Abstract
Although Snail factors promote cell survival in development and cancer, the tumor-suppressor p53 promotes apoptosis in response to stress. p53 and Snail2 act antagonistically to regulate p53 upregulated modulator of apoptosis (Puma) and cell death in hematopoietic progenitors following DNA damage. Here, we show that this relationship is conserved in the developing nervous system in which Snail genes are excluded from vertebrate neurons and they are substituted by Scratch, a related but independent neural-specific factor. The transcription of scratch2 is induced directly by p53 after DNA damage to repress puma, thereby antagonizing p53-mediated apoptosis. In addition, we show that scratch2 is required for newly differentiated neurons to survive by maintaining Puma levels low during normal embryonic development in the absence of damage. scratch2 knockdown in zebrafish embryos leads to neuronal death through the activation of the intrinsic and extrinsic apoptotic pathways. To compensate for neuronal loss, the proliferation of neuronal precursors increases in scratch2-deficient embryos, reminiscent of the activation of progenitor/stem cell proliferation after damage-induced apoptosis. Our data indicate that the regulatory loop linking p53/Puma with Scratch is active in the vertebrate nervous system, not only controlling cell death in response to damage but also during normal embryonic development.
Keywords: scratch, p53, puma, spinal cord, neuronal death, compensatory proliferation
When activated by cellular stress, p53 acts as a crucial tumor suppressor, acting through a very intricate signaling network involving multiple positive and negative regulatory feedback loops.1 Indeed, its activity is modulated by its own transcriptional targets, p53 upregulated modulator of apoptosis (Puma) and murine double minute gene 2 (Mdm2), which transduce the apoptotic response or trigger p53 degradation, respectively.2 Accordingly, irreparable damage results in apoptosis, whereas mild stress may elicit a p53 survival response associated with DNA repair.3 Hence, the effects of p53 are defined by its affinity for response elements (REs) in different target genes, the cell context and the nature and duration of the stress signal.
Scratch and Snail are transcriptional repressors from independent gene families that originated by duplication of the ancestral snail gene.4 They are prominently expressed during embryonic development, occupying complementary territories in vertebrates. As such, Snail genes are expressed in mesodermal and mesenchymal cells,5 whereas Scratch family members are neural specific.6, 7 Although Snail factors have received much attention in cancer research and developmental biology,8 the functional analyses of scratch genes in vivo are scarce9, 10 and still missing in vertebrates.
Although Snail factors drive the conversion of epithelial cells to migratory cells,8 scratch genes are excluded from epithelial and mesenchymal cells, which makes them unlikely to participate in epithelial–mesenchymal transition. However, members of the Snail family11, 12 and the Caenorhabditis elegans Scratch homolog, cell death specification 1 (ces-1), have been implicated in conferring resistance to cell death.10 Expression of the C. elegans puma homolog, egl-1 (egg laying defective 1), is repressed by ces-1,13 and Puma is the main effector of p53-induced cell death in vertebrates.2 Indeed, Snail2 can antagonize p53-mediated apoptosis by repressing Puma in hematopoietic precursors following DNA damage.12
The only Snail-like sequence found in the C. elegans genome is thought to encode a non-functional protein.4 Hence, we wondered whether ces-1, the sole Scratch homolog, had co-opted the role of Snail in repressing Puma in the nematode. Alternatively, Puma repression might be an ancestral function associated to both snail and scratch genes before their divergence. Therefore, we investigated the role of Scratch in vertebrate cell survival using zebrafish, a suitable model to study cell death and p53 signaling.14, 15, 16 We characterized three zebrafish scratch genes that were specifically expressed in the nervous system and we found that scratch2 acts as a survival factor in neurons in vivo. We show that scratch2 is directly upregulated by p53 and that it represses puma transcription after DNA damage. More importantly, we show that scratch2 protects neurons from apoptosis by maintaining Puma levels low during normal embryonic development.
Results
Expression of zebrafish scratch genes
Two Scratch genes exist in vertebrates, Scratch1 and Scratch2, and although an extra genome duplication gave rise to additional genes in the teleost lineage,17 only two scratch1 (scratch1a and 1b) and one scratch2 genes make up the scratch family in zebrafish.4 We obtained the three full-coding regions of zebrafish scratch genes that encode Scratch proteins with strong similarity to those predicted from other teleosts (Supplementary Figure 1) and vertebrates (Supplementary Figure 2). The three sequences contain the diagnostic Scratch domains: five Scratch-type C2H2 zinc-fingers and the Scratch and Snail1/GFI domains.18
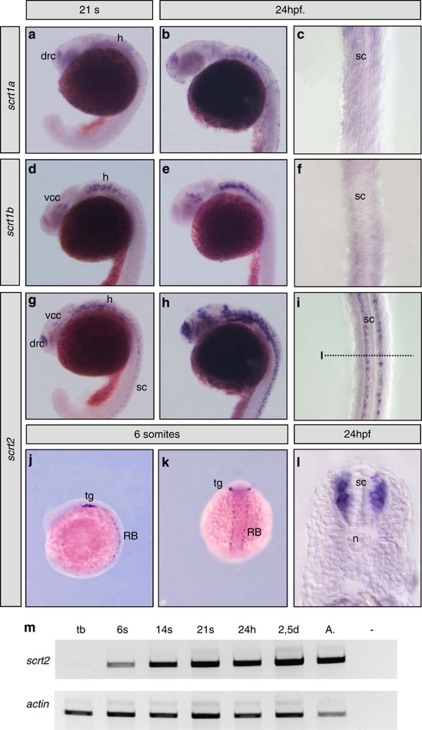
The scratch1 genes were first expressed at the 21 somite stage in the developing central nervous system of zebrafish embryos (Figures 1a and d). Although scratch1a transcripts were detected in the telencephalic dorsorostral cluster19 (drc) and hindbrain (h, Figure 1a), scratch1b was most strongly expressed in the hindbrain and diencephalic ventrocaudal cluster19 (vcc, Figure 1d). At 24 hours post-fertilization (h.p.f.), the expression of both genes was better defined in the same regions (Figures 1b and e). Similarly, scratch2 was expressed in both the drc and vcc, and in the hindbrain at the same stages, and at 24 h.p.f., it was also strongly expressed in the spinal cord (sc, Figures 1g–i), which was devoid of scratch1a and 1b transcripts (Figure 1c and f). scratch2 transcripts were detected in the spinal cord from the onset of its expression at the six-somite stage (Figure 1m), when it was also expressed in sensory Rohon-Beard neurons and in the placode of the trigeminal ganglion (tg, Figures 1j and k). However, this spinal cord expression was dynamic and transient, with no transcripts detected after 36 h.p.f. (data not shown). At 24 h.p.f., scratch2 transcripts were evident in the mantle layer of the spinal cord at different dorsoventral levels (Figure 1l) and not in the ventricular zone, indicating that it is expressed where postmitotic neurons are found. Hence, like the Drosophila,9 C. elegans10 and mouse scratch16, 7 and scratch27 homologs, the three zebrafish scratch genes are restricted to the nervous system, with scratch2 being expressed in the same territories as the two scratch1 genes, and in the spinal cord at the time of neuronal differentiation.
