Abstract
RNA regulators in bacteria are a heterogenous group of molecules that act by various mechanisms to modulate a wide range of physiological responses. One class comprises riboswitches, which are parts of the mRNAs they regulate. These leader sequences fold into structures amenable to conformational changes upon the binding of small molecules. Riboswitches thus sense and respond to the availability of various nutrients in the cell. Other small transcripts bind to proteins, among them global regulators, and antagonize their functions. The largest and most extensively studied set of small RNA regulators act through base pairing with RNAs, usually modulating the translation and stability of mRNAs. The majority of these small RNAs regulate responses to changes in environmental conditions. Finally, a recently discovered group of RNA regulators, known as the CRISPR RNAs, contain short regions of homology to bacteriophage and plasmid sequences. CRISPR RNAs interfere with bacteriophage infection and plasmid conjugation by targeting the homologous foreign DNA through an unknown mechanism. Here we discuss what is known about these RNA regulators, as well as the many intriguing questions that remain to be addressed.
RNA molecules that act as regulators were known in bacteria for years before the first microRNAs (miRNAs) and short interfering RNAs (siRNAs) were discovered in eukaryotes. In 1981, the ~108 nucleotide RNA I was found to block ColE1 plasmid replication by base pairing with the RNA that is cleaved to produce the replication primer (Stougaard et al., 1981; Tomizawa et al., 1981). This work was followed by the 1983 discovery of a ~70 nucleotide RNA which is transcribed from the pOUT promoter of the Tn10 transposon and represses transposition by preventing translation of the transposase mRNA (Simons and Kleckner, 1983). The first chromosomally-encoded small RNA regulator, reported in 1984, was the 174 nucleotide Escherichia coli MicF RNA, which inhibits translation of the mRNA encoding the major outer membrane porin OmpF (Mizuno et al., 1984). These first small RNA regulators, and a handful of others, were identified by gel analysis due to their abundance, by multicopy phenotypes, or by serendipity (reviewed in (Wassarman et al., 1999)).
While a few bacterial RNA regulators were identified early on, their prevalence and their contributions to numerous physiological responses were not initially appreciated. In 2001–2002, four groups reported the identification of many new small RNAs through systematic computational searches for conservation and orphan promoter and terminator sequences in the intergenic regions of E. coli (reviewed in (Livny and Waldor, 2007)). Additional RNAs were discovered by direct detection using cloning-based techniques or microarrays with probes in intergenic regions (reviewed in (Altuvia, 2007)). Variations of these approaches, aided by the availability of many new bacterial genome sequences, have led to the identification of regulatory RNAs in an ever-increasing number of bacteria. Enabled by recent technical advances, including multilayered computational searches (Livny et al., 2008; Weinberg et al., 2007), deep sequencing (Sittka et al., 2008), and tiled microarrays with full genome coverage (Landt et al., 2008), hundreds of candidate regulatory RNA genes in various bacteria have now been predicted. In E. coli alone, ~80 small transcripts have been verified, increasing the total number of genes identified for this organism by 2%.
In this review, we will focus our discussion on bacterial small RNAs that act as regulators. A limited number of small RNAs carry out specific housekeeping functions, namely the 4.5S RNA component of the signal recognition particle, the RNase P RNA responsible for processing of tRNAs and other RNAs, and tmRNA, which acts as both a tRNA and mRNA to tag incompletely translated proteins for degradation and to release stalled ribosomes (reviewed in (Holbrook, 2008; Kazantsev and Pace, 2006; Moore and Sauer, 2007)). We will not discuss these RNAs further, although their actions, as well as those of some tRNAs, can have regulatory consequences.
In addition, a few defining features are worthy of mention at the outset. Riboswitches are part of the mRNA they regulate, usually found within the 5’ untranslated region (5’-UTR), and hence act in cis. Most of the regulatory RNAs that act in trans by base pairing with other RNAs are synthesized as discrete transcripts with dedicated promoter and terminator sequences. Given that the longest of these RNAs, RNAIII of Staphylococcus aureus, is still only 514 nucleotides (reviewed in (Novick and Geisinger, 2008)), the RNAs are commonly referred to as small RNAs (sRNAs). We prefer this term to “noncoding RNA”, the term frequently used in eukaryotes, since a number of the sRNAs, including RNAIII, also encode proteins. In contrast to the base pairing sRNAs, some sRNAs that modulate protein activity as well as the CRISPR RNAs are processed out of longer transcripts.
Regulatory Functions of Bacterial RNAs
Regulatory RNAs can modulate transcription, translation, mRNA stability, and DNA maintenance or silencing. They achieve these diverse outcomes through a variety of mechanisms, including changes in RNA conformation, protein binding, base pairing with other RNAs, and interactions with DNA.
Riboswitches
Perhaps the simplest bacterial RNA regulatory elements are sequences at the 5’ end of mRNAs, or less frequently at the 3’ end, that can adopt different conformations in response to environmental signals, including stalled ribosomes, uncharged tRNAs, elevated temperatures, or small molecule ligands (reviewed in (Grundy and Henkin, 2006)). These elements were first described decades ago in elegant studies characterizing transcription attenuation. In this process, stalled ribosomes lead to changes in mRNA structure, affecting transcription elongation through the formation of terminator or antiterminator structures in the mRNA. Later studies showed that sequences found in transcripts encoding tRNA synthetases, termed “Tboxes”, bind the corresponding uncharged tRNAs, and that other leader sequences, known as “RNA thermometers”, fold in a manner that is sensitive to temperature. In both of these cases, the alternate structures lead to changes in the expression of the downstream gene.
More recently, it was found that leader sequences could bind small molecules and adopt different conformations in the presence or absence of metabolites (reviewed in (Mandal and Breaker, 2004; Montange and Batey, 2008; Nudler and Mironov, 2004)). These metabolite sensors, denoted “riboswitches”, directly regulate the genes involved in the uptake and use of the metabolite. In fact, in some cases, the presence of a riboswitch upstream of an uncharacterized or mis-annotated gene has helped to clarify the physiological role of the gene product. An ever-increasing number and variety of riboswitches are being identified in bacteria, as well as in some eukaryotes. For example, as many as 2% of all Bacillus subtilis genes are regulated by riboswitches which bind metabolites ranging from flavin mononucleotide (FMN) and thiamin pyrophosphate to S-adenosylmethionine, lysine and guanine.
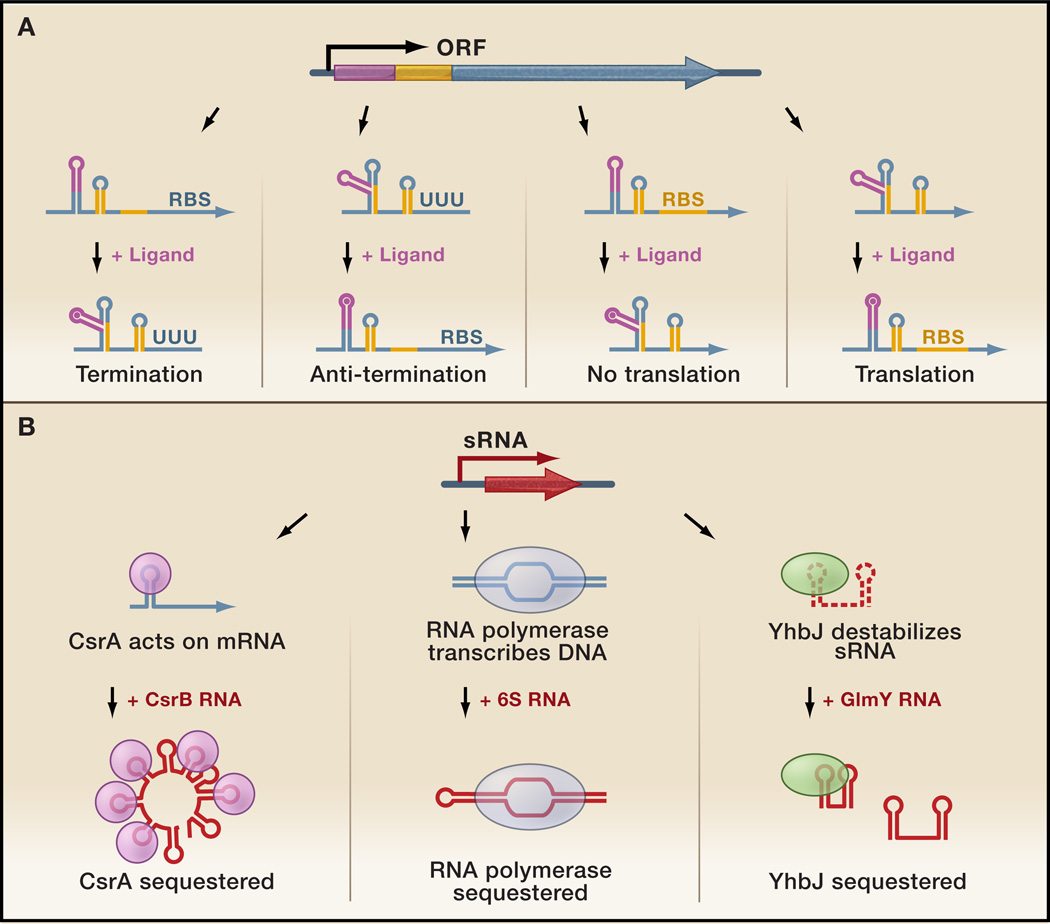
Riboswitches generally consist of two parts: the aptamer region, which binds the ligand, and the so-called expression platform, which regulates gene expression through alternative RNA structures that affect transcription or translation (reviewed in (Mandal and Breaker, 2004; Montange and Batey, 2008; Nudler and Mironov, 2004)) (Figure 1A). Upon binding of the ligand, the riboswitch changes conformation. These changes usually involve alternative hairpin structures which form or disrupt transcriptional terminators or antiterminators, or which occlude or expose ribosome binding sites (Figure 1A). In general, most riboswitches repress transcription or translation in the presence of the metabolite ligand; only a few riboswitches that activate gene expression have been characterized.
