Abstract
The Aeromonas hydrophila AH-3 WecP represents a new class of UDP-HexNAc:polyprenol-P HexNAc-1-P transferases. These enzymes use a membrane-associated polyprenol phosphate acceptor (undecaprenyl phosphate [Und-P]) and a cytoplasmic UDP-d-N-acetylhexosamine sugar nucleotide as the donor substrate. Until now, all the WecA enzymes tested were able to transfer UDP-GlcNAc to the Und-P. In this study, we present in vitro and in vivo proofs that A. hydrophila AH-3 WecP transfers GalNAc to Und-P and is unable to transfer GlcNAc to the same enzyme substrate. The molecular topology of WecP is more similar to that of WbaP (UDP-Gal polyprenol-P transferase) than to that of WecA (UDP-GlcNAc polyprenol-P transferase). WecP is the first UDP-HexNAc:polyprenol-P GalNAc-1-P transferase described.
INTRODUCTION
The lipopolysaccharide (LPS) is one of the major structural and immunodominant molecules of the outer membrane in Gram-negative bacteria. It consists of three moieties: lipid A, core oligosaccharide, and O-specific antigen or O side chain. The O antigen is the external component of LPS, and its structure consists of a polymer of oligosaccharide repeating units. The genetics of O-antigen biosynthesis in the Enterobacteriaceae have been intensively studied, and it has been shown that the wb* clusters usually contain genes involved in biosynthesis of activated sugars, glycosyl transferases, O-antigen polymerases, and O-antigen export (for a review, see reference 32). Despite the heterogeneity in the structures of the O antigens, only three pathways for the assembly of O antigens have been recognized: these are the Wzy-dependent pathway, the ABC transporter-dependent pathway, and the synthase-dependent pathway (14, 32). All of them initiate O-antigen LPS biosynthesis with an integral membrane protein that catalyzes the transfer of glucose (Glc)/galactose (Gal)-1-phosphate (WbaP) or N-acetylhexosamine (HexNAc)-1-phosphate onto undecaprenyl phosphate (Und-P) (38).
The UDP-HexNAc:polyprenol-P HexNAc-1-P transferase enzyme family, which includes both eukaryotic and prokaryotic transmembrane proteins, catalyzes the biosynthesis of polyprenol-linked oligosaccharides (26). These transferase-catalyzed reactions involve a membrane-associated polyprenol phosphate acceptor (Und-P) and a cytoplasmic UDP-d-N-acetylhexosamine sugar nucleotide as the donor substrate. Four subgroups of bacterial enzymes have been identified within the family, based on their specific substrate preference: these are MraY, WecA/TagO, WbcO/WbpL, and RgpG (5). MraY-type transferases are highly specific for UDP-N-acetylmuramate-pentapeptide (UDP-MurNAc-pp), whereas WecA proteins are selective for UDP-N-acetylglucosamine (UDP-GlcNAc) (31). The WbcO/WbpL substrate specificity has not yet been determined, but the structures of their biosynthetic end products imply that UDP-N-acetyl-d-fucosamine (UDP-FucNAc) and/or UDP-N-acetyl-d-quinovosamine (UDP-QuiNAc) is used (10, 41). Similar reasoning suggests that the RgpG subgroup is composed of relatively nonspecific transferases that can use either UDP-FucNAc or UDP- GlcNAc (40).
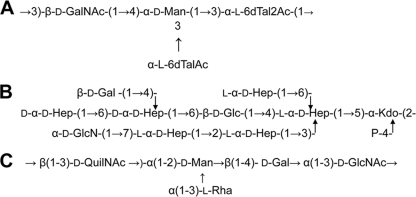
Mesophilic Aeromonas sp. serotype O34 strains have been recovered from moribund fish and from clinical specimens (17); O34 is the single most common Aeromonas serotype, accounting for 26.4% of all Aeromonas infections (18). Previous investigations have documented O34 strains as important causes of infections in humans. The Aeromonas hydrophila wbO34 gene cluster contains genes necessary for the production of O34-antigen LPS, and a role for the majority of these genes was previously attributed according to the chemical O34-antigen LPS structure obtained from strain AH-3 (Fig. 1) (20). By mutation and sequence homology, ORF15 from the A. hydrophila wbO34 gene cluster was putatively annotated as an undecaprenol-sugar-P-transferase (WecP, formerly WecA) able to initiate the assembly of the O34-antigen units, and the assembly of the O34 antigen was characterized as a Wzy-dependent pathway (20). In this study, we report the characterization of Aeromonas WecP, which represents a new class of UDP- HexNAc:polyprenol-P HexNAc-1-P transferases, according to its specific substrate preference.
Fig. 1.
Chemical structures of Aeromonas O34-antigen LPS (A) and LPS core (B) from A. hydrophila AH-3 and the O7-antigen LPS from E. coli strain VW187 (C).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are shown in Table 1. Aeromonas was grown in either tryptic soy broth (TSB) or tryptic soy agar (TSA), and Escherichia coli or Klebsiella pneumoniae was grown in Luria-Bertani (LB) Miller broth and LB Miller agar. Spectinomycin (50 μg/ml), tetracycline (20 μg/ml), chloramphenicol (25 μg/ml), gentamicin (20 μg/ml), kanamycin (50 μg/ml), or ampicillin (100 μg/ml) was added to the different media when required.
Table 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristic(s)b | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−endA hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 φ80dlacZΔM15 | 13 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44relA1 lac (F−proAB lacIqZDM15 Tn10) | Stratagene |
| S17-1 | hsdR pro recA, RP4-2 in chromosome; Km::Tn7 (Tc::Mu) | 20 |
| BL21(λD3) | F−ompThsdSB (rB− mB−) galdcm(λD3) | Novagen |
| VW187 | O7:K1; clinical isolate | M.A. Valvano |
| MV501 | VW187; wecA::Tn10 Tcr | M.A. Valvano |
| A. hydrophila strains | ||
| AH-3 | O34, wild type | 20 |
| AH-405 | AH-3, spontaneously Rifr | 20 |
| AH-3ΔwecPa | AH-3 wecP mutant in frame with pDM4 | 20 |
| S. enterica strains | ||
| LT2 | Wild type | M.A.Valvano |
| MSS2 | LT2, ΔwbaP::Cat, Cmr | M.A. Valvano |
| K. pneumoniae strains | ||
| 52145 | Wild-type O1:K2 | 30 |
| 52145ΔwecA | 51245 wecA mutant in frame with pKO3 | This study |
| Plasmids | ||
| pRK2073 | Helper plasmid, Spcr | 20 |
| pKO3 | CmrsacB temp-sensitive suicide vector | 27 |
| pGEMT-Gne | pGEM-T vector with complete gne of AH-3 | 8 |
| pBAD33 | Arabinose-inducible expression vector, Cmr | ATCC |
| pBAD33-Gm | pBAD33 vector with Gmr | 20 |
| pBAD33-WecPAh | pBAD33-Gm with A. hydrophila AH-3 wecA | This study |
| pBAD33-WecAEc | pBAD33-Gm with E. coli VW187 wecA | This study |
| pBAD33-WecAKp | pBAD33-Gm with K. pneumoniae 52145 wecA | This study |
| pBAD33-WbaPSe | pBAD33-Gm with S. enterica LT2 wbaP | This study |
| pET-30 Xa/LIC | IPTG-inducible expression vector, Kmr | Novagen |
| pET-30-WecPAh | pET-30 Xa/LIC with A. hydrophila AH-3 wecP | This study |
Formerly named AH-3ΔwecA.
