Abstract
Epigenetic modifications of chromatin, such as histone acetylation, are involved in repression of tumor antigens and multiple immune genes that are thought to facilitate tumor escape. The status of acetylation in a cell is determined by the balance of the activities of histone acetyltransferases and histone deacetylases. Inhibitors of histone deacetylase (HDACi) can enhance the expression of immunologically important molecules in tumor cells and HDACi treated tumor cells are able to induce immune responses in vitro and in vivo. Systemic HDACi are in clinical trails in cancer and also being used in several autoimmune disease models. To date, 18 HDACs have been reported in human cells and more than thirty HDACi identified, although only a few immune targets of these inhibitors have been identified. Here, we discuss the molecular pathways employed by HDACi and their potential role in inducing immune responses against tumors. We review data suggesting that selection of target specific HDACi and combinations with other agents and modalities, including those that activate stress pathways, may further enhance the efficacy of epigenetic therapies.
Keywords: Epigenetic gene regulation, Tumor vaccine, Histone deacetylase inhibitor, Immune response, Inflammatory cytokine
Introduction
Histone deacetylase inhibitors [HDACi] are a new generation of chemical agents being used to develop therapy against cancer and other diseases [1–3]. Many of these compounds, including trichostatin A [TSA] the first HDACi identified, were studied originally as differentiating agents. Several HDACi are in clinical trials based on their ability to inhibit cell growth and induce apoptosis and have shown significant activity against a spectrum of hematological and solid tumors [4, 5]. Monitoring gene effects following treatments with HDACi has furthered our understanding the role of epigenetic regulation in cancer. Many studies have identified numerous genes regulated epigenetically in cancer (reviewed in 6, 7) and several reviews have focused on the epigenetics of immune genes particularly in regulating T and B-cell differentiation [8–10]. Recent studies have also suggested that epigenetic silencing of immune genes in cancer may be as, or more, frequent a cause of gene repression as are mutations [11]. Here we consider additional mechanisms for the HDACi mediated effects in cancer cells and review evidence suggesting that cellular stress can enhance the expression of repressed immune genes elicited by HDACi in cancer cells. The first study showing the activation of silenced MHC genes in several tumor cells was performed with TSA [12]. This and other studies raised the possibility that systemic treatments with HDACi could potentially enhance the host immune responses by correcting the negative affects of the tumor on host immunity. We also discuss an epigenetically modified vaccine produced by treatment of tumor cells in vitro with HDACi. As will be emphasized, the HDACi currently in use affect numerous genes and pathways in tumors, as well as normal cells. Moreover, the effects of these agents may vary in different tumor types. We are just beginning to understand the complex epigenetic mechanisms involved in immune regulation and how HDACi can be most beneficially employed to modulate immune responses and the course of cancer.
Histone acetylation and cancer
In eukaryotic cells, complexes of genomic DNA and histones form the nucleosomal structures of chromatin, in which 146 base pairs of double-stranded DNA are wrapped around a central core of four basic histones (H3, H4, H2A and H2B). Each nucleosome has eight histone proteins arranged in a tripartite structure with one (H3H4) tetramer and two H2AH2B dimers. Nucleosomes are separated by linker DNA and compacted into higher-ordered structures by histone H1 [13]. While providing a mechanism of inserting several meters of DNA into a single nucleus, structural compactions can also restrict the access of regulatory proteins to DNA. N-terminal tails of all four histones protrude outward from the core histones and are more accessible to histone modifying enzymes [13]. Many studies over the past decade have shown that multiple covalent modifications (acetylation, phosphorylation, methylation, ubiquitination, sumoylation, ADP-ribosylation) occur on histone tail residues and, as more recent data demonstrates, also in the body of the histone proteins. The histone code hypothesis suggests that a dynamic constellation of these post-translational modifications determines the binding of chromatin remodeling factors to the nucleosome [14]. These factors, by altering chromatin structure, regulate the accessibility of transcription factors, cofactors, and the general transcriptional machinery to DNA, which ultimately determine gene expression. All of the epigenetic alterations currently recognized on histones are reversible and separate sets of enzymes for removing these marks have been identified [11]. The histone acetyltransferase [HAT] and histone deacetylase [HDAC] enzymes determine the status of nucleosome histone acetylation which is one of the most extensively studied of the chromatin modifications. It is the balance between the opposing activities of HATs and HDACs that determine chromatin structure at the gene level and, therefore, gene expression patterns. In addition, the relative level of total cellular HATs and HDACs determines the global status of acetylation in the genome which is important in regulating the cell’s response to endogenous and exogenous stimuli. Histone acetylation is generally correlated with gene activation and deacetylation with repression, although this is not always the case. For example, the ‘master regulator’ of MHC class II, CIITA is activated by STAT1 and this pathway requires HDACs [15], as does interferon stimulated innate immunity [16].
In humans, 18 HDACs have been identified which are divided into three classes based on sequence homology to yeast deacetylases [17]. Class I HDACs includes HDAC 1, 2, 3, and 8, which are mostly localized to the nucleus with ubiquitous distribution throughout human cell lines and tissues. Class II HDACs, divided into two subclasses IIa (HDAC 4, 5, 7 and 9) and IIb (HDAC 6 and 10) can shuttle between the cytoplasm and nucleus and have tissue-specific differences in distributions. Class III HDACs are the sirtuins (SIRT 1–7) and are structurally and mechanistically distinct from other HDACs. To generate acetate and deacetylated protein, class I and class II HDACs use a Zn2+-dependent mechanism, while class III HDACs requires NAD+. HDACs also modulate the acetylation of numerous other nonhistone proteins such as p53, cytoskeleton protein α-tubulin, and the molecular chaperone Hsp90 [18–20]. Direct acetylation of transcription factors and cofactors by HATs is another important mechanism by which acetylation regulates gene expression [21].
The transformation from normal to cancer cells involves multiple processes, which include genetic and epigenetic pathways. Chromatin modifications as well as DNA methylations are epigenetic changes that regulate gene expression, cell growth and differentiation without altering DNA sequences. Aberrant methylation of DNA, a well-characterized gene silencing mechanism, is an hallmark of cancer [6]. Dysregulation of histone modifications also lead to inappropriate expression or repression of genes, a key event in the pathogenesis of many forms of cancers [7]. Three major mechanisms are involved in the epigenetic modifications found in cancer: (1) activation of silenced genes, particularly oncogenes, (2) silencing of normally active tumor suppressors, and (3) silencing of certain immune genes involved in the antitumor response. There is substantial evidence that oncogenes and tumor suppressor genes are regulated by HDACs and that both HAT and HDAC mutations occur during carcinogenesis (reviewed in 22). Although it is established that tumor suppressor and immune genes are often silenced by HDAC in cancer cells, the events leading to epigenetic silencing are poorly understood. One possibility, suggested by recent data, is that micro-RNA [miRNA] often found at fragile sites in cancer cells are involved [23] in repression of gene at several levels including message stability, translation, and transcription [24]. We have found expression of many immune genes, including MHC and costimulatory molecules are regulated by miRNAs (Asirvatham AJ et al. manuscript submitted), but just how these small inhibitors regulate chromatin silencing in cancer cells is uncertain. One possibility is that miRNAs bind to DNA or nascent RNA transcripts and silence chromatin by recruiting HDACs as shown at heterochromatic foci [25]. Further studies are necessary to clarify this important issue.
