Abstract
CD1 activates T cells, but the functions and size of possible human T cell repertoires recognizing each of the CD1 antigen presenting molecules remain unknown. Using an experimental system that bypasses major histocompatibility complex (MHC) restriction and the requirement for defined antigens, we found that polyclonal T cells responded at higher rates to cells expressing CD1a compared to CD1b, CD1c or CD1d. Unlike invariant NKT cells, the CD1a-autoreactive repertoire contains diverse T cell receptors. Functionally, many CD1a-autoreactive T cells home to skin, where they produce interleukin 22 (IL-22) in response to CD1a on Langerhans cells. The strong and frequent responses among genetically diverse donors define CD1a-autoreactive cells as a normal part of the human T cell repertoire and CD1a as a target of TH22 cells.
Introduction
Four members of the human CD1 system, CD1a, CD1b, CD1c and CD1d, bind and present diverse lipids from mammalian cells and bacteria. It is therefore thought that the CD1 system allows T cells to survey CD1 expressing antigen presenting cells (APCs) for changes in lipid content caused by infection, inflammation or malignancy. This view is supported by several types of experimental studies. A particular subpopulation of CD1-restricted T cells, known as invariant NKT cells, was discovered based on their expression of conserved T cell receptors (TCRs) with limited Vβ 1 or Vα chains 2, 3. Therefore, using CD1d tetramers, the invariant TCRs of NKT cells serve as a defining surface marker that allows enumeration, tracking and measurement of effector functions on a single cell or population basis 4–7. Germ line deletion of CD1d or the joining (J) segment of the invariant Vα chain allows the selective deletion of NKT cells 8, 9, so that the influence of NKT cells in mouse models of autoimmunity, infection, tumor growth and other diseases has been broadly assessed 10. These approaches have led to the conclusion that invariant NKT cells are pre-primed memory T cells that secrete large amounts of interferon-γ (IFNγ) and work directly or through modulation of NK cells or dendritic cells to control infection and promote tumor regression 10. However, for the remainder of CD1d-restricted T cells that do not express the invariant TCR or for T cells that recognize the group 1 CD1 proteins (CD1a, CD1b and CD1c), there are no broadly useful cell surface markers or genetic means of deletion. Therefore, studying the immunological function of diverse CD1 restricted T cells outside the context of long term T cell clones remains a challenge.
The evolution of CD1 gene families in mammals suggest that human CD1a, CD1b and CD1c proteins and their non-human orthologs likely play important roles in the immune response. Although muroid rodents lack group 1 proteins, nearly all mammalian genomes harbor one or more CD1a genes, and no mammalian species is known to have survived without CD1 genes 11–13. The retention of large CD1 gene families in most mammals suggests that each type of CD1 gene has non-redundant and physiologically important roles14. This evolutionary hypothesis is supported by molecular and cellular evidence showing that CD1a, CD1b, CD1c and CD1d differ with regard to transcriptional regulation 15, patterns of tissue expression 16, subcellular trafficking 17 and antigen groove size 18.
Whether the clear differences in tissue distribution or cell biology of each CD1 protein translate into functional differences in the responding T cell populations is unknown. Two decades after the identification of human CD1-autoreactive T cells 19, nearly all insights into their possible functions are inferred from several long term T cell clones or lines rather than direct measurements of polyclonal T cells from blood and tissues. Clonal analysis has been successful for assigning certain unchanging aspects of phenotype, like TCR structure, CD4 and CD8 co-receptor expression 20, 21, antigens recognized 22 and molecular mechanisms of activation 18, 23. However, long term culture of human T cells promotes selective outgrowth of clones that have in vitro growth advantages and are unlikely to be representative of in vivo repertoires. This longstanding and basic gap in current knowledge persists due to limitations of even the best available reagents for tracking fresh polyclonal T cells, especially CD1 autoreactive T cells. Autoreactivity is a key feature of CD1-restricted T cells, which likely represents their ability to recognize endogenous lipid antigens 19, 24–27. Lipid self antigens have been identified 22, but the extent to which any one antigen can be used to track the larger pool of autoreactive T cells is unknown. In addition, tracking of autoreactive T cells with natural antigens loaded onto CD1 tetramers has been limited, because weak self agonists generally do not reliably confer adequate avidity to allow staining of TCRs 28.
We sought to overcome these problems by designing a system in which a cell type is used that provides a diverse pool of lipid antigens for loading onto CD1 expressed at high levels. For study of genetically diverse humans, a second design consideration was to minimize or remove the normally strong alloreactive responses to major histocompatibility complex (MHC) class I and II proteins that would confound CD1-reactive responses. We transfected human CD1 molecules into human myelogenous leukemia cells (K562 cells) that have low to absent levels of MHC 29, 30, resulting in APCs that can be universally used with MHC mismatched subjects. These cells presumably express a wide array of self lipid antigens, allowing the broad detection of CD1 autoreactivity without requiring prior knowledge of antigen structures. This method detected stronger and more frequent responses to CD1a than to any other human CD1 antigen presenting molecule. We found that CD1a autoreactive T cells are fundamentally different from invariant NKT cells based on their TCR patterns and effector functions. Further, CD1a-autoreactive T cells expressed skin homing markers and could be isolated from the skin and activated by epithelial CD1a-expressing Langerhans cells (LC). These studies identify CD1a-autoreactive cells as a subset of the human T cell repertoire and define CD1a as a target of TH22 cells, suggesting a new model of Langerhans cell – T cell interactions in epithelial homeostasis.
