Abstract
ClpXP is a two-component protease composed of ClpX, an ATP-dependent chaperone that recognizes and unfolds specific substrates, and ClpP, a serine protease. One ClpXP substrate in Escherichia coli is FtsZ, which is essential for cell division. FtsZ polymerizes and forms the FtsZ ring at midcell, where division occurs. To investigate the role of ClpXP in cell division, we examined the effects of clpX and clpP deletions in several strains that are defective for cell division. Together, our results suggested that ClpXP modulates cell division through degradation of FtsZ and possibly other cell division components that function downstream of FtsZ ring assembly. In the ftsZ84 strain, which is temperature sensitive for filamentation due to a mutation in ftsZ, we observed that deletion of clpX or clpP suppresses filamentation and reduces FtsZ84 degradation. These results are consistent with ClpXP playing a role in cell division by modulating the level of FtsZ through degradation. In another division-defective strain, ΔminC, the additional deletion of clpX or clpP delays cell division and exacerbates filamentation. Our results demonstrate that ClpXP modulates division in cells lacking MinC by a mechanism that requires ATP-dependent degradation. However, antibiotic chase experiments in vivo indicate that FtsZ degradation is slower in the ΔminC strain than in the wild type, suggesting there may be another cell division component degraded by ClpXP. Taken together these studies suggest that ClpXP may degrade multiple cell division proteins, thereby modulating the precise balance of the components required for division.
INTRODUCTION
The cell division machinery in Escherichia coli is responsible for the constriction and separation of a mother cell into two daughter cells. Many proteins spatially and temporally coordinate their activities to complete division in an efficient manner. The major structural component is FtsZ, a tubulin-like protein that assembles into a circumferential ring, termed the FtsZ ring, at midcell, the site of cell division (1). FtsZ polymerizes in a reaction requiring GTP binding but not hydrolysis. In vitro, FtsZ forms a range of structures, including single-stranded protofilaments, bundles, tubules, and sheets, depending on the experimental conditions used. However, the architecture of the FtsZ ring that is formed in vivo is not fully understood. Results from electron cryotomography studies of dividing cells suggest the FtsZ ring consists of short overlapping protofilaments rather than a continuous ring (27). In vitro and in vivo FtsZ polymers undergo dynamic disassembly and reassembly, which are coupled to GTP binding and hydrolysis by FtsZ (4, 12).
Proper placement of the FtsZ ring at midcell is regulated by several factors. One regulatory system in E. coli is the Min system, comprised of MinC, MinD, and MinE (28). The Min system promotes formation of the FtsZ ring at midcell by preventing its formation near the poles. MinC interacts directly with FtsZ and inhibits FtsZ polymerization (16). MinD, a membrane-associated ATPase, recruits MinC to the membrane, where it antagonizes FtsZ assembly. The function of MinC/MinD is controlled by MinE. MinE stimulates ATPase activity of MinD and promotes dissociation of MinD complexes from the membrane (25, 26). MinC and MinD are observed to oscillate between the two poles, inhibiting FtsZ ring formation near the poles and thereby preventing anucleate cells (29). In strains deleted for minC alone, or the full minCDE operon, cell division occurs; however, the FtsZ ring is misplaced, causing minicells, elongated cells, and short filaments to form (2, 17, 40). A second regulator of FtsZ ring position is a nucleoid occlusion factor, SlmA in E. coli (8). It prevents the assembly of the FtsZ ring over the nucleoid in vivo (8). In vitro, SlmA binds directly to FtsZ, likely altering the arrangement of FtsZ protofilaments (35).
An additional regulator of FtsZ that has been identified is ClpX (11, 34, 38). ClpX is a member of the AAA+ (ATPases associated with various cellular activities) family of ATPases and forms a complex with ClpP, a serine protease. ClpX recognizes substrates harboring ClpX recognition motifs or an SsrA tag and catalyzes ATP-dependent unfolding of the polypeptide and transfer of the unfolded protein into the cavity of ClpP for degradation (32). In Bacillus subtilis, the Levin group found that in vivo ClpX helps regulate cell division and that in vitro it inhibits FtsZ polymerization in an ATP- and ClpP-independent fashion (22, 38). Recently, it has been shown that ClpX also modulates cell division in Mycobacterium tuberculosis, possibly by an ATP-independent mechanism (18).
In E. coli, FtsZ was discovered to be associated with ClpXP protease in a proteomic study aimed at identifying ClpXP substrates (19). Subsequently, it was found that ClpXP degrades FtsZ in E. coli, but only 10 to 15% is turned over per cell cycle (11). Overproduction of both ClpX and ClpP expressed from the natural promoter by using a multicopy plasmid caused an increase in the rate of FtsZ degradation and a filamentous phenotype (11). The same study showed that ClpXP degrades both FtsZ monomers and polymers in vitro (11). In a subsequent study it was reported that overexpression of either full-length ClpX or the N-terminal substrate binding domain of ClpX to very high levels causes filamentation and perturbs formation of the FtsZ ring in a wild-type background (34). It was also reported that in vitro ClpX could inhibit FtsZ polymerization by an ATP- and ClpP-independent mechanism (34).
