Abstract
Trehalose accumulation is a common cell defense strategy against a variety of stressful conditions. In particular, our team detected high levels of trehalose in Propionibacterium freudenreichii in response to acid stress, a result that led to the idea that endowing Lactococcus lactis with the capacity to synthesize trehalose could improve the acid tolerance of this organism. To this end, we took advantage of the endogenous genes involved in the trehalose catabolic pathway of L. lactis, i.e., trePP and pgmB, encoding trehalose 6-phosphate phosphorylase and β-phosphoglucomutase, respectively, which enabled the synthesis of trehalose 6-phosphate. Given that L. lactis lacks trehalose 6-phosphate phosphatase, the respective gene, otsB, from the food-grade organism P. freudenreichii was used to provide the required activity. The trehalose yield was approximately 15% in resting cells and in mid-exponential-phase cells grown without pH control. The intracellular concentration of trehalose reached maximal values of approximately 170 mM, but at least 67% of the trehalose produced was found in the growth medium. The viability of mutant and control strains was examined after exposure to heat, cold or acid shock, and freeze-drying. The trehalose-producing strains showed improved tolerance (5- to 10-fold-higher survivability) to acid (pH 3) and cold shock (4°C); there was also a strong improvement in cell survival in response to heat shock (45°C), and no protection was rendered against dehydration. The insight provided by this work may help the design of food-grade strains optimized for the dairy industry as well as for oral drug delivery.
INTRODUCTION
Lactococcus lactis is a mesophilic homofermentative lactic acid bacterium used worldwide as a starter culture in food fermentations. In the dairy industry its primary function is the conversion of lactose to lactic acid, which provides an effective preservation of the fermented product. In addition, this organism contributes to the organoleptic and nutritional properties of the fermented foods. A wealth of information on lactococcal physiology has accumulated during the last decades, and a battery of tools for genetic manipulation is now available (reviewed in references 14 and 31). Hence, it is not surprising that the potential of this microorganism as a cell factory for the production of flavors, texturizers, and nutraceuticals has been explored to a great extent (27). L. lactis is also an excellent host for the production of heterologous proteins (37). Indeed, the ease of gene expression combined with the GRAS (generally regarded-as-safe) status of L. lactis prompted the use of this bacterium as a live vehicle for the delivery of antigens (live vaccines) or therapeutic proteins to mucosal surfaces (4, 60).
The use of L. lactis strains in starter cultures depends on functional properties (flavor and texture development) as well as growth performance and robustness. L. lactis grows optimally at pH values in the range of 6.3 to 6.9, but as a consequence of its metabolic activity, lactic acid accumulates, causing an acidification of the growth medium and, ultimately, growth arrest at a pH of around 4.3 (56). Acid stress has detrimental effects on the physiology of L. lactis, including cell membrane damage and the inhibition of metabolic reactions (35). Moreover, the usefulness of L. lactis as a live vehicle for the oral delivery of pharmaceuticals depends to a large extent on the ability of cells to endure the harsh acidic conditions in the upper gastrointestinal tract. Furthermore, during culture handling, storage, and product processing, lactic acid bacteria have to cope with dehydration (freeze-drying), elevated temperatures (≥41°C, e.g., in cheese processing), and cold stress (2°C to 6°C), among other stresses (56). Although lactococcal growth occurs in the range of 10°C to 40°C, cell viability is severely affected beyond these limits (61). In this context, it is clear that good performance in clinical and industrial applications depends largely on the ability of L. lactis to withstand various stresses, in particular acid stress.
Trehalose is a nonreducing disaccharide ubiquitously distributed in nature and is well known for its role in protecting cells against a variety of stresses (3, 28, 30, 49). Our team observed a substantial increase in the intracellular content of trehalose in Propionibacterium freudenreichii in response to acid stress (9). Inspired by this observation, we anticipated that the accumulation of trehalose could be a good strategy to improve the survival of L. lactis against acid stress. Therefore, we set out to engineer trehalose production in L. lactis by the de novo introduction of the P. freudenreichii trehalose biosynthetic pathway. In this organism trehalose is synthesized in two steps via the TpS-TpP pathway, the most widely used route for the synthesis of this disaccharide (8). First, glucose is transferred from NDP-glucose to glucose 6-phosphate (G6P) to yield trehalose 6-phosphate (Tre6P), in a reaction catalyzed by Tre6P synthase (TPS); subsequently, Tre6P is dephosphorylated to trehalose by the action of TPP, i.e., Tre6P phosphatase (8). Unfortunately, despite several attempts, the functional expression of the gene encoding P. freudenreichii TPS in L. lactis was not achieved (10). Concurrently, while characterizing glucose metabolism in an L. lactis CcpA (carbon catabolite protein A) deletion mutant, we observed a transient accumulation of Tre6P. In view of previous work on trehalose catabolism in L. lactis (2), we concluded that the synthesis of Tre6P occurred via the action of the Tre6P phosphorylase (TrePP) and β-phosphoglucomutase (β-PGM), which catalyze reversible steps (Fig. 1). Based on these results, we endeavored to produce trehalose in L. lactis using genes exclusively from food-grade organisms: the overexpression of the endogenous trePP and pgmB genes in addition to otsB from the dairy organism P. freudenreichii (Fig. 1). Meanwhile, a report appeared in the literature on the production of trehalose by a L. lactis construct overexpressing the Escherichia coli otsBA operon (53); therefore, this strain was also constructed and used for comparison.
Fig. 1.
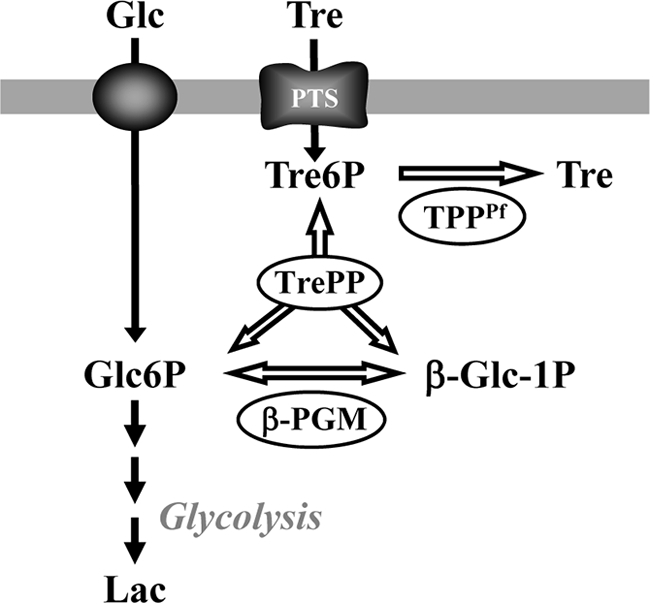
Scheme of the strategy followed to engineer Lactococcus lactis for the synthesis of trehalose. PTS, phosphotransferase system; TrePP, trehalose 6-phosphate phosphorylase; β-PGM, β-phosphoglucomutase; TPPPf, trehalose 6-phosphate phosphatase from Propionibacterium freudenreichii; Tre6P, trehalose 6-phosphate; Glc6P, glucose 6-phosphate; β-Glc1P, β-glucose 1-phosphate; Glc, glucose; Tre, trehalose; Lac, lactate. The genes encoding TrePP, β-PGM, and TPPPf were overexpressed in L. lactis NZ9000 with the purpose of channeling Glc6P for the synthesis of trehalose.
Here, we report the outcome of engineering L. lactis for trehalose synthesis. The intracellular and extracellular trehalose contents in the engineered strains were assessed; moreover, the abilities of these strains to survive stress associated with acid, cold, heat, and dehydration were examined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1 . For molecular biology procedures, L. lactis strains were grown as batch cultures (flasks) under static conditions in M17 medium (Difco) with 0.5% (wt/vol) glucose at 30°C. Propionibacterium freudenreichii cells were grown as described previously by Cardoso et al. (9). E. coli cells were grown aerobically at 37°C in LB medium. For physiological studies, L. lactis strains were grown in chemically defined medium (CDM) (44) with 1% glucose at 30°C under static conditions and without pH control (initial pH of 6.5) or in a 2-liter fermentor with pH controlled at 6.5. The pH was kept constant in the fermentor by the automatic addition of NaOH. Plasmid selection was achieved by the addition of chloramphenicol at a final concentration of 5 mg liter−1. Nisin (2 μg liter−1) was added when the optical density at 600 nm (OD600) reached approximately 0.4. Growth was monitored by measuring the OD600. Specific growth rates (μ) were calculated through linear regressions of the plots of ln(OD600) versus time during the exponential growth phase.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| MG1363 | Plasmid-free derivative of SH4109 | 22 |
| NZ9000 | MG1363; pepN::nisR nisK | 32 |
| NZ9000(pNZ8020) | Derivative of NZ9000 carrying pNZ8020 | This work |
| NZ9000ΔccpA | NZ9000 chromosomal deletion of ccpA | This work |
| NZ9000ΔccpA(pNZotsB) | Derivative of NZ9000ΔccpA carrying pNZotsB | This work |
| NZ9000ΔccpA(pNZtpo) | Derivative of NZ9000ΔccpA carrying pNZtpo | This work |
| NZ9000(pNZtpo) | Derivative of NZ9000 carrying pNZtpo | This work |
| NZ9000(pNZotsBA) | Derivative of NZ9000 carrying pNZotsBA | This work |
| Propionibacterium freudenreichii subsp. shermanii NIZO B365 | 9 | |
| E. coli DH5α | Amersham Biosciences | |
| Plasmids | ||
| pORI280::ΔccpA | pORI280 derivative carrying ccpA up- and downstream regions | 65 |
| pNZ8020 | Cmr; nisin-inducible PnisA | 13 |
| pNZotsB | pNZ8020 with P. freudenreichiiotsB cloned into the BamHI/EcoRI site; Cmr | This work |
| pNZtrePPpgmB | pNZ8020 with the lactococcal trePP and pgmB cloned into the SpeI/SacI site; Cmr | This work |
| pNZtpo | pNZtrePPpgmB with P. freudenreichiiotsB cloned into the SacI/XbaI site; Cmr | This work |
| pNZotsBA | pNZ8048 with E. coliotsB and otsA cloned into the NcoI/XbaI site; Cmr | This work |
Cmr, resistance to chloramphenicol.
