series, the 8.7 homolog grew 50 μm crystals. Much larger crystals, but of low diffraction quality, were obtained in 6.9 MAG. 7.8 MAG, by contrast, generated large, well formed crystals that diffracted to 2.8 Å.
3.3. Toward the Development of a Hosting Lipid Screen
The importance of having an assortment of MAGs, of the type described in this study, available for use in in meso crystallization trials is apparent from this study. The approach outlined in the current investigation was described as sequential screening where one MAG screen followed another in a rational manner. In practice, such hosting lipid screens should be run in parallel and be an integral part of the initial screening process. How many and which MAGs should be included in the hosting lipid screen is an open question. Certainly, at this juncture, monoolein ought to be the reference MAG and should be included in all hosting lipid screens. Based on the results of the current study, 7.8 MAG would also be a suitable candidate. Separately, we have found that this lipid produces crystals of other porins and of α-helical integral membrane proteins (unpublished work) and comes highly recommended. If space, time and materials permit 7.7 MAG should be included in the hosting lipid screen. It has produced crystals of a large, outer membrane protein (BtuB) and of an α-helical protein (bacteriorhodopsin).30 While it did not work with OprB, it has proven critical to the structure determination, using in meso-grown crystals, of a cytochrome c oxidase from Thermus thermophilus (unpublished work, J. Lyons, D. Aragão, T. Soulimane, M. Caffrey). Including a 16- and/or 17-carbon MAG is desirable but would not be considered essential for an initial hosting lipid screen.
3.4. How Hosting Lipid Affects on Crystallogenesis Might Come About
It is apparent that the acyl chain character of the MAG used to form the mesophase has a profound affect on the outcome of in meso crystallogenesis. To what can we ascribe this response? Reference has been made to the sensitivity of mesophase microstructure to lipid identity. Support for this statement is based on small-angle X-ray scattering measurements performed on the cubic phase prepared with an homologous series of MAGs of the type used in the current study.31–35 The data show expected behavior in that as chain length decreases so too does the thickness of the lipid layer that creates the apolar fabric of the cubic phase, when evaluated at a single temperature. Less intuitive perhaps is the finding that the aqueous channel diameter drops as chain length increases. This is consistent with a flattening and an attenuating curvature at the polar/apolar interface with the shorter-chain lipids. The response of the reconstituted protein to these affects will undoubtedly reveal itself in a differential crystallizability in meso, at the very least.
3.5. Expanding the Hosting Lipid Screen
The need for additional MAGs, beyond those used in the current study, with which to perform in meso crystallization trials is an obvious outcome of this work. With the exception of monoolein, all of the MAGs employed here were synthesized and purified in-house. A single research laboratory's resources are limited and not all possible MAGs can, or indeed, should be produced and tested. Nonetheless, in a proper hosting lipid screen MAGs having chain lengths in the 13- to 20-carbon range and with the olefinic bond positioned toward the middle of the chain (the so-called N.T MAG space) should be suitably sampled. With the recent successful application of different MAGs in in meso crystallization the need for a commercial supplier of such MAGs, of high quality and at an affordable price, is immediate.
Although lipid identity can be used to tailor phase behavior and microstructure, it is possible that the desired properties might not be accessible with a single lipid species in the temperature range of interest. In this case, it is possible to fine-tune by using mixtures of MAGs with different acyl chain lengths for which the mole ratio is adjusted to set the phase behavior and microstructure at the desired intermediate values.
The microstructure of the mesophase can be engineered over relatively wide limits to suit particular crystallogenesis needs by manipulating temperature and/or lipid identity and composition. However, the two metrics of the cubic phase - the polar and apolar compartment dimensions - are not independently adjustable and indeed are coupled tightly.35 Nonetheless, this feature of tunability is a valuable tool available to the crystal grower in search of a suitable lipid matrix with which to grow crystals. Thus, proteins with extramembranal domains that come in a variety of sizes can be accommodated as can those that originate from native membranes with different hydrophobic thicknesses.36
In addition to the need for a larger suite of MAGs with which to perform in meso crystallization, it is critically important to properly quantify the mesophase behavior and microstructure characteristics of these new lipids. As noted, this has been done most successfully to date using small-angle X-ray scatting.31–35 The information will be used to establish what is happening at a molecular level as the protein is reconstituted into the mesophase and as it migrates and phase separates in a process that ultimately leads to nucleation and crystal growth. With a mechanism in hand a more rational approach to in meso crystallogenesis, supported by increasingly reliable bioinformatics analyses of the target protein or complex, will become routine.
4. Conclusions
The data reported herein highlight the sensitive nature of the in meso crystallization process. Sensitive, that is, to the character of the acyl chain of the MAG that creates the bilayer of the hosting mesophase in and from which crystals grow. Not only are we witness to a profound crystal growth and quality response to acyl chain length but the position of the cis double bond along the chain matters too. Thus, a chain length disparity of just one carbon (in the 14- vs. 15-carbon homologs) meant either getting or not getting crystals of OprB. Further, in the case of the 15-carbon homologs, 8.7 MAG grew medium sized crystals, 6.9 MAG produced large crystals that diffracted poorly whilst 7.8 MAG generated large crystals diffracting to 2.8 Å.