An example of epigenetic repression of immune genes is seen in plasma cell tumors, which usually express MHC class I, but not class II, and this is associated with absence of mRNA for CIITA, a cofactor with HAT activity [12, 26]. It has been suggested that Blimp-1, a zinc-finger DNA-binding protein, recruits a corepressor complex containing HDAC to the CIITA promoter, and this is responsible for the failure of normal plasma cells and plasmacytomas to express class II [27]. HDAC1 and HDAC2 also inhibit the transcription of CIITA and the expression of MHC class II in human cervical cancer cell lines [28], as well as other tumor types [15]. MHC class II-antigen complexes are required for the recognition of tumor cells by CD4+ T-cells and antigen presentation via class II is critical for the activation of adaptive immune responses. Repressor complex containing HDAC1, HDAC2, and HDAC8 also mediate MHC class I downregulation in cervical cancer cells making them less susceptible to cytotoxic T-cells [CTL] [29]. Additionally, deacetylation of histone H3 and H4 is associated with the repression of the costimulatory molecules CD86 and ICAM-1 genes in AML cells [30] and HDAC mediated repression of MHC, CD40, and CD80 have been identified in various other human (e. g., neuroblastoma, squamous cell carcinoma, lymphoma) and mouse tumor cell lines (e.g., melanoma, adenocarcinoma, plasmacytoma) [12, 31–33]. Histone deacetylation and DNA methylation are also involved in silencing of MAGE antigen expression in different human cancer cells [34]. The quantitative levels of tumor antigens, as well as cell surface expression of MHC and costimulatory genes are important in determining T-cell activation, and epigenetic repression of these molecules in tumor cells provide a mechanism for tumor escape [11]. In tumor cells, the balance of histone acetylation and deacetylation is often disturbed and this is a rational for employing HDAC inhibitors in treatment protocols. Inhibition of HDAC shifts the balance toward acetylation with concomitant expression of repressed immune genes. Thus, treatments with HDACi could enhance antigen presentation as well as allow expression of cellular components required in the effector stages of antitumor immunity. However, the lack of an in-depth understanding of the specific function of different HDACs and their role in tumorigenesis is a major challenge in the selection of appropriate HDACi for therapies.
Inhibition of HDAC elicits immune genes in tumor cells
Inhibitors of class I and class II HDACs have been the focus of intense research in developing cancer therapy [5, 35]. Inhibitors of class III HDACs, such as the SIRT1 inhibitor Sirtinol, have recently been shown to induce senescence-like growth arrest in cancer cells [36], although little information is as yet available on immune gene expression patterns in senescence tumor cells. Naturally occurring and synthetic inhibitors of class I and class II HDAC can be divided into four main structural classes—hydroxamates, cyclic peptides, benzamides and aliphatic acids (selective examples in each class are shown in Table 1). Among these compounds, TSA—an antifungal agent isolated from Streptomyces hygroscopicus, is widely used in functional studies, because of its high reactivity and many synthetic HDACi has been modeled upon TSA. Recently newer generation of HDACi has been designed, such as (aryloxopropenyl)pyrrolyl hydroxyamides, with specific biological effects [37]. Many of the HDACi described have broad target specificity, such as SAHA an oral HDACi, which like TSA inhibits the activity of all class I and class II HDACs. However, some HDACi do in fact selectively target particular HDAC family members. For example, depsipeptide inhibits HDAC1 and HDAC2 (class I) but not HDAC 4 and HDAC6 (class II) [38]. MS-275 shows no inhibitory activity against HDAC8, but inhibits HDAC1 and HDAC3 [39]. In addition, (aryloxopropenyl)pyrrolyl hydroxyamides selectively inhibits HDAC4 but not HDAC1 [37]. Since, class I HDACs are well-known transcriptional corepressor and class IIa HDACs interact with DNA-binding transcription factors, as well as transcriptional corepressors [40], selective inhibition of specific HDAC could be targeted to induce genes important in activation of immune responses. However, it remains to be seen, whether selective inhibitors will be more effective and less toxic than the pan-HDACi in current use in inducing immune effector cells.
Table 1.
Different classes of HDAC inhibitors enhance immune molecules in tumor cells that may activate immune responses
| Inhibitors** | Target HDAC |
Immune genes enhanced by certain HDACi in tumor cells |
Effect on immune responses |
|---|---|---|---|
| Hydroxamates (e.g., TSA*, SAHA, LAQ824, LBH589, Pyridoxine, PXD101, CHAP) | Class I, IIa, IIb | MAGE 1–4, MHC class I | ↑ CTL recognition |
| MHC class I, class II, B7–1/2, CD40, ICAM1 | ↑ CD4+ and CD8+ T cell activation | ||
| Aliphatic acids (e.g., Butyrate, Valproic acid, AN-9) | Class I, IIa, | TAP1, TAP2, LMP2, LMP7, Tapasin | ↑Antigen processing and presentation |
| NKG2DL | ↑ Activation of NK and CD8+ T cells | ||
| Cyclic peptides (e.g., Depsipeptide*, Apicidin*) | Class I | ? | ? |
| Benzamides (e.g., CI-994, MS-275) | Class I | ? | ? |
| Nicotinamide, Sirtinol, Splitomicin | Class III | ? | ? |
DNA micro arrays of cells cultured with HDACi demonstrates that the expression of ~5% of the genes are altered depending on the cell type and the HDACi analyzed [5, 41]. TSA and valproic acid [VA] has been reported to enhance MHC class I expression in several tumor cells [12, 42, 43]. In addition to MHC class I, TSA treatment enhances the expression of molecules (TAP1, TAP2, LMP2, LMP7 and Tapasin) involved in antigen processing and presentation via class I pathway in melanoma cells. The TSA treated melanoma cells are capable of presenting class I restricted peptides and protein antigens directly to stimulate CD8+ T-cells in vitro (Khan ANH, manuscript submitted). VA treatment of cervical cancer cells enhances antigen-specific recognition of tumor cells by CTLs and the use of VA as an adjuvant has been suggested in cervical cancer patient [43]. Deficient expression of MHC class I and antigen processing components in tumor cell are well-established mechanisms of tumor escape from CTL mediated killing.