DNA techniques.
General molecular techniques were performed essentially as described elsewhere previously (48). Chromosomal and plasmid DNAs were isolated according the methods described previously by Johansen and Kibenich (29) and Birnboim and Doly (5), respectively. L. lactis was transformed with plasmid DNA by electroporation, as described previously by Holo and Nes (25). Restriction enzymes and T4 DNA ligase were obtained from New England BioLabs (Ipswich, MA), and Pwo polymerase and Taq polymerase were obtained from Roche Applied Science (Mannheim, Germany); all were used according to the suppliers' instructions. PCRs were performed by use of a MyCycler thermal cycler (Bio-Rad, Hercules, CA). The primers used are listed in Table S1 in the supplemental material and were purchased from Thermo Fisher Scientific (Waltham, MA).
Construction of strains and plasmids.
The deletion of ccpA in L. lactis NZ9000 was performed by using a two-step homologous recombination method as described previously by Zomer et al. (33, 65). The coding region of P. freudenreichii otsB (otsBPf) from P. freudenreichii B365 was amplified by PCR using primer pair tpp1-fw/tpp1-rev. The 0.89-kb BamHI/EcoRI fragment was digested with the indicated enzymes and cloned into BamHI/EcoRI-digested pNZ8020, yielding construct pNZotsB. The resulting construct was transformed into L. lactis strain NZ9000ΔccpA. The adjacent lactococcal trePP and pgmB genes (trehalose operon) and the otsB gene from P. freudenreichii were cloned and overexpressed in L. lactis NZ9000 as follows. The coding region of trePP-pgmB was amplified by PCR using primer pair trePPpgmB-fw/trePPpgmB-rev. The 3.14-kb SpeI/SacI fragment was digested with the indicated enzymes and cloned into SpeI/SacI-digested pNZ8020, yielding construct pNZ8020-trePPpgmB. The coding region of otsB was amplified by PCR using primer pair tpp-fw/tpp-rev. The 0.89-kb SacI/XbaI fragment was digested with the indicated enzymes and cloned into SacI/XbaI-digested pNZ8020-trePPpgmB, yielding constructs pNZ8020-trePPpgmBotsB, here designated pNZtpo. The resulting construct was transformed into L. lactis strains NZ9000 and NZ9000ΔccpA. L. lactis MG1363 DNA and P. freudenreichii B365 DNA were used as a template for the PCR amplification of trePP-pgmB and otsB inserts, respectively.
The coding region of otsBA from E. coli was amplified by PCR using primer pair otsBA-fw/otsBA-rev. The 3.811-kb NcoI/XbaI fragment was digested with the indicated enzymes and cloned into NcoI/XbaI-digested pNZ8048, yielding construct pNZotsBA. The resulting construct was transformed into L. lactis strain NZ9000. E. coli DH5α DNA was used as a template for the PCR amplification of the otsBA insert. The primer sequences used in this work are listed in Table S1 in the supplemental material.
Quantification of fermentation products during growth.
Culture samples (2 ml) were taken at different growth stages and centrifuged (2,000 × g for 5 min at 4°C); the supernatant solutions were stored at −20°C until they were analyzed by high-performance liquid chromatography (HPLC). Glucose, trehalose, acetate, ethanol, and lactate were quantified with a Dionex apparatus equipped with a refractive index detector (Shodex RI-101; Showa Denko K.K., Oita, Japan) using an HPX-87H anion-exchange column (Bio-Rad Laboratories, Inc., Richmond, CA) at 60°C, with 5 mM H2SO4 as the elution fluid and a flow rate of 0.5 ml min−1.
Enzyme activity measurements.
Cells were harvested during the exponential phase, washed twice, and suspended in 50 mM MES (morpholineethanesulfonic acid) buffer (pH 6.5). To measure β-PGM (EC 5.4.2.6) and TrePP (EC 2.4.1.216), cells were disrupted by using 0.5-g glass beads (diameter, 50 to 105 μm; Fischer Scientific BV, Den Bosch, Netherlands) using a Mini-BeadBeater-8 instrument (Biospec Products, Inc., Bartlesville, OK) with two 1-min pulses separated by 1 min of cooling down. TPP (EC 3.1.3.12) and TPS (EC 2.4.1.15) were assayed after the mechanical disruption of the cell suspension by passage through a French press (twice at 120 MPa). After cell disruption, the cell debris was pelleted, and activities were assayed at 30°C. One unit of enzyme activity is the amount of enzyme catalyzing the conversion of 1 μmol of substrate per minute under the experimental conditions used. Protein concentrations were determined by the method of Bradford (6).
The activity of β-PGM was measured as described previously by Qian et al. (45). The assay mixture (1 ml) contained 50 mM potassium phosphate (KPi) buffer (pH 7), 0.5 mM MgCl2, 1.75 U glucose 6-phosphate dehydrogenase, 0.5 mM NADP+, and 50 μM glucose 1,6-bisphosphate. Reactions were started by the addition of 1.5 mM β-glucose 1-phosphate.
The TrePP activity was measured according to a method described previously by Andersson et al. (2). The assay mixture (1 ml) contained 100 mM KPi buffer (pH 7.0), 3.75 U glucose 6-phosphate dehydrogenase, and 0.8 mM NADP+. Tre6P (0.67 mM) was used to start the reaction.
The TPP activity was assayed in a reaction mixture containing MES buffer (50 mM, pH 6.5) and 10 mM MgCl2. The reaction was initiated by the addition of 5 mM Tre6P to the mixture, the mixture was incubated at 30°C for different time periods, and the reaction was stopped by the addition of phosphate reagent (1 part of a 10% ascorbic acid solution and 6 parts of 0.42% ammonium molybdate in 1 N H2SO4) to the mixture. The A820 was proportional to the phosphate concentrations (1).
The TPS activity was determined with a reaction mixture containing MES buffer (50 mM, pH 6.5), 10 mM MgCl2, 15 mM glucose 6-phosphate, and 5% 2H2O. The reaction was initiated by the addition of 7.5 mM UDP-glucose to the mixture and was monitored online at 30°C by 31P nuclear magnetic resonance (NMR) spectroscopy. Spectra were acquired with a Bruker Avance II 500-MHz spectrometer (Bruker BioSpin GmbH, Karlsruhe, Germany) with a selective 5-mm-diameter probe head (SEX-P) by employing a pulse width of 8.7 μs (flip angle, 75°) and a recycle delay of 2.3 s. Chemical shifts are referenced to the resonance of external 85% H3PO4, designated at 0 ppm.
Reverse transcription assays: semiquantitative RT-PCR.
