Abstract
We propose the use of Desulfovibrio desulfuricans ND132 as a model species for understanding the mechanism of microbial Hg methylation. Strain ND132 is an anaerobic dissimilatory sulfate-reducing bacterium (DSRB), isolated from estuarine mid-Chesapeake Bay sediments. It was chosen for study because of its exceptionally high rates of Hg methylation in culture and its metabolic similarity to the lost strain D. desulfuricans LS, the only organism for which methylation pathways have been partially defined. Strain ND132 is an incomplete oxidizer of short-chain fatty acids. It is capable of respiratory growth using fumarate as an electron acceptor, supporting growth without sulfide production. We used enriched stable Hg isotopes to show that ND132 simultaneously produces and degrades methylmercury (MeHg) during growth but does not produce elemental Hg. MeHg produced by cells is mainly excreted, and no MeHg is produced in spent medium. Mass balances for Hg and MeHg during the growth of cultures, including the distribution between filterable and particulate phases, illustrate how medium chemistry and growth phase dramatically affect Hg solubility and availability for methylation. The available information on Hg methylation among strains in the genus Desulfovibrio is summarized, and we present methylation rates for several previously untested species. About 50% of Desulfovibrio strains tested to date have the ability to produce MeHg. Importantly, the ability to produce MeHg is constitutive and does not confer Hg resistance. A 16S rRNA-based alignment of the genus Desulfovibrio allows the very preliminary assessment that there may be some evolutionary basis for the ability to produce MeHg within this genus.
INTRODUCTION
Mercury methylation is a natural microbial process that converts inorganic Hg(II) to the bioaccumulative toxin methylmercury (MeHg). Methylmercury contamination of food webs causes significant risk to people and other organisms near the top of food webs worldwide (1, 67). Although the biogeochemistry of MeHg production in the environment has been studied in detail for more than 3 decades, the biochemical mechanism of methylation in bacterial cells remains poorly understood, especially relative to MeHg demethylation by the organomercury lyase pathway (3) or the redox transformations of metal contaminants like uranium (28, 69) and chromium (50). As of yet, no metabolic pathway or gene that is common to methylators but absent in nonmethylators has been identified.
Methylmercury production is an anaerobic process that occurs in saturated soils, wetlands, decaying periphyton mats, aquatic bottom sediments, and anaerobic bottom waters (5, 57). Studies at a variety of ecological scales show that MeHg production is intimately linked to the sulfur and iron cycles. Many studies have demonstrated sulfate stimulation of MeHg production in freshwater sediments and wetlands (e.g., references 12, 36, 44, and 70), and many have found that Hg methylation occurs most readily in zones of microbial sulfate or ferric iron reduction (e.g., references 21, 35, 42, and 48).
However, the ability to produce MeHg is not a common trait of dissimilatory sulfate-reducing bacteria (DSRB) or Fe(III)-reducing bacteria (FeRB). Only a subset of the sulfate- and Fe(III)-reducing bacterial species tested have the ability to methylate Hg. Overall, this capacity has been tested with fewer than 50 bacterial strains. The order Desulfovibrionales has been most extensively examined, and about half of the examined species have the ability to produce MeHg (18, 27, 37, 47, 51, 62). Mercury-methylating DSRB are also found within the Desulfobacterales (6, 13, 27, 47, 64). In addition, several species of Geobacter, FeRB in the order Desulfuromonales, produce MeHg (28, 46), as well as Desulfuromonas palmitatis SDBY1, in the same order. Limited testing for Hg methylation outside the Deltaproteobacteria has focused on FeRB and DSRB in the Gammaproteobacteria and in the Firmicutes, none of which methylate Hg (46, 62, 64). To summarize, most organisms tested for methylation have been sulfate- or Fe(III)-reducing Deltaproteobacteria, and only about half of those tested have the ability to produce MeHg. No organisms outside the Deltaproteobacteria have been shown to produce MeHg, but fewer than 15 have been tested.
The ability of certain organisms to produce MeHg could be linked to a specific methyl-transferase pathway, to a Hg-specific uptake pathway, or to the biochemistry of Hg binding and movement within cells. In the late 1980s and 1990s, Richard Bartha's group studied the metabolic pathways leading to MeHg, using an estuarine DSRB, Desulfovibrio desulfuricans LS, which was isolated from a brackish New Jersey marsh (18). They proposed that Hg methylation in this organism occurred via transfer of a methyl group from methyl-tetrahydrofolate via methylcobalamin (MeB12), with the methyl group originating either from C-3 of serine or from formate, via the acetyl-coenzyme A (CoA) synthase pathway (11, 15, 16). Since these pathways are not unique to D. desulfuricans LS, Bartha and colleagues proposed that the organism's ability to methylate mercury is most likely associated with the substrate specificity of its enzymes. Subsequent work confirmed that Hg methylation can occur independently of the acetyl-CoA pathway. Benoit et al. (6) demonstrated Hg methylation by Desulfobulbus propionicus, a DSRB that does not use the acetyl-CoA pathway. Ekstrom et al. (27) identified other Hg-methylating DSRB without the pathway. More recently, Ekstrom and Morel (26) showed that cobalt limitation inhibited MeHg production in Desulfococcus multivorans, an incomplete oxidizer that does use the acetyl-CoA pathway for major carbon metabolism. However, cobalt limitation did not affect Hg methylation in Desulfovibrio africanus (DSM 2603, strain Benghazi), an incomplete oxidizer that does not use that pathway, suggesting different methylation pathways in different organisms.
Differences in methylation rate among strains could also be due to differences in uptake pathways. The prevailing paradigm for Hg uptake by DSRB (5, 8, 23) is diffusion of small neutrally charged Hg complexes. However, Golding et al. (34) found that Hg uptake by Vibrio anguillarum and Escherichia coli strains modified with a mer-lux bioreporter system (which in this case did not include the Hg transport genes) was enhanced in the presence of a variety of small organic molecules, including amino acids. This result led to the hypothesis that Hg uptake may occur via a facilitated transport mechanism. Schaefer and Morel (66) showed that cysteine specifically enhanced Hg uptake and methylation in Geobacter sulfurreducens and proposed that Geobacter strains have a specific uptake mechanism for the Hg-cysteine complex.
Despite this progress, the mechanism of Hg uptake and methylation by bacteria remains unresolved. We have begun a concerted effort to identify the pathways and genes involved in MeHg production. Desulfovibrio desulfuricans ND132 was chosen for study because of its exceptionally high rates of Hg methylation in culture and its metabolic similarity to strain D. desulfuricans LS, the only organism for which methylation pathways have been partially defined. Before strain LS was lost, Pak and Bartha compared it to ND132 (58, 60). They noted that strain ND132 was similar in substrate utilization abilities but produced four times more MeHg than LS under the same lactate-sulfate culture conditions.
In developing a model organism for study, we hope to provide a well-characterized strain for further genetic and physiological study. Here, we describe the physiological characteristics of D. desulfuricans ND132, including antibiotic sensitivities important for genetic manipulation. We also provide basic information on ND132 MeHg production and degradation capabilities and explore how growth conditions affect these rates. Additionally, we explore Hg methylation within the genus Desulfovibrio in some detail. We examine the inducibility of Hg methylation and the role of methylation in Hg resistance among Desulfovibrio species. Finally, we summarize the available information on Hg methylation among Desulfovibrio species, including methylation rates for several new species. A potential evolutionary basis for methylation among Desulfovibrio species is explored through construction of a 16S rRNA gene-based phylogeny.
MATERIALS AND METHODS
Isolation of ND132.
Desulfovibrio desulfuricans ND132 was isolated from mid-Chesapeake Bay bottom sediments sampled in May 1985, as part of a study of tin methylation (29, 32). The sampling site (near station R64; 38°33.86′N, 76°26.38′W) was on the western slope of the main bay channel in about 20 m of water, which is often below the oxycline in summer. The sediment was soft black silt, with an average pore water sulfide (top 10 cm) concentration of 0.52 mM. Bottom water salinity was 23 ppt. Most-probable-number (MPN) estimates for sulfate-reducing bacteria (SRB) yielded >6,000 cells/g wet sediment on lactate and >500 cells/g on acetate (29). Sites near this station have been repeatedly occupied for a variety of research and monitoring work, including measurement of sulfate reduction rates and other biogeochemical parameters (29, 42, 55, 63). Significant rates of tin (32) and mercury (42) methylation have been measured at the site.
Isolation was done on agar plates from lactate-sulfate enrichments made from the top 10 cm of sediment. It is important to point out that enrichment and isolation of this strain were done in medium without added Hg or Sn. The strain was initially selected for its ability to produce MeSn3+ from inorganic tin. Since that time, the strain has been used to study the role of sulfur chemistry in Hg uptake and methylation (43), the uptake of thymidine by DSRB (30), and the potential for DSRB to reduce perrhenate (22).
PCR and sequencing.