IPTG, isopropyl-β-d-thiogalactopyranoside.
General DNA methods.
Standard DNA manipulations were done essentially as described previously (37). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers.
DNA sequencing and computer analysis of sequence data.
Double-stranded DNA sequencing was performed by using the dideoxy-chain termination method (39) with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). Oligonucleotides used for genomic DNA amplifications and DNA sequencing were purchased from Sigma-Aldrich. The DNA sequence was translated in all six frames, and all open reading frames (ORFs) were inspected. Deduced amino acid sequences were compared with those from DNA translated in all six frames from nonredundant GenBank and EMBL databases by using the BLAST (2) network service at the National Center for Biotechnology Information and the European Biotechnology Information. ClustalW was used for multiple-sequence alignments (6).
Mutant construction.
The K. pneumoniae 52145ΔwecA mutant was constructed by performing an in vitro in-frame deletion of the gene (27) as previously described (16). The mutant was constructed using 52145 chromosomal DNA and primers 52145wecA-A (5′-GAAGATCTGTATAATGGCGCCGGATAG-3′), 52145wecA-B (5′-CCCTACCACTAAACTTAAACACGAGGCGTGAAGGAATATAA-3′; bold type indicates restriction sites), 52145wecA-C (5′-TGTTTAAGTTTAGTGGATGGGGCGTTTGTCCTGATTACCC-3′), and 52145wecA-D (5′-GAAGATCTCGACTATCCAGCCTTTTCC-3′) in two sets of asymmetric PCRs to amplify DNA fragments AB and CD), respectively. DNA fragments AB and CD were annealed at their overlapping region (underlined letters in the 52145wecA-B and -C primer sequences) and amplified by PCR as a single fragment, using primers 52145wecA-A and -D. The fusion product was purified, BamHI digested, ligated into BamHI-digested and phosphatase-treated pKO3 vector, electroporated into E. coli DH5α, and plated on chloramphenicol LB agar plates at 30°C to obtain plasmid pKO3ΔwecAKp. The mutated gene was transferred to the chromosome by homologous recombination using the temperature-sensitive suicide plasmid pKO3, containing the counterselectable marker sacB. The plasmid with the engineered in-frame deletion (pKO3ΔwecAKp) was transferred into K. pneumoniae 52145 by transformation. Mutants were selected based on growth in LB agar containing 10 to 15% sucrose and loss of the chloramphenicol resistance marker of vector pKO3 (25). Mutation was confirmed by sequencing of the whole constructs in amplified PCR products.
Plasmid constructions and mutant complementation studies.
For complementation studies, the A. hydrophila AH-3 wecP, E. coli VW187 wecA, K. pneumoniae 52145 wecA, and Salmonella enterica LT2 wbaP genes were PCR amplified by using specific primer pairs and ligated to the plasmid pBAD33-Gm (22) (see the list of primers in Table SP1 in the supplemental material). The plasmid constructions were transformed into E. coli DH5α by electroporation, plated on gentamicin LB agar plates and incubated at 30°C. Plasmids with the amplified genes were independently transferred into the corresponding mutants by triparental mating using the mobilizing strain HB101/pRK2073 in Aeromonas or transformation by electroporation into E. coli, S. enterica, or K. pneumoniae. Transconjugants were selected on plates containing gentamicin (and rifampin for Aeromonas) and confirmed by PCR. Each gene was expressed from the arabinose-inducible and glucose-repressible pBAD33 promoter (PBAD). Repression from the araC promoter was achieved by growth in medium containing 0.2% (wt/vol) d-glucose, and induction was obtained by adding l-arabinose to a final concentration of 0.2% (wt/vol) (21).
Purification of A. hydrophila AH-3 His6-WecP.
For wecP overexpression, the pET-30 Xa/LIC vector (Novagen) and AccuPrime (Invitrogen) high-fidelity polymerase were used. The A. hydrophila AH-3 wecP was amplified from genomic DNA by using primers PET-A3wec-for (5′-GGTATTGAGGGTCGCATGCTTGCTGTGTGTTTACC-3′) and PET-A3wec-rev (5′-AGAGGAGAGTTAGAGCCATTCCTTTCTTCCCAAAGC-3′), and the PCR product was ligated into pET-30 Xa/LIC (Novagen) and electroporated into E. coli BL21(λDE3). The His6-WecP protein was overexpressed and cell lysates were obtained as previously reported for other proteins (8, 22). The total membrane fraction was obtained by ultracentrifugation (200,000 × g for 30 min at 10°C), the His6-WecP protein was solubilized and purified with a Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen) as previously reported (1). When needed, the His6-WecP protein was concentrated using a Centriplus 10-ml YM-30 centrifugal filter device (Amicon Bioseparations), and typical protein preparations contained yields of 0.07 to 0.08 g/ml as determined by the Bio-Rad Bradford assay.
LPS isolation and electrophoresis.
Cells were grown in LB, washed with water, and dehydrated by sequential washing with methanol-chloroform (1:1) (3 volumes), ethanol, acetone (2 volumes), and diethyl ether. The LPS was extracted from dehydrated cells after evaporation of the last dissolvent at room temperature. The phenol-chloroform/light petroleum ether method (11) was used for strains producing rough LPS (without O antigen), while the phenol-water procedure (48) was used for the strains producing the O-antigen domain (smooth LPS). For screening purposes, LPS was obtained after proteinase K digestion of whole cells (15). LPS samples were separated by SDS-PAGE or N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine (Tricine)-SDS-PAGE and visualized by silver staining as previously described (42).
Preparation of oligosaccharides.
The LPS preparations (20 mg) were hydrolyzed in 1% acetic acid (100°C for 120 min), and the precipitate was removed by centrifugation (8,000 × g for 30 min) and lyophilized to give lipid A. The supernatants were fractionated on a column (56 by 2.6 cm) of Sephadex G-50 in 0.05 M pyridinium acetate buffer, pH 4.5, with monitoring using a differential refractometer to obtain the oligosaccharide fractions.
GC-MS analysis.
