Abstract
Metastatic melanoma cells are highly adaptable to their in vivo microenvironment and can switch between protease-dependent mesenchymal and protease-independent amoeboid invasion to facilitate metastasis. Such adaptability can be visualized in vitro, when cells are cultured in conditions that recapitulate three-dimensional microenvironments. Using thick collagen layers in cell culture and in vivo extravasation assays, we found that pigment epithelium-derived factor (PEDF) suppressed lung extravasation of aggressive melanoma by coordinated regulation of cell shape and proteolysis. In cells grown on a thick collagen bed, PEDF overexpression and exogenous PEDF blocked the rapidly invasive, rounded morphology, and promoted an elongated, mesenchymal-like phenotype associated with reduced invasion. These changes in cell shape depended on decreased RhoA and increased Rac1 activation and were mediated by the up-regulation of Rac1-GEF, DOCK3 and down-regulation of Rac1-GAP, ARHGAP22. Surprisingly, we found that PEDF overexpression also blocked the trafficking of membrane-tethered, MT1-MMP to the cell surface through RhoA inhibition and Rac1 activation. In vivo, knockdown of Rac1 and DOCK3 or overexpression of MT1-MMP was sufficient to reverse the inhibitory effect of PEDF on extravasation. Using functional studies, we demonstrated that PEDF suppressed the rounded morphology and MT1-MMP surface localization through its antiangiongenic, 34-mer epitope and the recently identified PEDF receptor candidate, PNPLA2. Our findings unveil the coordinated regulation of cell shape and proteolysis and identify an unknown mechanism for PEDF's antimetastatic activity.
Introduction
During malignant progression, melanocytes acquire proinvasive characteristics that enable metastasis, the primary cause of cancer-associated death. Metastasis involves multiple events including detachment from the primary site, intravasation and survival in circulation, extravasation, and implantation at distant sites followed by proliferation [1]. Melanoma is a highly aggressive, undifferentiated lesion that rapidly acquires single-cell migration strategies allowing tissue invasion and colonization [2]. Moreover, depending on the microenvironment, it can switch from a slow, mesenchymal-like manner to a rapid, amoeboid mode of invasion [3]. The mesenchymal-like melanoma cells are characterized by elongated spindle shape and by actin-rich filopodia and lamellipodia at the leading edge [4]. Such cells are forming contacts with the extracellular matrix (ECM) and their motility requires integrin signaling, matrix metalloproteinases (MMPs), especially membrane type 1 matrix metalloprotease (MT1-MMP), and Rac1 function [4,5]. Integrins cluster at the focal adhesions and subsequently recruit MT1-MMP, which digests ECM and recruits and activates secreted MMP-2 and MMP-13 [6]. Amoeboid-like cancer cells appear rounded and rapidly change shape while migrating. They exhibit minimal protease activity; instead, they form bleb-like protrusions and move through existing spaces within the ECM. Their rapid deformability is dependent on RhoA and its downstream target Rho/ROCK kinase (ROCK), which phosphorylates myosin II light chain (MLC) and increases actomyosin contractility [7]. Extreme adaptability of the melanoma cells to diverse microenvironments depends on the switch from the mesenchymal-like to amoeboid morphology (mesenchymal-to-amoeboid transition, MAT) and back (amoeboid-to-mesenchymal transition, AMT) [8]. Such adaptability may explain poor efficacy of the broad-spectrum MMP inhibitors in clinical trials [9].
Pigment epithelium-derived factor (PEDF) is a ubiquitously expressed secreted protein that suppresses tumor growth and metastasis by targeting tumor cells and their microenvironment. In the microenvironment, PEDF induces apoptosis of the endothelial cells [10,11] through nuclear factor κB (NF-κB) and its downstream target Fas ligand (CD95L). Conversely, PEDF decreases endothelial cell survival by blocking the nuclear factor of activated T cells and, consequently, prosurvival FLICE-inhibitory protein (c-FLIP) [12,13]. We recently demonstrated that PEDF expression is high in melanocytes and poorly aggressive melanoma and is lost during the transition to a highly aggressive, metastatic state [14]. Our group and others showed that PEDF inhibits melanoma migration and invasion and significantly reduces metastasis [14,15], but the underlying molecular events remained unknown.
MT1-MMP is required for mesenchymal stem cell migration and invasion and is also critical for tumor invasion [16,17]. MT1-MMP is processed and activated in the trans-Golgi network before arrival at the plasma membrane [18,19]. Once tethered to the plasma membrane, MT1-MMP facilitates the activation and secretion of MMP-2 and MMP-13 and promotes local invasion in three-dimensional ECM environments [20–22]. MT1-MMP is enriched at the invadopodia, actin-based membrane protrusions typical of invading tumor cells [23], where it associates with β1-integrin clusters that interact with the ECM along the leading edge of the cell and facilitates traction and matrix degradation [4,6].
Few studies examine the connection between the regulation of tumor cell morphology and MT1-MMP activity. Here we show that PEDF, a potent antiangiogenic and antimetastatic protein, which is lost in highly aggressive, metastatic melanoma [14], suppressed extravasation and colonization of the lung parenchyma, in vivo, and link PEDF's antimetastatic action with a novel molecular function; PEDF inhibited the amoeboid and stimulated a mesenchymal-like morphology by blocking RhoA and inducing Rac1 activity, thus shifting the balance in favor of Rac1. Conversely, PEDF attenuated MT1-MMP function by preventing its distribution to the plasma membrane. Unexpectedly, PEDF-dependent blockade of MT1-MMP transport to cell surface was also mediated by the shift in RhoA/Rac1 balance. We conclude that PEDF's antimetastatic action has two distinct facets: the repression of amoeboid invasion and the restriction of proteolytic activity in mesenchymal-like melanoma cells. We also mapped PEDF's anti-metastatic effects on melanoma to its antiangiogenic, 34-mer peptide, and demonstrated the involvement of the candidate receptor, PNPLA2 (PEDF-R).
Materials and Methods
Cells
Human melanoma cell lines C81-61 and C8161 were from Dr Hendrix (Children's Memorial Hospital, Chicago, IL). SBcl2 line was provided by Dr Herlyn (Wistar Institute, Philadelphia, PA) and A375 was from ATCC (Manassas, VA). All cells were maintained as frozen stocks. After transfections or viral infections, only pooled clones were used to avoid clonal variations. The cells were cultured as previously described [24–26]. Human primary melanocytes and the medium were from Cascade Biologic (Invitrogen, Carlsbad, CA).
Expression and Silencing Vectors
For inducible expression, full-length PEDF was cloned into a multiple cloning site of the lentiviral vector, pLVX-Proteotuner (Clontech, Mountain View, CA), in-frame with the destabilization domain. The addition of (250 nM) Shield1 to the culture medium protects PEDF from proteasomal degradation causing rapid accumulation. PEDF and its functional epitopes, 34-mer and 44-mer, were cloned previously [27]; we subcloned them in a lentiviral vector, prrl.CMV.EGFP.wpre. SIN [14]. Primer sequences are given in Supplementary Methods. MT1-MMP cloned into a pcDNA3.1 (+) backbone was a gift from Dr Steve Weiss (University of Michigan, Ann Arbor, MI).