Results
CD1a-autoreactive T cells in peripheral blood
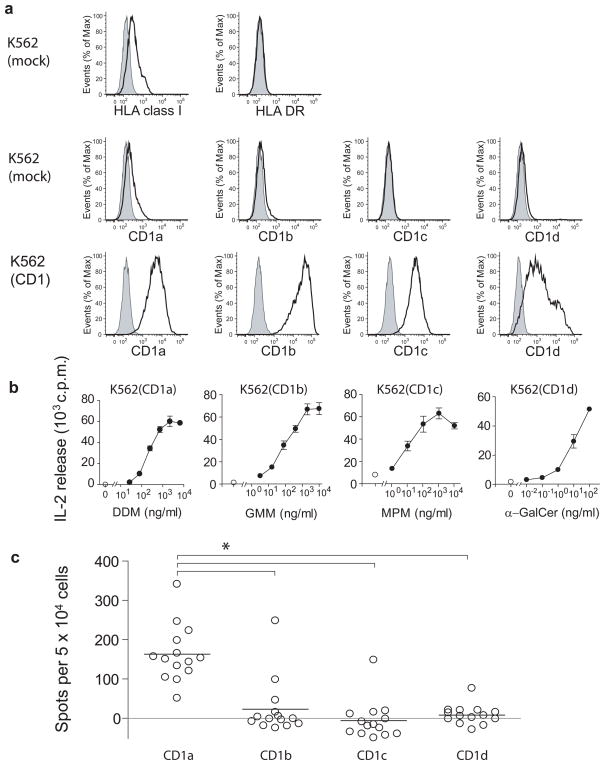
CD1 autoreactivity is defined as the specific recognition of CD1 in the absence of exogenous lipid antigen. Studies of CD1d-reactive NKT cells and CD1a-, CD1b- and CD1c-restricted T cell clones suggest that autoreactivity involves the recognition of an endogenous lipid antigen presented by CD119, 31, 32. To study human CD1-reactive T cells, we developed a system which can measure the CD1 autoreactivity of non-clonal T cell populations ex vivo. Using K562 cells with low surface density of MHC proteins, we transfected each of the genes encoding CD1a, CD1b, CD1c and CD1d, resulting in high surface expression of these molecules (Fig. 1a). APCs transfected with an empty vector control (mock) activated purified human T cells at frequencies of approximately 1 in 10,000 confirming the low alloreactive potential of these APC (Supplementary Fig. 1). The T cell activating function of each transfected CD1 molecule was confirmed by the presentation of known exogenous lipid antigens to CD1-restricted control T cell lines (Fig. 1b). To determine whether CD1-restricted T cells can be detected in human peripheral blood, we recruited 14 blood donors. After one or two in vitro stimulations with autologous dendritic cells (DCs) expressing CD1a, CD1b, CD1c and CD1d, the CD1 reactivity of polyclonal T cells was assessed by Elispot assays of IFN-γ secretion, using CD1 transfected K562 cells as APCs (Fig. 1c). A donor was considered to respond when the number of spots generated after contact with CD1-transfected K562 was higher than the background response to mock transfected K562 as assessed by the student’s t test (P<0.05). Using this criterion, three of 14 donors responded to CD1b and only one of 14 donors to CD1c or CD1d. The low responses were not due to the inability of K562 to activate responses through CD1b, CD1c or CD1d, because K562 cells efficiently stimulated CD1b and CD1c autoreactive T cell lines and CD1d autoreactive NKT cell clones (Supplementary Fig 2.) In addition, we found that K562-CD1b presented C80 GMM, an antigen presented only after endosomal recycling 33, indicating that K562 cells can survey endosomal antigens (data not shown). Remarkably, 14 out of 14 donors responded to K562-CD1a, and the mean responses were significantly higher than those against CD1b, CD1c or CD1d (P<0.01, Dunnett’s Multiple Comparison Test after one-way ANOVA).
Figure 1. A population study of CD1 autoreactive cells in blood of human donors.
(a) Surface expression of MHC and CD1 in K562 cells stably transfected with vector only (mock) or with vector containing the indicated human CD1 gene. Open histograms: MHC or CD1 staining, filled histograms: Isotype control. Results are representative of three or more experiments. (b) HT2 bioassay of IL-2 release in the supernatant of K562 cells incubated for 24 h with T cell lines recognizing CD1a and dideoxymycobactin (DDM), CD1b and glucose monomycolate (GMM), CD1c and mannosyl phosphomycoketide (MPM) or CD1d and α galactosyl ceramide (mean ± S.D.). Results are representative of three or more experiments. (c) Elispot of IFNγ release in polyclonal cell cultures from 14 donors stimulated in vitro with autologous DCs and analyzed for CD1 reactivity using K562-CD1 as APCs. IFNγ release is depicted as mean number of spots after subtraction of background spots formed in response to K562 transfected with vector alone. * (P<0.01) Dunnett’s Multiple Comparison Test after one-way ANOVA.
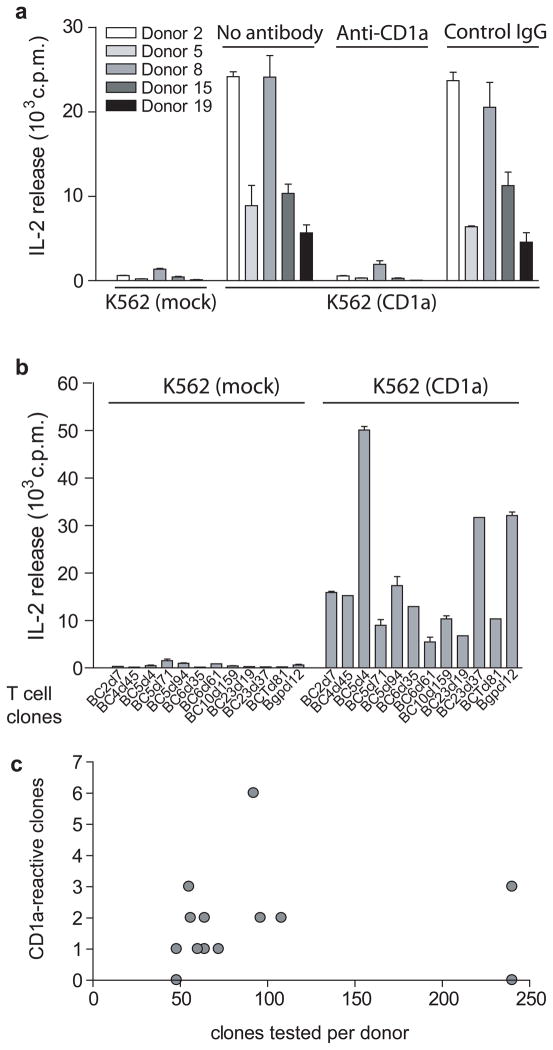
Considering that, unlike NKT cells, CD1a-autoreactive T cells have never been detected at the polyclonal level 4, 5, we undertook a series of analyses aimed at confirming the molecular target of recognition and more accurately measuring the frequency of responding cells. In a subset of five donors, we expanded responding T cells in the presence of dendritic cells and tested restriction of the resulting short term T cell lines with K562-CD1a in the presence or absence of an anti-CD1a blocking mAb. In all cases the responses required CD1a on the APC and were blocked to background level with anti-CD1a, indicating that the responses were dependent on CD1a and not on other cell surface determinants (Fig. 2a). Next, we performed a clonal precursor frequency analysis ex vivo in a separate group of 14 donors using autologous DCs as stimulator cells. We generated a panel of 1291 T cell clones detectably proliferating in response to autologous DCs and determined what percentage of autoreactive T cell clones secreted interleukin 2 (IL-2) in response to the K562-CD1a cells as compared to mock transfectants (Fig. 2b). One in every 50 autoreactive T cell clones, in samples from 12 out of 14 donors, was found to be CD1a-autoreactive (Fig. 2c). The first reported CD1a-autoreactive T cell clone expressed an αβ TCR, while lacking CD4 and CD8 coreceptor expression 19, but later examples of CD1a-autoreactive T cell clones expressed CD8 34. Among 12 clones that could be unambiguously analyzed for co-receptor expression, we found 11 CD4 single positive clones, one CD8 single positive clone and no CD4−CD8− clones.
