Abstract
The Chlamydia-specific hypothetical protein CT795 was dominantly recognized by human antisera produced during C. trachomatis infection but not by animal antisera raised against dead chlamydia organisms. The immundominant region recognized by the human antibodies was mapped to the N-terminal fragment T22-S69. The endogenous CT795 was detected in the cytoplasm of host cells during C. trachomatis infection and was highly enriched in the host cytosolic fraction but absent in the purified chlamydia organisms, suggesting that CT795 is synthesized and secreted into host cell cytoplasm without incorporation into the organisms. All C. trachomatis serovars tested secreted CT795. A predicted signal peptide of CT795 directed the mature PhoA to cross Escherichia coli inner membranes. The secretion of CT795 in Chlamydia-infected cells was inhibited by a C16 compound targeting signal peptidase I, but not by a C1 compound known to block the type III secretion pathway. These results suggest that CT795, like CPAF (a Chlamydia-secreted virulence factor), is secreted into the host cell cytoplasm via a sec-dependent mechanism and not by a type III secretion pathway. The above characterizations of CT795 have provided important information for further understanding the potential roles of CT795 in C. trachomatis pathogenesis.
INTRODUCTION
The Chlamydia trachomatis species consists of three human biovars that manifest different tissue tropisms, with the trachoma biovar (serovars A to C) infecting ocular epithelial cells, potentially resulting in trachoma (45), genital biovar (serovars D to K) targeting urogenital epithelial cells, leading to sexually transmitted diseases (29, 37), and the lymphogranuloma venerum (LGV) biovar, infecting colorectal tissues and causing disseminated infection (1, 38). Despite the differences in tissue tropisms, all C. trachomatis organism-induced diseases are characterized by inflammatory pathologies. However, the precise molecular mechanisms of chlamydial pathogenesis remain unknown, despite the tremendous amount of research efforts in the past half a century. It is proposed that the intracellular invasion and survival of the C. trachomatis organisms may mainly contribute to the pathogenesis. All C. trachomatis organisms undergo an intracellular growth cycle with distinct biphasic stages (15). The infection starts with invasion of an epithelial cell by an infectious elementary body (EB). The internalized EB rapidly develops into a noninfectious but metabolically active reticulate body (RB) for multiplication. The progeny RBs then differentiate back into EBs for exiting the infected host cells and spreading to nearby cells. All chlamydial biosynthesis and metabolism activities are restricted within a cytoplasmic vacuole known as the inclusion (15).
C. trachomatis organisms have evolved with the ability to secrete proteins into host cells for modifying host cellular processes and facilitating their own invasion, intracellular survival/replication, and spreading to new cells. For example, the EB-containing organisms can inject preexisting proteins into epithelial cells to induce endocytosis (7, 12), so that the EBs can rapidly enter host cells that are normally inefficient in taking up particles. Some of the injected proteins may further modulate host cell cytoskeletal structures and endocytic pathways (19) so that the chlamydia organism-laden vacuoles are not fused with host lysosomes (34). Once RBs are formed and initiate biosynthesis, some of the newly synthesized proteins are destined for the inclusion membrane (23, 32) and host cell cytoplasm (14, 41, 46). These newly secreted proteins may help the intracellular chlamydia organisms take up nutrients and energy from host cells (8, 16, 27, 39) and prevent the infected host cells from undergoing apoptosis or host immune detection and attack (46). For example, CPAF, a chlamydial protease/proteasome-like activity factor, is secreted into host cell cytosol (47). CPAF is a serine protease (4, 20) that can degrade a wide array of host proteins, including cytokeratins for chlamydial inclusion expansion (11, 22, 33), transcriptional factors required for major histocompatibility complex antigen expression for evading immune responses (48, 49), and BH3-only domain proteins for inhibiting apoptosis (13, 30).
Interestingly, some of the secretion proteins that are synthesized during live infection may not be (or may only minimally be) retained within the chlamydia organisms. For example, many proteins that are secreted into the inclusion membrane (called inclusion membrane proteins, or Incs) are highly enriched in the inclusion membrane with minimal association with the chlamydia organisms. The first Inc protein was identified by determining antigens preferentially recognized by antisera from animals infected with live chlamydia organisms and not sera from animals immunized with dead organisms (31). CPAF, which is actively secreted into the host cell cytosol during live chlamydia infection, is hardly detectable in the purified chlamydia organisms (47). As a result, humans or animals that are infected with live chlamydia organisms produce large amounts of anti-CPAF antibodies, while animals immunized with purified dead chlamydia organisms produce no anti-CPAF antibodies (35, 43). The proteins that are only synthesized during live infection without any significant retention in the organisms are designated infection-dependent antigens (43). Obviously, not all infection-dependent antigens are secretion proteins. Nevertheless, a comparison of antigen profiles recognized by antibodies produced during live infection versus those recognized by antisera induced by dead organisms may facilitate the identification of putative chlamydial secretion proteins.
Because searching for Chlamydia-secreted proteins has been a most productive approach for understanding the molecular mechanisms of chlamydial pathogenesis (2, 7, 9, 18, 19, 24, 28, 40–42, 47), we have developed various means for identifying chlamydial secretion proteins. When antigen profiles recognized by antisera from women urogenitally infected with C. trachomatis and those recognized by antibodies from rabbits immunized with dead chlamydia organisms were carefully compared, we found that the hypothetical protein CT795 was dominantly recognized by human but not rabbit antisera, suggesting that CT795 may be an infection-dependent antigen. When an antibody raised against a CT795 fusion protein was used to localize the endogenous protein, CT795 was detected in the cytoplasm of the C. trachomatis-infected cells, with a distribution pattern similar to CPAF, suggesting that CT795 is secreted into the host cell cytosol during chlamydial infection. The fact that CT795 was highly enriched in the cytosolic fraction but absent in the purified chlamydia organisms may explain why the dead chlamydia organisms failed to induce antibodies against CT795. Further characterization of the CT795 secretion showed that CT795 was secreted into the host cell cytosol 24 h after infection, and CT795 secretion seemed to be a common property of all C. trachomatis serovars. Since a predicted signal peptide of CT795 directed the mature PhoA to cross the Escherichia coli inner membrane and the secretion of CT795 in chlamydia-infected cells was inhibited by a C16 compound that targets signal peptidase I, but not by a C1 compound known to block the type III secretion pathway, we propose that CT795, like CPAF, is secreted via a sec-dependent but not type III secretion pathway. Since CPAF is a known bona fide virulence factor of C. trachomatis, the findings described in the current manuscript demonstrate that the sec-dependent secretion pathway may play important roles in C. trachomatis pathogenesis.
MATERIALS AND METHODS
Chlamydia infection.