To test culture purity and to help identify isolates, 16S rRNA gene sequences were obtained. DNA was amplified directly from cell cultures, with standard RedMix (Gene Choice, Frederick, MD) protocols with universal forward (27F) and reverse (1541R) primers (300 nM each) and a PTC-200 DNA Engine thermal cycler (Bio-Rad Laboratories, Hercules, CA). When necessary, PCR products were purified from agarose bands with GeneClean (MP Biomedicals, LLC) or from liquid with Qiagen (Qiagen, Inc., Valencia, CA) DNA cleanup kits. Sequencing chemistry used standard BigDye v3.1 reaction protocols (Applied Biosystems, Inc., Foster City, California). Sequencing was performed at the Smithsonian Laboratory of Analytical Biology, with an ABI 3100 automated capillary DNA sequencer (Applied Biosystems).
SEM preparation and observation.
Cells were prepared by fixing with 2.5% (vol/vol) anaerobic glutaraldehyde. After 2 h, fixed cells were placed onto a 0.22-μm-pore-sized Neopore filter. Filters were gently rinsed on a filter pad, 2 times with 0.1 M cacodylate buffer and 3 times with ultrapure water, and then covered with a second filter and dehydrated through an ethanol (EtOH) dehydration series. Dehydrated filters were placed between screens and critical point dried immediately (Tousimis Auto-Samdri; 815 Automatic Critical Point Dryer). The sandwiched membranes were then separated, mounted onto scanning electron microscopy (SEM) stubs and sputter coated (Emitech K575x Turbo Sputter Coater) with a thin layer of platinum on a rotating sample holder (20 kV, 45 s). The coated samples were then viewed with a Hitachi S-4700 field emission scanning electron microscope (FESEM) at a working distance of 5 to 6 mm, a voltage of 2.0 to 10.0 kV, and a magnification of approximately ×6,000.
Cultivation and maintenance of D. desulfuricans ND132.
Cultures were maintained in the Gilmour laboratory on SRM medium (without Hg) (6) containing 20 mM Na2SO4, 20 mM Na lactate, salts (170 mM NaCl, 1.4 mM NaH2PO4, 19 mM NH4Cl, 6.7 mM KCl, 1.5 mM MgCl2, and 1.5 mM CaCl2), 0.05% yeast extract (wt/vol), 25 nM selenate, 25 nM tungstate, 4.4 μM FeCl2, 10 mM MOPS (morpholinepropanesulfonic acid) buffer, trace metals (see Table S1 in the supplemental material), and vitamins (Table S2) at pH 7.2. The medium was usually reduced with 100 μM Ti-NTA (freshly made from TiCl3 and nitrilotriacetic acid [NTA] in saturated Na2CO3), and resazurin (0.001%) was used as a redox dye. Reductants were filter sterilized into medium tubes just before inoculation. Other reductants included thioglycolate-ascorbate (made from Na mercaptoacetic acid and Na ascorbate), and cysteine, both used at final concentrations of 100 μM.
In the Wall laboratory, strain ND132 was maintained on MOYLS4 medium (71) containing 40 mM Na lactate, 40 mM Na2SO4, 8 mM MgCl2, 20 mM NH4Cl, 0.6 mM CaCl2, 2 mM K2HPO4-NaH2PO4, 30 mM Tris-Cl, 0.1× Thauers vitamins, 0.5× trace elements, 0.1% (wt/vol) yeast extract, FeCl2-EDTA (0.06 mM and 0.12 mM, respectively) at pH 7.2 to 7.5. For ND132, the medium was typically modified to include 1% (wt/vol) NaCl, 1 mM cysteine and was reduced with 0.38 mM Ti-citrate. Prior to work described in this paper, purity was verified via single-colony isolation. To minimize phenotypic drift from repetitive culturing, all experiments used cells with fewer than three subcultures from frozen glycerol stocks. Cultures were routinely checked for aerobic contamination by streaking them on aerobic plates of complex medium containing glucose.
Metabolic by-product quantification.
Sulfide was analyzed by ion-specific electrode after preservation in freshly made sulfide antioxidant buffer (24). Sulfate was measured by ion chromatography (25).
Mercury methylation and demethylation measurements.
Methylmercury production and degradation in cultures were assessed using isotopically enriched stable Hg isotopes (39, 42, 46, 56). Throughout this report, the notation for individual Hg isotopes (e.g., 201Hg), is used as shorthand for the excess concentration of a single enriched isotope above its natural abundance. MeHg production in cultures was assayed by measuring the amount of Me201Hg produced from an inorganic 201HgCl2 spike, while demethylation was assayed by measuring the loss of Me199Hg spiked into a sample (38, 40, 53). Enriched Hg isotopes (199Hg [91.95%] and 201Hg [98.11%]) were obtained from Oak Ridge National Laboratory. Me199Hg was synthesized from 199HgCl2 using methylcobalamin (41). For most experiments, 201HgCl2 and Me199HgCl were added at 10 ng/ml of culture medium and tests were done in triplicate with uninoculated medium as a blank, unless specified otherwise. The stable isotope concentrations used were significantly above background Hg concentrations in the culture medium. For culture studies using 10 ng 201Hg/ml, detection limits for MeHg production were on the order of 0.1 pg/ml or 0.001% methylation, based on the method detection limit for excess Me201Hg. The average relative percent deviation (RPD) for duplicate analysis of methylation assays was 2%. For culture studies using a 10-ng-Me199Hg/ml spike, the detection limit for MeHg degradation was on the order of 3%, based on the average RPD of duplicate sample analyses. The amount of dissolved gaseous mercury (DGM) produced by cultures and controls was assessed by purging the sealed cultures onto gold traps. Mercury loss to bottle walls was measured directly at the end of the incubations by filling empty bottles with 0.5% (70:40 concentrated nitric-sulfuric) acid plus 1% BrCl and analyzing the digest acid after >24 h.
Induction of Hg methylation.
D. desulfuricans ND132 and another Chesapeake Bay Desulfovibrio isolate, T2, were grown through four batch culture cycles at four Hg concentrations (0, 0.5, 5, and 50 mg/liter) and then assayed for MeHg production. Mercury exposure and methylation assays were done in SRM medium (with lactate as the substrate and sulfate as the electron acceptor). Methylation was assayed during batch culture growth with 500 ng/ml added HgCl2.
Mercury toxicity.
Mercury toxicity was assessed during batch culture fermentative growth (without sulfate) with pyruvate as the carbon source. Mercury was added to medium as HgCl2 before inoculation. Toxicity was assessed by following the optical density (OD) of cultures through time.
Hg and MeHg analysis.
Hg and MeHg quantifications were carried out by isotope dilution inductively coupled plasma mass spectrometry (ICP-MS; PerkinElmer ElanDRC ICP-MS) coupled to standard digestion/distillation methods for trace-level Hg analysis and speciation, as previously described (42, 56). Briefly, total Hg samples were digested in 0.5% (vol/vol) digest acid (a mix of 70:40 concentrated nitric-sulfuric acids with 1% [wt/vol] BrCl). Digested samples were introduced into the ICP-MS following SnCl2 reduction in a flow injection/gas-stripping system. MeHg was separated from the sample matrix via atmospheric pressure water vapor distillation, followed by ethylation, purging with nitrogen gas, trapping on Tenax, thermal desorption, separation by gas chromatography, and detection with ICP-MS.
The concentrations of natural abundance Hg and MeHg and of excess enriched Hg stable isotopes were quantified via isotope dilution (39). Isotope dilution ICP-MS uses an enriched Hg stable isotope spike as an internal standard in every sample, significantly improving the accuracy and precision of these multistep analytical methods. Blanks and suitable certified reference materials were run with every batch.
Alignment and phylogenetic analyses of Hg methylation within Desulfovibrio.
A 16S rRNA gene-based phylogenetic tree of Desulfovibrio was constructed. The tree was based on the aligned sequences of 49 type strains of Desulfovibrio, plus Desulfobacter postgatei as the reference and outgroup taxon, all available from the Ribosomal Database Project (RDP) on 10 October 2010 (17). To these were added 13 additional species of Desulfovibrio that have been tested for mercury methylation ability and for which 16S rRNA gene sequences were available (see Tables S2 and S3 in the supplemental material). We sequenced available cultures for which sequences were not published. The RDP alignment was used as a reference for the addition of these taxa in GENEIOUS version 5.1, and the final alignment was manually corrected. The alignments were trimmed to a common region that included 1,384 aligned bases for all taxa, except in the cases of strain D. desulfuricans strain LS and D. africanus ADR13, for which only partial length sequences (1,183 and 540 bp, respectively) were available. The partial sequence for D. desulfuricans LS, done in the 1990s, was provided by R. Devereux. It lacks 200 bp at the 3′ end and has a number of gaps. A full list of the source of all sequences used is given in Table S2.