Strains NZ9000(pNZ8020), NZ9000(pNZotsBA), and NZ9000(pNZtpo) were grown as described above. Total RNA was isolated from cells at the mid-exponential phase of growth by using the SV total RNA isolation system (Promega), with the following modifications: incubation with lysozyme (5 mg ml−1 for 20 min at 37°C) preceded the first step of the kit protocol, and an additional incubation step with the DNase I in the kit (1.5 h at 23°C) was required to remove chromosomal DNA. Total RNA (1 μg), deoxynucleoside triphosphates (dNTPs) (final concentration of 0.5 mM), and random oligonucleotides (12 μg ml−1) (Invitrogen, Carlsbad, CA) were heated to 65°C for 5 min and chilled on ice. Dithiothreitol (final concentration, 5 mM), first-strand reverse transcription (RT) buffer, and Superscript III (1/20, vol/vol) (Invitrogen, Carlsbad, CA) were added, and samples were incubated for 5 min at 25°C, 60 min at 50°C, and 15 min at 70°C for enzyme inactivation. A parallel sample was treated in the same way, except for the addition of enzyme. cDNA was subsequently used at a dilution of 1/30 (vol/vol) in standard PCR mixtures. To test for the contamination of RNA with DNA, the RNA samples without reverse transcriptase were used as negative controls under all conditions tested. Chromosomal DNA from strains NZ9000(pNZ8020), NZ9000(pNZotsBA), and NZ9000(pNZtpo) were used as positive controls for the PCR. Primer pairs (in parentheses) were designed to amplify internal fragments of dnaK (dnaK_fwd/dnaK_rev), groEL (groEL_fwd/groEL_rev), recA (recA_fwd/recA_rev), clpP (clpP_fwd/clpP_rev), and tufA (tuf_fwd/tuf_rev). L. lactis tufA, a housekeeping gene coding for elongation factor Tu required for the continued translation of mRNA, was used as a control. RT-PCR was performed with RNA isolated from two or three independent cultures.
Extraction and quantification of intra- and extracellular trehalose during growth.
L. lactis strains were grown as described above, harvested at the exponential phase of growth, and rapidly pelleted by centrifugation (2,000 × g for 5 min at 4°C). The resulting supernatants were lyophilized, and the residues were suspended in 2H2O for the further quantification of extracellular trehalose. Ethanol cell extracts for the quantification of intracellular trehalose were prepared as described elsewhere previously (46). In brief, the cell pellets were suspended in 70% ethanol, and extraction was performed for 30 min with vigorous agitation in an ice bath. Debris was removed, ethanol was evaporated, and the residue was freeze-dried. The dried extracts were dissolved in 2H2O. The trehalose in supernatants or cell extracts was quantified by 1H-NMR spectroscopy. Formate was added as an internal concentration standard. 1H spectra were acquired with a Bruker Avance II 500-MHz spectrometer (Bruker BioSpin GmbH, Karlsruhe, Germany) with a broadband 5-mm-diameter probe head, with reverse detection employing a pulse width of 8 μs (flip angle, 90°) and a recycle delay of 2.5 s. The water resonance was suppressed with a presaturation pulse.
In vivo NMR studies with resting cells.
Cells were grown in CDM containing 1% glucose (wt/vol), and suspensions were prepared and made anaerobic as described elsewhere previously (39). In vivo NMR experiments were performed by using an online system and glucose specifically labeled with 13C on carbon 1 (40 mM) as a substrate (38, 39). In vivo 13C-NMR spectra were acquired at 125.77 MHz using a quadruple nucleus probe head at 30°C on a Bruker Avance II 500 MHz spectrometer (Bruker BioSpin GmbH, Karlsruhe, Germany) as described previously (39). Lactate was quantified in the NMR sample extract by 1H-NMR with a Bruker AMX300 instrument (Bruker BioSpin GmbH). The concentration of other metabolites was determined in fully relaxed 13C spectra of the NMR sample extracts as previously described (40). Each experiment was repeated at least twice, and the results were highly reproducible.
Quantification of trehalose 6-phosphate and trehalose in resting cells.
For the quantification of intracellular and extracellular pools of trehalose and trehalose 6-phosphate, cell suspensions were prepared as described above for in vivo NMR studies. After the addition of [1-13C]glucose (40 mM), 1-ml and 4-ml samples were taken independently for supernatants and total cell extracts, respectively, at different time points, as follows: for strain NZ(pNZtpo), samples were taken after 2.5, 5, 10, 15, 20, and 40 min, whereas for strain NZ(pNZotsBA), samples were collected at 2.5, 5, 7.5, 10, 20, and 40 min after the addition of glucose. For the quantification of extracellular trehalose, 1-ml samples were centrifuged (13,000 × g for 20 s at 4°C), and supernatants were stored at −20°C until further analysis. For the determination of the amounts of total trehalose and intracellular trehalose 6-phosphate, a cold solution of perchloric acid (final concentration, 0.6 M) was immediately added to the 4-ml samples. After stirring on ice for 20 min, the pH of the samples was adjusted to neutrality with 5 M KOH and centrifuged (30,000 × g for 20 min at 4°C). The resulting cell extracts were used for the quantification of trehalose and Tre6P. 13C-NMR spectra of supernatants and cell extracts were acquired with a 5-mm selective probe head using a pulse width corresponding to a 70° flip angle and a recycle delay of 1.5 s. Correction factors to take into account the incomplete relaxation of resonances were calculated by comparisons with spectra acquired under fully relaxed conditions (recycle delay, 60.5 s). Chemical shifts are referenced to the resonance of methanol in a glass capillary, designated at 49.3 ppm.
Cell viability assays.
L. lactis cells were grown in CDM at 30°C without pH control (initial pH of 6.5) until the exponential phase (OD600 about 1.3). After exposure to each stress, cell suspensions were adequately diluted in 50 mM KPi (pH 6.5), and the serial dilutions were plated onto M17 agar (1.5%) supplemented with 0.5% (wt/vol) glucose and 5 mg liter−1 chloramphenicol and incubated for approximately 36 h at 30°C for CFU counting. The viability was calculated as the ratio of CFU mg protein−1 of the sample exposed to stress for a given time period over the value determined at time zero. Values are average values from four to seven independent experiments and are given as percentages.
Acid shock.
Cultures (1 ml) were centrifuged (2,000 × g for 5 min at 25°C) and suspended in the same volume of 50 mM KPi (pH 3 [acidified with HCl] or pH 6.5 [control conditions]). Suspensions were incubated for defined time intervals (0, 10, 20, and 30 min) at 30°C, rapidly centrifuged (2,000 × g for 0.5 min at 25°C), and suspended in 50 mM KPi (pH 6.5) prior to plating.
Freeze-drying.
Cell samples (1 ml) were quickly frozen in liquid nitrogen and subsequently freeze-dried for 24 h. After lyophilization, the dried cells were reconstituted in 50 mM KPi (pH 6.5) and plated as described above.
Cold stress.
Cell cultures (40 ml) were placed into an ice bath during 5 min for rapid cooling down. Viability was assessed after 0, 1, 4, 8, and 14 days at 4°C. Day 0 (control condition) corresponds to cells plated immediately after the cooling-down step.
Heat shock.
Cell samples (1 ml) were transferred into a water bath at 45°C and incubated for different periods of time (0, 10, and 30 min).
Statistical analysis.
Statistical analyses of cell viability were performed by using the R Language for Statistical Computing, version 2.10.1 (R Development Core Team 2009). Prior to subjecting the data to 2-way analysis of variance (ANOVA) for the factors time and strain, we applied the Levene test for equality of variances. A rank transformation was applied in the case of inequality. For all stress conditions, a significant interaction between both factors was observed (P = 0.018 or lower for all stresses), indicating that the temporal development of viability was significantly different for each strain. We therefore proceeded with multiple testing for differences in the mean viabilities against the level at time zero (viability = 100%), as well as the mean viabilities between the strains, using the Welch t test and the Holm correction for multiple testing.
RESULTS
Detection of trehalose 6-phosphate accumulation in an L. lactis ΔccpA strain.
L. lactis subsp. cremoris NZ9000 cannot synthesize trehalose, but it possesses the enzymatic machinery to catabolize this disaccharide (2). The trehalose catabolic genes llmg_0453, llmg_0454, trePP, and pgmB, encoding the trehalose-PTS PTSTre, Tre6P phosphorylase, and β-phosphoglucomutase, are under the negative control of carbon catabolite protein A (CcpA) (59, 65). Curiously, an in vivo NMR study of glucose metabolism in resting cells of strain NZ9000ΔccpA revealed the transient accumulation of Tre6P (Fig. 2 A). Based on these data we designed the synthesis of trehalose in L. lactis by expressing an exogenous Tre6P phosphatase in the ccpA mutant. The otsB gene from P. freudenreichii (otsBPf) was cloned under a PnisA promoter, and the resulting plasmid, pNZotsB, was introduced into NZ9000ΔccpA.
Fig. 2.
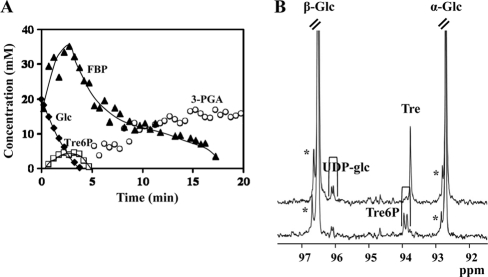
(A) Glucose metabolism in nongrowing cell suspensions of L. lactis strains with a deletion of the ccpA gene. Shown are the kinetics of [1-13C]glucose (20 mM) consumption and pools of intracellular metabolites in resting cells of L. lactis NZ9000ΔccpA at 30°C under anaerobic conditions with the pH controlled at 6.5. The maximal glucose consumption rate was 0.31 μmol min−1 mg protein−1. Symbols: closed diamonds, glucose; closed triangles, fructose 1,6-bisphosphate; open circles, 3-phosphoglycerate; open squares, trehalose 6-phosphate. The lines drawn in the graph are simple interpolations. (B) 13C-NMR spectra of perchloric acid extracts of nongrowing cell suspensions metabolizing [1-13C]glucose. Shown are data for strain NZ9000ΔccpA(pNZotsBPf) (top) and strain NZ9000ΔccpA (bottom). The resonance labeled with an asterisk is due to glucose 6-phosphate. Glc, glucose; FBP, fructose 1,6-bisphosphate; 3-PGA, 3-phosphoglycerate; Tre6P, trehalose 6-phosphate.