Several publications indicate that treatment with HDACi, such as TSA and VA, can also enhance expression of MHC class II and costimulatory molecules CD40, B7-1 and B7-2 in various human and mouse tumor cell lines [12, 30, 31, 33]. Additionally, TSA treated plasmacytoma and melanoma cells are capable of presenting antigens in the context of class II to stimulate IFNγ secretion and proliferation of CD4+ T-cells in vitro (15, Khan ANH et al. manuscript submitted). Expression of class II and costimulatory molecules on tumor cells involving gene transfection also enhances T-cell immunity in vivo [44]. The HDACi, sodium butyrate induces expression of ICAM-1 in different AML cell lines [30] and binding of ICAM-1 to LFA-1 on lymphocytes is crucial in activating T-cells [45]. Furthermore, TSA induces expression of ICAM-1 in tumor endothelial cells, which may modulate leukocyte infiltration into the tumor bed [46]. These studies collectively suggest that treatment of tumor cells with specific HDACi may alter their functional capability to induce T-cell mediated immune responses.
Our laboratory and others have shown that HDACi (TSA and VA) enhances the expression of NKG2D ligands [NKG2DL] on several different tumor cell types (Gregorie C et al. manuscript in preparation, 47, 48). NKG2DL on tumor cells can directly activate NK and CD8+ T-cells mediated responses against tumors [49, 50]. Activation of NK cells initiates a cascade of cytokine and chemokine expression leading to stimulation of macrophage, dendritic cells [DC] and other immune effectors [51, 52]. A review of chromatin changes in DNA damage suggests the possibility of an epigenetic code for DNA damage repair pathways [53]. In this scenario, NKG2DL also could be induced by the ATM/ATR DNA damage pathway in cancer cells [54]. However, whether the induction of NKG2DL by HDACi requires ATM/ATR or can occur by direct chromatin effects at the gene level is uncertain. It appears from the above data that acetylation by HDACi treatment of tumor cells can activate innate, as well as adaptive immunity.
Induction of immunity by epigenetic tumor vaccines
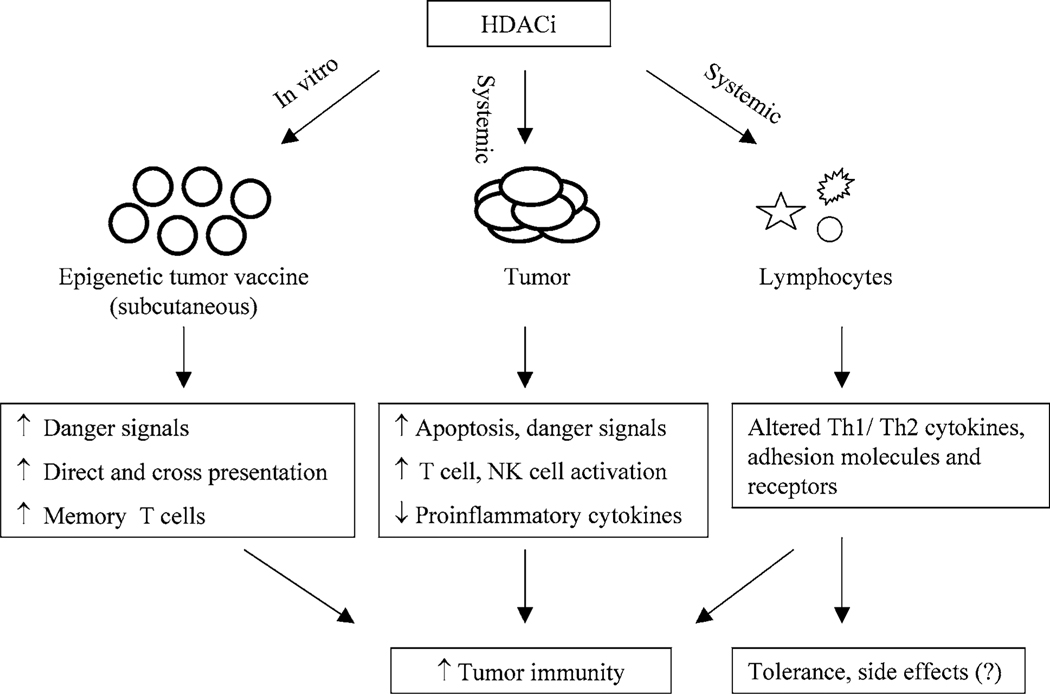
Activation of naïve CD4+ and CD8+ T-cells is achieved primarily through cross-presentation of tumor antigens by professional antigen presenting cells [APC] [55, 56]. Cross-presentation of antigen in absence of immunostimulatory signals can, however, induce tolerance. Direct antigen presentation by tumor cells could also activate naïve T-cells provided the tumor cells can deliver an MHC-restricted antigen specific signal together with appropriate costimulatory signals [43, 44]. We have shown in both murine plasmacytoma and melanoma models that effective antitumor immune responses can be achieved utilizing HDACi treated tumor cells that expressed MHC and costimulatory molecules CD40 and B7-1/2 (31, Khan ANH et al. manuscript submitted). In these studies, cytotoxic and IFNγ-producing T-cells were generated by vaccination with an optimal concentration of TSA treated tumor cells and a significant number of animals showed durable tumor specific immunity. Both cross and direct presentations appear to synergize in the activation of immune effector cells in these models. However, a remaining issue is the relative quantitative importance of direct versus cross-presentation in the immunity elicited by the epigenetic vaccine. Interestingly, NK cells, together with CD4+ and CD8+ T-cells, played an important role in initiation of immunity following HDACi treated tumor cell vaccination. Since HDACi treatment induces NKG2D ligand on tumor cells, we consider it likely that HDACi treated tumor cell vaccine may be capable of directly activating NK cell-mediated innate responses. These studies also show that apoptotic tumor cells produced by TSA and other DNA damaging agents vary in their ‘adjuvant’ effect in a vaccine setting, and vaccine inocula containing HDACi treated apoptotic cells (~50%) were the most effective (Khan ANH et al. manuscript submitted). Furthermore, apoptotic cell components can activate inflammatory regulators [57] and Toll-like receptors [TLR] [58] that potentially promote the antitumor immunity [59]. Although additional work is required to further dissect the mechanisms involved in epigenetic vaccination, these studies suggest that HDACi treated tumor cells are capable of activating both innate and adaptive immune responses in vivo (Fig. 1).
Fig. 1.