It makes sense therefore that integral to in meso crystallogenesis should be a hosting lipid screen that caters to the varied nature of membrane proteins entering crystallization trials. Ideally, the screen should be a part of the initial trials and should include 7.8 MAG and 9.9 MAG at a minimum. Based on considerable favorable experiences in the author's lab, 7.7 MAG should also be considered. Reducing the number of precipitant solutions used will facilitate more extensive host lipid screening. A reasonable approach toward this end is to remove replicate conditions from commercial screen kits (http://c6.csiro.au).37
In support of more extensive hosting lipid screening efforts a wide assortment of MAGs should be available such that N.T MAG space can be properly covered. Finer tuning of the mesophase can be done using MAGs in combination and by doping with additive lipids.
The ready availability, ideally commercially, of a suite of MAGs of high quality and reasonable price will enable their extensive testing in the crystallogenesis community and the identification of specific hosting lipids that work reliably. It may be that particular MAGs will emerge for distinct subsets of membrane proteins that, in turn, will lead to a more directed and rational approach to crystallogenesis and structure determination.
In parallel with on-going crystallogenesis trials must be a program devoted to the mesophase behavior and microstructure characterization of new MAGs. This information must then be correlated with crystallogenesis results and bioinformatics on the target proteins, pre- and post-structure determination, for a truly rational approach to crystallization with the ultimate objectives of solving structures, understanding interactions and function, and exploiting that information for the benefit of mankind.
Figure 3.
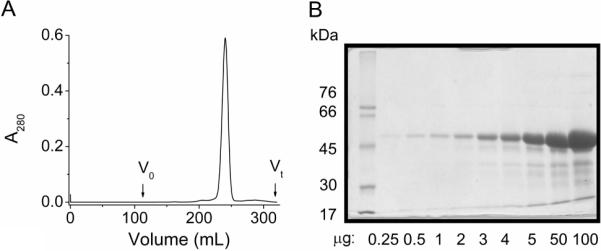
Purification of OprB. Size exclusion chromatographic analysis of OprB (A). Vo and Vt mark the void and total column volumes, respectively. The purity of the OprB used in crystallization trials is illustrated by SDS-PAGE analysis of a loading series (B). For PAGE analysis, samples were heated at 95 °C for 5 min in Laemmli buffer25 before loading on to a polyacrylamide gel composed of a 12 %(w/v) resolving and a 4 %(w/v) stacking gel. The latter was removed before staining with Coomassie Blue. Purity was estimated at >90 %.
Acknowledgements
The authors thank C. Arthur, J. Lyons and T. Smyth for important contributions to MAG synthesis, B. Sun for input regarding crystallogenesis, and V. Cherezov and D. Aragão for valuable assistance with macromolecular crystallography. This work was supported by grants from Science Foundation Ireland (07/IN.1/B1836), FP7 COST Action CM0902 and the National Institutes of Health (GM75915, P50GM073210). Diffraction data were collected at the GM/CA-CAT beamline, Advanced Photon Source. Use of the APS is supported by the US Department of Energy (DE-AC02-06CH11357). GM/CA-CAT is funded by the US National Institutes of Cancer (Y1-CO-1020) and General Medical Sciences (Y1-GM-1104).
Footnotes
The in meso crystallogenesis reported on herein makes use of MAGs containing cis-monounsaturated fatty acids. A shorthand system for describing the chemical constitution of these lipids is referred to as the N.T MAG notation.38 This is based on a simplistic view of the MAG molecule as an object consisting of a head, a neck, and a tail with the latter two joined by a trunk. In this representation, the head corresponds to the glycerol head group. It is in ester linkage to the neck corresponding to that part of the acyl chain extending from its carboxyl carbon to the first carbon of the olefin. The trunk is the cis - double bond. The tail extends from the second carbon of the olefin to the chain's methyl terminus. In the N.T MAG notation, N and T correspond to the number of carbon atoms in the neck and tail, respectively. The total number of carbon atoms in the chain is the sum of N and T. Thus, 11.7 MAG represents monovaccenin, a MAG with a fatty acyl chain 18 carbon atoms long where the cis - double bond resides between carbon atoms 11 and 12. It is an olefinic isomer of 9.9 MAG, commonly known as monoolein.
The bilayer thickness of the cubic phase formed by 9.9 MAG (monoolein) under conditions of full hydration reported here is for a sample temperature of 40 °C. The corresponding value at 20 °C is 35.6 Å.39 The reason for reporting bilayer thickness at such a high temperature is because this is the lowest temperature value available for 7.7 MAG 30 with which it is compared.