The following C. trachomatis organisms were used: serovars A/HAR-13, B/HAR-36, Ba/Ap-2, C/UW-1, D/UW-3/Cx, E/UW-5/CX, F/IC-Cal-3, G/UW-57/Cx, H/UW-43/Cx, I/UW-12/Ur, K/UW-31/Cx, L1/LGV-440, L2/LGV-434/Bu, and L3/LGV-404. All chlamydia organisms were either purchased from ATCC (Manassas, VA) or acquired from Harlan Caldwell at the Rocky Mountain Laboratory, NIAID, NIH (Hamilton, MT) or from Ted Kou at the University of Washington (Seattle, WA). The chlamydia organisms were propagated, purified, aliquoted, and stored as described previously (47). All chlamydia organisms were routinely checked for mycoplasma contamination. For infection, HeLa cells (human cervical carcinoma epithelial cells; ATCC catalog number CCL2) grown in either 24-well plates with coverslips or tissue flasks were inoculated with chlamydia organisms. The infected cultures were processed at various time points after infection for either immunofluorescence assays or Western blot analyses as described below. For some experiments, the cultures were treated with a C1 compound [N′-(3,5-dibromo-2-hydroxybenzylidene)-4-nitrobenzohydrazide; catalog number 5113023; ChemBridge, San Diego, CA], a small molecule known to inhibit the Yersinia type III secretion system (T3SS) and block chlamydial growth (44), or arylomycin C16, an inhibitor that targets signal peptidase I (5). The treated cultures were processed for immunofluorescence microscopy at 30 to 40 h after infection.
Chlamydia gene cloning, fusion protein expression, and antibody production.
The open reading frame (ORF) CT795 and its fragments from C. trachomatis serovar D were cloned into pGEX vectors (Amersham Pharmacia Biotech, Inc., Piscataway, NJ). The following primers were used for cloning the ct795 gene and its fragments: for ct795 full length (FL; without the first 21 codons, covering codons 22 to 163), forward primer 5′-CGC-GGATCC-(BamHI)-ACAGAGGATAAGCAGTGTCAA-3′ and ct795FL back primer 5′-TTTTCCTTTT-GCGGCCGC-(NotI)-CTACTCAACAAATTCAGGATTTA-3′; for ct795 fragment 1 (F1; codons 22 to 69), forward primer 5′-CGC-GGATCC-(BamHI)-ACAGAGGATAAGCAGTGTCAA-3′ and back primer 5′-TTTTCCTTTT-GCGGCCGC-(NotI)-TTAAGAAGTATCTAACAATACTACTAA-3′; for ct795F2 (codons 70 to 116), forward primer 5′-CGC-GGATCC-(BamHI)-GGGTATTCCTTCGAGACTCT-3′ and back primer 5′-TTTTCCTTTT-GCGGCCGC-(NotI)-TTAGCCAACCATAGGATCTGGAA-3′; for ct795F3 (codons 70 to 163), forward primer 5′-CGC-GGATCC-(BamHI)-GGGTATTCCTTCGAGACTCT-3′ and back primer 5′-TTTTCCTTTT-GCGGCCGC-(NotI)-CTACTCAACAAATTCAGGATTTA-3′; for ct795F4 (codons 117 to 163), forward primer 5′-CGC-GGATCC-(BamHI)-GAGATAGCGTTGTTCTTAGAAG-3′ and back primer 5′-TTTTCCTTTT-GCGGCCGC-(NotI)-CTACTCAACAAATTCAGGATTTA-3′. The gene ct795 and its fragments were expressed as fusion proteins with glutathione-S-transferase (GST) fused to the N terminus, as described previously (36). Expression of the fusion proteins was induced with isopropyl-beta-d-thiogalactoside (IPTG; Invitrogen, Carlsbad, CA), and the fusion proteins were extracted by lysing the bacteria via sonication in a Triton X-100 lysis buffer. The fusion protein-containing supernatants were directly used for enzyme-linked immunosorbent assays (ELISA), or fusion proteins were further purified using glutathione-conjugated agarose beads (Pharmacia) for raising antibodies in mice, as previously described (35, 36, 50). The fusion protein-specific antibodies after absorption with a bacterial lysate containing GST alone were used to localize endogenous CT795 in C. trachomatis-infected cells via an indirect immunofluorescence assay and to detect endogenous CT795 in a Western blot assay. In some experiments, the GST-fusion proteins bound to the glutathione-agarose beads were used to absorb the antibodies for confirming the antibody specificities. Production of various control GST-fusion proteins, including GST-CPAF (a chlamydia-secreted serine protease [10]), GST-Pgp3 (a chlamydial plasmid-encoded outer membrane and secretion protein [6, 24]), GST-cHSP60 (chlamydial HSP60), and GST-MOMP (the chlamydial major outer membrane protein from serovar D) have been described elsewhere (36).
Fusion protein microplate ELISA.
The fusion protein microplate ELISA was carried out as described previously (36). Briefly, the GST-fusion proteins in the form of bacterial lysates were applied to glutathione-coated 96-well microplates (catalog number 15140B; Pierce, Rockford, IL) and used to assay antibody reactivities. All primary antibodies were preabsorbed with a bacterial lysate containing GST alone before they were assayed on the ELISA plates. The human and rabbit antisera were obtained and produced as described previously (36, 43) and assayed at 1:500 dilutions of the original serum samples. The goat anti-human IgA-IgG-IgM or donkey anti-rabbit IgG secondary antibodies conjugated with horseradish peroxidase (HRP) (catalog numbers 109-035-064 and 711-035-152, respectively; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were used to probe the primary antibody binding. The soluble substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS; catalog number A1888-5G; Sigma) was used to visualize the reactions, and the reactivity was recorded as the absorbance (optical density [OD]) at 405 nm. A GST-alone bacterial lysate-coated well in each plate was used as a negative control, and the OD of the GST well was controlled at 0.05 or lower. Any wells with an OD equal to or greater than 4 times the OD value from the GST well were considered positive. The reactivity of a given antigen recognized by different antiserum samples was analyzed by comparing the raw OD values based on an analysis of variance (ANOVA) followed by Student's t test.
Immunofluorescence assay.
