Abstract
The protein tyrosine phosphatase PTPL1/PTPN13, whose activity is decreased through allelic loss, promoter methylation, or somatic mutations in some tumors, has been proposed as a tumor suppressor gene. Moreover, our recent clinical study identified PTPL1 expression level as an independent prognostic indicator of a favourable outcome for patients with breast cancer. However, how PTPL1 can affect tumor aggressiveness has not been characterized. Here, we first show that PTPL1 expression, assessed by Immunohistochemistry, is decreased in breast cancer and metastasis specimens compared with non-malignant tissues. Secondly, to evaluate whether PTPL1 plays a critical role in breast cancer progression, RNA interference experiments were performed in the poorly tumorigenic MCF-7 breast cancer cells. PTPL1 inhibition drastically increased tumor growth in athymic mice and also enhanced several parameters associated with tumor progression including cell proliferation on extracellular matrix components and cell invasion. Furthermore, the inhibition of Src kinase expression drastically blocked the effects of PTPL1 silencing on cell growth. In PTPL1 knockdown cells, the phosphorylation of Src on tyrosine 419 is increased, leading to the activation of its downstream substrates Fak and P130cas. Finally, substrate-trapping experiments revealed that Src tyrosine 419 is a direct target of the phosphatase.
Thus, by identification of PTPL1 as the first phosphatase able to inhibit Src through direct dephosphorylation in intact cells, we presently describe a new mechanism by which PTPL1 inhibits breast tumor aggressiveness.
Keywords: Animals; Blotting, Western; Breast Neoplasms; genetics; metabolism; pathology; Cell Adhesion; Cell Line, Tumor; Cell Movement; Cell Proliferation; Female; Fluorescent Antibody Technique; Humans; Immunoenzyme Techniques; Lymphatic Metastasis; Mice; Mice, Nude; Neoplasm Invasiveness; Phosphorylation; Protein Tyrosine Phosphatase, Non-Receptor Type 13; antagonists & inhibitors; genetics; metabolism; RNA, Small Interfering; pharmacology; Signal Transduction; Tissue Array Analysis; src-Family Kinases; antagonists & inhibitors; metabolism
Keywords: Tumor suppressor gene, breast cancer aggressiveness, Protein tyrosine phosphatase N13, Src, Signal transduction.
Introduction
PTPL1 (also called PTPN13, FAP-1, PTP-BAS, PTP1E), which is the nonreceptor type protein tyrosine phosphatase (PTP) with the highest molecular weight, 270 kDa, contains multiple interactive domains (1;2). Its physiological functions are poorly documented. PTP-BL (mouse homologue of PTPL1) KO mice present abnormal regulation of signal transducer and activator of transcription signaling in T cells (3), mice that lack the PTP-BL PTP activity show mild impairment of motor nerve repair (4) and we recently described the role of this phosphatase in adipocyte differentiation (5).
We reported the first evidence of the negative action of PTPL1 on cancer growth through our work on the anti-growth factor effect of antiestrogens in breast cancer (6–9). Other groups have confirmed that the PTPL1/PTPN13 gene presents the characteristics of a tumor suppressor gene (10;11). Its expression is frequently down-regulated or silenced through promoter hypermethylation within several tumor types (12;13). A mutational analysis of colorectal cancers identified different somatic mutations in PTPL1 (14). Additionally, the PTPL1/PTPN13 gene is located on chromosome 4q21, a region frequently deleted in ovarian and liver cancers (15). In agreement with these data, we recently showed that PTPL1 expression is an independent prognostic marker for increased overall survival in breast cancer, indicating that PTPL1 is an important regulatory element of human breast tumor aggressiveness (16).
A number of potential PTPL1-interacting partners point to a role for PTPL1 in several steps of tumor progression, such as modification of cell shape and motility, and indicate its potential role in cancer metastasis. These potential partners include PIP2 (1), TAPP1/2 (17), EphrinB1 (18), TRIP6/ZRP1 (19) and PARG1 (20), all of which are involved in the maintenance of the cytoskeleton.
In this study, we demonstrate that PTPL1 plays a critical role in breast cancer progression by acting on pathways dependent on cell-matrix interactions. We also delineate the underlying molecular mechanism of this effect, which involves a decrease of Src phosphorylation and the activation of Src substrates, FAK and p130cas. Using complementary substrate trapping, co-localization, and in cellulo dephosphorylation methods, we demonstrate that PTPL1 directly and specifically dephosphorylates Src on the activating tyrosine 419 (Y419). Our findings therefore provide a novel mechanism through direct Src dephosphorylation by which PTPL1 regulates breast cancer aggressiveness.
Materials and Methods
Immunohistochemistry
The tissue array containing selected areas of paraffin-embedded sections from primary breast cancers, benign breast tissues and lymph node metastases was obtained from SuperBioChips Laboratories. It was analyzed with anti-PTPL1(AC21 from AbCam) as previously described (21). Staining was revealed using a standard avidin-biotin enhanced immunoperoxidase technique (R.T.U. Vectastain Kit, Vector Labs). PTPL1 immunostaining was cytoplasmic. TMA was scanned with a Slide Scanner (Hamamatsu NANOZOOMER), and the cytoplasmic staining was evaluated with the Definiens developer (7.0) program (MRI, Montpellier).
Cell culture, plasmids and antibodies
HEK293, MDA MB 231 and MDA MB 436 cells were cultured in DMEM, MCF-7 and BT 549 cells in Dulbecco’s modified Eagle medium Ham’s F12/DMEM (50%/50%), T47D and ZR 75.1 cells in RPMI medium (Invitrogen), all supplemented with 10% FCS. The expression construct PTPL1 Wt was described previously as pHM6-PTPL1 (1). Mutants PTPL1-YF/DA and PTPL1-CS were obtained as described (8). All GST fusion proteins were constructed in pGEX-4T1 (Pharmacia Biotech) (8). Src and SrcY530F expression vectors were a gift of Dr S. Roche (CRBM, Montpellier, France). The following monoclonal and polyclonal antibodies were used: anti-HA (12CA5, Roche); anti-P130Cas (BD Biosciences); anti-phosphoTyrosine (4G10 and PY20) and anti-actin (Sigma); anti-PTPL1 (H300, Santa Cruz Biotechnology); anti-Fak, anti-Src, anti-phospho Src (Y419 and Y530) and anti-phospho Fak (Y397 and Y576/577) (Cell Signaling Technology).
Transfection and establishment of stable cell lines
Transient transfections were carried out using the jet PEI Cationic Polymer Transfection Reagent method (Qbiogene) with a ratio 1:10 of Src/PTPL1 Wt or mutant or empty pHM6 vector.