Figure 1. Gene Arrangement and Regulatory Functions of Ligand- and Protein-binding Regulatory RNAs.
(A) Riboswitches are composed of an aptamer region (pink) and an expression platform (orange) in the 5’ UTR of an mRNA (blue). Ligand binding can result in transcriptional regulation of mRNA synthesis or translational control of protein synthesis. (Left panel) In the absence of ligand, the expression platform assumes a conformation permissive of transcription—shown here as a stem-loop lacking a U-rich region—allowing synthesis of the entire mRNA. When the ligand binds the aptamer region, a conformational change leads to the disruption of this structure and the formation of an alternative hairpin followed by a string of U residues. This alternative hairpin acts as a transcriptional terminator, inhibiting gene expression. (Middle left panel) In the absence of ligand, the riboswitch initially forms a terminator. Upon ligand binding, this terminator is disrupted, allowing transcription to continue. (Middle right panel) In the absence of ligand, the ribosome binding site (RBS) is accessible, but upon ligand binding, is sequestered into an inhibitory stem-loop, preventing translation. (Right panel) In the absence of ligand, the expression platform forms a repressive secondary structure in which the ribosome binding site is occluded. When the ligand binds to the aptamer region, the ribosome binding site is released and translation can initiate. (B) Protein-binding sRNAs (red) that antagonize regulatory proteins. (Left panel) The CsrA protein (yellow circle) binds to GGA hairpins in mRNAs, altering expression from the transcripts. When CsrB RNA levels increase, the sRNA sequesters CsrA and prevents its regulatory effects. (Middle panel) Under conditions of low 6S abundance, σ70-RNA polymerase (blue oval) binds promoter DNA. When 6S levels increase, the sRNA titrates σ70- RNA polymerase away from some promoters, reducing transcription of certain housekeeping genes. (Right panel) When GlmY (shorter sRNA) levels are low, YhbJ (green oval) inactivates GlmZ (longer sRNA) by promoting its cleavage. When the GlmY competes with GlmZ for binding to YhbJ, GlmZ is stabilized.
Due to the modular nature of riboswitches, the same aptamer domain can mediate different regulatory outcomes or operate through distinct mechanisms in different contexts (reviewed in (Nudler and Mironov, 2004)). For example, the cobalamin riboswitch, which binds the coenzyme form of vitamin B12, operates by transcription termination for the btuB genes in Gram-positive bacteria but modulates translation initiation for the cob operons of Gram-negative bacteria. Some transcripts carry tandem riboswitches, which can integrate distinct physiological signals, and one notable riboswitch, the glmS leader sequence, even acts as a ribozyme to catalyze self-cleavage. Upon binding of its cofactor glucosamine-6-phosphate, the glmS riboswitch cleaves itself and inactivates the mRNA encoding the enzyme that generates glucosamine-6-phosphate, thus effecting a negative feedback loop for metabolite levels (Collins et al., 2007). In principle, riboswitches could be used in conjunction with any reaction associated with RNA, not just transcription, translation and RNA processing, but also RNA modification, localization or splicing.
Generally, the riboswitches in Gram-positive bacteria affect transcriptional attenuation, while the riboswitches in Gram-negative bacteria more frequently inhibit translation (reviewed in (Nudler and Mironov, 2004)). Possibly the preferential use of transcriptional termination in Gram-positive organisms is linked to the fact that genes are clustered together in larger biosynthetic operons where more resources would be wasted if the full-length transcript is synthesized. Gram-positive organisms also appear to rely more on cis-acting riboswitches than Gram-negative organisms, for which more trans-acting sRNA regulators are known. Research directions pursued in studies of the different organisms, however, may bias these generalizations.
sRNAs That Modulate Protein Activity
Three protein-binding sRNAs have intrinsic activity (RNase P) or contribute essential functions to a ribonucleoprotein particle (4.5S and tmRNA). In contrast, three other protein-binding sRNAs (CsrB, 6S, and GlmY) act in a regulatory fashion to antagonize the activities of their cognate proteins by mimicking the structures of other nucleic acids (Figure 1B).
The CsrB and CsrC RNAs of E. coli modulate the activity of CsrA, an RNA-binding protein that regulates carbon usage and bacterial motility upon entry into stationary phase and other nutrient-poor conditions (reviewed in (Babitzke and Romeo, 2007)). CsrA dimers bind to GGA motifs in the 5’ UTR of target mRNAs, thereby affecting the stability and/or translation of the mRNA. The CsrB and CsrC RNAs each contain multiple GGA binding sites, 22 and 13 respectively, for CsrA. Thus, when CsrB and CsrC levels increase, the sRNAs effectively sequester the CsrA protein away from mRNA leaders. Transcription of the csrB and csrC genes is induced by the BarA-UvrB two-component regulators when cells encounter nutrient poor growth conditions, though the signal for this induction is not known. The CsrB and CsrC RNAs also are regulated at the level of stability through the CsrD protein, a cyclic di-GMP binding protein, which recruits RNase E to degrade the sRNAs (Suzuki et al., 2006). CsrB and CsrC homologs (such as RsmY and RsmZ) have been found to antagonize the activities of CsrA homologs in a range of bacteria including Salmonella, Erwinia, Pseudomonas, and Vibrio where they impact secondary metabolism, quorum sensing and epithelial cell invasion (reviewed in (Lapouge et al., 2008; Lucchetti-Miganeh et al., 2008)).
The E. coli 6S RNA mimics an open promoter to bind to and sequester the σ70-containing RNA polymerase (reviewed in (Wassarman, 2007)). When 6S is abundant, especially in stationary phase, it is able to complex with much of the σ70-bound, housekeeping form of RNA polymerase, but is not associated with the σS-bound, stationary phase form of RNA polymerase (Trotochaud and Wassarman, 2005). The interaction between 6S and σ70-holoenzyme inhibits transcription from certain σ70 promoters and increases transcription from some σS regulated promoters, in part by altering the competition between σ70-and σS-holoenzyme binding to promoters. Interestingly, the 6S RNA can serve as a template for the transcription of 14–20 nucleotide product RNAs (pRNAs) by RNA polymerase, especially during outgrowth from stationary phase (Gildehaus et al., 2007; Wassarman and Saecker, 2006). In fact, it is thought that transcription from 6S when NTP concentrations increase may be a way to release σ70-RNA polymerase (Wassarman and Saecker, 2006). It is not known whether the pRNAs themselves have a function. The 6S RNA is processed out of a longer transcript and accumulates during stationary phase, but the details of this regulation have not been elucidated (reviewed in (Wassarman, 2007)). There are multiple 6S homologs in a number of organisms, including two in B. subtilis (Trotochaud and Wassarman, 2005). The roles of these homologs again are not known, but it is tempting to speculate that they inhibit the activities of alternative σ factor forms of RNA polymerase.
One additional sRNA, GlmY, has recently been proposed to have a protein-binding mode of action and is thought to function by titrating an RNA processing factor away from a homologous sRNA, GlmZ (reviewed in (Görke and Vogel, 2008)). Both GlmZ and GlmY promote accumulation of the GlmS glucosamine-6-phosphate synthase, however they do so by distinct mechanisms. The full-length GlmZ RNA base pairs with and activates translation of the glmS mRNA. Although the GlmY RNA is highly homologous to GlmZ in sequence and predicted secondary structure, GlmY lacks the region that is complementary to the glmS mRNA target and does not directly activate glmS translation. Instead, GlmY expression inhibits a GlmZ processing event that renders GlmZ unable to activate glmS translation. Although not yet conclusively shown, GlmY most likely stabilizes the full-length GlmZ by competing with GlmZ for binding to the YhbJ protein that targets GlmZ for processing. The GlmY RNA is also processed and its levels are negatively regulated by poly-adenylation (Reichenbach et al., 2008; Urban and Vogel, 2008).
CsrB RNA simulates an mRNA element, 6S imitates a DNA structure, and GlmY mimics another sRNA, raising the question as to what other molecules, nucleic acid or otherwise, might yet uncharacterized sRNAs mimic?