An immunofluorescence assay was carried out as described previously (50). Briefly, HeLa cells grown on coverslips were fixed with 2% paraformaldehyde (Sigma) dissolved in phosphate-buffered saline for 30 min at room temperature, followed by permeabilization with 2% saponin (Sigma) for an additional 30 min. After washing and blocking, the cell samples were subjected to antibody and chemical staining. Hoechst stain (blue; Sigma) was used to visualize DNA. A rabbit antichlamydia antibody (R1L2; raised with C. trachomatis L2 organisms [unpublished data]) or anti-IncA antibody (kindly provided by Ted Hackstadt, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, NIAID, NIH, Hamilton, MT [17]) plus a goat anti-rabbit IgG secondary antibody conjugated with Cy2 (green; Jackson ImmunoResearch Laboratories, Inc.) was used to visualize chlamydia organisms or the inclusion membrane. The various mouse antibodies plus a goat anti-mouse IgG conjugated with Cy3 (red; Jackson ImmunoResearch) were used to visualize the corresponding antigens. The mouse antibodies included polyclonal antibodies (pAbs) against GST-CT795 and GST-CT621 (both from the current study) and monoclonal antibody (MAb) 100a against CPAF (47). All primary antibodies were preabsorbed with a bacterial lysate containing GST alone before use. In addition, for some experiments, the primary antibodies were further absorbed with either the corresponding or heterologous fusion proteins immobilized onto glutathione-conjugated agarose beads (Pharmacia) prior to immunostaining in order to confirm the antibody-binding specificities. The immunofluorescence images were acquired using an Olympus AX-70 fluorescence microscope equipped with multiple filter sets and the Simple PCI imaging software (Olympus, Melville, NY) as described previously (13). A FluoView laser confocal microscope (Olympus, Center Valley, PA) was used to further analyze some of the immunofluorescence samples at the University of Texas Health Science Center at San Antonio institutional core facility as described previously (24). The images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA).
Western blot assay.
The Western blot assay was carried out as described elsewhere (49, 50). Briefly, HeLa cells with or without C. trachomatis infection and with or without fractionation, purified chlamydial RB and EB organisms, and various GST-fusion proteins were resolved on SDS-polyacrylamide gels, and the protein bands were transferred to nitrocellulose membranes for antibody detection. The primary antibodies were mouse pAb against CT795 (current study) and CT813 (an inclusion membrane protein [3]), MAb 100a against CPAF (47), MAb MC22 against the chlamydial MOMP (47), and MAb W27 against host cell HSP70 (catalog number sc-24; Santa Cruz Biotechnology, Santa Cruz, CA). All primary antibodies used in the current study were preabsorbed with an excess amount of bacterial lysates containing GST alone. The primary antibody binding was probed with an HRP-conjugated goat anti-mouse IgG secondary antibody (Jackson ImmunoResearch) and visualized by using an enhanced chemiluminescence (ECL) kit (Santa Cruz Biotechnology). Some of the C. trachomatis-infected HeLa cell (Ct-HeLa) samples were fractionated into a pellet fraction (containing host cell nuclei and chlamydial inclusions) and a cytosolic fraction (S100, containing chlamydia-secreted proteins) as described previously (24, 47). The chlamydia organisms were purified as described previously (6). The RB organisms were purified from 24-h cultures, while the EB organisms were from 40- to 50-h cultures.
BCIP assay.
To construct the plasmid pFLAG-CTC-CT795ss-′PhoA, a 63-bp DNA sequence coding for the CT795 signal peptide (M1-S21, designated CT795ss, with XhoI/BamHI restriction enzyme sites) was amplified from the Chlamydia trachomatis serovar D genome, and a 1,400-bp DNA sequence coding for ′PhoA (BamHI/KpnI) was amplified from the pFLAG-CTC-CPAFss-′PhoA plasmid; both 69-bp CT795 and 1,400-bp ′PhoA were inserted into the XhoI/KpnI sites of the plasmid pFLAG-CTC (catalog number E8408; Sigma) (′PhoA stands for mature PhoA without the signal peptide). The DH5α bacterial strain (Invitrogen, Carlsbad, CA) was used to express the three recombinant plasmids (pFLAG-PhoA, pFLAG-′PhoA, and pFLAG-CT795ss-′PhoA). For the 5-bromo-4-chloro-3-indolylphosphate (BCIP) assay, bacterial cells were grown in LB supplemented with the appropriate selection antibiotics at 37°C overnight. The overnight cultures were streaked onto LB agar containing the same selection antibiotics and 50 μg/ml BCIP (catalog number B6149; Sigma), and the plates were incubated at 30°C for 2 days. The bacterial colonies that were capable of exporting mature PhoA into the periplasm turned blue while the colonies incapable of doing so remained white.
Statistic analyses.
An ANOVA was used to compare OD values from multiple groups. Then, a two-tailed Student t test (for quantitative data) or chi-square test (for qualitative data) was used to compare the groups in the current study.
RESULTS
The hypothetical protein CT795 is dominantly recognized by antisera from women infected with live C. trachomatis but not from rabbits immunized with dead C. trachomatis organisms.
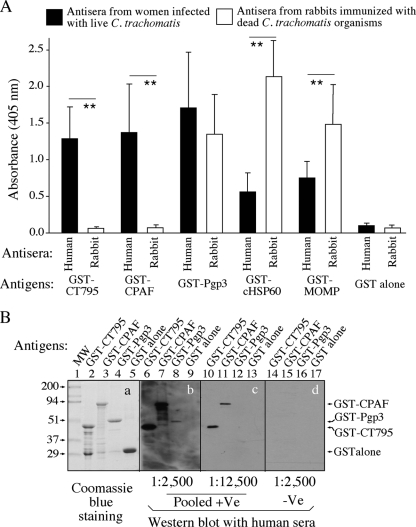
Since our previous studies demonstrated that CT795 is one of the immunodominant antigens recognized by antibodies from women urogenitally infected with C. trachomatis (36, 43), we further characterized the antigenicity of this hypothetical protein. When the reactivities of CT795 with antisera from 10 women infected with live chlamydia organisms versus antisera from 13 rabbits immunized with dead chlamydia organisms were carefully compared (Fig. 1A), we found that CT795 was recognized by all 10 human antiserum samples but not by any of the 13 rabbit serum samples. This recognition pattern is similar to that of CPAF but not those of Pgp3, cHSP60, or MOMP. Pgp3 was well recognized by both human and rabbit antisera, while cHSP60 and MOMP were preferentially recognized by the rabbit antisera. The antibody reactivities of these antigens correlated with the antigen localization in Chlamydia-infected cells, which is consistent with previous reports (31, 35, 43). CPAF is secreted into host cell cytosol but is absent in chlamydia organisms (47); thus, it is available for induction of antibody production during live chlamydia infection but not after dead chlamydia organism immunization. Pgp3 is both secreted into the host cell cytosol and associated with chlamydia organisms (6), while cHSP60 and MOMP are always associated with chlamydia organisms. Thus, immunization with dead organisms induced robust antibody responses to these antigens. The observation that CT795 was only dominantly recognized by human and not rabbit antisera suggested that CT795 might also be secreted out of the chlamydia organisms. In a Western blot assay, the human antibodies continued to dominantly recognize CPAF but not Pgp3 (Fig. 1B), which is consistent with various previous observations that human antisera can recognize denatured CPAF but not denatured Pgp3 (6, 25, 43). Interestingly, CT795 behaved similarly to CPAF. We further mapped the immunodominant region of CT795 to the N-terminal 48 amino acids (from residues T22 to S69) (Fig. 2).