Figure 1.
Expression of scratch genes in zebrafish embryos. The expression of scratch1a, scracth1b and scratch2 was analyzed at different developmental stages. (a–c) scratch1a is first expressed in the telencephalon and the hindbrain of 21-somite embryos, and this pattern is better appreciated at 24 h.p.f. (d–f) The onset of scratch1b expression is similar to that of scratch1a, although the expression in the hindbrain is more prominent and transcripts can also be detected in the diencepahalon. (g–i) scratch2 is prominently expressed in the regions of scratch1a and 1b expression and in the spinal cord.(j and k). scratch2 is expressed in the precursors of the trigeminal ganglion and in the sensory Rohon-Beard neurons from the six-somite stage (lateral and dorsal views, respectively). (l) scratch2 expression in the spinal cord at 24 h.p.f. is restricted to the mantle layer at all dorsoventral levels. (m) RT-PCR confirms the onset of scratch2 expression in six-somite stage embryo. h, hindbrain; n, notochord; drc, telencephalic dorsorostral cluster; RB, Rohon-Beard neurons; sc, spinal cord; tg, precursors of the trigeminal ganglion; vcc, diencephalic ventrocaudal cluster
scratch2 knockdown induces cell death in the spinal cord
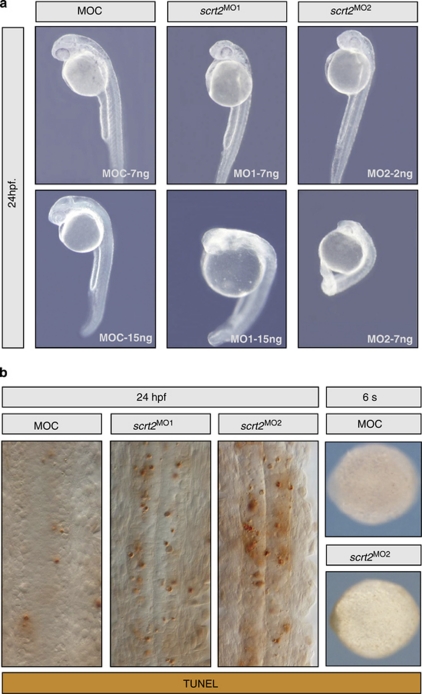
To study the function of scratch genes, we examined the effects of disrupting scratch2 activity in the spinal cord, which only expresses this scratch gene. Two morpholino oligonucleotides, one that overlapped the start codon (MO1) and another against the 5′ UTR (MO2, see Supplementary Figure 3), were injected into one-cell embryos. The survival of the embryos injected with MO1 was 86% (7 ng) and 70% (15 ng), whereas up to 84% (2 ng) and 50% (7 ng) of embryos survived when injected with MO2 (n>150 embryos per condition). Similar survival (84%) was evident after injection of a control morpholino (MOC), which did not produce any detectable phenotype at the concentrations used. In contrast, both MO1 and MO2 induced dose-dependent shortening of the embryos and opacity, characteristic of cell death at 24 h.p.f. (Figure 2a). At concentrations that produced similar embryo survival and opacity, injection of both MO's (7 ng of MO1 or 2 ng of MO2 per embryo) increased apoptosis in 94% of the embryos, when compared with the control MO (n=100; Figure 2b). Although distributed throughout the embryo (Supplementary Figure 3), the injected morpholinos should only affect regions that express target genes, and, indeed, apoptosis only increased in the nervous system. This cell death did not appear to be an off-target effect of morpholino injection (as described for around 20% of sequences20) because both MOs produced the same effect. Moreover, there was no morphological phenotype or additional apoptosis in injected embryos analyzed at the six-somite stage when scratch2 expression commences (Figure 2b). Indeed, when we co-injected MO2 (directed against the 5′ UTR sequences) with an mRNA containing the coding region of scratch2, morpholino-induced apoptosis was fully rescued in up to 85% of embryos (n=100; Figures 3g–i), reflecting the specificity of this effect. Hence, scratch2 might act as an anti-apoptotic factor in developing zebrafish embryos, although endogenous embryonic apoptosis persisted (Figures 3h and i) and scratch2 overexpression did not impede the endogenous cell death in the spinal cord (Figures 3j–l) and trigeminal ganglion (n=8 ganglia per condition) in which it is prominent at these stages (Supplementary Figure 4).
Figure 2.
scrtach2 knockdown induced cell death in zebrafish embryos at 24 h.p.f. (a) Two different morpholino antisense oligonucleotides were designed against adjacent sequences in the 5′ region of scratch2. scratch2 morpholino injections produced shortening and opacity, as assessed by morphology in embryos at 24 h.p.f. (b) A significant increase in cell death in the spinal cord was detected by TUNEL staining. Pictures show flat-mounted embryos highlighting dorsal views of the spinal cord. No morphological defects or significant cell death could be observed in embryos injected with control morpholino (MOC) or with scratch2 MOs analyzed as whole embryos at the six-somite stage (6s), when endogenous scratch2 begins to be expressed (anterior is to the left)
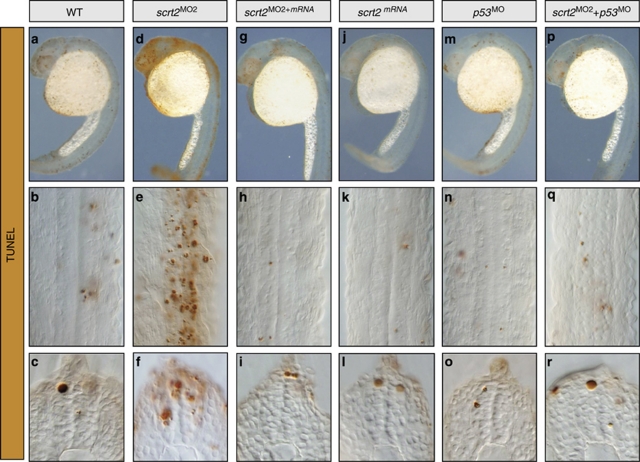
Figure 3.
