Abstract
A change in coreceptor preference from CCR5 to CXCR4 towards the end stage disease in some HIV-1 infected individuals has been well documented, but the reasons and mechanisms for this tropism switch remain elusive. It has been suggested that envelope structural constraints in accommodating amino acid changes required for CXCR4 usage is an obstacle to tropism switch, limiting the rate and pathways available for HIV-1 coreceptor switching. The present study was initiated in two R5 SHIVSF162P3N-infected rapid progressor macaques with coreceptor switch to test the hypothesis that an early step in the evolution of tropism switch is the adoption of a less constrained and more “open” envelope conformation for better CD4 usage, allowing greater structural flexibility to accommodate further mutational changes that confer CXCR4 utilization. We show that, prior to the time of coreceptor switch, R5 viruses in both macaques evolved to become increasingly sCD4-sensitive, suggestive of enhanced exposure of the CD4 binding site and an “open” envelope conformation, and this correlated with better gp120 binding to CD4 and with more efficient infection of CD4low cells such as primary macrophages. Moreover, significant changes in neutralization sensitivity to agents and antibodies directed against functional domains of gp120 and gp41 were seen for R5 viruses close to the time of X4 emergence, consistent with global changes in envelope configuration and structural plasticity. These observations in a simian model of R5-to-X4 evolution provide a mechanistic basis for the HIV-1 coreceptor switch.
Introduction
The human immunodeficiency virus (HIV) enters target cells via interaction of the viral glycoprotein with the cellular receptor CD4 and chemokine coreceptors, either CCR5 (R5 viruses) or CXCR4 (X4 viruses) [1]. Regardless of the route of transmission, R5 viruses account for most of the primary HIV-1 infections [2], [3]. With time, X4 variants arise and coexist with R5 viruses in ∼50% of subtype B infected individuals, and their emergence is associated with accelerated CD4+ T cell loss and disease progression [4]. The determinant of phenotypic change from R5 to X4 maps largely to the V3 loop of the envelope gp120 [5], [6], [7], requiring only a few amino acid substitutions in this region to expand or alter coreceptor preference [8], [9], [10]. Given the minimal requirement for V3 sequence change to confer the ability to use CXCR4, the high levels of virus replication and associated error rate [11], [12], [13], and the selective advantage of expanded target cell population in vivo [14], [15], it is surprising that the switch from R5 to X4 virus does not occur more rapidly and frequently in HIV-1 infected individuals. Although the mechanistic basis and blockade(s) for virus coreceptor switch remain ill-defined, several selective factors such as high viral load and evolutionary rate, CD4+CCR5+ target T cell limitation, and weakening of immune-driven pressures have been proposed as playing important roles [16], [17], [18].
We recently developed a simian model of coreceptor switching, based on infection of rhesus macaques with a pathogenic R5 SHIV isolate, SHIVSF162P3N [19], [20], [21]. The macaques infected intravenously or intrarectally with SHIVSF1623N in which X4 virus evolved and emerged were rapid progressors (RPs), with a clinical course that was characterized by extremely high levels of virus replication and weak or undetectable antiviral antibody and cellular immune responses. Sequence changes in the V3 loop of envelope gp120 were shown to determine the phenotypic change from R5 to X4 in macaques, and this process transitioned through dual-tropic (R5X4) variants capable of using both coreceptors, albeit with reduced efficiency [22]. Interestingly, while X4 appearance was associated with an accelerated drop in peripheral CD4+ T cell count, it followed rather than preceded the onset of precipitous CD4+ T cell loss in infected animals. The newly emerging R5X4 and X4 viruses were highly sensitive to neutralization with soluble CD4 (sCD4), and V3 sequence changes that confer CXCR4 usage are also sufficient to determine increase sCD4 sensitivity of the virus [22]. The conditions (e.g., extremely high levels of virus replication), genotypic requirements (i.e., V3 loop sequence changes) and pattern (e.g., emergence of neutralization sensitive X4 variants following the onset of CD4+ cell loss) for coreceptor switching in SHIVSF162P3N-infected macaques overlapped with those reported for HIV-1 infected humans [8], [9], [23], [24], [25], [26], [27], [28], supporting the use of this infection model to study the basis and underlying selection pressures for R5-to-X4 virus evolution in vivo.
In this respect, the findings in HIV-1-infected individuals and in SHIVSF162P3N-infected macaques that the emerging R5X4 and X4 variants were highly sensitive to sCD4 neutralization, and that the V3 sequence substitutions that altered coreceptor preference of the virus also determined its sCD4 sensitivity are noteworthy [19], [22], [27], [29], [30]. The former suggests that R5-to-X4 evolution is possible only when neutralization antibody selective pressure is absent or diminished with immune deterioration, while the latter implies that the early steps in the R5-to-X4 evolution process may require the same envelope conformation changes that render the virus sCD4 sensitive. Increased sCD4 sensitivity is indicative of enhanced CD4 binding and accessibility of the CD4 binding site, which is usually masked in the structure of the unliganded envelope glycoprotein of primary HIV-1 isolates in order to avoid the binding of potential neutralization antibodies [31], [32], [33], [34]. Since there is a diminished need to resist neutralizing antibodies in the rapidly progressing macaques, and perhaps in HIV-1 infected individuals towards end stage disease as well, when the immune system collapses, enhanced CD4 binding may be best achieved by adoption of an “open” envelope conformation to expose the CD4 binding site [31], [35]. As envelope structural constraints have been suggested to limit the pathways available for coreceptor switching [13], [36], [37], [38], an “open” envelope configuration can also release or minimize such constraints, allowing for greater flexibility in procuring the conformational transitions needed to confer CXCR4 utilization.
We tested this model for the R5-to-X4 phenotypic switch by assessing the sensitivity to sCD4 and a CCR5 antagonist of viruses pseudotyped with CCR5-using envelope gp160s (Envs) amplified over time from RP macaques with coreceptor switch, with these measurements serving as surrogate markers for CD4 and CCR5 utilization efficiencies, respectively [39], [40], [41], [42], [43], [44]. We also examined binding of soluble gp120 to CD4-Ig, as well as the ability of the R5 pseudoviruses to infect target cells that express low levels of the CD4 receptor. This is because, conceivably, the selection factor for viruses to expose the CD4 binding site and to bind CD4 better is to infect target cells that express low levels of the receptor more efficiently. Accordingly, HIV-1 R5 variants that can infect CD4low cells such as macrophages are frequently detected late in disease [45], [46], [47], [48], and macrophages are the major source of virus in SIV-infected RPs at end-stage disease [49]. Moreover, efficient infection of macrophages in vitro correlates with increased CD4 affinity, the capacity to use low CD4 levels, and with increased sensitivity to sCD4 [43], [50], [51], [52], [53], [54], [55], [56]. Lastly, susceptibility of the R5 pseudoviruses to neutralization with T20 and broadly reactive conformational antibodies was also determined, with broad changes in neutralization sensitivity interpreted as indicative of global rearrangements in glycoprotein structure and greater envelope plasticity [57], [58]. These studies suggest that adaptation of an “open” envelope conformation that binds CD4 more efficiently evolves in persisting R5 viruses, and is an early step in the pathway to the coreceptor switch in rhesus macaques.
Materials and Methods
Ethics Statement
This work used blood from SHIV infected macaques housed at the Tulane National Primate Research Center (TNPRC) in accordance with the Animal Welfare Act and Guide for the Care and Use of Laboratory Animals. TNPRC is accredited by the Association and Assessment and Accreditation of Laboratory Animal Care (AAALAC #000594). The OLAW animal welfare assurance number for TNPRC is A4499-01 and the USDA registration number is 72-R-0002. Care was provided by a faculty of 8 veterinarians, and 120 animal care technicians, veterinary technicians and enrichment staff. All procedures were performed on anesthetized animals and post-operative analgesics were administered as needed in accordance with IACUC approval. The Tulane University IACUC and the Division of Veterinary Medicine have established procedures to minimize pain and distress through several means. The use of preemptive and post procedural analgesia is required for procedures that would likely cause more than momentary pain or distress in humans undergoing the same procedure. Any deviation from the administration of analgesics according to this policy requires adequate scientific justification from the investigator and approval by the IACUC. Tulane also has a written endpoint policy to minimize potential pain and distress experienced by animals. If the animal becomes ill and/or meets the criteria for the IACUC approved endpoint policy, it will be euthanized using methods consistent with the recommendations of the American Veterinary Medical Association (AVMA) Panel on Euthanasia.
The Tulane IACUC specifically approved this study. And, in accordance with the recommendations of the Weatherall report “The use of non-human primates in research”, all steps were taken to protect animal welfare and to ameliorate suffering in all work involving non-human primates.
