Abstract
The development of lupus pathogenesis results from the integration of susceptibility and resistance genes. We have used a chronic graft-versus-host disease (cGVHD) model to characterize a suppressive locus at the telomeric end of the NZM2410-derived Sle2 susceptibility locus, which we named Sle2c2. cGVHD is induced normally in Sle2c2-expressing mice, but it is not sustained. The analysis of mixed bone marrow chimeras revealed that cGVHD resistance was eliminated by non-B non-T hematopoietic cells expressing the B6 allele, suggesting that resistance is mediated by this same cell type. Furthermore, Sle2c2 expression was associated with an increased number and activation of the CD11b+ GR-1+ subset of granulocytes before and in the early stage of cGVHD induction. We have mapped the Sle2c2 critical interval to a 6-Mb region that contains the Cfs3r gene, which encodes for the G-CSFR, and its NZM2410 allele carries a nonsynonymous mutation. The G-CSFR–G-CSF pathway has been previously implicated in the regulation of GVHD, and our functional data on Sle2c2 suppression suggest a novel regulation of T cell-induced systemic autoimmunity through myeloid-derived suppressor cells. The validation of Csf3r as the causative gene for Sle2c2 and the further characterization of the Sle2c2 MDSCs promise to unveil new mechanisms by which lupus pathogenesis is regulated.
Susceptibility to complex diseases such as systemic lupus erythematosus (SLE) results from the integration between susceptibility and resistance genes tipping in favor of susceptibility genes. Although most of the efforts have been devoted to the identification of susceptibility genes, the characterization of suppressor genes will arguably have a significant impact on our understanding of the disease process and holds substantial translational potentials. The NZM2410 mouse model of lupus is a recombinant inbred strain between the New Zealand Black (NZB) and New Zealand White (NZW) strains in which we have identified three strong susceptibility loci, Sle1, Sle2, and Sle3 (1). The expression of NZW-derived Sle1, which induces the loss of tolerance to chromatin (2), is necessary for disease induction (3). Linkage analysis has identified four Sle1 suppressor loci in the NZW strain, the strongest of which, H-2-linked Sles1, was sufficient to entirely suppress Sle1 induction of disease (4). Sles1 has now been mapped to a <1-Mb segment containing the class II MHC genes (5), and the identification of the corresponding gene promises to reveal a novel pathway by which tolerance to nuclear Ags is maintained. The three other Sle1-suppressor loci, Sles2, Sles3, and Sles4 on chromosomes 4, 3, and 9, respectively, have not been further characterized. Other lupus suppressor loci have been identified in other strains by linkage analysis, including one that overlaps with Sles3 in the BXSB/long-lived (6), MRL/lpr (7), and 129 (8) strains. In addition, a mutation in the Coronin 1A gene in a C57BL/6 substrain was shown to suppress disease in the MRL/lpr model (9).
Sle2 expression induces a number of B cell defects, including an expansion of the B1a cell compartment and an increased production of polyreactive IgM Abs (10). Using congenic recombinants, we have determined that the major contribution to the expansion of B1a cells mapped to the telomeric sublocus Sle2c (11), which corresponds to the only NZB portion of the NZM2410 Sle susceptibility loci. Polycongenic studies have shown that Sle2 expression increased fatal nephritis incidence from 41% in B6. Sle1.Sle3 to 98% in B6.Sle1.Sle2.Sle3 mice (3). Paradoxically, Sle2c expression decreased disease incidence in B6.Sle1.Sle3 mice (11). Fine mapping of the B1a cell expansion reduced the size of the Sle2c locus to a ~13-Mb region named Sle2c1 (11), and we have recently shown that Sle2c1 greatly enhanced the incidence and severity of lupus pathology when expressed on a Fas-deficient background (12). Interestingly, the disease severity was significantly more severe in B6.Sle2c1.lpr than in B6.Sle2.lpr mice. Overall, these data suggested the existence of a suppressor locus at the telomeric end of Sle2.
A chronic graft-versus-host disease (bm12-cGVHD) model, in which B6.C-H2bm12 lymphocytes are transferred into nonautoimmune H-2b B6 hosts, has been extensively used as an induced model of lupus (13). Within 3 wk after transfer, mice develop lupus-like phenotypes, including lymphocyte activation and antinuclear autoantibody production, which are dependent on cognate interactions between donor CD4+ T cells and host autoreactive B cells (14). cGVHD induction also requires host CD4+ T cells to nurture B cells through their development in a process that involves IL-4 and CD40L (15). We have shown that both Sle1c and Sle1a lupus-susceptibility loci enhanced lymphocyte activation and autoantibody production in the bm12-cGVHD model, indicating that these loci increase the frequency of autoreactive B cells that can be activated by cognate T cell help (16, 17). In this study, we have used the bm12-cGVHD model to demonstrate the presence of a suppressive locus at the telomeric end of Sle2, which we named Sle2c2. We showed that cGVHD induction initiates normally in Sle2c2-expressing mice, but it is not sustained. The analysis of mixed bone marrow (BM) chimeras revealed that non-B non-T hematopoietic cells from B6 origin were sufficient to break Sle2c2 cGVHD resistance. Furthermore, Sle2c2 expression is associated with an increased number and activation of the CD11b+ GR-1+ subset of granulocytes before and in the early stage of cGVHD induction. The Cfs3r gene, which encodes for G-CSFR, is located in the Sle2c2 interval, and its NZB allele carries a nonsynonymous mutation. G-CSFR has been shown recently to play a key role in regulating GVHD responses (18), and CD11b+ GR-1+ myeloid-derived suppressor cells (MDSCs) are potent suppressors of alloreactive T cell responses in a GVHD model (19). Overall, our results demonstrate that the Sle2c locus contains a potent lupus susceptibility gene, Sle2c1 (12), and a potent lupus suppressor gene (Sle2c2, this study) with ~15 Mb from each other. Our results suggest that Sle2c2 suppresses T cell-induced autoimmune responses through myeloid cells, and that Cfs3r is mediating this suppression through MDSCs. These suppressor cells have been largely associated with tumor and inflammation settings (20). Our results suggest that the Sle2c2 locus may serve as a model to investigate a novel involvement of MDSCs in suppressing T cell-mediated breach in tolerance to nuclear Ags.
Materials and Methods
Mice
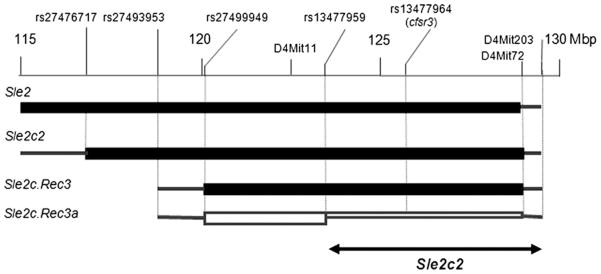
The Sle2c congenic interval located between D4Mit278 and D4Mit72 has been divided into the Sle2c1 and Sle2c2 intervals located between D4Mit278 and D4Mit37, and between D4Mit11 and D4Mit72, respectively, as shown in Fig. 2 (11). C57BL/6 (B6), B6.C-H2bm12/KhEg (bm12), B6.Rag1−/−, B6.Tcrβ−/−.Tcrδ−/− (B6.Tcr−/−), and B6.IgH6 were originally purchased from Jackson ImmunoResearch Laboratories. The B6. Sle2c.Rec3 recombinant strain was generated from B6.Sle2c, and the B6. Sle2c.Rec3a recombinant was generated from B6.Sle2c.Rec3. Fine mapping was performed by direct sequencing using single nucleotide polymorphism (SNP) markers that were polymorphic between B6 and NZB. All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Florida.