To investigate how protein degradation by ClpXP modulates cell division in E. coli, we examined the effects of clpX and clpP deletions in several strains defective in cell division at the level of the FtsZ ring. We observed that deletion of clpX and clpP suppresses temperature-sensitive filamentation of cells carrying the ftsZ84 allele and reduces FtsZ84 degradation, consistent with ClpXP playing a role in modulating the level of FtsZ. In a minC deletion strain, in which the majority of cells contain multiple FtsZ rings, the additional deletion of either clpX or clpP delays cell division and causes enhanced filamentation; however, FtsZ turnover is decreased in both ΔminC and ΔclpX ΔminC strains compared to the wild type. Our results show that ClpXP affects cell division by a mechanism that requires ATP-dependent degradation, suggesting that ClpXP modulates the balance of cell division proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) liquid broth at 30°C. E. coli strains containing full in-frame insertion-deletions for clpX, clpP, and minC were acquired from the Keio Collection (5). Deletion mutations were brought into MG1655 by P1 transduction. To construct multiple deletion sets, we first removed the kanamycin cassettes from our single-deletion strains by transformation with pCP20 to provide the FLP recombinase (13), and then strains were cured of pCP20 by a temperature upshift to 42°C. The cured strains were transduced again with P1 lysate prepared from a different deletion strain.
Table 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotypea | Source, reference, or constructionb |
|---|---|---|
| E. coli strains | ||
| BW25113 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ−rph-1 Δ(rhaD-rhaB)568 hsdR514 | 5 |
| JW1165 | BW25113 ΔminC765::kan | 5 |
| JW0428 | BW25113 ΔclpX724::kan | 5 |
| JW0427 | BW25113 ΔclpP723::kan | 5 |
| MG1655 | λ−rph-1 | 10 |
| JC0232 | MG1655 ΔminC::frt | P1(JW1165) × MG1655; pCP20 |
| JC0259 | MG1655 ΔclpX::kan | P1(JW0428) × MG1655 |
| JC0263 | MG1655 ΔclpP::kan | P1(JW0427) × MG1655 |
| JC0292 | MG1655 ΔminC::frt ΔclpX::frt | P1(JW0428) × JC0232; pCP20 |
| JC0302 | MG1655 ΔminC::frt ΔclpP::kan | P1(JW0427) × JC0232 |
| MCZ84 | [araD139] leu-260::Tn10 ftsZ84(Ts) Δ(argF-lac)169 λ−e14−flhD5301 Δ(fruK-yeiR)725(fruA) relA1 rpsL150(Strr) rbsR22 Δ(fimB-fimE)632(::IS1) deoC1 | 15 |
| JC0303 | MCZ84 ΔclpX::kan | P1(JW0428) × MCZ84 |
| JC0304 | MCZ84 ΔclpP::kan | P1(JW0427) × MCZ84 |
| Plasmids | ||
| pCP20 | amp flp recombinase | 13 |
| pBAD24 | amp (expression vector) | 21 |
| pClpX | amp Para::clpX | This study |
| pClpX(E185Q) | amp Para::clpX(E185Q) | This study |
| pClpP | amp Para::clpP | This study |
| pGfp-FtsZ | amp Para::gfp-ftsZ | This study |
| pClpXP | amp PclpP::clpPX | 11 |
The kan cassette is flanked by frt sites for removal by FLP recombinase. An frt scar remains after removal of the cassette by using FLP expressed from pCP20.
Strain constructions by P1 transduction are described as the P1(donor) × the recipient. Where indicated, removal of the Kan cassette (by use of pCP20) is noted.
The genes for clpX, clpP, and ftsZ were amplified by PCR and cloned into expression vector pBAD24. ftsZ was cloned into expression vector pBAD-GFP(uv) (24) at the 3′ end of the gfp sequence by using the restriction sites SacI and PstI to create pGfp-FtsZ. Site-directed mutagenesis of pClpX was performed to construct pClpX(E185Q) by using the QuikChange II kit (Stratagene). To test for complementation, plasmid-containing cells were grown overnight in LB medium with ampicillin (100 μg ml−1) and arabinose (0.05% or 0.1% for strains containing pClpX or pClpP, respectively). Cells were diluted into fresh medium to an optical density at 600 nm (OD600) of 0.01 and then grown at 30°C. At an OD600 of ∼0.4, cells were examined by microscopy. Expression levels of ClpX, ClpX(E185Q), and ClpP were verified by immunoblotting.
Microscopy and immunofluorescence.
Overnight cultures were diluted into LB broth to an OD600 of 0.01 and grown at 30°C for 3 h. Where indicated, cells from an overnight culture grown in LB were diluted into M63 minimal medium with 0.2% glucose to an OD600 of 0.01, and grown at 30°C for 6 h. Cells were collected by centrifugation at 600 × g for 5 min and washed in ice-cold phosphate-buffered saline (PBS) with 1 mM EDTA. Cells were applied to a glass slide, and a poly-l-lysine coated coverslip was added.