scratch2 protects from cell death by antagonizing the p53 pathway. Embryos at 24 h.p.f. were subjected to TUNEL staining and shown as whole embryos, flat mounts or in transverse sections through the spinal cord. (a–i). The cell death phenotype observed after scratch2 MO injection could be rescued by co-injection of scratch2 mRNA. (j–l) Injection of scratch2 mRNA alone does not protect from endogenous cell death. (m–o) p53 morphant embryos are indistinguishable from control embryos. (p–r) The co-injection of p53 MO prevents the death induced by scratch2 MO
scratch2 interacts with the p53 pathway to protect from cell death
As cell death augments in the developing spinal cord of scracth2 morphants, we wanted to determine the molecular pathway by which Scratch2 regulates apoptosis. Members of the p53 family are critical elements in the apoptotic program, p53 being the key family member in the zebrafish nervous system.21 An increase in p53 activity is associated with considerable apoptosis in the nervous system and growth arrest in embryos,14 a phenotype reminiscent to that observed in the scratch2 morphants. Hence, we assessed whether scratch2 and p53 might be acting in the same pathway. Injection of p53 MO alone did not induce any embryonic defects (98%, n=100; Figures 3m–o) as expected, as p53 is not essential for zebrafish (or mouse) embryonic development.14 Indeed, when MOs against scratch2 and p53 were co-injected into embryos, they were indistinguishable from control embryos (91%, n=77; Figures 3p–r). Hence, it appears that Scratch2 protects neurons from apoptosis and that the death induced by Scratch2 downregulation is mediated by p53.
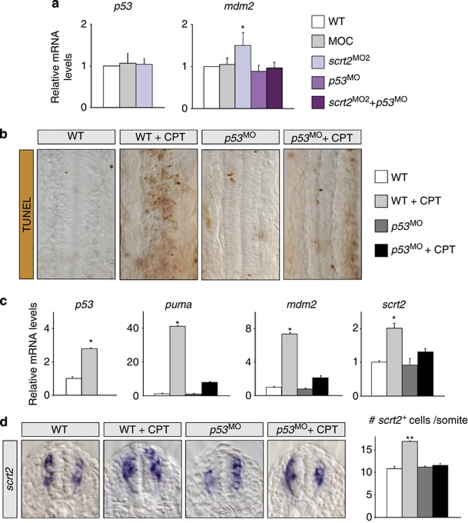
As Snail/Scratch proteins are transcriptional repressors,4 we assessed whether Scratch2 could protect cells from apoptosis by repressing p53 transcription. There were no significant changes in p53 expression in scratch2 morphant embryos (Figure 4a), indicating that Scratch2 does not act directly upstream of p53. However, Scratch2 downregulation activates p53, as it influences the expression of its main direct target, mdm2 (Figure 4a). Conversely, we determined whether activating p53 with camptothecin (CPT), an anti-cancer drug that promotes p53-mediated apoptosis in zebrafish, affects scratch2.14 CPT induced DNA damage and large-scale apoptosis in the spinal cord of 98% of embryos (n=100), which was impaired by injecting p53 MO (75% n=100). Hence, the apoptosis induced by CPT was mediated by p53 and it was concomitant with increased p53 transcription (Figures 4b and c). Upregulation of direct transcriptional targets of p53, mdm2 and the BH3-only puma further confirmed the functional activation of p53 (Figure 4c). Moreover, scratch2 transcription was also upregulated in CPT-treated embryos (Figure 4c), augmenting the number of cells expressing scratch2 in the spinal cord by 55% in a p53-dependent manner (from around 11 to 17 per somite in three independent embryos per condition, Figure 4d). The complement of cells expressing scratch2 was normal in p53-morphant embryos, indicating that p53 is not the endogenous inducer of scratch2. These results demonstrate that scratch2 is a novel p53 target activated after DNA damage.
Figure 4.
p53 activation induces scratch2 expression in the spinal cord. (a) Scratch2 does not regulate p53 transcription. Real-time RT-PCR from embryos injected with control (MOC), scratch2, p53 or scratch2 plus p53 MOs. The transcription of p53 is not affected by scratch2 knockdown, but its transcriptional activity, measured by the levels of its readout mdm2, is increased. (b) Flat-mounted spinal cords from 24 h.p.f. embryos showing the apoptosis induced by camptothecin (CPT)-mediated p53 activation.14 (c) Real-time RT-PCR to assess the transcription of p53, its targets puma and mdm2 and of scratch2 in embryos subjected to the treatments shown in b. scratch2 transcription is activated in response to CPT treatment. (d) scratch2 is activated in ectopic spinal cord cells on CPT treatment in a p53-dependent manner. Real-time RT-PCR data in a are means±S.E.M. of five independent experiments and c shows a representative experiment (n=3), with each point examined in triplicate in all cases. *P<0.05: **P<0.01
scratch2, a new direct target of p53
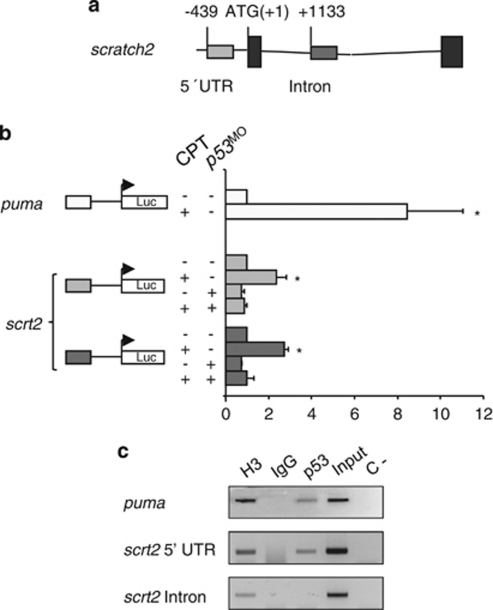
To determine whether p53 directly activates scratch2 transcription, we looked for putative p53-responsive elements in the scratch2 gene with the p53MH algorithm.22 We identified two putative p53-binding sites for which luciferase reporter constructs were generated, one in the 5′ UTR and another in the scratch2 intronic sequences (Figure 5a). A similar construct containing the p53 RE in puma was used as a positive control.12 The constructs were independently injected into one-cell zebrafish embryos and p53 was activated by treating the zebrafish embryos with CPT at the 21-somite stage. This activation of p53 transactivated each scratch2 reporter, and as this transactivation was prevented when p53 was knocked down, p53 appears to activate scratch2 transcription through the putative binding sites identified (Figure 5b). Moreover, in chromatin immunoprecipitation assays (ChIP) performed on embryos treated with CPT, p53 was specifically recruited to fragments containing the binding site in the scratch2 promoter (Figure 5c) and, hence, we consider scratch2 to be a novel direct target of p53.