Cells
293T cells and Hela TZM-bl cells expressing CD4, CCR5 and CXCR4 and containing integrated reporter genes for firefly luciferase and β-galactosidase under control of the HIV-1 LTR [59] were maintained in DMEM supplemented with 10% fetal bovine serum (FCS), 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM L-glutamine. RC49 and JC53 cells, which are clones of HeLa/CD4/CCR5 cells that express low and high levels of CD4 respectively [60], were maintained in the same media. Human peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll gradient centrifugation, stimulated with phytohemagglutinin (PHA, 3 µg/ml; Sigma, St. Louis, MO) in RPMI medium containing 10% FCS, penicillin, streptomycin, L-glutamine and 20 U/ml interleukin-2 (Norvatis, Emeryville, CA). Monocytes were enriched by centrifugation of PBMCs through a 40% percoll cushion followed by plastic adherence, and cultured in RPMI 1640 medium supplemented with 10% FCS and 5% human AB serum for 5–7 days to allow for differentiation into macrophages [61].
Plasmid constructs and pseudovirus production
For expression of envelope glycoproteins, full-length gp160 coding sequences were amplified from infected macaque PBMC or plasma RT products with primers SH43 (5′-AAGACAGAATTCATGAGAGTGAAGGGGATCAGGAAG-3′) and SH44 (5′-AGAGAGGGATCCTTATAGCAAAGCCCTTTCAAAGCCCT-3′), subcloned into the pCAGGS vector and sequenced for verification. To generate luciferase reporter viruses capable of only a single round of replication, envelope trans-complementation assay was used as previously described [62]. Briefly, Env expression plasmid and the NL4.3LucE-R+ vector were cotransfected with polyethylenimine (PEI, Polyscience, Warrington, PA) into 2.5×106 293T cells plated in 100 mm plate. Cell culture supernatants were harvested 72 hours later, filtered through 0.45-µm filters, and stored at −70°C in 1-ml aliquots. Pseudoviruses were quantified for p24 Gag content (Beckman Coulter, Fullerton, CA).
Virus infectivity
For assessment of Env infectivity and entry efficiency, 7×103 TZM-bl cells were seeded in 96-well plates 24 hours before use and infected, in triplicate, with 2 ng p24 Gag equivalent of the indicated pseudotyped viruses. Infected cells were cultured for 72 h at 37°C, at which time the cells were harvested, lysed and processed for luciferase activity according to the manufacturer's instructions (Luciferase Assay System; Promega, Madison, WI). Entry, as quantified by luciferase activity, was measured with an MLX microtiter plate luminometer (Dynex Technologies, Inc., Chantilly, VA). For RC49 and JC53 infections, 7×103 cells were seeded in each well of a 96-well plate on the day prior to infection. Infections were performed in duplicate with 2 ng p24 Gag equivalent of the indicated pseudoviruses, and cells harvested for quantitation of luciferase activity 72 hours later. For infection of primary cells, 106 and 105 cells of human PBMCs and macrophage respectively were infected in duplicate with 5 ng p24 Gag equivalent of the indicated pseudotyped viruses in each well of a 96-well plate. Infected cultures were harvested 72 hours later and processed for luciferase activity. To control for differences in Env entry efficiencies, infectivity for RC49 cells was expressed as a ratio of the infectivity for these cells compared to the infectivity in JC53 cells. Similarly, infectivity in macrophages was normalized to that achieved in peripheral blood mononuclear cells (PBMCs) from the same donor.
Receptor and coreceptor usage efficiency
For assessment of receptor usage efficiency, 2 ng p24 equivalent of the indicated pseudoviruses in 50 µl were incubated with equal 4-fold serial dilution volumes of the CD4-IgG2 fusion protein (sCD4; PRO 542, Progenics Pharmaceuticals, Tarrytown, NY) for 1 h at 37°C and then added to cells, in duplicate wells, for an additional 2 hours at 37°C. 100 µl of medium was then added to each well and the virus-protein cultures maintained for 72 hours. Control cultures received virus in the absence of sCD4. At the end of the culture period, the cells were lysed and processed for β-galactosidase activity (Galacto-Star System; Applied Biosystems, Bedford, MA). A neutralization curve was generated by plotting the percentage of neutralization vs sCD4 dilution, and 50% inhibitory concentrations (IC50) were determined using the Prism 4 software (GraphPad, San Diego, CA). For assessment of coreceptor usage efficiency, 7×103 TZM-bl cells per well of a 96-well plate were inoculated, in duplicate, with 2 ng p24 Gag antigen equivalent of the indicated pseudovirus in the absence or presence of 4-fold dilutions of the CCR5 antagonist PSC-RANTES. The cells were lysed after 72 hours at 37°C, processed for β-galactosidase activity, and IC50 determined using the Prism 4 software.
Soluble gp120 CD4-Ig binding
To examine CD4 binding, gp120 glycoproteins from 293T transfected cells were metabolically radiolabeled for 48 hours with 100 µCi/mL [35S]-methionine/cysteine ([35S] protein labeling mix; Perkin-Elmer, Waltham, Mass) in Dulbecco's modified Eagle's medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. Radiolabeled proteins released in the culture supernatant were incubated with either a mixture of sera from HIV-1 infected individuals or CD4-Ig [a fusion protein in which the N-terminal two domains of CD4 are linked to the Fc component of immunoglobulin G [63]] in the presence of 70 µl of 10% Protein A-Sepharose (American BioSciences Inc, Boulder, CO) for 2 hr at 37°C. The precipitates were analyzed on NuPAGE Novex Bis-Tris polyacrylamide gels (Invitrogen, Carlsbad, CA), followed by autoradiography and quantification with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Evaluation of spontaneous and sCD4-induced release of gp120
2.5×106 293T cells seeded in a 100-mm plate were transfected with Env expression plasmid by the polyethylenimine method. Transfected cells were collected 48 h later, washed twice, resuspended in phosphate-buffered saline and divided in half. sCD4 (0.5 µg/ml) was added to one of the two fractions, and both fractions were incubated for 2 h at 37°C. The supernatants and cells were subsequently collected, and the amounts of gp120 in each fraction were quantitated by ELISA according to the manufacturer's instructions (Advanced Biosciences Laboratories, Inc, Kensington, MD). Gp120 release was determined as a percentage of gp120 present in the supernatants compared to the total amount of Envs found in both the supernatants and the cell lysates. Results shown are expressed as the percentage difference in gp120 release in the presence of sCD4 relative to that seen in the absence of sCD4.
Neutralization assay
Virus neutralization was assessed using TZM-bl cells in 96-well plates. Briefly, equal volumes (50 µl) of pseudoviruses (2 ng p24 Gag equivalent) and 4-fold serial dilutions of IgG1b12, 447-52D and T20 were incubated for 1 h at 37°C and then added to cells, in duplicate wells, for an additional 2 hours at 37°C. 100 µl of medium was then added to each well and the virus-protein cultures maintained for 72 hours. Control cultures received virus in the absence of blocking agent. At the end of the culture period, the cells were lysed and processed for β-galactosidase activity. A neutralization curve was generated by plotting the percentage of neutralization vs agent dilution, and IC50 determined using the Prism 4 software.
Immunophenotyping of SHIV-infected cells
Identification of SHIV-infected macrophages was accomplished with double-label immunohistochemistry performed as previously described with modifications [64], [65]. Briefly, lymph node sections were deparaffinized in xylene and rehydrated through graded ethanol to tris-buffered saline (TBS) plus tween 20. Endogenous peroxidase activity was blocked by incubation in 3% H2O2 in PBS. Antigen retrieval was accomplished by microwave heating sections at 95°C for 20 minutes in citrate buffer (DAKO, Carpinteria, CA), followed by 20 minute cooling, and Dako protein block for 10 minutes. The blocked sections were incubated with SIVnef antibody (clone KK75, IgG1; 1∶200) overnight at 4°C then reacted with biotinylated secondary antibody (HAM-b, Dako, 1∶200) for 30 minutes. Sections were detected using standard avidin-biotin peroxidase complex technique (ABC Elite, Vector Laboratories, Burlingame, CA) and DAB chromagen (Dako). Sections were blocked again for 10 minutes with protein block (Dako) and incubated with Iba-1 antibody (Wako Chemicals, Richmond, VA, rabbit polyclonal, 019-19741, 1∶1000) for macrophages for 30 minutes at room temperature followed by biotinylated secondary antibody (GAR-b, Dako, 1∶200) for 30 minutes. Sections were detected using standard avidin-biotin alkaline phosphatase complex technique (Vectastain ABC-AP, Vector Laboratories, Burlingame, CA) and Permanent Red (Dako). Slides were counterstained with Mayer's hematoxylin, rinsed in tap water, coated with Clear Mount (Electron Microscopy Science, Hatfield, PA), air-dried overnight, then coverslipped.