To observe the location of FtsZ rings in live cells, we expressed Gfp-FtsZ in strains carrying the expression plasmid pGfp-FtsZ by growing the cells at 30°C in arabinose (0.015%) and ampicillin (100 μg ml−1). Log-phase cells were prepared as described above and examined immediately by fluorescence microscopy.
Cells were imaged by differential interference contrast (DIC) microscopy by using a Nomarski prism and by fluorescence microscopy using an Axio Imager M2 microscope (Zeiss) with an αPlan-Apochromat 100×/1.46-numerical aperture oil objective and the filter sets 4′,6-diamidino-2-phenylindole (DAPI) and fluorescein. Pictures were taken with an Axiocam camera. Images were processed using Adobe Photoshop CS3 and analyzed using NIH ImageJ software.
Protein turnover experiments.
E. coli strains were grown in LB at 30°C to the specified OD600 (0.3 to 0.5). To inhibit protein synthesis, spectinomycin was added to the cultures to a final concentration of 200 μg ml−1, and cell extracts were prepared at 1-h intervals after the addition of spectinomycin, as described previously (11). FtsZ turnover was monitored by immunoblotting with FtsZ antibodies as described previously (11). A minimum of three replicates was performed.
RESULTS AND DISCUSSION
Blocking protein degradation by ClpXP suppresses filamentation of ftsZ84.
Elevated levels of ClpXP increase FtsZ degradation in vivo and cause cellular filamentation (11), suggesting that ClpXP may influence cell division by modulating the intracellular concentration of FtsZ. The ftsZ84 strain carries a glycine-to-serine substitution mutation at amino acid position 105 of FtsZ (9). These cells divide normally at 28°C but form filaments at 37°C and are nonviable at 42°C (9). It has been shown that these phenotypes can be suppressed by increasing the intracellular FtsZ84 concentration (36). Therefore, to determine if protein degradation is also involved in limiting the amount of FtsZ84 available in the ftsZ84 temperature-sensitive strain (MCZ84), we deleted clpP and clpX and tested for suppression of temperature-sensitive filamentation.
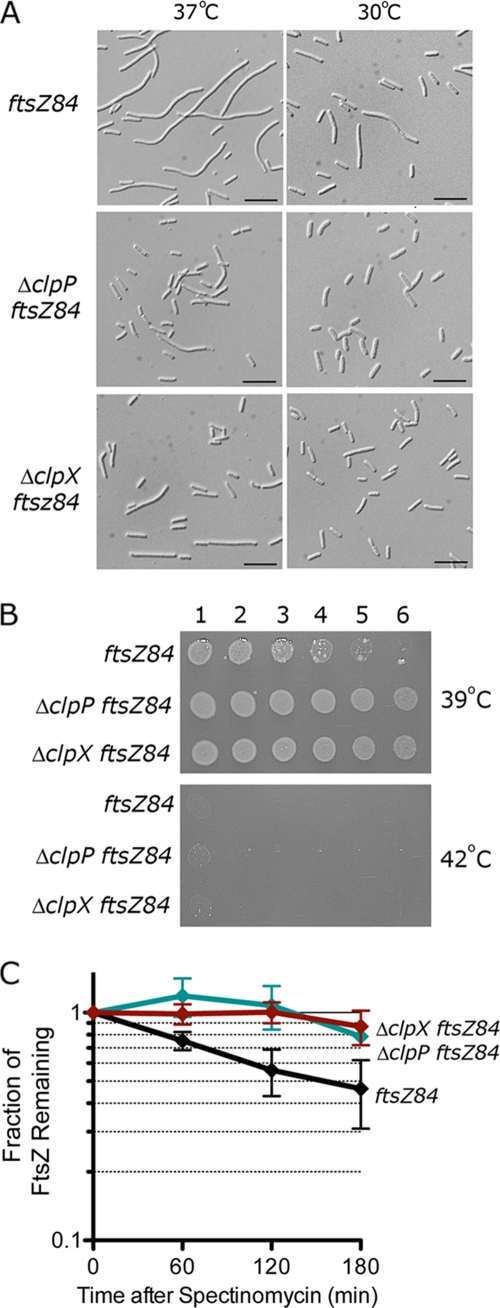
When we compared cellular morphology of ftsZ84 (MCZ84), ΔclpP ftsZ84 (JC0304), and ΔclpX ftsZ84 (JC0303) strains at 37°C, we saw that the ftsZ84 cells formed long filaments, as observed previously (Fig. 1A). In contrast, ΔclpP ftsZ84 and ΔclpX ftsZ84 cells formed elongated rods and short filaments at 37°C (Fig. 1A). A difference in cell length was also observed at 30°C between ftsZ84 cells and the double mutants, ΔclpP ftsZ84 and ΔclpX ftsZ84 (Fig. 1A); the cells of the double mutants were ∼20% shorter than ftsZ84 cells.
Fig. 1.