Figure 5.
scratch2 transcription is directly activated by p53. (a) p53-binding sites in scrtach2. Putative p53 response elements are found in the promoter and within the intron of scratch2 sequences (gray boxes, positions relative to the ATG). (b) Luciferase assays carried out in whole zebrafish embryos. CPT-mediated p53 activation correlates with reporter activity for puma-positive control constructs and scratch2 constructs containing p53-binding sites found either in the promoter or in the intron. Transactivation is prevented by the co-injection of p53 MO. *P<0.05 (c) Chromatin immunoprecipitation assays (ChIP) in embryos treated with CPT (see Materials and Methods). Input material was tested for each primer set. ChIP analyses were carried out with anti-p53 antibodies on 24 h.p.f. zebrafish embryos amplifying the p53 response element in puma intron as a positive control for immunoprecipitation. H3 and IgG are the positive and negative controls of the immunoprecipitate, respectively; C- is a PCR negative control. The data presented are representative of three independent experiments
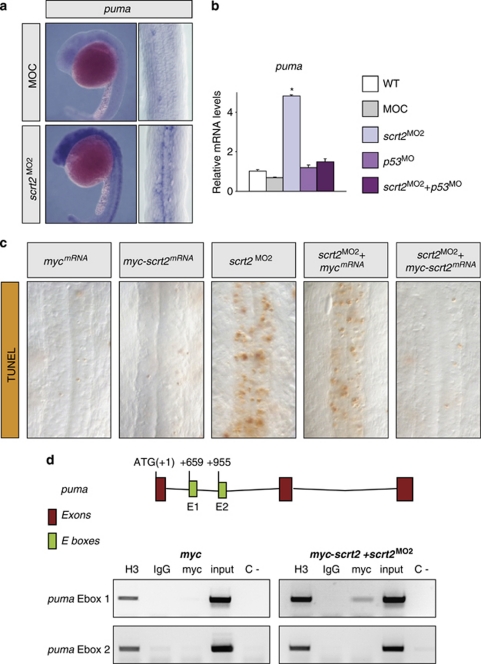
Scratch2 directly represses puma transcription
As scratch2 knockdown did not affect p53 transcription (Figure 4) and Snail/Scratch factors act as transcriptional repressors, Scratch2 could repress the transcription of pro-apoptotic genes downstream of p53. Hence, we assessed puma expression when Scratch2 function was compromised, one of the main mediators of apoptosis in the nervous system. Injection of MO2 strongly upregulated puma expression in zebrafish embryos at 24 h.p.f., an effect that was prevented by co-injection with p53 MO (Figures 6a and b). Thus, this increase in puma transcription appears to be mediated by p53, explaining the rescue of cell death observed when p53 MO was co-injected with scratch2 MO (Figures 3p–r). These data also support that Scratch2 could repress puma transcription. Similar to Snail, Scratch binds to E-boxes, and in murine hematopoietic cells, Snail2 has already been shown to bind to a consensus E-box in an intron of Puma.12 Indeed, we found two putative Scratch-binding sites in the first intron of zebrafish puma. As antibodies against Scratch are not available, we generated a myc-tagged Scratch2 fusion protein to perform ChIP assays. Moreover, as scratch2 overexpression is insufficient to protect against endogenous apoptosis (Figure 3), the ChiP experiment was performed in a scratch2-morphant background that can be rescued by injection of scratch2 mRNA (Figure 3). As expected, co-injection of scratch2 MO and myc-scratch2 mRNA prevented the apoptosis induced by scratch2 knockdown (75%, n=40; Figure 6c). Under these conditions, Scratch2 can bind to a fragment containing the first E-box in the puma intron (Figure 6d), demonstrating that Scratch2 can directly repress puma transcription, preventing embryonic cell death in the absence of damage.
Figure 6.
scratch2 antagonizes p53-mediated apoptosis by directly repressing puma transcription. (a) Increased puma transcription is observed by in situ hybridization in scratch2 morphants. (b) Real-time RT-PCR showing the increase in puma transcripts in scratch2-morphant embryos, which can be prevented by co-injection with p53 MO. *P<0.05. (c) A myc-scratch2 fusion mRNA is, similar to scratch2 mRNA, able to rescue the cell death induced by scratch2 MO. (d) Zebrafish Scratch2 is recruited to the E1-box in the first puma intron. Schematic representation of puma with E-boxes in green and positions relative to the ATG. Chromatin immunoprecipitation assays (ChIP) in embryos treated as in c. Input material was tested for each primer set and ChIP analyses were carried out with anti-myc antibody on 24 h.p.f. zebrafish embryos. An irrelevant sequence in scratch2 intron was amplified as a negative control of the immunoprecipitation with the anti-myc antibody (data not shown). H3 and IgG are the positive and negative controls of the immunoprecipitate, respectively. C- is a PCR negative control. The data presented are representative of three independent experiments
scratch2 downregulation activates intrinsic and extrinsic apoptotic pathways
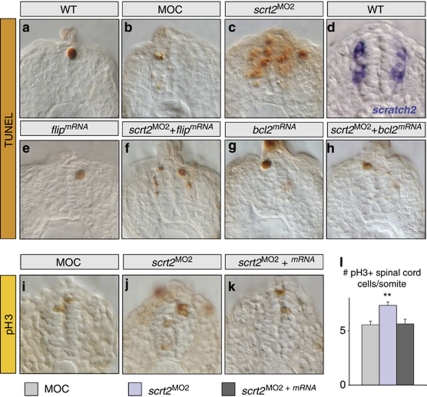
In the scratch2-morphant embryos studied here, apoptotic cells appeared in regions of endogenous scratch2 expression and in adjacent cells extending two or three cell layers, indicative of a non-cell-autonomous phenotype (Figures 3f and 7c). These apoptotic cells appear to account for about one-third of the total terminal deoxynuclotidyl transferase (TdT)-mediated dUTP nick end labelling (TUNEL)-positive cells. As this ectopic cell death was prevented by co-injection of scratch2 mRNA, it was considered a specific consequence of scratch2 knockdown. As a transcription factor, Scratch2 should act in a cell-autonomous manner and, thus, scratch2 MO should only produce effects in cells that normally express this factor. Nevertheless, these cells could produce non-cell-autonomous effects by activating an extrinsic apoptotic pathway. Accordingly, overexpression of a specific inhibitor of the extrinsic pathway (FLICE-inhibitory protein (FLIP))23 should block the apoptosis observed outside the endogenous domain of scratch2 expression. Injection of flip mRNA alone did not affect embryos, which were indistinguishable from controls (96%, n=30: Figures 7a, b and e), whereas there were fewer apoptotic cells in embryos co-injected with flip mRNA and scratch2 MO than when scratch2 was knocked down (89% n=37; compare Figure 7c with f). Interestingly, the remaining apoptotic cells were only found in the mantle layer of these embryos, within the limits of the endogenous scratch2 expression domain (compare Figures 7d with f). Hence, scratch2 knockdown leads to cell-autonomous activation of the intrinsic apoptotic pathway in cells that normally express scratch2, subsequently inducing cell death in adjacent cells through an extrinsic apoptotic pathway.15 Indeed, B-cell lymphoma 2 (bcl2) overexpression completely abolished the apoptotic phenotype in 75% of embryos co-injected with scratch2 MO (n=65; Figure 7h), both in endogenous territories of scratch2 expression and in adjacent cells.