All knockdowns were performed using the human lentiviral shRNAmir vector, GIPz (Open Biosystems, Huntsville, AL). For each target, we tested at least two short hairpin RNA (shRNA) constructs and chose the most efficient one (Supplementary Methods). All targets were silenced by more than 70% as was verified by quantitative reverse transcription-polymerase chain reaction (qRT-PCR; Figure W1). A nonsilencing (NS) sequence unable to target mammalian genes was used as a negative control. For shRNA sequences and testing, see Supplementary Methods.
Lentivirus Production, Transduction, and Plasmid Transfection
Lentiviruses were generated by cotransfecton of HEK-293T cells with lenti-X HT packaging system (Clontech) and the viral vector constructs. Culture supernatants were collected, concentrated, titrated, and used for viral transduction as described previously [14]. Infected cells were identified by GFP expression, enriched by cell sorting, and maintained in puromycin. Plasmid transfections were performed using Effectene reagent (Qiagen, Valencia, CA).
Western Blot Analysis and Antibodies
Western blot analysis was performed using a standard method with polyvinylidene fluoride membranes and the following antibodies: PEDF (BioProducts MD LLC, Middle Town, MD), RhoA and Cdc42 (Cell Signaling, Danvers, MA), Rac1 (Millipore, Billerica, MA), DOCK3 (AbCam, Cambridge, MA), p-MLC (Cell Signaling), MMP-2 (Cell Signaling), and MT1-MMP (Sigma-Aldrich, St Louis, MO). All Western blot analyses were performed three times with the exception of Rac1 pull-downs when DOCK3 and/or ARHGAP22 were silenced; in this case, pull-downs were performed as a functional control to verify knockdown (in addition to qRT-PCR). ImageJ software (National Institutes of Health, Bethesda, MD) was used to quantify Western blots.
RNA Extraction and qRT-PCR
Total messenger RNA (mRNA) was extracted with Qiagen RNeasy kit (Qiagen) and reverse-transcribed using SuperScript cDNA synthesis system (Promega, Madison, WI). Quantitative RT-PCR was performed with Perfecta SYBR Green SuperMix (Quanta Biosciences, Gaithersburg, MD) in Chromo4 thermal cycler (MJ Research, Waltham, MA; see Supplementary Methods for primer sequences). Each sample was tested in triplicate, and each experiment was repeated at least three times. Ribosomal Protein L19 was used as a reference mRNA in all qRT-PCR experiments. Representative experiments are shown.
Amoeboid/Mesenchymal Morphology Assays
Amoeboid/mesenchymal morphology assays were performed as previously described [28]. Twelve-well plates were coated with 700 µl of 1.7 mg/ml bovine type 1 collagen (GIBCO, Carlsbad, CA) in serumfree medium and incubated at 37°C for 1 hour. Twenty-five thousand cells were seeded in full medium atop the collagen layer. After 24 hours, the medium was replaced with low-serum (1%) or serum-free medium (SBcl2), and the cells were incubated an additional 16 hours. In the case of exogenous PEDF, the cells were incubated in low-serum medium (with PEDF) for 36 to 40 hours. Digital images of ten random fields (>500 cells treatment) were used to score the percent of elongated cells per field. Cells were classified as elongated when the length of longest process exceeded the length of the cell body by a factor of 2 or more.
Statistical Analysis
Statistical significance was determined using one-tailed Student's t test. All quantitative assays were performed in triplicate and reproduced three or more times.
Rho GTPase Pull-down Assays
Cells were serum-starved overnight in medium supplemented with 0.1% BSA and subsequently stimulated with EGF (100 ng/ml; PeproTech, Rocky Hill, NJ). Cell lysates were incubated 90 minutes on a rotating shaker at 4°C with either Rhotekin-RBD or PAK-GST beads (Cytoskeleton, Denver, CO) to detect active RhoA and Rac1/CdC42, respectively. The beads were washed in 50 mM Tris-HCl (pH 7.5) containing 1% Triton X-100, 150 mM NaCl, 10 mM MgCl2, protease inhibitor, and eluted with 2x sample buffer. An aliquot of total cell lysate was removed before pull-down to assess total Rho GTPase levels. Proteins were separated on 4% to 20% gradient polyacrylamide gels and transferred to polyvinylidene fluoride membranes, which were probed for RhoA, Rac1, or Cdc42, respectively.
Experimental Animals
Immunodeficient mice (Nu/Nu; Harlan, Indianapolis, IN) were maintained and handled at Northwestern University Center for Comparative Medicine according to the National Institutes of Health guidelines and treated according to the protocol approved by Northwestern University Animal Care and use Committee.
Lung Extravasation Assay
Lung extravasation assays were performed as previously described [29], with modifications. A375 melanoma cells were transduced with prrl. CMV.EGFP.wpre.SIN bicistronic lentiviral vector, control/empty (LV-C), or encoding PEDF (LV-PEDF) in combination with pGIPz LV-shRNAmir encoding shRNA-NS, DOCK3, or Rac1. All lentiviral vectors encoded GFP to facilitate imaging. The cells were injected into the tail vein of immunodeficient mice (Nu/Nu; Harlan). Three to five mice were used for each data point. The mice were killed 2 and 22 hours after injection, and the lungs were analyzed for GFP-positive tumor cells using Nikon AZ-100 epifluorescence macroscope (Nikon Instruments, Melville, NY) with a 5x objective.
Immunofluorescence
Cells grown on glass coverslips were fixed with 4% paraformaldehyde and permeabilized in 0.1% Triton X-100. Samples were blocked in 10% normal donkey serum, incubated overnight at 4°C with vinculin (AbCam) or MT1-MMP antibody (Sigma-Aldrich) and visualized with fluorophore-conjugated secondary antibody (Jackson Immunoresearch, West Grove, PA). Nuclei were counterstained with 4,6-diamidino-2-phenylindole, and the slides were mounted in Flouromount-G (Southern Biotech, Birmingham, AL). F-actin was visualized using conjugated phalloidin (Molecular Probes Invitrogen). Digital images were obtained using inverted epifluorescence microscope Eclipse TE200 (Nikon, Melville, NY).
Gelatin Zymography
The cells were starved overnight with 0.1% BSA, treated with 10 nM PEDF (where indicated), and conditioned medium was collected. In some experiments, MMPs were concentrated by incubation with gelatin-coated agarose beads (Sigma-Aldrich) at 4°C for 90 minutes. Samples were run on criterion zymogram gels containing gelatin substrate (Bio-Rad, Hercules, CA), renatured in 2.5% Triton, and developed at 37°C overnight. Gels were then fixed and stained with Coomassie Brilliant Blue. Activity was quantified using ImageJ software.