Small interfering RNA (siRNA) transfections were carried out using the Oligofectamine reagent method (Invitrogen). The two PTPL1-specific siRNAs (Si3:7313-GGAAAGAAGAGUUCGUUUA-7331 and Si4:1028-CAGAUCAGCUUUCCUGUAA-1046), the Src-specific siRNA (1403-GAAGCUGAGGCAUGAGAAG-1421) and the control non-targeting siRNA were from MWG.
For establishing clonal cell lines, MCF-7 cells were co-transfected with a 10-fold excess of pTER vector containing a short hairpin RNA (shRNA) control or shRNA-PTPL1 (Si3) over the pY3 hygromycin-resistance expression vector by the calcium phosphate-DNA precipitation technique. Colonies growing in the presence of hygromycin B (250 μg/mL, Invitrogen) were subsequently picked for expansion in cell lines, and PTPL1 expression was quantified by immunoblot. All experiments using transfected cells were performed at passage 4 +/− 1 and inhibition of PTPL1 expression was controlled over this period.
Cell invasion assay
Two days after siRNA transfection, cells were detached with trypsin-EDTA, and resuspended in 4% FBS Ham’s F12/DMEM. Cells (106/well) were seeded in triplicate in the upper chamber of a BD BioCoat Cell Culture Insert (12 μm diameter pore; BD Biosciences) that had been precoated with 30 μg of Matrigel (Becton Dickinson). The lower chamber contained Ham’s F12/DMEM with 10% FBS as the chemoattractant. After 48 h, the unmigrated cells in the upper chamber were gently scraped off the filter. The quantity of cells that had migrated through the filter was evaluated with the colorimetric MTT assay, as previously described (21), or by brilliant blue R staining and subsequent photography.
Preparation of GST fusion proteins
pGEX-4T1 Wt and DA were produced in Escherichia coli BL21 as previously described (8). After one wash with “Substrate-Trapping” buffer [20 mM Tris (pH 7.5), 100 mM NaCl, 1mM EDTA, 1% Triton X-100, 10% glycerol] containing 10 mM DTT, expression level and integrity of GST fusion proteins were verified by SDS-PAGE and Coomassie blue staining (data not shown).
Substrate trapping in vitro
Substrate trapping experiments were performed on cells treated with 10−4M pervanadate for 30 min (8). Briefly, cell lysates were incubated on ice for 30 min in the presence of 5 mM iodoacetic acid to irreversibly inactivate endogenous PTPs. After incubation, DTT was added for 10 min at 4°C to a final concentration of 10 mM, to inactivate the iodoacetic acid. Lysates were then incubated overnight at 4°C with 20 μL GST fusion protein–coupled beads (10 μg/μL). After seven washes affinity complexes were analyzed by immunoblot.
Growth assays
For Matrigel outgrowth assays, 20,000 cells were re-suspended in culture medium containing 10% FCS (0.5 mL) and added to 24-well plates that had been precoated with Matrigel (0.2 mL, 9.6 mg/mL) (BD Biosciences). Phase contrast optical photomicrographs were taken after five days.
Alternatively, 20,000 cells were plated in triplicate in 24-well dishes that had been coated with 10 μg/cm2 fibronectin (Becton Dickinson) in medium containing 10% FCS. Cells were fixed at various times with methanol, and their DNA content was determined by the diaminobenzoic acid fluorescence assay (22).
Cell attachment assay
For cell attachment assays 96-well dishes were coated with fibronectin, Matrigel or collagen 4 (Sigma) overnight (10 μg/cm2) and then blocked for 30 min with 0.5% BSA in PBS. Cells were trypsinized and resuspended at a final concentration of 105 cells/mL in culture medium containing 10% FCS. Two-hundred microliters of the cell suspension were added to the wells and the plates were incubated at 37°C for 30 min. Medium was then carefully suctioned out, and the wells were washed three times with culture medium. The colorimetric MTT-assay was used to determine the number of remaining cells (adherent cells).
Xenograft studies
In vivo experiments were performed in compliance with the French guidelines for experimental animal studies (Direction des Services Vétérinaires, Ministère de l’Agriculture, agreement no. B 34-45). Before cell implantation, a 1-cm-long silastic tube (silicone tube ID 0.062 in. and OD 0.095 in.) filled with a solid mixture of estradiol and cholesterol as a carrier (1:10) was implanted subcutaneously in the interscapular region of Swiss nu/nu (nude) mice (Charles River Laboratories International). Two days later, 107 cells were injected orthotopically into the thoracic mammary gland of athymic nude mice. Injected mice were examined every three or four days for tumor appearance. Tumor size was measured by digital caliper, and tumor volumes were estimated according to the formula for an ellipse: (short dimension)2 × (long dimension)/2.
Immunoprecipitation and Immunoblot analysis
Forty-eight hours after transient transfection, cells were washed twice in ice-cold PBS and lysed in lysis buffer [40 mM Tris (pH 8), 5 mM MgCl2, 40 mM Na4P207, 1% Triton x-100, 10 mM EDTA, 50 mM NaF, 100 μM Na3VO4, 1/250 aprotinin, 1 mM AEBSF]. Equal amounts of each cell lysate were immunoprecipitated with 2 μg antibodies overnight at 4°C and then incubated for 2 h with 40 μL protein A-Sepharose (6%) blocked in PBS, 4% BSA. Immune complexes were washed five times in lysis buffer before immunoblotting.
Immune complexes or 100 μg of total proteins were separated on 7.5 or 10% SDS/polyacrylamide gels before immunoblotting, as described (8), with the indicated antibody.
Immunofluorescence microscopy
Forty-eight hours after transient transfection, HEK293 cells that had been plated on coverslips were fixed with 3.7% formaldehyde in PBS for 15 min and permeabilized with 0.5% Triton X-100 in PBS for 10 min. Immunolabeling was performed as described (8) with FITC or Texas red–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Coverslips were mounted with mounting medium and visualized with a Bio-Rad 1024 CLSM system using a 60X (1.4 NA) planapochromatic objective (Nikon). A series of optical sections were collected and projected onto a single image plane using the laser sharp 1024 software and processing system.
Statistical analysis
The results of the cell proliferation, migration, adhesion and tumor growth assays were assessed with the student’s t test. For immunohistochemistry study, the statistical significance of differences between groups was evaluated by the Mann-Whitney test or the Wilcoxon rank sum test. All statistical tests were two sided, and P values less than 0.05 were considered to be statistically significant.