Figure 7.
scratch2 downregulation activates both the intrinsic and extrinsic apoptotic pathways in the spinal cord, concomitant with compensatory proliferation. Embryos at 24 h.p.f. were TUNEL stained (a–c and e–h) or subjected to in situ hybridization for scratch2 (d), and transverse sections through the spinal cord are shown. (a–d) After scratch2 MO injection cell death is observed in regions of endogenous scratch2 expression and in adjacent cells. Compare c with d. (e–f) Co-injection of scratch2 MO and mRNA for the specific inhibitor of extrinsic apoptotic pathway flip prevents the death induced outside the scratch2 expression domain, confirming the activation of this pathway after scratch2 downregulation. (g and h) The co-injection of scrt2 MO and bcl2 mRNA prevents the activation of both the intrinsic and the extrinsic death pathways induced by scratch2 MO. (i–l) pH3 immunoreactivity and cell quantification reveals that cell death in scratch2 morphants induce compensatory proliferation in the ventricular zone (compare j with i, and l). Co-injection of scratch2 MO and mRNA rescues this phenotype (k and l). **P<0.01
The apoptosis in the scratch2 morphants did not significantly reduce the size of the spinal cord, perhaps because compensatory proliferation can be triggered in adjacent cells, as in Drosophila and mice.24, 25, 26 Indeed, the apoptosis observed in scratch2 morphants was followed by an increase in mitosis in the ventricular zone, assessed by phosphohistone 3 (pH3) staining (Figures 7i–l). This increased proliferation was specifically related to scratch2 knockdown, as the normal number of mitotic cells were observed when the morpholino was co-injected with scratch2 mRNA (Figures 7k and l). Hence, the apoptosis induced by loss of Scratch2 is associated with compensatory proliferation that attempts to heal the tissue.
Discussion
By characterizing the zebrafish scratch genes, we show that, similar to their homologs in other species, they are specifically expressed in the nervous system during early development.6, 7, 9, 10 The three scratch genes are expressed in overlapping and specific patterns in the developing brain and scratch2 alone is expressed in the embryonic spinal cord. This specific expression of scratch2 allowed us to analyze vertebrate Scratch function in vivo for the first time, avoiding the problems of compensation due to co-expression of multiple family members. Accordingly, we pinpoint a role for Scratch in neuron survival during normal embryonic development, repressing Puma expression in the absence of DNA damage.
Scratch2 promotes neuronal survival by suppressing Puma expression during normal development
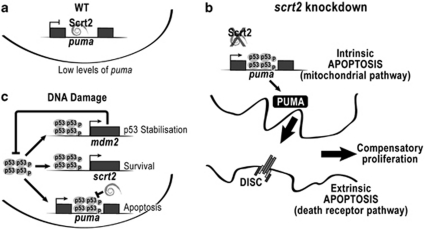
The induction of Puma expression by stress can provoke indiscriminate death of mature cells, whereas in deregulated systems (e.g., tumors), the loss of Puma may be sufficient to overcome apoptosis.27 Thus, maintaining Puma levels low is essential for cell survival. Snail2 can protect hematopoietic progenitor cells from cell death induced by DNA damage by repressing Puma transcription.12 In the nervous system, scratch2 knockdown de-represses puma and provokes neuronal death, even in the absence of DNA damage. This indicates that Scratch2 is required for neuron survival during normal development, and that Puma must be actively repressed, even in the absence of stress signals. Newly differentiated spinal cord neurons need an effective way to repress Puma activity to survive, and this can be achieved during embryonic development through Scratch2 (Figure 8a).
Figure 8.
Role of Scratch2 in neuron survival during development. (a) In the wild-type embryo, Scratch2 maintains puma transcription repressed, allowing neuronal differentiation and survival. (b) Scratch2 downregulation de-represses puma, permitting p53 binding to its response element in the intron and resulting in neuronal cell death. Apoptosis mediated by the intrinsic pathway in these cells induces the extrinsic apoptotic program in the neighboring cells. Dying cells induce progenitor cells in the ventricular zone to increase proliferation in an attempt to compensate for cell loss. (c) On DNA damage, p53 induces the transcription of several target genes, including mdm2, puma and scratch2. p53 protein levels are controlled by degradation induced by Mdm2. Cells expressing high levels of Scratch2 can interfere with the p53 apoptotic program through the repression of puma, leading to cell survival. A long-lasting or strong damage induces apoptosis as the final outcome of persistent p53 activity
The knockdown of scratch2 augments puma expression and neuronal death in a p53-dependent manner (Figure 8b), even though p53 expression remains unaffected. As p53 is constitutively transcribed during development, its activity is tightly regulated through proteasomal degradation triggered by its own target, Mdm2,28 and by its sequestration by anti-apoptotic members of the Bcl-2 family.29 Puma is a common target of p53 and Scratch2 for activation or repression, respectively. Indeed, Scratch2 binds to an E-box located very close to the p53 consensus RE in a puma intron, suggesting that they might compete for binding, as previously postulated for Snail2.30 In the absence of damage, low levels of p53 enable Scratch2 to bind to the intron of puma and prevent its transcription (Figure 8a). Conversely, p53 can activate puma transcription in the absence of Scratch2 (Figure 8b). Interestingly, the p53/Bcl-XL equilibrium is disrupted by Puma, releasing p53 into the cytoplasm.31 Thus, newly expressed Puma may augment the p53 cytoplasmic pool, reinforcing Puma expression and cell death.
After DNA damage, p53 activation directly increases scratch2 transcription in the zebrafish, establishing a negative feedback loop similar to that described for Mdm2, but through direct repression of Puma rather than p53 degradation (Figure 8c). Similarly, p53 upregulates the transcription of other prosurvival genes, including Snail family members.12 Weak Scratch expression permits Puma transcription, whereas strong Scratch expression would prevent p53 from acting on Puma, probably by interfering with the assembly of the transcriptional complex.
Scratch2 is required for newly differentiated neurons to survive: evolutionary considerations
Evolution has favored the need for Scratch to promote neuron survival. Increased scratch (ces-1) expression in C. elegans leads to the appearance of supernumerary neurons, although it is not, in principle, required for neuron survival.32 However, scratch2 knockdown in zebrafish is sufficient to promote neuronal death, while its overexpression does not promote the survival of additional neurons, which also seems to be true in the chick embryo (our unpublished results). Interestingly, vertebrate evolution has implemented robust mechanisms to regulate cell number and to eliminate supernumerary neurons,33 which suggests the need for a system ensuring controlled neuronal survival. Although in tissues with little proliferation, mature cells need to be protected, mature cells are dispensable in those with high proliferation rates, as long as there are sufficient progenitors.12 For example, the progenitors in the hematopoietic system express Snail2 and, hence, they are protected from p53-induced cell death after DNA damage.12 Other adult tissues have poor proliferative capacity and they must protect mature cells, as also seems to be the case in the developing nervous system in which differentiated neurons express scratch to protect them from cell death.