Results
PEDF Suppresses Amoeboid Morphology
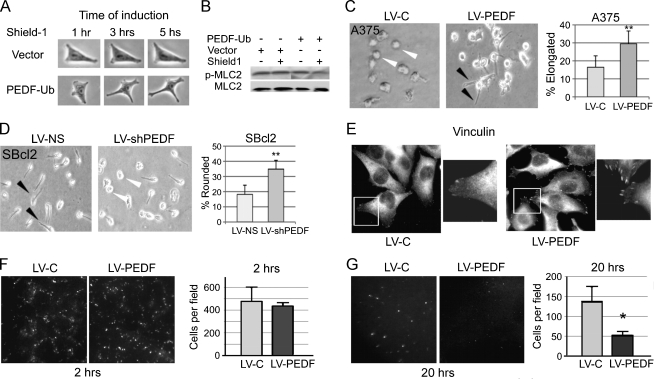
We previously showed that PEDF inhibited human melanoma metastasis in mice [13,14]. Because the amoeboid morphology is often associated with rapid invasion and increasingly metastatic behavior in melanoma [5], we sought to determine how PEDF affects melanoma morphology. In vitro, we used an inducible lentiviral expression system that allows rapid PEDF expression (iPEDF). Briefly, PEDF was tagged with proteasome-targeting degradation domain, which is masked by a small-molecule inhibitor, shield1. On addition of shield1, PEDF was stabilized and accumulated at high levels within 30 minutes (Figure W2A). We transduced A375 melanoma, a cell line that expresses no detectable PEDF, with iPEDF or control vector and evaluated cell morphology in the presence and in the absence of shield1. Within 3 hours of shield1 addition, PEDF-expressing cells assumed an elongated shape and protrusions consistent with mesenchymal-like morphology (Figure 1A). In agreement, we observed that PEDF knockdown in primary human melanocytes, which express high endogenous PEDF levels, reduced the number of processes (Figure W3). Furthermore, iPEDF expression in A375 melanoma decreased the levels of phosphorylated myosin light chain, a marker of actomyosin contractility and amoeboid phenotype (Figure 1B).
Figure 1.
PEDF promotes mesenchymal morphology and blocks lung extravasation. (A and B) A375 cells expressing inducible PEDF (iPEDF) were treated with shield1 (250 nM) to induce PEDF expression, and images were obtained at indicated time points (A). The lysates were collected 30 minutes after induction, and MLC phosphorylation was measured by Western blot (B). (C) A375 cells constitutively expressing PEDF (LV-PEDF) or vector control (LV-C) were seeded on a thick collagen bed where LV-PEDF showed a significantly higher proportion of mesenchymal cells. The data are expressed as percent elongated or rounded cells per field (10 random fields and >500 cells were examined per experiment). **P < .01. Error bars indicate standard deviation. (D) SBcl2 melanoma, expressing high endogenous PEDF infected with nonsilencing control, LV-NS, or LV-shPEDF were grown on collagen; PEDF knockdown significantly increased the rounded (amoeboid) cell population. **P < .01. (E) In A375 cells transduced with the lentiviral vector expressing PEDF (LV-PEDF) or control virus (LV-C), focal contacts were visualized by vinculin immunofluorescence. (F and G) Extravasation assay was performed with A375 (LV-C) or A375 (LV-PEDF) cells injected into the tail vein of nude mice. Lung surfaces were examined for GFP-expressing tumor cells 2 (F) and 20 hours (G) after injection. GFP-positive colonies were imaged using Nikon AZ-100 and analyzed using Metamorph software; SEM values are shown. *P < .05.
We therefore tested if PEDF could suppress the amoeboid phenotype of cells grown on a thick collagen layer, representative of a three-dimensional matrix. Constitutive PEDF overexpression in A375 using lentiviral vector (Figure W2, B and C; LV-PEDF) or treatment with exogenous recombinant PEDF (rPEDF; Figure W4) significantly increased the percentage of elongated cells compared with the vector control (LV-C) (Figure 1C). In agreement, PEDF knockdown in poorly aggressive cell line, SBcl2, which expresses high PEDF and possesses a large percentage of elongated (mesenchymal-like) cells, significantly increased the amoeboid population compared with the nonsilencing (LV-NS) control (Figures 1D and W2C). Finally, PEDF dramatically increased the number of focal adhesions, indicative of a mesenchymal, adherent phenotype, as was demonstrated by immunofluorescence for vinculin (Figure 1E). Therefore, in melanoma, PEDF promoted a mesenchymal-like morphology and its loss enabled the transition to an amoeboid phenotype.
PEDF Inhibits Extravasation to the Lung
Invasion is a key requirement for tumor cell extravasation from the blood vessels and colonization at remote sites. Because rapid extravasation is associated with the amoeboid morphology [5,29], we tested whether PEDF can block extravasation from the lung vasculature. In a short-term lung colonization assay, we injected the mouse tail vein with A375 melanoma overexpressing PEDF (LV-PEDF) or infected with control lentivirus (LV-C). Two hours after injection, equal numbers of PEDF-positive (LV-PEDF) and PEDF-null (LV-C) cells could be seen at the lung surface in a pattern following that of the lung vasculature (Figure 1F). Later, 22 to 24 hours after injection, only GFP-positive cells that escaped from circulation remained visible at the lung surface; extravasation of PEDF-positive cells was significantly lower than in control, suggesting that PEDF opposes lung extravasation and rapid invasion—a process associated with amoeboid morphology [29] (Figure 1G).
PEDF Suppresses Amoeboid Phenotype by Increasing Rac1 Activation
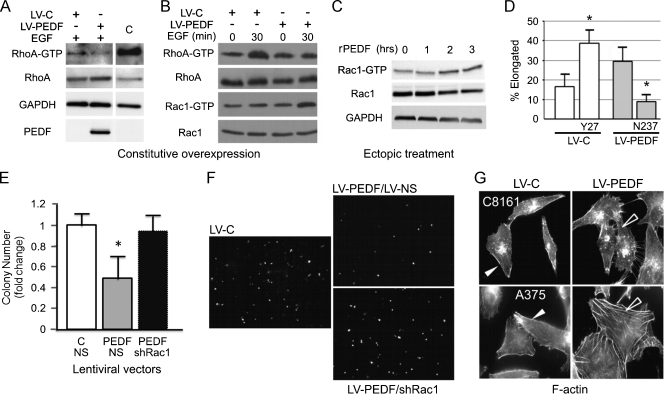
Cytoskeletal rearrangements critical for migration and invasion are regulated by the Rho GTPases, RhoA, Rac1, and Cdc42 [30]. Rho and Rac both support migration but exert opposing effects on cell shape; RhoA promotes an amoeboid morphology and rapid invasion, whereas Rac1 supports an adherent, elongated phenotype and slow invasion [31–36]. We found that PEDF overexpression reduced RhoA activity in A375 melanoma cells (Figure 2, A and B). In contrast, PEDF overexpression or treatment with recombinant PEDF (rPEDF) increased active Rac1-GTP (Figure 2, B and C). In agreement, Rac1 inhibitor, NSC23766, abrogated the increase in mesenchymal-like population caused by PEDF overexpression in A375 cells (Figure 2D). Further, blocking the Rho pathway with the ROCK inhibitor, Y27632, mimicked PEDF's ability to stimulate an elongated morphology (Figure 2D). Thus, the shift of the Rho/Rac balance in favor of Rac1 was critical for the decrease of the amoeboid population. In vivo, Rac1 silencing using lentiviral shRNA (Figure W2D; LV-shRac1) reversed the ability of PEDF to suppress extravasation (Figure 2, E and F). Finally, PEDF overexpression in A375 and C8161 (highly aggressive and PEDF-negative melanoma) cells dramatically altered their F-actin profile by reducing cortical actin, typical of the amoeboid morphology, and stimulating the formation of stress fibers, a characteristic of protease-dependent, mesenchymal-like invasion (Figure 2G). Taken together, our data show that PEDF shifts the balance between RhoA and Rac1 in favor of Rac1 and promotes mesenchymal-like state associated with a significant decrease in extravasation in vivo.