Results
PTPL1 regulates breast cancer cell aggressiveness in vitro and in vivo
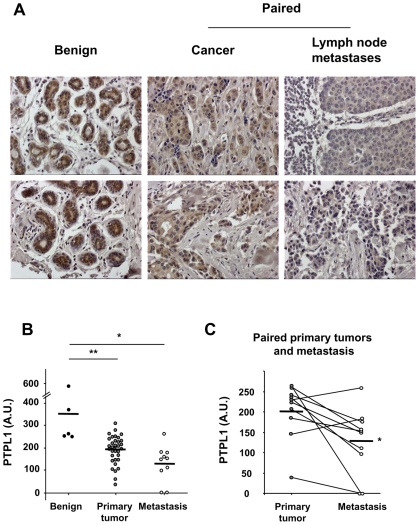
We have previously shown that the PTPL1 mRNA expression level is negatively correlated with breast tumor aggressiveness (16). In this study, we compared PTPL1 expression at the protein level by immunohistochemical analysis of tissue microarray that contained 5 benign breast tissues adjacent to breast cancer, 34 primary infiltrating ductal carcinomas and 10 matched pairs of primary tumors and lymph node metastases. Although the specimens were heterogeneous, PTPL1 expression in cancers was significantly lower than in benign tissues (Fig. 1A and B). Furthermore, lymph node metastases showed significantly lower levels of PTPL1 expression than paired primary tumors (Fig. 1A and C). In agreement with this result, highly invasive breast cancer cell lines (MDA MB 231, MDA MB 436 and BT 459) expressed lower PTPL1 levels than poorly invasive breast cancer cell lines (MCF7, T47D, ZR 75.1) (Fig. S1).
Figure 1. Evaluation of PTPL1 expression in breast cancer specimens.
A microarray of tissue sections from 5 benign tissues, 24 primary cancers and 10 paired primary cancers and lymph node metastases was immunostained with an antibody against PTPL1. A, Representative staining of two benign tissues and two paired primary cancers and lymph node metastases (magnification Gx200). B, Differences in the expression of PTPL1 between benign and cancer or metastasis groups were assessed using the Mann-Whitney test. (P=0.0004 and P=0.0013, respectively) C, Differences in the expression of PTPL1 between cancer and metastasis groups were assessed using the Wilcoxon signed rank test (P=0.014).
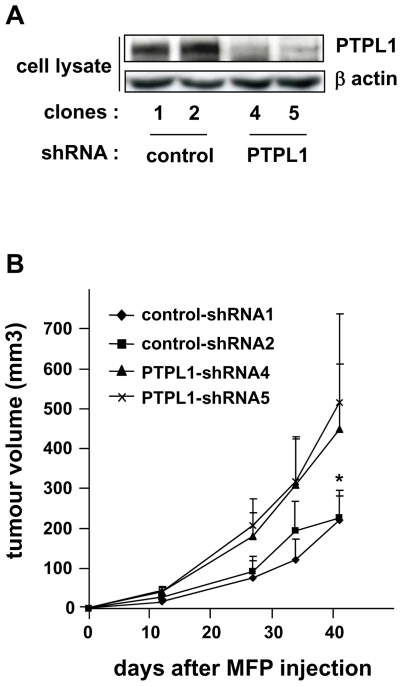
To directly address the role of PTPL1 in tumor aggressiveness, we engineered, from the non-invasive and poorly tumorigenic MCF-7 breast cancer cells, stable clones expressing a shRNA directed against PTPL1. PTPL1 expression was inhibited by approximately 95% in two different shRNA-PTPL1 clones (Fig. 2A). We investigated the effect of PTPL1 extinction on xenograft growth in nude mouse mammary fat pads. Tumor growth from the MCF-7 shRNA-PTPL1 clones was markedly higher than that of tumors from control clones (Fig. 2B), suggesting that PTPL1 actively contributes to a less aggressive phenotype.
Figure 2. Implication of PTPL1 in breast cancer cells aggressiveness.
A, Expression of PTPL1 in stable cell clones was monitored by western blot using anti-PTPL1 antibodies. The loading control was obtained by re-probing the membrane with anti-actin. B, Six-week-old mice were injected with clonal cell lines in the mammary glands. Tumor volume was measured at the indicated time, and the mean ± 95% confidence interval is shown (n=11 per group). *P<0.0012 for mean of PTPL1-shRNAs versus mean of control-shRNAs (Student’s t-test).
PTPL1 affects cell growth on extracellular matrix components, cell invasiveness and adhesion
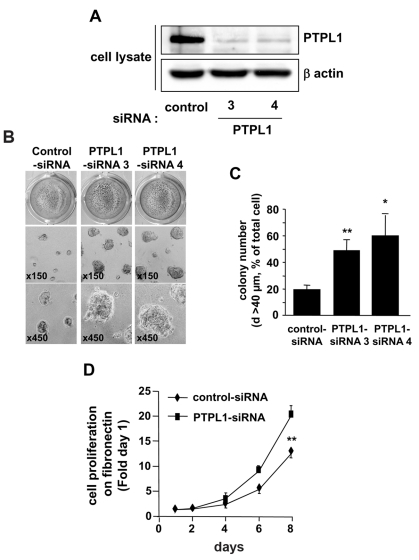
To test biological parameters associated with tumor cell aggressiveness which were quantifiable in rapid in vitro assays, we chose to downregulate PTPL1 expression by siRNA to avoid clone divergence. Five days after transfection in MCF-7 cells, PTPL1-siRNAs efficiently decreased PTPL1 expression by about 80% (Fig. 3A). We first studied the growth of cells as cell monolayers on plastic and no differences were observed between cells transfected with PTPL1-siRNAs and control cells (data not shown). Considering that PTPL1 extinction is effective in vivo, we tested its effect on MCF-7 cells grown under more physiological conditions. Inhibition of PTPL1 expression increased the size of the round colonies formed by MCF-7 cells seeded on Matrigel (a reconstituted basement membrane) (Fig. 3B). Indeed, quantification of the large colonies showed comparable increases, 2.5-fold and 3-fold, by PTPL1-siRNA3 and PTPL1-siRNA4, respectively (Fig. 3C). Furthermore, time-course experiments on cells plated on fibronectin, a basement membrane component, revealed that cells expressing PTPL1-siRNA grew exponentially with a doubling time of 28 h, which was significantly lower than that of control cells (41 h) (Fig. 3D). Similar results were obtained with two other poorly tumorigenic breast cancer cell lines (Fig. S2).
Figure 3. Cell growth on extra-cellular matrices of siRNA-PTPL1 transfected cells.
A, Five days after siRNA transfection, MCF-7 cells were lysed and cell extracts were analyzed by immunoblotting with anti-PTPL1 and anti-actin antibodies. B–C, Mock-transfected (control-siRNA) and PTPL1-siRNA (PTPL1-siRNA) cell lines were coated on Matrigel. B, Phase contrast optical photomicrographs after five days of culture and photographs after p-iodonitrotetrazolium violet staining. C, Colonies on Matrigel with a diameter greater than 40 μm were counted on four different fields per well. The data given are the mean ± s.d. of quadruplet samples. *P=0.003 and **P<0.001 versus control-siRNA. One representative experiment out of four is shown. D, One day after siRNA transfection, cells were plated on fibronectin coated wells and the DNA was quantified at different days, as described in the Materials and Methods. Cell growth was expressed relative to day one (mean ± s.d. of six wells from two independent experiments out of four). **P=0.0001 versus control-siRNA.