Modulation of antitumor immune responses by treatment with HDACi. Vaccination with HDACi treated tumor cells can induce danger signals, T-cell, and NK cell-mediated immunity. Systemic HDACi potentially activate adaptive and innate immune responses through induction of immune genes on tumor cells (e.g., MHC, CD40, B7-1/2, ICAM-1, NKG2DL), by enhancing antigen availability and promoting apoptosis of tumor cells. HDACi may also suppress proinflammatory cytokines in tumor microenvironment. Furthermore, effects of systemic HDACi on lymphocytes (T-cells, macrophages and DC) may augment immunity by secretion of cytokines (e.g., IL-2, IFNγ) and activating adhesion molecules (e.g., ICAM-1) and receptors (e.g. TLR). However, in certain situation HDACi mediated inhibition of Th1 effector cell function may alter the Th1 and Th2 balance and promote tolerance instead of immunity
Role of systemic HDAC inhibitors in activating immune responses
Systemic HDACi have shown significant antitumor effects in preclinical models and some are currently in clinical trials [11, 60]. A new drug application has recently been approved for SAHA treatment of T-cell lymphoma [60]. However, the precise molecular pathways involved in the antitumor effects have not been fully determined. It has been shown that upregulation of p21waf, p16, and p27 and downregulation of Cyclin A, Cyclin D, CDK4 and dephosphorylation of pRb are common features of HDAC inhibition and are necessary for cell cycle arrest and growth arrest of tumor cells [4]. Altered expression of genes involved in the apoptotic pathways is also associated with HDACi treatment [61]. Additionally, HDACi are thought to induce tumor regression through inhibition of angiogenesis [62]. Although current systemic treatments have not been specifically designed to target the immune system, as discussed above, optimal concentrations of HDACi may upregulate repressed tumor immune genes, activate T-cells and allow host effector responses. Systemic HDACi, while altering tumor associated antigens, MHC and costimulatory molecule expression and antigen presentation, may also affect other components in tumor cells, including those that represent ‘danger signals’ (i.e., heat-shock proteins, HMGB1) or inflammatory mediators (i.e., cytokines, chemokines) as discussed below. Furthermore, much recent evidence suggests a ‘continuum’ between innate and adaptive immune responses [63] and these immune cell populations may also be target of HDACi to generate distinct patterns of immune responsiveness. Thus, in addition to direct differentiating, cytostatic, and cytotoxic effects, systemic HDACi may modify immunity through multiple host and tumor pathways to augment the efficacy of the antitumor therapy (Table 1 and Fig. 1).
Modulation of inflammatory cytokines by HDACi
In addition to their anticancer activities, several HDACi are effective in selected immune disease models. For example, treatment with SAHA inhibits TNFα and IL-6 production by stimulated MRL-lpr mesangial cells in vitro and blocks renal disease progression in MRL-lpr mice, a murine model of systemic lupus erythematosus [SLE] [64]. SAHA treatment also significantly reduces experimental colitis and this correlates with suppression of colonic proinflammatory cytokines [65]. TSA treatment can suppress adjuvant-induced rheumatoid arthritis in rats [66] and reduce clinical symptoms in a murine model of multiple sclerosis [67]. SAHA and TSA, at lower concentration, also inhibit the production of Th1 (IL-12 and IFNγ), as well as Th2 (IL-6 and IL-10) cytokines by MRL-lpr splenocytes [68, 69]. Butyrate, when administered topically, can effectively treat ulcerative colitis [70]. In addition, systemic treatment with SAHA suppresses serum levels of inflammatory cytokines and reduces graft-versus-host disease in a murine bone marrow transplant model [71]. It has been suggested that inhibition of proinflammatory cytokine production (IL-1, IL-6, IL-8, IL-10, IL-12, IFNγ and TNFα) is correlated with reduced disease progression after HDACi treatment [3, 72]. However, the cellular and molecular mechanisms governing the effects of HDACi in these disease models are not well-understood. Furthermore, as discussed below, the relation of inflammation to cancer progression and immunity is far from certain. While additional investigations are needed, these studies do suggest that suppression of inflammatory cytokines by HDACi can play a role in the treatment of autoimmune diseases.
With regard to tumors, much data now suggests that chronic inflammation may be etiologically related to the initiation and progression of cancer [73, 74]. In fact, antiinflammatory agents, such as aspirin and NSAIDs have been reported to prevent cancer, for example cancer of the colon and the progression of Barrett’s esophagus to esophageal cancer [75, 76]. It has been suggested, as outlined above for autoimmune disorders, that HDACi enhance regression of tumors via inhibition of NF-κB and proinflammatory cytokines levels in malignant cells, as well as macrophages present in the tumor microenvironment [72]. Therefore, the inhibition of proinflammatory cytokines by HDACi might be expected to be beneficial in the treatment of tumors, which are associated with chronic inflammation (Fig. 1). However, in the acute situation the role of ‘good’ versus ‘bad’ inflammation is uncertain and likely depends on the type and extent of the inflammatory process, its location, the genetic setting, associated diseases and other complex and incompletely understood determinants [73, 77]. Importantly, the initiation of the immune response depends on inflammation, and adjuvants are well-established immune promoters. Thus, some ‘controlled’ inflammation is beneficial and required in antitumor immunity and likely as suggested in developing better vaccines [57]. Many adjuvants (CpG, IL-12, CD40L etc.) employed in combination with tumor vaccines elicit inflammation and promote immunity [78] while others, including radiation and chemotherapy, induce apoptotic cells which can also enhance inflammation. Moreover, in contrast to the antiinflammatory effects discussed above, HDACi can enhance proinflammatory cytokines in certain situation and cell types. For example, TSA and SAHA have been shown to increase the LPS-induced expression of IL-6, TNFα, and MIP-2 in microglial cells [79]. Treatment with TSA also enhances IL-8 production, but not IL-12, by SV-40-transformed lung epithelial cells [80]. In addition, TSA treatment enhances TLR4 expression in embryonic stem cells and this is correlated with increased IL-6 and TNFα expression in stimulated cells [81]. These data, in sum, suggest that enhanced production of certain inflammatory cytokines by HDACi may be important in the activation of both innate immune effector cells and adaptive responses. However, whether HDACi alone, or in association with other agents, could be used to augment inflammatory cytokine mediated immune responses in the tumor microenvironment remains to be determined. Inflammatory infiltrates differ in composition, and it will be necessary in future work to identify those elements of inflammation that boost tumor immunity versus those that are involved in tumor progression.