Figure 2.
PEDF regulates lung extravasation by modulating the balance of active RhoA and Rac1. (A and B) Rho and Rac pull-down assays were performed on A375 melanoma cells constitutively expressing PEDF (LV-PEDF) or vector control (LV-C): “C” indicates positive control provided with the kit. (C) Time course of Rac1 activation using 10 nM recombinant human PEDF (rPEDF). (D) The effects of RhoA and Rac1 on cell morphology: A375 LV-PEDF and A375 LV-C cells were grown on a thick collagen layer. Rho inhibitor, Y27632 (Y27), and Rac1 inhibitor, NSC23766 (N237) were added where indicated and incubated overnight. (E and F) Lung extravasation of A375 melanoma cells transduced with the indicated combinations of LV-C and LV-PEDF with LV-NS (nonsilencing control) or LVshRac1. Pictures shown are representative images taken 20 hours after injection. *P < .05. (G) F-actin profile of A375 and C8161 (HA) melanoma cells transduced with LV-C or LV-PEDF and stained with fluorescence-labeled phalloidin.
PEDF Requires DOCK3 and WAVE2 to Promote an Elongated Morphology
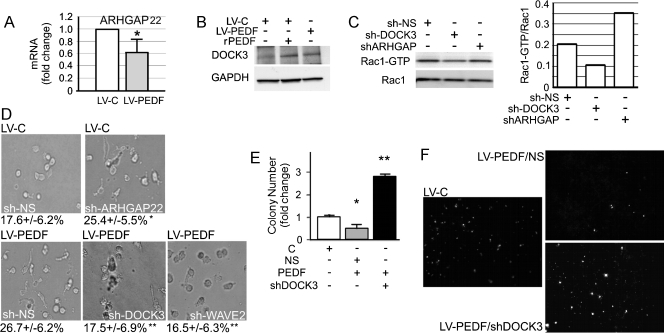
The guanine nucleotide exchange factors (GEFs), DOCK3 and DOCK10, and the GTPase-activating protein (GAP), ARHGAP22, modify Rho GTPase functions that determine elongated or rounded phenotypes [5,28]. We measured their expression in A375 control and PEDF-expressing cells (LV-PEDF) by qRT-PCR. PEDF did not have a clear effect on DOCK10, the GEF that regulates Cdc42 [28]; in agreement, PEDF had no effect on Cdc42 activity (Figure W5). In contrast, PEDF increased the expression of Rac1-specific GEF, DOCK3 (Figure 3B), and suppressed a Rho-dependent inhibitor of Rac1, ARHGAP22 (Figure 3A). Treatment of parental A375 cells with rPEDF also reduced ARHGAP22 (Figure W6) and increased DOCK3 (Figure 3B). We used lentiviral vectors encoding shRNA for DOCK3, ARHGAP22, and Rac1 downstream target, WAVE2, which caused at least 70% knockdown of their respective targets as was measured by qRT-PCR (Figure W1). Furthermore, Rac1 pull-down assay showed that DOCK3 silencing decreased active Rac1, whereas ARHGAP22 knockdown modestly increased Rac1 activation (Figure 3C). To determine whether DOCK3, WAVE2, or ARHGAP22 contributes to the PEDF-dependent AMT, we assessed cell morphology in a three-dimensional collagen matrix. In the absence of DOCK3 or Rac1 downstream effector, WAVE2, PEDF-expressing A375 cells (LV-PEDF) retained a rounded, amoeboid morphology, and ARHGAP22 silencing was sufficient to promote an elongated morphology in control (LV-C) A375 cells (Figure 3D). Together, these results suggest that PEDF activates DOCK3 GEF and inactivates ARHGAP22 to cause the shift of the Rho/Rac balance in favor of Rac1 and subsequent accumulation of adherent, mesenchymal-like cells. PEDF's requirement for DOCK3 to suppress A375 extravasation in vivo (Figure 3, E and F) further supported Rac1 involvement.
Figure 3.
PEDF attenuates ARHGAP22 expression and requires DOCK3 and WAVE2 to stimulate mesenchymal morphology. (A) Suppression of ARHGAP22 expression by PEDF (qRT-PCR). (B) DOCK3 protein expression in A375 cells infected with LV-C or LV-PEDF or treated overnight with 10 nM recombinant PEDF (rPEDF). (C) Rac1 pull-down assay in A375 cells after DOCK3 (LV-shDOCK3) or ARHGAP22 (LV-ARHGAP22) knockdown and quantification with ImageJ software. (D) Amoeboid/mesenchymal morphology of A375 LV-C or A375 LV-PEDF transduced with LV-shARHGAP22, LV-shDOCK3, LV-shWAVE2, or control LV-NS and grown on collagen. Data are shown as percent elongated cells per field. **P < .01. (E and F) Lung extravasation of A375 melanoma cells transduced with the indicated combinations of LV-C and LV-PEDF with LV-NS and LV-shDOCK3. Quantitative analysis of the experiment: highly significant increase in lung colonies formed when PEDF is overexpressed in the absence of shDOCK3 when compared with LV-PEDF. *P < .05. **P < .01.
PEDF Blocks MT1-MMP Cell Surface Localization by Blocking RhoA and Activating Rac1
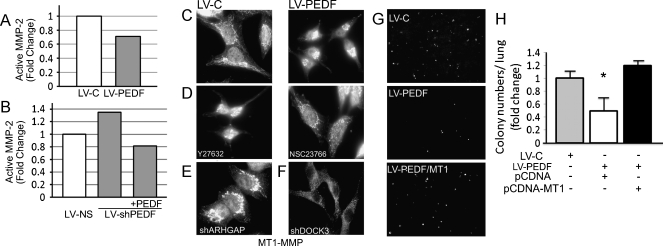
MT1-MMP activity is critical for migration and invasion of adherent cells [4]. Given that PEDF promotes cell spreading and suppresses migration, invasion, and metastasis [37], we investigated MT1-MMP function and distribution in melanoma cells overexpressing PEDF. We first tested and registered a moderate inhibition of MMP-2 activity by gelatin zymography in A375 LV-PEDF cells compared with LV-C but were unable to detect a difference in MMP-2 protein levels by Western blot (Figures 4A and W7, A and B). However, PEDF knockdown (LV-shPEDF) in SBcl2 cells, which express high levels of PEDF and are poorly aggressive, increased MMP-2 activity by gelatin zymography (Figures 4, A and B, and W7B).