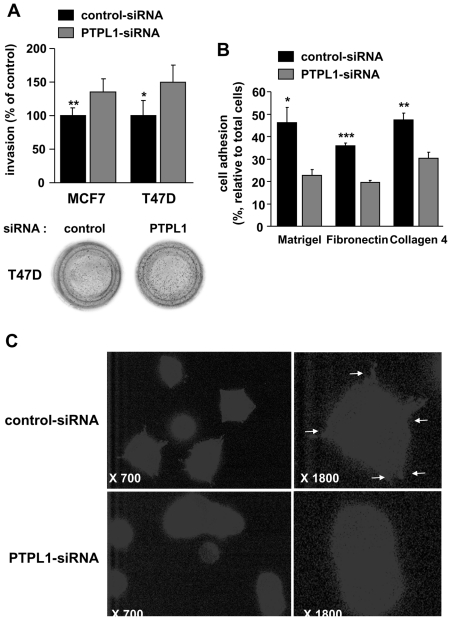
These growth effects require coating with extracellular matrix components, suggesting that PTPL1 could play a role in the signaling pathways initiated by cell-matrix interactions. We then analyzed cell adhesion and invasion, which are closely related to cell-matrix interactions. The invasive activity of MCF-7 cells was significantly increased in cells transfected with PTPL1 siRNA compared to control cells (Fig. 4A, left bars), suggesting an inhibitory role of PTPL1 on cell invasion. Because basal MCF-7 cell invasiveness is very low, we confirmed the inhibitory role of PTPL1 on another breast cancer cell line, T47D. PTPL1 extinction increased the invasiveness of these cells by 50% (Fig. 4A, right bars). We next examined the influence of PTPL1 on the adhesion of MCF-7 cells on fibronectin, Matrigel or collagen 4. PTPL1 siRNA significantly reduced the number of adherent cells by about 40% depending on the substrate (Fig. 4B). Furthermore, immunofluorescence experiments on MCF-7 cells plated on fibronectin demonstrated a drastic decrease in the number of focal adhesions after PTPL1 extinction, corroborating the influence of PTPL1 on the cell adhesion process (Fig. 4C).
Figure 4. Effect of PTPL1 downregulation on cell invasion and adhesion.
A, Two days after transfection, cells were plated onto the upper well of a Matrigel coated Transwell Boyden Chamber and allowed to migrate toward the chemoattractant 10% FBS for 48 h. The percentage of cells that migrated through Matrigel-coated filters was quantified relative to the total number of seeded cells. The data represent the percentage of cells that migrated to the lower side of the filter relative to control-siRNA cells. The results are the means ± s.d of triplicate samples from two independent experiments (MCF-7; n=6) or quadruplet samples from one experiment (T47D). *P=0.036, **P=0.003. Photographs are representative brilliant blue stained filters. B, Five days after siRNA transfection, MCF-7 cells were plated in triplicate on Matrigel, collagen 4 or fibronectin for 30min. Data represent the percentage of adherent cells compared to the total seeded cells. The results are means ± s.d of triplicate. *P=0.011, **P=0.004, ***P=0.0001. One representative experiment out of four is shown. C, Immunofluorescence of cells plated on fibronectin was performed using a phosphotyrosine antibody. Representative cells from one experiment out of two. For control and PTPL1 siRNA transfected cells, left panels correspond to a 700-fold magnification and right panels correspond to an 1800-fold magnification.
Src kinase is essential for PTPL1 biological activity and knockdown of PTPL1 stimulates the Src kinase pathway
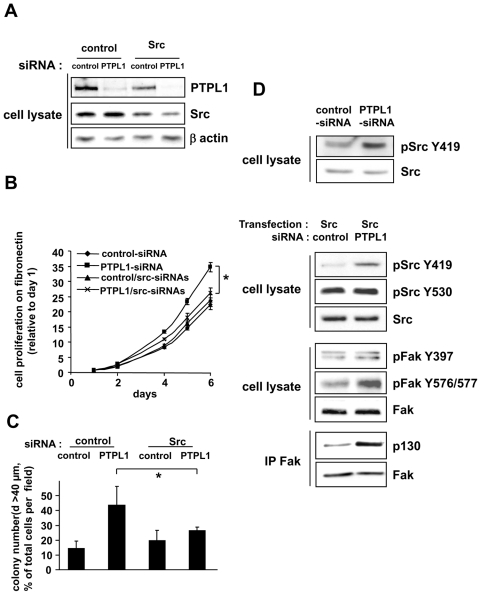
Next, we sought to determine the mechanism of these effects. Some experiments have suggested that PTPL1 might inactivate Src kinase activity in vitro (18;23). Moreover, the Src kinase family plays a major role in the regulation of focal adhesion and cell growth induced by cell-matrix interactions in normal and malignant cells (24;25). Thus, we hypothesized that PTPL1 regulates breast cancer cells aggressiveness through inactivation of the Src kinase pathway. To test this hypothesis, we downregulated Src expression in PTPL1-siRNA MCF-7 cells (Fig. 5A) and analyzed their capacity to grow on Matrigel and fibronectin. Although the silencing of Src protein did not affect cell growth, it strongly inhibited the positive effect of PTPL1 knockdown (Fig. 5B and C), suggesting that PTPL1 expression inhibits Src signaling in basal conditions.
Figure 5. Implication of Src kinase activity for PTPL1 biological activity.
A, Five days after siRNA transfection, MCF-7 cells were lysed, and cell extracts were analyzed by immunoblot using anti-Src, anti-PTPL1 and anti-actin antibodies. B–C, One day after siRNA transfection, cells were plated on B) fibronectin and C) Matrigel. Outgrowth of transfected cells was monitored as described in Figure 2 C–D. The data given are the mean ± s.d. of B) triplicate C) quadruplet samples. One representative experiment out of three is shown. B) *P≤0.003. C)*P≤0.035. D, One day after siRNA transfection, MCF-7 cells were (second, third and fourth panel) or were not (first panel) transiently transfected with the Src expression vector. The phosphorylation state of specific tyrosine residues in Src (top blot) or FAK (middle blot) was monitored by western blot using anti-pSrcY419 and anti-pSrcY530 or anti-pFakY397 and anti-pFak576/577 antibodies. Equivalent amounts of Src or FAK expression were confirmed by re-probing the blots with anti-Src and anti-FAK antibodies. For Fak/P130 association (bottom blot), lysates were immunoprecipitated with anti-Fak antibody and immunoblotted with anti-P130cas. An equal immunoprecipitation level of FAK was confirmed by direct immunoblotting of the membrane after stripping with anti-Fak antibody.