Effects of HDACi on immune cells
The HDACi given systemically in clinical trials are generally well tolerated with minimal hematological effects [82, 83], but accumulation of acetylated histones in immune cells, such as T-cells and antigen presenting cells could alter their functional capacity depending on the dose, route and specific drug. A recent in vitro study shows that LAQ824 alters TLR4-dependent activation and function of macrophages and DC, which leads to activation of Th2 but not Th1 effector cells [84]. Treatment with TSA abrogates IL-2 production and CD28 expression by CD4+ T–cells, but enhances the expression of HSP70, TGFβ1 and ICAM1 genes in T-cells [85]. TSA also inhibits CD4+ expression in stimulated T-cells, but not in unstimulated T-cells suggesting that certain HDACi may induce factors responsible for negative regulation of CD4+ T-cells [86]. While low concentration of TSA treatment enhances IFNγ production by normal T-cells and PMA/ Ionomycin stimulated SLE T-cells [87], high concentrations of TSA and butyrate reduce IFNγ production by normal T-cells [88]. Since, T-cells are a major source of IFNγ, a key cytokine in tumor immunity, these findings should be considered in systemic HDACi therapies. Moreover, as mentioned above, SAHA has antiinflammatory properties and suppresses LPS-induced secretion of TNFα, IL-1 and IL-12 by human PBMC [68]. It is difficult at present to rationalize the contrasting effects of HDACi on cytokine and T-cells. They may be related not only to the type of HDACi and its dosage but also to the stage of T-cell differentiation. Complex patterns of epigenetic modifications are causally associated with cytokine gene activation and repression in different T-cell lineages [89]. For example, differentiation of naïve CD4+ Th0 cells into mature Th2 cells is associated with hyperacetylation of histone H3 at the IL-4 gene, whereas differentiation into Th1 cells is associated with hypoacetylation of histone H3 at the IFNγ gene [90]. Additional factors might be direct acetylation of nonhistone proteins, such as NF-κB, which regulate cytokine expression in lymphocytes. Although many details of the epigenetic regulation of lymphocytes are still undefined, these studies do suggest that systemic HDACi therapy may alter immune homeostasis and tumor immunity (Fig. 1).
The transcription factor NF-κB, an important regulator of immune and inflammatory responses, is predominantly located in the cytoplasm of unstimulated cells. Various extracellular stimuli result in nuclear translocation of NF-κB and activation of target genes in lymphocytes and cancer cells [91]. Histone modification plays a critical role in determining expression of several NF-κB dependent genes, including both pro and antiapoptotic genes in various cells [92, 93]. HDACi can suppress NF-κB-dependent gene expression through different mechanisms. For example, TSA and butyrate can prevent the nuclear translocation of NF-κB, as well as NF-κB-dependent promoter activity in macrophage cell lines [94]. HDACi mediated stabilization of I-κB is associated with reduced gene transcription in LPS-stimulated macrophage cells [95]. In addition, HDACi suppress NF-κB-regulated gene expression in mesangial cells by inhibiting the HDAC2 - NF-κB interaction at the promoter [96], and in a colon cell line by reducing the proteasome-dependent degradation of I-κB [97]. However, the effects of HDACi may induce NF-κB in certain situation. For example, HDACi can enhance acetylation of NF-κB in the cytoplasm, impair I-κB binding, promote nuclear translocation of NF-κB [98] and NF-κB activation by HDACi can induce transcription of inflammatory genes in certain cells [72]. Therefore, as discussed above, the cell type, the specificity of the HDAC and the type of external stimuli appear to determine, whether HDACi exerts inhibitory or inducing effects on NF-κB.
HDACi may also affect other signaling pathways that regulate immune genes. Importantly, the mitogen activated protein kinases [MAPK] pathways activated by stress stimuli can directly phosphorylate histone H3 on serine 10 and 28, and this substantially enhances histone acetylation by HDACi [99, 100]. The marked enhancement of acetylation and gene expression by combinations of HDACi and MAPK activators has therapeutic implications. In addition, as mentioned, STAT a major signaling molecule downstream of many cytokine receptors depend on HDAC for transcription regulation and propagation of inflammatory signals. Recent studies have shown that induction of CIITA and MHC class II by IFNγ can be enhanced or inhibited by HDACi depending on the order of treatment, and that both acetylation and deacetylation can be associated with enhanced transcription [15, 16, 101]. One possibility for the requirement of HDAC in transcription is that the necessity of establishing repressed chromatin after RNA Pol II passage during transcriptional elongation. By removing acetylation, HDACs provide the repressive state required to prevent spurious transcription from intragenic sites behind the elongation complex [102].
Conclusions
As we understand more of the complex mechanisms of epigenetic regulation, it may be possible to select specific HDACi or combinations of HDACi with other epigenetic agents (e.g., DNA methyltransferase inhibitor, histone methyltransferase inhibitor) to activate optimal immune responses against tumors. Newer HDACi are now being explored that show specificity for particular classes of HDAC [37]. The additional specificity will hopefully provide selectivity in the subset of genes activated and more effective, less-toxic epigenetic tumor therapies. As signal transduction pathways that impact epigenetic modifications are further characterized, activators or inhibitors of these signaling pathways (e.g., MAPK, NF-κB, PI3K/AKT and others) may prove more efficacious than current HDACi and/or could be used in combination with HDACi to enhance their activity.
Since, epigenetic regulation is central to many processes of development and differentiation, we consider it likely that HDACi therapy will modify gene expression in normal tissues [15, 85, 103]. Therefore, it will be important to evaluate the impact of HDACi therapy on MHC, costimulatory molecule, NKG2DL and other immune genes in normal cells as well as tumors obtained from systemically treated patient. Moreover, we emphasis, as discussed above, that HDACi treatment may alter cytokine production, as well as T-cell proliferation and differentiation, and could potentially interfere with certain antitumor immune responses. Thus, HDACi therapies, which we feel have great potential in cancer treatment, must be carefully examined as to target specificity, dosage and combinations with other agents in order to maximize immunity and minimize adverse effects. In addition, antiinflammatory effects of specific HDACi may be useful for the treatment of certain inflammatory diseases and inflammation associated cancers. However, we stress that the effects of HDACi on inflammation are complex, variable and at present poorly understood.
Autologus tumor cell vaccines generated by HDACi in vitro, as described above, involves tumor-associated antigens, unique to the individual tumor and would be applicable without prior knowledge of the tumor antigens. While this vaccination approach would also have the advantage of being histocompatible with the host and capable of generating tumor specific durable immunity, clearly there are challenges and potential problems in adapting this type of vaccine for the cancer treatments. For example, the design of methods to kill tumor cells following the HDACi treatment and yet maintain immunogenicity will be important in future work. Additionally, activation of immune suppressor cells in tumor microenvironment, such as Treg, may promote tolerance [104]. In this regard, use of different HDACi in combination with anti-Treg and other adjuvant treatments may generate more effective tumor cell vaccines. Furthermore, vaccination approaches utilizing tumor-derived exosomes, which are membrane bound vesicles secreted by the normal cellular endocytic pathway, have demonstrated immunogenicity in murine tumor models [105, 106]. Treatments of tumor cells with HDACi, which enhance immune molecules including tumor antigens, may be reflected in the exosomes produced by the HDACi treated cells. Thus, exosomes could be designed to maximize tumor immunogenicity while avoiding many of the issues associated with whole cell vaccination. Finally, with the advent of additional data on the role of miRNA in cancer immunity and methods of delivering RNAi to cells, new prevention and treatment approaches based on small RNAs could be imagined. For example, tumor vaccines produced by derepressing the inhibitory effects of miRNA on immune genes (a dicer vaccine) or even systemic treatments using anti-miRNAs delivered specifically to tumors.