Partially methylated alditol acetates and methyl glycoside acetates were analyzed on an Agilent Technologies 5973N mass spectrometry (MS) instrument equipped with a 6850A gas chromatograph (GC) and an RTX-5 capillary column (30-m by 0.25-mm inner diameter [i.d.]; flow rate, 1 ml/min; helium as the carrier gas; Restek). Acetylated methyl glycoside analysis was performed with the following temperature program: 150°C for 5 min, increase from 150°C to 250°C at 3°C/min, and 250°C for 10 min. For partially methylated alditol acetates, the temperature program was 90°C for 1 min, increase from 90°C to 140°C at 25°C/min, increase from 140°C to 200°C at 5°C/min, increase from 200°C to 280°C at 10°C/min, and 280°C for 10 min.
Isolation of cell envelope fractions.
The cultures were grown on TSB at 30°C. Cell envelopes were prepared by lysis of whole cells in a French press at 16,000 lb/in2. Unbroken cells were removed by centrifugation at 10,000 × g for 10 min, and the envelope fraction (membrane fraction) was collected by centrifugation at 100,000 × g for 2 h.
WecP transferase assay.
The reaction mixture used for the in vitro transferase assay contained approximately 50 μg of the total protein membrane fraction from A. hydrophila AH-3 or A. hydrophila AH-3ΔwecP(pBAD33-WecPAh) (providing the enzyme and the endogenous Und-P acceptor), 10 mM MgCl2, and 131.4 pmol of UDP-[14C]GalNAc (237 mCi/mmol) or UDP-[14C]GlcNAc (230 mCi/mmol) (NEN Life Science), in 250 μl of 10 mM Tris-HCl (pH 8.5). The mixture was incubated for 30 min at 37°C, and the lipid fraction was extracted twice with the same volume of 1-butanol. The extracts were washed at least three times with the same volume of distilled water, and the amount of radioactivity incorporated into the lipid fraction was measured in a scintillation counter. Radioactive counts were normalized to the background value by using 50 μg of the total protein membrane fraction from the AH-3ΔwecP mutant (lacking the enzyme) instead of the wild-type membrane fraction. The assay was also performed using purified His6-WecP (0 to 10 ng) and 50 μg of the total protein membrane fraction from A. hydrophila AH-3ΔwecP (providing the endogenous Und-P acceptor). One unit of enzyme activity was defined as 10−3 pmol of GalNAc/GlcNAc incorporated into the lipid fraction per min per mg of protein in the reaction.
RESULTS
A. hydrophila WecP characterization.
A BLAST-X analysis of A. hydrophila AH-3 WecP revealed similar proteins (identities around 45 to 50% and similarities around 60 to 70%) in a number of bacteria like Leptospira or Oceanobacter spp. besides the genus Aeromonas (>90% similarity). However, the well-characterized Enterobacteriaceae WecA protein showed low-level similarities with A. hydrophila WecP (identities below 30% and similarities below 50%) (data not shown).
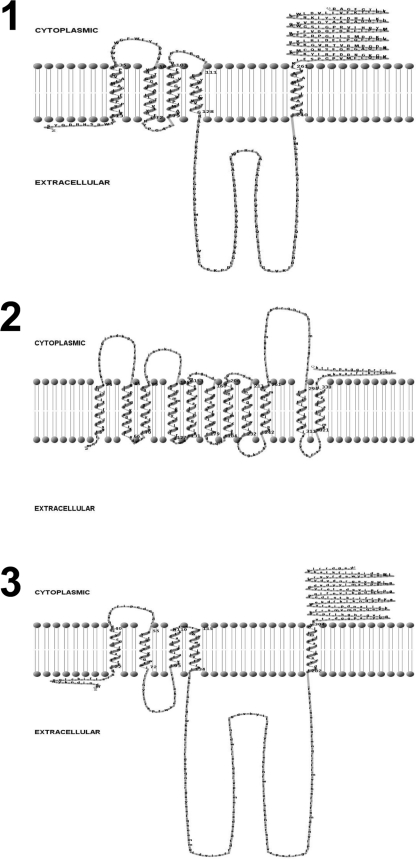
Computer programs such as TMHHM 2.0 (prediction of transmembrane helices in proteins, from the Center for Biological Sequence Analysis, Denmark [http://www.cbs.dtu.dk/services/TMHMM/]) have been used to predict transmembrane helices and membrane topology models for proteins involved in the transfer of sugar-phosphate residues to undecaprenol phosphate (Und-P), such as WecA (25) and WbaP (36). The same computer program was used to model the A. hydrophila AH-3 WecP molecular topology, revealing three predicted domains that could be clearly delineated: these were an N-terminal region containing a cluster of four transmembrane helices, a large central periplasmic loop, and a C-terminal cytosolic tail (Fig. 2). The A. hydrophila AH-3 WecP molecular topology model resembles that of Salmonella enterica WbaP, which catalyzes the transfer of Gal-1-P from UDP-Gal onto Und-P (34). The WbaP membrane topology model (Fig. 2) also consists of an N-terminal region containing a cluster of four transmembrane helices, a large central periplasmic loop, and a C-terminal cytosolic tail (36). Previous work (45) has shown that the C-terminal half of WbaP carries the Gal-1-P transferase activity. In contrast, the topological model for E. coli WecA (Fig. 2), which is responsible for the transfer of GlcNAc-1-P from UDP-GlcNAc to polyprenol-P, consists of 11 transmembrane helices, five cytosolic loops, and five periplasmic loops (25).
Fig. 2.
Membrane topological models for S. enterica LT2 WbaP (panel 1), E. coli VW187 WecA (panel 2), and A. hydrophila AH-3 WecP (panel 3).
Because of the differences between Aeromonas WecP and Enterobacteriaceae WecA, we suggest the name of WecP for this protein in Aeromonas (Wec for being a putative UDP-HexNAc:polyprenol-P HexNAc-1-P transferase and P for the similarity in the molecular topology to WbaP). Note that the names WecB, -C, and -D are already used (29). In order to identify the sugar donor for WecP, we decided to introduce well-known sugar-Und-P transferases into an A. hydrophila wecP mutant as well as this WecP protein into well-characterized wecA and wbaP mutants.
Complementation studies of the AH-3ΔwecP mutant.