We resorted to 13C-NMR analyses of cell extracts obtained during the metabolism of [1-13C]glucose by resting cells to monitor trehalose and Tre6P. A resonance at 93.70 ppm assigned to the C-1/C-1′ atoms of trehalose was detected in 13C spectra of strain NZ9000ΔccpA(pNZotsB) (C-1/C-1′ refers to the carbon atoms at position 1 of the two glucose molecules in trehalose); in contrast, in spectra of strain NZ9000ΔccpA the resonances (at 93.75 and 93.85 ppm) due to the two anomeric carbon atoms of Tre6P were observed, while the resonance due to trehalose was absent (Fig. 2B).
L. lactis strains engineered for trehalose synthesis.
Capitalizing on our previous data, we devised a strategy to obtain an L. lactis trehalose producer using genes only from GRAS organisms by overexpressing simultaneously the lactococcal trePP and pgmB genes together with otsBPf from P. freudenreichii. The trePP and pgmB genes were cloned under the PnisA promoter in pNZ8020, creating plasmid pNZ8020-trePPpgmB. The cloning of otsBPf downstream of pgmB rendered plasmid pNZtpo in host strain NZ9000, here designated NZ9000(pNZtpo); pNZtpo was subsequently transformed into NZ9000ΔccpA. While this work was in progress, Termont et al. (53) reported the production of trehalose in an engineered L. lactis strain overexpressing the otsBA operon (E. coli TPP [TPPEc] and TPSEc) from E. coli. Therefore, we decided to construct a similar strain and compare the two constructs. The E. coli otsBA genes were cloned under the control of PnisA, resulting in plasmid pNZotsBA, yielding strain NZ9000(pNZotsBA). The NZ9000 strain harboring pNZ8020, NZ9000(pNZ8020), was used as a control.
To evaluate the functional expression of the cloned gene products, cell extracts were obtained from the mid-exponential phase of nisin-induced cultures (2 μg liter−1), and the relevant activities were assayed. In NZ9000(pNZtpo), the activity of TrePP was 0.30 ± 0.01 μmol min−1 mg protein−1, 298-fold higher than that for control strain NZ9000(pNZ8020); β-PGM was 22-times overexpressed, showing a specific activity of 1.11 ± 0.19 μmol min−1 mg protein−1 (about 0.05 μmol min−1 mg protein−1 in the control); and the activity of the heterologous TPPPf was 0.027 ± 0.001 μmol min−1 mg protein−1. In strain NZ9000(pNZotsBA), the heterologous activity of TPPEc was 0.26 μmol min−1 mg protein−1, and the TPS activity was about 10-fold lower. A similar activity profile was previously reported for the expression of the E. coli otsBA genes in Corynebacterium glutamicum (42).
Trehalose produced by engineered strains grown with pH control.
All strains were grown in CDM supplemented with 1% glucose and with the pH controlled at 6.5 under anaerobic conditions and induced with nisin (2 μg liter−1) at an OD600 of 0.5. During the mid-exponential (OD600 of 2.2) and stationary (OD600 of approximately 5) phases of growth, samples were collected and rapidly centrifuged to remove the growth medium. The cell pellets were subjected to ethanol extraction for the quantification of intracellular trehalose, while extracellular trehalose was measured in the supernatant solutions (growth medium) (Table 2).
Table 2.
Trehalose contents in ethanol extracts (intracellular trehalose) or in the growth medium (extracellular trehalose) of L. lactis cells collected during mid-exponential and stationary phases of growtha
| Strain | Mean extracellular trehalose concn (mM) ± SD |
Mean intracellular trehalose concn (mM) ± SD |
Mean total trehalose (μmol/mg protein) |
|||
|---|---|---|---|---|---|---|
| Exp | Stat | Exp | Stat | Exp | Stat | |
| NZ9000ΔccpA(pNZtpo) | 1.2b | 0.4b | 170.8b | 6.2b | 3.9b | 0.4b |
| NZ9000(pNZtpo) | 0.8 ± 0.1 | 2.1 ± 0.1 | 167.0 ± 9.2 | 123.1 ± 7.4 | 2.2 ± 0.2 | 2.6 ± 0.3 |
| NZ9000(pNZotsBA) | 0.4 ± 0.1 | 1.2 ± 0.04 | 79.2 ± 18.1 | 75.7 ± 1.3 | 1.2 ± 0.3 | 1.4 ± 0.04 |
Cultures were grown with the pH controlled at 6.5. Trehalose was quantified by proton NMR. Exp, mid-exponential phase; Stat, stationary phase.
A single experiment was performed.
The three engineered strains excreted trehalose into the medium. In strain NZ9000ΔccpA(pNZtpo) the levels of intracellular and extracellular trehalose decreased by about 30- and 3-fold, respectively, from the mid-exponential phase to the stationary phase of growth (Table 2). These results show that this strain utilizes the produced trehalose efficiently, most likely due to the derepression of the trehalose operon triggered by the ccpA deletion. In view of this nondesired feature, strain NZ9000ΔccpA(pNZtpo) was not considered further in this study. In contrast, for strains NZ9000(pNZtpo) and NZ9000(pNZotsBA) the levels of extracellular trehalose were higher during the stationary phase, and the level of intracellular trehalose was only slightly reduced.
Growth profiles of recombinant and control strains without pH control.
Strains NZ9000(pNZtpo) and NZ9000(pNZotsBA) and control strain NZ9000(pNZ8020) were grown in CDM without pH control (initial pH of 6.5) and induced with 2 μg liter−1 nisin at an OD600 of 0.4 (Fig. 3). The maximal biomass was identical for all strains, but the specific growth rate of NZ9000(pNZtpo) was only 70% of that of the control strain, while NZ9000(pNZotsBA) exhibited a growth profile identical to that of the control (Fig. 3).
Fig. 3.

Growth profiles of L. lactis strains engineered for trehalose synthesis. Growth was performed with CDM with 1% (wt/vol) glucose at 30°C without pH control (initial pH of 6.5). The growth rates (μ) were 0.69 ± 0.006 h−1 for the control strain, 0.49 ± 0.006 h−1 for NZ9000(pNZtpo), and 0.70 ± 0.026 h−1 for NZ9000(pNZotsBA). Symbols: diamonds, control strain; squares, NZ9000(pNZtpo); triangles, NZ9000(pNZotsBA). Closed symbols indicate OD600 values; open symbols indicate pH values during growth. Data shown are representative of data from three identical experiments.
Despite trehalose production, the major end product of NZ9000(pNZtpo) was lactate, accounting for 83.5% of the glucose consumed, and a similar fermentation pattern was observed for strain NZ9000(pNZotsBA). As expected, the control strain was fully homolactic and unable to produce trehalose (Table 3).
Table 3.
Major end products from the metabolism of glucose in L. lactis strains NZ9000(pNZtpo) and NZ90000(pNZotsBA) and control strain NZ9000(pNZ8020) during growth without pH controla
| OD600 of sampling (phase) | Concn (mM) |
|||||
|---|---|---|---|---|---|---|
| NZ9000(pNZtpo) |
NZ9000(pNZotsBA) |
NZ9000(pNZ8020) |
||||
| Lactate | Trehalose | Lactate | Trehalose | Lactate | Trehalose | |
| 0.4 (induction) | 12.2 | ND | 9.0 | ND | 9.9 | ND |
| 1.3 (mid-exponential) | 25.2 | 0.3 | 20.8 | 0.2 | 30.0 | ND |
| 2.0–2.2 (late exponential) | 47.2 | 1.1 | 47.7 | 1.0 | 63.4 | ND |
| >2.4 (stationary) | 75.1 | 2.1 | 77.6 | 2.8 | 83.1 | ND |
| % from glucose | 83.5 | 16.8b | 82.9 | 8.5b | 92.8 | ND |
Nisin (2 μg liter−1) was added at an OD600 of 0.4. Metabolites in supernatant solutions of culture samples collected at different time points during growth were measured by HPLC. Initial and final glucose concentrations were 65.6 and 20.6 mM for NZ9000(pNZtpo), 63.0 and 16.2 mM for NZ9000(pNZotsBA), and 62.9 and 18.1 mM for the control strain, respectively. Values are for a representative growth out of two experiments. Induction means the time of nisin addition. ND, below the detection limit.