Acknowledgment
This work was supported by the National Institutes of Health grant HD 17013.
Contributor Information
A. Nazmul H. Khan, Email: anmnazmul.khan@roswellpark.org, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm & Carlton Streets, Buffalo, NY 14263, USA.
Thomas B. Tomasi, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm & Carlton Streets, Buffalo, NY 14263, USA Departments of Medicine and Microbiology & Immunology, School of Medicine and Biomedical Sciences, State University of New York, Buffalo, NY 14214, USA.
References
- 1.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Conley BA, Wright JJ, Kummar S. Targeting epigenetic abnormalities with histone deacetylase inhibitors. Cancer. 2006;107:832–840. doi: 10.1002/cncr.22064. [DOI] [PubMed] [Google Scholar]
- 3.Huang L. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases. J Cell Physiol. 2006;209:611–616. doi: 10.1002/jcp.20781. [DOI] [PubMed] [Google Scholar]
- 4.Marks PA, Dokmanovic M. Histone deacetylase inhibitors: discovery and development as anticancer agents. Expert Opin Investig Drugs. 2005;14:1497–1511. doi: 10.1517/13543784.14.12.1497. [DOI] [PubMed] [Google Scholar]
- 5.Bhalla KN. Epigenetic and chromatin modifiers as targeted therapy of hematological malignancies. J Clin Oncol. 2005;23:3971–3993. doi: 10.1200/JCO.2005.16.600. [DOI] [PubMed] [Google Scholar]
- 6.Laird PW. Cancer epigenetics. Human Mol Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 7.Ducasse M, Brown MA. Epigenetic aberrations and cancer. Mol Cancer. 2006 doi: 10.1186/1476-4598-5-60. ( www.molecular-cancer.com/content/5/1/60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nature Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 9.Bergman Y, Cedar H. A stepwise epigenetic process controls immunoglobulin allelic exclusion. Nature Rev Immunol. 2004;4:753–761. doi: 10.1038/nri1458. [DOI] [PubMed] [Google Scholar]
- 10.Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu Rev Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 11.Tomasi TB, Magner WJ, Khan ANH. Epigenetic regulation of immune escape genes in cancer. Cancer Immunol Immunother. 2006;55:1159–1184. doi: 10.1007/s00262-006-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magner WJ, Kazim AL, Stewart C, et al. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol. 2000;165:7017–7024. doi: 10.4049/jimmunol.165.12.7017. [DOI] [PubMed] [Google Scholar]
- 13.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 14.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 15.Chou S-D, Khan ANH, Magner WJ, Tomasi TB. Histone acetylation regulates the cell type specific CIITA promoters, MHC class II expression and antigen presentation in tumor cells. Int Immunol. 2005;17:1483–1494. doi: 10.1093/intimm/dxh326. [DOI] [PubMed] [Google Scholar]
- 16.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeRuijten AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 19.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacs JJ, Murphy PJ, Gaillard S, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Moll Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 22.Hake SB, Xiao A, Allis CD. Linking the epigenetic language of covalent histone modifications to cancer. Br J Cancer. 2004;90:761–769. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 25.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh N, Gyory I, Wright G, Wood J, Wright KL. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J Biol Chem. 2001;276:15264–15268. doi: 10.1074/jbc.M100862200. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by Blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20:2592–2603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zika E, Greer SF, Zhu X-S, Ting JP-Y. Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and Major Histocompatibility Complex class II enhanceosome formation. Mol Cell Biol. 2003;23:3091–3102. doi: 10.1128/MCB.23.9.3091-3102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Ou X, Xiong J, Wang T. HPV16E7 mediates HDAC chromatin repression and downregulation of MHC class I genes in HPV16 tumorigenic cells through interaction with an MHC class I promoter. Biochem Biophys Res Commun. 2006;349:1315–1321. doi: 10.1016/j.bbrc.2006.08.182. [DOI] [PubMed] [Google Scholar]
- 30.Maeda T, Towatari M, Kosugi H, Saito H. Up-regulation of costimulatory/adhesion molecules by histone deacetylase inhibitors in acute myeloid leukemia cells. Blood. 2000;96:3847–3856. [PubMed] [Google Scholar]
- 31.Khan ANH, Magner WJ, Tomasi TB. An epigenetically altered tumor cell vaccine. Cancer Immunol Immunother. 2004;53:748–754. doi: 10.1007/s00262-004-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gialitakis M, Kretsovali A, Spilianakis C, et al. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin A. Nucleic Acid Res. 2006;34:765–772. doi: 10.1093/nar/gkj462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanaseki T, Ikeda H, Takamura Y, et al. Histone deacetylation, but not hypermethylation, modifies class II transactivator and MHC class II gene expression in squamous cell carcinoma. J Immunol. 2003;170:4980–4985. doi: 10.4049/jimmunol.170.10.4980. [DOI] [PubMed] [Google Scholar]
- 34.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4:339–349. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 35.Acharya MR, Sparreboom A, Venitz J, Figg WD. Rational development of histone deacetylase inhibitors as anticancer agents: a review. Mol Pharm. 2005;68:917–932. doi: 10.1124/mol.105.014167. [DOI] [PubMed] [Google Scholar]
- 36.Ota H, Tokunaga E, Chang K, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 37.Mai A, Massa S, Pezzi R, et al. Class II (IIa)-selective histone deacetylase inhibitors.1. synthesis and biological evaluation of novel (Aryloxopropenyl)pyrrolyl Hydroxyamides. J Med Chem. 2005;48:3344–3353. doi: 10.1021/jm049002a. [DOI] [PubMed] [Google Scholar]
- 38.Furumai R, Matsuyama A, Kobashi N, et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- 39.Hu E, Dul E, Sung C-M, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- 40.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 41.Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–4572. [PubMed] [Google Scholar]
- 42.Bubenik J. Prospects for immunotherapy of MHC class I deficient tumors. Folia Biol (Praha) 2003;49:95–99. [PubMed] [Google Scholar]