The alignment was analyzed through a Bayesian phylogenetic approach as implemented in MrBayes version 3.1 (65). The program was set to use a HKY85 substitution model and a gamma rate distribution, to run in six parallel chains, and to save every 100th tree. A total of 2,100,000 generations of the chains in the Markov chain Monte Carlo (MCMC) simulation procedure were conducted. The average standard deviation (SD) of split frequencies between the MCMC chain runs was <0.01 upon termination of the analysis. Trees from the initial 100,000 generations were discarded as burn-in of the chains, and the remaining 2,000,000 are summarized in a 50% majority rule consensus tree with the branch posterior probabilities shown.
RESULTS
Description and morphology.
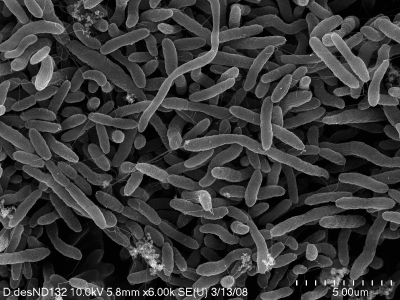
D. desulfuricans strain ND132 cells are Gram-negative, slightly curved rods with a relatively constant diameter of ∼0.75 μm but variable lengths, averaging ∼4 μm (Fig. 1). The strain is motile and produces bisulfate reductase (desulfoviridin) (61). It is nonsporulating based on visual observation of old cultures and was unable to grow after exposure to 90°C heat for 10 min. Phylogenetic analysis based on the 16S rRNA gene sequence (HQ693571) assigned the strain to the genus Desulfovibrio (Deltaproteobacteria → Desulfovibrionaceae).
Fig. 1.
Scanning electron microscopy image of D. desulfuricans ND132 showing a rod shape of variable length that has a tendency to curve.
Electron donors and acceptors.
D. desulfuricans strain ND132 is an incomplete oxidizer, with limited metabolic flexibility (Table 1). The most robust growth was observed during sulfate respiration with either lactate or pyruvate as the carbon source. Fermentative growth occurred on pyruvate in the absence of sulfate, but with lower cell yields than during respiratory growth with pyruvate and sulfate. The organism was not able to grow on lactate alone. It was also able to utilize fumarate or formate during sulfate respiration. Growth on fumarate alone occurred after a long lag but achieved nearly the same cell densities as growth with pyruvate alone. All media containing formate also included 20 mM acetate as a carbon source, since Desulfovibrio species require an organic C2 compound in addition to CO2 for cell synthesis. Pyruvate synthesis by a reductive carboxylation of the activated acetate, acetyl-CoA, has been shown to be the first step in cell synthesis when cells were grown on hydrogen or formate (2, 68).
Table 1.
Electron donors and acceptors supporting the growth of D. desulfuricans ND132a
| Substrate | Electron acceptor | Growthb |
|---|---|---|
| Respiration | ||
| Lactate | Sulfate | +++ |
| Pyruvate | Sulfate | +++ |
| Malate | Sulfate | − |
| Fumarate | Sulfate | ++ |
| Succinate | Sulfate | − |
| Proline | Sulfate | − |
| EtOH | Sulfate | − |
| Cysteine | Sulfate | − |
| Alanine | Sulfate | − |
| Glycinebetaine | Sulfate | − |
| Choline | Sulfate | − |
| Formate-acetate | Sulfate | + |
| Lactate | Sulfite | ++ |
| Pyruvate | Sulfite | + |
| Formate-acetate | Sulfite | − |
| Lactate | Nitrate | − |
| Lactate | Fumarate | + |
| Pyruvate | Fumarate | +++ |
| Formate-acetate | Fumarate | + |
| Lactate | Fe(III) | −* |
| Pyruvate | Fe(III) | −* |
| Pyruvate-acetate | Fe(III) | −* |
| Acetate | Fe(III) | −* |
| Fermentation | ||
| Lactate | − | |
| Pyruvate | + | |
| Malate | − | |
| Fumarate | + | |
| Succinate | − | |
| Proline | − | |
| EtOH | − | |
| Cysteine | − | |
| Alanine | − | |
| Glycinebetaine | − | |
| Choline | − | |
| Formate-acetate | − |
Growth in modified SRM (6) or MOY (71) medium, with potential organic substrates added at 30 to 60 mM concentrations, was tested. For formate, 20 mM Na-acetate was also added as the carbon source. Sulfate, sulfite, fumarate, and Fe(III) citrate (50 mM) were tested as potential electron acceptors. All media contained yeast extract. All tests were done in at least triplicate, and positive cultures were subcultured again in the same medium. Growth was measured once cells reached stationary phase.
+++, OD660 ≥ 1.0; ++, OD660 ≥ 0.5; +, OD660 ≥ 0.1; *, no reduction of Fe(III).
Unlike for the well-studied Desulfovibrio vulgaris (61), growth of ND132 on formate was poor. ND132 was unable to respire sulfate in the presence of other citric acid intermediates, amino acids, and other common DSRB substrates listed in Table 1. ND132 did not reduce Fe(III) chelated with citrate, given a variety of electron donors. As is typical of most Desulfovibrio species, ND132 did not grow on nitrate with lactate as the electron donor (52).
Importantly, ND132 was able to grow well using fumarate as an electron acceptor and pyruvate as the electron donor. At comparable substrate concentrations, ND132 growing on pyruvate-fumarate achieved an optical density about two-thirds of that of lactate-sulfate-grown cells. This growth mode allows experimentation with minimal generation of sulfide, a strong inhibitor of Hg methylation (6). At 32°C, in modified MOYLS4 medium, the optimal conditions for growth rate and yield were 40 mM (each) pyruvate and fumarate and 100 μM Ti-NTA as a reductant. Higher concentrations of pyruvate and fumarate did not enhance growth, nor did different ratios of pyruvate-fumarate (see Fig. S1 in the supplemental material).
Optimal growth parameters.
D. desulfuricans strain ND132 is a salt-tolerant mesophile. The optimal NaCl concentration for growth was about 2% (wt/vol), although good growth was achieved over a range of 0% to 3% (see Fig. S2 in the supplemental material). This wide range of salt tolerance reflects the estuarine habitat from which the strain was isolated. The strain grew well over a pH range of 6.8 to 8.2, with an optimum pH of 7.8 (Fig. S2). The optimal growth temperature was 32°C. Growth rate was only slightly less at 30 and 37°C, but no substantial growth occurred at 45°C. Thiogylcolate-ascorbate, Ti-NTA, and cysteine (all 100 μM) were adequate reductants for growth on either lactate-sulfate or pyruvate-fumarate in liquid medium. Strain ND132 appears to be extremely sensitive to O2. Single colonies failed to form in the anaerobic glove bag unless the agar plates were additionally placed in sealed containers with additional palladium catalyst.
Antibiotic resistance.
Determination of antibiotic sensitivity profiles for a bacterium may offer insight into the genetic complement of the organism and give a useful foundation for future genetic studies. Strain ND132 was resistant to ampicillin (up to 200 μg/ml), whereas it was sensitive to all other antibiotics tested (the aminoglycosides, kanamycin, spectinomycin, gentamicin, and G418 or genetisin) within the chosen concentration ranges (see Fig. S3 in the supplemental material). Additionally, chloramphenicol (50 μg/ml) has previously been shown to reduce leucine uptake and MeHg production by this strain (37), demonstrating sensitivity to this antibiotic and suggesting involvement of new protein synthesis for MeHg production.
Methylmercury production and degradation.
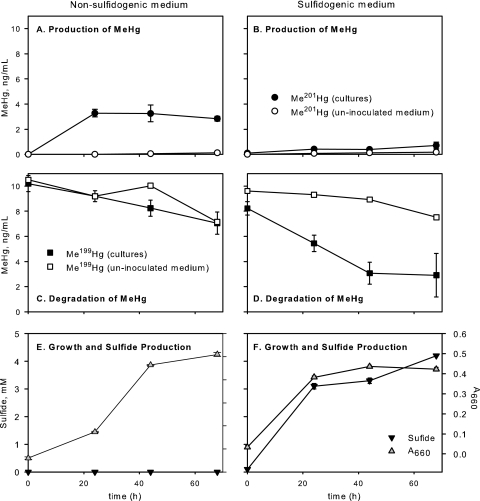
MeHg production and degradation by strain ND132 were measured simultaneously in batch culture by the addition of isotopically enriched Me199Hg and 201HgCl2 to culture medium prior to inoculation, each at 10 ng/ml (Fig. 2). The experiment was done with both sulfidogenic and nonsulfidogenic media.
Fig. 2.
Methylmercury production (A, B) and degradation (C, D) during batch culture growth of strain ND132 on pyruvate-fumarate medium (A, C, E) and lactate-sulfate medium (B, D, F). Media were amended with enriched 201HgCl2 and Me199HgCl before inoculation, both to 10 ng/ml. In panels A and B, Me201Hg concentrations represent MeHg production, while in panels C and D, Me199Hg concentrations show simultaneous MeHg degradation. Error bars represent the standard deviations of results from three replicate cultures. Optical density and sulfide concentrations are shown for cultures in panels E and F.