Statistical Analysis
Differences in susceptibility to sCD4, IgG1b12, 447-52D and T20, as well as infection of CD4low cells and binding of gp120 to CD4-Ig between the acute (w2 for BR24 and w1 for CA28) and the evolving R5 viruses were examined using Mann-Whitney U test. P-values<0.05 were considered statistically significant.
Results
Increased sCD4 sensitivity of evolving R5 viruses in a RP with coreceptor switch
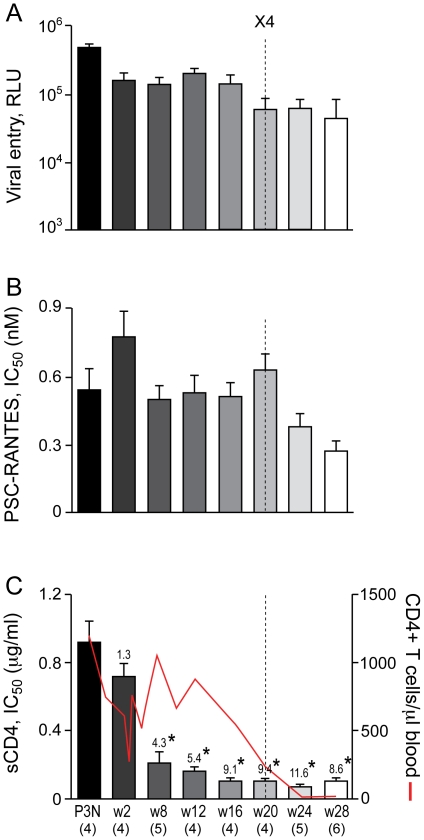
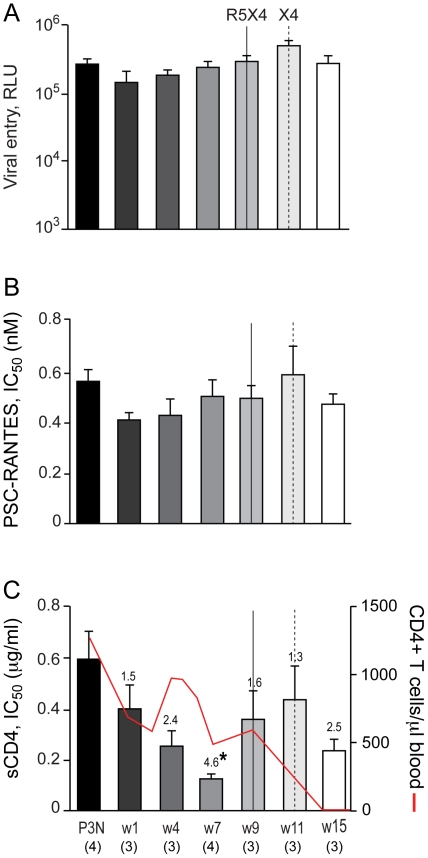
We first determined if R5 viruses evolved over time to be more sCD4 sensitive in BR24, the initial reported case of coreceptor switching in R5 SHIVSF162P3N-infected RP macaques [19]. BR24 sustained high viremia and progressed to disease at 28 week post-infection (wpi) in the absence of seroconversion, with tropism switch documented at 20 wpi. We obtained multiple CCR5-using full-length envelope gp160 (Env) at 2, 8, 12, 16, 20, 24 and 28 wpi and generated single-round replication-competent luciferase reporter viruses for functional characterization. Four randomly selected functional Env clones from the SHIVSF162P3N inoculum were also characterized for comparison. We found no significant difference in the entry efficiency of R5 viruses bearing Envs amplified from macaque BR24 at 2–16 wpi, when measured in CD4hi CCR5hi TZM-bl cells, but R5 viruses present during and following the time of X4 emergence at 20 wpi infected TZM-bl cells less efficiently (2–3 fold reduction in RLU; Figure 1A ). There was also no significant change in the ability of the evolving R5 viruses to use the CCR5 coreceptor up to the time of switch, as indicated by similar IC50 inhibitory dose with the CCR5 inhibitor PSC-RANTES ( Figure 1B ). However, viruses evolving following the time of switch showed a 1.5- to 2-fold increase in susceptibility to PSC-RANTES inhibition, suggesting that they used the CCR5 coreceptor less efficiently. These findings of reduced entry fitness and decreased CCR5 use for R5 viruses that coexist with emerging X4 viruses in BR24 towards end-stage disease at 20, 24 and 28 wpi contrast with reports of increased replication and efficacy of CCR5 usage with disease progression in HIV-1 infected individuals with R5 viruses only [44], [66], [67], [68], [69], [70], [71], [72], but are consistent with results for late R5 viruses from HIV-1 infected individuals with detectable CXCR4-using variants [73], [74], [75].
Figure 1. Entry efficiency, PSC-RANTES and sCD4 sensitivity of R5 viruses evolving over time in BR24.
Entry of luciferase reporter viruses expressing CCR5-using envelopes into TZM-bl cells expressed as relative light unit (RLU)(A), and susceptibility of the reporter viruses to neutralization with PSC-RANTES (B) and sCD4 (C) were determined. The dashed vertical line indicates time of tropism switch in BR24 (20 wpi), and the numbers in the brackets indicate the number of clones analyzed at each time point. Envelope clones from the SHIVSF162P3N inoculum (P3N) were also included in the characterization for comparison. Absolute CD4+ T-cell count in the animal over the course of infection is shown in (C) for reference, and values above the bars indicate fold increase in sCD4 sensitivity relative to that of the w2 viruses. * P<0.05 (Mann-Whitney U test). Data are representative of 2–3 independent experiments (error bars, s.d.).
In contrast, R5 viruses in BR24 evolved prior to the time of coreceptor switch to become increasingly susceptible to inhibition with CD4-IgG2, a tetrameric soluble CD4 (sCD4) construct based on IgG ( Figure 1C ). There was little difference in the concentrations of sCD4 needed to achieve 50% neutralization (IC50) of the week 2 (w2) and the inoculating P3N viruses (0.92 and 0.72 µg/ml, respectively), but a statistically significant 4.3-fold increase in sCD4 sensitivity was evident for viruses present six weeks later (w8; IC50 of 0.21 µg/ml). sCD4 sensitivity continued to increase significantly for R5 viruses in BR24, with a 5.4-fold increase seen for the w12 viruses (IC50 of 0.17 µg/ml), and a 9.1–9.4 fold increase for viruses present at 16 and 20 wpi (IC50 of ∼0.1 µg/ml) that is suggestive of increased accessibility of oligomeric gp120 to CD4 prior to and during the time of coreceptor switch. Notably, acquisition of increased sCD4 sensitivity of the R5 viruses took place in the presence of a high CD4+ T cell count (>500 CD4+ T cells per µl blood at 16 wpi), suggesting that paucity of CD4+ target T cells is not the driving force for viruses to expose their CD4 binding site. Increase in sCD4 sensitivity continued for R5 viruses evolving following the time of coreceptor switch in BR24. Compared to the w2 viruses, the w24 and w28 viruses exhibited 11.6- and 8.6-fold increases in sensitivity, respectively.
The increase in sCD4 sensitivity of early R5 viruses in BR24 correlates with better CD4 binding and with infection of CD4low cells, but this association dissipates near the time of coreceptor switch
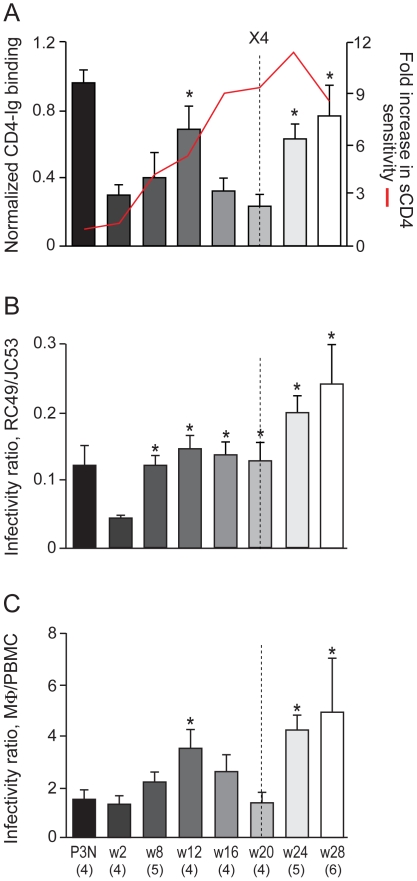
We next sought to establish, for the evolving R5 viruses in BR24, an association between sCD4 sensitivity, soluble gp120 (sgp120) binding to CD4-Ig, infection of primary macrophages and HeLa RC49 cells, the latter having been used as an indicator of macrophage-tropism and ability to utilize low levels of CD4 for infection [43], [76]. Results showed that the 4.3 and 5.4 fold increase in sCD4 sensitivity of the w8 and w12 viruses as compared to the w2 viruses respectively was accompanied by a 2–3 fold increase in binding of the w8 and w12 sgp120s to CD4-Ig ( Figure 2A ), and with a corresponding fold increase in the ability of the viruses to infect RC49 cells ( Figure 2B ) and primary macrophages ( Figure 2C ). The increase in the ability of the w12 viruses to infect CD4low cells and to bind CD4-Ig is statistically significant. Thus, viruses in BR24 are evolving early to adopt an “open” Env conformation in order to bind CD4 more efficiently for infection of CD4low cells.