Cis-encoded Base Pairing sRNAs
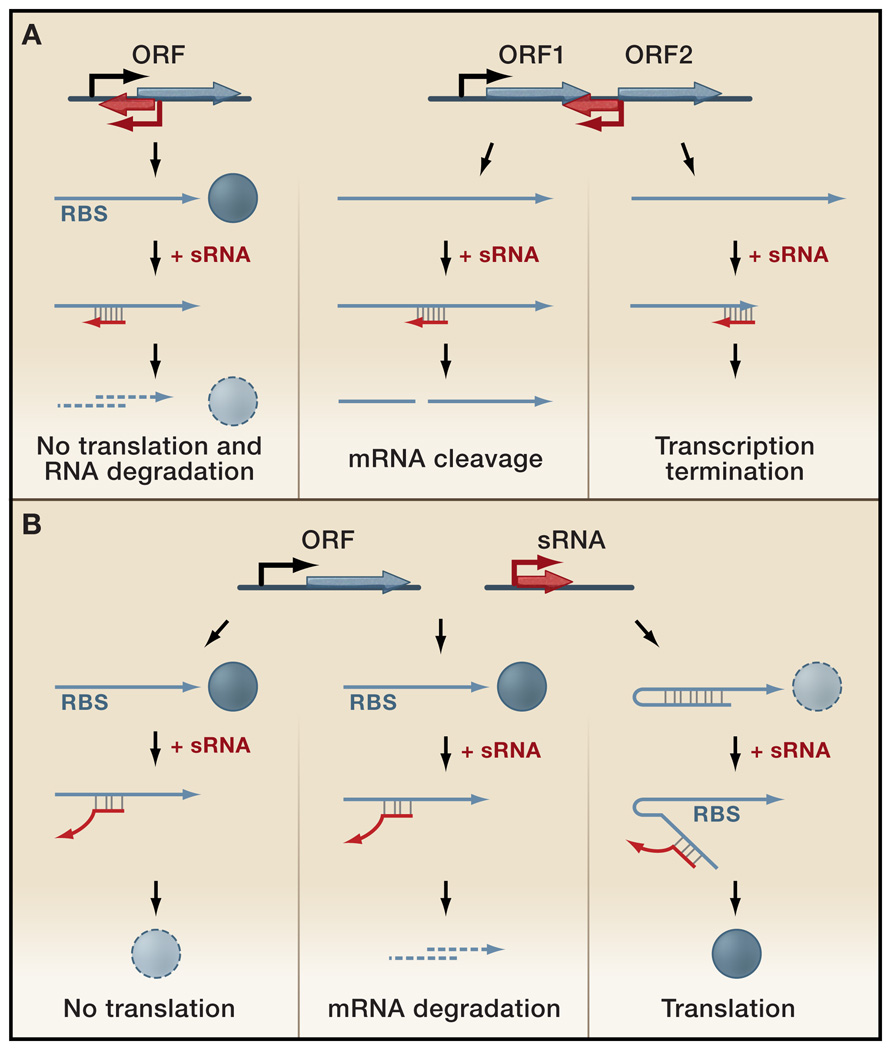
In contrast to the few known protein-binding sRNAs, most characterized sRNAs regulate gene expression by base pairing with mRNAs and fall into two broad classes: those having extensive potential for base pairing with their target RNA (Figure 2A) and those with more limited complementarity (Figure 2B). We will first focus on sRNAs that are encoded in cis on the DNA strand opposite the target RNA and share extended regions of complete complementarity with their target, often 75 nucleotides or more (Figure 2A) (reviewed in (Brantl, 2007; Wagner et al., 2002)). While the two transcripts are encoded in the same region of DNA, they are transcribed from opposite strands as discrete RNA species and function in trans as diffusible molecules. For the few cases where it has been examined, the initial interaction between the sRNA and target RNA involves only limited pairing, though the duplex can subsequently be extended. The most well-studied examples of cis-encoded antisense sRNAs reside on plasmids or other mobile genetic elements, however chromosomal versions of these sRNAs increasingly are being found.
Figure 2. Gene Arrangement and Regulatory Functions of Base Pairing Regulatory RNAs.
(A) Two possible configurations of cis-encoded antisense sRNAs (red) and their target RNAs (blue) which share extensive complementarity. (Left panel) An sRNA encoded opposite to the 5’ UTR of its target mRNA. Base pairing inhibits ribosome binding and often leads to target mRNA degradation. (Right panels) An sRNA encoded opposite to the sequence separating two genes in an operon. Base pairing of the sRNA can target RNases to the region and cause mRNA cleavage, with various regulatory effects, or the sRNA can cause transcriptional termination, leading to reduced levels of downstream genes.
(B) Genes encoding trans-encoded antisense sRNAs (red) are located separate from the genes encoding their target RNAs (blue) and only have limited complementarity. Trans-encoded sRNA can act negatively by base pairing with the 5’ UTR and blocking ribosome binding (left panel) and/or targeting the sRNA-mRNA duplex for degradation by RNases (middle panel). Trans-encoded sRNA can act positively by preventing the formation of an inhibitory structure, which sequesters the ribosome binding site (RBS) (right panel).
Most of the cis-encoded antisense sRNAs expressed from bacteriophage, plasmids and transposons function to maintain the appropriate copy number of the mobile element (reviewed in (Brantl, 2007; Wagner et al., 2002)). They achieve this through a variety of mechanisms, including inhibition of replication primer formation and transposase translation, as mentioned for plasmid ColE1 RNA I and Tn10 pOUT RNA, respectively. Another common group act as antitoxins to repress the translation of toxic proteins that kill cells from which the mobile element has been lost.
In general, the physiological roles of the cis-encoded antisense sRNAs expressed from bacterial chromosomes are less well understood. A subset promote degradation and/or repress translation of mRNAs encoding proteins that are toxic at high levels (reviewed in (Fozo et al., 2008a; Gerdes and Wagner, 2007)). In E. coli, there are also two sRNAs, IstR and OhsC, that are encoded directly adjacent to genes encoding potentially toxic proteins. Although these sRNAs are not true antisense RNAs, they do contain extended regions of perfect complementarity (19 and 23 nucleotides) with the toxin mRNAs. Interestingly, most of these sRNAs appear to be expressed constitutively. Some of the chromosomal antitoxin sRNAs are homologous to plasmid antitoxin sRNAs (for example, the Hok/Sok loci present in the E. coli chromosome) or are located in regions acquired from mobile elements (for example, the RatA RNA of B. subtilis found in a remnant of a cryptic prophage). These observations indicate that the antitoxin sRNA and corresponding toxin genes might have been acquired by horizontal transfer. The chromosomal versions may simply be non-functional remnants. However, some cis-encoded antisense antitoxin sRNAs do not have known homologs on mobile elements. In addition, given that bacteria have multiple copies of several loci, all of which are expressed in the cases examined, it is tempting to speculate that the antitoxin sRNAs-toxin proteins encoded on the chromosome provide beneficial functions (Fozo et al., 2008b). Although high levels of the toxins kill cells, more moderate levels produced from single-copy loci under inducing conditions may only slow growth. Thus one model proposes that chromosomal toxin-antitoxin modules induce slow growth or stasis under conditions of stress to allow cells time to repair damage or otherwise adjust to their environment (Kawano et al., 2007; Unoson and Wagner, 2008). Another possibility is that certain modules may be retained in bacterial chromosomes as a defense against plasmids bearing homologous modules, assuming that the chromosomal antisense sRNA can repress the expression of the plasmid-encoded toxin.
Another group of cis-encoded antisense sRNAs modulates the expression of genes in an operon. Some of these sRNAs are encoded in regions complementary to intervening sequence between ORFs (Figure 2A). For example, in E. coli, base pairing between the stationary phase-induced GadY antisense sRNA and the gadXW mRNA leads to cleavage of the duplex between the gadX and gadW genes and increased levels of a gadX transcript (Opdyke et al., 2004; Tramonti et al., 2008). For the virulence plasmid pJM1 of Vibrio anguillarum, the interaction between the RNAβ antisense sRNA and the fatDCBAangRT mRNA leads to transcription termination after the fatA gene, thus reducing expression of the downstream angRT genes (Stork et al., 2007). In Synechocystis, the iron-stress repressed IsrR antisense sRNA base pairs with sequences within isiA coding region of the isiAB transcript and leads to decreased levels of an isiA transcript (Dühring et al., 2006). In this case, it is not known whether isiB expression is also affected.
The list of cis-encoded antisense sRNAs is far from complete, especially for chromosomal versions, and other mechanisms of action are sure to be found.
Trans-encoded Base Pairing sRNAs
Another class of base pairing sRNAs is the trans-encoded sRNAs, which, in contrast to the cis-encoded antisense sRNAs, share only limited complementarity with their target mRNAs. These sRNAs regulate the translation and/or stability of target mRNAs and are, in many respects, functionally analogous to eukaryotic miRNAs (reviewed in (Aiba, 2007; Gottesman, 2005)).
The majority of the regulation by the known trans-encoded sRNAs is negative (reviewed in (Aiba, 2007; Gottesman, 2005)). Base pairing between the sRNA and its target mRNA usually leads to repression of protein levels through translational inhibition, mRNA degradation, or both (Figure 2B). The bacterial sRNAs characterized to date primarily bind to the 5’ UTR of mRNAs and most often occlude the ribosome binding site, though some sRNAs such as GcvB and RyhB inhibit translation through base pairing far upstream of the AUG of the repressed gene (Sharma et al., 2007; Vecerek et al., 2007). The sRNA-mRNA duplex is then frequently subject to degradation by RNase E. For the few characterized sRNA-mRNA interactions, the inhibition of ribosome binding is the main contributor to reduced protein levels, while the subsequent degradation of the sRNA-mRNA duplex is thought to increase the robustness of the repression and make the regulation irreversible (Morita et al., 2006). However, sRNAs can also activate expression of their target mRNAs through an anti-antisense mechanism whereby base pairing of the sRNA disrupts an inhibitory secondary structure which sequesters the ribosome binding site ((Hammer and Bassler, 2007; Prévost et al., 2007; Urban and Vogel, 2008) and reviewed in (Gottesman, 2005; Prévost et al., 2007)) (Figure 2B). Theoretically, base pairing between a trans-encoded sRNA and its target could promote transcription termination or antitermination, as has been found for some cis-encoded sRNAs, or alter mRNA stability through changes in poly-adenylation.