Figure 2. Autoreactive T cells in the blood recognize the CD1a protein.
(a) Polyclonal T cell cultures were incubated with K562 cells transfected with vector only (K562 mock), K562-CD1a cells or K562-CD1a cells pretreated with anti-CD1a blocking antibody or isotype control. After 24 hrs supernatant was analyzed for IL-2 release using HT2 bioassay (mean ± S.D.) Results are representative of three or more experiments for each T cell line. (b) T cell clones were incubated with K562 cells transfected with vector only (K562 mock) or K562-CD1a for 24 hrs and IL-2 release was measured using HT2 bioassay. Each clone was tested in two or more experiments. (c) 1291 T cell clones were generated from 14 donors by ex vivo limiting dilution and tested for CD1a response as in (b) or by incubating with K562-CD1a cells pretreated with anti-CD1a blocking antibody or isotype control and enumerating responding cells by IFN-γ Elispot assay.
Invariant NKT cells recognizing CD1d are defined by Vα24Jα18 containing TCRα, but it is unknown whether T cells recognizing CD1a are conserved or diverse because only two Vα and Vβ pairs have been reported 27. Sequencing of the TCRα and β chains showed that none of the CD1a-reactive clones expressed the invariant Vα24 (TRAV10) or Vβ11 (TRBV25) found on human NKT cells. Although several CD1a-autoreactive clones shared the same variable regions and even joining regions in the TCRα or β, none of the clones expressed identical CDR3 sequences (data not shown). We conclude that CD1a-autoreactive T cells are common in the peripheral blood of humans and do not show the highly conserved TCR sequences seen in invariant NKT cells.
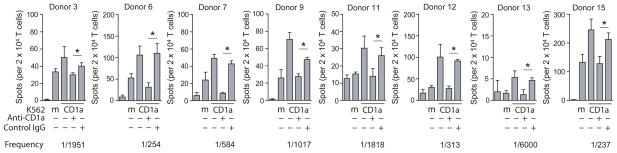
Because CD1a-autoreactive T cells were abundant in these cultures, we hypothesized that they might be a pre-primed T cell population that is detectable without expansion by autologous DCs. Using IFNγ Elispot assays we detected responses to K562-CD1a in purified CD45RO+ memory T cells in eight out of eight random donors (Fig. 3), and antibody blocking experiments showed that CD1a was necessary and sufficient for the response in all cases. Based on the fraction of the response that was blocked by anti-CD1a compared to control IgG, the frequency of CD1a autoreactive T cells among peripheral blood memory T cells is estimated to be between 0.02 and 0.4%. However, this could be an underestimate, considering that not all CD1a-reactive T cells may produce IFNγ. In agreement with the prior analysis using DC stimulated PBMC (Fig. 1c), the direct ex vivo analysis showed that T cell responses against CD1a were of greater magnitude than those against other CD1 isoforms (Supplementary Fig. 3). Collectively, these results show that CD1a-autoreactive T cells are abundant in the blood without immune stimulation and present in most or all donors, so that they can be considered to constitute a subset of the normal humanαβ T cell repertoire.
Figure 3. Quantitative detection of CD1a-autoreactive memory T cells.
CD45RO+ T cells from eight blood bank donors were incubated with K562 cells transfected with CD1a or vector only (mock, m). K562-CD1a cells were pre incubated with anti-CD1a or control IgG for 1 h before adding T cells and IFNγ-secreting cells were enumerated by Elispot assay (mean ± S.D.). * (P<0.05, Student’s t-test, two-tailed)
CD1a-autoreactive T cells home to the skin
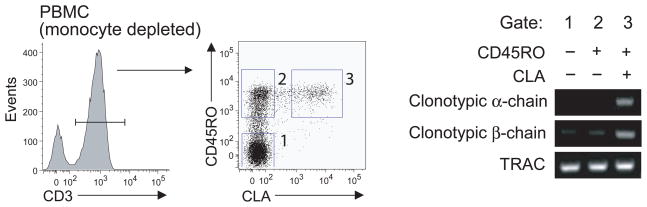
The expression of CD1a in humans is restricted to thymocytes, myeloid DCs and Langerhans cells (LCs). In the periphery, CD1a is found predominantly in the skin, where epidermal LCs express CD1a at extremely high cell surface density 35. Owing to high cellular density and large surface area 36, LCs form a nearly contiguous network of high density CD1a-containing membranes in the epidermis 37. The localization of CD1a in the skin led us to hypothesize that CD1a-autoreactive T cells in the peripheral circulation might normally home to and localize within the skin. We first determined whether CD1a-autoreactive T cells recovered from the blood express skin-homing markers. Because surface expression of homing markers are rapidly altered during in vitro culture 38, peripheral blood T cells were sorted directly ex vivo based on the expression of the CD45RO memory marker and cutaneous lymphocyte antigen (CLA), a carbohydrate epitope that identifies T cells that participate in immune responses in the skin39.
For this experiment we took advantage of the serendipitous finding that donor 8 harbored a CD1a-autoreactive T cell clone that was isolated from three independent samples, which indicates that these cells had undergone clonal expansion in vivo. The clonality was confirmed by the expression of identical TCRα and TCRβ chains in each of the isolates, and also because this clone had two rearranged TCRα chains, one of which had a CDR3 out of frame (data not shown). After validating clonotypic primers for the α and β chain of this TCR, we could measure the presence of this CD1a-specific clone within the naïve (CD45RO− CLA−), memory (CD45RO+ CLA−) and skin-homing memory (CD45RO+ CLA+) T cell fractions of the peripheral blood. PCR products for the clonotypic primers were almost exclusively found in the CD45RO+ CLA+ fraction (Fig. 4), indicating that, in this donor, CD1a-reactive T cells are CLA+ memory T cells, and further suggesting that CD1a-reactive T cells can be part of the skin homing T cell subset.
Figure 4. CD1a-autoreactive T cells in the blood express skin homing markers.
RNA was extracted from CD45RO−CLA−, CD45RO+CLA−, CD45RO+CLA+ T cells and analyzed by RT PCR with primers validated to be specific for the CDR3 regions of TCR α and β chains. TCRα constant region primers (TRAC) were used as a control for input cDNA.
CD1a dependent IL-22 production
Previous studies have described CD1a-autoreactive T cell clones mainly in the context of pathologic conditions associated with autoimmunity 31, 32 or allergy 40. However, detection of polyclonal T cells recognizing CD1a among many human blood donors suggests that they fulfill some physiological function without immune provocation or dysregulation. Lacking any precedent for possible effector functions associated with CD1a-autoreactive T cells, we had initially screened for the possible existence of a repertoire using IFN-γ (Fig. 1) and IL-2 (Fig. 2) because these are somewhat promiscuously produced by T cell subsets and so increased the chance of discovering the repertoire. We next screened a broader panel of cytokines that might provide hints at specific effector functions. We tested for candidate cytokines associated with TH1, TH2 and TH17 subsets of the MHC-restricted repertoire as well as cytokines as well as cytokines produced by CD1d restricted NKT cells. The dominant cytokine normally produced by NKT cells activated with foreign antigens is IFNγ. However, autoreactive NKT cell responses are associated with low levels of IFNγ and interleukin 4 (IL-4) and instead produce high levels of granulocyte-macrophage colony stimulating factor (GM-CSF) and interleukin 13 (IL-13) in the absence of IL-2 10, 41.