Figure 2. sgp120 CD4-Ig binding and infection of CD4low cells with BR24 viruses.
The binding of sgp120 to CD4-Ig together with the fold-increase in sCD4 sensitivity (A), infectivity of HeLa RC49 cells (B) and primary macrophages (mΦ; C) that express low levels of CD4 with pseudotyped viruses bearing CCR5-using Envs amplified over time from BR24 were determined. Properties of four envelope clones in the SHIVSF162P3N inoculum (P3N) were also determined and shown for reference. sgp120 binding to CD4-Ig (A) was normalized to that of sgp120 binding to polyclonal serum from HIV-1 infected individuals. Infectivity in RC49 cells (B) and macrophages (C) that express low levels of CD4 was expressed as a ratio of infectivity in JC53 cells and autologous PBMCs that express high levels of CD4 and CCR5, respectively. The dashed vertical line indicates time of tropism switch. For sgp120 CD4-Ig binding, data are the means and standard deviations from at least two independent experiments. For infection of CD4low cells, data are representative of at least 3 independent experiments (error bars, s.d.). * above the bars indicates normalized CD4-Ig binding and CD4low cell infectivity ratios that are statistically different between the acute (w2) and the evolving R5 viruses.
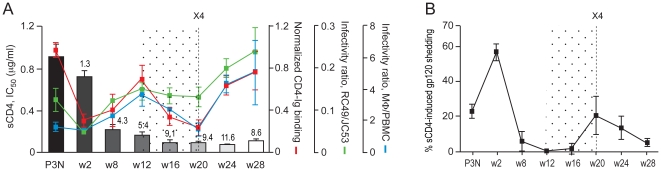
The association between sCD4 sensitivity, sgp120 CD4-Ig binding and infection of CD4low cells was also seen for R5 viruses evolving following the time of coreceptor switch at 20 wpi in BR24. The late w24 and w28 viruses were highly susceptible to sCD4 inhibition (IC50 of 0.08 and 0.11 µg/ml, respectively), and bound CD4-Ig and infected CD4low cells with great efficiencies. The notable exceptions were R5 viruses present close to (16 wpi) and at the time of switch (20 wpi). As illustrated in Figure 3A , despite an increase in sCD4 sensitivity when compared to the w12 viruses, the w16 and w20 sgp120s exhibited decreased CD4-Ig binding. Moreover, infectivity of the w16 and w20 viruses for CD4low cells was either comparable to that of the w12 viruses (in RC49 cells) or reduced (in primary macrophages).
Figure 3. Relationship between sCD4 sensitivity, CD4-Ig binding, infection of CD4low cells and sCD4-induced gp120 release of BR24 viruses.
(A) The relationship between sgp120 binding to CD4-Ig, sCD4 sensitivity, infection of RC49 cells and primary macrophages (mΦ) of BR24 dervied viruses is illustrated. Values above the bars indicate fold increase in sCD4 sensitivity of BR24 viruses compared to viruses in the SHIVSF162P3N inoculum (P3N). (B) Extent of sCD4-induced gp120 from surface of 293T cells transiently expressing BR24-derived envelope glycoproteins. Percentage difference in gp120 release in the presence of sCD4 relative to that in the absence of sCD4 is shown. The data are the means and standard deviations of two independent experiments. The vertical dashed line in (A) and (B) indicates the time of coreceptor switching, and the dotted area highlights the time when the relationship between sCD4 sensitivity, sgp120 binding to CD4-Ig and infection of CD4low cells dissipates.
To understand the disconnect between sCD4 neutralization and CD4 binding of the w16 and w20 viruses, we assessed sCD4-induced gp120 release from the surface of 293T cells transiently expressing their envelope glycoprotein trimers, as this had been shown to be a mechanism by which sCD4 neutralizes HIV-1 infection [77], [78], [79], [80], [81]. Results showed that the extent of sCD4-induced gp120 detachment from the surface of Env-expressing 293T cells was negligible for the w8, w12 and w16 Envs ( Figure 3B ). sCD4-induced gp120 release increased for the w20, w24 and w28 viruses as compared to the w16 virus, but was lower than that of the sCD4-resistant inoculating and w2 viruses. These findings are consistent with previous studies with HIV-1 primary isolates, showing a lack of correlation between sCD4 inhibition and the degree of sCD4-dependent gp120 release [80], [82]. Thus, we conclude that high sCD4 sensitivity of the w16 and w20 R5 viruses in BR24 cannot be explained by increased affinity of the envelope glycoprotein complex for CD4, or by increased sCD4-induced gp120 shedding.
Global changes in envelope glycoprotein structure of R5 viruses near and at the time of coreceptor switch in macaque BR24
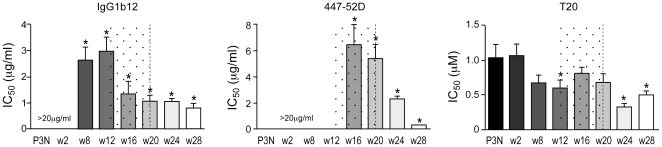
Binding to CD4 induces major conformational changes in the envelope glycoprotein that play key roles in Env-mediated fusion. Among these are exposure of the V3 loop and formation of the coreceptor-binding site on gp120 [83], [84], [85], [86], [87], and of a triple-stranded coiled coil activated fusion intermediate structure composed of the N-terminal heptad repeat (HR1) region of gp41 [88], [89], [90], [91], [92], [93]. Structural alterations in the gp120 CD4 binding site or in the V3 domain for BR24 viruses present near or at the time of coreceptor switch, therefore, could have affected their sCD4 susceptibility. Furthermore, recent studies showed that induction of an activated state in the HIV-1 Env that rapidly decays into functionally inactive forms could also mediate sCD4 sensitivity [94]. Accordingly, we assessed neutralizing antibody and T20 sensitivity of the w16 and w20 viruses to probe the conformational state of their envelope glycoproteins. The antibodies used were the broadly neutralizing antibody IgG1b12 directed against the CD4 binding site, and the anti-V3 loop MAb 447-52D [95], [96]. The fusion inhibitor T20 (also known as Fuzeon or enfuvirtide) binds to the hydrophobic groove on the surface of the coiled coil formed by HR1 [88], [97], and sensitivity to T20 has been shown to be modulated by gp120 interactions with the coreceptor as well as the half-life of the HR-1 groove [94], [98], [99], [100].
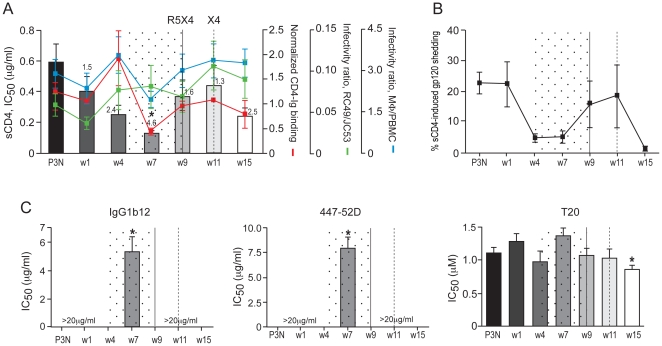
Statistically significant changes in virus sensitivity to b12 and 447-52D were evident beginning at 8 and 16 wpi, respectively ( Figure 4 ). Notably, there was an increase in b12 sensitivity that may be indicative of structural changes in CD4 binding site conformation or accessibility. 50% inhibition of the w16 and w20 viruses was achieved with ∼1 µg/ml of the antibody compared to 3 and >20 µg/ml needed for the earlier (w8 and w12) and acute (w2) viruses, respectively. The w16 and w20 viruses were also more sensitive to 447-52D neutralization as compared to the earlier R5 viruses (IC50 of 5–6 µg/ml compared to >20 µg/ml, respectively), with continued increase in sensitivity for viruses following the time of switch, perhaps suggestive of increased exposure of the V3 loop. Furthermore, the w16 viruses were slightly more resistant to T20 neutralization (IC50 of 0.8 µg/ml) compared to the early (w8 and w12; IC50, ∼0.6 µg/ml) as well as the late (w24 and w28; IC50, 0.3–0.5 µg/ml) viruses; one possible explanation is that exposure of the gp41 HR1 groove on envelope glycoproteins of w16 viruses decays faster. Together, these findings are in support of significant changes in structure or accessibility of the CD4 and the V3 loop, and easier induction of a metastable activated state of the envelope glycoprotein which could account for the increase in sCD4 sensitivity of the w16 and w20 R5 viruses.