FIGURE 2.
Genetic map of the Sle2c2 locus. The Sle2 region located between 115 and 130 Mb is shown, as well as the three Sle2c congenic intervals used in this study. The NZM2410 (NZB)-derived homozygous intervals are shown by black boxes flanked on each side by a line indicating the area of recombination with the B6 genome in between the indicated markers. The B6. Sle2c.Rec3a interval is defined by a recombination event that occurred on one chromosome between rs13477959 and rs13477964. The region between rs13477964 and D4Mit72 is heterozygous in that strain. The filled rectangles indicate the strains that showed cGVHD resistance, and the open rectangle indicates the strain that showed a B6-like cGVHD. This mapped the Sle2c2 critical interval between rs13477959 and D4Mit203.
cGVHD induction
bm12-cGVHD was induced according to an established protocol (14) Briefly, 50–80 × 106 bm12 splenocytes were injected i.p. into 2- to 4-mo-old mice. In some experiments, 2 × 107 magnetic bead-purified CFSE-labeled bm12 CD4+ T cells were injected i.v. into B6 or B6.Sle2c2 mice. Groups of at least five recipient mice were sacrificed 2, 7, and 21 d after transfer, when splenocytes were assessed by flow cytometry and serum autoantibodies by ELISA, as previously described (17).
The T cell-induced GVHD in BM-reconstituted lethally irradiated hosts (BM-GVHD) was induced according to a previously published protocol (21). Briefly, lethally irradiated (1000 rad) B6 and B6.Sle2c2 mice (10 mice per strain) were reconstituted by i.v. injection with 107 BM cells combined with 107 purified splenic T cells (mouse pan-T Dynabeads; Invitrogen) from BALB/c mice. Survival and skin clinical scores were monitored for 50 d following induction. Skin involvement was scored on a 0–4 scale, as previously described (22).
The parent-into-F1 (PF1) model of acute GVHD (aGVHD) was induced according to a previously published protocol (23). Briefly, 5 × 107 splenocytes from either B6 or B6.Sle2c2 mice were i.p. injected into (B6 × DBA/2)F1 mice (BDF1). The BDF1 recipients were sacrificed 2 wk later and analyzed for spleen lymphocyte numbers and activation.
Flow cytometry
Briefly, cells were first blocked with anti-CD16/CD32 (2.4G2), and then stained with pretitrated amounts of the following FITC-, PE-, allophycocyanin-, or biotin-conjugated Abs: CD4 (RM4-5), CD69 (H1.2F3), CD44 (IM7), CD62L (MEL-14) B220 (RA3-6B2), CD86 (GL1), CD80 (16-10A1), CD22.2 (Cy34.1), I-ab (AF6-120.1), CD11b (M1/70), CD11c (HL3), L6C/G (GR-1/RB6-8C5), or isotype controls, all from BD Biosciences (San Diego, CA). Biotin-conjugated Abs were revealed by streptavidin-PercP-Cy5a. Cell staining was analyzed using a FACSCalibur (BD Biosciences). At least 30,000 events were acquired per sample, and dead cells were excluded based on scatter characteristics.
Dendritic cell phenotypes and MLR
The number and activation phenotypes of CD11b+CD11c+ dendritic cells (DCs) were evaluated in the spleen of 10-wk-old mice after collagenase digestion, as previously described (24). Splenic DCs and CD4+ T cells were isolated by positive and negative selection, respectively, according to the manufacturer’s instructions (Miltenyi Biotec). The purity averaged 75% for DCs and 91% for CD4+ T cells. B6 or B6.Sle2c2 DCs were mixed with bm12 CD4+ T cells (106 cells/ml/well) at either 1:5 or 1:10 ratio, and cultured in RPMI 1640 medium supplemented with 10% FBS for 72 h. Proliferation in the last 24 h was measured by the addition of 10 μM BrdU.
BM chimeras
BM chimeras were performed, as previously described (25). Briefly, irradiated (1000 rad) B6.Sle2c2 recipient mice received 107 T cell-depleted BM cells from each of two donor strains, as shown in Table I (2 × 107 cells in group 2 with a single donor strain). cGVHD was induced in the chimeras with 7 × 107 splenocytes 2 mo after reconstitution, and the mice were analyzed 3 wk later.
Table I.
Mixed BM chimera groups used in this study showing the BM donor partner and the resulting origin of T, B, and myeloid cells for each group
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | |
|---|---|---|---|---|---|
| BM donor 1 | B6 | B6.Rag1−/− | B6.Tcr−/− | B6.IgH6 | |
| BM donor 2 | Sle2c2 | Sle2c2 | Sle2c2 | Sle2c2 | Sle2c2 |
| T cells | Mixed | Sle2c2 | Sle2c2 | Sle2c2 | Mixed |