The role of Scratch2 in the survival of zebrafish and C. elegans neurons (refs. 10, 13 and data herein) indicates that resistance to cell death is an ancestral function associated with the Scratch superfamily. Snail proteins have also been implicated in promoting cell survival on different apoptotic stimuli,8, 11 indicating that cell survival is an ancestral function associated with both families before diploblasts and bilateria diverged.4 Accordingly, the better-known role of Snail factors in regulating cell adhesion and movement was probably co-opted later by Snail genes alone. It is intriguing that evolutionary patterns also show that p53 genes existed before the appearance of cancer.34 Therefore, the crucial tumor suppressor functions associated with this family were not ancestral but rather, similar to Snail and Scratch, the ancestral function appears to be the regulation of cell death, which existed even before the regulation of the cell cycle.35 Whether the Snail, Scratch and p53 families evolved hand-in-hand to establish the p53 regulatory networks during evolution might be an interesting topic to explore in the future.
Scratch2 downregulation, activation of intrinsic and extrinsic apoptotic pathways and compensatory proliferation
During embryonic development, puma is activated after scratch2 knockdown to stimulate the mitochondrial apoptotic pathway in the zebrafish neural tube. Scratch2 deficiency provokes cell-autonomous death of differentiated neurons that otherwise express scratch2. Similarly, it activates non-autonomous cell death through an extrinsic apoptotic pathway in progenitor cells that do not normally express scratch2, revealing a role for the extrinsic apoptotic pathway in the zebrafish nervous system15 (Figure 8b). Similar cell death of both proliferative and differentiated cells occurs in the intestine of young mice deficient for Mdm2.26 Interestingly, fish and mice respond similarly to the loss of a p53-induced negative regulator, Scratch2 or Mdm2, not only in terms of cell death but also by inducing compensatory proliferation (ref. 26 and data herein) Indeed, although the apoptosis induced by Scratch2 downregulation could reduce the size of the spinal cord, increased proliferation of neural progenitors in the ventricular zone appears to prevent this from occurring. Hence, this tissue seems to activate mechanisms to overcome neuronal loss and maintain tissue homeostasis, similar to the compensatory mechanisms described in Drosophila.24, 25
The apoptosis and compensatory proliferation observed in our scratch2 morphants, is reminiscent of the massive leukocyte cell death and compensatory proliferation, which repopulates damaged tissue following irradiation of wild-type mice and which leads to secondary malignancies appearing because of the damage to progenitor/stem cell pools.36, 37 Conversely, the lack of cell death and of subsequent compensatory proliferation in Puma-deficient mice protects them from developing secondary tumors.36, 37
In summary, although both p53 and Puma seem to be dispensable for normal embryonic development, they can respond rapidly to damage; thus, inhibitors must be available to tightly control their activity. Here, we describe one such inhibitor, Scratch2, which acts directly on the main readout of p53 signaling, puma. When scratch2 is downregulated, puma levels increase, promoting neuronal cell death by activating both intrinsic and extrinsic apoptotic pathways even in the absence of DNA damage. Neuronal loss activates compensatory progenitor cell proliferation, a physiological mechanism at play during development to maintain tissue homeostasis, which may be corrupted in circumstances of previous DNA damage, to provoke the appearance of cancer.
Materials and Methods
Fish maintenance
AB and Tup-Lof wild-type Zebrafish strains maintained at 28°C under standard conditions were used in all experiments. The embryos were staged, as indicated elsewhere.38
Isolation of zebrafish cDNAs
Total RNA was extracted with Trizol (Invitrogen, Paisley, UK) from 24-h.p.f. zebrafish embryos and used to synthesize cDNA using SuperScript III reverse transcriptase (Invitrogen) and oligodT as a primer. Subsequent PCR amplification was carried out using primers directed to the 5′- and 3′ UTR sequences as follows: scratch1a: 5′-ATGGATCCGGATAACGTGCTGGAAGAGG-3′and 3′-AATCTAGAGAAGTCACGCCTCAATGGAC-5′ scratch1b: 5′-CGGGATCCCGGACGAGTGTTTTCTCCTT-3′ and 3′-GCTCTAGATGCGATTAATCCTCGGTGTT-5′ scratch2: 5′-GGATCCTCTGTGCGACCGTGAACC-3′ and 3′-GCCTCGAGGGATTTTTGCGAGCATTAAA-5′. Amplified fragments were subcloned into pTOPO and pCS2+. Zebrafish actin was amplified as a control for scratch2 expression during developmental stages with the following primers: 5′-CTGGTTGTTGACAACGGATCCG-3′ and 5′-CAGACTCATCGTACTCCTGCTTGC-3′. Zebrafish puma (DQ860151), flip (NM_194399) and bcl2 (ENSDART00000105690) coding sequences were amplified with the following sets of oligonucleotide primers: puma, 5′-CACGATTCTGGGATAACATCAA-3′ and 5′-ACCCGTGTGTTCATCTGAGG-3′ flip, 5′-GGATCCGGTGAAATGGCAGATGGATT-3′ and 5′-GGATCCGGAGTGGTTTGTGTTGTGTTG-3′ bcl2, 5′-GGATCCGCGCGTTTCTATCGTGATTT-3′ and 5′-TCTAGAGAGGCTGTCACTTCTGAGCA-3′. In all cases, sequences represent forward and reverse primers, respectively. The puma DNA fragments were subcloned into the pGEM.TE plasmid and those from flip and bcl2 into the pCS2+ plasmid. Myc-tagged scratch2 was subcloned in pCS2+-NLS-MT (kindly provided by JL Gómez-Skarmeta) after amplification with the following primers: Forward 5′-GAATTCCATGCCTCGCTCGTTTCT-3′ and reverse, 5′-CTCGAGTTAATTCTCTGAGCCAGATTCG-3′.