Figure 4. Changes in neutralization sensitivity of R5 viruses evolving over time in macaque BR24.
Susceptibility of BR24 R5 pseudoviruses to neutralization with b12, 447-52D and T20 was determined, with sensitivity of variants from the inoculating virus SHIVSF162P3N (P3N) shown for reference. The vertical dashed line indicates the time of coreceptor switching, and the dotted area designates the period of marked envelope conformational changes. Data are representative of at least two independent experiments (error bars, s.d.). * above the bars indicate IC50 values that are statistically different between the acute (w2) and the evolving R5 viruses.
Similar early events for tropism switch in another SHIVSF162P3N infected RP macaque
To corroborate the above findings, we investigated if the early events of envelope evolution prior to the time of switch in BR24 are recapitulated in CA28, another RP macaque with coreceptor switch. Peak and set-point viremia were one-log higher in CA28 than in BR24, and the animal progressed to AIDS at 15 wpi, with transient seroconversion at 7 wpi [20]. We previously documented X4 emergence in CA28 at 11 wpi, but more recent studies revealed the presence of another R5-to-X4 evolutionary pathway that led to the emergence of a distinct dual-tropic virus at 9 wpi [101]. We amplified CCR5-using Envs from CA28 at w1, 4, 7, 9, 11 and 15 wpi and found them to mediate comparable entry into TZM-bl cells ( Figure 5A ). There was no notable difference in PSC-RANTES sensitivity of the evolving R5 viruses (<2-fold, Figure 5B ), but consistent with findings in BR24, R5 viruses in CA28 prior to the time of tropism switch were more sensitive to sCD4 neutralization ( Figure 5C ). Compared to the w1 viruses, the w7 viruses from CA28 were significantly more sensitive to sCD4 neutralization. And, as was observed in BR24, the increase in sCD4 sensitivity was acquired in the presence of high CD4+ T cell numbers (∼500 CD4+ T cells per µl blood at 7 wpi). sCD4 sensitivity however decreased for viruses during the time of switch in CA28 (w9 and w11 viruses). The transient development of anti-SHIV antibody at 7 wpi in this animal could be a contributing factor.
Figure 5. Entry efficiency, PSC-RANTES and sCD4 sensitivity of R5 viruses evolving over time in CA28.
Entry of luciferase reporter viruses expressing CCR5-using envelopes into TZM-bl cells (A), and susceptibility of the reporter viruses to neutralization with PSC-RANTES (B) and sCD4 (C) were determined. The solid and dashed vertical lines indicate the two switch events in CA28 leading to the emergence of distinct dual-tropic and X4 viruses, respectively. The numbers in the brackets denote the number of envelope clones analyzed at each time point. Absolute CD4+ T-cell count in the animal over the course of infection is shown in (C), and values above the bars indicate fold increase in sCD4 sensitivity of CA28 viruses compared to viruses in the SHIVSF162P3N inoculum (P3N). *P<0.05 (Mann-Whitney U test). Data are representative of at least three independent experiments (error bars, s.d.).
The 2.4-fold increase in sCD4 sensitivity of the w4 viruses correlated with a corresponding fold increase in sgp120 binding to CD4-Ig and with enhanced infection of RC49 cells and primary macrophages ( Figure 6A ), but this association dissipated at w7, two weeks prior to the first switch event in this animal. Importantly, and consistent with what was observed for BR24 w16 viruses, the dissociation between sCD4 neutralization and CD4 binding of the CA28 w7 viruses cannot be explained by greater sCD4-induced gp120 release ( Figure 6B ), but by antigenic change in the receptor binding site and the V3 loop ( Figure 6C ). The extent of sCD4-induced gp120 release was comparable for the w4 and w7 viruses, but 50% neutralization of the w7 viruses was achieved with ∼5 µg/ml of the anti-CD4BS antibody b12 and ∼8 µg/ml of the anti-V3 mAb 447-52D as compared to >20 µg/ml for the w4 and viruses present at the other time points examined. The increase in sensitivity to b12 and 447-52D neutralization of the w7 viruses is statistically significant. Furthermore, the w7 viruses showed a modest increase in T20 resistance (IC50 of 1.4 µg/ml in comparison to ∼1 µg/ml for the other viruses). Collectively, the similarities in increase sCD4 sensitivity that is associated with better CD4 binding of the early R5 viruses, and changes in envelope configuration for R5 viruses close to the time of switch in BR24 and CA28 support similar mechanism(s) and selective pressures for change in coreceptor preference in the two RP macaques.
Figure 6. Structure and function of R5 viruses evolving over time in macaque CA28.
The relationship between sCD4 sensitivity, binding of sgp120 to CD4-Ig, infectivity of HeLa RC49 cells and primary macrophages (mΦ)(A), the extent of sCD4-induced gp120 release (B), and neutralization susceptibility (C) of pseudoviruses bearing CCR5-using Envs amplified over time from CA28 is shown. The solid and dashed vertical lines indicate time of the two switch events in CA28, and the dotted area marks the time when correlation between sCD4 sensitivity, sgp120 binding to CD4-Ig and infectivity of CD4low cells dissipates (A), and period of notable envelope conformational change (B and C). Data are representative of at least two independent experiments (error bars, s.d.). * above bars indicate differences in sCD4 sensitivity, CD4-Ig binding and susceptibility to agents and antibodies between the acute (w2) and the evolving R5 viruses that are statistically significant.
Macrophages are the predominant virus-producing cells at end-stage disease in macaques BR24 and CA28
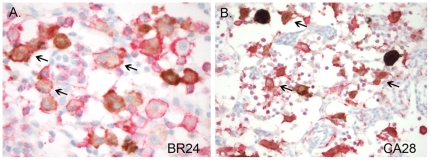
To determine if viruses are evolving in BR24 and CA28 for infection of macrophages in vivo, double labeled immunohistochemical staining for SIV nef (brown) and the macrophage marker lba-1 (red) was used to identify SHIV-expressing cells in the mesenteric lymph node at time of euthanasia. Based on coexpression of lba-1, the majority of SHIV–expressing cells in the lymph node of BR24 and CA28 were found to be macrophages ( Figure 7 ). Thus, similar to findings in SIV-infected RPs at end-stage disease [49], macrophage infection is responsible for sustaining virus replication in the two R5 SHIVSF162P3N-infected RP macaques at very late stages of disease.
Figure 7. SHIV-infected macrophages identified with double-label SIVnef and Iba-1 immunohistochemistry.
Tissue macrophages are the primary SHIV infected cells at end stage disease in BR24 (A) and CA28 (B). Double-labeled immunohistochemical staining for SIVnef (brown) and the macrophage marker lba-1 (red) was performed. Arrows mark representative double-positive cells.
Discussion
A change in coreceptor preference from CCR5 to CXCR4 late in infection has been well documented in some HIV-1 infected individuals since the early days of the AIDS epidemics, but the reasons and mechanisms for this tropism switch remain elusive. Because X4 emergence is strongly associated with rapid CD4+ T–cell loss and disease progression, and concerns that the introduction of CCR5 entry inhibitors as anti-HIV therapeutics could facilitate X4 emergence and exacerbate disease, there is an increasing need to improve our understanding of the selection pressures which favor CCR5-to-CXCR4 switch. Using a simian model of HIV-1 coreceptor switch, we tested in this study the hypothesis that an early selective force in the evolutionary pathway of tropism switch is the need for viruses to increase the efficiency of CD4 binding for infection of CD4low-expressing cells such as tissue macrophages. The adoption of a less constrained and more “open” envelope conformation that exposes the CD4 binding site for enhanced CD4 binding, in turn, releases or reduces envelope structural constraints that have been suggested to limit the pathways available for change in coreceptor preference. We show that R5 viruses evolved early in two rapid progressor macaques to become sCD4-sensitive, and this correlated with better gp120 binding to CD4 and with efficient infection of CD4low cells such as primary macrophages and the HeLa RC49 cells. Furthermore, significant changes in neutralization sensitivity to agents and antibodies directed against functional domains of both gp120 and gp41, including the V3 loop that is important for coreceptor binding were seen for R5 viruses present close to the time of X4 emergence in these rapid progressing macaques, consistent with global changes in envelope conformation and structural plasticity that facilitate the remodeling needed to expand or switch to CXCR4 usage. These observations in two R5 SHIVSF162P3N-infected macaques therefore support our proposed mechanistic model for coreceptor switching.