We, therefore, determined whether Src expression or activity was regulated by PTPL1 silencing. Src expression and Src Y530 phosphorylation, which locks human Src in an inactive conformation, were detectable and unaffected by PTPL1 silencing in native MCF-7 cells. SrcY419 phosphorylation, which is necessary for full Src catalytic activity, was low and only detectable after important protein loading and long E.C.L. exposure. In these conditions, SrcY419 phosphorylation was increased in the presence of PTPL1 siRNA (Fig. 5D first panel), indicating that PTPL1 inhibits Src activation. Then, we used MCF-7 cells transfected with a low amount of Src expression vector (Fig. 5D) for further experiments on Src phosphorylation and signaling. The effect of PTPL1 extinction on cell growth on Matrigel was comparable in cells overexpressing Src and in native MCF-7 cells (Fig. S3 and Fig. 3C). Whereas SrcY530 was unaffected, SrcY419 phosphorylation was strongly increased in the presence of PTPL1 siRNA (Fig. 5D), corroborating the finding that the phosphatase counteracts Src-induced effects by inhibiting Src activation. We next analyzed whether PTPL1 also affected the activation of downstream Src substrates. Upon integrin stimulation, Fak is phosphorylated on tyrosine 397 (Y397), generating a high affinity binding site for Src (25). Then, Src phosphorylates Fak on tyrosine residues, which are required for the full activation of Fak. Two of these residues are Y576/577. As expected, Fak-Y576/577 phosphorylation was increased when PTPL1 was silenced (Fig. 5D), although no changes were observed on Y397. The activated Fak/Src complex recruits proteins involved in cell adhesion, actin dynamics, proliferation and survival, such as p130cas. The proportion of p130cas co-precipitating with FAK was clearly increased in PTPL1 silenced cells (Fig. 5D).
These results suggest that, through Src activation, PTPL1 silencing increases the tyrosine phosphorylation of critical proteins involved in integrin signaling.
Src is a direct target of PTPL1
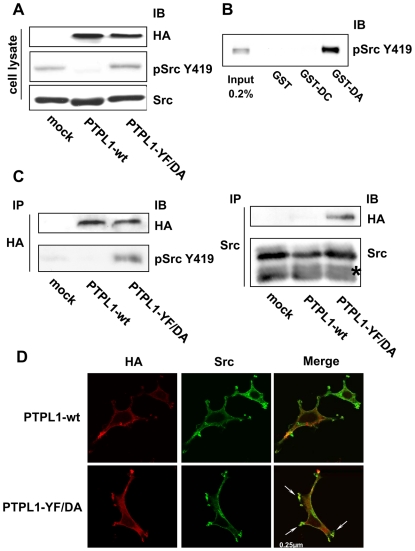
Up to now, the effect of PTPL1 on Src phosphorylation has only been described in vitro (18;23). Next, we tested whether PTPL1 overexpression was sufficient to change Src phosphorylation in cellulo. As Src kinase is mainly expressed as an inactive form in cells, we assessed PTPL1-induced phosphorylation changes in HEK293 cells overexpressing Src (to increase the amount of Src phosphorylated at Y419). Cells were cotransfected by expression vectors coding for wt PTPL1 or PTPL1-YF/DA, a catalytically inactive mutant of PTPL1. In the absence of PTPL1, Src was highly phosphorylated at Y419. This phosphorylation was abolished by the overexpression of wt PTPL1, whereas it was unaffected when the catalytically inactive PTPL1-YF/DA mutant was introduced in the cells (Fig. 6A). This result suggested that PTPL1 phosphatase activity was essential for Src hypophosphorylation.
Figure 6. Phosphorylation and localization of Src kinase in PTPL1 transfected cells.
A, HEK293 cells were transiently cotransfected with Src and HA-tagged wt PTPL1 or PTPL1-YF/DA expression vectors. Total lysates were analyzed by direct immunoblotting with anti-pSrcY419 antibody (middle). Equivalent amounts of PTPL1 and Src were confirmed by re-probing the blots with anti-HA (top) and anti-Src (bottom) antibodies. B, HEK293 cells overexpressing the Src Y530F expression vector were serum-starved and stimulated with 10 mM pervanadate for 30 min. Cells were lysed and proteins were resolved by SDS-PAGE (7.5% gel; input) or incubated with the indicated GST fusion proteins immobilized on beads. Bound materials were immunoblotted with anti-pSrcY419 antibody. C, PTPL1 was immunoprecipitated from lysates prepared from HEK293 cells transfected as described in A) using an anti-HA antibody and immunoblotted for the presence of Src tyrosine-phosphorylated protein at residue 419 with anti-pSrcY419 antibody (bottom-left). Equivalent amounts of PTPL1 were confirmed by re-probing the blots with an anti-HA antibody (top-left). Src was immunoprecipitated from the same lysates using an anti-Src antibody and immunoblotted for the presence of PTPL1 using an anti-HA antibody (top-right). An equal expression level of Src was confirmed by direct immunoblotting of lysate with anti-Src antibody (bottom-right). Asterisks indicate IgG bands. D, Cells were treated for indirect FITC localization of PTPL1 constructs with anti-HA antibody (left) and indirect TRITC localization of Src (middle). Images represent merges of horizontal confocal sections (right). Bar, 250 nm.
Today, “substrate trapping” is currently used to identify PTP substrates (26). The in vitro assay was, therefore, performed on pervanadate-treated HEK293 cell lysates overexpressing a constitutively active form of Src, SrcY530F. Lysates were incubated with either GST alone, a recombinant PTPL1 catalytic domain fused to GST (GST-wt) or the substrate trapping fusion protein GST-DA, which forms stable complexes with substrates. Whereas no protein retained by GST or GST-wt was detectable (Fig. 6B, lane 2–3), the GST-DA trapping fusion protein specifically bound Y419-phosphorylated Src (Fig. 6B, lane 4). We next analyzed whether PTPL1 forms a complex with Src in cells. In cellulo substrate trapping was done in HEK293 cells expressing the constitutively active form of Src. Src was readily coimmunoprecipitated with the substrate trapping mutant PTPL1-YF/DA (Fig. 6C, left panel lane 3). However, no complex formation was detected with wt PTPL1 or control transfectants lacking PTPL1 (Fig. 6C, left panel lane 1–2). This specific interaction between the PTPL1 substrate trapping mutant and Src was then confirmed in the reverse experiment in which the same lysates were immunoprecipitated with an anti-Src antibody. The substrate trapping mutant complexed with Src, whereas wild type phosphatase did not co-precipitate (Fig. 6C, right panel). Finally, we also studied the localization of PTPL1 and Src by confocal microscopy in HEK293 cells overexpressing Src and wt PTPL1 or PTPL1-YF/DA. PTPL1 and PTPL1-YF/DA both localized to the plasma membrane, as described previously (1). In cells overexpressing wt PTPL1, Src was detected in the cytoplasm and at the plasma membrane, leading to a partial colocalization with wt PTPL1 (Fig. 6D). In cells overexpressing PTPL1-YF/DA, Src was strongly delocalized to plasma membrane protrusions and an intense colocalization with PTPL1 was observed (Fig. 6D, arrows). These data clearly demonstrate that PTPL1 directly regulates SrcY419 phosphorylation in intact cells.