Figure 4.
PEDF restricts MT1-MMP surface localization by suppressing ARHGAP22. (A and B) MMP-2 activity measured by gelatin zymography in A375 LV-C and A375 LV-PEDF (A) and in SBcl2 LV-NS and SBcl2 LV-shPEDF cells (B). LV-C and LV-NS were used as controls. (C) MT1-MMP localization in A375 LV-PEDF and control (LV-C) cells. (D) Role of Rac1 in PEDF's effect on MT1-MMP: the distribution from the perinuclear area to the cell periphery is reversed by treatment with Rac1 inhibitor, NSC23766. Note similar effect on A375 LV-C. Rho/ROCK inhibitor, Y27632, mimics perinuclear localization of MT1-MMP caused by PEDF. (E and F) MT1-MMP distribution in A375 LV-C cells after ARHGAP22 silencing with LV-shARHGAP22 (E) and in A375 LV-PEDF after DOCK3 silencing with LVshDOCK (F). (G and H) Decreased A375 extravasation caused by LV-PEDF was overcome by overexpression of MT1-MMP. **P < .01. Error bars, SEM.
Importantly, we found that forced PEDF expression or exogenous PEDF (Figure W7C) blocked MT1-MMP localization to the cell surface and confined it to the perinuclear region, a pattern not observed in LV-C controls (Figure 4C). Treatment of A375 LV-PEDF cells with Rac1 inhibitor, NSC23766, reversed PEDF's effect by causing MT1-MMP redistribution to the cell periphery. In contrast, Rho/ROCK inhibitor, Y27632, caused MT1-MMP perinuclear localization in A375 LV-C cells (Figure 4D). These results suggest that, similar to AMT, PEDF regulates MT1-MMP distribution through RhoA/Rac1 balance. To identify other molecular mediators, we silenced ARHGAP22 or DOCK3. ARHGAP22 knockdown phenocopied the MT1-MMP perinuclear localization caused by PEDF (Figure 4E). In contrast, LV-shDOCK3 restored MT1-MMP distribution in A375 LV-PEDF cells (Figure 4F). MT1-MMP overexpression in A375 LV-PEDF cells also restored their ability to extravasate in vivo in the short-term lung colonization assay (Figures 4, G and H, and W8).
PEDF Drives AMT through the 34-mer Functional Epitope and Cell Surface Receptor, PEDF-R
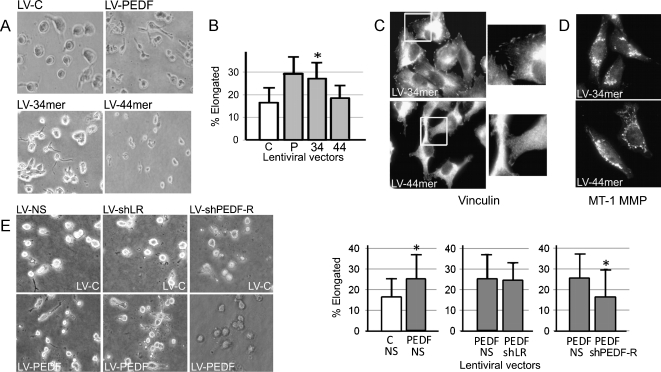
Studies by our group and others ascribed PEDF's antiangiogenic and neurotrophic activities to the distinct N-terminal peptides, the 34-mer and 44-mer, respectively [27,38,39]. To determine which fragment causes AMT, we generated lentiviral vectors encoding the 34-mer (LV-34mer) and the 44-mer (LV-44mer) peptides (Figure W9). The ability to induce AMT in the three-dimensional matrix was maintained by the 34-mer but not by the 44-mer (Figure 5, A and B). In concert, the 34-mer, but not the 44-mer, suppressed MT1-MMP distribution and increased the number of focal contacts as was measured by vinculin immunofluorescence (Figure 5, C and D). Therefore, PEDF's antimetastatic functions, including the ability to suppress amoeboid invasion and block MT1-MMP, reside in the 34-mer, PEDF fragment, previously identified as antiangiogenic.
Figure 5.
PEDF regulates AMT through the 34-mer epitope and PEDF-R cell surface receptor. (A and B) We generated A375 cells with lentiviral overexpression of PEDF, the 34-mer, and the 44-mer (LV-PEDF, LV-34mer, LV-44mer) and analyzed their morphology on collagen gels. A representative of three independent experiments is shown. *P < .05. (C) Focal adhesions were visualized by vinculin immunostaining. (D) MT1-MMP staining of A375 LV-34mer or A375 LV-44mer. (E) A375 LV-C and A375 LV-PEDF were infected with lentiviral vectors encoding shRNA targeting the two known PEDF receptors, PEDF-R (LV-shPEDF-R) or LR (LV-shLR), and morphology was analyzed by collagen assay. Data are presented as the percent elongated cells per field. *P < .05. Error bars, SD per field.
Currently, two PEDF binding partners and putative surface receptors are known, PNPLA2 (PEDF-R) and nonintegrin 37/67-kDa laminin receptor (LR) [40,41]. We silenced each candidate receptor in A375 melanoma using lentiviral vectors encoding the respective shRNA (Figure W1) and counted cells with amoeboid and mesenchymal-like morphology when grown on a thick layer of collagen. Despite the fact that LR is expressed at higher levels than PEDF-R in A375 melanoma (Figure W10), we found that PEDF-R knockdown eliminated the increase in the elongated population due to PEDF, whereas LR silencing had no effect (Figure 5E). Concurrently, we found that PEDF-R is required for the 34-mer to promote elongated morphology (Figure W11).
Discussion
Cell invasion is a critical component of metastasis and can occur by mesenchymal or amoeboid invasion strategies. Spindle-shaped mesenchymal cells with a rigid cytoskeleton rely on proteolysis to generate de novo tunnels in surrounding matrix, whereas rounded amoeboid cells possess highly contractile cortical actin that allow rapid shape deformability and penetration through existing gaps in a protease-independent manner [17,42,43]. Plasticity, the ability to rapidly switch between mesenchymal and amoeboid invasion, provides highly aggressive tumors with extreme adaptability to diverse tissue microenvironments. Thus far, it was unclear if the shape and proteolytic activity of the tumor cells are controlled independently or through related signaling pathways. Our study, for the first time, identifies an extracellular factor and its cognate receptor, PEDF-R, which promotes the mesenchymal-like phenotype and restricts surface distribution and function of MT1-MMP, a key protease for the invasion of adherent tumor cells.