Several mechanisms can explain sCD4 sensitivity of HIV/SIV. For the early R5 viruses in macaques BR24 (w8, w12) and CA28 (w4), we showed that increase sCD4 sensitivity correlated with better CD4-Ig binding ( Figures 3A and 6A ), consistent with exposure of the CD4 binding site and adoption of an “open” envelope conformation. For R5 viruses close to the time of switch (w16 for BR24 and w7 for CA28), however, changes in the CD4 binding site and/or alteration in the conformational changes induced by CD4 binding appeared to be the underlying basis ( Figures 4 and 6C ). Interestingly, we observed, in both macaques, that sCD4-induced gp120 shedding decreased for Envs evolving prior to the time of switch ( Figures 3B and 6B ), suggesting that a tighter interaction between the gp120 and gp41 may be necessary during the process of envelope remodeling to acquire CXCR4 use. Alternatively, it has been proposed that an increased number of virion-associated Env complexes available for receptor interaction might facilitate infection of CD4low cells [54], [102], [103]. Thus, it is conceivable that a more stable gp120-gp41 interaction, in particular for BR24 w8 and w12 and CA28 w4 Envs, increases gp120 retention by Env complexes for infection of CD4low cells. Genetic studies to determine if virion-gp120 retention and infection of CD4low cells of these early viruses in BR24 and CA28 are linked will be required to examine this latter possibility.
We show that acquisition of increased sCD4 sensitivity occurred in the presence of high amounts of CD4+ T cells, implying that paucity of CD4+ target T cells is not the driving force for viruses to expose their CD4 binding site and to increase CD4 binding. Moreover, we recently reported that viruses did not evolve early to become sCD4 sensitive in macaques that were depleted of B cells to abrogate or diminish antiviral antibody responses prior to infection with SHIVSF162P3N, implying that the reduced antibody-driven pressure in the RPs was also not sufficient to select for viruses with an “open” Env conformation [104]. Rather, the tight association between CD4 binding and infection of CD4low cells of the evolving R5 viruses in both BR24 and CA28, and the finding that primary macrophages are the principle virus-producing cells at end-stage disease in these two macaques with coreceptor switch suggest that adoption of an “open” Env is in response to the need to use low levels of CD4 receptor more efficiently. However, increased sCD4 sensitivity and CD4 binding were seen as early as 4–8 wpi, a time when CD4+ T cells and not tissue macrophages are the preferred targets of HIV/SIV infection [105], [106], [107]. This then raises the intriguing possibility that a selective pressure for altered CD4 affinity of the early R5 viruses in BR24 and CA28 could be decreasing CD4 expression levels on target T cells. Although direct evidence in support is lacking, infectivity of HIV-1 primary isolates in vitro is strongly dependent on the level of CD4 expression [40], [108], [109]. Moreover, transmitted and founder viruses in acute HIV-1 infection have been reported to replicate poorly in monocyte-derived macrophages [3], [110] and to require high receptor levels for entry [76]. Our finding that the ability of the acute viruses (w2 for BR24 and w1 for CA28) to bind CD4 and to infect CD4low cells in both macaques is decreased is consistent with these reports in human, and suggests that CD4+ T cells expressing high amounts of the receptor may be the earliest and preferred targets of virus infection and depletion in vivo, leaving only cells with lower CD4 levels available during the post-acute phase of infection. Nevertheless, CD4 and CCR5 concentration requirements for R5 HIV-1 infections in vitro have been shown to be interdependent, with viruses being highly dependent on the CD4 concentrations or strength of the initial virus-CD4 bond when cell surface CCR5 density is low [60]. Thus, it is possible that the selection factor for better CD4 usage we observed in the RP macaques following acute R5 SHIVSF162P3N infection could be due to initially low CCR5 and not CD4 expressions on T lymphocytes. Studies to monitor variations in CD4 and CCR5 cell surface densities on target T cells during the course of SHIVSF162P3N infection and to examine their relationship to macrophage infection and tropism switch in RP macaques will be needed to more clearly address the selection factors for viruses to evolve early to use low levels of the CD4 receptor more efficiently.
Because most HIV-1-infected individuals have developed neutralizing antibodies, less constrained and “open” envelopes are selected against and not commonly found. This then raises the question as to what extent the observed changes associated with the coreceptor switch in rapid progressor macaques that did not develop or maintain a strong antiviral antibody response reflect what occurs in humans. In this regard, it is noteworthy that X4 dominance is seen only towards end-stage disease in HIV-1 infected individuals, when the immune system is impaired [25], [28], [111], [112]. And, although rare, rapid progressor status has been documented in HIV-1 infected individuals [73], [113], [114], [115], [116], [117], [118], [119], with phenotypic switch reported in cases where this was examined [73], [119]. Moreover, emergence of sCD4 neutralization-sensitive X4 viruses in the presence of neutralizing antibodies has been reported [27], suggesting that X4 virus evolution is in anatomical compartments with lower antibody pressure than in the plasma, and/or that these viruses spread via cell-cell, a mode of transmission that is less susceptible to antibody neutralization. Indeed, we have shown that peripheral lymph nodes that are enriched in target cells for X4 viruses are the preferred sites of their evolution and amplification [21], [22], and the syncytium-inducing/fusion capacity of X4 viruses has been well documented [120]. Thus, the observations made in the SHIV-infected macaques studied here are likely to represent an important step toward our understanding of HIV-1 coreceptor switch in humans.
In summary, our findings provide evidence that adoption of an “open” Env by R5 viruses in response to the selection pressure for better CD4 usage and infection of CD4low cells represents an early step in the chain of events leading to R5-to-X4 evolution, allowing other selection factors such as virus replication-associated mutational events that are required for tropism switch, but which usually come with costs to viral fitness because of structural constraints, to be manifested. Studies of coreceptor switch in RPs are useful for they allow examination of the process of R5 envelope evolution required for a generalized switch uncomplicated by the selection pressure of antiviral antibody responses. Although our studies were limited with respect to the number of animals, the similarity of the evolutionary pattern in structure and function of R5 envelope variants seen in the two outbred RP macaques that differed in the kinetics and levels of virus replication prior to the time of coreceptor switch support a shared mechanism and selective pressure(s) for the change in coreceptor preference. Further research will be required to determine if acquisition of an “open” Env conformation to increase CD4 affinity is a property unique to the early R5 viruses in R5-SHIVSF162P3N-infected RPs with coreceptor switch, and how broadly our findings in the SHIV-rhesus model relate to HIV infection of humans. Additionally, it will be of interest to examine coreceptor switching in SHIVSF162P3N-infected macaques that have developed a neutralizing antibody response, to discern the impact of humoral immune selection forces on the tempo and molecular pathways available for tropism switch.