| B cells | Mixed | Sle2c2 | Sle2c2 | Mixed | Sle2c2 |
| Myeloid cells | Mixed | Sle2c2 | Mixed | Mixed | Mixed |
Statistics
Statistical analyses were performed with GraphPad Prism4, using two-tailed unpaired t tests or Bonferroni’s multiple comparison tests when several groups were compared. Nonparametric statitistics were also performed and yielded the same results. Graphs show means and SEMs, and statistical significance is represented as *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
The telomeric region of Sle2 contains a suppressive locus to alloreactive T cell-induced cGVHD
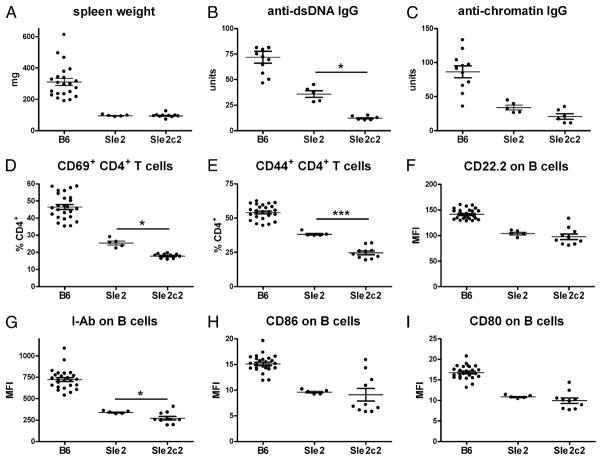
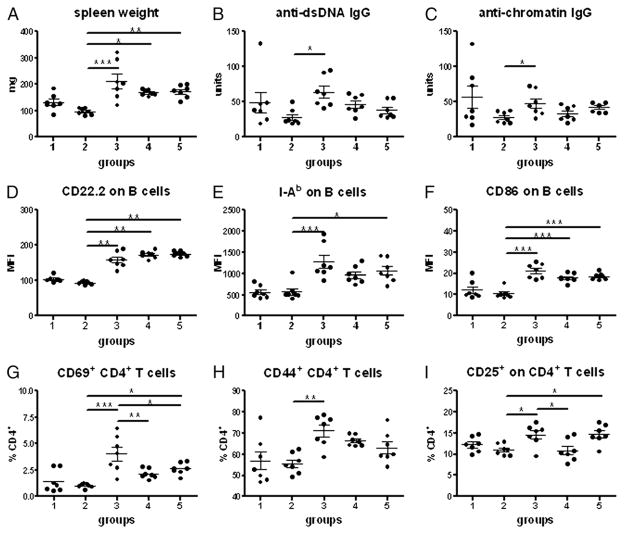
When bm12-cGVHD was compared between B6.Sle2 and B6 mice, we found unexpectedly that all cGVHD phenotypes were significantly lower in B6.Sle2 than in B6 mice 3 wk after bm12 splenocyte transfers. This included splenic expansion (Fig. 1A), serum anti-dsDNA, and antichromatin IgG (Fig. 1B, 1C), CD4+ T cell (Fig. 1D, 1E), and B cell (Fig. 1F–I) activation. Previous studies with Sle2 subcongenic intervals have suggested the existence of a suppressor locus in the telomeric portion of Sle2 (11, 12). Accordingly, we assessed the two telomeric B6.Sle2c subcongenic strains, B6.Sle2c1 and B6.Sle2c2, for cGVHD responses. The most centromeric Sle2c1 interval that is associated with B1a cell expansion (11) produced cGVHD responses identical to B6 (data not shown). In contrast, B6.Sle2c2 mice produced cGVHD responses that were significantly lower than B6 for all phenotypes, and even significantly lower than B6.Sle2 for autoantibody production (Fig. 1B, 1C), CD4+ T cell activation (Fig. 1D, 1E), and I-Ab expression on B cells (Fig. 1G). These data showed that the NZB-derived Sle2c2 locus (Fig. 2) prevents lymphocyte activation and autoantibody production induced by alloreactive T cells.
FIGURE 1.
B6.Sle2 and B6.Sle2c2 mice are resistant to bm12-cGVHD induction. A, Spleen weight. Anti-dsDNA (B) and antichromatin (C) IgG. Percentage of CD69+ (D)- and CD44+ (E)-expressing CD4+ T cells. CD22.2 (F), I-Ab (G), CD86 (H), and CD80 (I) mean fluorescence intensity (MFI) on B220+ B cells. Measurements were performed 3 wk after cGVHD induction. The graphs show means and SEMs. All phenotypes were significantly different between B6 and either B6.Sle2 or B6.Sle2c2. p < 0.001. Differences between B6.Sle2 and B6.Sle2c2 mice are indicated when significant. *p < 0.05, ***p < 0.001.
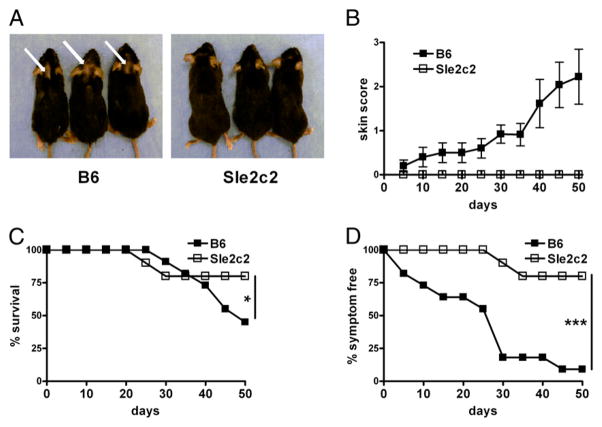
We investigated whether the suppressive activity of Sle2c2 was restricted to the bm12-cGVHD model by adapting another GVHD model (BM-cGVHD) that was induced by allotypic BALB/c T cells introduced into lethally irradiated mice reconstituted with BALB/c BM (21). As expected, B6 developed alopecia, but none of the B6.Sle2c2 mice did (Fig. 3A, 3B). Furthermore, survival and the overall percentage of symptom-free mice were significantly lower in B6 than in B6.Sle2c2 mice (Fig. 3C, 3D). Overall, these results demonstrate that Sle2c2 strongly suppresses cGVHD induced by alloreactive T cells.
FIGURE 3.
The B6.Sle2c2 strain is resistant to BM-cGVHD induction. A, Representative B6 (left) and B6.Sle2c2 mice 20 d after receiving BALB/c BM and T cells. The arrows point to alopecia-affected areas. B, Progression of the skin semiquantitative clinical scores following induction. Percentage of survival (C) and percentage of symptom-free mice combining skin involvement and survival (D). The graphs show means and SEMs. The statistical significance shown in graphs C and D corresponds to χ2 tests performed on values at the last day of observation. *p < 0.05, ***p < 0.001.
cGVHD resistance occurs late in the response of B6.Sle2c2 mice
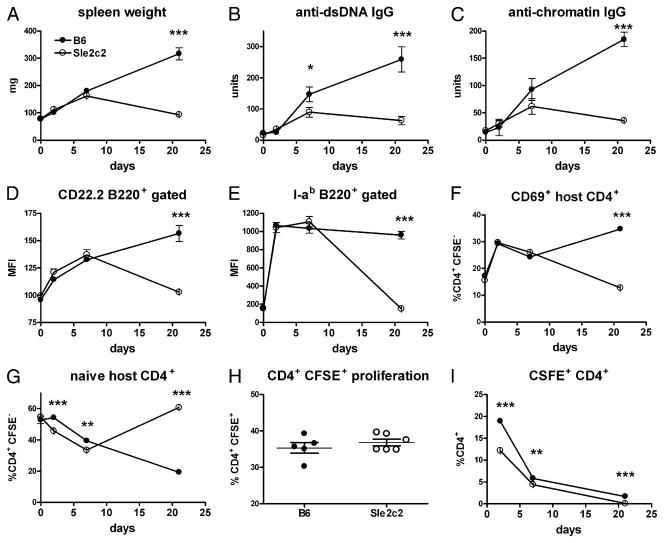
To better dissect the mechanisms of Sle2c2 resistance, cGVHD was induced with purified CFSE-labeled bm12 CD4+ T cells. As shown in Fig. 4A–G, bm12-CD4+ T cells induced the expected cGVHD phenotypes 3 wk after transfer in B6 mice, but not in B6. Sle2c2 mice, with differences similar to what was observed with transfer of whole splenocytes. This indicated that Sle2c2 suppresses CD4+ T cell alloreactivity or its consequences, without the requirement of other bm12 cell types.