Morpholino oligonucleotides and mRNA injections
All morpholino antisense oligonucleotides were obtained from Gene Tools LLC (Philomath, OR, USA) and used as described by Nasevicius and Ekker.39 Two antisense morpholinos (MOs) were used. MO1, 5′-GAGGCATGGTTCACGGTCGCACAGA-3′, overlapping the start codon and injected from 2 to 15 ng per embryo, and MO2, 5′-CGCACCAGAAAATCCTTCACATCGG-3′, located at the 5′ UTR and injected from 1 to 7 ng per embryo (Supplementary Figure 3). The sequence of p53 MO was described previously.20 The standard control morpholino from Gene Tools was used for control injections. All oligonucleotides were injected into the yolk of one- or two-cell embryos using a pressure injector (PicoSpritzer, Parker, Fairfield, NJ, USA). MO1 was fluoresceinated to confirm that the morpholino was distributed to all cells after injection and during embryo development (Supplementary Figure 4).
For rescue and overexpression experiments, mRNA was synthesized and capped using the mMessage mMachine Kit (Ambion, Austin, TX, USA). Rescue analyses were performed with 75 pg per embryo of scratch2 mRNA lacking the 5′ UTR region to which MO2 binds. For overexpression analyses, 75–100 pg per embryo of myc-scratch2, flip and control RNAs or 200 pg per embryo of bcl2 mRNA were used.
In situ hybridization and immunohistochemistry
In situ hybridization and immunohistochemistry of whole-mount zebrafish embryos for scratch2, puma and pH3 (Rabbit anti-pH3 antibody, 1:1000, Upstate Biotechnology, Lake Placid, NY, USA) were performed as described previously.40 Embryos were then fixed and embedded in gelatin to obtain vibratome sections (30 μm). Embryos were photographed using an Olympus DP70 digital camera (Olympus, Hamburg, Germany) on either a Leica M10 dissecting microscope or a Leica DMR microscope (Leica, Wetzlar, Germany). Flat-mounted 24-h.p.f. embryo spinal cords are shown anterior-up, unless otherwise indicated.
Quantitative reverse transcription PCR
Quantitative reverse transcription PCR was performed using the Step One Plus (Applied Biosystems, Carlsbad, CA, USA) sequence detection system and the SYBR Green method (Applied Biosystems). The data show one representative experiment. Statistics correspond to n=3 unless otherwise indicated, with each point examined in triplicate.
Transcript levels were calculated using the comparative Ct method normalized to GAPDH. The final results were expressed relative to the control condition using the 2−(ΔΔCt)±S.D. formula. The primers used are listed below:
Gapdh (AY818346), 5′-GCAGAAAAGCCAGACCATTC-3′ and 5′-CTCGTAGGTGGGAACAGGAA-3′ scrt2, 5′-ACTACGAGTCGGCCTGCTT-3′ and 5′-GGGATTTTTGCGAGCATTAAA-3′ puma (DQ860151), 5′-ACGCTGTCTTCCTTCAGAGG-3′ and 5′-GGTAGAGGGCATTGATGGTG-3′ p53 (DRU60804), 5′-CAGTCTGGCACAGCAAAATC-3′ and 5′-ATTTGAACGGGGCAAGTTTT-3′ and mdm2 (AF356346), 5′-ATTGTTCACGGAAGGACTGG-3′ and 5′-CCACTGACTGAATGGGCTCT-3′.
CPT treatment and TUNEL assay
CPT (Calbiochem) treatment was performed as described.14 A 50 μM stock of CPT in DMSO was diluted in Danieau's buffer to obtain a 500 nM working solution that was applied to 21-somite stage embryos at 28°C until the stage of analysis (24 h.p.f.). Embryos used for luciferase assays and ChIP experiments were treated with CPT for the first 2 h and kept in embryo medium until they reached 24 h.p.f. Control embryos were mock treated with Danieau's buffer containing the same concentration of DMSO. Apoptosis was analyzed with the TUNEL kit (Roche, Basel, Switzerland) according to manufacturer's instructions.
Cell quantification
For the analyses of scrt2- and pH3-positive cells, 30 μm consecutive sections corresponding to five consecutive somites (s5–s9) were analyzed in embryos at 24 h.p.f. Quantification of TUNEL-positive cells in trigeminal ganglia was carried out in flat-mounted 24 h.p.f. embryos. At least three embryos were counted per condition and marker (n=3 for scrt2, n=6 for pH3 and n=8 for trigeminal ganglia).
Luciferase assays
The p53MH algorithm22 was used to identify putative p53 REs present in scratch2 and puma genomic sequences. To generate the luciferase reporter plasmids, various fragments containing the p53 putative REs were subcloned in pGL3Basic (Promega, Madison, WI, USA). PCR amplification was performed on adult zebrafish genomic DNA as template with the following sets of oligonucleotide primers:
scrt2 5′ UTR, 5′-CATCATGGTACCTGATCTCACTCCCACCGT-3′ and 5′-CATCATGCTAGCTCAGCACTGGATAGCTCC-3′ scrt2 intron, 5′-CATCATGCTAGCTCCCACCTGTTACGAAAA-3′ and 5′-CATCATCTCGAGTGATGCTTAAACCATTCAT-3′ and puma intron, 5′-CATCATACGCGTTGCATTAATTTCTGACAAGT-3′ and 5′-CATCATCCCGGGACCAGCGACAGGCTAT-3′.
Luciferase assays were performed as previously described.41 Briefly, the reporter plasmid of the firefly luciferase gene, under the control of the different p53 REs obtained for scratch2 (12.5 pg per embryo) and puma (2.5 pg per embryo), was injected into one-cell stage zebrafish embryos, together with 1.25 pg per embryo of a plasmid containing the Renilla luciferase gene under the control of the Tk promoter. The 21-somite stage embryos were treated with 500 nM CPT for 2 h to promote p53 activation and the embryos were then kept in embryo medium for 2 more hours. Subsequently, 20 embryos of each experimental condition were lysed in 100 μl lysis buffer (Passive lysis buffer, Promega) and assays were carried out on 10 μl of the embryo lysate with a Dual-Luciferase Reporter Assay System (Promega). The data are represented as the mean±S.D. of three different experiments.
Chromatin immunoprecipitation
ChIP experiments were carried out following the Upstate cell signaling solutions protocol (Upstate cell signaling solutions, Billerica, MA, USA). Briefly, embryos were treated with CPT (see above) and then 30 embryos were sonicated in 300 μl lysis buffer per condition in each experiment. The chromatin was immunoprecipitated with the anti-p53 antibodies, kindly provided by Professor G Del Sal (University of Trieste, Trieste, Italy) and goat anti-myc (Abcam, Cambridge, UK). The antibodies used as negative and positive controls were Rabbit anti-IgG (Diagenode, Liège, Belgium) and Rabbit anti-H3 (Abcam), respectively. The oligonucleotide primers used for PCR detection were: scrt2 5′ UTR, 5′-TGATCTCACTCCCACCGT-3′ and 5′-CTCAGCACTGGATAGCTCC-3′ scrt2 intron, 5′-TCCCACCTGTTACGAAAA-3′ and 5′-TGATGCTTAAACCATTCAT-3′ puma p53 RE and E-box1, 5′-TGCATTAATTTCTGACAAGT-3′ and 5′-ACCAGCGACAGGCTATCAA-3′ puma E-box2, 5′-TCTGCTGACACTCCTCCTCA-3′ and 5′-GCAACAGCCTGAGCTGAAAT-3′. The data presented are representative of three independent experiments.