Values calculated for total trehalose (intracellular plus extracellular) determined by NMR with cells collected during the mid-exponential phase.
In the mid-exponential phase of growth (OD600 of 1.3), the concentration of trehalose inside the cells was determined by NMR as described in Materials and Methods (Table 3). Strain NZ9000(pNZtpo) accumulated 150 ± 7 mM trehalose, while NZ9000(pNZotsBA) accumulated 92 ± 2 mM. Taking into consideration the total amount of trehalose produced (intracellular plus extracellular trehalose), we estimated that 15.8% ± 1.4% of the glucose supplied was converted to trehalose in NZ9000(pNZtpo), whereas only 8.5% ± 0.2% of glucose was directed toward trehalose production in NZ9000(pNZotsBA) (Table 4).
Table 4.
Yield of trehalose and percentage of trehalose excreted in strains NZ9000(pNZtpo) and NZ9000(pNZotsBA)a
| Culture type | Mean % trehalose yield ± SD |
Mean % trehalose excreted ± SD |
||
|---|---|---|---|---|
| NZ9000(pNZtpo) | NZ9000(pNZotsBA) | NZ9000(pNZtpo) | NZ9000(pNZotsBA) | |
| Resting cells at pH 6.5 | 14.9b | 10.7b | 26.8b | 21.6b |
| Growing cells | ||||
| pH controlled at 6.5 | 6.5 ± 2.0 (Exp), 11.4 ± 4.0 (Stat) | 5.4 (Exp), 5.2 (Stat)c | 78.0 ± 3.9 (Exp), 86.9 ± 0.6 (Stat) | 80.5 ± 1.3 (Exp), 84.2 ± 0.3 (Stat) |
| Without pH control | 15.8 ± 1.4 (Exp) | 8.5 ± 0.2 (Exp) | 66.7 ± 8.0 (Exp) | 80.7 ± 1.2 (Exp) |
The trehalose produced and glucose consumed were quantified by NMR (see Materials and Methods). In growing cells, levels of intracellular and extracellular trehalose were determined for cell extracts and supernatants derived from cultures harvested during mid-exponential (Exp) and stationary (Stat) phases of growth, respectively. Unless stated otherwise, the values are averages of data from at least two independent experiments.
Values refer to the experiment shown in Fig. 4.
A single experiment was performed.
Dynamics of trehalose and trehalose 6-phosphate in resting cells of engineered strains.
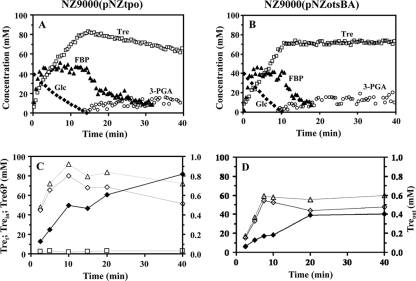
The metabolism of [1-13C]glucose (40 mM) in strains NZ9000(pNZtpo) and NZ9000(pNZotsBA) was studied by in vivo 13C-NMR in suspensions of nongrowing cells under an argon atmosphere and at a constant pH of 6.5 (Fig. 4). Maximal glucose consumption rates of 0.33 ± 0.01 and 0.39 ± 0.01 μmol min−1 mg protein−1 were determined for strains NZ9000(pNZtpo) and NZ9000(pNZotsBA), respectively. These values should be compared with the 0.37 ± 0.01 μmol min−1 mg protein−1 determined for the control strain (not shown). Both recombinant strains produced lactate as a major end product, which accounted for approximately 83% of the supplied glucose. In the engineered strains, the profile of accumulation of fructose 1,6-bisphosphate (FBP) resembled that of the wild-type and control strains (38, 47); in brief, FBP accumulated transiently, and levels started to decline at the onset of glucose depletion (Fig. 4A and B). Trehalose was detected immediately after the addition of glucose; in strain NZ9000(pNZtpo) the buildup of total trehalose plus Tre6P was very fast during the first 2 min and continued at a lower rate, reaching a maximal level of 83 mM (calculated on the basis that all trehalose was inside the cells). Therefore, approximately 15% of the glucose supplied was channeled toward trehalose synthesis. Once glucose was exhausted, trehalose was consumed at a low rate (0.04 μmol min−1 mg protein−1). In strain NZ9000(pNZotsBA), the level of trehalose increased steadily while glucose was available, leveling off at 68 mM (calculated as though the total trehalose was intracellular). In this strain 10.7% of the glucose supplied was directed toward trehalose synthesis. It is impossible to distinguish between the NMR signals of trehalose and those of Tre6P in spectra of living cells due to extensive line broadening and overlapping. Moreover, extracellular trehalose and intracellular trehalose cannot be distinguished by NMR. To quantify intra- and extracellular trehalose and discriminate between trehalose and Tre6P, we analyzed the cell extracts and supernatant solutions derived from samples taken during the metabolism of glucose by 13C-NMR (Fig. 4C and D). Trehalose 6-phosphate was detected in strain NZ9000(pNZtpo) (up to 4 mM) but not in strain NZ9000(pNZotsBA). In strain NZ9000(pNZtpo) the concentration of trehalose increased concomitantly with glucose consumption in both intra- and extracellular compartments. Upon glucose depletion the concentration of intracellular trehalose decreased, while the concentration of extracellular trehalose moderately increased. It is curious that while considerable amounts of glucose 6-phosphate (G6P) accumulated in strain NZ9000(pNZotsBA) (similarly to wild-type strains [38]), only traces were detected in NZ9000(pNZtpo) (not shown). The lack of an accumulation of the intermediate metabolite G6P is probably related to the lower glycolytic flux in this strain (Fig. 4A).
Fig. 4.
(A and B) Kinetics of [1-13C]glucose (40 mM) consumption and pools of metabolites in resting cells of L. lactis strains engineered for the synthesis of trehalose: NZ9000(pNZtpo) (A) and NZ9000(pNZotsBA) (B). The experiments were monitored online by in vivo 13C-NMR and carried out at 30°C under anaerobic conditions and with the pH controlled at 6.5. Maximal glucose consumption rates (μmol min−1 mg protein−1) were 0.33 (A) and 0.39 (B). Symbols: closed diamonds, glucose; closed triangles, fructose 1,6-bisphosphate; open circles, 3-phosphoglycerate; open squares, total trehalose plus trehalose 6-phosphate expressed as an intracellular concentration. (C and D) Parallel experiments were run to study the kinetics of trehalose 6-phosphate and trehalose (intracellular and extracellular pools) in NZ9000(pNZtpo) (C) and NZ9000(pNZotsBA) (D). The extracellular trehalose concentration was determined in cell supernatants, while intracellular trehalose and trehalose 6-phosphate concentrations were determined in perchloric acid extracts. These metabolites were quantified by proton NMR. Symbols: closed diamonds, extracellular trehalose; open diamonds, intracellular trehalose; open triangles, total trehalose expressed as an intracellular concentration; open squares, trehalose 6-phosphate. Each type of experiment was performed twice, with good reproducibility.
Data on the percentage of trehalose that was secreted into the external medium as well as the trehalose yield are summarized in Table 4 for the various experimental conditions examined.
Synthesis of trehalose improves acid tolerance of L. lactis.
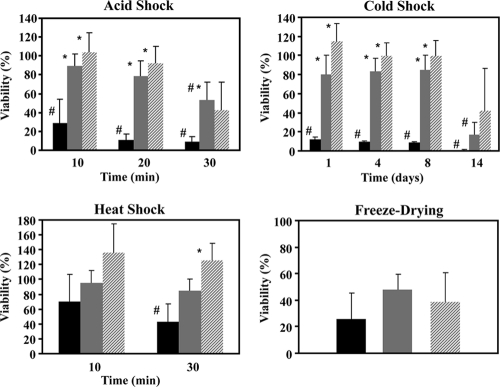
The survival of engineered and control strains when exposed to pH 3 for time periods of up to 30 min was evaluated. To perform these tests, cell suspensions were harvested and centrifuged, and the supernatants were discarded to remove extracellular trehalose. Cells were then suspended in 50 mM KPi acidified with HCl to pH 3 or in 50 mM KPi at pH 6.5 (reference conditions). After incubation, cells were quickly centrifuged to remove acid and suspended in buffer with an optimal pH (KPi at pH 6.5). For each strain examined, the survival rates of cells in KPi at pH 6.5 (reference conditions) were similar for all the incubation times examined. The numbers of viable cells per mg of protein were 2.83 × 1010 ± 0.81 × 1010 for the control strain, 3.14 × 108 ± 0.42 × 108 for NZ9000(pNZtpo), and 2.50 × 109 ± 0.55 × 109 for NZ9000(pNZotsBA). In contrast, exposure to pH 3.0 reduced considerably the viability of the control strain in a time-dependent manner (Fig. 5). Noteworthy is the fast decay (about a 72% reduction; P = 0.012) within the first 10 min. Conversely, the trehalose producer NZ9000(pNZtpo) showed no significant loss of viability (P = 0.296) during the first 20 min of exposure to pH 3.0. Only after 30 min of exposure was a significant decrease in survival of about 45% observed (P = 0.011). However, this survival rate (∼55%) was significantly higher (P = 0.002) than that of the control strain (∼8%). The performance of strain NZ9000(pNZotsBA) at a low pH was comparable to that of NZ9000(pNZtpo), as differences were not significant at any time point (P > 0.5 for all times).