D. desulfuricans ND132 produced MeHg in both media, primarily during exponential growth (Fig. 2A and B). The level of MeHg production was higher during growth in nonsulfidogenic (pyruvate-fumarate) medium. In this medium, roughly 35% of the Hg added as enriched 201HgCl2 was converted to Me201Hg during batch growth (Fig. 2A). In lactate-sulfate medium, when millimolar concentrations of sulfide were generated, less than 10% of the added 201Hg(II) was converted to Me201Hg (Fig. 2B). Uninoculated culture medium was used to assess abiotic methylation from culture medium components. In both uninoculated controls, less than 1% of the added 201HgCl2was converted to MeHg over 68 h. MeHg production was also measured in cell-free spent medium by adding 201HgCl2 to filtrates from active cultures. Me201Hg production was similar to that observed in uninoculated medium, confirming that extracellular metabolites do not directly methylate Hg.
Methylmercury was also substantially degraded during batch culture growth and in uninoculated controls (Fig. 2C and D). About two-thirds of the added Me199Hg added was lost during batch culture growth on lactate-sulfate medium (Fig. 3D), while about 20% was lost in the corresponding uninoculated control. On pyruvate-fumarate medium, MeHg was degraded to the same extent (about 30%) in both cultures and uninoculated controls (Fig. 2C). The degradation of MeHg in uninoculated culture medium, in the dark, highlights the well-known lability of this compound and the need for appropriate controls when assessing demethylation in cultures and in the environment. It is notable that the sulfidogenic cultures degraded more MeHg than did uninoculated controls, while the nonsulfidogenic cultures did not. Chemical disproportionation of MeHg by sulfide (19, 69) is one potential explanation for the loss of MeHg in sulfidogenic cultures. We conducted a separate, matched experiment to test for abiotic MeHg degradation by sulfide in these media and found <3% MeHg degradation in uninoculated medium in the presence of up to 10 mM sulfide over 32 h at 27°C (see Fig. S4 in the supplemental material). We interpret these results to mean that demethylation is at least in part an active microbial process in this strain. However, the mechanism and extent of MeHg degradation by DSRB need further attention.
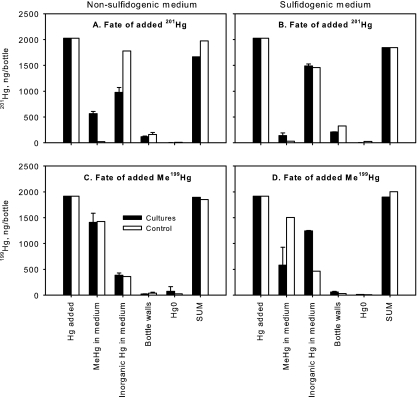
Fig. 3.
Mass balance of Hg and MeHg in strain ND132 culture bottles after 68 h. Panels A and C are from pyruvate-fumarate cultures; panels B and D are from lactate-sulfate cultures. Panels A and B show the fate of inorganic 201Hg added to cultures; panels C and D show the fate of Me199Hg added to cultures. Data correspond to the last time point in Fig. 2. Closed bars represent averages of results from triplicate cultures, with standard deviations. Open bars represent uninoculated medium controls (single bottles). Each bar represents the total amount of Hg or MeHg in each phase in the 200-ml culture bottles. Left to right: total Hg or MeHg added to bottles, MeHg in culture medium (unfiltered), inorganic Hg in culture medium (unfiltered), total Hg on bottle walls, Hg0, and the sum of all measured 201Hg or 199Hg at 68 h.
To evaluate the products of methylation and demethylation, and to ensure that the sampling design captured all of the Hg and MeHg in the experiments, we determined the mass balances for inorganic Hg and MeHg in the cultures (Fig. 3). We took filtered and unfiltered samples from the cultures and controls for Hg and MeHg analysis over time. At the end of the incubations, we purged the remaining medium in each bottle onto a gold column to trap any gaseous Hg that had formed. After the culture bottles were emptied, Hg that had sorbed to the glass bottle walls was measured by adding digestion acid to the empty bottles and then determining Hg in the acid.
Figure 3A and B show the mass balances for Hg added to cultures and controls as inorganic 201Hg. About 2 μg (as Hg) of 201HgCl2 was added to each 200-ml culture bottle prior to inoculation (leftmost bars in each panel). The remaining bars show the distribution of 201Hg or MeHg after 68 h of incubation (corresponding to the last data point in Fig. 2). Other than conversion to MeHg, the Hg added as inorganic 201Hg remained mainly as inorganic Hg in culture medium in both sulfidogenic and nonsulfidogenic media. In this figure, the bars showing Hg and MeHg in culture medium include both the filterable and the particulate phases. Between 5 and 15% of the added 201Hg(II) had sorbed to the glass bottle walls (fourth set of bars). Less than 1% of 201Hg was converted to DGM in either medium and in uninoculated controls. After 68 h growth, the sum of all measured fractions was within 10% of the amount of added 201Hg in all but one of the bottles (rightmost bar in each panel).
Figure 3C and D show the mass balances for Me199Hg added to cultures and controls. About 2 μg (as Hg) of Me199HgCl2 was added to each 200-ml culture bottle prior to inoculation (leftmost bars in each panel). Me199Hg was degraded to inorganic 199Hg(II), with less than 5% of Me199Hg converted to DGM in either medium. After 68 h of growth, the sum of all measured fractions was within 5% of the added Me199Hg in all of the culture bottles. Although dimethylmercury was not explicitly measured, it would have been included in the MeHg fraction, since our samples were preserved in HCl acid, which readily converts dimethylmercury to MeHg in aqueous solution (4). This mass balance helps confirm that the declines in MeHg concentration were due to degradation (biotic and abiotic) and not to other loss mechanisms (like sorption to bottle walls or reduction to elemental Hg0).
We also measured the distribution of Hg and MeHg between filterable and particulate phases within the culture medium (Fig. 4). These data provide information on bioavailability, sorption of Hg and MeHg to cells, fate of MeHg formed by cells, and chemical precipitation of Hg in uninoculated control medium. Even in uninoculated medium, more than half of the inorganic 201Hg was initially found in the particulate phase (Fig. 4C and D), although there was no visible precipitate (Fig. 4C and D). In cultures, less than 10% of inorganic Hg remained in the filterable phase. Interestingly, this was true of both sulfidic and nonsulfidogenic cultures, suggesting that sulfide precipitation is not the driving mechanism. We interpret this to mean that there was rapid precipitation of inorganic Hg in the culture medium and, additionally, rapid association of inorganic Hg with cells. Potential Hg-containing precipitates include Ca and Mg phosphates and any TiO2 formed from the Ti-NTA medium reductant. Initially, almost all of the Me201Hg produced from inorganic 201Hg by strain ND132 was filterable (Fig. 4E and F), demonstrating that most MeHg produced by cells is excreted. Over time, the Me201Hg produced by cultures moved slowly into a particulate phase. The Me199Hg added to cultures remained primarily in the filterable phase throughout the time course (data not shown).
Fig. 4.
Distribution of Hg and MeHg between the filterable and particulate phases in cultures and controls over time in batch cultures. “Filterable” refers to any Hg or MeHg that passes a 0.2-μm-pore-sized filter and is therefore not adsorbed to cells or bottle walls. Data correspond to the experiments represented in Fig. 2 and 3. (A and B) Inorganic 201Hg in cultures. (C and D) Inorganic 201Hg in uninoculated control medium. (E and F) Me201Hg formed from inorganic 201Hg in cultures. Data points show the averages of results from triplicate cultures, with standard deviations, or from single controls. Hg and MeHg were measured explicitly in the filtered and unfiltered samples; particulate data were obtained by subtraction. The level for inorganic Hg was calculated as the difference between the levels for total and MeHg.
Role of growth mode and reductants in methylation.
We examined the role of substrates, electron acceptors, and reductants in MeHg production by measuring MeHg production under different culture conditions (Fig. 5). The choice of organic substrate had limited effect on MeHg production. However, as expected (6, 7), sulfidogenic growth resulted in an almost 10-fold decrease in MeHg production relative to the level for growth without sulfide production. The choice of reductant had little impact on net MeHg production, at least during batch culture growth (Fig. 5, right).
Fig. 5.
Influence of substrates, electron acceptors, and medium reductants on growth and MeHg production by D. desulfuricans ND132. Data were collected at the end of batch growth on 40 mM substrates. Left panels show growth with a range of electron donors and acceptors, all reduced with 100 μM Ti-NTA; “S” represents sulfate (40 mM). Right panels show growth on sulfate-lactate (S-lac), sulfate-pyruvate (S-pyr), sulfate-formate-aceate (Sfor-ace), pyruvate (pyr), fumarate (fum), fumarate-pyruvate (fum-pyr), or fumarate-formate (fum-for) medium with Ti-NTA, thioglycolate-ascorbate ([Thio/asc]), or cysteine ([cys]) as the medium reductant (all added to give 100 μM). Error bars represent the standard deviations of results from three replicate growth tubes, with levels for blanks (uninoculated culture medium) subtracted where appropriate.