Discussion
We have recently showed that PTPL1 expression level is an independent prognostic indicator of favorable outcome for patients with breast cancer (16). In this study, we demonstrated that PTPL1 expression is decreased in breast cancers and metastases compared with benign breast tissues. Moreover, using in vitro and animal models, we evaluated the impact of PTPL1 in breast cancer cell aggressiveness and delineated the molecular mechanism responsible for this effect. We found that PTPL1 regulates the aggressiveness of a poorly tumorigenic mammary cell line because we observed that silencing of the phosphatase led to increased tumor growth in athymic mice. In line with our findings, Spanos and collaborators (27) have recently demonstrated that PTPL1 inhibition, induced either by the presence of HPV type 16 E6 or by a short hairpin RNA strategy, induces tumor growth of murine tonsil epithelial cells when it was associated with the oncogene Rasv12 (27).
Here, we show that PTPL1 induces phenotypic changes of breast cancer cells that are dependent on the cell-matrix interactions, suggesting an involvement of the signaling pathways initiated by these interactions such as integrin/Fak/Src kinase signaling (25). Silencing of PTPL1 expression is associated with increased cell proliferation when the cells are coated on extracellular matrix components, although the increase is not observed when cells are plated on plastic. We also show enhanced invasiveness associated with decreased cell adhesion of PTPL1 knockdown cells. PTPL1 associates with a number of actin-associated molecules (2), and it may play a general role in modulating cell invasion through the binding and/or dephosphorylation of substrates involved in cell adhesion and migration. Indeed, PTPL1 has been implicated in the regulation of cell invasion in ovarian cancer cells. In these studies, PTPL1 regulates cell motility by negatively acting on the phosphorylation of either TRIP6, after lysophosphatidic acid stimulation (28), or HER- 2 receptor in the HER-2-overexpressing SKOV3 cell line (29). On the other hand, the non-receptor tyrosine kinase Src alters the actin cytoskeleton and the adhesion networks that control cell migration and also transmits signals that regulate proliferation and cell survival (24;25). Thus, in human cancers, elevated Src activity has been shown to increase growth rate (30) and migratory and metastatic potential (31), while inhibiting cell-cell adhesion (32). In human breast cancers, Src activity is reported to be elevated (33). Interaction of Src with numerous breast cancer-associated growth factors and signaling pathways, such as prolactin (34), EGFR (35), ERK1/2, PI3-kinase (36) and estrogen receptor (37), supports the notion that Src activity contributes to the growth and survival of breast cancer cells. Therefore, we investigated the implication of Src signaling in the PTPL1 biological effects. We demonstrated that in MCF-7 cells interruption of Src expression inhibited PTPL1 knockdown-dependent growth induction and Src activity was increased in PTPL1 knockdown cells. There are number of candidate phosphatases for Src Y530 dephosphorylation, including cytoplasmic PTP1B (38), Shp1 (39), Shp2 (40) and transmembrane enzymes PTPα (41), PTPε (42) and PTPλ (43). Conversely, no phosphatase was shown to dephosphorylate Src Y419 in intact cells, whereas some experiments have suggested that PTPL1 might inactivate Src kinase activity in vitro (18;23). In this study, we use in cellulo dephosphorylation, co-localization and substrate trapping experiments both in vitro and in cellulo to identify the PTPL1 substrate in Src signaling. We demonstrated that PTPL1 partially co-localizes with Src and specifically dephosphorylates Src on Y419. Recently, two studies have reported interacting proteins necessary for PTPL1 action on HER2 and Src signaling. Necl-2 forms a complex with PTPL1 and HER2 allowing silencing of HER-2/HER3 signaling by PTPL1 (44). RIL forms a complex with PTPL1 and Src inducing a PTPL1-dependent Src dephosphorylation (45). In the latter case, experiments were performed in colon cancer cells in which inactivation of Src activity requires the presence of both proteins, PTPL1 and RIL (45). In agreement with Boumber et al. (46) who have shown that RIL transcription is suppressed in multiple types of human cancer cells, including breast cancer cells such as MCF-7 and T47D, no RIL expression was detected in the MCF-7 cells used in our experiments (data not shown), indicating that RIL was not implicated in PTPL1 action in our model. Moreover, the absence of interaction between Src and wild type PTPL1 in substrate trapping experiments indicated that PTPL1 and Src did not complex with any other partners, such as RIL or Necl-2, in our cell culture conditions.
In estrogen receptor positive (ER-positive) breast cancer cells, acquisition of tamoxifen resistance is associated with increased levels of Src kinase activity and a more motile and invasive phenotype (47;48). Pharmacological inhibition of Src strongly enhances the inhibitory effects of tamoxifen or estrogen deprivation on cell growth (49). Moreover, treating ER-positive breast cancer cells with a combination of a Src kinase inhibitor and tamoxifen suppresses growth and invasion and prevents the acquisition of tamoxifen-resistant growth (50). In ER-positive breast cancer cells, we have shown that PTPL1 is up-regulated by antiestrogens and has a major role in the negative effect of tamoxifen on growth factor signaling (6). Our data link PTPL1 to Src in the regulation of breast cancer aggressiveness and corroborate the hypothesis that the metastatic spread and progression of this disease can be mitigated by blocking Src activation in tamoxifen-resistant breast cancer.
In summary, by identifying Src as the PTPL1 target in signaling pathways dependent on cell-matrix interactions, we presently describe a new mechanism by which PTPL1 inhibits tumor progression that may explain the increased aggressiveness of breast tumors expressing a low level of PTPL1 (Fig. S2). This interplay between Src and PTPL1 points to the necessity for further studies on the mechanisms by which the catalytic activity of this phosphatase is regulated and reinforces the interest in new therapeutic routes combining antiestrogens and Src inhibitors.
Supplementary Material
Acknowledgments
We thank Imade Ait Arsa (IRCM, Montpellier, France) for technical assistance, Jean-Yves Cance (IRCM, Montpellier, France) for photographs and Chamroeun Sar (INM-MRI, Montpellier, France) for IHC analysis.