Fig. 5.
Effects of different stresses on the survival of L. lactis strains engineered for trehalose synthesis. Black bars, control strain; gray bars, NZ9000(pNZtpo); dashed bars, NZ9000(pNZotsBA). Viability was calculated as the number of CFU mg protein−1 of cells exposed to stress as a percentage of the number of CFU mg protein−1 of nonstressed cells (for details, see Materials and Methods). For acid stress, cells harvested during the mid-exponential phase were exposed to pH 3.0 (50 mM KPi acidified with HCl) for defined time periods (10, 20, and 30 min). Values are the means of data from at least five independent experiments. For cold stress, survival at 4°C after 1, 4, 8, and 14 days was determined. Values are the means of data from at least four independent experiments. For heat stress, survival was determined upon exposure to 45°C for different time periods. Values are the means of data from at least four independent experiments. For freeze-drying of cells, the rate of survival of cells subjected to one-cycle freeze-drying for 24 h was determined. Values are the means of data from at least five independent experiments. Error bars indicate standard deviations. The asterisk designates statistically significant differences (P < 0.05) between the survival of the engineered strain and that of the control strain. The sharp symbol designates significant differences (P < 0.05) compared to the viability at the 100% level.
Trehalose protects L. lactis against cold shock.
The experimental design consisted of rapidly transferring and incubating the cultures on ice for 5 min; subsequently, the cultures were moved to a chamber at 4°C. One day at 4°C sufficed to significantly reduce the viability of the control strain to survival rates of around 12% (P = 0.022) (Fig. 5). In contrast, the viability of strain NZ9000(pNZtpo) after 8 days at 4°C was approximately 80% and was significantly reduced (P = 0.001) only after 14 days at this temperature. Strain NZ9000(pNZotsBA) showed apparently higher survival rates than did NZ9000(pNZtpo) upon cold exposure, but the differences were not significant (P = 0.146 or higher for all times).
Trehalose confers tolerance to heat shock in L. lactis.
In this study, the viability of strain NZ9000(pNZtpo) was not significantly affected after 10 or 30 min of incubation at 45°C (P = 0.495 and P = 0.307, respectively), whereas the percentage of survival of the control strain significantly decreased to 40% after 30 min at 45°C (P = 0.033) (Fig. 5). Although no significant differences were found between the survival rates of strains NZ9000(pNZtpo) and NZ9000(pNZotsBA) when exposed to 45°C (P = 0.161 and P = 0.092 at 10 and 30 min, respectively), it is noteworthy that the latter strain consistently presented higher survival rates.
Effect of trehalose accumulation on cell survival in response to freeze-drying.
The survival rates of the trehalose producers NZ9000(pNZtpo) and NZ9000(pNZotsBA) and that of the control strain were assessed after one cycle of freeze-drying. The viability of the control strain was reduced by 75%, while strains NZ9000(pNZtpo) and NZ9000(pNZotsBA) showed viability reductions of nearly 50 and 60%, respectively (Fig. 5). The differences with respect to the control strain, however, were not significant (P = 0.17 and P = 0.20); hence, trehalose did not protect L. lactis from the stress imposed by freezing and dehydration.
Stress genes are not induced by heterologous expression of biosynthetic genes.
To confirm that the beneficial effects of trehalose synthesis on the stress resistance of engineered strains were not caused by an unintended overexpression of stress genes, we performed semiquantitative RT-PCR assays. The transcription levels of four genes known to be involved in the stress response of L. lactis were assessed. The dnaK and groEL genes encode a chaperone and a chaperonin, respectively, both overproduced in response to acid and heat stress conditions (7, 61, 63). The recA gene mediates the stress response upon DNA damage, and its involvement in the acid shock response was reported previously (7). The clpP gene encodes a heat shock protein also involved in the acid shock response (19).
The results showed that these stress response genes are equally transcribed in control strain NZ9000(pNZ8020) and in the trehalose-producing strains NZ9000(pNZotsBA) and NZ9000(pNZtpo) (see Fig. S1 in the supplemental material). We conclude that the observed improved stress resistance of the engineered strains is associated directly with the presence of trehalose.
DISCUSSION
Lactococcus lactis is an important industrial organism widely used for dairy fermentations. In addition to this traditional use, L. lactis was recently proposed to be an efficient producer of heterologous proteins and vehicle for drug delivery. The commercial importance of these applications underlies the demand for the development of robust strains that are able to perform well even under adverse bioprocess conditions such as extremes of temperature or pH, high concentrations of weak acids, and dehydration. In view of the use of L. lactis in the food industry as well as for oral drug delivery, we envisaged the construction of a food-grade strain with improved tolerance to stress, in particular to acid stress. To this end, the synthesis of trehalose, a solute widespread in stress responses, was engineered in L. lactis using genes derived exclusively from food-grade organisms.
The growth rate of strain NZ9000(pNZtpo) was approximately 30% lower than those of the control and the mutant overexpressing otsBA, and a similar trend was observed for the specific glucose consumption rate and the cell viability in the absence of stress. We noticed that the two recombinant strains were clearly different with respect to Tre6P accumulation: in resting cells of NZ9000(pNZtpo) the concentration of Tre6P was 3 mM, whereas in NZ9000(pNZotsBA) the level of this metabolite was below the detection level of the NMR technique (Fig. 4C). Furthermore, Tre6P was also detected (thin-layer chromatography assays) in cell extracts derived from mid-exponential-phase growing cultures of NZ9000(pNZtpo) but not in L. lactis cells overexpressing otsBA (data not shown). In view of these results it is tempting to suggest the involvement of Tre6P in the mechanisms leading to the impairment of the growth rate, glucose consumption, and viability of NZ9000(pNZtpo). The toxicity ascribed to sugar-phosphate accumulation is often invoked to justify impaired or arrested growth, but a comprehensive explanation remains elusive in many cases (17, 58). However, it is known that the accumulation of Tre6P in Saccharomyces cerevisiae causes a strong reduction in the glycolytic flux because Tre6P inhibits hexokinase in this organism (54). Also, the deleterious effect of the Tre6P accumulation on cell viability was demonstrated previously with a TPP-deficient mutant of S. cerevisiae (16). To our knowledge, the effect of Tre6P on the regulation of the glycolytic enzymes of L. lactis has not been studied, and our data on metabolite dynamics during glucose metabolism (Fig. 4) do not provide a clue for a putative glycolytic target. Therefore, the hypothetical toxic effect of Tre6P in L. lactis requires further investigation.
In the present work, the production of trehalose was achieved by overexpressing the trePP and pgmB genes from L. lactis (trehalose operon) and the otsB gene from P. freudenreichii. Growing cultures and resting cells of the resulting trehalose-producing strain, NZ9000(pNZtpo), converted a maximum of 16.8% of the glucose supplied into trehalose, a value far from the theoretical maximum of 66.7%. It is conceivable that the low activity of TPPPf (0.027 μmol min−1 mg protein−1) could limit the synthesis of trehalose. In fact, this view is strongly supported by the accumulation in this strain of Tre6P, the substrate of TPP (Fig. 4). Therefore, the enhancement of the TPP activity should be a primary goal of future strategies aimed at improving the yield of trehalose in L. lactis.
Curiously, the strain engineered with the trehalose pathway of E. coli, NZ9000(pNZotsBA), showed even lower yields of trehalose production despite a 10-fold-higher TPP activity (0.26 μmol min−1 mg protein−1). In this case, the metabolic bottleneck for trehalose synthesis is probably at the level of the reaction catalyzed by TPS, whose activity was very low (0.02 μmol min−1 mg protein−1). Likewise, a defective TPS activity was previously evoked as the main reason for the poor trehalose production in Corynebacterium glutamicum engineered for trehalose overproduction by the expression of otsBA from E. coli (43).
Altogether, the results with the two recombinant strains strongly indicate that a “pull strategy” should be followed to drive G6P away from glycolysis and direct it toward the synthesis of trehalose; this implies a high level of activity of the enzymes that use G6P (TrePP or TPS), combined with TPP activity of a similar magnitude. This type of approach has proven highly effective for the optimization of mannitol production in L. lactis (21). Combining high activities of mannitol 1-phosphate dehydrogenase and mannitol 1-phosphate phosphatase led to an efficient channeling of fructose 6-phosphate and a mannitol yield close to the theoretical maximum.