FIGURE 4.
cGVHD resistance occurs late in the response of B6.Sle2c2 mice. Phenotypes were analyzed in cohorts of five mice sacrificed 2, 7, and 21 d after cGVHD induction with purified CFSE-labeled bm12 CD4+ T cells and compared with age-matched unmanipulated mice. A, Spleen weight. B, Serum anti-dsDNA IgG. C, Serum antichromatin IgG. Activation markers CD22.2 (D) and I-ab (E) MFI on B cells. CSFEnegative host CD4+ T cell activation as measured by CD69 (F) and naive markers CD62L+CD44− (G) expression. H, Percentage of CFSE+ bm12 CD4+ T cells that have proliferated at day 2. I, CFSE+ bm12 CD4+ T cells expressed as the percentage of total CD4+ T cells in host mice. The graphs show means and SEMs. **p < 0.01, ***p < 0.001.
To determine whether cGVHD induction fails in B6.Sle2c2 mice, or alternatively, if it initiates normally, but is not sustained, we assessed the time course of the bm12-cGVHD response. Cohorts of five mice per strain were evaluated 2, 7, and 21 d after transfer of bm12-purified CD4+ T cells. No difference was observed for any of the cGVHD phenotypes in unmanipulated age-matched mice (Fig. 4). Spleen weight significantly increased in both strains by day 2 (p < 0.001) and continued to increase in B6, but dipped to pre-cGVHD weight by day 21 in B6.Sle2c2 mice (Fig. 4A). Anti-dsDNA and antichromatin IgG were produced by both strains at day 7, although to a lower level in B6.Sle2c2 mice, but continued to increase at day 21 only in B6 mice (Fig. 4B, 4C). Activation of recipient B cells, indicated by CD22.2 and I-ab upregulation, increased similarly in both strains at days 2 and 7, but diverged significantly by day 21 to pre-cGVHD levels in B6. Sle2c2 mice (Fig. 4D, 4E). The same results were obtained for CD86 (data not shown). The activation of host CD4+ T cells followed the same pattern, with significantly lower levels of CD69 expression in B6.Sle2c2 mice only at day 21 (Fig. 4F). The percentage of CD62L+CD44− naive recipient CD4+ T cells decreased progressively in both strains from day 2 to 7, although to significantly lower levels in B6.Sle2c2 mice, and then reverted in that strain to a level that was even higher than in unmanipulated mice by day 21 (Fig. 4G). The proliferation of bm12 CD4+ T cells was similar in both hosts at d 2 (Fig. 4H). However, the percentage of the bm12 CFSE+ CD4+ T cells was significantly lower in B6. Sle2c2 than in B6 recipients (Fig. 4I). This indicated that allorecognition occurred to a similar extent in both host strains, but alloreactive T cells represented a smaller proportion of the total T cells in B6.Sle2c2 mice, either due to their decreased survival or, conversely, to a proportionally greater expansion of B6 host CD4+ T cells.
To confirm that alloreactive T cell responses were intact in B6. Sle2c2 mice, we used the PF1 model of aGVHD. In this model, naive parental strain T cells transferred into F1 mice differentiate into antihost CTL specific for host MHC class I that eliminate host lymphocytes, particularly splenic B cells (23). Splenocytes from either B6 or B6.Sle2c2 mice induced similar aGVHD in BDF1 mice, as measured 2 wk after transfer by an elevated number of T cells, an elimination of the B cells, and an increased CD4+ and B cell activation (Supplemental Fig. 1). These results indicate that the alloreactive CTL response is intact in B6.Sle2c2 mice. Overall, these results showed that B6 and B6.Sle2c2 mice have identical lymphocyte activation patterns before cGVHD induction, and that bm12 CD4+ T cells trigger alloreactive responses in both strains, which are not sustained in B6.Sle2c2 mice.
cGVHD resistance requires Sle2c2 expression in non-B non-T BM-derived cells
To determine which cell type is responsible for Sle2c2 cGVHD resistance, we produced mixed BM chimeras in which the expression of the B6 and Sle2c2 alleles was controlled in B cells, T cells, or non-B non-T BM-derived cells (Table I). Groups 1 and 2 were chimera controls reconstituted with BM-derived cells from either B6 and B6.Sle2c2, or B6.Sle2c2 mice only, respectively. In group 3 B6 alleles were expressed only in non-B non-T BM-derived cells, whereas in group 4 B6 alleles were expressed in B cells and in non-B non-T BM-derived cells, and in group 5 B6 alleles were expressed in T cells and in non-B non-T BM-derived cells. bm12-cGVHD was induced in these chimeras 2 mo after BM reconstitution, and phenotypes were examined 3 wk later. As shown in Fig. 5, the expression of B6 alleles in non-B non-T BM-derived cells was sufficient to break the resistance to cGVHD and significantly increase all phenotypes examined as compared with chimeras controls in which all BM-derived cells express only the Sle2c2 alleles (compare groups 2 and 3). The expression of B6 alleles in B or T cells did not amplify the cGVHD phenotypes conferred by the expression of B6 alleles in only non-B non-T BM-derived cells. Indeed, the amplitude of the cGVDH response in either group 4 or 5 did not reach that of group 3, and T cell activation was actually significantly lower in group 4 than 3 (Fig. 5G, 5I). Our data even suggest that the B6 allele of Sle2c2 expressed in either B or T cells may have a weak protective effect, that is stronger when expressed in both B and T cells, which would explain the weak cGVHD phenotypes obtained in group 1 mice (Fig. 5). This experiment demonstrates that expression of the NZB allele of Sle2c2 in non-B non-T BM-derived cells, which are largely myeloid cells, controls cGVHD resistance in B6.Sle2c2 mice.
FIGURE 5.
cGVHD resistance is maintained by non-B non-T BM-derived cells. cGVHD was induced by bm12 splenocytes transferred into five groups of mixed BM chimeras described in Table I (1, B6 + Sle2c2; 2, Sle2c2; 3, B6.Rag−/− + Sle2c2; 4, B6.Tcr−/− + Sle2c2; 5, B6.Igh6 + Sle2c2). Phenotypes were analyzed 3 wk later. A, Spleen weight. B, Serum anti-dsDNA IgG. C, Serum antichromatin IgG. Surface activation marker expression on splenic B cells (D–F) and CD4+ T cells (G–I). The graphs show means and SEMs. *p < 0.05, **p < 0.01, ***p < 0.001.