Funding :This work was supported by the «Institut National de la Santé et de la Recherche Médicale», the «Ligue Nationale Contre le Cancer-comité des Pyrénées Orientales», and by the «Institut National du Cancer» (grant number 0611-3D1019-34/valo, 0610-3D1616-118PL2006). M. Dromard and M. Glondu-Lassis were supported by grants from “Ligue Nationale Contre le Cancer” and the “Institut National du Cancer”, respectively.
References
- 1.Bompard G, Martin M, Roy C, Vignon F, Freiss G. Membrane targeting of protein tyrosine phosphatase PTPL1 through its FERM domain via binding to phosphatidylinositol 4,5-biphosphate. J Cell Sci. 2003;116:2519–30. doi: 10.1242/jcs.00448. [DOI] [PubMed] [Google Scholar]
- 2.Abaan OD, Toretsky JA. PTPL1: a large phosphatase with a split personality. Cancer Metastasis Rev. 2008;27:205–14. doi: 10.1007/s10555-008-9114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakahira M, Tanaka T, Robson BE, Mizgerd JP, Grusby MJ. Regulation of signal transducer and activator of transcription signaling by the tyrosine phosphatase PTP-BL. Immunity. 2007;26:163–76. doi: 10.1016/j.immuni.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Wansink DG, Peters W, Schaafsma I, et al. Mild impairment of motor nerve repair in mice lacking PTP-BL tyrosine phosphatase activity. Physiol Genomics. 2004;19:50–60. doi: 10.1152/physiolgenomics.00079.2004. [DOI] [PubMed] [Google Scholar]
- 5.Glondu-Lassis M, Dromard M, Chavey C, et al. Downregulation of protein tyrosine phosphatase PTP-BL represses adipogenesis. Int J Biochem Cell Biol. 2009;41:2173–80. doi: 10.1016/j.biocel.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Freiss G, Puech C, Vignon F. Extinction of insulin-like growth factor-I mitogenic signaling by antiestrogen-stimulated Fas-associated protein tyrosine phosphatase-1 in human breast cancer cells. Mol Endocrinol. 1998;12:568–79. doi: 10.1210/mend.12.4.0088. [DOI] [PubMed] [Google Scholar]
- 7.Bompard G, Puech C, Prebois C, Vignon F, Freiss G. Protein-tyrosine phosphatase PTPL1/FAP-1 triggers apoptosis in human breast cancer cells. J Biol Chem. 2002;277:47861–9. doi: 10.1074/jbc.M208950200. [DOI] [PubMed] [Google Scholar]
- 8.Dromard M, Bompard G, Glondu-Lassis M, et al. The Putative Tumor Suppressor Gene PTPN13/PTPL1 Induces Apoptosis through Insulin Receptor Substrate-1 Dephosphorylation. Cancer Res. 2007;67:6806–13. doi: 10.1158/0008-5472.CAN-07-0513. [DOI] [PubMed] [Google Scholar]
- 9.Freiss G, Vignon F. Protein tyrosine phosphatases and breast cancer. Crit Rev Oncol Hematol. 2004;52:9–17. doi: 10.1016/j.critrevonc.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 11.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 12.Yeh SH, Wu DC, Tsai CY, et al. Genetic characterization of fas-associated phosphatase-1 as a putative tumor suppressor gene on chromosome 4q21. 3 in hepatocellular carcinoma. Clin Cancer Res. 2006;12:1097–108. doi: 10.1158/1078-0432.CCR-05-1383. [DOI] [PubMed] [Google Scholar]
- 13.Ying J, Li H, Cui Y, et al. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20:1173–5. doi: 10.1038/sj.leu.2404193. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 15.Inazawa J, Ariyama T, Abe T, et al. PTPN13, a fas-associated protein tyrosine phosphatase, is located on the long arm of chromosome 4 at band q21. 3. Genomics. 1996;31:240–2. doi: 10.1006/geno.1996.0039. [DOI] [PubMed] [Google Scholar]
- 16.Revillion F, Puech C, Rabenoelina F, et al. Expression of the putative tumor suppressor gene PTPN13/PTPL1 is an independent prognostic marker for overall survival in breast cancer. Int J Cancer. 2009;124:638–43. doi: 10.1002/ijc.23989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimber WA, Deak M, Prescott AR, Alessi DR. Interaction of the protein tyrosine phosphatase PTPL1 with the PtdIns(3,4)P 2 binding adaptor protein TAPP1. Biochem J. 2003;(Pt) doi: 10.1042/BJ20031154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer A, Zimmer M, Erdmann KS, et al. EphrinB phosphorylation and reverse signaling: regulation by Src kinases and PTP-BL phosphatase. Mol Cell. 2002;9:725–37. doi: 10.1016/s1097-2765(02)00488-4. [DOI] [PubMed] [Google Scholar]
- 19.Murthy KK, Clark K, Fortin Y, Shen SH, Banville D. ZRP-1, a zyxin-related protein, interacts with the second PDZ domain of the cytosolic protein tyrosine phosphatase hPTP1E. J Biol Chem. 1999;274:20679–87. doi: 10.1074/jbc.274.29.20679. [DOI] [PubMed] [Google Scholar]
- 20.Saras J, Franzen P, Aspenstrom P, et al. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J Biol Chem. 1997;272:24333–8. doi: 10.1074/jbc.272.39.24333. [DOI] [PubMed] [Google Scholar]
- 21.Glondu M, Liaudet-Coopman E, Derocq D, et al. Down-regulation of cathepsin-D expression by antisense gene transfer inhibits tumor growth and experimental lung metastasis of human breast cancer cells. Oncogene. 2002;21:5127–34. doi: 10.1038/sj.onc.1205657. [DOI] [PubMed] [Google Scholar]
- 22.Vignon F, Capony F, Chambon M, et al. Autocrine growth stimulation of the MCF 7 breast cancer cells by the estrogen-regulated 52 K protein. Endocrinology. 1986;118:1537–45. doi: 10.1210/endo-118-4-1537. [DOI] [PubMed] [Google Scholar]
- 23.Superti-Furga G, Jonsson K, Courtneidge SA. A functional screen in yeast for regulators and antagonizers of heterologous protein tyrosine kinases. Nat Biotechnol. 1996;14:600–5. doi: 10.1038/nbt0596-600. [DOI] [PubMed] [Google Scholar]
- 24.Frame MC. Newest findings on the oldest oncogene; how activated src does it. J Cell Sci. 2004;117:989–98. doi: 10.1242/jcs.01111. [DOI] [PubMed] [Google Scholar]