Database search
NCBI and EMBL-EBI/WellcomeTrust Sanger Institute databases were searched through the BLAST family of programs (http://www.ncbi.nlm.nih.gov/BLAST) and http://www.ensembl.org, respectively. Sequence alignments were carried out using Clustal W (http://www.ebi.ac.uk/clustalw; ref. 42) and corrected by visual inspection.
Acknowledgments
We are very grateful to Elisa Guida and Mireille Tora for their help in generating some constructs; to Jose Luis Gomez-Skarmeta for providing the pCS2+-NLS-MT plasmid; to Alejandro Barrallo-Gimeno for his help and training in the use of the zebrafish; to Giannino del Sal for providing the anti-p53 antibody; to Diana Abad for excellent care of our zebrafish facility; to Isabel Fabregat and to Elisa Guida and Joan Galcerán among other lab members for helpful discussions and to Stuart B Ingham for his help with Figure 8. Work in the lab is supported by grants from the Spanish Ministry of Science and Innovation (BFU2008-01042, CONSOLIDER-INGENIO 2010 CSD2007-00017 and CSD2007-00023) and the Generalitat Valenciana (Prometeo 2008/049) to MAN.
Glossary
- Puma
p53 upregulated modulator of apoptosis
- Mdm2
murine double minute gene 2
- ces-1
cell death specification 1
- h.p.f.
hours post-fertilization
- TUNEL
terminal deoxynuclotidyl transferase (TdT)-mediated dUTP nick end labelling
- CPT
Camptothecin
- ChIP
chromatin immunoprecipitation
- RT-PCR
reverse transcription PCR
- FLIP
FLICE-inhibitory protein
- bcl2
B-cell lymphoma 2
- pH3
phosphohistone 3
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by L Greene
Supplementary Material
References
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. Evolutionary history of the Snail/Scratch superfamily. Trends Genet. 2009;25:248–252. doi: 10.1016/j.tig.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Nakakura EK, Watkins DN, Schuebel KE, Sriuranpong V, Borges MW, Nelkin BD, et al. Mammalian Scratch: a neural-specific Snail family transcriptional repressor. Proc Natl Acad Sci USA. 2001;98:4010–4015. doi: 10.1073/pnas.051014098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Nieto MA. The expression of Scratch genes in the developing and adult brain. Dev Dyn. 2006;235:2586–2591. doi: 10.1002/dvdy.20869. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Roark M, Sturtevant MA, Emery J, Vaessin H, Grell E, Bier E. scratch, a pan-neural gene encoding a zinc finger protein related to snail, promotes neuronal development. Genes Dev. 1995;9:2384–2398. doi: 10.1101/gad.9.19.2384. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Horvitz HR. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol Cell. 1999;4:309–319. doi: 10.1016/s1097-2765(00)80333-0. [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Thellmann M, Hatzold J, Conradt B. The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development. 2003;130:4057–4071. doi: 10.1242/dev.00597. [DOI] [PubMed] [Google Scholar]
- Langheinrich U, Hennen E, Stott G, Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol. 2002;12:2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- Eimon PM, Ashkenazi A. The zebrafish as a model orgnaims for study of apoptosis. Apoptosis. 2010;15:331–349. doi: 10.1007/s10495-009-0432-9. [DOI] [PubMed] [Google Scholar]
- Kratz E, Eimon PM, Mukhyala K, Stern H, Zha J, Strasser A, et al. Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ. 2006;13:1631–1640. doi: 10.1038/sj.cdd.4402016. [DOI] [PubMed] [Google Scholar]
- Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, et al. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares M, Locascio A, Nieto MA. The increasing complexity of the Snail gene superfamily in metazoan evolution. Trends Genet. 2001;17:178–181. doi: 10.1016/s0168-9525(01)02232-6. [DOI] [PubMed] [Google Scholar]
- Ross LS, Parrett T, Easter SS., Jr Axonogenesis and morphogenesis in the embryonic zebrafish brain. J Neurosci. 1992;12:467–482. doi: 10.1523/JNEUROSCI.12-02-00467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. p53 family in development. Mech Dev. 2008;125:919–931. doi: 10.1016/j.mod.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The cell is dead Long live the cell! Trends Cell Biol. 2008;18:467–473. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FA, Perez-Garijo A, Morata G. Apoptosis in Drosophila: compensatory proliferation and undead cells. Int J Dev Biol. 2009;53:1341–1347. doi: 10.1387/ijdb.072447fm. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Okano H, Lozano G. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 2008;15:1772–1781. doi: 10.1038/cdd.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- Endo H, Kamada H, Nito C, Nishi T, Chan PH. Mitochondrial translocation of p53 mediates release of cytochrome c and hippocampal CA1 neuronal death after transient global cerebral ischemia in rats. J Neurosci. 2006;26:7974–7983. doi: 10.1523/JNEUROSCI.0897-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S, Alsheich-Bartok O, Haupt Y. Clues from worms: a Slug at Puma promotes the survival of blood progenitors. Cell Death Differ. 2006;13:913–915. doi: 10.1038/sj.cdd.4401906. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- Nikoletopoulou V, Lickert H, Frade JM, Rencurel C, Giallonardo P, Zhang L, et al. Neurotrophin receptors TrkA and TrkC cause neuronal death whereas TrkB does not. Nature. 2010;467:59–63. doi: 10.1038/nature09336. [DOI] [PubMed] [Google Scholar]
- Aranda-Anzaldo A, Dent MA. Reassessing the role of p53 in cancer and ageing from an evolutionary perspective. Mech Ageing Dev. 2007;128:293–302. doi: 10.1016/j.mad.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- Labi V, Erlacher M, Krumschnabel G, Manzl C, Tzankov A, Pinon J, et al. Apoptosis of leukocytes triggered by acute DNA damage promotes lymphoma formation. Genes Dev. 2010;24:1602–1607. doi: 10.1101/gad.1940210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Vandenberg CJ, Delbridge AR, Wu L, Scott CL, Adams JM, et al. Apoptosis-promoted tumorigenesis: gamma-irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev. 2010;24:1608–1613. doi: 10.1101/gad.1940110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown' in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Patel K, Wilkinson DG. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
- Alcaraz-Perez F, Mulero V, Cayuela ML. Application of the dual-luciferase reporter assay to the analysis of promoter activity in Zebrafish embryos. BMC Biotechnol. 2008;8:81. doi: 10.1186/1472-6750-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.