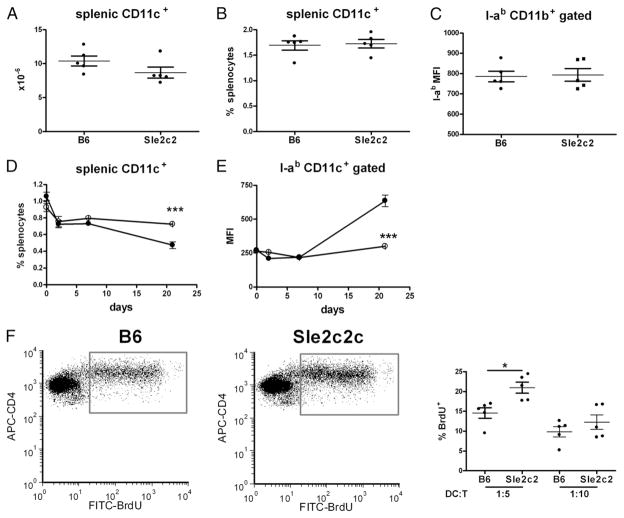
To evaluate whether among the non-B non-T cells, dendritic cells (DCs) mediated the bm12-cGVHD resistance, we first determined their number and activation after cGVHD induction with whole splenocytes. There was no difference between the number, percentage, or activation of B6 and B6.Sle2c2 DCs 3 wk after induction (Fig. 6A–C). Because donor DCs could not be distinguished from host DCs in that experiment, we evaluated the DC compartment following induction with purified CD4+ T cells. In both B6 and B6.Sle2c2 mice, the percentage of splenic CD11c+ CD11b+ DCs initially decreased, but whereas it continued decreasing in B6 mice, it stabilized in B6.Sle2c2 mice (Fig. 6D). In contrast, DC activation levels remained fairly constant in B6. Sle2c2 mice, whereas they increased in B6 mice, resulting in a significant difference 21 d after induction (Fig. 6E). Similar results were obtained with CD11b+CD11c− GR-1− macrophages (data not shown). B6 or B6.Sle2c2 DC function was evaluated by their ability to induce bm12 CD4+ T cells to proliferate in vitro (Fig. 6F). At a 1:5 DC:T cell ratio, bm12 T cells proliferated significantly more in response to B6.Sle2c2 than to B6 DCs, whereas there was no difference at a 1:10 ratio. These results suggest that DC hyporesponsiveness is unlikely to be responsible for cGVHD resistance in B6.Sle2c2 mice.
FIGURE 6.
cGVHD resistance in B6.Sle2c2 mice is not due to DC hyporesponsivess to alloreactive T cells. Number (A), percentage (B), and I-ab expression (C) of CD11c+CD11b+ splenocytes 3 wk after cGVHD induction with bm12 splenocytes. Time course of the percentage of CD11c+CD11b+ splenocytes (D) and their I-ab expression (E) after transfer of bm12 CD4+ T cells. F, bm12 CD4+ T cell proliferation induced by either B6 or B6.Sle2c2 DCs in 72-h cocultures at a 1:5 and 1:10 DC:T cell ratio. Representative FACS plot of CD4+ gated BrdU+ cells is shown on the left. The graphs show means and SEMs. *p < 0.05, ***p < 0.001.
cGVHD resistance maps to a 6-Mb interval on the telomeric end of Sle2
We have generated two recombinants within the Sle2c2 interval to further map the bm12-cGVHD resistance (Fig. 2). The Sle2c.Rec3 interval resulting from a centromeric recombination event between rs27493953 and rs27499949 was bred to homozygozity. Sle2c.Rec3a was obtained from a recombination event on one chromosome of Sle2c.Rec3 between rs13477959 and rs13477964. It was therefore NZB/B6 heterozygous between rs13477964 and D4Mit72 and centromeric to rs13477964 up to rs13477959.
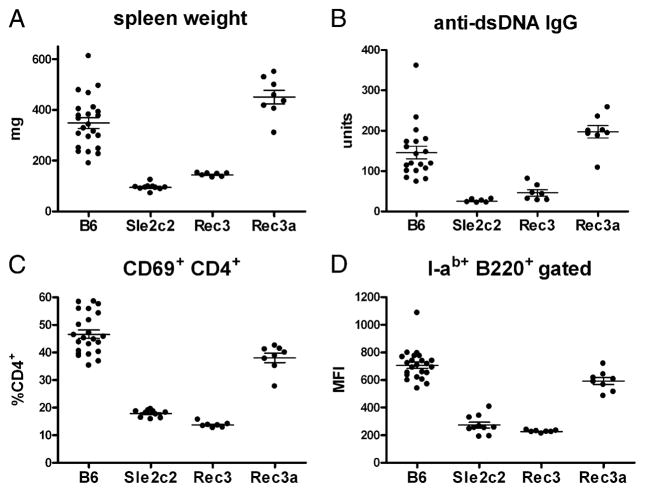
bm12-cGVHD was induced in B6.Sle2c.Rec3 and B6.Sle2c. Rec3a mice comparatively to B6 and B6.Sle2c2 controls. As shown in Fig. 7, the results were unambiguous with the B6.Sle2. Rec3 mice failing to show splenic expansion (Fig. 7A), produce anti-dsDNA IgG (Fig. 7B), or upregulate CD69 on CD4+ T cells and I-ab on B cells (Fig. 7C, 7D). The same results were obtained for CD44 expression on CD4+ T cells; CD22.2, CD80, and CD86 expression on B cells; and antichromatin IgG (data not shown). This located the Sle2c2 critical interval containing the gene responsible for cGVHD resistance between rs13477959 and D4Mit72. This interval is gene rich with 106 RefSeq genes. Among these genes, Csf3r encodes for the G-CSFR and rs13477964 corresponds to a nonsynonymous G > A SNP that results in a Ser379 Asn amino acid change in exon 10. A recent study has shown that the expression level of the G-CSFR regulates GVHD in mice (18). None of the other genes has a known function related to either autoimmunity of GVHD, making Csf3r the top candidate gene for Sle2c2. Interestingly, the Csf3r gene region corresponds to a ~30-Kb haplotype block of identity between B6 and NZW (including rs13477964), but contains 16 polymorphic SNPs between B6 and NZB (http://phenome.jax.org/SNP/). We found that the level of Csf3r transcripts was similar between B6 and B6.Sle2c2 spleens (data not shown), which corroborates the absence of published polymorphisms between NZB and B6 in the promoter region.
FIGURE 7.
cGVHD resistance maps to the ~6 Mb at the telomeric end of the Sle2c2 interval. cGVHD was induced in the B6.Sle2c.Rec3 and B6.Sle2c.Rec3a strains with B6 and B6.Sle2c2 as negative and positive controls, respectively. Spleen weight (A), serum anti-dsDNA IgG production (B), CD69 expression on CD4+ T cells (C), and I-Ab MFI on B220+ B cells (D) 3 wk after induction. For each phenotype, Bonferroni’s multiple comparison tests showed a significant difference between B6.Sle2c2 and either B6 or B6.Sle2c. Rec3a, but not with B6.Sle2c.Rec3 values. p < 0.01. The graphs show means and SEMs.