For trans-encoded sRNAs, there is little correlation between the chromosomal location of the sRNA gene and the target mRNA gene. In fact, each trans-encoded sRNA typically base pairs with multiple mRNAs (reviewed in (Gottesman, 2005; Prévost et al., 2007)). The capacity for multiple base pairing interactions results from the fact that trans-encoded sRNAs make more limited contacts with their target mRNAs in discontinuous patches, rather than extended stretches of perfect complementarity, as for cis-encoded antisense sRNAs. The region of potential base pairing between trans-encoded sRNAs and target mRNAs typically encompasses ~10–25 nucleotides, but in all cases where it has been examined only a core of the nucleotides seem to be critical for regulation. For example, although the SgrS sRNA has the potential to form 23 base pairs with the ptsG mRNA across a stretch of 32 nucleotides, only four single mutations in SgrS significantly affected downregulation of ptsG (Kawamoto et al., 2006).
In many cases, the RNA chaperone Hfq is required for trans-encoded sRNA-mediated regulation, presumably to facilitate RNA-RNA interactions due to limited complementarity between the sRNA and target mRNA (reviewed in (Aiba, 2007; Brennan and Link, 2007; Valentin-Hansen et al., 2004)). The hexameric Hfq ring, which is homologous to Sm and Sm-like proteins involved in splicing and mRNA decay in eukaryotes, may actively remodel the RNAs to melt inhibitory secondary structures. Hfq also may serve passively as a platform to allow sRNAs and mRNAs to sample potential complementarity, effectively increasing the local concentrations of sRNAs and mRNAs. It should be noted that when the E. coli SgrS RNA is pre-annealed with the ptsG mRNA in vitro, the Hfq protein is no longer required (Maki et al., 2008). However, in vivo in E. coli, sRNAs no longer regulate their target mRNAs in hfq mutant strains, and all trans-encoded base pairing sRNAs examined to date co-immunoprecipitate with Hfq. In fact, enrichment of sRNAs by co-immunoprecipitation with Hfq proved to be a fruitful approach to identify and validate novel sRNAs in E. coli (Zhang et al., 2003) and has been extended to other bacteria, such as S. typhimurium (Sittka et al., 2008).
Beyond facilitating base pairing, Hfq contributes to sRNA regulation through modulating sRNA levels (reviewed in (Aiba, 2007; Brennan and Link, 2007; Valentin-Hansen et al., 2004)). Somewhat counterintuitively, most E. coli sRNAs are less stable in the absence of Hfq, presumably because Hfq protects sRNAs from degradation in the absence of base pairing with mRNAs. Once base paired with target mRNAs, many of the known sRNA-mRNA pairs are subject to degradation by RNase E, and Hfq may also serve to recruit RNA degradation machinery through its interactions with RNase E and other components of the degradosome. In addition, competition between sRNAs for binding to Hfq may be a factor controlling sRNA activity in vivo.
Although all characterized E. coli trans-encoded sRNAs require Hfq for regulation of their targets, the need for an RNA chaperone may not be universal. For example, VrrA RNA repression of OmpA protein expression in V. cholerae is not eliminated in hfq mutant cells, though the extent of repression is higher in cells expressing Hfq (Song et al., 2008). In general, longer stretches of base pairing, as is the case for the cis-encoded antisense sRNAs that usually do not require Hfq for function, and/or high concentrations of the sRNA may obviate a chaperone requirement.
In contrast to cis-encoded sRNAs, several of which are expressed constitutively, most of the trans-encoded sRNAs are synthesized under very specific growth conditions. In E. coli for example, these regulatory RNAs are induced by low iron (Fur-repressed RyhB), oxidative stress (OxyR-activated OxyS), outer membrane stress (σE-induced MicA and RybB), elevated glycine (GcvA-induced GcvB), changes in glucose concentration (CRP-repressed Spot42 and CRP-activated CyaR), and elevated glucose-phosphate levels (SgrR-activated SgrS) ((De Lay and Gottesman, 2008; Johansen et al., 2008; Urbanowski et al., 2000) and reviewed in (Görke and Vogel, 2008; Gottesman, 2005)). In fact, it is possible that every major transcription factor in E. coli may control the expression of one or more sRNA regulators. It is also noteworthy that a number of the sRNAs are encoded adjacent to the gene encoding their transcription regulator, including E. coli OxyR-OxyS, GcvA-GcvB, and SgrR-SgrS.
The fact that a given base pairing sRNA often regulates multiple targets means that a single sRNA can globally modulate a particular physiological response, in much the same manner as a transcription factor, but at the post-transcriptional level (reviewed in (Bejerano-Sagie and Xavier, 2007; Massé et al., 2007; Valentin-Hansen et al., 2007)). Well-characterized regulatory effects of these sRNAs include the down regulation of iron-sulfur cluster containing enzymes under conditions of low iron (E. coli RyhB), repression of outer membrane porin proteins under conditions of membrane stress (E. coli MicA and RybB), and repression of quorum sensing at low cell density (Vibrio Qrr). The fact that direct or indirect negative feedback regulation is observed for a number of sRNAs emphasizes that sRNAs are integrated into regulatory circuits. In E. coli for example, ryhB is repressed when iron is released after RyhB down-regulates iron-sulfur enzymes (Massé et al., 2005), and micA and rybB are repressed when membrane stress is relieved upon their down-regulation of outer membrane porins (Johansen et al., 2006; Thompson et al., 2007). As another example, the Qrr sRNAs in Vibrio base pair with and inhibit expression of the mRNAs encoding the transcription factors responsible for the activation of the qrr genes (Svenningsen et al., 2008; Tu et al., 2008).
CRISPR RNAs
A unique class of recently discovered regulatory RNAs is the CRISPR RNAs, which provide resistance to bacteriophage (reviewed in (Sorek et al., 2008)) and prevent plasmid conjugation (Marraffini and Sontheimer, 2008). CRISPR systems share certain similarities with eukaryotic siRNA-driven gene silencing, although they exhibit distinct features as well, and present an exciting new arena of RNA research. The CRISPR sequences have been found in ~40% of bacteria and ~90% of archaea sequenced to date (Sorek et al., 2008), emphasizing their wide-ranging importance.
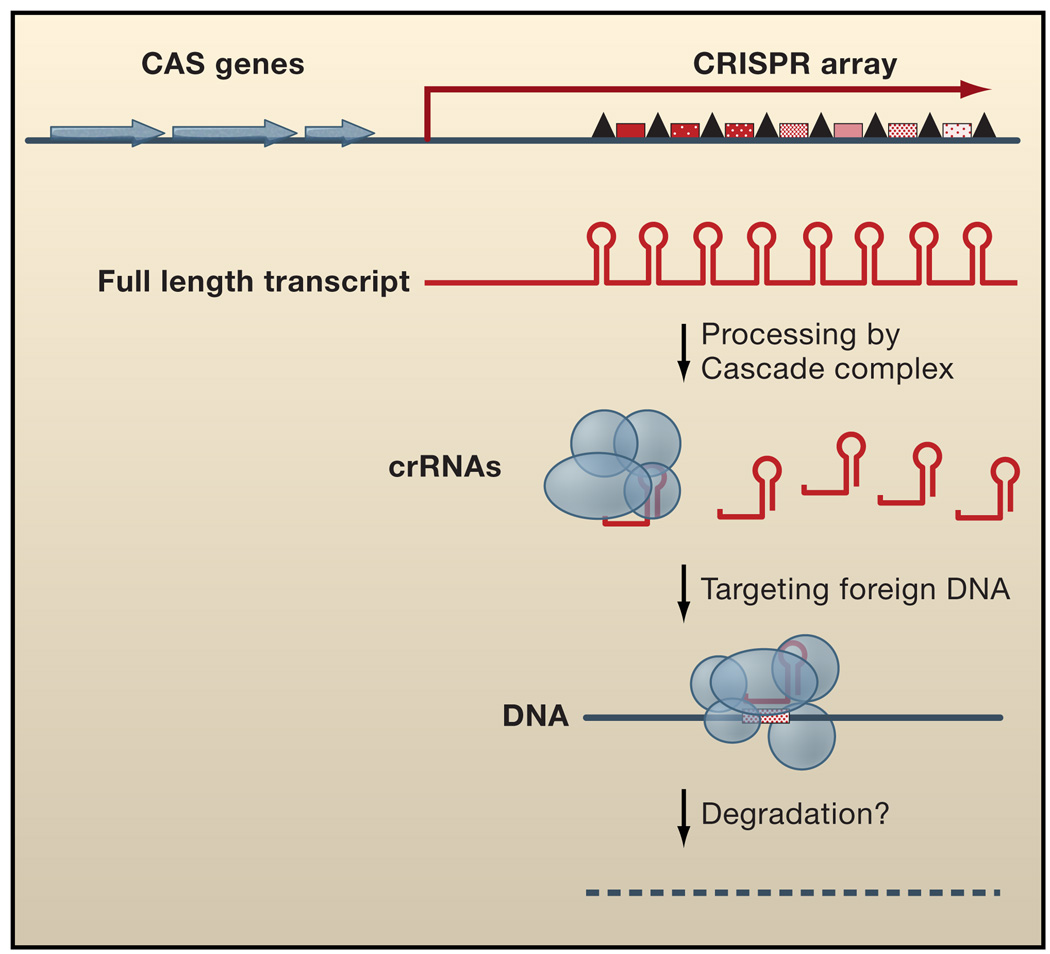
CRISPR sequences (Clustered Regularly Interspaced Short Palindromic Repeats) are highly variable DNA regions which consist of a ~550 bp leader sequence followed by a series of repeat-spacer units (Figure 3) (reviewed in (Sorek et al., 2008)). The repeated DNA can vary from 24 to 47 base pairs, but the same repeat sequence usually appears in each unit in a given CRISPR array, and is repeated two to 249 times. The repeat sequences diverge significantly between bacteria, but can be grouped into 12 major types and often contain a short 5–7 base pair palindrome. Unlike other repeated sequences in bacterial chromosomes, the CRISPR repeats are regularly interspersed with unique spacers of 26 to 72 base pairs; these spacers are not typically repeated in a given CRISPR array. Although the repeats can be similar between species, the spacers between the repeats are not conserved at all, often varying even between strains, and are most often found to be homologous to DNA from phages and plasmids, an observation that was initially perplexing.