Mercury concentration dependence of MeHg production.
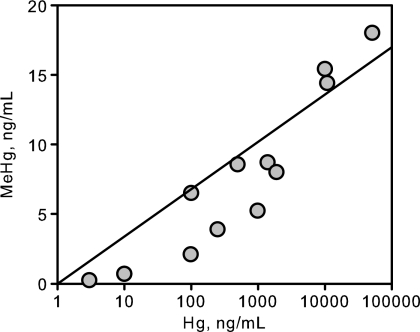
We also explored the relationship between Hg concentration in culture medium and net MeHg production. A concentration dependence experiment was done with batch cultures, and MeHg concentration was examined once cultures reached stationary phase. Under these test conditions, net MeHg production by strain ND132 growing on lactate-sulfate medium was a strong positive function of the log of the total added Hg concentration (Fig. 6).
Fig. 6.
Concentration dependence of MeHg production on inorganic Hg(II) added to medium by strain ND132 growing sulfidogenically on lactate-sulfate medium. All data were taken from batch cultures once cells had reached stationary phase. Mercury was added to culture medium prior to inoculation. The final optical densities (A660) of cultures ranged from 0.4 to 0.7. The regression line fit to the data forced the intercept through zero: MeHg = 3 × (log Hg concentration).
Hg methylation is not inducible.
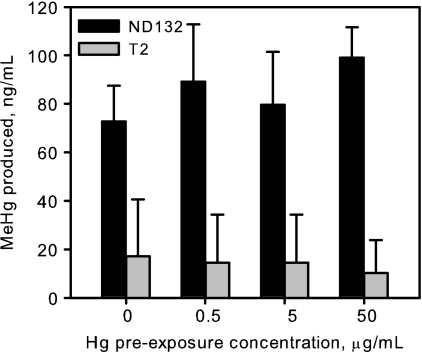
We tested the inducibility of methylation in two Hg-methylating DSRB isolates, D. desulfuricans ND132 and another Chesapeake Bay Desulfovibrio isolate, T2. Cultures were exposed to four different Hg concentrations for multiple generations, after which time MeHg production rates were measured at a fixed mercury concentration. Preexposure to Hg did not affect rates of MeHg production by the strains tested (Fig. 7). For both strains, neither MeHg production nor growth was affected by preexposure to any of the Hg concentrations tested (analysis of variance [ANOVA]; single factor, alpha <0.05). Mercury methylation is a constitutive activity in these organisms. This small study also highlights the resistance of DSRB to Hg.
Fig. 7.
Preexposure of two DSRB cultures to mercury did not increase MeHg production rates. D. desulfuricans ND132 and D. desulfuricans T2 were exposed to 0, 0.5, 5, or 50 μg Hg/ml through four batch culture cycles and then assayed for MeHg production from batch cultures with 500 ng/ml added HgCl2. Bars show the amount of MeHg produced by cells preexposed to different Hg concentrations. Each bar represents the average and standard deviation of results from 4 methylation assays. Cell growth was not affected by preexposure to Hg, as assessed by the final optical density and the growth rate in the methylation assays (data not shown).
The ability to produce MeHg does not confer Hg resistance.
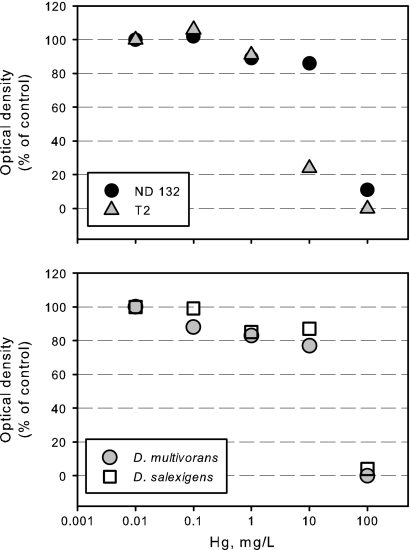
To test whether the ability to produce MeHg confers Hg resistance on DSRB species, we compared the toxicities of Hg(II) to four estuarine DSRB species, two of which are capable of MeHg production and two of which are not. D. desulfuricans ND132 was no more or less resistant to inorganic Hg (based on growth) than two other estuarine Desulfovibrio strains that are not capable of Hg methylation (Fig. 8). One of the Hg-methylating strains tested (T2) was more sensitive to Hg than were the two nonmethylating strains tested. A study in which the aqueous concentration and complexation of Hg were controlled and modeled would provide stronger evidence, but these results suggest that the ability to produce MeHg does not confer Hg resistance. This makes sense given that MeHg is more toxic to bacteria than is inorganic Hg (45).
Fig. 8.
The ability to produce MeHg does not confer Hg resistance. Mercury toxicity, as assessed by optical density at the end of batch culture growth, is shown for four estuarine DSRB species. The strains in the top panel are capable of MeHg production, whereas the strains in the bottom panel are not. All cultures were grown fermentatively on pyruvate. Each data point represents the average of results from three separate cultures. The standard deviation for each point averaged about 5%. OD660 readings were corrected for OD in medium blanks with matched Hg concentrations.
Mercury methylation capability among Desulfovibrio strains.
In order to better understand the distribution of strains capable of producing MeHg within the genus Desulfovibrio, the Hg methylation ability of several Desulfovibrio strains was tested (Table 2). The Hg methylation capability of many of these organisms has not previously been published, with the exceptions of Desulfovibrio desulfuricans subsp. desulfuricans ATCC 13541 (47), D. africanus DSM 2603 (27) and D. vulgaris DSM 644 (62). Among the strains from culture collections, only D. africanus DSM 2603 produced MeHg concentrations significantly above that of uninoculated medium controls. Both Chesapeake Bay strains produced MeHg, although at very different rates.
Table 2.
Mercury methylation capabilities of several Desulfovibrio species
| Species or strain | Source | Medium | MeHg produced (ng/ml)a |
|
|---|---|---|---|---|
| Avg (SD) | Abiotic control | |||
| D. desulfuricans aestuariiSylt3 | ATCC 29578 | Sulfate-lactate | 0.040 (0.02) | 0.020 |
| D. desulfuricans subsp. desulfuricans Essex 6 | DSM 642 | Sulfate-lactate | 0.001 (0.000) | 0.001 |
| D. desulfuricans subsp. desulfuricans El Agheila Z | DSM 1926 | Sulfate-lactate | 0.001 (0.001) | 0.001 |
| D. desulfuricans G20 | J. Wall | Sulfate-lactate | 0.001 (0.001) | 0.001 |
| D. gigas | DSM 1382 | Sulfate-lactate | 0.000 (0.001) | 0.020 |
| D. salexigens | DSM 2638 | Sulfate-lactate | 0.24 (0.32) | 0.08 |
| D. africanus | DSM 2603 | Sulfate-lactate | 0.51 (0.052) | 0.02 |
| D. africanus | DSM 2603 | Pyruvate-fumarate | 3.4 (0.79) | 0.003 |
| D. vulgaris Hildenborough | DSM 644 | Sulfate-lactate | 0.03 (0.02) | 0.01 |
| Desulfovibrio sp. strain T2 | Chesapeake sediments | Sulfate-lactate | 1.5 (0.3) | 0.02 |
| Desulfovibrio sp. strain X2 | Chesapeake sediments | Sulfate-lactate | 0.35 (0.1) | 0.10 |
| D. desulfuricans ND132 | Chesapeake sediments | Sulfate-lactate | 0.70 (0.26) | 0.16 |
| D. desulfuricans ND132 | Chesapeake sediments | Pyruvate-fumarate | 3.3 (0.30) | 0.05 |
Methylation was assayed by measuring MeHg at the end of batch growth, i.e., once cultures reached stationary phase, on medium containing 10 ng Hg/ml, added as HgCl2 prior to inoculation. All assays were performed in triplicate, and averages are presented with standard deviations (SD).
It can be difficult to compare reported methylation rates among organisms, because medium chemistry, growth phase, and mercury concentration all affect methylation rates, as described here for strain ND132. However, King et al. (47) showed that MeHg production rates differ among Hg-methylating bacterial species. We also found significant difference among strains grown on the same medium (Table 2).
With the addition of the strains listed in Table 2, 19 Desulfovibrio strains have been tested for their ability to produce MeHg. Of those, about half (nine) do so. More Desulfovibrio strains have been tested than strains in any other single genus. Table S3 in the supplemental material provides a compilation of literature reports of Hg methylation by Desulfovibrio strains.
Phylogenetic distribution of the ability to produce MeHg.