In L. lactis, trehalose was detected not only inside the cells but also in the extracellular medium both during growth and under nongrowing conditions. In fact, when mid-exponential-phase cultures of NZ9000(pNZtpo) were analyzed for trehalose production, at least 67% of the total trehalose was found in the extracellular medium. In resting cells, however, only around 20% of the trehalose produced was exported to the medium (Table 4). Fairly similar results were observed with strain NZ9000(pNZotsBA). The ability of many organisms to synthesize and accumulate trehalose, and other compatible solutes, in response to osmotic stress has been extensively documented (49, 64). Regrettably, the assessment of solute excretion is rarely performed, probably because it seems counterintuitive that a protecting compound is synthesized and then lost to the medium, apparently without a sound physiological reason. The excretion of trehalose is well known for C. glutamicum and E. coli (23, 52). In the latter bacterium, the cytoplasmic trehalose level is regulated by a futile cycle involving the overproduction, excretion, and reutilization of this sugar (52), and a similar excretion/reutilization cycle was proposed previously for ectoine in Halomonas elongata (24); in fact, this hypothesis was validated by the disruption of the uptake system in H. elongata, which led to a beneficial 20% increase in ectoine productivity. In view of these reports, the excretion of trehalose observed for the two L. lactis trehalose-producing strains was not unexpected. It is known that lactic acid bacteria in general, and L. lactis in particular, have a limited capacity to synthesize compatible solutes but are able to import glycine betaine, carnitine, or proline to counterbalance the external osmotic pressure (36, 62). Therefore, it is conceivable that trehalose, a compatible solute extraneous to L. lactis, is released via mechanosensitive channels that respond to an increase in cell turgor pressure (18). Subsequently, the excreted trehalose can be taken up via the specific phosphotransferase system described in the literature (2) and further metabolized. This convoluted pathway, involving the excretion and uptake of trehalose, probably provides the only route to catabolize intracellular trehalose in the engineered trehalose-producing L. lactis, since trehalase or trehalose phosphorylase activities have not been detected in this organism (2).
The trehalose-producing strains engineered in this work showed a remarkable tolerance to acid, cold, and heat shocks. In fact, the survival of strains NZ9000(pNZtpo) and NZ9000(pNZotsBA) was not significantly affected by severe insults such as up to 20 min of exposure to pH 3 for 8 days at 4°C or 30 min at 45°C (Fig. 5). The role of trehalose in the protection of L. lactis during stress emerges from the sharp contrast between the high tolerance of trehalose-accumulating cells and the poor performance of the control strain. However, the possible contribution of stress proteins should be considered. As is the case for many other organisms, L. lactis develops adaptation strategies when the environmental conditions are shifted far away from the optimal parameters. This is called the stress response and involves the induction of the synthesis of several proteins whose role is to prevent cell death by counteracting the damage provoked by harsh conditions. The response of L. lactis to acid, osmotic, cold, and heat stress has been extensively investigated, and many stress proteins were identified (7, 19, 61, 63). The primary role of trehalose in the protection of L. lactis from acid and cold stress is clearly demonstrated by our results, since the strain lacking trehalose was severely affected by these stresses, in contrast with the excellent performance of the trehalose-producing strains. Therefore, the intrinsic stress response alone did not provide sufficient protection. However, there is an apparent synergism between the actions of trehalose and the heat stress response, specially evident for strain NZ9000(pNZotsBA), which exhibited survival rates that were consistently higher than those in the absence of heat shock.
The engineering strategy used in this work involved the overexpression in L. lactis of heterologous genes encoding trehalose biosynthetic enzymes. Therefore, one could argue that the observed increased stress resistance could, at least in part, result from the induction of stress proteins. This hypothesis was ruled out by semiquantitative RT-PCR experiments; the transcript levels of four selected genes encoding stress proteins, i.e., dnaK, recA, groEL, and clpP, were assessed, and the results showed no evidence for alterations in the expressions of these genes (see Fig. S1 in the supplemental material). We conclude that the stress resistance phenotype derives primarily from the presence of trehalose.
The mechanisms underlying the protecting effect of trehalose are not clearly understood, and it is especially intriguing that this compound can protect cells from a variety of stresses (cold, heat, low pH, high osmolarity, free radicals, and desiccation). We reported the involvement of trehalose in the acid stress response (9), and a similar behavior was recently found in Rhizopus oryzae (55). On the other hand, the role of trehalose in protection against cold stress has been extensively studied in plants and the model organisms E. coli and S. cerevisiae (20, 30, 50). Interestingly, trehalose also increases the life span of the nematode Caenorhabditis elegans, as demonstrated recently (26). Additionally, the ability of trehalose to stabilize the protein structure against heat denaturation and to prevent concomitant protein aggregation has been extensively documented both in vitro and in vivo (28, 51). Trehalose also acts as a scavenger of reactive oxygen species, which accumulate under several stressful conditions, thereby protecting proteins and other cell components from damage caused by strong oxidizing agents (3).
Despite the limited knowledge on the mode of action of trehalose, or, indeed, of any other protecting osmolyte, a potential stabilizing effect of trehalose on cell membranes has frequently been proposed (30, 34, 57). At low temperatures, trehalose would counteract the decrease in membrane fluidity (12). At a low pH, the extrastabilization conferred by trehalose would result in decreased permeability to hydrogen ions, a highly beneficial trait when membranes are subjected to strong pH gradients. The latter view stemmed mainly from the huge trehalose levels observed for Sulfolobus solfataricus, a thermoacidophilic archaeon whose cell membranes can stand gradients as high as 5 pH units (41).
Trehalose-producing L. lactis showed no significantly improved survival when exposed to a cycle of freeze-drying, despite the high level of intracellular trehalose (in the 100 mM range). This result is not surprising, as protection against dehydration is provided largely by exogenous trehalose, although this effect is amplified by the presence of intracellular trehalose (15). Upon the addition of 0.1 to 0.5 M trehalose, enhanced survival in response to freeze-drying was observed for different bacterial cultures, e.g., E. coli, Bacillus thuringiensis, and Lactobacillus acidophilus (11, 34). Here, the concentration of trehalose typically found in the external medium of L. lactis cultures (around 1 mM) was too low to provide effective protection. Our results contrast with those described previously by Termont et al. (53), who reported a 100% retention of cell viability after the freeze-drying of NZ9000(pNZotsBA). The reasons for this discrepancy probably arise from the different conditions used by those authors for the induction of trehalose synthesis, namely, highly toxic levels of nisin (400 μg liter−1) and aerobic conditions.
In summary, we demonstrated that trehalose, a compound unrelated to wild-type L. lactis strains, plays a definite role in the protection of this bacterium against damage caused by acid, cold, or heat shock. Moreover, this work represents a proof of concept for the development of robust, food-grade L. lactis strains able to perform under demanding working conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Oscar Kuipers (Groningen University) for providing plasmid pORI280::ΔccpA. We thank Rui Neves for the construction of strain NZ9000ΔccpA.