- 43.Mora-García ML, Duenas-González A, Hernández-Montes J, et al. Up-regulation of HLA class-I antigen expression and antigen-specific CTL response in cervical cancer cells by the demethylating agent hydralazine and the histone deacetylase inhibitor valproic acid. J Transl Med. 2006 doi: 10.1186/1479-5876-4-55. ( www.translational-medicine.com/content/4/1/55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pulaski BA, Ostrand-Rosenberg S. Reduction of established spontaneous mammary carcinoma metastases following immunotherapy with major histocompatibility complex class II and B7.1 cell-based tumor vaccines. Cancer Res. 1998;58:1486–1493. [PubMed] [Google Scholar]
- 45.Kim JJ, Tsai A, Nottingham LK, et al. Intracellular adhesion molecule-1 modulates β-chemokines and directly costimulates T cells in vivo. J Clin Invest. 1999;103:869–877. doi: 10.1172/JCI6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hellebrekers DM, Castermans K, Viré E, et al. Epigenetic regulation of tumor endothelial cell anergy: silencing of intercellular adhesion molecule-1 by histone modifications. Cancer Res. 2006;66:10770–10777. doi: 10.1158/0008-5472.CAN-06-1609. [DOI] [PubMed] [Google Scholar]
- 47.Skov S, Pedersen MT, Andresen L, et al. Cancer cells become susceptible to natural killer cell killing after exposure to histone deacetylase inhibitors due to glycogen synthase kinase-3-dependent expression of MHC class I-related chain A and B. Cancer Res. 2005;65:11136–11145. doi: 10.1158/0008-5472.CAN-05-0599. [DOI] [PubMed] [Google Scholar]
- 48.Armeanu S, Bitzer M, Lauer UM, et al. Natural killer cell-mediated lysis of hepatoma cells via specific induction of NKG2D ligands by the histone deacetylase inhibitor sodium valproate. Cancer Res. 2005;65:6321–6329. doi: 10.1158/0008-5472.CAN-04-4252. [DOI] [PubMed] [Google Scholar]
- 49.Zhou H, Luo Y, Lo FJ, et al. DNA-based vaccines activate innate and adaptive antitumor immunity by engaging the NKG2D receptor. Proc Natl Acad Sci USA. 2005;102:10846–10851. doi: 10.1073/pnas.0502208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamiryo Y, Yajima T, Saito K, et al. Soluble branched (1,4)-β-d-glucans from Acetobacter species enhances anti-tumor activities against MHC class I-negative and –positive malignant melanoma through augmented NK activity and cytotoxic T cell response. Int J Cancer. 2005;115:769–776. doi: 10.1002/ijc.20934. [DOI] [PubMed] [Google Scholar]
- 51.Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take center stage. Nature Rev Immunol. 2005;5:112–124. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 52.Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr Opin Immunol. 2005;17:29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Hassa PO, Hottiger MO. An epigenetic code for DNA damage repair pathways? Biochem Cell Biol. 2005;83:270–285. doi: 10.1139/o05-034. [DOI] [PubMed] [Google Scholar]
- 54.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heath WR, Carbone FR. Cross-presentation, dendritic cells, tolerance and immunity. Annu Rev Immunol. 2001;19:47–64. doi: 10.1146/annurev.immunol.19.1.47. [DOI] [PubMed] [Google Scholar]
- 56.Thomas AM, Santarsiero LM, Lutz ER, et al. Mesothelin-specific CD8+ T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med. 2004;200:297–306. doi: 10.1084/jem.20031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Restifo NP. Building better vaccines: how apoptotic cell death can induce inflammation and activate innate and adaptive immunity. Curr Opin Immunol. 2000;12:597–603. doi: 10.1016/s0952-7915(00)00148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albert ML. Death-defying immunity: do apoptotic cells influence antigen processing and presentation. Nature Rev Immunol. 2004;4:223–230. doi: 10.1038/nri11308. [DOI] [PubMed] [Google Scholar]
- 59.Gamrekelashvili J, Krüger C, von Wasielewski R, et al. Necrotic tumor cell death in vivo impairs tumor-specific immune responses. J Immunol. 2007;178:1573–1580. doi: 10.4049/jimmunol.178.3.1573. [DOI] [PubMed] [Google Scholar]
- 60.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 61.Moore PS, Barbi S, Donadelli M, et al. Gene expression profiling after treatment with histone deacetylase inhibitor trichostatin A reveals altered expression of both pro- and anti-apoptotic genes in pancreatic adenocarcinoma cells. Biochem Biophy Acta. 2004;1693:167–176. doi: 10.1016/j.bbamcr.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Qian DZ, Kato Y, Shabbeer S, et al. Targeting tumor angiogenesis with histone deacetylase inhibitors: the hydroxamic acid derivative LBH589. Clin Cancer Res. 2006;12:634–642. doi: 10.1158/1078-0432.CCR-05-1132. [DOI] [PubMed] [Google Scholar]
- 63.Borghesi L, Milcarek C. Innate versus Adaptive Immunity: A Paradigm Past Its Prime? Cancer Res. 2007;67:3989–3993. doi: 10.1158/0008-5472.CAN-07-0182. [DOI] [PubMed] [Google Scholar]
- 64.Reilly CM, Mishra N, Miller JM, et al. Modulation of renal disease in MRL/lpr mice by suberoylanilide hydroxamic acid. J Immunol. 2004;173:4171–4178. doi: 10.4049/jimmunol.173.6.4171. [DOI] [PubMed] [Google Scholar]
- 65.Glauben R, Batra A, Fedke I, et al. Histone hyperacetylation is associated with amelioration of experimental colitis in mice. J Immunol. 2006;176:5015–5022. doi: 10.4049/jimmunol.176.8.5015. [DOI] [PubMed] [Google Scholar]
- 66.Chung YL, Lee MY, Wang AJ, Yao LF. A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol Ther. 2003;8:707–717. doi: 10.1016/s1525-0016(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 67.Camelo S, Iglesias A, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 68.Leoni F, Zaliani A, Bertolini G, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99:2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra N, Reilly CM, Brown DR, et al. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111:539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vernia P, Annese V, Bresci G, et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur J Clin Invest. 2003;33:244–248. doi: 10.1046/j.1365-2362.2003.01130.x. [DOI] [PubMed] [Google Scholar]
- 71.Reddy P, Maeda Y, Hotary K, et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc Natl Acad Sci USA. 2004;101:3921–3926. doi: 10.1073/pnas.0400380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blanchard F, Chipoy C. Histone deacetylase inhibitors: new drugs for the treatment of inflammatory diseases? Drug Discov Today. 2005;10:197–204. doi: 10.1016/S1359-6446(04)03309-4. [DOI] [PubMed] [Google Scholar]