Figure 3. Gene Arrangement and Regulatory Functions of CRISPR RNAs.
CRISPR arrays are comprised of DNA repeats (black triangles) separated by unique spacers (red hatched boxes). CAS genes (blue) which encode proteins that function in CRISPR RNA processing and/or DNA silencing are located nearby. The CRISPR arrays are initially transcribed as a long RNA, which is subsequently processed by the Cascade complex (blue circles and ovals) to individual repeat-spacer units, called crRNAs. These crRNAs target foreign DNA through an unknown mechanism likely involving other CAS proteins and the degradation of the exogenous DNA.
Adjacent to the CRISPR DNA array are several CRISPR-associated (CAS) genes (reviewed in (Sorek et al., 2008)). Two to six core CAS genes seem to be associated with most CRISPR systems, but different CRISPR subtypes also have specific CAS genes encoded in the flanking region. Other CAS genes, that are never present in strains lacking the repeats, may be found in genomic locations distant from the CRISPR region(s). The molecular functions of the CAS proteins are still mostly obscure, but they often contain RNA- or DNA-binding domains, helicase motifs, and endo- or exonuclease domains.
After the initial report of CRISPR sequences in 1989, several different hypotheses were advanced as to possible functions of these repeats (reviewed in (Sorek et al., 2008)). The proposal that CRISPRs confer resistance to phages came in 2005 with findings that the spacers often contain homology to phage or plasmids. Another major advance was the discovery that the CRISPR DNA arrays are transcribed in bacteria (Brouns et al., 2008) and archaea (Tang et al., 2002; Tang et al., 2005). The full-length CRISPR RNA initially extends the length of the entire array, but is subsequently processed into shorter fragments the size of a single repeat-spacer unit. Recently, it was shown that the E. coli K12 CasA-E proteins associate to form a complex termed Cascade, for CRISPR-Associated Complex for Antiviral Defense (Brouns et al., 2008). The CasE protein within the Cascade complex is responsible for processing of the full-length CRISPR RNA transcript.
Importantly, it was demonstrated that new spacers corresponding to phage sequences are integrated into existing CRISPR arrays during phage infection and that these new spacers confer resistance to subsequent infections with the cognate phage, or other phage bearing the same sequence (Barrangou et al., 2007). The new spacers are inserted at the beginning of the array, such that the 5’ end of the CRISPR region is hypervariable between strains and conveys information about the most recent phage infections, while the 3’ end spacers are consequences of more ancient infections. Single nucleotide point mutations in the bacterial spacers or the phage genome abolish phage resistance and, further, introduction of novel phage sequences as spacers in engineered CRISPR arrays provides de novo immunity to bacteria that have never encountered this phage. Similar observations were recently made for spacers found to correspond to sequences present on conjugative plasmids (Marraffini and Sontheimer, 2008).
These findings, together with the observation that some CAS genes encode proteins with functions potentially analagous to eukaryotic RNAi enzymes (Makarova et al., 2006), have led to a model for CRISPR RNA function (Figure 3). The CRISPR DNA array is transcribed into a long RNA, which is processed by the Cascade complex of CAS proteins into a single repeat-spacer unit known as a crRNA (Brouns et al., 2008). The crRNAs, which are single-stranded unlike double-stranded siRNAs, are retained in the Cascade complex (Brouns et al., 2008). By analogy with eukaryotic RNAi systems, Cascade or other CAS effector proteins may then direct base pairing of the crRNA spacer sequence with phage or plasmid nucleic acid targets. Until recently, it was not known whether the crRNAs would target DNA or RNA, but CRISPR spacers generated from both strands of phage genes can effectively confer phage resistance (Barrangou et al., 2007; Brouns et al., 2008). In addition, the insertion of an intron into the target gene DNA in a conjugative plasmid abolishes interference by crRNAs, even though the uninterrupted target sequence is regenerated in the spliced mRNA (Marraffini and Sontheimer, 2008). These results all point to DNA as the direct target, but how the crRNAs interact with the DNA and what occurs subsequently are still unknown. Further studies addressing the details of the molecular mechanism behind CRISPR RNA-mediated “silencing” of foreign DNA and how new spacers are selected and then acquired are eagerly anticipated and will provide further insight into the similarities and differences with the eukaryotic RNAi machinery.
The CRISPR system has broad evolutionary implications. The extreme variability of CRISPR arrays between organisms and even strains of the same species provides useful tools for researchers to genotype strains and to study horizontal gene transfer and micro-evolution (reviewed in (Sorek et al., 2008)). The CRISPR loci record the history of recent phage infection and allow differentiation between strains of the same species. This property can be used to identify pathogenic bacterial strains and track disease progression world-wide, as well as to monitor the population dynamics of non-pathogenic bacteria (Horvath et al., 2008). Additionally, the presence of phage sequences within the CRISPR arrays that confer resistance against infection provide a strong selective pressure for the mutation of phage genomes and may partially underlie the rapid phage mutation rate (Andersson and Banfield, 2008).
Dual Function RNAs
The distinctions between some of the categories of RNA regulators discussed above as well as between the RNA regulators and other RNAs can be blurry. For example, a few of the trans-encoded base pairing sRNAs encode proteins in addition to base pairing with target mRNAs. The S. aureus RNAIII has been shown to base pair with mRNAs encoding virulence factors and a transcription factor (Boisset et al., 2007), but also encodes a 26 amino acid δ-hemolysin peptide. Similarly, the E. coli SgrS RNA, which blocks translation of the ptsG mRNA encoding a sugar-phosphate transporter, is translated to produce the 43 amino acid SgrT protein (Wadler and Vanderpool, 2007). In this case, the SgrT protein is thought to reinforce the regulation exerted by SgrS by independently down-regulating glucose uptake through direct or indirect inhibition of the PtsG protein. We predict that other regulatory sRNAs will be found to encode small proteins and that conversely some mRNAs encoding small proteins will be found to have additional roles as sRNA regulators. It also deserves mention that some of the cis-encoded antisense sRNAs, in addition to regulating their cognate sense mRNA, may base pair with other mRNAs via limited complementarity or, in independent roles, bind proteins to affect other functions. Similarly, while riboswitches are synthesized as part of an mRNA, the small transcripts that are generated by transcription attenuation or autocleavage potentially could go on to perform other functions as their own entities.
Factors Influencing Regulation by RNAs
While there has been a great explosion in the discovery and characterization of RNA regulators in the past ten years, a number of critical questions about their regulatory mechanisms remain to be answered.
RNA Structures, Levels, and Localization
What are the structures of the RNAs and how do they impact ligand, protein and mRNA binding? Three-dimensional structures for several riboswitches, both in the presence and absence of their respective ligands, have been solved in recent years (Montange and Batey, 2008). These studies have shown that some riboswitches have a single, localized ligand-binding pocket. In these cases, the conformational changes induced by ligand binding are confined to a small region. In other riboswitches, the ligand-binding site is comprised of at least two distinct sites, such that ligand binding results in more substantial changes in the global tertiary structure. In contrast, no three-dimensional structures have been solved for bacterial sRNAs. In fact, the secondary structures for only a limited number of sRNAs have been probed experimentally. Another generally unknown quantity, which has important implications for how an RNA interacts with other molecules, is the concentration of the RNA. After induction, the OxyS RNA has been estimated to be present at 4,500 molecules per cell (Altuvia et al., 1997), but it is not known whether this is typical for other sRNAs and whether all of the sRNA molecules are active. Do nucleotide modifications or metabolite binding alter the abundance or activities of any of the sRNAs? It is also intriguing to ask whether any of the regulatory RNAs show specific subcellular localization or are even secreted. In eukaryotes, localization of regulatory RNAs to specific subcellular structures, such as P bodies and Cajal bodies, is connected to their functions (reviewed in (Pontes and Pikaard, 2008)). It is plausible that subcellular localization similarly impacts regulatory RNA function in bacteria. In support of this idea, RNase E has been found to bind membranes in vitro (Khemici et al., 2008), and membrane targeting of the ptsG mRNA-encoded protein is required for efficient SgrS sRNA repression of this transcript (Kawamoto et al., 2005). Another attractive, but untested, hypothesis is that bacterial RNAs might be secreted into a host cell where they could modulate eukaryotic cell functions.
Proteins Involved
What proteins are associated with regulatory RNAs and how do the proteins impact the actions of the RNAs? So far much of the attention has been focused on the RNA chaperone Hfq. Even so, the details of how this protein binds to sRNAs and impacts their functions are murky. For example, structural and mutational studies indicate that both faces of the donut-like Hfq hexamer can make contacts with RNA (reviewed in (Aiba, 2007; Brennan and Link, 2007)), but it is not clear whether the sRNA and mRNA bind both faces simultaneously, whether the sRNA and mRNA bind particular faces, and whether base pairing is facilitated by changes in RNA structure or proximity between the two RNAs or both. The Hfq protein has been shown to copurify with the ribosomal protein S1, components of the RNase E degradosome, and polynucleotide phosphorylase (Mohanty et al., 2004; Morita et al., 2005; Sukhodolets and Garges, 2003), among others, but these are all abundant RNA-binding proteins and the in vivo relevance of these interactions is poorly understood. In addition, only half of all sequenced Gram-negative and Gram-positive species and one archaeon have Hfq homologs (reviewed in (Valentin-Hansen et al., 2004)). Do other proteins substitute for Hfq in the organisms that do not have homologs, or does base pairing between sRNAs and their target mRNAs not require an RNA chaperone in these cases?