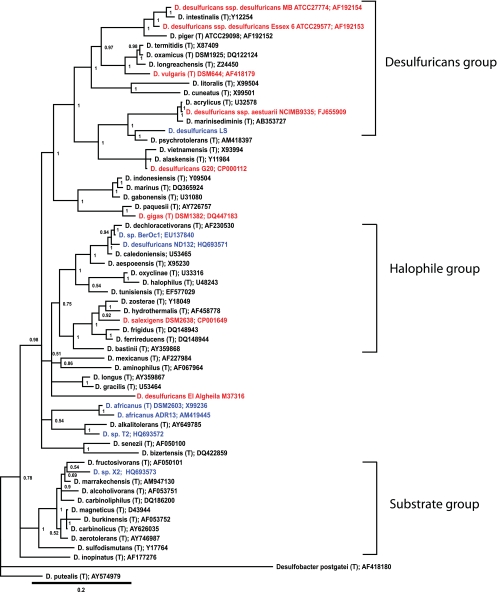
We constructed a 16S rRNA gene-based phylogeny of the genus Desulfovibrio in order to explore the evolutionary basis for MeHg production within this group (Fig. 9). Desulfovibrio is a large diverse genus, with 49 type strains listed in the RDP database (17). Our alignment included the 15 strains for which Hg methylation has been tested and for which sequences were also available (including strains presented here and those in the literature), plus all of the Desulfovibrio type strains. The analysis yielded four well-supported deep-branching main clades plus a number of less-well-supported smaller groups.
Fig. 9.
16S rRNA gene phylogeny for the genus Desulfovibrio. The tree was based on the aligned gene sequences of 16S rRNA from 49 type species of Desulfovibrio, plus 13 additional species of Desulfovibrio that have been tested for mercury methylation ability (Table 2; see also Table S2 in the supplemental material). The tree contains all Desulfovibrio strains for which Hg methylation ability has been reported to date, including those previously reported by other investigators (where sequences were available) (Table S2) and those reported here (Table 2). Type strains are indicated by “(T).” Strains able to produce MeHg are shown in blue, those unable to do so are in red, and strains that have not been tested are black. Positive methylation was defined as MeHg production significantly in excess of available (usually abiotic) controls. Alignment and phylogenetic methods are given in detail in the text. Desulfobacter postgatei was used as a reference and outgroup taxon.
Most of the well-studied Desulfovibrio desulfuricans type strains, including D. vulgaris Hildenborough and D. desulfuricans strains Essex 6 and G20, fall into two related groups, labeled “Desulfuricans” groups 1 and 2 in Fig. 9. Interestingly, D. desulfuricans LS also aligned in this clade. However, only a partial (<1,200-bp) and discontinuous sequence is available for this lost strain, and its placement should be interpreted with some caution. Most of the strains tested for methylation to date fall in this group, but other than strain LS, there are no identified Hg methylators.
Strain ND132 falls into a group that is dominated by marine and halophilic strains (labeled the “Halophile” group in Fig. 9). This group includes another closely related methylator, Desulfovibrio strain BerOc1, isolated from a Mediterranean estuary (62). However, the group also includes a strain without the ability to produce MeHg. Weakly related to this group is D. africanus. Both the type strain and strain ADR13, isolated from a French estuary (62), are Hg methylators. They are both weakly related to Hg-methylating strain T2 from Chesapeake Bay.
A group of species with the ability to utilize a broader set of substrates than is common in other Desulfovibrio species clusters loosely together in the “Substrate” group. This group contains Chesapeake Bay Hg-methylating strain X2. No other species in the group have been tested.
DISCUSSION
Phylogenetic distribution of Hg-methylating organisms.
About half of the bacterial species known to produce MeHg are in the genus Desulfovibrio, but only about half of the Desulfovibrio species tested so far have the ability to produce MeHg. All of the other known Hg methylators are also Deltaproteobacteria. Ranchou-Peyruse et al. (62) constructed a 16S rRNA-based phylogeny of Hg methylators in the Deltaproteobacteria and concluded that, within the Deltaproteobacteria, there is no obvious taxonomic structure associated with the ability to methylate Hg.
Because so much bacterial diversity is unaccounted for in testable pure cultures, and since all of the known type strains of the genus have not been tested, a fully statistically supported ancestral state reconstruction for Hg methylation (e.g., using ModelTest or other methods) was not possible. However, our more specific alignment of Hg methylators within the genus Desulfovibrio suggests a potential, but unidentified, evolutionary basis for Hg methylation. Most of the commonly studied Desulfovibrio strains, including those for which full genomic sequences are available, aligned into a clade that contains few (if any) Hg methylators, while most of the strains with the ability to methylate Hg fall, at least loosely, into a group that is dominated by halophiles. This finding merits further investigation but must be interpreted with caution since several of the lineages within our analysis have relatively weak support, and the fraction of Desulfovibrio strains tested for Hg methylation remains low. As more Desulfovibrio genome sequences become available, it will be interesting to see how our alignment holds up with multiple-gene phylogenies. Further, since few organisms outside the Deltaproteobacteria have been tested for methylation, it is premature to assume that this ability is confined to a few clades within Deltaproteobacteria. To summarize, most of the Hg-methylating bacteria identified to date fall within a few clades in the Deltaproteobacteria, including two clades in the genera Desulfovibrio and Geobacte r plus a few scattered species in the Desulfobacteraceae and the Desulfovibrionaceae.
Mercury methylation and MeHg demethylation by ND132.
D. desulfuricans ND132 has the ability to produce MeHg and to degrade it, as first reported by Pak and Bartha (58, 59). The net rate of MeHg production depends on the ratio of rate constants for each process and the substrate pools. This is the same balance observed in most anaerobic sediments, where demethylation rate constants generally exceed methylation rate constants, but the substrate pool for methylation is much larger than the MeHg concentration, so that on balance a small percentage of the total Hg pool is found as MeHg (e.g., references 42 and 57).
As expected, gross methylation rates were substantially lower in the presence of sulfide. The strong inhibition of net MeHg production by sulfide has repeatedly been demonstrated to occur in cultures and natural sediments (9, 20, 31, 35, 54). Interestingly, the demethylation rates observed in strain ND132 were much higher in sulfidogenic cultures. Thus, the net MeHg production level is lower in sulfidogenic cultures because of both lower production and higher degradation. Abiotic controls containing sulfide did not significantly demethylate MeHg, suggesting no direct effect of sulfide on demethylation. The mechanism of MeHg degradation by species that do not contain the mer operon is unknown. Although sulfide-mediated abiotic degradation has been proposed, our data suggest that demethylation is a metabolic process, albeit one that is significantly affected by medium chemistry and MeHg bioavailability.
We observed that MeHg production by strain ND132, under otherwise fixed conditions, was a function of the log Hg concentration (Fig. 6). Put another way, the fraction of Hg converted to MeHg decreases significantly as Hg concentrations increase. King et al. (49) also found a nonlinear relationship in estuarine sediment slurries and attributed it to first-order Michaelis-Menten kinetics. They hypothesized that the relationship was due to saturation of methylating enzymes in SRB. However, this relationship does not necessarily mean that Hg methylation follows Michaelis-Menten kinetics or that Hg uptake becomes saturated at higher Hg concentrations (48). It is more plausible to attribute the relationship to the solubility and speciation of inorganic Hg in the culture medium (10). Many researchers have hypothesized that Hg is available only to cells from the aqueous phase (8, 56) or perhaps bound to small organics that are taken up by cells (3, 33, 34, 66). Mercury is highly reactive to surfaces and particles, and the concentration in solution is not usually a linear function of the total Hg concentration.
Status of understanding the Hg methylation process.
The ability to produce MeHg is constitutive, i.e., not inducible by prior exposure to Hg. However, the rate of methylation by any organism depends heavily on the bioavailability of Hg in culture medium for uptake. Like Desulfobulbus propionicus (6, 37), strain ND132 is an example of a DSRB that can produce MeHg when growing on a variety of electron acceptors. In our hands, all DSRB strains with the ability to methylate Hg have done so under all culture conditions tested, while strains that did not methylate Hg could not be induced to do so by changing conditions. From these results, we infer that methylation is not inducible nor is it linked directly to the sulfate electron transport pathway.
Our data show that Hg methylation occurs inside cells but that MeHg is rapidly exported out of cells. Our tests with spent culture medium confirm that extracellular metabolites do not methylate Hg. Mercury complexation dramatically influences uptake and methylation. In order to assess and compare Hg methylation rates among bacterial species and across growth conditions, it is necessary to know how Hg has partitioned among cells, medium, and bottle walls. Some measure of important aqueous Hg ligands, including thiols, dissolved organic matter, and sulfide, is also critical. Although it is clear that Hg complexation affects Hg uptake by DSRB, competing hypotheses for Hg uptake mechanisms by Deltaproteobacteria have not been resolved; indeed, uptake mechanisms may differ among Hg-methylating organisms (8, 66).