Fig. 1.
Recognition of the hypothetical protein CT795 by human and animal antibodies. (A) Reactivities were measured in an ELISA of various GST-fusion proteins, including CT795, CPAF (a Chlamydia-secreted serine protease), Pgp3 (a chlamydial outer membrane protein that is also secreted into host cell cytosol), chlamydial HSP60 (cHSP60), and MOMP, as well as GST alone, as indicated along the x axis, with antisera from 10 women who were urogenitally infected with live C. trachomatis organisms and 13 rabbits immunized with dead chlamydia organisms. The GST-fusion proteins were immobilized onto glutathione-coated microplates, and all antisera were used at a 1:500 dilution. The goat anti-human and rabbit IgG conjugates were used to detect primary binding, and the reactivities of the fusion proteins with the antisera were recorded as the absorbance at 405 nm (OD values displayed along the y axis). Note that GST-CT795 and GST-CPAF were preferentially recognized by human but not rabbit antisera, while the rest of the antigens were recognized by both human and rabbit antisera. (B) Pooled positive antisera (pooled +Ve; the 10 human antisera described for panel A pooled at an equal ratio) or pooled negative sera (pooled -Ve) from 8 women without C. trachomatis infection were used at various dilutions (as indicated at the bottom of the images) and reacted with GST-CT795, GST-CPAF, GST-Pgp3, or GST alone in a Western blot assay. Although the amounts of full-length fusion proteins loaded into the corresponding lanes were similar (lanes 2 to 5 in gel a), the human antibodies recognized both the denatured GST-CT795 (lanes 6 and 10) and GST-CPAF (lanes 7 and 11) better than the denatured GST-Pgp3 (lanes 8 and 12). Lane 1 was loaded with a prestained molecular weight marker (MW). **, P < 0.01 (ANOVA followed by Student's t test).
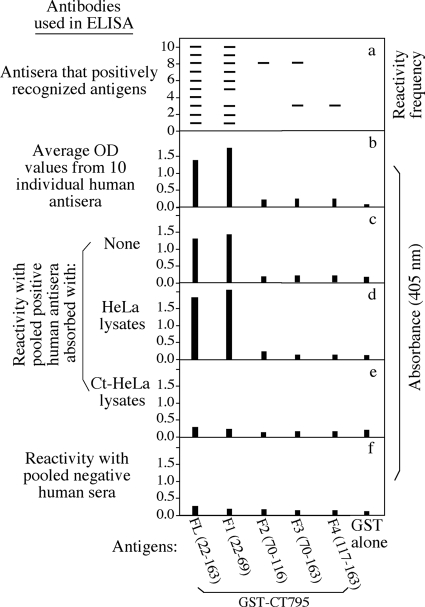
Fig. 2.
Map of the immunodominant regions of CT795. The full-length (FL) CT795 (covering amino acid residues 22 to 163 [the putative signal peptide M1-S21 was left out]) and its fragments, including F1 (residues 22 to 69), F2 (residues 70 to 116), F3 (residues 70 to 163), and F4 (residues 117 163) expressed as GST-fusion proteins (displayed along the x axis), were reacted with each of the 10 positive human antisera in an ELISA as described for Fig. 1. A positive reactivity between a given antigen and a given antiserum was defined as an OD value equal to or greater than 4 times the OD value obtained from the GST-alone-coated well in the same microplate. (a) Each positive reactivity is represented by a horizontal bar. For example, serum 3 positively recognized antigens FL, F1, F3, and F4, and antigen FL was positively recognized by all 10 antisera. (b) Average OD values for each antigen based on its reactivity with the 10 antisera. (c to e) OD values for each antigen recognized by the pooled positive antisera without (c) or with absorption with the normal HeLa cell lysates (d) or Chlamydia-infected HeLa cell (Ct-HeLa) lysates (e). (f) Reactivity of each antigen with the pooled negative antisera, as described for Fig. 1. Note that the human antisera recognized F1 as dominantly as the full-length CT795, without much reactivity with any other fragments.
Localization of the hypothetical protein CT795 in host cell cytosol.
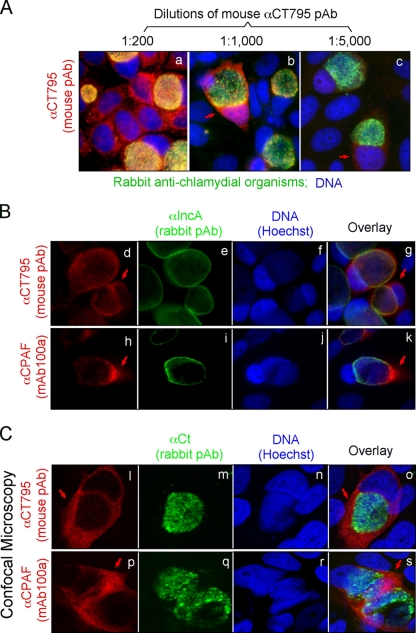
Given the similarities in antigenicities between CT795 and CPAF, we carefully characterized the endogenous CT795 protein. When a mouse antiserum raised with a GST-CT795 fusion protein was used to localize the endogenous protein in C. trachomatis-infected HeLa cells in an immunofluorescence assay, we found that the vast majority of CT795 was localized in the host cell cytosol, and the cytosolic signal became clearer as the dilution of the antiserum increased (Fig. 3A), suggesting that most antibodies or the antibodies with highest affinity detected the antigens in the host cell cytosol. The cytosolic labeling with the anti-CT795 antiserum was confirmed when the infected cells were colabeled with a rabbit anti-IncA antibody (Fig. 3B). Most anti-CT795 labeling was localized outside the inclusion membrane, a pattern similar to CPAF (a chlamydial serine protease known to be secreted into the host cell cytosol). Furthermore, under a confocal microscope, there was little overlap between the anti-CT795 and anti-chlamydia organism labeling (Fig. 3C), which suggests that most CT795 proteins may not be associated with the chlamydia organisms at all, again, a pattern similar to CPAF.
Fig. 3.