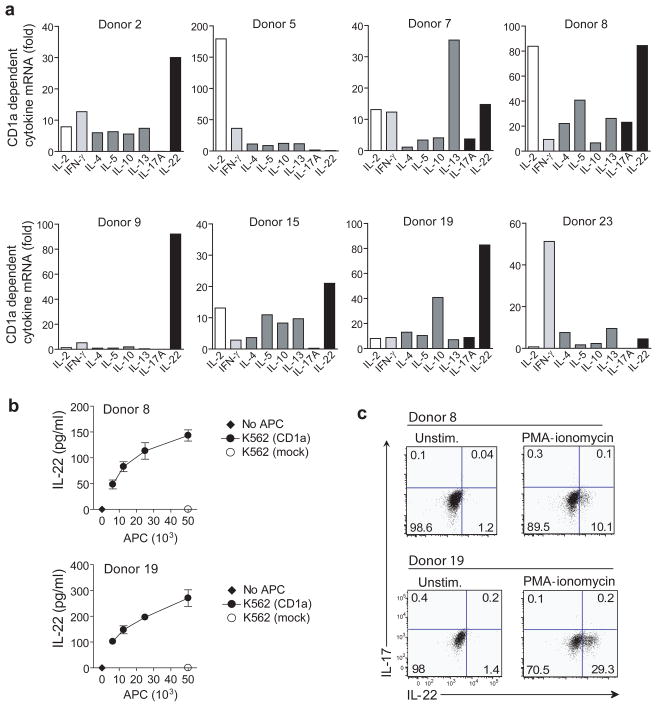
CD1a reactive T cell lines were stimulated with K562-CD1a cells, and cytokine mRNAs were measured by real time PCR. To ensure that CD1a was both necessary and sufficient to induce measured responses, we compared the mRNA levels in response to K562-CD1a treated with control IgG to K562-CD1a treated with anti-CD1a blocking antibody. Two out of eight donors tested showed a dominant upregulation of IFNγ or IFNγ and IL-2, whereas five other donors showed an unexpected pattern, characterized by a strong upregulation of interleukin 22 (IL-22) (Fig. 5a). IL-22 protein secretion was also detected in a dose-dependent manner in response to CD1a (Fig. 5b).
Figure 5. CD1a-dependent IL-22 production.
(a) K562-CD1a cells were preincubated with anti-CD1a or control IgG and then co-cultured with polyclonal CD1a-autoreactive T cell cultures for 6 hrs at a K562 to T cell ratio of 1 : 10. CD1a dependent cytokine gene upregulation was measured by real-time PCR. Samples were normalized to β-actin. (b) T cell lines from two donors that showed upregulation of IL-22 transcripts were incubated with K562 vector only (mock) or increasing numbers of K562-CD1a. After 24 h the supernatant was analyzed for IL-22 protein by ELISA (mean ± S.D.). Results are representative of two independent experiments for each T cell line. (c) T cell lines from the same donors were analyzed for intracellular IL-17 and IL-22 staining in response to stimulation with PMA-ionomycin.
We considered that IL-22 might be an important effector function of the CD1a-autoreactive repertoire because this cytokine is normally restricted to a limited subset of T cells, and both IL-22 and CD1a were previously suspected to play roles in skin immunity. IL-22 is a member of the IL-10 cytokine family and acts on non-hematopoietic epithelial cells, such as keratinocytes of the skin 42, where it triggers the production of antimicrobial peptides and expression of genes involved in cellular differentiation and survival 43, 44. It is therefore thought to be involved in early host defense against microbial pathogens and in the homeostasis of epithelia. In mice, IL-22 is often detected in TH17 cells 45, but human T cells known as ‘IL-22 only’ or TH22 cells can produce IL-22 in the absence of IL-1746–49. In our polyclonal analysis, the lack of significant CD1a-dependent upregulation of IL-17 mRNA in most donors suggested that these two cytokines are not coordinately upregulated in human CD1a-autoreactive T cells (Fig. 5a). Single cell level intracellular cytokine staining revealed that even a strong stimulus like PMA-ionomycin does not induce significant production of IL-17 in two IL-22 producing CD1a autoreactive T cell lines (Fig. 5c), while the same stimulus induces production of both cytokines in the T cell fraction of total PBMC (data not shown). These data support the idea that IL-17 and IL-22 represent effector molecules produced by distinct T cell population in humans, and that CD1a-autoreactive cells fall into the IL-22 producing subset.
IL-22 producing T cell populations sometimes show polyfunctional cytokine response, producing not only IL-17, but also IL-13, IL-2 and in some cases IFN-γ 47–49. Although not as prominent as IL-22, IFN-γ was present in CD1a-autoreactive populations (Fig. 1 and 5), which might have been produced either by TH1-like CD1a-restricted cells, or from clones that produce both IFN-γ and IL-22 47–49. A set of CD1a-autoreactive clones, selected on the basis of their production of IFN-γ and validated for clonality by Vβ PCR analysis, expressed either IFN-γ alone or IFN-γ and IL-22 in varied ratios in response to CD1a (Supplementary Fig. 4). This provides direct evidence for dual cytokine producing CD1a-autoreactive T cells at the clonal level, and suggests that our earlier measurements of CD1a-dependent IFN-γ responses at the polyclonal level, likely reflect both TH1 and TH22/TH1 cells 49.