Amino acid 379 of the G-CSFR is not conserved between primates and rodents (Supplemental Fig. 2A) and is located outside of the solved structure in both the human (Brookhaven Protein Data Bank [PDB]: 2D9Q-B) and mouse (PDB: 1CD9-B) proteins (26). According to the Conserved Domain Database (27), the I337 to E429 region of G-CSFR contains a putative fibronectin III domain (FNIII; Supplemental Fig. 2B), which is a structure found in ~2% of all animal extracellular and intracellular proteins, including membrane-spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules (28). When blasted against the primary protein sequences available in the PDB, the G-CSFR’s FNIII domain showed 30% sequence similarity to the FNIII domain of the IL-6R gp130 (PDB: 3L5H). To investigate the potential role of the Ser379Asn in the function of the G-CSF3R, we constructed a homology model of the receptor using the SWISS-MODEL web service (29), which suggested that aa 379 is exposed to the solvent and may interact with the receptor ligand, other parts of the G-CSFR, or other surface proteins (Supplemental Fig. 2C). An in silico glycosylation analysis performed with NetNGlyc 1.0 (www.cbs.dtu.dk/services/NetNGlyc) predicted Asn379 is not glycosylated; however, the homology model showed that it is in close proximity to a highly probable glycosylation site at aa 408 (Supplemental Fig. 2B). Overall, these data do not predict an obvious functional change associated with this nonsynonymous mutation, but do not exclude that rs13477964 or other yet unidentified polymorphisms in this gene are responsible for the NZB allele cGVHD resistance.
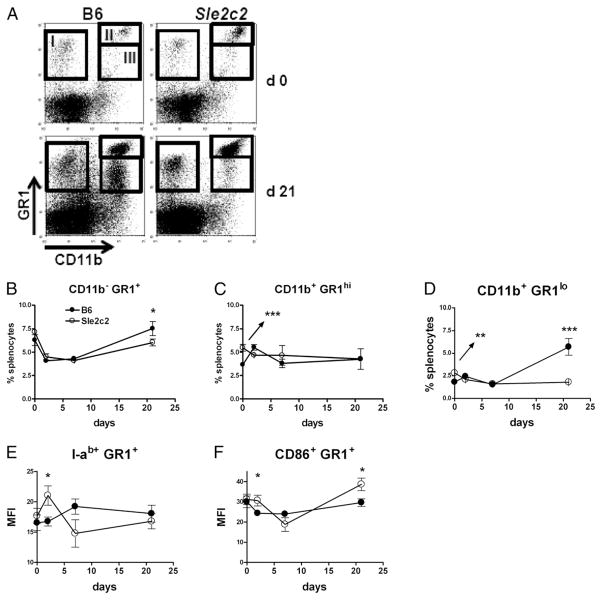
G-CSF is a growth factor for GR-1+ (Ly6G/C) granulocytes, and a recent study has shown that CD11b+ GR-1+ MDSCs suppressed GVHD (19). We therefore compared the status of the GR-1+ cell populations (Fig. 8A) relative to cGVHD induction between B6 and B6.Sle2c2 mice. The CD11b− GR-1+ cells followed a similar pattern between the two strains, initially decreasing and going back to pre-cGVHD levels by day 21 after induction in B6, but not in B6.Sle2c2 mice (Fig. 8B). The percentage of granulocytic CD11b+ GR1high MDSCs was significantly higher in B6.Sle2c2 than in B6 mice before cGVHD induction, but normalized to similar levels in both strains after induction (Fig. 8C). The percentage of monocytic CD11b+ GR1low MDSCs was also significantly higher in B6.Sle2c2 than in B6 mice before cGVHD induction, but dramatically increased in B6, but not in B6.Sle2c2 mice (Fig. 8D). Finally, we observed a higher level of activation, as indicated by I-ab and CD86 expression, in GR-1+ cells in B6. Sle2c2 mice, most notably at day 2 after cGVHD induction (Fig. 8E, 8F). Overall, these results show differences between B6 and B6.Sle2c2 GR1+ cells before or early in the cGVHD response.
FIGURE 8.
Differential response of GR-1+ cells to cGVHD in B6 and B6.Sle2c2 mice. A, Representative FACS plots showing the GR-1 and CD11b stains on B6 and B6.Sle2c2 splenocytes before and at day 21 after bm12 CD4+ T cell transfer. The three populations evaluated in the study are shown as gates I–III. Percentage of CD11b− GR-1+ (B, gate I), CD11b+ GR-1high (C, gate II), and CD11b+ GR-1low (D, gate III) splenocytes. I-ab (E) and CD86 (F) expression on GR-1+ splenocytes. Filled symbols represent B6 values, and open symbols represent B6.Sle2c2 values (mean and SEM of five mice per strain per time point). Statistical significance values are between the two strains at specific time points. *p < 0.05, ***p < 0.001.
Discussion
This study demonstrates the existence of a NZB-derived suppressor locus at the telomeric end of the Sle2 lupus susceptibility locus. We used an induced model of lupus to demonstrate the suppressive activity of Sle2c2. Our previous results involving this region crossed either to the Sle1/Sle3 combination (11) or to Fas deficiency (12) strongly suggest that this suppressor locus is also effective in spontaneous lupus. These results predict that the combination of Sle1/Sle2/Sle3 could be rendered more pathogenic by the elimination of Sle2c2, which we are currently testing. A close proximity of susceptibility and suppressive loci, such as the one that we have revealed for Sle2c1 and Sle2c2, has been previously described as gene masking in the NOD model of type I diabetes (30), and is likely to represent a frequent occurrence in the genetic architecture of complex traits. Other suppressive loci have been identified in various mouse models of lupus (4, 7, 8). With exception of the Coronin 1A gene, which affects the strength of the T cell synapse (9), and Sles1, which has been mapped to a small H-2–linked region, very little has been achieved toward the identification of the suppressor genes and the mechanisms by which they confer protection.
Sle2c2 suppressed not only the lupus-like phenotypes in the classic bm12-cGVHD model, but also in the fully allotypic T cell-induced BM-cGVDH. This indicates that Sle2c2 suppressed either the induction or maintenance of alloreactive T cell responses or their consequences. Our time course analysis of cGVHD responses and in vitro MLR results showed that alloreactive responses are initiated normally in Sle2c2-expressing mice, but that they are not maintained. Our BM-chimera experiments showed that the expression of B6 alleles in BM-derived nonlymphoid cells was sufficient to restore normal cGVHD responses, and suggest that Sle2c2 resistance is mediated by the same type of cells in which the B6 allele is dominant. To our knowledge, these results establish for the first time the involvement of nonlymphoid cell population in the bm12-cGVHD models. We have ruled out macrophages and DCs to be the most likely cell population responsible for Sle2c2 resistance first because their number and activation status is not affected by Sle2c2 expression before and in the early stage of cGVHD induction, and second because bm12 CD4+ T cells strongly respond to Sle2c2-expressing DCs. We propose that MDSCs are strong candidates for mediating Sle2c2 expression. In addition to the BM nonlymphoid origin of the cells in which Sle2c2 is functionally expressed, several lines of evidence support this hypothesis. First, MDSCs expanded in vitro with G-CSF or GM-CSF have potent immunosuppressive functions of alloreactive T cells and suppress GVHD in vivo (19). Second, we have found that contrary to lymphocytes, Sle2c2 expression was associated with an expanded population of MDSCs in unmanipulated mice and with a different response of this cell compartment to cGVDH. The greatest difference was found for monocytic GR-1low MDSCs, which have been recently associated with the greatest suppressive activity of the proliferation of allogenic CD8+ T cells in a tumor model (31) and syngeneic CD4+ T cells in the lupus-prone MRL/lpr mice (32). The respective roles of GR-1high and GR-1low MDSCs in Sle2c2-mediated cGVHD resistance will have to be addressed directly with adoptive transfers. Third, Csf3r, the gene encoding for the G-CSFR, maps to Sle2c2, and its NZB allele carries multiple known mutations, including a nonsynonymous mutation in the extracellular domain. The NZB genome is poorly characterized, and genomic sequencing of the NZB allele of Csf3r may reveal additional mutations. Altered G-CSFR function could affect either the size or the function of MDSCs. The potential role of MDSCs in autoimmunity is still largely unexplored. A recent study reported an increased percentage of MDSCs in the blood and kidneys of MRL/ lpr mice with disease progression (32). This corroborates our findings of an expanded GR-1low MDSC population in B6 mice with established cGVHD, and the hypothesis that inflammation drives the expansion of MDSCs (20). We hypothesize that Sle2c2 cGVHD resistance is mediated through the MDSC population existing at the initiation of the CD4+ T cell alloreactive response, and therefore, the increase seen in B6 mice with the cGVHD progression is a secondary effect of the inflammatory process.