- 25.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–23. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–5. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spanos WC, Hoover A, Harris GF, et al. The PDZ binding motif of human papillomavirus type 16 E6 induces PTPN13 loss, which allows anchorage-independent growth and synergizes with ras for invasive growth. J Virol. 2008;82:2493–500. doi: 10.1128/JVI.02188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai YJ, Lin WC, Lin FT. PTPL1/FAP-1 negatively regulates TRIP6 function in lysophosphatidic acid-induced cell migration. J Biol Chem. 2007;282:24381–7. doi: 10.1074/jbc.M701499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu JH, Chen R, Yi W, et al. Protein tyrosine phosphatase PTPN13 negatively regulates Her2/ErbB2 malignant signaling. Oncogene. 2008;27:2525–31. doi: 10.1038/sj.onc.1210922. [DOI] [PubMed] [Google Scholar]
- 30.Irby R, Mao W, Coppola D, et al. Overexpression of normal c-Src in poorly metastatic human colon cancer cells enhances primary tumor growth but not metastatic potential. Cell Growth Differ. 1997;8:1287–95. [PubMed] [Google Scholar]
- 31.Frame MC. Src in cancer: deregulation and consequences for cell behaviour. Biochim Biophys Acta. 2002;1602:114–30. doi: 10.1016/s0304-419x(02)00040-9. [DOI] [PubMed] [Google Scholar]
- 32.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–80. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 33.Reissig D, Clement J, Sanger J, et al. Elevated activity and expression of Src-family kinases in human breast carcinoma tissue versus matched non-tumor tissue. J Cancer Res Clin Oncol. 2001;127:226–30. doi: 10.1007/s004320000197. [DOI] [PubMed] [Google Scholar]
- 34.Acosta JJ, Munoz RM, Gonzalez L, et al. Src mediates prolactin-dependent proliferation of T47D and MCF7 cells via the activation of focal adhesion kinase/Erk1/2 and phosphatidylinositol 3-kinase pathways. Mol Endocrinol. 2003;17:2268–82. doi: 10.1210/me.2002-0422. [DOI] [PubMed] [Google Scholar]
- 35.Biscardi JS, Ishizawar RC, Silva CM, Parsons SJ. Tyrosine kinase signalling in breast cancer: epidermal growth factor receptor and c-Src interactions in breast cancer. Breast Cancer Res. 2000;2:203–10. doi: 10.1186/bcr55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaben AM, Saucier C, Bedin M, Redeuilh G, Mester J. Mitogenic activity of estrogens in human breast cancer cells does not rely on direct induction of mitogen-activated protein kinase/extracellularly regulated kinase or phosphatidylinositol 3-kinase. Mol Endocrinol. 2004;18:2700–13. doi: 10.1210/me.2003-0133. [DOI] [PubMed] [Google Scholar]
- 37.Migliaccio A, Di Domenico M, Castoria G, et al. Steroid receptor regulation of epidermal growth factor signaling through Src in breast and prostate cancer cells: steroid antagonist action. Cancer Res. 2005;65:10585–93. doi: 10.1158/0008-5472.CAN-05-0912. [DOI] [PubMed] [Google Scholar]
- 38.Bjorge JD, Pang A, Fujita DJ. Identification of protein-tyrosine phosphatase 1B as the major tyrosine phosphatase activity capable of dephosphorylating and activating c-Src in several human breast cancer cell lines. J Biol Chem. 2000;275:41439–46. doi: 10.1074/jbc.M004852200. [DOI] [PubMed] [Google Scholar]
- 39.Frank C, Burkhardt C, Imhof D, et al. Effective dephosphorylation of Src substrates by SHP-1. J Biol Chem. 2004;279:11375–83. doi: 10.1074/jbc.M309096200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang SQ, Yang W, Kontaridis MI, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13:341–55. doi: 10.1016/s1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 41.Ponniah S, Wang DZ, Lim KL, Pallen CJ. Targeted disruption of the tyrosine phosphatase PTPalpha leads to constitutive downregulation of the kinases Src and Fyn. Curr Biol. 1999;9:535–8. doi: 10.1016/s0960-9822(99)80238-3. [DOI] [PubMed] [Google Scholar]
- 42.Gil-Henn H, Elson A. Tyrosine phosphatase-epsilon activates Src and supports the transformed phenotype of Neu-induced mammary tumor cells. J Biol Chem. 2003;278:15579–86. doi: 10.1074/jbc.M210273200. [DOI] [PubMed] [Google Scholar]
- 43.Fang KS, Sabe H, Saito H, Hanafusa H. Comparative study of three protein-tyrosine phosphatases. Chicken protein-tyrosine phosphatase lambda dephosphorylates c-Src tyrosine 527. J Biol Chem. 1994;269:20194–200. [PubMed] [Google Scholar]
- 44.Kawano S, Ikeda W, Kishimoto M, Ogita H, Takai Y. Silencing of ErbB3/ErbB2 signaling by immunoglobulin-like Necl-2. J Biol Chem. 2009 doi: 10.1074/jbc.M109.025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Tu Y, Zhao J, Chen K, Wu C. Reversion-induced LIM interaction with Src reveals a novel Src inactivation cycle. J Cell Biol. 2009;184:785–92. doi: 10.1083/jcb.200810155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boumber YA, Kondo Y, Chen X, et al. RIL, a LIM gene on 5q31, is silenced by methylation in cancer and sensitizes cancer cells to apoptosis. Cancer Res. 2007;67:1997–2005. doi: 10.1158/0008-5472.CAN-06-3093. [DOI] [PubMed] [Google Scholar]
- 47.Planas-Silva MD, Bruggeman RD, Grenko RT, Stanley SJ. Role of c-Src and focal adhesion kinase in progression and metastasis of estrogen receptor-positive breast cancer. Biochem Biophys Res Commun. 2006;341:73–81. doi: 10.1016/j.bbrc.2005.12.164. [DOI] [PubMed] [Google Scholar]
- 48.Hiscox S, Morgan L, Green TP, et al. Elevated Src activity promotes cellular invasion and motility in tamoxifen resistant breast cancer cells. Breast Cancer Res Treat. 2006;97:263–74. doi: 10.1007/s10549-005-9120-9. [DOI] [PubMed] [Google Scholar]
- 49.Planas-Silva MD, Hamilton KN. Targeting c-Src kinase enhances tamoxifen’s inhibitory effect on cell growth by modulating expression of cell cycle and survival proteins. Cancer Chemother Pharmacol. 2007;60:535–43. doi: 10.1007/s00280-006-0398-z. [DOI] [PubMed] [Google Scholar]
- 50.Hiscox S, Jordan NJ, Smith C, et al. Dual targeting of Src and ER prevents acquired antihormone resistance in breast cancer cells. Breast Cancer Res Treat. 2009;115:57–67. doi: 10.1007/s10549-008-0058-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.