Detection of CT795 in the cytosol of C. trachomatis-infected cells. HeLa cells infected with C. trachomatis organisms were processed for immunolabeling with mouse antibodies visualized with a goat anti-mouse IgG conjugated with Cy3 (red), rabbit antibodies visualized with a Cy2-conjugated goat anti-rabbit IgG (green), and the DNA Hoechst stain (blue). (A) A mouse antibody raised against the GST-CT795 fusion protein was costained at various dilutions with a rabbit anti-chlamydia organism antibody. As the dilution of the anti-GST-CT795 antiserum increased, a unique cytosolic signal became clear (images b and c; red arrows). (B) The anti-GST-CT795 antibody (at 1:1,000) was costained with a rabbit anti-IncA antibody, and the CT795 signal (images d to g; red arrows) was detected outside the inclusion with a distribution pattern similar to that of CPAF (images h to k). (C) The costaining of the anti-CT795 antibody with the anti-chlamydial organism antibody was visualized under a confocal microscope. The CT795 signal (images l and o; red arrows) did not overlap with the organism signal (images m and o; green) at all. As a positive control, the CPAF-specific signal was also distinct from the organism signal in parallel cultures (images p to s).
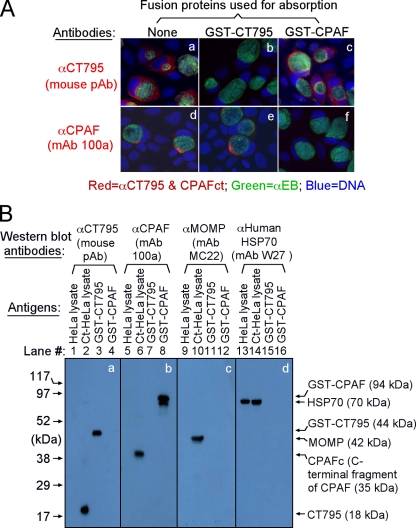
We next confirmed the antibody labeling specificity by using an absorption procedure (Fig. 4A). The labeling in host cell cytosol by the anti-CT795 antiserum was removed by absorption with the fusion protein GST-CT795 but not by GST-CPAF, while anti-CPAF labeling was blocked by GST-CPAF but not by GST-CT795. These results demonstrated that the anti-CT795 and anti-CPAF antibodies specifically labeled the corresponding endogenous proteins without cross-reacting with each other. In a Western blot assay (Fig. 4B), these antibodies only recognized the protein bands migrating at the molecular weights expected for the corresponding endogenous proteins from the C. trachomatis-infected cell and GST-fusion protein samples. As loading controls, the anti-MOMP antibody detected MOMP in the infected cell sample while the anti-mammalian HSP70 antibody recognized HSP70 in both normal and infected HeLa samples. These results further confirmed that both anti-CT795 and anti-CPAF antibodies only specifically detected the corresponding endogenous proteins without cross-reacting with any other chlamydial or host cell proteins. Thus, we can conclude that the cytosolic signals labeled by the anti-CT795 and anti-CPAF antibodies revealed by immunofluorescence microscopy represent the corresponding endogenous proteins.
Fig. 4.
The anti-GST-CT795 fusion protein antiserum specifically detected endogenous CT795 protein. (A) The anti-GST-CT795 antibody and the anti-CPAF MAb 100a with or without preabsorption with the corresponding or heterologous GST-fusion proteins were used to detect the endogenous proteins in C. trachomatis-infected cells. Note that the cytosolic signals detected by the anti-CT795 and anti-CPAF antibodies were removed by preabsorption with the corresponding but not heterologous fusion proteins. (B) Antigens, including HeLa lysates (lanes 1, 5, 9, and 13) and chlamydia-infected HeLa (Ct-HeLa) lysates (lanes 2, 6, 10, and 14) or GST-CT795 (lanes 3, 7, 11, and 15) or GST-CPAF (lanes 4, 8, 12, and 16) fusion proteins, were resolved on SDS-polyacrylamide gels and blotted onto nitrocellulose membranes for detection with antibodies against CT795 (mouse polyclonal antibody [image a]), CPAF (MAb 100a [image b]), MOMP (MAb MC22 [image c]), and human HSP70 (MAb W27 [image d]). Note that the mouse anti-GST-CT795 antibody only reacted with GST-CT795, without cross-reacting with GST-CPAF fusion proteins, and specifically detected the endogenous CT795 in the chlamydia-infected cell sample without cross-reacting with any other chlamydial or host proteins at the concentration used. The three control MAbs only detected the corresponding recombinant and/or endogenous antigens. As stated in Materials and Methods, all antibodies used in the current study were preabsorbed with bacterial lysates containing GST alone. CPAFc represents the C-terminal fragment of CPAF, generated as a result of CPAF processing occurring in chlamydia-infected cells. The epitope recognized by MAb 100a is located in the C-terminal fragment.
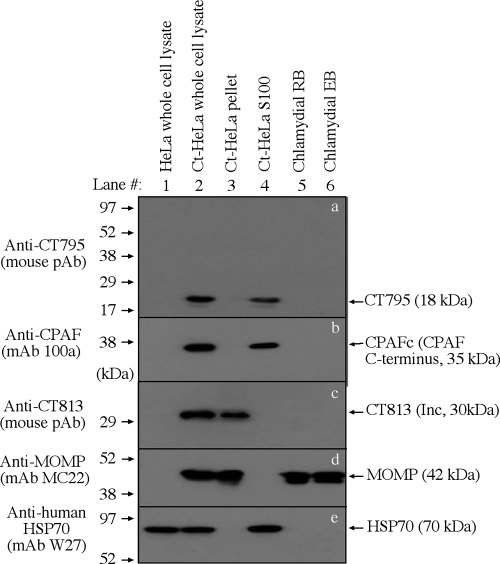
To directly visualize the molecular basis of the anti-CT795 antibody-labeled cytosolic signals in C. trachomatis-infected cells, the infected cells were fractionated into cytosolic and nuclear/inclusion (pellet) fractions. CT795 and the various control proteins were detected in each fraction in a Western blot assay (Fig. 5). CPAF was only detected in either the C. trachomatis-infected whole-cell lysate (Ct-HeLa) or the cytosolic fraction (Ct-HeLa S100) samples and not in other samples, including the purified C. trachomatis RB and EB organisms (Fig. 5b), which is consistent with what has been described previously (47). Interestingly, CT795 displayed a distribution pattern similar to that of CPAF, indicating that CT795 was also secreted into the host cell cytosol. To monitor the quality of the fractionation, the anti-CT813 antibody (a known inclusion membrane protein) and anti-MOMP antibody were used to indicate the pellet fraction containing the chlamydial inclusions, while an anti-human HSP70 antibody was used to indicate the host cell cytosolic fraction containing the C. trachomatis-secreted proteins. Detection with these antibodies revealed no cross-contamination between the pellet and cytosolic fractions. In addition, detection with the anti-MOMP antibody also showed that the amounts of chlamydia organisms in the C. trachomatis-infected HeLa whole-cell lysate, the pellet fraction, and purified EB and RB samples were equivalent. These results together have independently confirmed that, like CPAF, CT795 is secreted into the cytoplasm of C. trachomatis-infected cells without associating with RBs harvested at 24 h or EBs at 48 h postinfection.