References
- (1).Landau EM, Rosenbusch JP. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Caffrey M. Annu. Rev. Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- (3).Raman P, Cherezov V, Caffrey M. Cell. Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- (5).Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chien EYT, Liu W, Zhao Q, Katritch V, Won Han G, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Caffrey M, Cherezov V. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hancock RE, Carey AM. FEMS Microbiol. Lett. 1980;8:105–109. [Google Scholar]
- (11).Wylie JL, Worobec EA. J. Bacteriol. 1995;177:3021–3026. doi: 10.1128/jb.177.11.3021-3026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Trias J, Rosenberg EY, Nikaido H. Biochim. Biophys. Acta. 1988;938:493–496. doi: 10.1016/0005-2736(88)90148-4. [DOI] [PubMed] [Google Scholar]
- (13).Wylie JL, Bernegger-Egli C, O'Neil JD, Worobec EA. J. Bioenerg. Biomembr. 1993;25:547–556. doi: 10.1007/BF01108411. [DOI] [PubMed] [Google Scholar]
- (14).Cherezov V, Yamashita E, Liu W, Zhalnina M, Cramer WA, Caffrey M. J. Mol. Biol. 2006;364:716–734. doi: 10.1016/j.jmb.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Cherezov V, Liu W, Derrick JP, Luan B, Aksimentiev A, Katritch V, Caffrey M. Proteins. 2008;71:24–34. doi: 10.1002/prot.21841. [DOI] [PubMed] [Google Scholar]
- (16).Coleman BE, Cwynar V, Hart DJ, Havas F, Mohan JM, Patterson S, Ridenour S, Schmidt M, Smith E, Wells AJ. Synlett. 2004;2004:1339–1342. [Google Scholar]
- (17).Caffrey M, Lyons J, Smyth T, Hart DJ. In: Current Topics in Membranes. Chapter 4. Larry D, editor. Vol. 63. Academic Press; 2009. pp. 83–108. [Google Scholar]
- (18).Cheng A, Hummel B, Qiu H, Caffrey M. Chem. Phys. Lipids. 1998;95:11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- (19).Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- (20).Hofer N, Aragao D, Caffrey M. Biophys. J. 2010;99:23–25. doi: 10.1016/j.bpj.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Fischetti RF, Xu S, Yoder DW, Becker M, Nagarajan V, Sanishvili R, Hilgart MC, Stepanov S, Makarov O, Smith JL. J. Synchrotron Radiat. 2009;16:217–225. doi: 10.1107/S0909049508040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. Nucleic Acids Res. 2004;32:400–404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Strop P, Brunger AT. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Herrmann KW. J. Phys. Chem. 1962;66:295–300. [Google Scholar]
- (25).Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- (26).Wang YF, Dutzler R, Rizkallah PJ, Rosenbusch JP, Schirmer T. J. Mol. Biol. 1997;272:56–63. doi: 10.1006/jmbi.1997.1224. [DOI] [PubMed] [Google Scholar]
- (27).Schirmer T, Keller TA, Wang YF, Rosenbusch JP. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- (28).Meyer JE, Hofnung M, Schulz GE. J. Mol. Biol. 1997;266:761–775. doi: 10.1006/jmbi.1996.0823. [DOI] [PubMed] [Google Scholar]
- (29).Forst D, Welte W, Wacker T, Diederichs K. Nat. Struct. Biol. 1998;5:37–46. doi: 10.1038/nsb0198-37. [DOI] [PubMed] [Google Scholar]
- (30).Misquitta LV, Misquitta Y, Cherezov V, Slattery O, Mohan JM, Hart D, Zhalnina M, Cramer WA, Caffrey M. Structure. 2004;12:2113–2124. doi: 10.1016/j.str.2004.09.020. [DOI] [PubMed] [Google Scholar]
- (31).Briggs Briggs, J. PhD Thesis. The Ohio State Univ; 1994. [Google Scholar]
- (32).Briggs J, Caffrey M. Biophys. J. 1994;67:1594–1602. doi: 10.1016/S0006-3495(94)80632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Briggs J, Caffrey M. Biophys. J. 1994;66:573–587. doi: 10.1016/s0006-3495(94)80847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Briggs J, Chung H, Caffrey M. J. Phys. II France. 1996;6:723–751. [Google Scholar]
- (35).Caffrey M. J. Struct. Biol. 2003;142:108–132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]
- (36).Sharpe HJ, Stevens TJ, Munro S. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Newman J, Fazio VJ, Lawson B, Peat TS. Cryst. Growth Des. 2010;10:2785–2792. [Google Scholar]
- (38).Misquitta Y, Caffrey M. Biophys. J. 2001;81:1047–1058. doi: 10.1016/S0006-3495(01)75762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Liu W, Caffrey M. J. Struct. Biol. 2005;150:23–40. doi: 10.1016/j.jsb.2004.12.007. [DOI] [PubMed] [Google Scholar]