The ability to produce MeHg does not confer Hg resistance. Thus, Hg methylation did not evolve as a mechanism to protect cells from Hg toxicity. Rather, Hg methylation is most likely a metabolic mistake. Choi et al. (16) showed that MeB12 is the proximate methylating agent for Hg in Desulfovibrio strain LS, with the methyl group likely originating through the acetyl-CoA pathway. However, we now know that this pathway is not common to all Hg methylators, so small differences in enzymes within this pathway probably do not control Hg methylation ability among strains. We remain ignorant of whether differences in methylation ability among bacterial species are driven by differences in methyl transfer pathways, uptake mechanisms, metal-thiol housekeeping within cells, or something we have not yet suspected.
Desulfovibrio desulfuricans ND132 as a model organism.
D. desulfuricans ND132 makes an ideal organism for the study of Hg methylation because it exhibits exceptionally high rates of MeHg production but otherwise appears to be a relatively typical anaerobic, mesophilic estuarine Desulfovibrio strain. Phylogenetically, ND132 falls into a large group of salt-tolerant Desulfovibrio strains that are relatively distant from the best-studied strains in the genus. The organism has a wide range of salt and pH tolerance. Unlike many DSRB strains, ND132 has the ability to grow well using fumarate as an alternative electron acceptor to sulfate, allowing study of methylation during rapid growth while avoiding sulfide inhibition of methylation. The newly finished genome for strain ND132 (14) is the first complete genome that we are aware of for a Desulfovibrio strain that generates MeHg. The genome sequence will allow comparison with the many available full-genome sequences of other Desulfovibrio species and other DSRB and FeRB and present an unparalleled opportunity for follow-on studies that might include comparative transcriptomic and proteomic studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Georgia Riedel for MeHg analyses, Sarah Werner and the Smithsonian Laboratory of Analytical Biology for 16S rRNA analysis, and Taylan Morcol and Tyler Bell for culture work with ND132. R. Devereux provided the partial 16S rRNA sequence from D. desulfuricans LS.
This work was supported by the U.S. Department of Energy under the Subsurface Biogeochemical Research Program (SBR), Office of Biological and Environmental Research, Office of Science, in part through the Mercury Science Focus Area Program at Oak Ridge National Laboratory (ORNL), through DE-FG02-07ER64396 (J.D.W.), by National Science Foundation grant DEB0351050 (C.C.G.), and by the SERC Research Experience for Undergraduates program. ORNL is managed by University of Tennessee UT—Battelle, LLC, for the Department of Energy under contract no. DE-AC05-00OR22725.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Axelrad D. A., Bellinger D. C., Ryan L. M., Woodruff T. J. 2007. Dose-response relationship of prenatal mercury exposure and IQ: an integrative analysis of epidemiologic data. Environ. Health Perspect. 115:609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Badziong W., Ditter B., Thauer R. K. 1979. Acetate and carbon-dioxide assimilation by Desulfovibrio vulgaris (Marburg), growing on hydrogen and sulfate as sole energy-source. Arch. Microbiol. 123:301–305 [Google Scholar]
- 3. Barkay T., Gillman M., Turner R. R. 1997. Effects of dissolved organic carbon and salinity on bioavailability of mercury. Appl. Environ. Microbiol. 63:4267–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barone V., Bencini A., Totti F., Uytterhoeven M. G. 1996. Theoretical characterization of the mechanism of Hg-C bond cleavage by halogenic acids. Organometallics 15:1465–1469 [Google Scholar]
- 5. Benoit J. M., Gilmour C. C., Heyes A., Mason R. P., Miller C. L. 2003. Geochemical and biological controls over methylmercury production and degradation in aquatic ecosystems, p. 262–297 In Cai Y., Braids O. C.(ed.), Biogeochemistry of environmentally important trace elements. ACS Symposium Series, vol. 835. American Chemical Society, Washington, DC [Google Scholar]
- 6. Benoit J. M., Gilmour C. C., Mason R. P. 2001. Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3). Appl. Environ. Microbiol. 67:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benoit J. M., Gilmour C. C., Mason R. P. 2001. The influence of sulfide on solid phase mercury bioavailability for methylation by pure cultures of Desulfobulbus propionicus (1pr3). Environ. Sci. Technol. 35:127–132 [DOI] [PubMed] [Google Scholar]
- 8. Benoit J. M., Gilmour C. C., Mason R. P., Heyes A. 1999. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ. Sci. Technol. 33:951–957 [Google Scholar]
- 9. Benoit J. M., Gilmour C. C., Mason R. P., Riedel G. S., Riedel G. F. 1998. Behavior of mercury in the Patuxent River estuary. Biogeochemistry 40:249–265 [Google Scholar]
- 10. Benoit J. M., Mason R. P., Gilmour C. C. 1999. Estimation of mercury-sulfide speciation in sediment pore waters using octanol-water partitioning and implications for availability to methylating bacteria. Environ. Toxicol. Chem. 18:2138–2141 [DOI] [PubMed] [Google Scholar]
- 11. Berman M., Chase T., Bartha R. 1990. Carbon flow in mercury biomethylation by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 56:298–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Branfireun B. A., Roulet N. T., Kelly C. A., Rudd J. W. M. 1999. In situ sulphate stimulation of mercury methylation in a boreal peatland: toward a link between acid rain and methylmercury contamination in remote environments. Global Biogeochem. Cycles 13:743–750 [Google Scholar]
- 13. Bridou R., Monperrus M., Gonzalez P. R., Guyoneaud R., Amouroux D. 2011. Simultaneous determination of mercury methylation and demethylation capacities of various sulfate-reducing bacteria using species-specific isotopic tracers. Environ. Toxicol. Chem. 30:337–344 [DOI] [PubMed] [Google Scholar]
- 14. Brown S. D., et al. 2011. Genome sequence of the mercury-methylating strain Desulfovibrio desulfuricans ND132. J. Bacteriol. 193:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi S. C., Chase T., Bartha R. 1994. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 60:1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi S. C., Chase T., Bartha R. 1994. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 60:4072–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cole J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Compeau G. C., Bartha R. 1985. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 50:498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Craig P. J., Moreton P. A. 1984. The role of sulfide in the formation of dimethyl mercury in river and estuary sediments. Mar. Pollut. Bull. 15:406–408 [Google Scholar]
- 20. Craig P. J., Moreton P. A. 1986. Total mercury, methylmercury and sulfide levels in British estuarine sediments. Water Res. 20:1111–1118 [Google Scholar]
- 21. Creswell J. E., et al. 2008. Factors controlling temporal and spatial distribution of total mercury and methylmercury in hyporheic sediments of the Allequash Creek wetland, northern Wisconsin. J. Geophys. Res. Biogeosci. 113:9 [Google Scholar]
- 22. Dolor M. K., Gilmour C. C., Helz G. R. 2009. Distinct microbial behavior of Re compared to Tc: evidence against microbial Re fixation in aquatic sediments. Geomicrobiol. J. 26:470–483 [Google Scholar]
- 23. Drott A., Lambertsson L., Bjorn E., Skyllberg U. 2007. Importance of dissolved neutral mercury sulfides for methyl mercury production in contaminated sediments. Environ. Sci. Technol. 41:2270–2276 [DOI] [PubMed] [Google Scholar]
- 24. Eaton A. D., Clesceri L. S., Rice E. W., Greenberg A. E., Franson M. A. H. (ed.). 2005. Method 4500. Sulfide. G. Ion-selective electrode method. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC [Google Scholar]
- 25. Eaton A. D., Clesceri L. S., Rice E. W., Greenberg A. E., Franson M. A. H. 2005. Method 4110. Determination of anions by ion chromatography. B. Ion chromatography with chemical suppression of eluent conductivity. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC [Google Scholar]
- 26. Ekstrom E. B., Morel F. M. M. 2008. Cobalt limitation of growth and mercury methylation in sulfate-reducing bacteria. Environ. Sci. Technol. 42:93–99 [DOI] [PubMed] [Google Scholar]
- 27. Ekstrom E. B., Morel F. M. M., Benoit J. M. 2003. Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria. Appl. Environ. Microbiol. 69:5414–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fleming E. J., Mack E. E., Green P. G., Nelson D. C. 2006. Mercury methylation from unexpected sources: molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Appl. Environ. Microbiol. 72:457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilmour C. 1985. Estuarine methylation of tin and its relationship to the microbial sulfur cycle. University of Maryland, College Park, MD [Google Scholar]