As we previously published, the AH-3ΔwecP mutant was completely unable to biosynthesize the O34-antigen LPS (20). The mutant harboring plasmid pBAD33-WecPAh (carrying the gene from strain AH-3) showed an LPS profile identical to that of the wild-type strain, while no changes could be observed when the mutant carried the plasmid vector alone or plasmid pBAD33-WbaPSe carrying the Salmonella enterica LT2 wbaP (Fig. 3). When plasmid pBAD33-WecAEc (carrying the E. coli VW187 [O7] wecA) was introduced in the mutant and expressed with arabinose, we could see two bands on LPS gels (the band corresponding to the mutant plus an additional band with reduced motility) (Fig. 3). Isolated LPS of the AH-3ΔwecP mutant with plasmid pBAD33-WecAEc grown under expressing conditions (with arabinose) was devoid of high-molecular-mass O-antigen polysaccharide (O-antigen PS). Sugar analysis of LPS (see Materials and Methods) from the AH-3ΔwecP mutant renders the following monosaccharides: glucose (Glc), Gal, glucosamine (GlcN), l-glycero-d-manno-heptopyranose and d-glycero-d-manno-heptopyranose (l,d-Hep and d,d-Hep, respectively), and 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo), corresponding to the LPS core (Fig. 1). However, the sugar analysis of LPS from AH-3ΔwecP mutant with plasmid pBAD33-WecAEc grown under expressing conditions (with arabinose) showed the same monosaccharides as described above plus 6-deoxy-talose (l-6dTal) and mannose (d-Man), in agreement with the reported O34-antigen LPS (Fig. 1) (20, 23), but with GlcNAc instead of N-acetyl-galactosamine (GalNAc), which is the other sugar from the O34-antigen LPS in the wild-type strain (Fig. 1). The amount of GlcNAc is similar to the amounts of d-Man and d-Glc and about half of the amount of l-6dTal. From these results, it seems that the additional band with reduced motility observed in the LPS gels for the AH-3ΔwecP mutant with plasmid pBAD33-WecAEc grown under expressing conditions (with arabinose) corresponds to an LPS molecule with a single O-antigen repeating unit. No GlcNAc was detected in the LPS of AH-3ΔwecP mutant alone or with plasmid vector pBAD33.
Fig. 3.
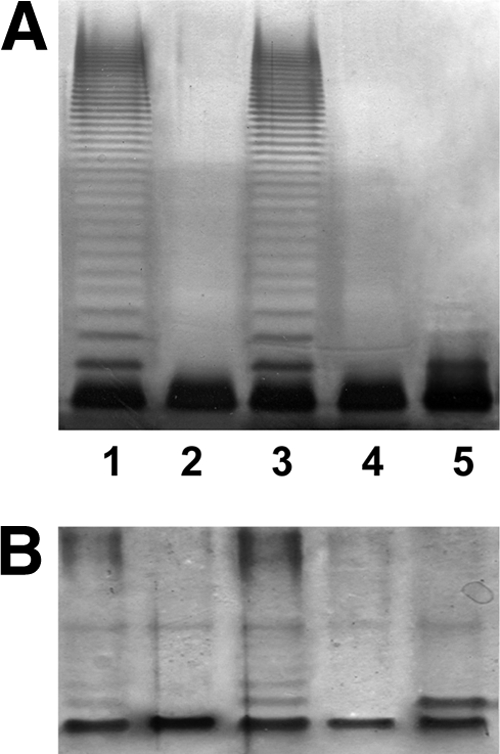
Polyacrylamide gels showing the migration of LPS from AH-3ΔwecA and its complementation. The LPS samples were separated on SDS-PAGE (A) and SDS-Tricine-PAGE (B) and visualized by silver staining. Shown are LPS samples from A. hydrophila AH-3 (wild type [WT]) (lanes 1), the AH-3ΔwecP mutant (lanes 2), AH-3ΔwecP(pBAD33-WecPAh) (lanes 3), AH-3ΔwecP(pBAD33-WbaPSe) (lanes 4), and AH-3ΔwecP(pBAD33-WecAEc) (lanes 5).
The AH-3ΔwecP mutant was complemented by different pBAD33 plasmids with several Aeromonas wecP homologues from strains like A. hydrophila PPD134/91 (20), A. caviae Sch3N (12), A. veronii biovar Sobria AH-41 (34), and A. salmonicida subsp. salmonicida A449 (21), according to their LPS profiles (data not shown).
Complementation studies of E. coli MV501 (WecA) and S. enterica MSS2 (WbaP) mutants.
Plasmid pBAD33-WecPAh (carrying the gene from strain AH-3) was unable to change the S. enterica MSS2 or E. coli MV501 LPS profile (Fig. 4). One possible reason the LPS profile not to change could be the absence of UDP-GalNAc in S. enterica strain MSS2 or E. coli strain MV501. Generation of UDP-GalNAc from UDP- GlcNAc requires the presence of a Gne (UDP-GalNAc 4-epimerase) homologue. Thus, we introduced the A. hydrophila gne-containing plasmid pGEMT-Gne (7) into S. enterica strain MSS2 and E. coli strain MV501. No changes in LPS profile were observed in S. enterica strain MSS2 carrying A. hydrophila AH-3 wecP and gne. However, when both genes were introduced into the E. coli MV501 mutant, we could see an LPS profile showing two bands (one with the same motility as the mutant and the other one with a reduced motility) (Fig. 4A and B, lanes 5). LPS isolated from the E. coli MV501 mutant with plasmids pBAD33-WecPAh and pGEMT-Gne together grown under expressing conditions (with arabinose) was sugar analyzed. Among the different sugars (Kdo, l,d-Hep, Glc, Gal, GlcNAc, Man, rhamnose [Rha], and QuiNAc), GalNAc was detected in ratios similar to Man (a unique sugar in the reported E. coli O7-antigen LPS structure [Fig. 1] [28, 47]). The presence of Man could not be detected in isolated LPS of E. coli MV501, LPS of E. coli MV501 carrying pGEMT-Gne alone, or LPS of E. coli MV501 carrying pBAD33-WecPAh alone. GalNAc has not been described as being present in LPS of E. coli strains 32 with O7-antigen LPS (Fig. 1) (25, 47) or LPS core (4) and was not detected in the LPS of E. coli MV501 with or without plasmid vector pBAD33, pBAD33-WecPAh, or pGEMT-Gne. Because it is only a single additional band in the LPS profile of the E. coli MV501 mutant with plasmids pBAD33-WecPAh and pGEMT-Gne together grown under expressing conditions (with arabinose) and the Man or GalNAc values for this LPS are always about three times less than that of Glc (a monosaccharide from the LPS core but not from the O7-antigen LPS), this additional band could account for LPS molecules with a single O-antigen repeating unit.
Fig. 4.