It is likely still other proteins that act on or in conjuction with the regulatory RNAs remain to be discovered. The RNase E and RNase III endonucleases are known to cleave base pairing sRNAs and their targets (Viegas et al., 2007), but these may not be the only ribonucleases to degrade the RNAs. Pull-down experiments with tagged sRNAs indicate that other proteins, such as RNA polymerase (Windbichler et al., 2008), also bind the RNA regulators, but again the physiological relevance of this interaction is not known. In addition, genetic studies hint at the involvement of proteins such as YhbJ, which antagonizes GlmY and GlmZ activity, though the activity of this protein is still mysterious (Kalamorz et al., 2007; Urban and Vogel, 2008).
Requirements for Productive Base Pairing
What are the rules for productive base pairing? Trans-encoded sRNAs bind to their target mRNAs using discontiguous and imperfect base pairing, of which often only a core set of interactions is essential, stimulating questions as to how specificity between sRNAs and mRNAs is imparted and how such limited pairing can cause translation inhibition or RNA degradation. Several algorithms for the predictions of base pairing targets for trans-encoded sRNAs have been developed ((Tjaden, 2008) and reviewed in (Pichon and Felden, 2008; Vogel and Wagner, 2007)). However, the accuracy of these predictions has been varied. For some sRNAs, such as RyhB and GcvB, there are distinct conserved single-stranded regions, which appear to be required for base pairing with most targets and are associated with more accurate predictions (Sharma et al., 2007; Tjaden et al., 2006). For other sRNAs such as OmrA and OmrB, few known targets were predicted in initial searches (Tjaden et al., 2006). Mutational studies to define the base pairing interactions with known OmrA and OmrB targets (Guillier and Gottesman, 2008) highlight possible impediments to computational predictions. These can include the lack of knowledge about the sRNA domains required for base pairing, limited base pairing interactions, and base pairing to mRNA regions outside the immediate vicinity of the ribosome binding site. Recent systematic analysis indicates sRNAs can block translation by pairing with sequences in the coding region, as far downstream as the fifth codon (Bouvier et al., 2008). Other factors such as the position of Hfq binding and the secondary structures of both the mRNA and sRNA are also likely to impact base pairing in ways that have not been formalized. In vitro studies exploring the role of Hfq in facilitating the pairing between the RprA and DsrA RNAs and the rpoS mRNA show that binding between Hfq, the mRNA and the sRNAs is clearly influenced by what portion of the rpoS 5’ leader is assayed (Soper and Woodson, 2008; Updegrove et al., 2008). With an increasing number of validated targets that can serve as training sets, the ability to accurately predict targets should significantly improve.
As with eukaryotic miRNAs and siRNAs, there may be mechanistic differences between the trans- and cis-encoded base pairing sRNAs based on their different properties. Trans-encoded sRNAs, which have imperfect base pairing with their targets like miRNAs, often interact with Hfq. In contrast, cis-encoded sRNAs, which have complete complementarity with targets like siRNAs, do not appear to require Hfq, but tend to be more structured and may use other factors to aid in base pairing. These differences may have broader implications for the types of targets regulated, the nature of the proteins required, as well as the mechanistic details of base pairing.
New Mechanisms of Action
What novel mechanisms of action remain to be uncovered? Most sRNAs characterized to date base pair in the 5’ UTR of target mRNAs near the ribosome binding site, however other locations for base pairing and consequent mechanisms of regulation are possible. Only a few bacterial ribozymes have been described. Will other sRNAs or riboswitches be found to have enzymatic activity? As already alluded to, the mechanism of crRNA action in targeting and interfering with DNA is not understood. Completely novel mechanisms may be revealed by further studies of the CRISPR sequences. Finally, nearly a third of the E. coli sRNAs identified to date, and the vast majority of those in other organisms, have yet to be characterized in significant detail. These too may have unanticipated roles and modes of action.
Physiological Roles of Regulatory RNAs
In addition to further exploring the mechanisms by which riboswitches, sRNAs and crRNAs act, it is worth reflecting on what is known, as well as what is not understood, about the physiological roles of these regulators.
Association with Specific Responses
A number of themes are emerging with respect to the physiological roles of riboswitches and sRNAs. In general terms, riboswitches, protein binding sRNAs, trans-encoded base pairing sRNAs and some cis-encoding base pairing sRNAs mediate responses to changing environmental conditions by modulating metabolic pathways or stress responses. Riboswitches and T-boxes tend to regulate biosynthetic genes, as these elements directly sense the concentrations of various metabolites, while some RNA thermometers, such as the 5’-UTR of the mRNA encoding the heat shock sigma factor σ32 (Morita et al., 1999), control transcriptional regulators. The CsrB and 6S families of sRNAs also control the expression of large numbers of genes in response to decreases in nutrient availability by repressing the activities of global regulators. The trans-encoded base pairing sRNAs mostly contribute to the ability to survive various environmental insults by modulating the translation of regulators or repressing the synthesis of unneeded proteins. In particular, it is intriguing that a disproportionate number of trans-encoded sRNAs regulate outer membrane proteins (MicA, MicC, MicF, RybB, CyaR, OmrA and OmrB) or transporters (SgrS, RydC, GcvB). Other pervasive themes include RNA-mediated regulation of iron metabolism, not only in bacteria but also in eukaryotes, as well as RNA regulators of quorum sensing.
Pathogenesis presents a set of behaviors one might expect to be regulated by sRNAs since bacterial infections involve multiple rounds of rapid and coordinated responses to changing conditions. The central role of sRNAs in modulating the levels of outer membrane proteins, which are key targets for the immune system, as well as other responses important for survival under conditions found in host cells, such as altered iron levels, also implicates these RNA regulators in bacterial survival in host cells. Indeed, although these studies are still at the early stages, several sRNAs have been shown to alter infection. These include members of the CsrB family of sRNAs in Salmonella, Erwinia, Yersinia, Vibrio and Pseudomonads which bind to and antagonize CsrA family proteins that are global regulators of virulence genes; RyhB of Shigella which represses a transcriptional activator of virulence genes; RNAIII of Staphylococcus which both base pairs with mRNAs encoding virulence factors and encodes the δ-hemolysin peptide; and the Qrr sRNAs of Vibrio which regulate quorum sensing ((Heroven et al., 2008; Murphy and Payne, 2007) and reviewed in (Romby et al., 2006; Toledo-Arana et al., 2007)). hfq mutants of a wide range of bacteria also show reduced virulence (reviewed in (Romby et al., 2006; Toledo-Arana et al., 2007)). Some sRNAs, such as a number of sRNAs encoded in Salmonella and Staphylococcus pathogenicity islands, show differential expression under pathogenic conditions (Padalon-Brauch et al., 2008; Pfeiffer et al., 2007; Pichon and Felden, 2005). Other sRNAs, such as five in Listeria monocytogenes, are specific to pathogenic strains (Mandin et al., 2007). Finally, thermosensors and riboswitches can have roles in as regulators of pathogenesis, upregulating virulence genes upon increased temperature encountered in host cells or upon binding signals such as the “second messenger” cyclic di-GMP (Johansson et al., 2002; Sudarsan et al., 2008). Further studies of these and other pathogenesis-associated regulatory RNAs could lead to opportunities for interfering with disease.
A subset of the cis-encoded antisense sRNAs expressed from bacterial chromosomes act as antitoxins but their physiological roles are not clear. They may also be involved in altering cell metabolism in response to various stresses enabling survival. Alternatively, they may play a role in protecting against foreign DNA. This is clearly the function of CRISPR RNAs, which have been demonstrated to repress bacteriophage and plasmid entry into the cell, and in principle could be used to silence genes from other mobile elements.