Acknowledgments
We are grateful to William Olsen for PRO 542, Oliver Hartley for PSC-RANTES, David Kabat for the RC49 and JC53 cell lines, Dennis Burton and Susan Zolla-Pazner for the IgG1b12 and 447-52D antibody, respectively, Shibo Jiang for T20, and Hiroshi Mohri for help in Statistical Analysis. The TZM-bl cell line (catalog no. 8129 from Dr. John C. Kappes, Xiaoyun Wu and Tranzyme, Inc.) was obtained through the NIH AIDS Research and Reagent Program, Division of AIDS, NIAID, NIH; and the SIVnef antibody (from Dr. K. Kent and Ms. C. Arnold) was obtained from the NIBSC Centralized Facility for AIDS Reagents, supported by EU Programme EVA contract (BMH4 97/2515) and the UK Medical Research Council.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was supported by NIH grants RO1AI46980 and R37AI41945. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Berger EA. HIV entry and tropism: the chemoking receptor connection. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 2.van't Wout AB, Kootstra NA, Mulder-Kampinga GA, Albrecht-van Lent N, Scherpbier HJ, Veenstra J, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. Journal of Clinical Investigation. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 5.Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, et al. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 6.Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 7.Shioda T, Levy JA, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 8.Fouchier RA, Groenink M, Kootstra NA, Tersmette M, Huisman HG, et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen MA, van 't Wout AB. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev. 2003;5:104–112. [PubMed] [Google Scholar]
- 11.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 12.Malim MH, Emerman M. HIV-1 sequence variation: drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 13.Overbaugh J, Bangham CR. Selection forces and constraints on retroviral sequence variation. Science. 2001;292:1106–1109. doi: 10.1126/science.1059128. [DOI] [PubMed] [Google Scholar]
- 14.Berkowitz RD, Alexander S, Bare C, Linquist-Stepps V, Bogan M, et al. CCR5- and CXCR4-utilizing strains of HIV-1 exhibit differential tropism and pathogenesis in vivo. Journal of Virology. 1998;72:10108–10117. doi: 10.1128/jvi.72.12.10108-10117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaak H, van't Wout AB, Brouwer M, Hooibrink B, Hovenkamp E, et al. In vivo HIV-1 infection of CD45RA(+)CD4(+) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(+) T cell decline. Proc Natl Acad Sci U S A. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors–central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 17.Regoes RR, Bonhoeffer S. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol. 2005;13:269–277. doi: 10.1016/j.tim.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Miedema F, Tersmette M, van Lier RA. AIDS pathogenesis: a dynamic interaction between HIV and the immune system. Immunol Today. 1990;11:293–297. doi: 10.1016/0167-5699(90)90116-q. [DOI] [PubMed] [Google Scholar]
- 19.Ho SH, Tasca S, Shek L, Li A, Gettie A, et al. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J Virol. 2007;81:8621–8633. doi: 10.1128/JVI.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho SH, Trunova N, Gettie A, Blanchard J, Cheng-Mayer C. Different mutational pathways to CXCR4 coreceptor switch of CCR5-using simian-human immunodeficiency virus. J Virol. 2008;82:5653–5656. doi: 10.1128/JVI.00145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren W, Tasca S, Zhuang K, Gettie A, Blanchard J, et al. Different tempo and anatomic location of dual-tropic and X4 virus emergence in a model of R5 simian-human immunodeficiency virus infection. J Virol. 2010;84:340–351. doi: 10.1128/JVI.01865-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tasca S, Ho SH, Cheng-Mayer C. R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch. J Virol. 2008;82:7089–7099. doi: 10.1128/JVI.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 24.Moyle GJ, Wildfire A, Mandalia S, Mayer H, Goodrich J, et al. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis. 2005;191:866–872. doi: 10.1086/428096. [DOI] [PubMed] [Google Scholar]
- 25.van Rij RP, Hazenberg MD, van Benthem BH, Otto SA, Prins M, et al. Early viral load and CD4+ T cell count, but not percentage of CCR5+ or CXCR4+ CD4+ T cells, are associated with R5-to-X4 HIV type 1 virus evolution. AIDS Res Hum Retroviruses. 2003;19:389–398. doi: 10.1089/088922203765551737. [DOI] [PubMed] [Google Scholar]
- 26.Kassaye S, Johnston E, McColgan B, Kantor R, Zijenah L, et al. Envelope coreceptor tropism, drug resistance, and viral evolution among subtype C HIV-1-infected individuals receiving nonsuppressive antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;50:9–18. doi: 10.1097/QAI.0b013e31818ffdff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunnik EM, Quakkelaar ED, van Nuenen AC, Boeser-Nunnink B, Schuitemaker H. Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J Virol. 2007;81:525–531. doi: 10.1128/JVI.01983-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casper C, Naver L, Clevestig P, Belfrage E, Leitner T, et al. Coreceptor change appears after immune deficiency is established in children infected with different HIV-1 subtypes. AIDS Res Hum Retroviruses. 2002;18:343–352. doi: 10.1089/088922202753519124. [DOI] [PubMed] [Google Scholar]
- 29.Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science. 1992;257:535–537. doi: 10.1126/science.1636088. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien WA, Chen IS, Ho DD, Daar ES. Mapping genetic determinants for human immunodeficiency virus type 1 resistance to soluble CD4. J Virol. 1992;66:3125–3130. doi: 10.1128/jvi.66.5.3125-3130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 32.Bou-Habib DC, Roderiquez G, Oravecz T, Berman PW, Lusso P, et al. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lusso P, Earl PL, Sironi F, Santoro F, Ripamonti C, et al. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J Virol. 2005;79:6957–6968. doi: 10.1128/JVI.79.11.6957-6968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 35.Poignard P, Klasse PJ, Sattentau QJ. Antibody neutralization of HIV-1. Immunol Today. 1996;17:239–246. doi: 10.1016/0167-5699(96)10007-4. [DOI] [PubMed] [Google Scholar]
- 36.Kuiken CL, de Jong JJ, Baan E, Keulen W, Tersmette M, et al. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastore C, Ramos A, Mosier DE. Intrinsic obstacles to human immunodeficiency virus type 1 coreceptor switching. J Virol. 2004;78:7565–7574. doi: 10.1128/JVI.78.14.7565-7574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van't Wout AB, Blaak H, Ran LJ, Brouwer M, Kuiken C, et al. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during course of human immunodeficiency virus infection. Journal of Virology. 1998;72:5099–5107. doi: 10.1128/jvi.72.6.5099-5107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, et al. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci U S A. 2002;99:16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak SL, Platt EJ, Madani N, Ferro FE, Jr, Peden K, et al. CD4, CXCR-4, and CCR5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. Journal of Virology. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platt EJ, Madani N, Kozak SL, Kabat D. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J Virol. 1997;71:883–890. doi: 10.1128/jvi.71.2.883-890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters PJ, Duenas-Decamp MJ, Sullivan WM, Brown R, Ankghuambom C, et al. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology. 2008;5:5. doi: 10.1186/1742-4690-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Etemad B, Fellows A, Kwambana B, Kamat A, Feng Y, et al. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. J Virol. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray L, Sterjovski J, Churchill M, Ellery P, Nasr N, et al. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology. 2005;337:384–398. doi: 10.1016/j.virol.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Juarez J, Alali M, Dwyer D, Collman R, et al. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999;73:9741–9755. doi: 10.1128/jvi.73.12.9741-9755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuttle DL, Anders CB, Aquino-De Jesus MJ, Poole PP, Lamers SL, et al. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res Hum Retroviruses. 2002;18:353–362. doi: 10.1089/088922202753519133. [DOI] [PubMed] [Google Scholar]
- 48.Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276:1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 49.Brown CR, Czapiga M, Kabat J, Dang Q, Ourmanov I, et al. Unique pathology in simian immunodeficiency virus-infected rapid progressor macaques is consistent with a pathogenesis distinct from that of classical AIDS. J Virol. 2007;81:5594–5606. doi: 10.1128/JVI.00202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol. 2000;74:10984–10993. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mori K, Rosenzweig M, Desrosiers RC. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Means RE, Matthews T, Hoxie JA, Malim MH, Kodama T, et al. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J Virol. 2001;75:3903–3915. doi: 10.1128/JVI.75.8.3903-3915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, et al. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J Virol. 2002;76:2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, et al. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78:6915–6926. doi: 10.1128/JVI.78.13.6915-6926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, et al. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J Virol. 2005;79:4828–4837. doi: 10.1128/JVI.79.8.4828-4837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, et al. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology. 2007;360:105–119. doi: 10.1016/j.virol.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pacheco B, Basmaciogullari S, Labonte JA, Xiang SH, Sodroski J. Adaptation of the human immunodeficiency virus type 1 envelope glycoproteins to new world monkey receptors. J Virol. 2008;82:346–357. doi: 10.1128/JVI.01299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho SH, Martin F, Higginbottom A, Partridge LJ, Parthasarathy V, et al. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J Virol. 2006;80:6487–6496. doi: 10.1128/JVI.02539-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 63.Chowdhury IH, Koyanagi Y, Takamatsu K, Yoshida O, Kobayashi S, et al. Evaluation of anti-human immunodeficiency virus effect of recombinant CD4-immunoglobulin in vitro: a good candidate for AIDS treatment. Med Microbiol Immunol. 1991;180:183–192. doi: 10.1007/BF00215247. [DOI] [PubMed] [Google Scholar]