Fig. 5.
CT795 is enriched in the cytosolic fraction of chlamydia-infected HeLa cells. HeLa cells infected with C. trachomatis organisms (Ct-HeLa) were fractionated into nuclear (Ct-HeLa pellet, containing chlamydial inclusions; lane 3) and cytosolic (Ct-HeLa S100, containing chlamydia-secreted proteins; lane 4) portions. The cellular fractions along with total cell lysates (normal HeLa, lane 1, and Ct-HeLa, lane 2) and purified chlamydial RB (lane 5) and EB (lane 6) organisms, as listed at the top, were resolved on SDS-polyacrylamide gels. The resolved protein bands were blotted onto nitrocellulose membranes for reacting with antibodies (listed on the left) against CT795 (a), CPAF (b), CT813 (c; an inclusion membrane protein), MOMP (d), and human HSP70 (e). All antibodies detected their corresponding proteins in the HeLa-L2 whole-cell lysate sample (lane 2) and other corresponding samples (as indicated on the right). Note that both CT795 and CPAF were highly enriched in the cytosolic fraction (lane 4 in panels a and b) without any significant association with the purified organisms (lanes 5 and 6 in panels a and b).
Characterization of CT795 secretion.
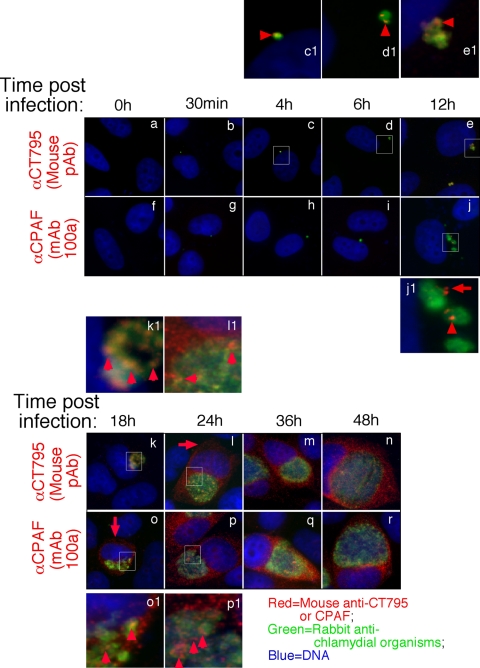
We further used the specific anti-CT795 antibody to monitor the biosynthesis and secretion of CT795 at the single-cell level (Fig. 6). CT795 was first detected as early as 4 h postinfection. Clear secretion into host cell cytosol was only detected 24 h after infection. In contrast, CPAF was synthesized at 12 h after infection, and secretion of CPAF took place simultaneously and became obvious at 18 h after infection. Although there was a delay in CT795 secretion into host cell cytosol, many CT795 molecules appeared to be secreted by the chlamydia organisms after biosynthesis, since most of the CT795-postive granules, although still inside the inclusions, did not overlap with the chlamydia organisms. CT795 may first reach the inclusion lumen and then enter the host cell cytosol.
Fig. 6.
Time course expression of CT795 protein during C. trachomatis infection. The C. tracomatis-infected culture samples were processed at various times after infection (as indicated on the top) for immunofluorescence staining as described for Fig. 1. The mouse anti-CT795 (images a to e and k to n) and anti-CPAF (MAb 100a; panels f to j and o to r) antibodies were visualized with a goat anti-mouse IgG conjugated with Cy3 (red), while the chlamydia organisms were visualized with a rabbit antichlamydia antibody plus a goat anti-rabbit IgG-Cy2 conjugate (green). Areas amplified in separate images are marked with white squares, and the corresponding amplified images are labeled with the same letters followed by the number 1. Note that CT795 was first detected associated with the chlamydial inclusions at 4 h after infection (panels c and c1; red arrowheads), and CPAF was first detected at 12 h after infection (panels j and j1; the red arrow indicates CPAF dissociated from the inclusion, while the arrowhead indicates CPAF still associated with the inclusion). Significant cytosolic signal was first detected at 24 h postinfection for CT795 (l; red arrows) and at 18 h postinfection for CPAF (o; red arrow). Red arrowheads were used to mark signals still associated with the inclusions.
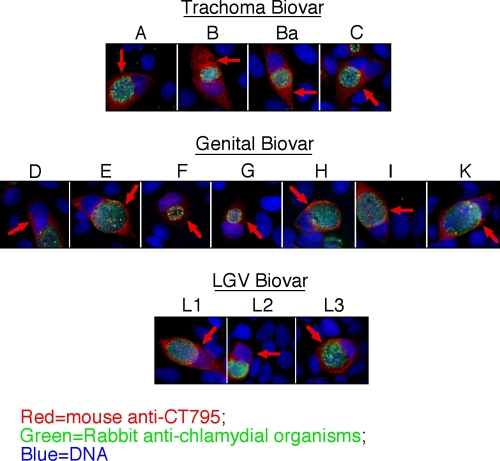
CT795 is a Chlamydia-specific hypothetical protein. It is highly conserved between the C. trachomatis serovars A, B, D, and L2, with >99% amino acid sequence identity. However, the amino acid sequence identity of CT795 with its homologs in other chlamydial species is much lower: 67% for C. muridarum organisms (MoPn, Nigg strain) and <40% for the remaining Chlamydia species (http://stdgen.northwestern.edu/) (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The anti-CT795 antibody detected cytosolic signals in cells infected with C. trachomatis serovars from all three biovars, including trachoma serovars (A, B, B1, and C), genital serovars (D to G, H, I, and K), and LGV (L1 to L3) (Fig. 7). These results demonstrated that secretion of CT795 is a common property of all C. trachomatis organisms but not a strain- or serovar-specific phenomenon.
Fig. 7.
Secretion of CT795 into host cell cytosol is a common feature of all C. trachomatis serovars tested. HeLa cells infected with C. trachomatis trachoma biovar (serovars A, B, Ba, and C), genital biovar (D, E, F, G, H, I, and K), and LGV biovar (L1 to L3) as indicated on the top of each image were processed 40 h after infection for immunofluorescence labeling as described for Fig. 1. The CT795 antibody detected significant signals in the cytosol of host cells infected with all serovars from the Q3 biovars. Red arrows mark the CT795-sepcific signals in the cytosol of the chlamydia-infected cells.