Physiological Roles of Multiple Copies
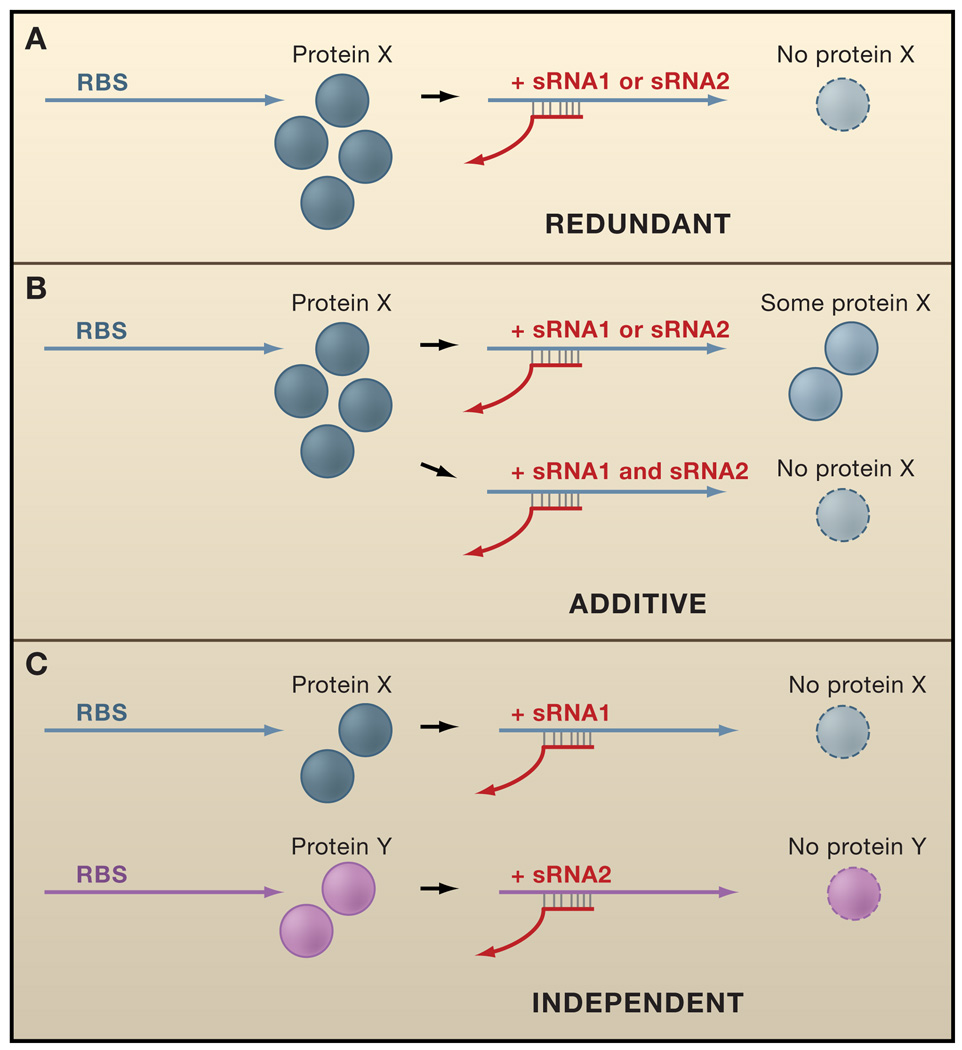
Some sRNAs including OmrA/OmrB, Prr1/Prr2, Qrr1–5, 6S homologs, CsrB homologs, GlmY/GlmZ, and several toxin-antitoxin modules are present in multiple copies in a given bacterium. Although the physiological advantages of the repeated sRNA genes are only understood in a subset of cases, multiple copies can have several different roles (Figure 4).
Figure 4. Possible Roles of Duplicated RNA Genes.
Two homologous sRNAs (red) can act in different manners to regulate mRNAs (blue and purple) and correspondingly alter protein levels (blue and purple circles).
(A) Redundant functions. Homologous sRNAs may target the same mRNAs.
(B) Additive functions. Multiple sRNAs may precisely control the levels of regulated proteins by each binding to a subset of the target mRNAs. Since relative levels of sRNAs and mRNAs are critical to the effectiveness of regulation, altering the amount of repeated sRNAs can finetune the stability and/or translation of the target mRNAs.
(C) Independent functions. Similar sRNAs may use unique sequences to regulate distinct mRNA targets.
Firstly, homologous RNAs can act redundantly, serving as back ups in critical pathways or to increase the sensitivity of a response. In V. cholerae, any single Qrr RNA is sufficient to repress quorum sensing by down regulating the HapR transcription factor, and the deletion of all four qrr genes is required to constitutively activate the quorum sensing behaviors (Lenz et al., 2004). Since the effectiveness of sRNA regulation is directly related to their abundance relative to mRNA targets, this redundancy has been proposed to permit an ultrasensitive, switch-like response for quorum sensing in V. cholerae and may help amplify a small input signal to achieve a large output.
Secondly, repeated RNAs can act additively, as in the case of the V. harveyi Qrr sRNAs (Tu and Bassler, 2007). In this case, the five qrr genes have divergent promoter regions and are differentially expressed, suggesting each Qrr sRNA may respond to different metabolic indicators to integrate various environmental signals. Deletion of individual Qrr genes affects the extent of quorum sensing behaviors, indicating they do not act redundantly. Rather, the total amount of Qrr sRNAs in V. harveyi produces distinct levels of regulated genes, such that altering the abundance of any given Qrr sRNA changes the extent of the response. This additive regulation is thought to allow fine-tuning of luxR levels across a gradient of expression, leading to precise, tailored amounts of gene expression. It is surprising that within the same quorum sensing system in two related species of Vibrio, the multiple Qrr sRNAs operate according to two distinct mechanisms. While the reason for this is not clear, the difference illustrates the evolvability of RNA regulators and the regulatory nuances that can be provided by having multiple copies.
A third possibility is that the duplicated RNAs can act independently of each other. This could occur in several ways. For base pairing sRNAs, each sRNA could regulate a different set of genes, most likely in a somewhat overlapping manner. For protein-binding sRNAs, different homologs could interact with distinct proteins, giving rise to variations in the core complexes. As mentioned above, various B. subtilis 6S isoforms could repress RNA polymerase bound to different σ factors. Homologous RNA species also can employ very different mechanisms of action, as observed for the E. coli GlmY and GlmZ RNAs (Urban and Vogel, 2008). GlmZ functions by base pairing, while GlmY likely acts as a mimic to titrate away YhbJ and other factors that inactive GlmZ.
In some cases it is still perplexing why multiple copies are maintained. One example is the toxin-antitoxin modules, which are not only encoded by multiple genes in E. coli chromosomes, but which can vary in gene number even within the same species (reviewed in (Fozo et al., 2008a)). Redundant RNAs may simply indicate a recent evolutionary event, which has not yet undergone variation to select new functions. Alternatively, additional genes may be selected by the pressure to maintain at least one copy across a population. Complete answers to the question of why various regulatory RNA genes are duplicated await more characterization of each set of RNAs.
Advantages of Regulatory RNAs
RNA regulators have several advantages over protein regulators. They are less costly to the cell and can be faster to produce, since they are shorter than most mRNAs (~100–200 nucleotides compared to 1,000 nucleotides for the average ~350 amino acid E. coli protein) and do not require the extra step of translation.
The effects of the RNA regulators themselves also can be very fast. For cis-acting riboswitches, the coupling of a sensor directly to an mRNA allows a cell to respond to the signal in an extremely rapid and sensitive manner. Similarly, since sRNAs are faster to produce than proteins and act post-transcriptionally, it was anticipated that, in the short term, they could shut off or turn on expression more rapidly than protein-based transcription factors. Indeed this expectation is supported by some dynamic simulations (Mehta et al., 2008; Shimoni et al., 2007). Other unique aspects of sRNA regulation revealed by recent modeling studies are related to the threshold linear response provided by sRNAs, in contrast to the straight linear response provided by transcription factors (Legewie et al., 2008; Levine et al., 2007; Mehta et al., 2008). Most sRNAs characterized thus far act stoichiometrically through the noncatalytic mechanisms of mRNA degradation or competitive inhibition of translation, reactions in which the relative concentrations of the sRNA and mRNA are critical. Thus for negatively-acting sRNAs, when [sRNA] ≫ [mRNA], gene expression is tightly shut off, but when [mRNA] ≫ [sRNA], the sRNA has little effect on expression. This threshold property of sRNA repression suggests that sRNAs are not generally as effective as proteins at transducing small or transient input signals. In contrast, when input signals are large and persistent, sRNAs are hypothesized to be better than transcription factors at strongly and reliably repressing proteins levels, as well as at filtering noise. Moreover, sRNA-based regulation is thought to be ultra-sensitive to changes in sRNA and mRNA levels around the critical threshold, especially in the case of multiple, redundant sRNAs as in the V. cholerae Qrr quorum sensing system, which is proposed to lead to switch-like “all or nothing” behavior (Lenz et al., 2004).
Additional features of different subsets of the RNA regulators provide other advantages. Some riboswitches lead to transcription termination or self-cleavage and some base pairing sRNAs direct the cleavage of their targets, rendering their regulatory effects irreversible. For the cis-encoded antisense sRNAs and the CRISPR RNAs, the extensive complementarity with the target nucleic acids imparts extremely high specificity. In contrast, the ability of trans-encoded sRNAs to regulate many different genes allows these sRNAs to control entire physiological networks with varying degrees of stringency and outcomes. The extent and quality of base pairing with sRNAs can prioritize target mRNAs for differential regulation and could be used by cells to integrate different states into gene expression programs (Mitarai et al., 2007). In addition, when multiple target mRNAs of a given sRNA are expressed in a cell, their relative abundance and binding affinities can strongly influence expression of each other through cross-talk (Levine et al., 2007; Mehta et al., 2008; Shimoni et al., 2007). Conversely, competition between different sRNAs for Hfq or a specific mRNA is likely to alter dynamics within a regulatory network. Finally, base pairing flexibility presumably also allows rapid evolution of sRNAs and mRNA targets.
Moreover, while not an advantage per se, RNA regulators usually act at a level complementary to protein regulators, most often functioning at the post-transcriptional level as opposed to transcription factors that act before sRNAs or enzymes such as kinases or proteases that act after sRNAs. Different combinations of these protein and RNA regulators can provide a variety of regulatory outcomes, such as extremely tight repression, an expansion in the genes regulated in response to a single signal or conversely an increase in the number of signals sensed by a given gene (Shimoni et al., 2007).
Evolution of Regulatory RNAs
We do not yet know whether all bacteria contain regulatory RNAs or whether we are coming close to having identified all sRNAs and riboswitches in well-studied bacteria. Given the redundancy in the sRNAs being found, the searches for certain classes of sRNAs, in particular sRNAs encoded in intergenic regions and expressed under typical laboratory conditions, appear near saturation in E. coli. However, other types of sRNAs, such as cis-encoded antisense sRNAs and sRNAs whose expression is tightly regulated, may still be missing from the lists of identified RNA regulators.