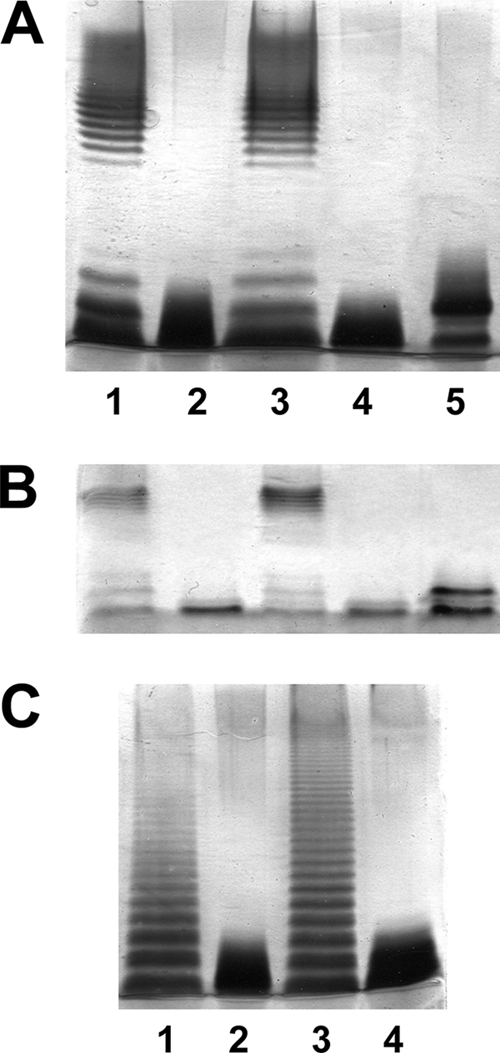
Polyacrylamide gels showing the migration of LPS from E. coli WecA (MV501) and S. enterica WbaP (MSS2) mutants and its complementation. The LPS samples were separated on SDS-PAGE (A and C) and SDS-Tricine-PAGE (B) and visualized by silver staining. (A and B) Shown are LPS samples from E. coli VW187 (WT) (lanes 1), MV501 (lanes 2), MV501(pBAD33-WecAEc) (lanes 3), MV501(pBAD33-WecPAh) (lanes 4), and MV501(pBAD33-WecPAh)(pGEMT-Gne) (lanes 5). (C) Shown are LPS samples from S. enterica LT2 (WT) (lane 1), MSS2 (lane 2), MSS2(pBAD33-WbaPSe) (lane 3), and MSS2(pBAD33-WecPAh)(pGEMT-Gne) (lane 4).
All these results suggest that A. hydrophila WecP is a UDP-HexNAc:polyprenol-P GalNAc-1-P transferase in a Wzy-dependent pathway of O-antigen assembly.
K. pneumoniae 52145 (O1:K2) WecA mutant and complementation.
WecA initiates O-unit synthesis in many Enterobacteriaceae, Klebsiella pneumoniae O1 being one of them (9). Also, this O1 antigen is an example of those using an ABC transporter-dependent pathway for assembly (38). By using the methodology described in Materials and Methods, we constructed an in-frame wecA mutant of strain 52145 (O1:K2) (30), which was devoid of the O1-antigen LPS banding pattern in gels (Fig. 5). The LPS of the 52145ΔwecA mutant harboring plasmid pBAD33-WecAKp or pBAD33-WecAEc showed in gels the same banding pattern exhibited by the wild-type LPS. In contrast, the LPS of the 52145ΔwecA mutant harboring the plasmid vector alone (data not shown), with plasmid pBAD33-WecPAh from A. hydrophila AH-3, or with pBAD33-WbaPSe from S. enterica LT2 did not show changes in its LPS profile (Fig. 5). In K. pneumoniae O1:K2, no generation of UDP-GalNAc from UDP-GlcNAc is possible, due to the lack of the epimerase enzyme, like in E. coli VW187 (O7). Therefore, when we included plasmid pGEMT-Gne together with plasmid pBAD33-WecPAh from A. hydrophila AH-3 in the 52145ΔwecA mutant grown under inducing conditions, the O1-antigen banding pattern was rescued, according to SDS-PAGE (Fig. 5). As has been previously described (34) for the LPSs of the strains using the ABC-transporter dependent pathway, like in K. pneumoniae O1, the initial sugar for the O-antigen LPS is retained in their structure. The initial sugar for the wild-type strain is GlcNAc, as reported for K. pneumoniae (43, 44). When we examined the sugar composition of the LPS from 52145ΔwecA mutant with plasmids pBAD33-WecPAh and pGEMT-Gne together grown under inducing conditions, we observed a large amount of Gal (in agreement with the reported O1-antigen LPS structure [9]) and minor GalNAc content. No GlcNAc could be detected in this LPS sample. As previously reported for strain 52145, no GalNAc was present either in the LPS core or in the K. pneumoniae O1 antigen (9, 33, 43, 44). All these results prompted us to study the in vitro enzymatic activity of WecP.
Fig. 5.
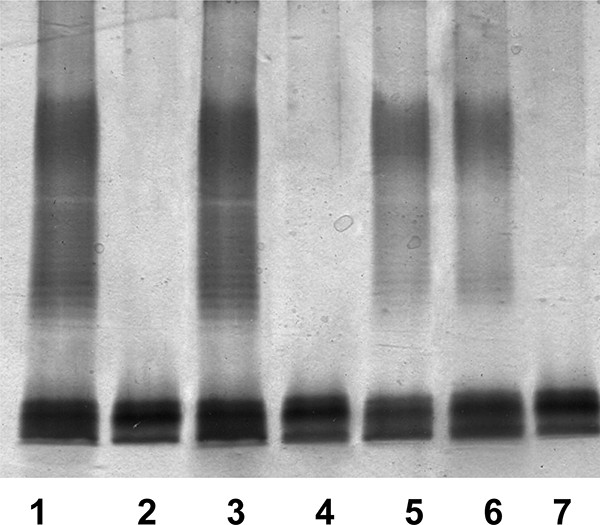
Polyacrylamide gels showing the migration of LPS from the 52145ΔwecA mutant and its complementation. The LPS samples were separated on SDS-PAGE and visualized by silver staining. Shown are LPS samples from K. pneumoniae 52145 (lane 1), 52145ΔwecA (lane 2), 52145ΔwecA(pBAD33-WecAKp) (lane 3), 52145ΔwecA(pBAD33-WecPAh) (lane 4), 52145ΔwecA(pBAD33-WecPAh)(pGEMT-Gne) (lane 5), 52145ΔwecA(pBAD33-WecAEc) (lane 6), and 52145ΔwecA(pBAD33-WbaPSe) (lane 7).
UDP-HexNAc:polyprenol-P HexNAc-1-P transferase assay.
The assay was performed as described in Materials and Methods by using the total membrane fractions of A. hydrophila AH-3 and A. hydrophila AH-3ΔwecP(pBAD33-WecPAh) as the source of both enzyme and the endogenous Und-P acceptor. As controls, the membrane fractions of A. hydrophila AH-3ΔwecP and A. hydrophila AH-3ΔwecP (pBAD33) were used. The membrane fraction from A. hydrophila AH-3 catalyzed the incorporation of GalNAc-1-P from UDP-GalNAc with an specific activity similar to that obtained when using the membrane fraction of A. hydrophila AH-3ΔwecP(pBAD33-WecPAh) grown under noninducing conditions (Table 2). Membrane fractions obtained from AH-3ΔwecP(pBAD33-WecPAh) grown in the presence of l-arabinose (inducing conditions for PBAD) resulted in about a 10-fold increase in specific activity (Table 2). None of the membrane preparations described above was able to catalyze the transfer of GlcNAc-1-P from UDP-GlcNAc at significant levels. As expected, experiments performed using a membrane preparation from A. hydrophila AH-3ΔwecP or from AH-3ΔwecA(pBAD33), both under inducing and noninducing conditions, did not catalyze the incorporation of GalNAc or GlcNAc.
Table 2.