Deletion of clpP or clpX suppresses temperature-sensitive filamentation in cells carrying the FtsZ84 mutation. (A) ftsZ84 (MCZ84), ΔclpP ftsZ84 (JC0304), and ΔclpX ftsZ84 (JC0303) cells were grown at 30°C and 37°C to an OD600 of 0.5 and analyzed by DIC microscopy. Median cell lengths (n = 200) at 30°C for each strain were 3.6 μm ± 0.1 (ΔclpP ftsZ84), 3.5 μm ± 0.1 (ΔclpX ftsZ84), and 4.4 μm (ftsZ84). Bars, 10 μm. (B) Log dilutions of cultures grown overnight at 30°C that were then spotted onto LB plates. Plates were incubated overnight at 39°C and 42°C. (C) FtsZ84 turnover was monitored in cultures of ftsZ84, ΔclpP ftsZ84, and ΔclpX ftsZ84 strains grown at 30°C in LB after addition of spectinomycin. The relative change in FtsZ84 band intensity was monitored by immunoblotting with FtsZ antibodies.
To determine if deletion of either clpP or clpX restored viability of ftsZ84 at 42°C, the nonpermissive temperature, we grew these strains in liquid LB at 30°C and then plated the cultures onto LB plates. While none of these strains grows at 42°C, deletion of either clpP or clpX enhanced viability at 39°C (Fig. 1B). These results indicate that deletion of either clpP or clpX partially suppresses the temperature-sensitive phenotype caused by the chromosomal ftsZ84 mutation.
We next tested if degradation of FtsZ84 is reduced in ΔclpP ftsZ84 and ΔclpX ftsZ84 strains by monitoring FtsZ84 turnover by immunoblotting after halting protein synthesis. We calculated that the half-life of FtsZ84 is ∼150 min, and deletion of clpP or clpX prevents degradation of FtsZ84 (Fig. 1C). Our results suggest that deletion of clpP or clpX suppresses temperature-sensitive filamentation by decreasing FtsZ84 protein turnover.
Aberrant division in clp min double mutants.
Degradation of FtsZ modulates cell division in wild-type (11) and ftsZ84 (Fig. 1) strains. ClpX, with and without ClpP, functions as a negative regulator of FtsZ assembly in vitro, yet ClpP and ClpX are not essential proteins in E. coli (20). We hypothesized that in the absence of the main negative regulator of FtsZ assembly in the cell, MinC, ClpXP may play a more important role in cell division. To address this we constructed double deletions of either clpP or clpX with minC and examined cell length by microscopy.
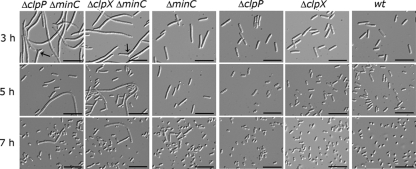
As previously seen (17), we observed that deletion of minC caused cells to divide asymmetrically, resulting in short filaments, elongated rods, and minicells (Fig. 2). Short filaments produced by the ΔminC strain were approximately 2- to 3-fold longer than cells produced by the wild-type strain. When we examined cells from exponentially growing cultures (OD600, ∼0.3; 3 h of growth) of both the clpP minC (JC0302) and the clpX minC (JC0292) double deletion strains, we observed severe defects in cell division (Fig. 2). Cultures of both ΔclpP ΔminC and ΔclpX ΔminC strains contained very long filamentous cells, in contrast to the short filaments and elongated cells of the ΔminC culture. Numerous long filaments observed in the double deletion strains were ∼10-fold longer than those of a wild-type E. coli cell, and some filaments were branched (Fig. 2). Two percent of the filaments in cultures of ΔclpP ΔminC and ΔclpX ΔminC strains were branched when cells were grown in LB broth (Fig. 2). Branches were not observed in cultures of wild-type cells or any of the strains carrying a single deletion. Branching has previously been observed in cells carrying the minCDE triple deletion under certain conditions and in combination with ftsZ84 (3, 39). The number of minicells in all strains carrying the minC deletion was similar (∼25%) with respect to the total number of cells in each culture, regardless of length. Deletion of either clpP (JC0263) or clpX alone (JC0259) did not induce filamentation, and these cells appeared similar to wild-type cells (MG1655), as previously reported (11, 20) (Fig. 2). Our results suggest that in the absence of ClpXP protease, a minC deletion causes extensive cellular filamentation.
Fig. 2.
Deletion of clpP or clpX in the minC deletion strain exacerbates cellular filamentation. ΔclpP ΔminC (JC0302), ΔclpX ΔminC (JC0292), ΔminC (JC0232), ΔclpP (JC0263), ΔclpX (JC0259), and wild-type (wt; MG1655) cells were grown at 30°C. Live cells were analyzed by DIC microscopy after growth for 3 h, 5 h, and 7 h as described in Materials and Methods. Arrows point to branches. Bars, 10 μm.