Increased G-CSFR expression on DCs induced by total body irradiation exacerbated GVHD through an amplification loop involving IFN-γ production by NKT cells (18). In contrast, the anti-inflammatory properties of G-CSF in regulating T cell tolerance have now been well documented (33), with G-CSF treatment preventing or ameliorating several mouse models of immune-related diseases. Among these studies, it was shown that pre-treatment of B6 mice with G-CSF significantly reduced the severity of PF1 GVHD (34). Moreover, preventive treatment with high doses of G-CSF reduced autoantibody production and renal disease in MRL/lpr mice (35). Interestingly, low-dose treatment greatly accelerated the disease process in the same strain. Overall, these studies strongly support Csf3r as the prime candidate gene for Sle2c2, but cannot predict whether the NZB allele confers a gain or loss of function because an increased G-CSF/G-CSFR signal has been associated with both increased and decreased immune pathogenesis.
We have mapped Sles2, a NZW-derived suppressor of Sle1, to the same general region as Sle2c2 (4). The quantitative trait locus mapping of Sles2 was performed at a low resolution, and our work with the Sle1 locus (36) has shown that multiple loci can be closely linked. Therefore, we cannot predict at this time whether Sles2 and Sle2c2 are allelic. If Csf3r is the gene corresponding to Sle2c2, it is unlikely that it also corresponds to Sles2, as the region containing this gene is highly conserved between B6 and NZW. Our results with the BM chimeras and with the B6.Sle2c.Rec3a strain suggest that Sle2c2 suppression is recessive, as coexpression of the B6 and NZB alleles was sufficient to restore B6-like cGVHD. Because Sles2 is also recessive (4), complementation experiments should be able to determine whether these two suppressive loci correspond to the same gene.
In conclusion, we have identified a NZB-derived suppressor locus that prevents T cell-induced autoimmune and cGVHD manifestations. We have mapped this locus to a small region that contains the Csf3r gene, with a nonsynonymous mutation in the NZB allele, and shown that suppression is not mediated by either B or T cells. The fact that this gene has recently been implicated in the regulation of GVHD and our functional data on Sle2c2 suppression suggest a novel regulation of T cell-induced systemic autoimmunity through myeloid-derived suppressor cells. The validation of Csf3r as the causative gene for Sle2c2 and the further characterization of the Sle2c2 MDSCs promise to unveil new mechanisms by which lupus pathogenesis is regulated.
Supplementary Material
Acknowledgments
We thank Leilani Zeumer for excellent technical help and Xuekun Su for outstanding animal care.
This work was supported by National Institutes of Health Grants RO1 AI068965 (to L.M.) and K01AR056725 (to Z.X.).
Abbreviations used in this article
- aGVHD
acute graft-versus-host disease
- BM
bone marrow
- cGVHD
chronic graft-versus-host disease
- DC
dendritic cell
- FNIII
fibronectin III domain
- GVHD
graft-versus-host disease
- MDSC
myeloid-derived suppressor cell
- MFI
mean fluorescence intensity
- NZB
New Zealand Black
- NZW
New Zealand White
- PDB
Brookhaven Protein Data Bank
- PF1
parent-into-F1
- SLE
systemic lupus erythematosus
- SNP
single nucleotide polymorphism
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J, Wakeland EK. Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity. 1994;1:219–229. doi: 10.1016/1074-7613(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 2.Mohan C, Alas E, Morel L, Yang P, Wakeland EK. Genetic dissection of SLE pathogenesis: Sle1 on murine chromosome 1 leads to a selective loss of tolerance to H2A/H2B/DNA subnucleosomes. J Clin Invest. 1998;101:1362–1372. doi: 10.1172/JCI728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morel L, Croker BP, Blenman KR, Mohan C, Huang G, Gilkeson G, Wakeland EK. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci USA. 2000;97:6670–6675. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morel L, Tian XH, Croker BP, Wakeland EK. Epistatic modifiers of autoimmunity in a murine model of lupus nephritis. Immunity. 1999;11:131–139. doi: 10.1016/s1074-7613(00)80088-6. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian S, Yim YS, Liu K, Tus K, Zhou XJ, Wakeland EK. Epistatic suppression of systemic lupus erythematosus: fine mapping of Sles1 to less than 1 mb. J Immunol. 2005;175:1062–1072. doi: 10.4049/jimmunol.175.2.1062. [DOI] [PubMed] [Google Scholar]
- 6.Haywood MEK, Gabriel L, Rose SJ, Rogers NJ, Izui S, Morley BJ. BXSB/long-lived is a recombinant inbred strain containing powerful disease suppressor loci. J Immunol. 2007;179:2428–2434. doi: 10.4049/jimmunol.179.4.2428. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Nose M, Kamoto T, Nishimura M, Hiai H. Host modifier genes affect mouse autoimmunity induced by the lpr gene. Am J Pathol. 1997;151:1791–1798. [PMC free article] [PubMed] [Google Scholar]
- 8.Carlucci F, Fossati-Jimack L, Dumitriu IE, Heidari Y, Walport MJ, Szajna M, Baruah P, Garden OA, Cook HT, Botto M. Identification and characterization of a lupus suppressor 129 locus on chromosome 3. J Immunol. 2010;184:6256–6265. doi: 10.4049/jimmunol.0901463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haraldsson MK, Louis-Dit-Sully CA, Lawson BR, Sternik G, Santiago-Raber ML, Gascoigne NR, Theofilopoulos AN, Kono DH. The lupus-related Lmb3 locus contains a disease-suppressing Coronin-1A gene mutation. Immunity. 2008;28:40–51. doi: 10.1016/j.immuni.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohan C, Morel L, Yang P, Wakeland EK. Genetic dissection of systemic lupus erythematosus pathogenesis: Sle2 on murine chromosome 4 leads to B cell hyperactivity. J Immunol. 1997;159:454–465. [PubMed] [Google Scholar]