PEDF is ubiquitously expressed and has potent antiangiogenic activity; it inhibits migration and invasion of the endothelial and tumor cells and halts melanoma metastasis [14,15]. We previously demonstrated that PEDF is expressed at high levels by normal melanocytes and is lost during the transition to a highly invasive, aggressive melanoma [14]. Here, we identify key cellular functions and signaling pathways regulated by PEDF to suppress tumor metastasis (Figure 6).
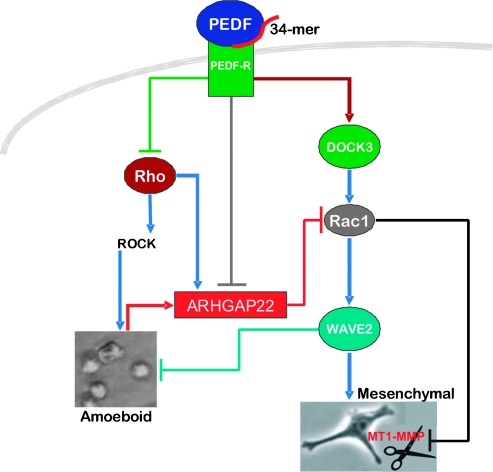
Figure 6.
Summary of the signaling pathways altered by PEDF to suppress amoeboid morphology and proteolysis, extravasation, and metastasis. PEDF, via the 34-mer epitope, activates PEDF-R, which blocks RhoA activation and represses ARHGAP22, an inhibitor of Rac1 activity. At the same time, PEDF stimulates DOCK3 expression, which drives Rac1 activity and, consequently, WAVE2 signaling. The net result is increased active Rac1 and a consequent shift in the balance of active Rac1/RhoA in favor of Rac1, which drives AMT and the accumulation of the mesenchymal-like cells. At the same time, PEDF, through the 34-mer epitope, restricts MT1-MMP (denoted as scissors) surface localization and consequent signaling. Therefore, endogenous PEDF blocks melanoma lung extravasation by “locking” melanoma cells in a mesenchymal state and simultaneously inhibiting MT1-MMP, a critical player for mesenchymal-mediated extravasation.
We found that PEDF promoted an elongated, adherent phenotype in melanoma, as was evidenced by the decreased MLC phosphorylation, increased number of elongated cells, focal contacts, and stress fiber formation. Given that PEDF blocks vasculogenic mimicry in melanoma [14] and promotes neuronal differentiation [44,45], it is not unexpected that it also regulates cellular morphology in melanoma, a disease of melanocytes—the cells of neural crest origin. We demonstrate that PEDF promotes AMT through a previously identified mechanism, pushing the balance between active RhoA and Rac1 in favor of Rac1, and that Rac1-GEF, DOCK3, and the Rac1 inhibitor, ARHGAP22, are critical regulators of this balance. We discovered that PEDF blocked proteolysis, which is required for the mesenchymal-like cells to invade, by causing MT1-MMP redistribution to the perinuclear region. Surprisingly, the redistribution of MT1-MMP was also mediated through the alteration of the RhoA/Rac1 balance, where the Rho inhibitor mimicked perinuclear accumulation of MT1-MMP by PEDF and the Rac1 inhibitor restored MT1-MMP membrane localization when PEDF was overexpressed. The evidence in favor of Rac1 precluding MT1-MMP trafficking to the cell surface was further solidified when ARHGAP22 silencing increased perinuclear MT1-MMP and DOCK3 knockdown restored MT1-MMP distribution to the cell membrane. Together, our findings clearly demonstrate that the balance between active Rac1 and RhoA regulate MT1-MMP subcellular localization and therefore activity. Our study pinpoints a relationship between the signaling components in control of cellular morphology and MT1-MMP distribution in the tumor context.
Several studies highlight the significance of MT1-MMP signaling and function in organ development and in tumor progression and demonstrate its critical role in mesenchymal movement [17]. In mammary epithelial tubule model, cells sort according to MT1-MMP expression and Rho kinase signaling [46]. Moreover, Cdc42 and MT1-MMP are essential for the single-cell invasion in three-dimensional cultures; it has been proposed that coordinated Cdc42 and MT1-MMP activity allow the formation of single-cell invasion tunnels through which fibrosarcoma cells subsequently invade [43]. Bone marrow myeloid progenitor cells require MT1-MMP to maintain typical cell morphology, lamellipodia formation, and motility [47]. By overexpressing MT1-MMP, we were able to override PEDF's antimetastatic effect and restore extravasation; thus, MT1-MMP blockade and Rac1 activation are critical drivers of PEDF's antimetastatic function.
While significant progress has been made in defining intracellular events that control tumor cell morphology, the extracellular cues that drive this transition remained unknown. Matrix stiffness is one important determinant of the modes in which macrophage-derived blood monocytes move [48]. PEDF is a secreted, ubiquitously expressed protein, which can be incorporated into the ECM, and belongs to a class of matricellular proteins, which also includes thrombospondin [49]. We identified PEDF as a paracrine factor that controls the reversible transition between mesenchymal-like and amoeboid morphology and blocks MT1-MMP function. Furthermore, we identified an active PEDF peptide, the 34-mer, and cell surface receptor, PEDF-R, that transmits this signal. In support of the fact that PEDF-R drives Rac1, we have preliminary data demonstrating that PEDF promotes melanosome dispersion and melanogenesis, a pathway stimulated by a secreted phospholipase A2 [50], in melanocytes and B16F10 melanoma by increasing cAMP, a robust inducer of Rac1.
In conclusion, we identified a previously unknown function of PEDF, where it suppresses melanoma metastasis by locking the cells in a mesenchymal-like state while abrogating MT1-MMP and proteolysis. Further, we outline a molecular mechanism through which PEDF elicits its antimetastatic effects. Given that we previously identified an 18-amino acid active peptide that maintains the 34-mer functional activity [51], this study could yield a new drug for prevention of metastasis.
Supplementary Methods
shRNAmir Constructs
All constructs were purchased from Open Biosystems (underlined clones refer to the most efficient vector to knockdown each respective transcript).