A.L.C. and F.S.C. held fellowships SFRH/BD/30419/2006 and SFRH/BD/5080/2001 from the Fundação para a Ciência e a Tecnologia (FCT). The NMR spectrometers are part of the National NMR Network (REDE/1517/RMN/2005), supported by the Programa Operacional Ciência e Inovação (POCTI) 2010 and the FCT.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Ames B. N. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8:115–118 [Google Scholar]
- 2. Andersson U., Levander F., Radstrom P. 2001. Trehalose-6-phosphate phosphorylase is part of a novel metabolic pathway for trehalose utilization in Lactococcus lactis. J. Biol. Chem. 276:42707–42713 [DOI] [PubMed] [Google Scholar]
- 3. Benaroudj N., Lee D. H., Goldberg A. L. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276:24261–24267 [DOI] [PubMed] [Google Scholar]
- 4. Bermudez-Humaran L. G. 2009. Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Hum. Vaccin. 5:264–267 [DOI] [PubMed] [Google Scholar]
- 5. Birnboim H. C., Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 7. Budin-Verneuil A., Pichereau V., Auffray Y., Ehrlich D. S., Maguin E. 2005. Proteomic characterization of the acid tolerance response in Lactococcus lactis MG1363. Proteomics 5:4794–4807 [DOI] [PubMed] [Google Scholar]
- 8. Cardoso F. S., Castro R. F., Borges N., Santos H. 2007. Biochemical and genetic characterization of the pathways for trehalose metabolism in Propionibacterium freudenreichii, and their role in stress response. Microbiology 153:270–280 [DOI] [PubMed] [Google Scholar]
- 9. Cardoso F. S., Gaspar P., Hugenholtz J., Ramos A., Santos H. 2004. Enhancement of trehalose production in dairy propionibacteria through manipulation of environmental conditions. Int. J. Food Microbiol. 91:195–204 [DOI] [PubMed] [Google Scholar]
- 10. Carvalho A. L., et al. 2006. Towards the production of trehalose in Lactococcus lactis, abstr. P051. Abstr. 10th Int. Symp. Genet. Ind. Microorgan., Prague, Czech Republic [Google Scholar]
- 11. Conrad P. B., Miller D. P., Cielenski P. R., de Pablo J. J. 2000. Stabilization and preservation of Lactobacillus acidophilus in saccharide matrices. Cryobiology 41:17–24 [DOI] [PubMed] [Google Scholar]
- 12. Crowe J. H., Crowe L. M., Chapman D. 1984. Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science 223:701–703 [DOI] [PubMed] [Google Scholar]
- 13. de Ruyter P. G., Kuipers O. P., de Vos W. M. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Vos W. M., Hugenholtz J. 2004. Engineering metabolic highways in lactococci and other lactic acid bacteria. Trends Biotechnol. 22:72–79 [DOI] [PubMed] [Google Scholar]
- 15. Diniz-Mendes L., Bernardes E., de Araujo P. S., Panek A. D., Paschoalin V. M. 1999. Preservation of frozen yeast cells by trehalose. Biotechnol. Bioeng. 65:572–578 [DOI] [PubMed] [Google Scholar]
- 16. Elliott B., Haltiwanger R. S., Futcher B. 1996. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics 144:923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Englesberg E., Anderson R. L., Weinberg R., Lee N., Hoffee P., Huttenhauer G., Boyer H. 1962. l-Arabinose-sensitive, l-ribulose 5-phosphate 4-epimerase-deficient mutants of Escherichia coli. J. Bacteriol. 84:137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Folgering J. H., Moe P. C., Schuurman-Wolters G. K., Blount P., Poolman B. 2005. Lactococcus lactis uses MscL as its principal mechanosensitive channel. J. Biol. Chem. 280:8784–8792 [DOI] [PubMed] [Google Scholar]
- 19. Frees D., Vogensen F. K., Ingmer H. 2003. Identification of proteins induced at low pH in Lactococcus lactis. Int. J. Food Microbiol. 87:293–300 [DOI] [PubMed] [Google Scholar]
- 20. Garg A. K., et al. 2002. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. U. S. A. 99:15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaspar P. 2008. Metabolic engineering of Lactococcus lactis for mannitol production. Ph.D. thesis. ITQB, Universidade Nova de Lisboa, Oeiras, Portugal [Google Scholar]
- 22. Gasson M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gourdon P., Lindley N. D. 1999. Metabolic analysis of glutamate production by Corynebacterium glutamicum. Metab. Eng. 1:224–231 [DOI] [PubMed] [Google Scholar]
- 24. Grammann K., Volke A., Kunte H. J. 2002. New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581(T). J. Bacteriol. 184:3078–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holo H., Nes I. F. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195–199 [DOI] [PubMed] [Google Scholar]
- 26. Honda Y., Tanaka M., Honda S. 2010. Trehalose extends longevity in the nematode Caenorhabditis elegans. Aging Cell 9:558–569 [DOI] [PubMed] [Google Scholar]
- 27. Hugenholtz J., et al. 2002. Metabolic engineering of lactic acid bacteria for the production of nutraceuticals. Antonie Van Leeuwenhoek 82:217–235 [PubMed] [Google Scholar]
- 28. Jain N. K., Roy I. 2009. Effect of trehalose on protein structure. Protein Sci. 18:24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johansen E., Kibenich A. 1992. Isolation and characterization of IS1165, an insertion sequence of Leuconostoc mesenteroides subsp. cremoris and other lactic acid bacteria. Plasmid 27:200–206 [DOI] [PubMed] [Google Scholar]
- 30. Kandror O., DeLeon A., Goldberg A. L. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. U. S. A. 99:9727–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleerebezem M., Hugenholtz J. 2003. Metabolic pathway engineering in lactic acid bacteria. Curr. Opin. Biotechnol. 14:232–237 [DOI] [PubMed] [Google Scholar]
- 32. Kuipers O. P., De Ruyter P. G. G. A., Kleerebezem M., de Vos W. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 33. Leenhouts K., et al. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217–224 [DOI] [PubMed] [Google Scholar]
- 34. Leslie S. B., Israeli E., Lighthart B., Crowe J. H., Crowe L. M. 1995. Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl. Environ. Microbiol. 61:3592–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mercade M., Lindley N. D., Loubiere P. 2000. Metabolism of Lactococcus lactis subsp. cremoris MG 1363 in acid stress conditions. Int. J. Food Microbiol. 55:161–165 [DOI] [PubMed] [Google Scholar]
- 36. Molenaar D., Hagting A., Alkema H., Driessen A. J., Konings W. N. 1993. Characteristics and osmoregulatory roles of uptake systems for proline and glycine betaine in Lactococcus lactis. J. Bacteriol. 175:5438–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morello E., et al. 2008. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 14:48–58 [DOI] [PubMed] [Google Scholar]
- 38. Neves A. R., et al. 2002. Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular metabolite pools determined by in vivo nuclear magnetic resonance. Appl. Environ. Microbiol. 68:6332–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neves A. R., et al. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200–212 [DOI] [PubMed] [Google Scholar]
- 40. Neves A. R., et al. 2002. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD(+) and NADH pools determined in vivo by 13C NMR. J. Biol. Chem. 277:28088–28098 [DOI] [PubMed] [Google Scholar]
- 41. Nicolaus B., et al. 1988. Trehalose in Archaebacteria. Syst. Appl. Microbiol. 10:215–217 [Google Scholar]
- 42. Padilla L., Kramer R., Stephanopoulos G., Agosin E. 2004. Overproduction of trehalose: heterologous expression of Escherichia coli trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in Corynebacterium glutamicum. Appl. Environ. Microbiol. 70:370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Padilla L., Morbach S., Kramer R., Agosin E. 2004. Impact of heterologous expression of Escherichia coli UDP-glucose pyrophosphorylase on trehalose and glycogen synthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 70:3845–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poolman B., Konings W. N. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qian N., Stanley G. A., Hahn-Hagerdal B., Radstrom P. 1994. Purification and characterization of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells. J. Bacteriol. 176:5304–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramos A., Boels I. C., de Vos W. M., Santos H. 2001. Relationship between glycolysis and exopolysaccharide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 67:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramos A., et al. 2004. Effect of pyruvate kinase overproduction on glucose metabolism of Lactococcus lactis. Microbiology 150:1103–1111 [DOI] [PubMed] [Google Scholar]
- 48. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Santos H., da Costa M. S. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501–509 [DOI] [PubMed] [Google Scholar]
- 50. Schade B., Jansen G., Whiteway M., Entian K. D., Thomas D. Y. 2004. Cold adaptation in budding yeast. Mol. Biol. Cell 15:5492–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singer M. A., Lindquist S. 1998. Thermotolerance in Saccharomyces cerevisiae: the yin and yang of trehalose. Trends Biotechnol. 16:460–468 [DOI] [PubMed] [Google Scholar]
- 52. Styrvold O. B., Strom A. R. 1991. Synthesis, accumulation, and excretion of trehalose in osmotically stressed Escherichia coli K-12 strains: influence of amber suppressors and function of the periplasmic trehalase. J. Bacteriol. 173:1187–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Termont S., et al. 2006. Intracellular accumulation of trehalose protects Lactococcus lactis from freeze-drying damage and bile toxicity and increases gastric acid resistance. Appl. Environ. Microbiol. 72:7694–7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thevelein J. M., Hohmann S. 1995. Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem. Sci. 20:3–10 [DOI] [PubMed] [Google Scholar]
- 55. Uyar E. O., Hamamci H., Turkel S. 2010. Effect of different stresses on trehalose levels in Rhizopus oryzae. J. Basic Microbiol. 50:368–372 [DOI] [PubMed] [Google Scholar]
- 56. van de Guchte M., et al. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187–216 [PubMed] [Google Scholar]
- 57. van den Bogaart B. G., Hermans N., Krasnikov V., de Vries A. H., Poolman B. 2007. On the decrease in lateral mobility of phospholipids by sugars. Biophys. J. 92:1598–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vanderpool C. K. 2007. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 10:146–151 [DOI] [PubMed] [Google Scholar]
- 59. Wegmann U., et al. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wells J. M., Robinson K., Chamberlain L. M., Schofield K. M., Le Page R. W. 1996. Lactic acid bacteria as vaccine delivery vehicles. Antonie Van Leeuwenhoek 70:317–330 [DOI] [PubMed] [Google Scholar]
- 61. Whitaker R. D., Batt C. A. 1991. Characterization of the heat shock response in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 57:1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolters J. C., et al. 2010. Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA. PLoS One 5:e10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xie Y., Chou L. S., Cutler A., Weimer B. 2004. DNA macroarray profiling of Lactococcus lactis subsp. lactis IL1403 gene expression during environmental stresses. Appl. Environ. Microbiol. 70:6738–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yancey P. H. 2005. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208:2819–2830 [DOI] [PubMed] [Google Scholar]
- 65. Zomer A. L., Buist G., Larsen R., Kok J., Kuipers O. P. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.