- 11.Xu Z, Duan B, Croker BP, Wakeland EK, Morel L. Genetic dissection of the murine lupus susceptibility locus Sle2: contributions to increased peritoneal B-1a cells and lupus nephritis map to different loci. J Immunol. 2005;175:936–943. doi: 10.4049/jimmunol.175.2.936. [DOI] [PubMed] [Google Scholar]
- 12.Xu Z, Cuda CM, Croker BP, Morel L. The NZM2410-derived lupus susceptibility locus Sle2c1 increases TH17 polarization and induces nephritis in Fas-deficient mice. Arthritis Rheum. 2010 doi: 10.1002/art.30146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenberg R. Mechanisms of systemic autoimmunity in murine models of SLE. Immunol Res. 1998;17:41–47. doi: 10.1007/BF02786429. [DOI] [PubMed] [Google Scholar]
- 14.Morris SC, Cheek RL, Cohen PL, Eisenberg RA. Autoantibodies in chronic graft versus host result from cognate T-B interactions. J Exp Med. 1990;171:503–517. doi: 10.1084/jem.171.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury A, Cohen PL, Eisenberg RA. B cells require “nurturing” by CD4 T cells during development in order to respond in chronic graft-versus-host model of systemic lupus erythematosus. Clin Immunol. 2010;136:105–115. doi: 10.1016/j.clim.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Cuda C, Morel L. Genetic determination of T cell help in loss of tolerance to nuclear antigens. J Immunol. 2005;174:7692–7702. doi: 10.4049/jimmunol.174.12.7692. [DOI] [PubMed] [Google Scholar]
- 17.Cuda CM, Zeumer L, Sobel ES, Croker BP, Morel L. Murine lupus susceptibility locus Sle1a requires the expression of two sub-loci to induce inflammatory T cells. Genes Immun. 2010;11:542–553. doi: 10.1038/gene.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris ES, MacDonald KPA, Kuns RD, Morris HM, Banovic T, Don ALJ, Rowe V, Wilson YA, Raffelt NC, Engwerda CR, et al. Induction of natural killer T cell-dependent alloreactivity by administration of granulocyte colony-stimulating factor after bone marrow transplantation. Nat Med. 2009;15:436–441. doi: 10.1038/nm.1948. [DOI] [PubMed] [Google Scholar]
- 19.Highfill SL, Rodriguez PC, Zhou Q, Goetz CA, Koehn BH, Veenstra R, Taylor PA, Panoskaltsis-Mortari A, Serody JS, Munn DH, et al. Bone marrow myeloid-derived suppressor cells (MDSC) inhibit graft-versus-host (GVHD) disease via an arginase-1 dependent mechanism that is upregulated by IL-13. Blood. 2010;116:5738–5747. doi: 10.1182/blood-2010-06-287839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen JL, Trenado A, Vasey D, Klatzmann D, Salomon BL. CD4(+)CD25(+) immunoregulatory T cells: new therapeutics for graft-versus-host disease. J Exp Med. 2002;196:401–406. doi: 10.1084/jem.20020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson BE, McNiff JM, Matte C, Athanasiadis I, Shlomchik WD, Shlomchik MJ. Recipient CD4+ T cells that survive irradiation regulate chronic graft-versus-host disease. Blood. 2004;104:1565–1573. doi: 10.1182/blood-2004-01-0328. [DOI] [PubMed] [Google Scholar]
- 23.Puliaev R, Nguyen P, Finkelman FD, Via CS. Differential requirement for IFN-gamma in CTL maturation in acute murine graft-versus-host disease. J Immunol. 2004;173:910–919. doi: 10.4049/jimmunol.173.2.910. [DOI] [PubMed] [Google Scholar]
- 24.Vremec D, O’Keeffe M, Hochrein H, Fuchsberger M, Caminschi I, Lahoud M, Shortman K. Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood. 2007;109:1165–1173. doi: 10.1182/blood-2006-05-015354. [DOI] [PubMed] [Google Scholar]
- 25.Sobel ES, Satoh M, Chen Y, Wakeland EK, Morel L. The major murine systemic lupus erythematosus susceptibility locus Sle1 results in abnormal functions of both B and T cells. J Immunol. 2002;169:2694–2700. doi: 10.4049/jimmunol.169.5.2694. [DOI] [PubMed] [Google Scholar]
- 26.Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, Tokunaga M, Kuroki R. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. Proc Natl Acad Sci USA. 2006;103:3135–3140. doi: 10.1073/pnas.0511264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol. 2006;359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 29.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37(Database issue):D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridgway WM, Peterson LB, Todd JA, Rainbow DB, Healy B, Burren OS, Wicker LS. Gene-gene interactions in the NOD mouse model of type 1 diabetes, Ch. 6. In: Emil RUaH., editor. Advances in Immunology Immunopathogenesis of Type 1 Diabetes Mellitus. Academic Press; New York, NY: 2008. p. 151. [DOI] [PubMed] [Google Scholar]
- 31.Dolcetti L, Peranzoni E, Ugel S, Marigo I, Fernandez Gomez A, Mesa C, Geilich M, Winkels G, Traggiai E, Casati A, et al. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. Eur J Immunol. 2010;40:22–35. doi: 10.1002/eji.200939903. [DOI] [PubMed] [Google Scholar]
- 32.Iwata Y, Furuichi K, Kitagawa K, Hara A, Okumura T, Kokubo S, Shimizu K, Sakai N, Sagara A, Kurokawa Y, et al. Involvement of CD11b+ GR-1low cells in autoimmune disorder in MRL-Faslpr mouse. Clin Exp Nephrol. 2010;14:411–417. doi: 10.1007/s10157-010-0309-9. [DOI] [PubMed] [Google Scholar]
- 33.Rutella S, Zavala F, Danese S, Kared H, Leone G. Granulocyte colony-stimulating factor: a novel mediator of T cell tolerance. J Immunol. 2005;175:7085–7091. doi: 10.4049/jimmunol.175.11.7085. [DOI] [PubMed] [Google Scholar]
- 34.Pan L, Delmonte JJ, Jr, Jalonen CK, Ferrara JL. Pretreatment of donor mice with granulocyte colony-stimulating factor polarizes donor T lymphocytes toward type-2 cytokine production and reduces severity of experimental graft-versus-host disease. Blood. 1995;86:4422–4429. [PubMed] [Google Scholar]
- 35.Zavala F, Masson A, Hadaya K, Ezine S, Schneider E, Babin O, Bach JF. Granulocyte-colony stimulating factor treatment of lupus autoimmune disease in MRL-lpr/lpr mice. J Immunol. 1999;163:5125–5132. [PubMed] [Google Scholar]
- 36.Morel L. Genetics of SLE: evidence from mouse models. Nat Rev Rheumatol. 2010;6:348–357. doi: 10.1038/nrrheum.2010.63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.