PEDF:
VL2HS_221662 shRNAmir
PEDF-R:
V2LHS_215390 shRNAmir
V2LHS_216192 shRNAmir
Laminin receptor:
V2LHS_271522 shRNAmir
V2LHS_133908 shRNAmir
DOCK3:
V2LHS_64763 shRNAmir
V2LHS_64761 shRNAmir
WAVE2:
V2LHS_95116 shRNAmir
V2LHS_269093 shRNAmir
ARHGAP22:
V3LHS_393447 shRNAmir
V3LHS_393444 shRNAmir
Primer Sequences for Real-time PCR
DOCK3:
Forward: 5′-ACA GAG AAG TAC GGC CTG TTG GTT-3′
Reverse: 5′-ACG TGT CCA TCA CAG GTC GAA AGT-3′
ARHGAP22:
Forward 5′-AGA AGG GAG TGC TGA CCT GAG AAA-3′
Reverse: 5′-ACT CCT CCA TTT CCC TCT GCA ACA-3′
DOCK10:
Forward: 5′-AGC AGT TCT GCA CCA CTC TCA GAA-3′
Reverse: 5′-AAC TGA CGT TTC CAG AGC CTC CTT-3′
WAVE2:
Forward: 5′-CCA AAC CTT TCC CTG AAG CAA CCA-3′
Reverse: 5′-ACA GGC ACT TGA AGG AAA GAG GGA-3′
PEDF-R
Forward: 5′-GCA GTT TCC TGC TGA AGG TC-3′
Reverse: 5′-GCT CGT CCT TGG AGT TGA-3′
LR
Forward: 5′-AGA AGG CAG TGA CCA AGG AGG AAT-3′
Reverse: 5′-ATT GCT GAA TAG GCA CAG AGG GCA-3′
Cloning Strategy for PEDF Fragments
N-terminal fragment (N-Ter). Target sequence was amplified from a previously described clone, pcDNA4/TO/myc-His expressing the PEDF N-Terminus [1] using the following primer sequences:
DNA: pcDNA4/TO/myc-His-N-Ter
Forward: 5′-CGGTCTAGAAGGATGCAGGCCCTGGTG-3′
Reverse: 5′-CCCTCGAGACTCAATGGTGATGGTGATGGTGGGTACCATGGATG-3′
34-Mer. The 34-mer sequence was amplified from pcDNA4/TO/myc-His-34-mer [1] using the following primer sequences:
DNA: pcDNA4/TO/myc-His-34-mer [1] Forward: 5-GGGCACAGCAGCTGCGATCCTTTCTTCAAAG-3 Reverse: 5′-CCCTCGAGACTCAATGGTGATGGTGATGGTGGTTGGTCGTGGG-3′
44-Mer. The 44-mer PEDF epitope was amplified from pcDNA4/TO/myc-His-44-mer [1] using:
DNA: pcDNA4/TO/myc-His-44-mer [1]
Forward: 5′-GGGCACAGCAGCTGCGTGCTCCTGTCTCCTC-3′
Reverse: 5′-CCCTCGAGACTCAATGGTGATGGTGATGGTGGGTACCATGGATG-3
The resultant PCR product(s) were then inserted into the lentiviral vector, prrl.CMV.EGFP.wpre.SIN [2] obtained from L. Naldini (San Raffaele - Telethon Institute for Gene Therapy, Milan, Italy) and previously used by our group [3].
Acknowledgments
The authors thank Steve Weiss at the University of Michigan for donating MT1-MMP construct and Vladimir Gelfand (Northwestern University) for helpful discussion.
Abbreviations
- PEDF
pigment epithelium-derived factor
- MT1-MMP
membrane type-1 matrix metalloproteinase
- PEDF-R
PNPLA2
- LR
laminin receptor
- AMT
amoeboid-to-mesenchymal transition
Footnotes
This work was supported by the National Institutes of Health (R01 HL68033 and R01HL077471 to O.V.) and the Ministerio de Ciencia e Innovación (SAF2007-6292 and SAF2010-19256 to B.J.). O.L. was supported by a National Institutes of Health/National Cancer Institute training grant (T32CA009560) and by the Malkin Scholars Program. J.L.O was supported by a Ministerio de Ciencia e Innovación fellowship and a SAF2007-62292 contract.
This article refers to supplementary materials, which are designated by Figures W1 to W11 and are available online at www.neoplasia.com.
References
- 1.Chaffer C, Weinberg R. A perspective on cancer cell metastasis. Science. 2011;6024:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hegerfeldt Y, Tusch M, Brocker EB, Friedl P. Collective cell movement in primary melanoma explants: plasticity of cell-cell interaction, β1-integrin function, and migration strategies. Cancer Res. 2002;62:2125–2130. [PubMed] [Google Scholar]
- 4.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 6.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 7.Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 8.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 9.Dove A. MMP inhibitors: glimmers of hope amidst clinical failures. Nat Med. 2002;8:95. doi: 10.1038/nm0202-95. [DOI] [PubMed] [Google Scholar]
- 10.Dawson DW, Volpert OV, Gillis P, Crawford SE, Xu H, Benedict W, Bouck NP. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 11.Volpert OV, Zaichuk T, Zhou W, Reiher F, Ferguson TA, Stuart PM, Amin M, Bouck NP. Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium-derived factor. Nat Med. 2002;8:349–357. doi: 10.1038/nm0402-349. [DOI] [PubMed] [Google Scholar]
- 12.Aurora AB, Biyashev D, Mirochnik Y, Zaichuk TA, Sanchez-Martinez C, Renault MA, Losordo D, Volpert OV. NF-κB balances vascular regression and angiogenesis via chromatin remodeling and NFAT displacement. Blood. 2010;116:475–484. doi: 10.1182/blood-2009-07-232132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaichuk TA, Shroff EH, Emmanuel R, Filleur S, Nelius T, Volpert OV. Nuclear factor of activated T cells balances angiogenesis activation and inhibition. J Exp Med. 2004;199:1513–1522. doi: 10.1084/jem.20040474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orgaz JL, Ladhani O, Hoek KS, Fernandez-Barral A, Mihic D, Aguilera O, Seftor EA, Bernad A, Rodriguez-Peralto JL, Hendrix MJ, et al. Loss of pigment epithelium-derived factor enables migration, invasion and metastatic spread of human melanoma. Oncogene. 2009;28:4147–4161. doi: 10.1038/onc.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia M, Fernandez-Garcia NI, Rivas V, Carretero M, Escamez MJ, Gonzalez-Martin A, Medrano EE, Volpert O, Jorcano JL, Jimenez B, et al. Inhibition of xenografted human melanoma growth and prevention of metastasis development by dual antiangiogenic/antitumor activities of pigment epithelium-derived factor. Cancer Res. 2004;64:5632–5642. doi: 10.1158/0008-5472.CAN-04-0230. [DOI] [PubMed] [Google Scholar]
- 16.Lu C, Li XY, Hu Y, Rowe RG, Weiss SJ. MT1-MMP controls human mesenchymal stem cell trafficking and differentiation. Blood. 2010;115:221–229. doi: 10.1182/blood-2009-06-228494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzone M, Baldassarre M, Beznoussenko G, Giacchetti G, Cao J, Zucker S, Luini A, Buccione R. Intracellular processing and activation of membrane type 1 matrix metalloprotease depends on its partitioning into lipid domains. J Cell Sci. 2004;117:6275–6287. doi: 10.1242/jcs.01563. [DOI] [PubMed] [Google Scholar]
- 19.Yana I, Weiss SJ. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol Biol Cell. 2000;11:2387–2401. doi: 10.1091/mbc.11.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler GS, Butler MJ, Atkinson SJ, Will H, Tamura T, Schade van Westrum S, Crabbe T, Clements J. The TIMP2 membrane type 1 metalloproteinase “receptor” regulates the concentration and efficient activation of progelatinase A. A kinetic study. 'Ortho MP, Murphy G, (1998;J Biol Chem 273:871–880. doi: 10.1074/jbc.273.2.871. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, Sato H, Okada A, Ohuchi E, Imai K, Okada Y, Seiki M. TIMP-2 promotes activation of progelatinase A by membrane-type 1 matrix metalloproteinase immobilized on agarose beads. J Biol Chem. 1998;273:16098–16103. doi: 10.1074/jbc.273.26.16098. [DOI] [PubMed] [Google Scholar]