In vitro WecP transferase assay using UDP-[14C]GlcNAc or UDP-[14C]GalNAcd
| Membrane fraction or protein | Incorporation (U/mg/min) ofa: |
|
|---|---|---|
| GlcNAc | GalNAc | |
| A. hydrophila AH-3 (wild type) | <1 | 8 ± 1 |
| AH-3ΔwecP mutant | ND | ND |
| AH-3ΔwecP mutant + pBAD33-WecPAh | ||
| Noninducedb | <1 | 5 ± 2 |
| Inducedc | <1 | 76 ± 6 |
| AH-3ΔwecP mutant + pBAD33 | ||
| Noninducedb | <1 | <1 |
| Inducedc | <1 | <1 |
| His6-WecP (ng) | ||
| 0 | <1 | <1 |
| 5 | <1 | 150 ± 12 |
| 10 | <1 | 224 ± 17 |
Standard deviations shown were obtained from three independent experiments. ND, not detected.
Cultures were grown in the absence of l-arabinose, not inducing the PBAD promoter.
Cultures were grown in the presence of l-arabinose, inducing the PBAD promoter.
Reaction mixtures used for the in vitro transferase assay contained approximately 50 μg of the total protein membrane fraction (providing the enzyme and the endogenous Und-P acceptor), 10 mM MgCl2, and the corresponding radiolabeled UDP-HexNAc in 10 mM Tris-HCl (pH 8.5). In assays using purified His6-WecP, 50 μg of the total protein membrane fraction from A. hydrophila AH-3ΔwecP was used as the source of Und-P. The mixture was incubated for 30 min at 37°C, and the lipid fraction was extracted twice with the same volume of 1-butanol. The extracts were washed at least three times with the same volume of distilled water, and the amount of radioactivity incorporated into the lipid fraction was measured in a scintillation counter. All the values are normalized to the background obtained using the lipid extract from the membrane fraction of AH-3ΔwecP mutant.
To prove the role of WecP, we amplified the wecP gene from A. hydrophila AH-3 and cloned it into pET-30 Xa/LIC (see Materials and Methods). The construction was made to obtain a WecP protein with an extra amino-terminal sequence of 47 amino acid residues, with 6 histidine residues in positions 2 to 7. This His6-WecP was overproduced in E. coli BL21(λD3) harboring pET-30-WecPAh and purified from the membrane fraction as previously reported for WecA (1). The transferase activity was determined as described above by using the total membrane fraction of A. hydrophila AH-3ΔwecP as the source of the endogenous Und-P acceptor and by using 1 to 10 ng of purified His6-WecP. In control experiments without added His6-WecP, neither GlcNAc-1-P nor GalNAc-1-P was incorporated from UDP-GlcNAc or UDP-GalNAc. In contrast, in the presence of His6-WecP, GalNAc-1-P was incorporated from UDP-GalNAc but no GlcNAc-1-P was incorporated when using UDP-GlcNAc. The specific activity of GalNAc-1-P incorporation was dependent on the amount of His6-WecP added to the reaction (Table 2). These results strongly suggest that the WecP protein is indeed a UDP-GalNAc:polyprenol-P GalNAc-1-P transferase.
DISCUSSION
The A. hydrophila AH-3 WecP represents a new class of UDP-HexNAc:polyprenol-P HexNAc-1-P transferases. These transferase-catalyzed reactions involve a membrane-associated polyprenol phosphate acceptor and a cytoplasmic UDP-d-N-acetylhexosamine sugar nucleotide as the donor substrate. Four subgroups of bacterial enzymes have been identified based on their specific substrate preference (31). However, until now, all the WecA enzymes tested were able to transfer UDP-GlcNAc to the Und-P. It has been indicated that WecA can also transfer N-acetylgalactosamine-1-phosphate (25), as it is also essential for the synthesis of O antigens containing N-acetylgalactosamine (46). However, reference 46 only suggested that in a study of the E. coli O157 wb* cluster, WecA transfers GalNAc-phosphate to Und-P to initiate O-unit synthesis as well as initiating enterobacterial common-antigen synthesis by transfer of GlcNAc-phosphate. Assuming this suggestion, they subsequently identified the genes for all the required steps for synthesis of the O157 O-antigen (46). In no case is there any biochemical proof for the transfer of GalNAc. Furthermore, recently, a novel biosynthetic pathway for the production of GalNAc-P-P-Und by epimerization after the formation of GlcNAc-P-P-Und by WecA in E. coli O157 has been described (35).
In this study, we present in vitro and in vivo proofs that A. hydrophila AH-3 WecP transfers GalNAc to Und-P and is unable to transfer GlcNAc to the same enzyme substrate. The in vitro assays clearly indicated that [14C]GalNAc but not [14C]GlcNAc is incorporated in the lipid fraction of the protein membrane (endogenous Und-P acceptor) by A. hydrophila AH-3 WecP.
The LPS profiles in gels together with the sugar analysis of the mutants used in this study carrying well-defined sugar-Und-P transferases clearly indicate the in vivo transfer of GalNAc to the Und-P by WecP. In these studies, it is important to point out that in A. hydrophila AH-3 or E. coli O7, assembly of O antigen is through the Wzy-dependent pathway, while in the case of K. pneumoniae O1, it is through the ABC transporter-dependent pathway (32). In the first case for synthesis of O antigens, monomers are assembled on a lipid carrier (Und-P) by enzymes encoded in the wb gene cluster before their incorporation into the LPS molecule through the O-antigen polymerase and the flippase activity (Wzy and Wzx). In the second case, the complete O-antigen LPS is assembled on a lipid carrier (Und-P) before its incorporation into the LPS molecule through an ABC-2-type pathway (Wzm-Wzt). These differences could explain why we found a partial complementation in LPS gels in A. hydrophila AH-3ΔwecP carrying E. coli WecA or in E. coli MV501 (WecA mutant) carrying A. hydrophila WecP plus Gne, while the complementation was fully achieved, based on LPS gels, in K. pneumoniae 52145ΔwecA carrying A. hydrophila WecP plus Gne. The presence of GalNAc in the sugar analysis of LPS from E. coli or K. pneumoniae complemented strains, and the presence of GlcNAc in LPS from AH-3ΔwecP carrying E. coli O7 WecA instead of GalNAc, fully supported in vivo the transfer of GalNAc to the Und-P by WecP.