We continued to monitor cell length as cells progressed through exponential phase to determine if the ΔclpP ΔminC and ΔclpX ΔminC cells were blocked or delayed for cell division. Compared to the large number of filamentous cells seen during exponential growth of ΔclpP ΔminC and ΔclpX ΔminC cells, many fewer filaments were seen in the cultures after 5 h of growth, as cells approached stationary phase (Fig. 2; see also Fig. S1A in the supplemental material). After 7 h, the majority of cells in both cultures began to acquire a smaller round appearance typical of stationary-phase wild-type and ΔminC cells (Fig. 2). By this time, only a few relatively short filaments remained in the ΔclpP ΔminC and ΔclpX ΔminC strains. Since colony formation of all strains was similar after cells reached stationary phase (see Fig. S1B), our results suggest that the filaments seen in exponential phase undergo division, resulting in viable cells.
Cell division and cell growth are coordinately regulated based on nutrient availability (37). Slowing cell growth can overcome certain cell division delays, allowing cells more time to complete a division cycle. Culturing cells in minimal medium suppresses filamentation of ΔslmA ΔminCDE cells (8). Therefore, to determine if slow cell growth also suppresses filamentation of ΔclpX ΔminC and ΔclpX ΔminC cells, we grew these strains in M63 minimal medium with glucose (0.2%). Cultures of ΔclpX ΔminC and ΔclpP ΔminC cells grown in minimal medium were not filamentous and were similar in length to ΔminC cells (see Fig. S2 in the supplemental material).
Our results indicate that in the absence of MinC and either ClpX or ClpP, there is a profound delay in the time required to complete division when cells are grown in nutrient-rich medium. They further suggest that in the absence of MinC, ClpXP promotes FtsZ ring constriction. The observation that deletion of minC is more detrimental in a ΔclpX or ΔclpP strain implies that MinC and ClpXP have an overlapping function during division, although the mechanisms by which they carry out this function would undoubtedly be different.
ATP-dependent protein unfolding and proteolysis by ClpXP are essential for promoting FtsZ ring constriction in the minC mutant.
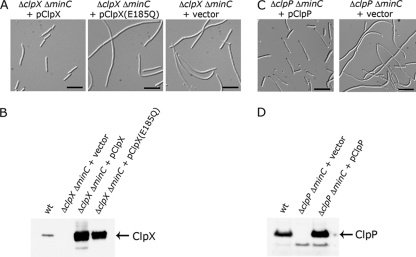
The previous result suggested that ClpXP may perform an overlapping function with MinC to promote cell division. To determine if the filamentous phenotype in the ΔclpX ΔminC strain could be rescued by the addition of ClpX, we expressed clpX from a plasmid under the control of the arabinose-inducible promoter and observed the cells by microscopy. The majority of ΔclpX ΔminC cells expressing clpX from a plasmid were short filaments (Fig. 3A and B), similar to the short filaments observed for the ΔminC strain (Fig. 2). The majority of ΔclpX ΔminC cells carrying the control vector were very long filaments (Fig. 3A). In control experiments, we observed that the overexpression of ClpX driven by the arabinose-inducible promoter did not cause filamentation in the wild-type E. coli strain MG1655 (see Fig. S3 in the supplemental material). Together, the results show that the lack of ClpX is responsible for the cell division delay in the ΔclpX ΔminC strain.
Fig. 3.
Suppression of long filaments in the ΔclpX ΔminC and ΔclpP ΔminC strains. (A) ΔclpX ΔminC (JC0292) cells containing pClpX, pClpX(E185Q), or control vector pBAD24 were grown at 30°C to an OD600 of ∼0.4 in the presence of arabinose (0.05%) and ampicillin (100 μg ml−1) and then visualized by DIC microscopy. Bars, 10 μm. (B) Extracts of wild-type (MG1655) or ΔclpX ΔminC cells expressing ClpX or ClpX(E185Q) were immunoblotted with antibodies to ClpX. (C) ΔclpP ΔminC (JC0302) cells containing pClpP or pBAD24 were grown and visualized as described above except the medium contained 0.1% arabinose. Bars, 10 μm. (D) Extracts of wild-type (MG1655) or ΔclpP ΔminC cells expressing ClpP were immunoblotted with antibodies to ClpP.
We next tested whether the ATP-dependent protein unfolding activity of ClpX is required to prevent the formation of long filaments in the ΔminC strain. To do this we analyzed cells following expression of a ClpX mutant with an amino acid substitution in the Walker B nucleotide binding motif, ClpX(E185Q). This mutation blocks the ability of ClpX to hydrolyze ATP, thus preventing ATP-dependent unfolding by ClpX (23). When plasmid-encoded ClpX(E185Q) was expressed in the ΔclpX ΔminC double deletion strain, we observed long filamentous cells similar to those seen in the double deletion strain carrying the empty vector (Fig. 3A). Expression of ClpX(E185Q) was verified by immunoblot analysis (Fig. 3B). Importantly, these results demonstrated that ATP hydrolysis by ClpX is required for the function of ClpX during cell division in ΔminC cells.