- 22.Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 23.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- 24.Seftor EA, Brown KM, Chin L, Kirschmann DA, Wheaton WW, Protopopov A, Feng B, Balagurunathan Y, Trent JM, Nickoloff BJ, et al. Epigenetic transdifferentiation of normal melanocytes by a metastatic melanoma microenvironment. Cancer Res. 2005;65:10164–10169. doi: 10.1158/0008-5472.CAN-05-2497. [DOI] [PubMed] [Google Scholar]
- 25.Hendrix MJ, Seftor EA, Chu YW, Seftor RE, Nagle RB, McDaniel KM, Leong SP, Yohem KH, Leibovitz AM, Meyskens FL, Jr, et al. Coexpression of vimentin and keratins by human melanoma tumor cells: correlation with invasive and metastatic potential. J Natl Cancer Inst. 1992;84:165–174. doi: 10.1093/jnci/84.3.165. [DOI] [PubMed] [Google Scholar]
- 26.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819–3825. [PubMed] [Google Scholar]
- 27.Filleur S, Volz K, Nelius T, Mirochnik Y, Huang H, Zaichuk TA, Aymerich MS, Becerra SP, Yap R, Veliceasa D, et al. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 2005;65:5144–5152. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- 28.Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr Biol. 2008;18:1456–1465. doi: 10.1016/j.cub.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 29.Pinner S, Sahai E. PDK1 regulates cancer cell motility by antagonising inhibition of ROCK1 by RhoE. Nat Cell Biol. 2008;10:127–137. doi: 10.1038/ncb1675. [DOI] [PubMed] [Google Scholar]
- 30.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 31.Smith A, Bracke M, Leitinger B, Porter JC, Hogg N. LFA-1-induced T cell migration on ICAM-1 involves regulation of MLCK-mediated attachment and ROCK-dependent detachment. J Cell Sci. 2003;116:3123–3133. doi: 10.1242/jcs.00606. [DOI] [PubMed] [Google Scholar]
- 32.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 33.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 34.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worthylake RA, Lemoine S, Watson JM, Burridge K. RhoA is required for monocyte tail retraction during transendothelial migration. J Cell Biol. 2001;154:147–160. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keely PJ, Westwick JK, Whitehead IP, Der CJ, Parise LV. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- 37.Dass CR, Ek ET, Choong PF. PEDF as an emerging therapeutic candidate for osteosarcoma. Curr Cancer Drug Targets. 2008;8:683–690. doi: 10.2174/156800908786733487. [DOI] [PubMed] [Google Scholar]
- 38.Meyer C, Notari L, Becerra SP. Mapping the type I collagen-binding site on pigment epithelium-derived factor. Implications for its anti-angiogenic activity. J Biol Chem. 2002;277:45400–45407. doi: 10.1074/jbc.M208339200. [DOI] [PubMed] [Google Scholar]
- 39.Bilak MM, Becerra SP, Vincent AM, Moss BH, Aymerich MS, Kuncl RW. Identification of the neuroprotective molecular region of pigment epithelium-derived factor and its binding sites on motor neurons. J Neurosci. 2002;22:9378–9386. doi: 10.1523/JNEUROSCI.22-21-09378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard A, Gao-Li J, Franco CA, Bouceba T, Huet A, Li Z. Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor. J Biol Chem. 2009;284:10480–10490. doi: 10.1074/jbc.M809259200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notari L, Baladron V, Aroca-Aguilar JD, Balko N, Heredia R, Meyer C, Notario PM, Saravanamuthu S, Nueda ML, Sanchez-Sanchez F, et al. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006;281:38022–38037. doi: 10.1074/jbc.M600353200. [DOI] [PubMed] [Google Scholar]
- 42.Friedl P, Wolf K. Proteolytic and non-proteolytic migration of tumour cells and leucocytes. Biochem Soc Symp. 2003:277–285. doi: 10.1042/bss0700277. [DOI] [PubMed] [Google Scholar]
- 43.Fisher KE, Sacharidou A, Stratman AN, Mayo AM, Fisher SB, Mahan RD, Davis MJ, Davis GE. MT1-MMP- and Cdc42-dependent signaling co-regulate cell invasion and tunnel formation in 3D collagen matrices. J Cell Sci. 2009;122:4558–4569. doi: 10.1242/jcs.050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith ND, Schulze-Hoepfner FT, Veliceasa D, Filleur S, Shareef S, Huang L, Huang XM, Volpert OV. Pigment epithelium-derived factor and interleukin-6 control prostate neuroendocrine differentiation via feed-forward mechanism. J Urol. 2008;179:2427–2434. doi: 10.1016/j.juro.2008.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Garcia NI, Volpert OV, Jimenez B. Pigment epithelium-derived factor as a multifunctional antitumor factor. J Mol Med. 2007;85:15–22. doi: 10.1007/s00109-006-0111-z. [DOI] [PubMed] [Google Scholar]
- 46.Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM. Self-organization of engineered epithelial tubules by differential cellular motility. Proc Natl Acad Sci USA. 2009;106:14890–14895. doi: 10.1073/pnas.0901269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalo P, Guadamillas MC, Hernandez-Riquer MV, Pollan A, Grande-Garcia A, Bartolome RA, Vasanji A, Ambrogio C, Chiarle R, Teixido J, et al. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev Cell. 2010;18:77–89. doi: 10.1016/j.devcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V. Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J Immunol. 2010;184:1049–1061. doi: 10.4049/jimmunol.0902223. [DOI] [PubMed] [Google Scholar]
- 49.Bornstein P. Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J Cell Biol. 1995;130:503–506. doi: 10.1083/jcb.130.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon S, Kim NH, Koo BS, Lee HJ, Lee AY. Bee venom stimulates human melanocyte proliferation, melanogenesis, dendricity and migration. Exp Mol Med. 2007;39:603–613. doi: 10.1038/emm.2007.66. [DOI] [PubMed] [Google Scholar]
- 51.Mirochnik Y, Aurora A, Schulze-Hoepfner FT, Deabes A, Shifrin V, Beckmann R, Polsky C, Volpert OV. Short pigment epithelial-derived factor-derived peptide inhibits angiogenesis and tumor growth. Clin Cancer Res. 2009;15:1655–1663. doi: 10.1158/1078-0432.CCR-08-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Filleur S, Volz K, Nelius T, et al. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 2005;65:5144–5152. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- 2.Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 3.Orgaz JL, Ladhani O, Hoek KS, et al. Loss of pigment epithelium-derived factor enables migration, invasion and metastatic spread of human melanoma. Oncogene. 2009;28:4147–4161. doi: 10.1038/onc.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.