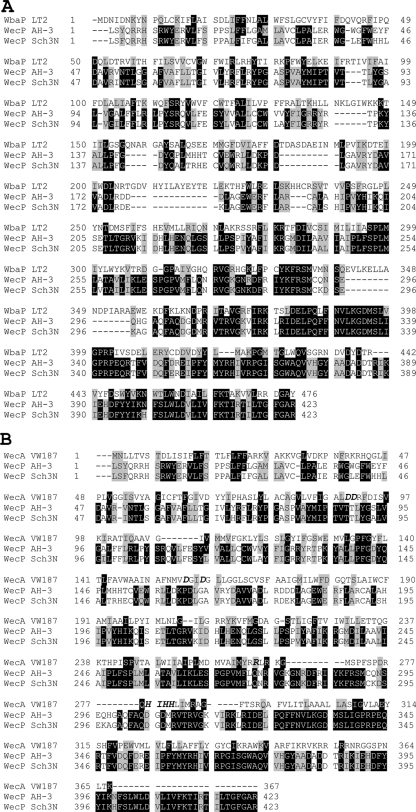
The WecP enzyme (UDP-GalNAc:polyprenol-P GalNAc-1-P transferase) differs from WecA (UDP-GlcNAc:polyprenol-P GlcNAc-1-P transferase) in membrane topology. The WecP protein membrane topology model resembles that of the WbaP enzyme (UDP-Gal:polyprenol-P Gal-1-P transferase), with the two enzymes sharing three predicted domains: an N-terminal region containing four transmembrane helices, a central periplasmic loop, and a C-terminal domain containing the last transmembrane helix and a large cytoplasmic tail which carries the sugar-phosphate transferase activity. In addition, the C-terminal putative catalytic domain of WecP showed a considerable degree of conservation with that of WbaP (Fig. 6A), in agreement with the fact that WbaP showed this conservation with a broad spectrum of bacterial species (36). The other two WbaP domains showed a low degree of conservation compared to WecP (Fig. 6B), in agreement with the reduced homology to a number of proteins previously described (36).
Fig. 6.
Multiple-sequence alignments of S. enterica strain LT2 WbaP with A. hydrophila AH-3 WecP and A. caviae Sch3N (A) and E. coli strain VW187 WecA with the same Aeromonas WecP proteins mentioned above (B). Identical (black) and similar (gray) residue conservations are shown. Amino acids of functional importance in the transfer of GlcNAc-1-P to Und-P (26) are italic and labeled in boldface for E. coli strain VW187 WecA (B).
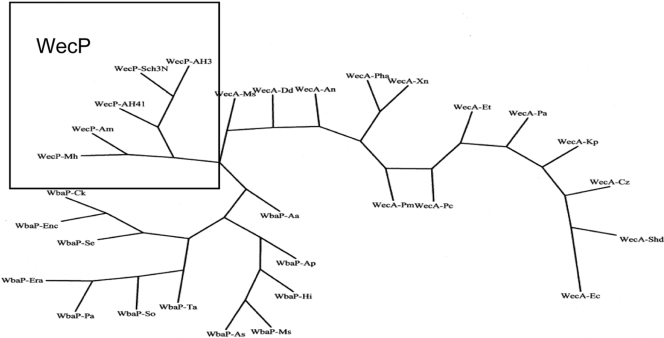
WecA is localized in discrete regions within the membrane, and five cytosolic loops have been determined using computer programs and some experimental data (25). Specific amino acid residues of functional importance in the transfer of GlcNAc-1-P to Und-P have been identified in cytosolic loops 2, 3, and 5 of WecA (25). Replacement of Asp90 and Asp91 in cytosolic loop 2 and Asp156 and Asp159 in cytosolic loop 3 with other amino acids affected the function of WecA, as demonstrated by the lack of in vitro enzymatic activity or reduced in vitro enzymatic activity of the mutated proteins, which could not mediate O-antigen LPS production in vivo (3). Alignment between WecP and WecA did not show Asp residues equivalent to Asp90, Asp91, Asp156, and Asp159 (Fig. 6B). The differences in substrate specificity and membrane topology between WecP and WecA suggest that their phylogenetic relationship is not very close. This was checked by running the phylogenetic inference software Protpars from the PHYLIP package (J. Felsenstein, PHYLIP [Phylogeny Inference Package] version 3.5c, Department of Genetics, University of Washington, Seattle, WA, 1993), using WecP, WecA, and WbaP homologues (Fig. 7). This approach suggests that each of these enzyme classes constitutes a different phylogenetic branch.
Fig. 7.
Phylogenetic tree of selected WecP, WecA, and WbaP homologues. The tree was generated by using the Protpars program (parsimony method) in the PHYLIP package, using default parameters and bootstrap values. WecP proteins shown are from A. hydrophila strains AH-3 (WecP-AH3) (EU274663) and AH-41 (WecP-AH41), A. caviae Sch3N (WecP-Sch3N), Actinobacillus minor 202 (WecP-Am) (ZP_03611743), and Mannheimia haemolytica PHL213 (WecP-Mh) (ZP_04978354). WecA proteins shown are from E. coli VW187 (WecA-Ec), K. pneumoniae 52145 (WecA-Kp), Shigella dysenteriae Sd 197 (WecA-Shd) (YP_405383), Cronobacter sakazakii ATCC BAA-894 (WecA-Cz) (YP_001439802), Pantoea annanatis LMG 620103 (WecA-Pa) (YP_003518442), Xenorhabdus nematophila ATCC 19061 (WecA-Xn) (YP_003710698), Photorhabdus asymbiotica (WecA-Pha) (YP_003042971), Arsenophonus nasoniae (WecA-An) (CBA71837), Proteus mirabilis HI4320 (WecA-Pm) (YP_002153001), Pectobacterium carotovorum WP114 (WecA-Pc) (ZP_03829856), Edwardsiella tarda ATCC 23685 (WecA-Et) (ZP_06716201), Dickeya dadantii 3937 (WecA-Dd) (YP_003885016), and Mannheimia succiniciproducens MBEL55E (WecA-Ms) (YP_088103). WbaP proteins shown are from S. enterica serovar Typhimurium LT2 (WbaP-Se) (NP_461027), Enterobacter cloacae ATCC 13047 (WbaP-Enc) (YP_003613836), Citrobacter koseri ATCC BAA-895 (WbaP-Ck), Erwinia amylovora ATCC BAA-2158 (WbaP-Era) (CBX81132), Pantoea annanatis LMG 620103 (WbaP-Pa) (YP_003520801), Serratia odorifera 4Rx13 (WbaP-So) (ZP_06189355), Tolumonas auensis DSM 9187 (WbaP-Ta) (YP_002893229), Actinobacillus succinogenes 130Z (WbaP-As) (YP_001344127), Mannheimia succiniciproducens MBEL55E (WbaP-Ms) (YP_087854), Haemophilus influenzae RdKW20 (WbaP-Hi) (NP_439033), Actinobacillus pleuropneumoniae L20 (WbaP-Ap) (YP_0010541601), and Aggregatibacter aphrophilus NJ8700 (WbaP-Aa) (YP_002893229) (designations following the protein names represent GenBank accession numbers).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Plan Nacional de I + D + i and FIS grants (Ministerio de Educación, Ciencia y Deporte and Ministerio de Sanidad, Spain) and by the Generalitat de Catalunya (Centre de Referència en Biotecnologia).