To determine whether ClpP suppresses the filamentous phenotype in the ΔclpP ΔminC double mutant, we expressed clpP from a plasmid under the control of the arabinose-inducible promoter and examined the cells by microscopy (Fig. 3C). We observed that the majority of cells expressing ClpP from a plasmid were short filaments or elongated rod-shaped cells, in contrast to the long filamentous cells carrying the control vector, indicating that the long filament phenotype in the ΔclpP ΔminC strain is suppressed by plasmid-encoded ClpP. Expression of ClpP was verified by immunoblot analysis (Fig. 3D). Taken together, our results show that both ClpX and ClpP are important for promoting cell division in the ΔminC strain. The results suggest that ATP-dependent degradation by ClpXP modulates cell division in ΔminC cells.
FtsZ localizes to rings, but the rings are slow to constrict in ΔclpP ΔminC and ΔclpX ΔminC cells.
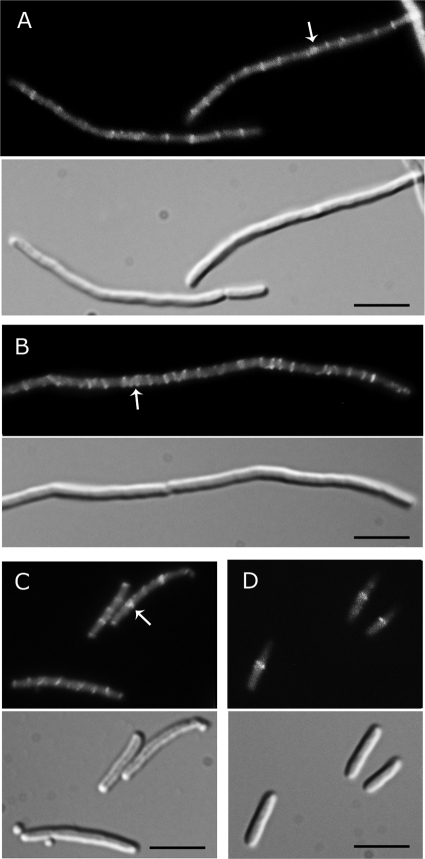
A cell division delay suggests that ΔclpP ΔminC and ΔclpX ΔminC cells may be defective in assembly of the FtsZ ring, constriction, or septation. To visualize the ring, we expressed Gfp-FtsZ from a vector under the control of the arabinose promoter to a level that was approximately 30% of the total cellular FtsZ (see Fig. S4 in the supplemental material) and observed live cells by fluorescence microscopy. ΔclpX ΔminC and ΔclpP ΔminC cells showed fluorescent FtsZ rings that appeared similar to the FtsZ rings in ΔminC cells (Fig. 4). Similar results were obtained by immunofluorescence detection of native FtsZ (see Fig. S5 in the supplemental material). We occasionally observed thick or double FtsZ rings in all strains deleted for minC (Fig. 4; see also Fig. S5). We also observed variations in FtsZ ring intensity and frequency along the length of the filament in all cells deleted for minC; a similar observation was made previously in filamentous cells deleted for minCDE (40). The ΔclpP ΔminC and the ΔclpX ΔminC strains as well as the ΔminC strain had approximately twice as many FtsZ rings per unit length as the wild-type cells (see Fig. S5), consistent with previously reported results in ΔminCDE cells (8). DNA staining with DAPI showed that FtsZ rings are positioned between nucleoids in ΔminC, ΔclpP ΔminC, and ΔclpX ΔminC cells (see Fig. S5). Our results show that although the ΔclpX ΔminC and ΔclpP ΔminC filaments are delayed for division, FtsZ rings form at intervals poised to participate in dividing the filament into many rod-shaped cells.
Fig. 4.
FtsZ rings are present in long filaments of ΔclpX ΔminC and ΔclpP ΔminC cells. Cells expressing plasmid-encoded Gfp-FtsZ were prepared as described in Materials and Methods. FtsZ rings were visible by fluorescence (upper panel) and DIC (lower panel) microscopy in all strains: ΔclpX ΔminC (A), ΔclpP ΔminC (B), ΔminC (C), and wild-type MG1655 (D). Arrows indicate positions of thick FtsZ bands or adjacent rings. Bars, 5 μm.
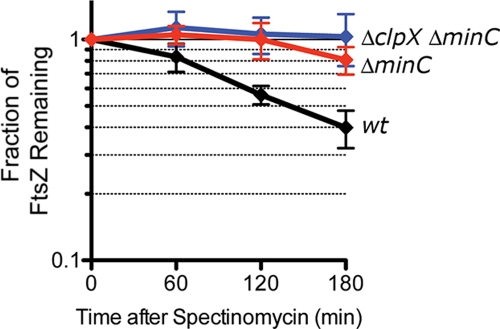
FtsZ degradation is slowed in ΔminC cells.
Since we previously observed that ClpXP degrades FtsZ in vivo (11), we compared FtsZ turnover rates in the wild-type, ΔminC, and ΔclpX ΔminC strains. We performed antibiotic chase experiments and calculated that the half-life of FtsZ in the wild-type strain grown at 30°C is ∼140 min (Fig. 5). Unexpectedly, we observed that FtsZ turnover was slower in a ΔminC strain than in a wild-type strain; in the 3 h following addition of spectinomycin, only ∼20% of the cellular FtsZ was degraded in the ΔminC strain, compared with ∼60% in the wild-type strain. FtsZ turnover was not significantly slower in the ΔclpX ΔminC strain than in the ΔminC strain (Fig. 5). These results suggest that MinC promotes degradation of FtsZ by ClpXP either directly or indirectly.