Skin-homing TH22 cells contain CD1a-autoreactive cells
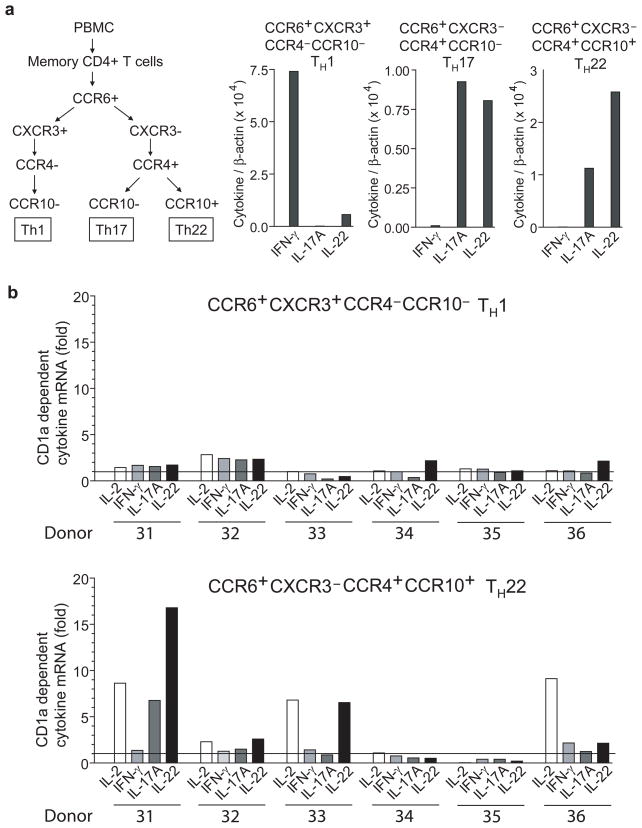
Production of IL-22 is only one feature of an emerging definition of a T helper cell subset, TH22, which is also characterized by expression of the aryl-hydrocarbon receptor 50 and expression of the chemokine receptors CCR6, CCR4 and CCR10, which promote skin homing 47, 48. Although antigen presenting molecules or other molecular targets recognized by TH22 cells are not well characterized, CD1a represents a good candidate because it is localized in skin, where TH22 cells normally home. We found that CD1a-restricted T cell lines express the aryl hydrocarbon receptor (Supplementary Fig. 5). To directly address if CD1a-autoreactive T cells represent a part of the TH22 subset and their relative abundance in this subset as compared to TH1 or TH17 cells, we sorted memory CD4+ T cells into the following fractions: CCR6+CXCR3+CCR4−CCR10−, CCR6+CXCR3−CCR4+CCR10− and CCR6+CXCR3− CCR4+CCR10+. The cytokine profiles measured by RT-PCR matched prior results obtained with ELISA 47, 48 and confirmed that these three sorted populations are enriched for TH1, TH17 and TH22 cells, respectively (Fig. 6a). After a single in vitro expansion with DCs, we detected CD1a-dependent upregulation of cytokine mRNA (fold increase >2) in five out of six donors, and the fraction associated with TH22 cells (CCR6+CCR4+CCR10+) showed the strongest and most frequent CD1a responses (Fig. 6b and data not shown). In addition, similar to our previous analysis of CD1a-autoreactive T cell cultures (Fig. 5a), IL-2 and IL-22 were the most dominantly upregulated cytokines in CCR4+CCR6+CCR10+ CD4+ T cells. Thus, CD1a-autoreactive T cells are found in the human TH22 subset, which defines CD1a as a target for TH22 cells.
Figure 6. TH22 subset contains CD1a-autoreactive T cells.
(a) Memory CD4+ T cells from six blood donors were sorted into CCR6+CXCR3+CCR4−CCR10−, CCR6+CXCR3−CCR4+CCR10−, and CCR6+CXCR3−CCR4+CCR10+ fractions and stimulated for 6 h with OKT3 to assess the cytokine profiles by real time PCR. (b) After a single in vitro expansion with DCs, sorted T cell fractions were co-cultured for 6 h with K562-CD1a preincubated with anti-CD1a or control IgG at a K562 to T cell ratio of 1 : 10. CD1a dependent cytokine gene upregulation was measured by real-time PCR. Samples were normalized to β-actin.
CD1a-autoreactive responses from skin
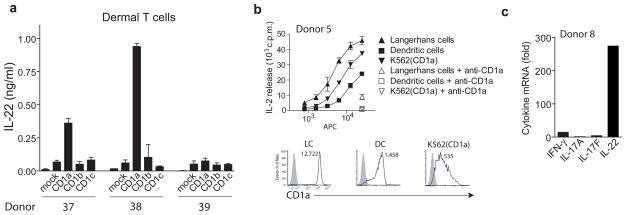
The expression of CLA and skin-homing chemokine receptors on the surface of CD1a-autoreactive T cells predicts that these cells would be present in the skin. To assess this, we isolated T cells from human skin biopsy specimens and tested for CD1a recognition 51. Similar to results found from blood derived T cells (Fig. 2c), initial screens isolated skin T cell clones whose activation was blocked by anti-CD1a (data not shown). We next tested for CD1a-dependent IL-22 production in polyclonal T cells (Fig. 7a). In two out of three samples, lymphocytes isolated and expanded from normal human dermis showed a significant IL-22 production in response to CD1a transfected, but not against CD1b or CD1c transfected K562 cells, demonstrating the dominance of CD1a-mediated responses in healthy human skin and their ability to produce IL-22.
Figure 7. IL-22 producing CD1a-autoreactive T cells in human skin.
(a) Lymphocytes from three normal human dermis samples, expanded in vitro with DC, were incubated with CD1 transfected K562 cells for 24 h after which the IL-22 release was measured by ELISA (mean ± S.D.). (b) Freshly isolated Langerhans cells, monocyte-derived DCs and K562-CD1a were incubated with a CD1a-autoreactive T cell line for 24 hrs after which supernatant was analyzed for IL-2 by HT2 bioassay (mean ± S.D.). The results are representative of separate experiments performed for three different T cell lines. The mean fluorescence intensity of CD1a on the antigen presenting cells was determined by flow cytometry. Open histograms: CD1a staining, filled histograms: Isotype control. (c) Freshly isolated Langerhans cells were pre-incubated with control IgG or anti-CD1a and the incubated with a CD1a-reactive T cell line for 18 hrs after which RNA was extracted and after reverse transcription the CD1a dependent upregulation of cytokine mRNA was measured by real time PCR. Results are representative of two experiments with natural CD1a expressing APC.
Expanding on the notion that CD1a-autoreactive T cells might play a role in skin immunity, we measured the ability of CD1a-autoreactive T cells to recognize natural CD1a-expressing antigen presenting cells. We isolated Langerhans cells (LC) from epidermal sheets of human skin and compared them to K562-CD1a transfectants as well as monocyte-derived DCs, which mimic many features of myeloid DCs. All three cell types activated T cell lines in a CD1a-dependent manner, with the highest potency of activation seen against LCs (Fig. 7b). Similar to K562-CD1a cells, LCs induced strong upregulation of IL-22, in the absence of significant upregulation of either IFNγ or IL-17 (Fig. 7c), indicating that the pattern of CD1a-dependent cytokine gene induction is similar when either K562 or freshly isolated Langerhans cells were used as stimulator cells.
Discussion
More so than other organs, skin resident T cell repertoires differ fundamentally between mice and humans. For example, mouse skin contains large numbers ofγδ T cells with Vγ5+ Vδ1+ invariant TCRs known as dendritic epidermal T cells (DETC), whose development requires skint1, a immunoglobulin-like dermal protein 52. Human skin lacks skint1 and DETC, and instead contains predominantly T cells that express rearranged αβ TCRs 53, 54. During the evolution of muroid rodents, a break in chromosome 1 resulted in a loss of group 1 CD1 genes, so human, but not mouse skin, contains Langerhans cells that express very high density CD1a 13. These general observations highlight two key reasons for developing experimental systems for studying skin T cell immunity in humans. First, the natural functions of CD1a cannot be studied in mice. Second, CD1a and other immune receptors, which naturally differ between these two species, represent candidate molecules for controlling the dichotomously different organization of skin T cell populations seen in these species.