We thank Maite Polo for her technical assistance and Miguel A. Valvano for kindly providing E. coli strains (VW187 and MV501) and S. enterica strains (LT2 and MSS2).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Al-Dabbagh B., Mengin-Lecreux D., Bouhss A. 2008. Purification and characterization of the bacterial UDP-GlcNAC:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J. Bacteriol. 190:7141–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amer A. O., Valvano M. A. 2002. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148:571–582 [DOI] [PubMed] [Google Scholar]
- 4. Amor K., et al. 2000. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 68:1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson M. S., Eveland S. S., Price N. P. 2000. Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family. FEMS Microbiol. Lett. 191:169–175 [DOI] [PubMed] [Google Scholar]
- 6. Bateman A., et al. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Canals R., et al. 2006. A gene (uridine diphosphate N-acetylgalactosamine 4-epimerase) is essential for mesophilic Aeromonas serotype O:34 virulence. Infect. Immun. 74:537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canals R., et al. 2007. The role of Gne and GalE in the virulence of Aeromonas hydrophila serotype O34. J. Bacteriol. 189:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clarke B. R., et al. 1995. Role of Rfe and RfbF in the initiation of biosynthesis of D-galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae serotype O1. J. Bacteriol. 177:5411–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DiGiandomenico A., et al. 2002. Glycosylation of Pseudomonas aeruginosa 1244 pilin: glycan substrate specificity. Mol. Microbiol. 46:519–530 [DOI] [PubMed] [Google Scholar]
- 11. Galanos C., Lüderitz O., Westphal O. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245–249 [DOI] [PubMed] [Google Scholar]
- 12. Gryllos I., Shaw J. G., Gavín R., Merino S., Tomás J. M. 2001. Role of flm locus in mesophilic Aeromonas species adherence. Infect. Immun. 69:65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 14. Heinrichs D. E., Yethon J. A., Whitfield C. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221–232 [DOI] [PubMed] [Google Scholar]
- 15. Hitchcock P. J., Brown T. M. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izquierdo L., et al. 2003. The Klebsiella pneumoniae wabG gene: their role in the biosynthesis of the core lipopolysaccharide and virulence. J. Bacteriol. 185:7213–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janda J. M., Abbott S. L. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332–344 [DOI] [PubMed] [Google Scholar]
- 18. Janda J. M., Abbott S. L., Khashe S. G., Kellogg H., Shimada T. 1996. Further studies on biochemical characteristics and serologic properties of the genus Aeromonas. J. Clin. Microbiol. 34:1930–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reference deleted.
- 20. Jiménez N., et al. 2008. The Aeromonas hydrophila wb*O34 gene cluster: genetics and temperature regulation. J. Bacteriol. 190:4198–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiménez N., et al. 2009. Genetics and proteomics of Aeromonas salmonicida lipopolysaccharide core biosynthesis. J. Bacteriol. 191:2228–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiménez N., et al. 2009. A bifunctional enzyme in a single gene catalyzes the incorporation of GlcN into the Aeromonas core LPS. J. Biol. Chem. 284:32995–33005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knirel Y. A., Shaskov A. S., Senchenkova S. N., Merino S., Tomás J. M. 2002. Structure of the O-polysaccharide of Aeromonas hydrophila O:34: a case of random O-acetylation of 6-deoxy-L-talose. Carbohydr. Res. 337:1381–1386 [DOI] [PubMed] [Google Scholar]
- 24. Reference deleted.
- 25. Lehrer J., Vigeant K. A., Tatar L. D., Valvano M. A. 2007. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J. Bacteriol. 189:2618–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lehrman M. A. 1994. A family of UDP-GlcNAc/MurNAc:polyisoprenol-P GlcNAc/MurNAc-1-P transferases. Glycobiology 4:768–771 [DOI] [PubMed] [Google Scholar]
- 27. Link A. J., Phillips D., Church G. M. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. L'vov V. L., et al. 1984. Structural studies of the O-specific side chain of the lipopolysaccharide from Escherichia coli O7. Carbohydr. Res. 126:249–259 [DOI] [PubMed] [Google Scholar]
- 29. Meier-Dieter U., Barr K., Starman R., Hatch L., Rick P. D. 1992. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J. Biol. Chem. 267:746–753 [PubMed] [Google Scholar]
- 30. Nassif X., Fournier J. M., Arondel J., Sansonetti P. J. 1989. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 57:546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price N. P., Momany F. A. 2005. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology 15:29–42 [DOI] [PubMed] [Google Scholar]
- 32. Raetz C. R., Withfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Regué M., et al. 2005. A second outer-core region in Klebsiella pneumoniae lipopolysaccharide. J. Bacteriol. 187:4198–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rubires X., Merino S., Aguilar A., Nogueras M. M., Tomás J. M. 1998. Isolation of three different bacteriophage from mesophilic Aeromonas sp. that use different types of monopolar flagella as their primary receptor. FEMS Microbiol. Lett. 161:53–57 [DOI] [PubMed] [Google Scholar]
- 35. Rush J. S., Alaimo C., Robbiani R., Wacker M., Waechter C. J. 2010. A novel epimerase that converts GlcNAc-P-P-undecaprenol to GalNAc-P-P-undecaprenol in Escherichia coli O157. J. Biol. Chem. 285:1671–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saldías M. S., et al. 2008. Distinct functional domains of the Salmonella enterica WbaP transferase that is involved in the initiation reaction for synthesis of the O antigen subunit. Microbiology 154:440–453 [DOI] [PubMed] [Google Scholar]
- 37. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 38. Samuel G., Reeves P. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503–2519 [DOI] [PubMed] [Google Scholar]
- 39. Sanger F., Nicklen S., Coulson A. R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shibata Y., Yamashita Y., Ozaki K., Nakano Y., Koga T. 2002. Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect. Immun. 70:2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skurnik M., Bengoechea J. A. 2003. The biosynthesis and biological role of lipopolysaccharide O-antigens of pathogenic Yersiniae. Carbohydr. Res. 338:2521–2529 [DOI] [PubMed] [Google Scholar]
- 42. Tsai C. M., Frasch C. E. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115–119 [DOI] [PubMed] [Google Scholar]
- 43. Vinogradov E., Perry M. B. 2001. Structural analysis of the core region of the lipopolysaccharides from eight serotypes of Klebsiella pneumoniae. Carbohydr. Res. 335:291–296 [DOI] [PubMed] [Google Scholar]
- 44. Vinogradov E., et al. 2002. Structures of lipopolysaccharides from Klebsiella pneumoniae. Elucidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the O chains. J. Biol. Chem. 277:25070–25081 [DOI] [PubMed] [Google Scholar]
- 45. Wang L., Liu D., Reeves P. R. 1996. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J. Bacteriol. 178:2598–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang L., Reeves P. R. 1998. Organization of the Escherichia coli O157 O antigen cluster and identification of its specific genes. Infect. Immun. 66:3545–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., et al. 2007. Biochemical characterization of dTDP-D-Qui4N and dTDP-D-Qui4NAc biosynthetic pathways in Shigella dysenteriae type 7 and Escherichia coli O7. J. Bacteriol. 189:8626–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Westphal O., Jann K. 1965. Bacterial lipopolysaccharide extraction with phenol-water and further application of the procedure. Methods Carbohydr. Chem. 5:83–91 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.