Fig. 5.
Deletion of minC reduces the rate of FtsZ degradation by ClpXP. FtsZ turnover was calculated by comparing relative FtsZ levels in each strain after addition of spectinomycin and immunoblotting as described in Materials and Methods. Strains tested include wild-type (wt) MG1655, ΔminC, and ΔclpX ΔminC.
We compared global levels of FtsZ from cultures in log phase without halting protein synthesis and found no significant differences in the levels of FtsZ in any of the strains tested (see Fig. S6 in the supplemental material). This result is in agreement with previously published work indicating that FtsZ levels do not change significantly in min mutants (39).
Since there is slower turnover of FtsZ in ΔminC and ΔminC ΔclpX strains than in a wild-type strain, one possible explanation for synthetic filamentation in the clp min double mutant is the persistence of a cell division inhibitor, which is a substrate for ClpXP. Failure to degrade an inhibitor due to deletion of clpX or clpP could lead to division delay, which may be more pronounced in a minC deletion strain, where FtsZ is diluted among multiple ring structures. A second possibility to explain these results is that ClpXP may degrade a subpopulation of FtsZ that is dependent on MinC for localization.
Next, we tested whether increasing the intracellular concentration of FtsZ by expressing FtsZ from a plasmid could suppress the formation of long filaments in ΔclpX ΔminC and ΔclpP ΔminC cultures. We observed that overexpression of FtsZ alone, or overexpression of FtsZ, FtsA, and FtsQ from the pZAQ plasmid (data not shown), reduced overall cell length and suppressed the long filament phenotype of ΔclpX ΔminC and ΔclpP ΔminC strains (see Fig. S7 in the supplemental material). This result is consistent with the above suggestion that the availability of functional FtsZ is limited in the ΔclpX ΔminC and ΔclpP ΔminC strains. However, since multiple cell division abnormalities can be suppressed by overexpression of FtsZ, including those that are synthetic with a min deletion, such as envC and slmA, as well as several cell shape mutants (6–8), the explanation may be more complex.
Model for ClpXP function during cell division.
We have shown that prevention of degradation by ClpXP partially suppresses temperature-sensitive filamentation of ftsZ84 cells and prevents FtsZ84 turnover, suggesting that ClpXP may modulate cell division through FtsZ degradation. We have also shown that deletion of clpX or clpP in combination with deletion of minC causes cell division defects. However, FtsZ degradation is reduced in the ΔminC strain compared to the wild-type strain and is not significantly more reduced in the ΔclpX ΔminC mutant, suggesting that ClpXP may act in cell division by degrading a cell division component other than FtsZ. Together, our results suggest that ClpXP may degrade multiple cell division proteins.
Any model to explain the observations must include a role for ClpX that incorporates ATP-dependent protein unfolding activity and a role for ClpP. Thus, the mechanism of action of ClpX in B. subtilis cell division, which involves ATP-independent and ClpP-independent inhibition of FtsZ polymerization (22, 38), is not easily applicable to E. coli. However, under certain conditions for E. coli where ClpX is greatly overexpressed independent of ClpP, cell division is perturbed (34), and the mechanism is likely through direct binding of ClpX to FtsZ and prevention of FtsZ polymerization.
One model to explain how ClpXP might modulate cell division, consistent with ClpXP acting directly on FtsZ (11) and FtsZ84 (Fig. 1), is that ClpXP degrades FtsZ, thereby destabilizing the FtsZ ring and allowing FtsZ polymers to undergo necessary remodeling and conformational changes associated with ring constriction and septation. This might occur as the result of FtsZ polymer severing directly through extraction and degradation of FtsZ protomers from the fibers followed by remodeling of the shorter fibers. ClpXP may also degrade free FtsZ protomers, thereby driving the FtsZ monomer-polymer equilibrium in the direction of polymer disassembly. Similar mechanisms of action have been proposed for other Clp and Clp-like proteins, such as disassembly of amyloid fibers by Hsp104 (33) and severing of microtubules by spastin (31). A notable aspect of this model is that very few cycles of FtsZ protomer degradation are required to promote polymer fragmentation, which is consistent with the relatively small amount (10 to 15% per generation) of FtsZ degradation seen in vivo (11).
Our data with minC double mutants with clpX and clpP are consistent with a model in which ClpXP promotes degradation of another cell division protein that functions downstream of FtsZ ring assembly, possibly participating in FtsZ ring remodeling, the timing of constriction or the insertion of cell wall components. For example, this unidentified cell division protein could be a division inhibitor that, in the absence of ClpXP, causes a cell division delay, which is more pronounced in a minC deletion strain. Alternatively, this unidentified protein might be a cell division component that becomes out of balance with the other components when ClpXP is not present. For example, the levels of FtsZ and FtsA are in balance; increasing FtsA causes cell division inhibition, and the inhibition is relieved by excess FtsZ (14). If FtsA turnover were controlled in a ClpXP-dependent manner, as suggested by a proteomic study (30), then deletion of clpX or clpP would cause excess FtsA, leading to division inhibition. More complicated models that involve degradation of a subpopulation of FtsZ cannot be ruled out at this time.