In mice, IL-22 is typically co-secreted with IL-17 and is usually understood as part of the TH17 phenotype 55, although overexpression of Notch results in production of IL-22 in the absence of IL-17 56. Human αβ T cells producing IL-22 in the absence of IL-17 have been frequently observed and are now emerging as a recognized T helper subset, TH22. TH22 cells express the aryl hydrocarbon receptor 50, skin-homing chemokine receptors (CCR4, CCR10) 47,48, and localize to healthy and diseased skin 49. Although the antigen presenting molecules or any molecular targets recognized by TH22 cells are currently unknown, CD1a represents a good candidate because it is absent in mice and highly expressed in tissues that are enriched in TH22 cells. Here we show that CD1a autoreactive T cells have all of the known properties of TH22 cells, thereby identifying CD1a as molecular target of TH22 cells. Determining the relative contribution of CD1a and MHC antigen presenting molecules in TH22 cell activation will be important for understanding and intervening in skin immunopathology caused by IL-22 in psoriasis and other skin diseases 46, 49. More generally, our data show that populations of CD1a-autoreactive T cells are abundantly present in the blood and skin of most human donors tested and so we conclude that they are a normal part of the humanαβ T cell repertoire.
The experimental system we have generated is unbiased in the sense that it is able to detect responses against any CD1 protein, but T cell activation by CD1a dominated in magnitude of response and the percentage of individuals responding in both blood and skin-derived T cells. An obvious possibility to explain these findings is that CD1a-autoreactive T cells may be as or more common than T cells recognizing other CD1 isoforms in unchallenged human hosts. The estimated precursor frequency for CD1a-autoreactive T cells among memory T cells in the blood measured here with CD1a expressing cells (0.02 to 0.4 %) is similar to that found for human NKT cells measured with CD1d-α-galactosylceramide tetramers (undetectable to 1.0 %) 4, 5, 7. Tetramer based detection of NKT cells is direct, but necessarily requires a priori knowledge of antigens, measures responses to only one antigen and typically uses a synthetic superagonist, α-galactosyl ceramide. The detection system reported here differs in that it is presumably measuring responses to diverse and natural cellular self antigens, a speculation supported by data showing that lipid extracts of K562 cells activate CD1a autoreactive T cells (not shown). Given the incomplete current understanding of the diversity of self antigens for CD1 proteins, the features of this detection system were not only desirable, but also necessary to make the first global measurements of CD1 autoreactivity ex vivo in humans.
However, because this detection method requires an activation response, the number of cells detected depends on activation thresholds. Therefore, the second possibility for the increased detection of CD1a-autoreactive cells is that they have a lower threshold of activation than T cells recognizing other CD1 proteins. Because CD1a is generally absent or low in peripheral blood and tissues other than skin, CD1a-autoreactive T cell contact with CD1a-expressing APCs is not expected to be a common event in the blood and most other tissues. This situation contrasts to CD1d, which is expressed on the peripheral blood B cells, monocytes and other cells in blood and tissues 16. These considerations suggest a model in which CD1a-reactive T cell activation might have lower intrinsic resistance to activation and that their function is controlled by their ability to gain direct access to CD1a-expressing APCs in certain tissue environments.
Three related observations suggest that CD1a recognition occurs as a mechanism of skin homeostasis: restriction of CD1a expression to the skin, homing receptors (CLA, CCR6, CCR4, CCR10) that drive T cells to the skin and linkage of CD1a autoreactivity to the TH22 phenotype. Rather than stimulating other hematopoietically derived immune cells, IL-22 acts on heterodimers of IL-10R2 and IL-22R1 expressed on epithelial cells, including keratinocytes 42. Therefore, IL-22 secretion from chronic interactions with CD1a autoreactive TCRs on LCs might promote epithelial matrix remodeling in a baseline state without necessarily provoking broader immune activation or immunopathology 44. However, IL-22 over-production promotes acanthosis observed in psoriasis 57, and triggered release of the less abundant cytokines detected in CD1a autoreactive cells (IL-2, IL-13, IFN-γ) can promote immunopathology. These considerations drive ongoing efforts to understand regulation of the CD1a autoreactivity. Our preliminary data suggest that CD1a autoreactive T cells are present in dermis (Fig. 7 and data not shown), yet LCs mainly localize to the epidermis. Therefore, one model is that these near neighbors cross the dermal-epidermal junction to contact each other. Of course, skin is a rich source of tissue-specific oils and waxes that might function as CD1a presented lipid autoantigens.
Methods
Antibodies
Details of antibodies used in flow cytometry and neutralization assays are provided in Supplementary Table 1.
CD1 expressing antigen presenting cells
Peripheral blood mononuclear cells (PBMC) were obtained from blood donations at Massachusetts General Hospital, as approved by the Partners Healthcare Institutional Review Board, and were isolated from buffycoats by density centrifugation over Ficoll Hypaque. Monocyte-derived DC expressing CD1 were isolated by adherence to plastic and treatment with 300 IU/ml GM-CSF and 200 IU/ml Interleukin-4 for 72 hrs, followed by γ-irradiation (5000 Rad).
K562 cells were transfected with pcDNA3.1 or pcDNA3 vector containing: no insert, CD1a, CD1b, CD1c or CD1d cDNA. The CD1a, CD1c and CD1d constructs contained cDNA encoding the heavy chain gene only, whereas the CD1b construct contained the CD1 heavy chain linked to human β-2 microglobulin. K562 clones were selected for high CD1 expression and low expression of HLA class I. HLA DR surface expression was undetectable on all clones.
Polyclonal and clonal CD1-autoreactive T cell cultures
Polyclonal T cell cultures
Autologous monocyte-derived DC expressing CD1 were cultured with non-adherent cells (at a ratio of 1:5) in 96 well plates (105 cells / well) in RPMI supplemented with 10% fetal bovine serum (Hyclone) and 2% human AB serum (Gemini), essential and non-essential amino acids (Gibco), penicillin/streptomycin (Gibco), and β-mercaptoethanol (complete media). On day 2 and 7, IL-2 (Chiron, 0.2 nM) and IL-15 (Peprotech, 5 ng/ml) were added to the cultures and they were split if the wells became full. After 10–14 days the cells were tested for reactivity against CD1 transfected K562 cells. CD1-autoreactive T cells were maintained in culture by restimulation with autologous dendritic cells.