- 73.de Visser KE, Coussens LM. The interplay between innate and adaptive immunity regulates cancer development. Cancer Immunol Immunother. 2005;54:1143–1152. doi: 10.1007/s00262-005-0702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nature Rev cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rigas B. The use of nitric oxide-donating nonsteroidal anti-inflammatory drugs in the chemoprevention of colorectal neoplasia. Curr Opin Gastroenterol. 2007;23:55–59. doi: 10.1097/MOG.0b013e32801145b0. [DOI] [PubMed] [Google Scholar]
- 76.Anderson LA, Johnston BT, Watson RG, et al. Nonsteroidal anti-inflammatory drugs and the esophageal inflammation-metaplasia-adenocarcinoma sequence. Cancer Res. 2006;66:4975–4982. doi: 10.1158/0008-5472.CAN-05-4253. [DOI] [PubMed] [Google Scholar]
- 77.Karin M. Inflammation and cancer: the long reach of Ras. Nature Med. 2005;11:20–21. doi: 10.1038/nm0105-20. [DOI] [PubMed] [Google Scholar]
- 78.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nature Med. 2004;10:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suuronen T, Huuskonen J, Pihlaja R, Kyrylenko S, Salminen A. Regulation of microglial inflammatory response by histone deacetylase inhibitors. J Neurochem. 2003;87:407–416. doi: 10.1046/j.1471-4159.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- 80.Iwata K, Tomita K, Sano H, et al. Trichostatin A, a histone deacetylase inhibitor, down-regulates interleukin-12 transcription in SV-40-transformed lung epithelial cells. Cell Immunol. 2002;218:26–33. doi: 10.1016/s0008-8749(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 81.Zampetaki A, Xiao Q, Zeng L, et al. TLR4 expression in mouse embryonic stem cells and in stem cell-derived vascular cells is regulated by epigenetic modifications. Biochem Biophys Res Commun. 2006;347:89–99. doi: 10.1016/j.bbrc.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 82.Sandor V, Bakke S, Robey RW, et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res. 2002;8:718–728. [PubMed] [Google Scholar]
- 83.O’Connor OA, Heaney ML, Schwartz L, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J Clin Oncol. 2006;24:166–173. doi: 10.1200/JCO.2005.01.9679. [DOI] [PubMed] [Google Scholar]
- 84.Brogdon JL, Xu Y, Szabo SJ, et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood. 2007;109:1123–1130. doi: 10.1182/blood-2006-04-019711. [DOI] [PubMed] [Google Scholar]
- 85.Moreira JM, Scheipers P, Sorensen P. The histone deacetylase inhibitor Trichostatin A modulates CD4+ T cell responses. BMC Cancer. 2003 doi: 10.1186/1471-2407-3-30. ( www.biomedcentral.com/1471–2407/3/30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kozlowska A, Jagodzinski PP. Effect of Trichostatin A on CD4 surface density in peripheral blood T cells. Folia Histochem Cytobiol. 2006;44:259–262. [PubMed] [Google Scholar]
- 87.Mishra N, Brown DR, Olorenshaw IM, Kammer GM. Trichostatin A reverses skewed expression of CD154, interleukin-10, and interferon-gamma gene and protein expression in lupus T cells. Proc Natl Acad Sci USA. 2001;98:2628–2633. doi: 10.1073/pnas.051507098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dangond F, Gullans SR. Differential expression of human histone deacetylase mRNAs in response to immune cell apoptosis induction by trichostatin A and butyrate. Biochem Biophys Res Commun. 1998;247:833–837. doi: 10.1006/bbrc.1998.8891. [DOI] [PubMed] [Google Scholar]
- 89.Reiner SL. Epigenetic control in the immune response. Hum Mol Genet. 2005;14(Spec 1):R41–R46. doi: 10.1093/hmg/ddi115. [DOI] [PubMed] [Google Scholar]
- 90.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 91.Greten FR, Karin M. The IKK/NF-kappaB activation pathway-a target for prevention and treatment of cancer. Cancer Lett. 2004;206:193–199. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 92.Graham B, Gibson SB. The two faces of NFkappaB in cell survival responses. Cell Cycle. 2005;4:1342–1345. doi: 10.4161/cc.4.10.2047. [DOI] [PubMed] [Google Scholar]
- 93.Berghe WV, Ndlovu MN, Hoya-Arias R, et al. Keeping up NF-κB appearances: Epigenetic control of immunity or inflammation-triggered epigenetics. Biochem Pharmacol. 2006;72:1114–1131. doi: 10.1016/j.bcp.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 94.Rahman MM, Kukita A, Kukita T, et al. Two histone deacetylase inhibitors, trichostatin A and sodium butyrate, suppress differentiation into osteoclasts but not into macrophages. Blood. 2003;101:3451–3459. doi: 10.1182/blood-2002-08-2622. [DOI] [PubMed] [Google Scholar]
- 95.Chakravorrty D, Koide N, Kato Y, et al. The inhibitory action of butyrate on lipopolysaccharide-induced nitric oxide production in RAW 264.7 murine macrophage cells. J Endotoxin Res. 2000;6:243–247. [PubMed] [Google Scholar]
- 96.Yu Z, Zhang W, Kone BC. Histone deacetylases augment cytokine induction of the iNOS gene. J Am Soc Nephrol. 2002;13:2009–2017. doi: 10.1097/01.asn.0000024253.59665.f1. [DOI] [PubMed] [Google Scholar]
- 97.Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J Bio Chem. 2001;276:44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]
- 98.Chen LF, Greene WC. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J Mol Med. 2003;81:549–557. doi: 10.1007/s00109-003-0469-0. [DOI] [PubMed] [Google Scholar]
- 99.Soloaga A, Thomson S, Wiggin GR, et al. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546:51–58. doi: 10.1016/s0014-5793(03)00451-4. [DOI] [PubMed] [Google Scholar]
- 101.Klampfer L, Huang J, Swaby L-A, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J Biol Chem. 2004;279:30358–30368. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 102.Carrozza MJ, Li B, Florens L, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 103.Sanderson L, Tylor GW, Aboayge EO, et al. Plasma pharmacokinetics and metabolism of the histone deacetylase inhibitor Trichostatin A after intraperitoneal administration to mice. Drug Metab Dispos. 2004;32:1132–1138. doi: 10.1124/dmd.104.000638. [DOI] [PubMed] [Google Scholar]
- 104.Zou W. Immunosuppressive networks in the tumor environment and their therapeutic relevance. Nature Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 105.Altieri SL, Khan ANH, Tomasi TB. Exosomes from plasmacytoma cells as a tumor vaccine. J Immunother. 2004;27:282–288. doi: 10.1097/00002371-200407000-00004. [DOI] [PubMed] [Google Scholar]
- 106.Chaput N, Taieb J, Schartz N, et al. The potential of exosomes in immunotherapy of cancer. Blood Cells Mol Dis. 2005;35:111–115. doi: 10.1016/j.bcmd.2005.05.009. [DOI] [PubMed] [Google Scholar]