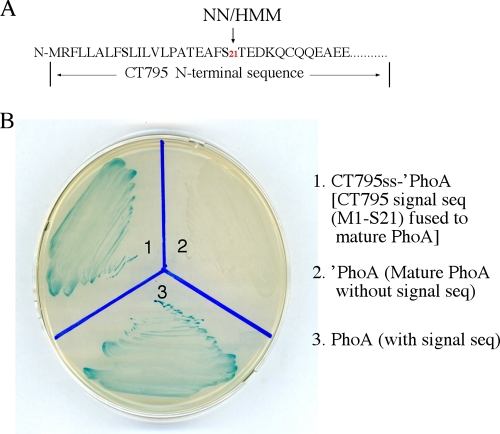
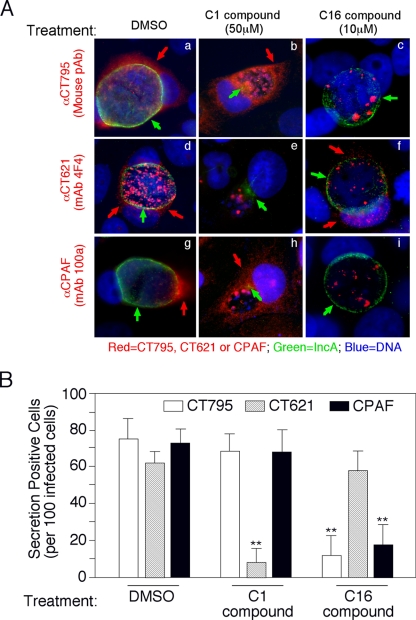
The next question was how CT795 is secreted into host cell cytosol. When the CT795 sequence was analyzed using the program SignalP version 3.0 with the NN (neural network) and HMM (hidden Markov model) algorithms (www.expasy.ch), a putative bacterial signal peptide covering residues M1 to S21 was identified (Fig. 8A). We then evaluated the functionality of the signal sequence in a bacterium-based phoA gene fusion system. This assay system exploits two characteristics of PhoA: the enzyme is only active after translocation into the bacterial periplasm, and the phosphatase activity can be conveniently monitored with the chromogenic substrate BCIP (26). The predicted CT795 signal sequence (CT795ss), when fused to the N terminus of the mature PhoA (′PhoA), directed translocation of ′PhoA to cross the bacterial inner membrane into the periplasmic space, as evidenced by the observation that the periplasmic PhoA changed the BCIP medium to blue (Fig. 8B). As controls, the medium for bacteria that expressed ′PhoA alone stayed white, while the bacteria expressing the full-length PhoA turned the medium blue. These results show that the putative CT795 signal sequence is functional and may be able to direct CT795 across the chlamydial inner membrane to enter the periplasmic region. Most importantly, the secretion of both CT795 and CPAF into the host cell cytosol was inhibited by a C16 compound that is known to target signal peptidase I, but not by a C1 compound known to block the type III secretion pathway, while under the same conditions, the secretion levels of IncA and CT621 (both known chlamydia type III secretion substrates) were inhibited by the C1 but not the C16 compound (Fig. 9). The above results together demonstrate that secretion of CT795 into the host cell cytosol is likely dependent on a sec-dependent but not type III secretion pathway, although the precise mechanism as to how the periplasmic CT795 reaches the host cell cytosol remains to be addressed.
Fig. 8.
Prediction and evaluation of secretion signal sequences in CT795. (A) SignalP software was used to identify type II secretion signal peptides. Both the NN and HMM algorithms predicted the N-terminal region covering the first 21 amino acids as a signal sequence. The DNA coding for the CT795 signal sequence, CT795ss, was then used to replace the native signal peptide-coding region of the PhoA gene. The chimeric CT795ss-′PhoA construct was transformed into E. coli in order to detect the translocation of PhoA into the periplasmic space, where PhoA enzymatic activity can be measured by using BCIP-containing plates. Blue indicates that PhoA has crossed the inner membrane and reached the periplasm. Note that the bacteria transformed with the full-length PhoA (plate slot 1) but not ′PhoA missing the secretion sequence (slot 2) turned blue. The CT795ss-′PhoA chimeric construct-transformed bacteria also turned blue (slot 3).
Fig. 9.
Secretion of CT795 is inhibited by a C16 compound that targets signal peptidase I. (A) HeLa cells infected with C. trachomatis were treated with dimethyl sulfoxide (DMSO) (panels a, d, and g), 50 μM C1 compound (b, e, and h), or 10 μM C16 compound (c, f, and i) at 6 h postinfection and processed for immunofluorescence microscopy analysis 30 h after treatment. The mouse antibodies (red) against CT795 (mouse pAb; panels a, b, and c), CT621 (MAb 4F4; panels d, e, and f), or CPAF (MAb 100a; panels g, h, and i) were costained with a rabbit anti-IncA antibody (green) and the DNA dye Hoechst stain (blue). The IncA protein, whether localized in the inclusion membrane (a, c, d, f, g, and i) or sequestered in the inclusion lumen (b, e, and h) is indicated with green arrows, while secreted CT795 (a and b), CT621 (d and f), and CPAF (g and h) are indicated with red arrows. (B) The secretion-positive cells (red arrows) were quantitated by counting 100 infected cells randomly from each coverslip, and results are expressed as the percentage of the total infected cells. The data presented came from three independent experiments. Note that the number of CT621 secretion-positive cells was significantly lower in C1-treated samples than in DMSO- or C16-treated samples (**, P < 0.01), while the number of CT795 secretion-positive cells was significantly lower in C16-treated samples than in DMSO- or C1-treated samples (P < 0.01), as was also true for the cells positive for secretion of the control CPAF protein.
DISCUSSION
C. trachomatis organisms have adapted an obligate intracellular lifestyle, which allows the organisms to hide from extracellular host defense mechanisms and also requires that the organisms evolve strategies to deal with the hostile intracellular environments. One of the chlamydial strategies is to secrete chlamydial proteins into host cell cytosol. Thus, searching for C. trachomatis-secreted proteins has been a hot subject within this field. We have used various approaches to identify C. trachomatis-secreted proteins, and in the current study we have presented convincing evidence that the hypothetical protein CT795 is secreted into the host cell cytosol. First, CT795 was dominantly recognized by antisera from C. trachomatis-infected humans but not from dead organism-immunized rabbits, suggesting that CT795 may be synthesized during live infection but not packaged back into the mature organisms. Second, fractionation of C. trachomatis-infected cells revealed that CT795 was highly enriched in the host cell cytosol and not present in the purified EB or RB organisms, confirming that CT795 is secreted into the host cell cytosol. Although direct detection of chlamydial protein expression in humans has been difficult, the above observations together have demonstrated that CT795 is expressed and may even be secreted during chlamydial infection in humans. Third, a significant amount of CT795 was visualized in the cytosol of C. trachomatis-infected cells, starting at 24 h after infection, and all C. trachomatis serovars tested were found to secrete CT795, indicating that CT795 secretion is a common property of all C. trachomatis organisms. Fourth, the putative signal sequence of CT795 was functional in directing a heterologous protein to cross the bacterial inner membrane and secretion of CT795 into host cell cytosol was inhibited by a C16 compound but not by a C1 compound, suggesting that CT795 may reach the host cell cytosol via specific mechanisms involving a sec-dependent secretion pathway. Finally, we have extensively demonstrated that the detection of CT795 in the host cell cytosol is specific. The antibody specificity was confirmed in multiple independent assays. Thus, CT795, like CPAF, is another bona fide secretion protein of C. trachomatis.