Are RNA regulators remnants of the RNA world or are the genes recent additions to bacterial genomes? We propose that the answer to this question is both. Some of the regulators such as riboswitches and CRISPR systems, which are very broadly conserved, are likely to have ancient evolutionary origins. In contrast, while regulation by base pairing may long have been in existence, individual antisense regulators, both cis- and trans-encoded sRNAs may be recently acquired and rapidly evolving. This is exemplified by the poor conservation of sRNA sequences across bacteria. For example, the Prr RNAs of Pseudomonas bear almost no resemblance to the equivalent RyhB sRNA of E. coli although both are repressed by Fur and act on similar targets (Wilderman et al., 2004). One might imagine that the expression of a spurious transcript, either antisense or with limited complementarity to a bona fide mRNA, which provides some selective advantage could easily be fixed in a population.
It is intriguing to note that distinct RNA regulators have been used to solve specific regulatory problems, emphasizing the pervasiveness and adaptability of RNA-mediated regulation. For example, in B. subtilis, the glmS mRNA is inactivated by the self-cleavage of the glucosamine-6-phosphate-responsive cis-acting riboswitch (Collins et al., 2007), whereas in E. coli, the glmS mRNA is positively regulated by the two trans-acting sRNAs GlmY and GlmZ (Urban and Vogel, 2008). As another example, RyhB-like trans-encoded sRNAs repress the expression of iron-containing enzymes during iron starvation in various bacteria, while the cis-encoded IsiR sRNA of Synechocystis represses expression of the IsiA protein, a light harvesting antenna, under iron replete conditions (reviewed in (Massé et al., 2007)).
Applications of Regulatory RNAs
The central roles played by RNA regulators in cellular physiology make them attractive for use as tools to serve as biosensors or to control bacterial growth either positively or negatively. Endogenous RNAs could serve as signals of the environmental status of the cell. For example, the levels of the RyhB and OxyS sRNAs, respectively, are powerful indicators of the iron status and hydrogen peroxide concentration in a cell (Altuvia et al., 1997; Massé and Gottesman, 2002). CRISPR sequences provide insights into the history of the extracellular DNA encountered by the bacteria and have been used to genotype strains during infectious disease outbreaks (reviewed in (Hebert et al., 2008; Sorek et al., 2008)). Regarding the control of bacterial cell growth, one can imagine how riboswitches might be exploited as drug targets given their potential to bind a wide variety of compounds (reviewed in (Blount and Breaker, 2006)). Similarly, since interference with the functions of some of the sRNAs is detrimental to growth and several sRNAs contribute to virulence, these regulators and their interacting proteins also could be targeted by antibacterial therapies. Alternatively, ectopic expression of specific regulatory RNAs might be used to increase stress resistance and facilitate bacterial survival in various industrial or ecological settings.
RNA also presents a powerful system for rational design as it is modular, easily synthesized and manipulated, and can attain an enormous diversity of sequence, structure, and function. Although less developed than in eukaryotes, the application of synthetic RNAs is being explored in bacteria (reviewed in (Hebert et al., 2008; Isaacs et al., 2006)). For example, riboswitch elements have been engineered to use novel ligands, and sRNAs have been designed to base pair with novel transcripts. Engineered CRISPR repeats present an obvious mechanism by which to repress uptake of specific DNA sequences. Limitations to these approaches include incomplete repression observed for the synthetic riboswitches and base pairing sRNAs thus far, off target effects, as well as problems in delivering the RNA regulators into cells where they might be of greatest utility. Nevertheless, synthetic RNAs have potential to provide a variety of useful tools and therapeutics in the future.
Perspectives
In summary, RNA molecules serve a wide range of regulatory functions in bacteria and modulate almost every aspect of cell metabolism. Examples of these RNA regulators were known long before the discovery of similar regulators in eukaryotes, though the large numbers of riboswitches, sRNAs, and CRISPR RNAs, as well as their correspondingly large importance to cellular physiology and defense mechanisms, were not anticipated. Many bacteria are facile experimental systems and have small genomes, which aid computational predictions and robust model development. In addition, hundreds of bacterial genome sequences, representing a broad diversity of species with a variety of lifestyles and ecological niches, are available. These factors make bacteria an ideal system in which to delve deeply into mechanistic, physiological and evolutionary questions regarding regulatory RNAs.
ACKNOWLEDGMENTS
We regret that we were only able to cite a subset of the most recent publications due to the broad scope of the review. Readers are referred to numerous reviews for more in depth coverage of specific topics. We appreciate the comments from so many of our colleagues. Supported by the Intramural Research Program of NICHD and a fellowship from the PRAT program of NIGMS (L.S.W.).
REFERENCES
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:45–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Xavier KB. The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 2007;10:189–198. doi: 10.1016/j.mib.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5' mRNA coding region inhibits translation initiation. Mol. Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. The Crp-activated sRNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 2008 doi: 10.1128/JB.01157-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. USA. 2006;103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008a;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008b;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Wagner EGH. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Gildehaus N, Neusser T, Wurm R, Wagner R. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res. 2007;35:1885–1896. doi: 10.1093/nar/gkm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2925. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit. Rev. Biochem. Mol. Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. The 5' end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 2007;104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert CG, Valdes JJ, Bentley WE. Beyond silencing--engineering applications of RNA interference and antisense technology for altering cellular phenotype. Curr Opin Biotechnol. 2008;19:500–505. doi: 10.1016/j.copbio.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Heroven AK, Böhme K, Rohde M, Dersch P. Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol Microbiol. 2008A;68:1179–1195. doi: 10.1111/j.1365-2958.2008.06218.x. [DOI] [PubMed] [Google Scholar]
- Holbrook SR. Structural principles from large RNAs. Annu. Rev. Biochem. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P. Down-regulation of outer membrane proteins by non-coding RNAs: unraveling the cAMP-CRP- and σE-dependent CyaR-ompX regulatory case. J. Mol. Biol. 2008;383:1–9. doi: 10.1016/j.jmb.2008.06.058. [DOI] [PubMed] [Google Scholar]
- Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Kalamorz F, Reichenbach B, März W, Rak B, Görke B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 2007;65:1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of posttranscriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Pace NR. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol. Microbiol. 2008;68:600–614. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Legewie S, Dienst D, Wilde A, Herzel H, Axmann IM. Small RNAs establish delays and temporal thresholds in gene expression. Biophys J. 2008;95:3232–3238. doi: 10.1529/biophysj.108.133819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;6:e5. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Teonadi H, Livny M, Waldor MK. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE. 2008;3:e3197. doi: 10.1371/journal.pone.0003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology. 2008;154:16–29. doi: 10.1099/mic.0.2007/012286-0. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki K, Uno K, Morita T, Aiba H. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc. Natl. Acad. Sci. USA. 2008;105:10332–10337. doi: 10.1073/pnas.0803106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Goyal S, Wingreen NS. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol. Syst. Biol. 2008;4:221. doi: 10.1038/msb.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitarai N, Andersson AM, Krishna S, Semsey S, Sneppen K. Efficient degradation and expression prioritization with small RNAs. Phys. Biol. 2007;4:164–171. doi: 10.1088/1478-3975/4/3/003. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc. Natl. Acad. Sci. USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor s32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl. Acad. Sci. USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ER, Payne SM. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 2007;75:3470–3477. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Kang J-G, Storz G. GadY, a small RNA reglator of the acid response in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol. 2007A;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon C, Felden B. Small RNA gene identification and mRNA target predictions in Bacteria. Bioinformatics. 2008;24:2807–2813. doi: 10.1093/bioinformatics/btn560. [DOI] [PubMed] [Google Scholar]
- Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr. Opin. Genet. Dev. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Görke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acid Res. 2008;36:2570–2580. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romby P, Vandenesch F, Wagner EGH. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol. Syst. Biol. 2007;3:138. doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Kleckner N. Translational control of IS10 transposition. Cell. 1983;34:683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JCD, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000163. e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, Wai SN. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Stork M, Di Lorenzo M, Welch TJ, Crosa JH. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J. Bacteriol. 2007;189:3479–3488. doi: 10.1128/JB.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard P, Molin S, Nordström K. RNAs involved in copy-number control and incompatibility of plasmid R1. Proc. Natl. Acad. Sci. USA. 1981;78:6008–6012. doi: 10.1073/pnas.78.10.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Waters CM, Bassler BL. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev. 2008;22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Hüttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- Thompson KM, Rhodius VA, Gottesman S. sE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 2007;189:4243–4256. doi: 10.1128/JB.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B. TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res. 2008;36:W109–W113. doi: 10.1093/nar/gkn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Tomizawa J, Itoh T, Selzer G, Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc. Natl. Acad. Sci. USA. 1981;78 doi: 10.1073/pnas.78.3.1421. 1421-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42-bp GadX/GadW-specific binding sites. Mol. Microbiol. 2008;70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Waters CM, Svenningsen SL, Bassler BL. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol. Microbiol. 2008;70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoson C, Wagner EGH. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008;70:258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]
- Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5' rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Johansen J, Rasmussen AA. Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 2007;10:152–155. doi: 10.1016/j.mib.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Vecerek B, Moll I, Bläsi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Wagner EGH. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EGH, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–1603. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Zhang A, Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N, von Pelchrzim F, Mayer O, Csaszar E, Schroeder R. Isolation of small RNA-binding proteins from E. coli: evidence for frequent interaction of RNAs with RNA polymerase. RNA Biol. 2008;5:30–40. doi: 10.4161/rna.5.1.5694. [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]