Perhaps it is more likely that ClpXP influences cell division by degrading multiple components, thereby modulating the finely balanced interplay of the many factors required for cell division. In E. coli there are several cell division components and regulators among the large and diverse set of proteins that have been implicated as potential ClpXP substrates, including MinD and penicillin binding proteins 5 and 6, in addition to FtsZ and FtsA (19, 30).
Supplementary Material
ACKNOWLEDGMENTS
We thank Susan Gottesman, Kumaran Ramamurthi, and Maria Sandkvist for helpful discussions, Nadim Majdalani for technical assistance, Joe Lutkenhaus for the pZAQ expression plasmid, and Matt Chenoweth for construction of pFtsZ. We are grateful to the reviewers for their insightful comments. We also thank Shannon Doyle, Danielle Johnston, Marie-Caroline Miot, and Olivier Genest for critical reading of the manuscript and helpful discussions.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute Center for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 11 February 2011.
REFERENCES
- 1. Adams D. W., Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 [DOI] [PubMed] [Google Scholar]
- 2. Akerlund T., Bernander R., Nordstrom K. 1992. Cell division in Escherichia coli minB mutants. Mol. Microbiol. 6:2073–2083 [DOI] [PubMed] [Google Scholar]
- 3. Akerlund T., Nordstrom K., Bernander R. 1993. Branched Escherichia coli cells. Mol. Microbiol. 10:849–858 [DOI] [PubMed] [Google Scholar]
- 4. Anderson D. E., Gueiros-Filho F. J., Erickson H. P. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bendezu F. O., de Boer P. A. 2008. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J. Bacteriol. 190:1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernhardt T. G., de Boer P. A. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52:1255–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernhardt T. G., de Boer P. A. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bi E., Lutkenhaus J. 1990. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). J. Bacteriol. 172:5602–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 11. Camberg J. L., Hoskins J. R., Wickner S. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U. S. A. 106:10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y., Erickson H. P. 2005. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem. 280:22549–22554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cherepanov P. P., Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 14. Dai K., Lutkenhaus J. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai K., Xu Y., Lutkenhaus J. 1993. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts). J. Bacteriol. 175:3790–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dajkovic A., Lan G., Sun S. X., Wirtz D., Lutkenhaus J. 2008. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18:235–244 [DOI] [PubMed] [Google Scholar]
- 17. de Boer P. A., Crossley R. E., Rothfield L. I. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–649 [DOI] [PubMed] [Google Scholar]
- 18. Dziedzic R., et al. 2010. Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS One 5:e11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., Baker T. A. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671–683 [DOI] [PubMed] [Google Scholar]
- 20. Gottesman S., Clark W. P., de Crecy-Lagard V., Maurizi M. R. 1993. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J. Biol. Chem. 268:22618–22626 [PubMed] [Google Scholar]
- 21. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haeusser D. P., Lee A. H., Weart R. B., Levin P. A. 2009. ClpX inhibits FtsZ assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity. J. Bacteriol. 191:1986–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hersch G. L., Burton R. E., Bolon D. N., Baker T. A., Sauer R. T. 2005. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell 121:1017–1027 [DOI] [PubMed] [Google Scholar]
- 24. Hoskins J. R., Singh S. K., Maurizi M. R., Wickner S. 2000. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. U. S. A. 97:8892–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Z., Lutkenhaus J. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337–1343 [DOI] [PubMed] [Google Scholar]
- 26. Ivanov V., Mizuuchi K. 2010. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc. Natl. Acad. Sci. U. S. A. 107:8071–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Z., Trimble M. J., Brun Y. V., Jensen G. J. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lutkenhaus J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539–562 [DOI] [PubMed] [Google Scholar]
- 29. Meinhardt H., de Boer P. A. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. U. S. A. 98:14202–14207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neher S. B., et al. 2006. Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol. Cell 22:193–204 [DOI] [PubMed] [Google Scholar]
- 31. Roll-Mecak A., Vale R. D. 2008. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 451:363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sauer R. T., et al. 2004. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shorter J., Lindquist S. 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304:1793–1797 [DOI] [PubMed] [Google Scholar]
- 34. Sugimoto S., et al. 2010. AAA+ chaperone ClpX regulates dynamics of prokaryotic cytoskeletal protein FtsZ. J. Biol. Chem. 285:6648–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tonthat N. K., et al. 2011. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 30:154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X. D., de Boer P. A., Rothfield L. I. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10:3363–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weart R. B., Levin P. A. 2003. Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 185:2826–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weart R. B., Nakano S., Lane B. E., Zuber P., Levin P. A. 2005. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol. Microbiol. 57:238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu X. C., Margolin W. 2000. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J. Bacteriol. 182:6203–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu X. C., Margolin W. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315–326 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.