- 30. Gilmour C. C., Leavitt M. E., Shiaris M. P. 1990. Evidence against incorporation of exogenous thymidine by sulfate-reducing bacteria. Limnol. Oceanogr. 35:1401–1409 [Google Scholar]
- 31. Gilmour C. C., et al. 1998. Methylmercury concentrations and production rates across a trophic gradient in the northern Everglades. Biogeochemistry 40:327–345 [Google Scholar]
- 32. Gilmour C. C., Tuttle J. H., Means J. C. 1987. Anaerobic microbial methylation of inorganic tin in estuarine sediment slurries. Microb. Ecol. 14:233–242 [DOI] [PubMed] [Google Scholar]
- 33. Golding G. R., Kelly C. A., Sparling R., Loewen P. C., Barkay T. 2007. Evaluation of mercury toxicity as a predictor of mercury bioavailability. Environ. Sci. Technol. 41:5685–5692 [DOI] [PubMed] [Google Scholar]
- 34. Golding G. R., et al. 2002. Evidence for facilitated uptake of Hg(II) by Vibrio anguillarum and Escherichia coli under anaerobic and aerobic conditions. Limnol. Oceanogr. 47:967–975 [Google Scholar]
- 35. Hammerschmidt C. R., Fitzgerald W. F. 2004. Geochemical controls on the production and distribution of methylmercury in near-shore marine sediments. Environ. Sci. Technol. 38:1487–1495 [DOI] [PubMed] [Google Scholar]
- 36. Harmon S. M., King J. K., Gladden J. B., Chandler G. T., Newman L. A. 2004. Methylmercury formation in a wetland mesocosm amended with sulfate. Environ. Sci. Technol. 38:650–656 [DOI] [PubMed] [Google Scholar]
- 37. Henry E. 1992. The role of sulfate-reducing bacteria in environmental mercury methylation. Harvard University, Cambridge, MA [Google Scholar]
- 38. Heyes A., Mason R. P., Kim E. H., Sunderland E. 2006. Mercury methylation in estuaries: Insights from using measuring rates using stable mercury isotopes. Mar. Chem. 102:134–147 [Google Scholar]
- 39. Hintelmann H., Evans R. D. 1997. Application of stable isotopes in environmental tracer studies—measurement of monomethylmercury (CH3Hg+) by isotope dilution ICP-MS and detection of species transformation. Fresenius J. Anal. Chem. 358:378–385 [Google Scholar]
- 40. Hintelmann H., Keppel-Jones K., Evans R. D. 2000. Constants of mercury methylation and demethylation rates in sediments and comparison of tracer and ambient mercury availability. Environ. Toxicol. Chem. 19:2204–2211 [Google Scholar]
- 41. Hintelmann H., Ogrinc N. 2003. Determination of stable mercury isotopes by ICP/MS and their application in environmental studies, p. 321–338 In Cai Y., Braids O. C. (ed.), Biogeochemistry of environmentally important trace elements . ACS Symposium Series, vol. 835. American Chemical Society, Washington, DC [Google Scholar]
- 42. Hollweg T. A., Gilmour C. C., Mason R. P. 2009. Methylmercury production in sediments of Chesapeake Bay and the mid-Atlantic continental margin. Mar. Chem. 114:86–101 [Google Scholar]
- 43. Jay J. A., et al. 2002. Mercury methylation by Desulfovibrio desulfuricans ND132 in the presence of polysulfides. Appl. Environ. Microbiol. 68:5741–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeremiason J. D., et al. 2006. Sulfate addition increases methylmercury production in an experimental wetland. Environ. Sci. Technol. 40:3800–3806 [DOI] [PubMed] [Google Scholar]
- 45. Jonas R. B., Gilmour C. C., Stoner D. L., Weir M. M., Tuttle J. H. 1984. Comparison of methods to measure acute metal and organometal toxicity to natural aquatic microbial communities. Appl. Environ. Microbiol. 47:1005–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kerin E. J., et al. 2006. Mercury methylation by dissimilatory iron-reducing bacteria. Appl. Environ. Microbiol. 72:7919–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. King J. K., Kostka J. E., Frischer M. E., Saunders F. M. 2000. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl. Environ. Microbiol. 66:2430–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. King J. K., Kostka J. E., Frischer M. E., Saunders F. M., Jahnke R. A. 2001. A quantitative relationship that remonstrates mercury methylation rates in marine sediments are based on the community composition and activity of sulfate-reducing bacteria. Environ. Sci. Technol. 35:2491–2496 [DOI] [PubMed] [Google Scholar]
- 49. King J. K., Saunders F. M., Lee R. F., Jahnke R. A. 1999. Coupling mercury methylation rates to sulfate reduction rates in marine sediments. Environ. Toxicol. Chem. 18:1362–1369 [Google Scholar]
- 50. Klonowska A., et al. 2008. Hexavalent chromium reduction in Desulfovibrio vulgaris Hildenborough causes transitory inhibition of sulfate reduction and cell growth. Appl. Microbiol. Biotechnol. 78:1007–1016 [DOI] [PubMed] [Google Scholar]
- 51. Lin C. C., Jay J. A. 2007. Mercury methylation by planktonic and biofilm cultures of Desulfovibrio desulfuricans. Environ. Sci. Technol. 41:6691–6697 [DOI] [PubMed] [Google Scholar]
- 52. Marietou A., Griffiths L., Cole J. 2009. Preferential reduction of the thermodynamically less favorable electron acceptor, sulfate, by a nitrate-reducing strain of the sulfate-reducing bacterium Desulfovibrio desulfuricans 27774. J. Bacteriol. 191:882–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martin-Doimeadios R. C., et al. 2004. Mercury methylation/demethylation and volatilization pathways in estuarine sediment slurries using species-specific enriched stable isotopes. Mar. Chem. 90:107–123 [Google Scholar]
- 54. Marvin-DiPasquale M., Agee J. L. 2003. Microbial mercury cycling in sediments of the San Francisco Bay-Delta. Estuaries 26:1517–1528 [Google Scholar]
- 55. Marvin-DiPasquale M. C., Capone D. G. 1998. Benthic sulfate reduction along the Chesapeake Bay central channel. I. Spatial trends and controls. Mar. Ecol. Prog. Ser. 168:213–228 [Google Scholar]
- 56. Mitchell C. P. J., Gilmour C. C. 2008. Methylmercury production in a Chesapeake Bay salt marsh. J. Geophys. Res. Biogeosci. 113:G00C04doi: 10.1029/2008JG000765 [DOI] [Google Scholar]
- 57. Munthe J., et al. 2007. Recovery of mercury-contaminated fisheries. Ambio 36:33–44 [DOI] [PubMed] [Google Scholar]
- 58. Pak K., Bartha R. 1998. Products of mercury demethylation by sulfidogens and methanogens. Bull. Environ. Contam. Toxicol. 61:690–694 [DOI] [PubMed] [Google Scholar]
- 59. Pak K. R., Bartha R. 1998. Mercury methylation and demethylation in anoxic lake sediments and by strictly anaerobic bacteria. Appl. Environ. Microbiol. 64:1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pak K. R., Bartha R. 1998. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl. Environ. Microbiol. 64:1987–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Postgate J. R., Kent H. M., Robson R. L., Chesshyre J. A. 1984. The genomes of Desulfovibrio gigas and Desulfovibrio vulgaris. J. Gen. Microbiol. 130:1597–1601 [DOI] [PubMed] [Google Scholar]
- 62. Ranchou-Peyruse M., et al. 2009. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiology J. 26:1–8 [Google Scholar]
- 63. Roden E. E., Tuttle J. H. 1993. Inorganic sulfur cycling in mid and lower Chesapeake Bay sediments. Mar. Ecol. Prog. Ser. 93:101–118 [Google Scholar]
- 64. Rodriguez-Gonzalez P., et al. 2009. Species-specific stable isotope fractionation of mercury during Hg(II) methylation by an anaerobic bacteria (Desulfobulbus propionicus) under dark conditions. Environ. Sci. Technol. 43:9183–9188 [DOI] [PubMed] [Google Scholar]
- 65. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 66. Schaefer J. K., Morel F. M. M. 2009. High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat. Geosci. 2:123–126 [Google Scholar]
- 67. Scheulhammer A. M., Meyer M. W., Sandheinrich M. B., Murray M. W. 2007. Effects of environmental methylmercury on the health of wild birds, mammals, and fish. Ambio 36:12–18 [DOI] [PubMed] [Google Scholar]
- 68. Sorokin Y. I. 1966. Role of carbon dioxide and acetate in biosynthesis by sulphate-reducing bacteria. Nature 210:551–552 [DOI] [PubMed] [Google Scholar]
- 69. Wallschlager D., Hintelmann H., Evans R. D., Wilken R. D. 1995. Volatilization of dimethylmercury and elemental mercury from River Elbe floodplain soils. Water Air Soil Pollut. 80:1325–1329 [Google Scholar]
- 70. Wiener J. G., et al. 2006. Mercury in soils, lakes, and fish in Voyageurs National Park (Minnesota): importance of atmospheric deposition and ecosystem factors. Environ. Sci. Technol. 40:6261–6268 [DOI] [PubMed] [Google Scholar]
- 71. Zane G. M., Yen H. C. B., Wall J. D. 2010. Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 76:5500–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.