- 64.Schwartz AJ, Alvarez X, Lackner AA. Distribution and immunophenotype of DC-SIGN-expressing cells in SIV-infected and uninfected macaques. AIDS Res Hum Retroviruses. 2002;18:1021–1029. doi: 10.1089/08892220260235380. [DOI] [PubMed] [Google Scholar]
- 65.Yearley JH, Kanagy S, Anderson DC, Dalecki K, Pauley DR, et al. Tissue-specific reduction in DC-SIGN expression correlates with progression of pathogenic simian immunodeficiency virus infection. Dev Comp Immunol. 2008;32:1510–1521. doi: 10.1016/j.dci.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, et al. Sensitivity to inhibition by B-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proceedings of the National Academy of Science. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van't Wout AB, Ran LJ, Kuiken CL, Kootstra NA, Pals ST, et al. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72:488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scoggins RM, Taylor JR, Jr, Patrie J, van't Wout AB, Schuitemaker H, et al. Pathogenesis of primary R5 human immunodeficiency virus type 1 clones in SCID-hu mice. J Virol. 2000;74:3205–3216. doi: 10.1128/jvi.74.7.3205-3216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koning FA, Kwa D, Boeser-Nunnink B, Dekker J, Vingerhoed J, et al. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 2003;188:864–872. doi: 10.1086/377105. [DOI] [PubMed] [Google Scholar]
- 70.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 71.Karlsson I, Antonsson L, Shi Y, Oberg M, Karlsson A, et al. Coevolution of RANTES sensitivity and mode of CCR5 receptor use by human immunodeficiency virus type 1 of the R5 phenotype. J Virol. 2004;78:11807–11815. doi: 10.1128/JVI.78.21.11807-11815.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Repits J, Oberg M, Esbjornsson J, Medstrand P, Karlsson A, et al. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J Gen Virol. 2005;86:2859–2869. doi: 10.1099/vir.0.81111-0. [DOI] [PubMed] [Google Scholar]
- 73.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine -mediated suppression. Nature Medicine. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 74.Low AJ, Marchant D, Brumme CJ, Brumme ZL, Dong W, et al. CD4-dependent characteristics of coreceptor use and HIV type 1 V3 sequence in a large population of therapy-naive individuals. AIDS Res Hum Retroviruses. 2008;24:219–228. doi: 10.1089/aid.2007.0140. [DOI] [PubMed] [Google Scholar]
- 75.Coetzer M, Nedellec R, Salkowitz J, McLaughlin S, Liu Y, et al. Evolution of CCR5 use before and during coreceptor switching. J Virol. 2008;82:11758–11766. doi: 10.1128/JVI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alexander M, Lynch R, Mulenga J, Allen S, Derdeyn CA, et al. Donor and recipient envs from heterosexual human immunodeficiency virus subtype C transmission pairs require high receptor levels for entry. J Virol. 2010;84:4100–4104. doi: 10.1128/JVI.02068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Orloff SL, Kennedy MS, Belperron AA, Maddon PJ, McDougal JS. Two mechanisms of soluble CD4 (sCD4)-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) infectivity and their relation to primary HIV-1 isolates with reduced sensitivity to sCD4. J Virol. 1993;67:1461–1471. doi: 10.1128/jvi.67.3.1461-1471.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 79.Hart TK, Kirsh R, Ellens H, Sweet RW, Lambert DM, et al. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc Natl Acad Sci U S A. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thali M, Furman C, Helseth E, Repke H, Sodroski J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J Virol. 1992;66:5516–5524. doi: 10.1128/jvi.66.9.5516-5524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore JP, McKeating JA, Huang YX, Ashkenazi A, Ho DD. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Groenink M, Moore JP, Broersen S, Schuitemaker H. Equal levels of gp120 retention and neutralization resistance of phenotypically distinct primary human immunodeficiency virus type 1 variants upon soluble CD4 treatment. J Virol. 1995;69:523–527. doi: 10.1128/jvi.69.1.523-527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 85.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 86.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 87.Sattentau QJ, Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci U S A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang S, Lin K, Strick N, Neurath AR. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 90.Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 91.Koshiba T, Chan DC. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J Biol Chem. 2003;278:7573–7579. doi: 10.1074/jbc.M211154200. [DOI] [PubMed] [Google Scholar]
- 92.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 93.Markovic I, Clouse KA. Recent advances in understanding the molecular mechanisms of HIV-1 entry and fusion: revisiting current targets and considering new options for therapeutic intervention. Curr HIV Res. 2004;2:223–234. doi: 10.2174/1570162043351327. [DOI] [PubMed] [Google Scholar]
- 94.Haim H, Si Z, Madani N, Wang L, Courter JR, et al. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 96.Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- 97.Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 98.Reeves JD, Lee FH, Miamidian JL, Jabara CB, Juntilla MM, et al. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J Virol. 2005;79:4991–4999. doi: 10.1128/JVI.79.8.4991-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Platt EJ, Durnin JP, Kabat D. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J Virol. 2005;79:4347–4356. doi: 10.1128/JVI.79.7.4347-4356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Derdeyn CA, Decker JM, Sfakianos JN, Zhang Z, O'Brien WA, et al. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J Virol. 2001;75:8605–8614. doi: 10.1128/JVI.75.18.8605-8614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shakirzyanova M, Ren W, Zhuang K, Tasca S, Cheng-Mayer C. Fitness disadvantage of transitional intermediates contributes to dynamic change in the infecting-virus population during coreceptor switch in R5 simian/human immunodeficiency virus-infected macaques. J Virol. 2010;84:12862–12871. doi: 10.1128/JVI.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miller ED, Duus KM, Townsend M, Yi Y, Collman R, et al. Human immunodeficiency virus type 1 IIIB selected for replication in vivo exhibits increased envelope glycoproteins in virions without alteration in coreceptor usage: separation of in vivo replication from macrophage tropism. J Virol. 2001;75:8498–8506. doi: 10.1128/JVI.75.18.8498-8506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, et al. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology. 2004;321:8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 104.Tasca S, Zhuang K, Gettie A, Knight H, Blanchard J, et al. Effect of B-cell-depletion on coreceptor switching in R5 SHIVinfection of rhesus macaques. J Virol. 2011 doi: 10.1128/JVI.02150-10. doi: 10.1128/JVI.021510-021510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 106.Mattapallil JJ, Smit-McBride Z, McChesney M, Dandekar S. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1B expression and display antiviral cytotoxic activity despite severe CD4+ T cell depletion in primary SIV infection. Journal of Virology. 1998;72:6421–6429. doi: 10.1128/jvi.72.8.6421-6429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kabat D, Kozak SL, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Platt EJ, Kozak SL, Kabat D. Critical role of enhanced CD4 affinity in laboratory adaptation of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000;16:871–882. doi: 10.1089/08892220050042819. [DOI] [PubMed] [Google Scholar]
- 110.Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, et al. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol. 2009;83:8208–8220. doi: 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Roda Husman AM, Koot M, Cornelissen M, Keet IP, Brouwer M, et al. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann Intern Med. 1997;127:882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- 112.Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Demarest JF, Jack N, Cleghorn FR, Greenberg ML, Hoffman TL, et al. Immunologic and virologic analyses of an acutely HIV type 1-infected patient with extremely rapid disease progression. AIDS Res Hum Retroviruses. 2001;17:1333–1344. doi: 10.1089/08892220152596597. [DOI] [PubMed] [Google Scholar]
- 114.Martin-Rico P, Pedersen C, Skinhoj P, Nielsen C, Lindhardt BO. Rapid development of AIDS in an HIV-1-antibody-negative homosexual man. AIDS. 1995;9:95–96. [PubMed] [Google Scholar]
- 115.Ganeshan S, Dickover RE, Korber BT, Bryson YJ, Wolinsky SM. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu S-L, Schacker T, Musey L, Shriner D, McElrath MJ, et al. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. Journal of Virology. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Michael NL, Brown AE, Voigt RF, Frankel SS, Mascola JR, et al. Rapid disease progression without seroconversion following primary human immunodeficiency virus type 1 infection–evidence for highly susceptible human hosts. J Infect Dis. 1997;175:1352–1359. doi: 10.1086/516467. [DOI] [PubMed] [Google Scholar]
- 118.Montagnier L, Brenner C, Chamaret S, Guetard D, Blanchard A, et al. Human immunodeficiency virus infection and AIDS in a person with negative serology. J Infect Dis. 1997;175:955–959. doi: 10.1086/513999. [DOI] [PubMed] [Google Scholar]
- 119.Oka S, Ida S, Shioda T, Takebe Y, Kobayashi N, et al. Genetic analysis of HIV-1 during rapid progression to AIDS in an apparently healthy man. AIDS Res Hum Retroviruses. 1994;10:271–277. doi: 10.1089/aid.1994.10.271. [DOI] [PubMed] [Google Scholar]
- 120.Schuitemaker H, Kootstra NA, de Goede RE, de Wolf F, Miedema F, et al. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]