CD1a-autoreactive T cell clones by ex vivo limiting dilution
96-well plates were seeded with 100 autologous DCs and 100 non-adherent cells per well. After 24 h, 105 autologous irradiated PBMC were added per well in complete media containing IL-2 (0.5 nM) and IL-15 (2 ng/ml). The plates were screened for clones after 14 days, and all clones were tested for CD1a reactivity. The number of replicates per clone depended on the available amount of cells. Clonality of the cell cultures was determined by PCR for T cell receptor α and β chains using a primer panel covering the T cell receptor variable regions (International Immunogenetics, IMGT, primer database) combined with constant region primers.
CD1a-autoreactive T cell clones from skin by ex vivo limiting dilution
Skin T cell clones were generated by the method described above, with the difference that allogeneic DCs were used as APC. DCs were pretreated with anti-HLA class I and anti-HLA DR 20 μg/ml prior to incubation with T cells.
CD1a-autoreactive T cell clones from polyclonal cultures
CD1a-autoreactive T cell clones were derived from polyclonal cultures by limiting dilution. T cells were seeded at a concentration of 1 cell / well in 96 wells plates in 100 μl of cell suspension containing irradiated B-LCL (105 / ml), PBMC (106/ml) and K562 CD1a (5×105/ml), IL-2 (0.5 nM), and IL-15 (2 ng/ml) and PHA (Sigma-Aldrich 0.5 μg/ml). After 14 days the plates were screened and all clones were tested for CD1a reactivity.
Cell purification and sorting
Memory CD4+ T cell were purified from fresh PBMC using CD4+ T cell Isolation Kit (Miltenyi Biotech), and T-helper subsets were sorted after staining with CXCR3, CCR6, CCR4, CCR10 antibodies. Cell sorting was performed on a FACSAria (Becton Dickinson), 11-color flow cytometer. Total memory and CLA+ T cells were sorted after staining with CD3 and CD45RO and CLA antibodies.
Normal human skin samples were obtained as discarded material after cosmetic surgery under rules of the Institutional Review Board (Partners Human Research Committee, Boston, MA). The isolation of Langerhans cells and dermal lymphocytes was performed as published previously 58. Total skin T cells were isolated from skin biopsies by a method published previously 51.
Cellular assays
Measuring CD1 autoreactivity
T cell cultures were analyzed for CD1-autoreactivity by co-culture with APC (K562-mock, K562-CD1, DC or LC) at an APC : T cell ratio between 1:1 and 1:5 (1:10 for real-time PCR experiments). To confirm the CD1 dependence of the response, K562 CD1 cells were pre-incubated for one hour at 37°C with 10 μg/ml of blocking antibody against the corresponding CD1 molecule or control IgG.
IL-2 Bioassay
IL-2 was determined using an HT2 bioassay. Supernatants from T cell assays were added to wells containing 104 IL-2 dependent HT2 cells in 100μl T cell media, which were cultured for 24 h before adding 1 μCi 3H-thymidine for an additional 6 hrs of culture, followed by harvesting and counting β-emissions. Assays were performed in triplicate and reported as the mean ± standard deviation.
IFNγ Elispot
APC and T cell co-cultures incubated 16 hrs in a Multiscreen-IP filter plate (96 wells, Millipore) coated with anti-IFNγ antibody (1-D1-K) according to manufacturer’s instructions (Mabtech, Sweden).
Intracellular cytokine staining
T cells were stimulated with PMA (10 ng/ml) and Ionomycin (250 ng/ml). After two hours Brefeldin A (Golgiplug, BDbiosciences, 1:1000) was added and incubated for 6 hrs, when the cells were fixed with 4% paraformaldehyde and permeabilized using Cytoperm/wash buffer (BD Biosciences) and stained for intracellular IL-17 and IL-22.
Interleukin-22 ELISA
Measurement of IL-22 protein in cell culture supernatants was performed by IL-22 ELISA (Peprotech) according to manufacturer’s instructions.
Polymerase Chain Reaction (PCR)
RNA was isolated from cells using RNeasy kit (Qiagen) and cDNA was synthesized using Quantitect reverse transcription kit (Qiagen), including a genomic DNA removal step. Sequences of the CDR3 clonotypic primers were TCAACGTTGCTGAAGGGAATCCTC (Vα forward), CCTGCACTCTCCTGGGGGA (CDR3α reverse), GCAGGGTCCAGGTCAGGACCCCCA (Vβ forward primer), CTCCCGCTATCCATCCCAGGC (CDR3β). These primers were validated for specificity using cDNA from the specific T cell clone and showed minimal or no annealing with cDNA from random donor PBMC.
Cytokine analysis by real-time PCR
K562-CD1a cells were preincubated with anti-CD1a or control IgG and then co-cultured with CD1a-autoreactive T cell cultures for 6 hrs at a K562 to T cell ratio of 1:10. RNA was extracted, and after reverse transcription, real-time PCR was used to determine the CD1a dependent cytokine gene upregulation.
Primer pair sequences for cytokine mRNA were derived from primerbank (http://pga.mgh.harvard.edu/primerbank/) and were validated for specificity (melt-curve analysis) and priming efficiency. Real-time PCR was run on a Stratagene Mx3000P cycler using PerfeCTa SYB®RGreen Supermix (Quanta Biosciences). Background transcript levels for K562-CD1a were determined, and were >100 fold lower than background transcript in unstimulated T cells for all cytokines tested.
Statistical Analysis
Unpaired two-sided student’s t test was performed to determine if a response to CD1 transfected APCs was significantly different from that to mock transfected control. Responses to CD1a, CD1b, CD1c and CD1d transfected APC were compared by one-way ANOVA, and Dunnett’s Multiple Comparison Test.
Supplementary Material
Acknowledgments
The authors acknowledge D.C. Barral, M. Brenner, M. Relloso and M. Sugita for providing CD1 plasmid constructs, J. Gumperz and M. Brigl for providing human NKT cell lines, G. Losyev for cell sorting, and K. Magalhães, S. Huang, I.C. Ho and R. Grenningloh for technical advice. This work was supported by grants from the NIAMS (048632 to D.B.M. AR056720 to R.A.C), NIAID (AI054456 and AI056299), the Damon Runyon Clinical Investigator Award (to R.A.C.), the Burroughs Wellcome Fund for Translational Research (to D.B.M.) and the National Psoriasis Foundation, USA (to A.J).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Author contributions
A.J. designed and performed the experiments, A.J. and D.B.M. prepared the manuscript. D.B.M. supervised the experiments. V.P.C. isolated Langerhans cells from human epidermis and lymphocytes from the dermis. T.Y.C performed T cell culture and Western blot analysis. I.V.R. assisted in experiments and R.A.C. provided T cells isolated from human skin biopsies.
References
- 1.Fowlkes BJ, et al. A novel population of T-cell receptor alpha beta-bearing thymocytes which predominantly expresses a single V beta gene family. Nature. 1987;329:251–254. doi: 10.1038/329251a0. [DOI] [PubMed] [Google Scholar]
- 2.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4–8− T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8− alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195:625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karadimitris A, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci USA. 2001;98:3294–3298. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 9.Cui J, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 10.Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–1388. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Rhijn I, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 12.Looringh van Beeck FA, et al. Two canine CD1a proteins are differentially expressed in skin. Immunogenetics. 2008;60:315–324. doi: 10.1007/s00251-008-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- 14.Kasmar A, Van Rhijn I, Moody DB. The evolved functions of CD1 during infection. Curr Opin Immunol. 2009;21:397–403. doi: 10.1016/j.coi.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roura-Mir C, et al. Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR-2. J Immunol. 2005;175:1758–1766. doi: 10.4049/jimmunol.175.3.1758. [DOI] [PubMed] [Google Scholar]
- 16.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol. 2007;314:113–141. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]
- 17.Sugita M, Cernadas M, Brenner MB. New insights into pathways for CD1-mediated antigen presentation. Curr Opin Immunol. 2004;16:90–95. doi: 10.1016/j.coi.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 19.Porcelli S, et al. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 20.Rosat JP, et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. J Immunol. 1999;162:366–371. [PubMed] [Google Scholar]
- 21.Sieling PA, et al. Evidence for human CD4+ T cells in the CD1-restricted repertoire: derivation of mycobacteria-reactive T cells from leprosy lesions. J Immunol. 2000;164:4790–4796. doi: 10.4049/jimmunol.164.9.4790. [DOI] [PubMed] [Google Scholar]
- 22.Moody DB. The surprising diversity of lipid antigens for CD1-restricted T cells. Adv Immunol. 2006;89:87–139. doi: 10.1016/S0065-2776(05)89003-0. [DOI] [PubMed] [Google Scholar]
- 23.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 24.Shamshiev A, et al. The alphabeta T cell response to self-glycolipids shows a novel mechanism of CD1b loading and a requirement for complex oligosaccharides. Immunity. 2000;13:255–264. doi: 10.1016/s1074-7613(00)00025-x. [DOI] [PubMed] [Google Scholar]
- 25.Shamshiev A, et al. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieling PA, et al. Human double-negative T cells in systemic lupus erythematosus provide help for IgG and are restricted by CD1c. J Immunol. 2000;165:5338–5344. doi: 10.4049/jimmunol.165.9.5338. [DOI] [PubMed] [Google Scholar]
- 27.Vincent MS, Xiong X, Grant EP, Peng W, Brenner MB. CD1a-, b-, and c-restricted TCRs recognize both self and foreign antigens. J Immunol. 2005;175:6344–6351. doi: 10.4049/jimmunol.175.10.6344. [DOI] [PubMed] [Google Scholar]
- 28.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 29.Klein E, et al. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer. 1976;18:421–431. doi: 10.1002/ijc.2910180405. [DOI] [PubMed] [Google Scholar]
- 30.Britten CM, et al. The use of HLA-A*0201-transfected K562 as standard antigen-presenting cells for CD8(+) T lymphocytes in IFN-gamma ELISPOT assays. J Immunol Methods. 2002;259:95–110. doi: 10.1016/s0022-1759(01)00499-9. [DOI] [PubMed] [Google Scholar]
- 31.Shamshiev A, et al. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667–1675. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Roura-Mir C, et al. CD1a and CD1c activate intrathyroidal T cells during Graves’ disease and Hashimoto’s thyroiditis. J Immunol. 2005;174:3773–3780. doi: 10.4049/jimmunol.174.6.3773. [DOI] [PubMed] [Google Scholar]
- 33.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 34.Vincent MS, et al. CD1-dependent dendritic cell instruction. Nat Immunol. 2002;3:1163–1168. doi: 10.1038/ni851. [DOI] [PubMed] [Google Scholar]
- 35.Meunier L, et al. Quantification of CD1a, HLA-DR, and HLA class I expression on viable human Langerhans cells and keratinocytes. Cytometry. 1996;26:260–264. doi: 10.1002/(SICI)1097-0320(19961215)26:4<260::AID-CYTO4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 36.Yu RC, Abrams DC, Alaibac M, Chu AC. Morphological and quantitative analyses of normal epidermal Langerhans cells using confocal scanning laser microscopy. Br J Dermatol. 1994;131:843–848. doi: 10.1111/j.1365-2133.1994.tb08587.x. [DOI] [PubMed] [Google Scholar]
- 37.Chu A, et al. Immunoelectron microscopic identification of Langerhans cells using a new antigenic marker. J Invest Dermatol. 1982;78:177–180. doi: 10.1111/1523-1747.ep12506352. [DOI] [PubMed] [Google Scholar]
- 38.Armerding D, Kupper TS. Functional cutaneous lymphocyte antigen can be induced in essentially all peripheral blood T lymphocytes. Int Arch Allergy Immunol. 1999;119:212–222. doi: 10.1159/000024197. [DOI] [PubMed] [Google Scholar]
- 39.Fuhlbrigge RC, Kieffer JD, Armerding D, Kupper TS. Cutaneous lymphocyte antigen is a specialized form of PSGL-1 expressed on skin-homing T cells. Nature. 1997;389:978–981. doi: 10.1038/40166. [DOI] [PubMed] [Google Scholar]
- 40.Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, et al. Natural killer T-cell autoreactivity leads to a specialized activation state. Blood. 2008;112:4128–4138. doi: 10.1182/blood-2008-05-157529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolk K, et al. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Boniface K, et al. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174:3695–3702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- 44.Wolk K, Sabat R. Interleukin-22: a novel T- and NK-cell derived cytokine that regulates the biology of tissue cells. Cytokine Growth Factor Rev. 2006;17:367–380. doi: 10.1016/j.cytogfr.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nograles KE, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol. 2009;123:1244–1252. e1242. doi: 10.1016/j.jaci.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 48.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 49.Eyerich S, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–3585. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 51.Clark RA, et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J Clin Invest. 2006;126:1059–1070. doi: 10.1038/sj.jid.5700199. [DOI] [PubMed] [Google Scholar]
- 52.Strid J, Tigelaar RE, Hayday AC. Skin immune surveillance by T cells--a new order? Semin Immunol. 2009;21:110–120. doi: 10.1016/j.smim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Foster CA, et al. Human epidermal T cells predominantly belong to the lineage expressing alpha/beta T cell receptor. J Exp Med. 1990;171:997–1013. doi: 10.1084/jem.171.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark RA, et al. The vast majority of CLA+ T cells are resident in normal skin. J Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 55.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Ann Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 56.Alam MS, et al. Notch signaling drives IL-22 secretion in CD4+ T cells by stimulating the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 107:5943–5948. doi: 10.1073/pnas.0911755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boniface K, et al. A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol. 2007;150:407–415. doi: 10.1111/j.1365-2249.2007.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pena-Cruz V, et al. Extraction of human Langerhans cells: a method for isolation of epidermis-resident dendritic cells. J Immunol Methods. 2001;255:83–91. doi: 10.1016/s0022-1759(01)00432-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.