Despite the overwhelming evidence that CT795 is secreted into the host cell cytosol during chlamydia infection in cell culture and possibly during human infection, the complete molecular pathways that chlamydia organisms use to export CT795 are still unclear. The findings that the CT795 N-terminal leader sequence directed the translocation of mature PhoA into E. coli periplasm and the secretion was inhibited by a signal peptidase I inhibitor but not a type III secretion pathway inhibitor suggest that a sec-dependent export mechanism may play an important role in the secretion of CT795 into the host cell cytosol. Indeed, the chlamydial genome encodes many homologs of the key components required for a functional sec-dependent pathway (http://stdgen.northwestern.edu/). The chlamydial homolog of the essential translocase subunit SecY is encoded by the C. trachomatis ORF CT510, while the homolog of SecF is CT448. Although there is no SecB homolog, the Chlamydia genome does encode many general chaperones, including three GroELs (CT110, CT604, and CT755 [21]). Some of these chaperones may help secretion by interacting with two chlamydial homologs of SecA (CT141 and CT701) to bring the substrate proteins to the chlamydial SecYEG translocon. In addition, the chlamydial FtsY (an SRP receptor-like protein) may act as a GTP-dependent chaperone that promotes interaction between the substrate protein and the SecYEG translocon. The chlamydial homologs of signal peptidases LepB and LspA are CT020 and CT408, respectively. Thus, it is possible that the CT795 molecules secreted into the chlamydial inclusion lumen and host cell cytosol are from the periplasmic pool. However, at this moment, it remains unknown how the periplasmic CT795 molecules pass through the outer membrane to enter the chlamydial inclusion lumen and further into the host cell cytosol. The C. trachomatis organisms do encode homologs of the general secretion proteins (Gsps) in the outer membrane, including CT570 to -572 for GspF, -E, and -D, respectively. Although these outer membrane secretion proteins can export periplasmic proteins out of the organisms, it is hard to imagine how soluble proteins secreted into the inclusion lumen cross the inclusion membrane to enter host cell cytosol. Previous studies have shown that secretion of CPAF is also dependent on a sec-dependent secretion pathway (5). However, it remains unclear how the periplasmic CPAF molecules cross the outer membrane and enter the host cell cytoplasm. An outer membrane vesicular budding model has been proposed for CPAF secretion (5), and this model may also be suitable for the secretion of CT795. This is because CT795-containing granules/vesicles that are free of the organisms were detected in the chlamydial inclusions, which was especially true at the time points before CT795 was detected in the host cell cytosol (Fig. 6). Although we still don't know exactly how CT795 or CPAF is secreted into the host cell cytosol, as more effector molecules are identified, more tools will be available for figuring out the precise secretion pathways in chlamydia organisms that have evolved for exporting virulence factors.
The next question is what roles secreted CT795 may play in chlamydial pathogenesis. Since CT795 is not retained in the chlamydia organisms in any appreciable amount, it is reasonable to hypothesize that CT795 may mainly target host cell components. Previous studies showed that CPAF, a Chlamydia-secreted serine protease, can target various host proteins for degradation to benefit chlamydial intracellular replication and survival (20, 30, 47). However, CT795 has neither significant sequence homology with any proteases nor any detectable proteolytic activities (data not shown). Thus, CT795 may fulfill a distinct function from CPAF, which is consistent with the observation that the time course of CT795 expression and secretion significantly differed from that of CPAF (Fig. 6). Amino acid sequence analysis revealed no known protein function domains in CT795. The immunoreactivity of CT795 with human antibodies was highly concentrated in the N-terminal fragment covering residues T22 to S69, suggesting that the T22-S69 region may be surface exposed and involved in interactions with other molecules. Experimental approaches are under way to identify the CT795 interaction partners and to unravel the functionality of the secreted CT795. Regardless of which cellular components may be targeted by CT795 and what roles the secreted CT795 may play in chlamydial pathogenesis, the finding that CT795 is secreted into the host cell cytosol has laid a solid foundation for further understanding the chlamydia pathogenic mechanisms contributed by the hypothetical CT795.
ACKNOWLEDGMENT
This work was supported in part by grants (to G. Zhong) from the U.S. National Institutes of Health.
Footnotes
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Bauwens J. E., et al. 2002. Epidemic lymphogranuloma venereum during epidemics of crack cocaine use and HIV infection in the Bahamas. Sex. Transm Dis. 29:253–259 [DOI] [PubMed] [Google Scholar]
- 2. Chellas-Gery B., Linton C. N., Fields K. A. 2007. Human GCIP interacts with CT847, a novel Chlamydia trachomatis type III secretion substrate, and is degraded in a tissue-culture infection model. Cell. Microbiol. 9:2417–2430 [DOI] [PubMed] [Google Scholar]
- 3. Chen C., et al. 2006. The hypothetical protein CT813 is localized in the Chlamydia trachomatis inclusion membrane and is immunogenic in women urogenitally infected with C. trachomatis. Infect. Immun. 74:4826–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen D., Chai J., Hart P. J., Zhong G. 2009. Identifying catalytic residues in CPAF, a Chlamydia-secreted protease. Arch. Biochem. Biophys. 485:16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen D., et al. 2010. Secretion of the chlamydial virulence factor CPAF requires sec-dependent pathway. Microbiology 156:3031–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen D., et al. 2010. Characterization of Pgp3, a Chlamydia trachomatis plasmid-encoded immunodominant antigen. J. Bacteriol. 192:6017–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clifton D. R., et al. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cocchiaro J. L., Kumar Y., Fischer E. R., Hackstadt T., Valdivia R. H. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong F., et al. 2006. Localization of the hypothetical protein Cpn0797 in the cytoplasm of Chlamydia pneumoniae-infected host cells. Infect. Immun. 74:6479–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong F., Pirbhai M., Zhong Y., Zhong G. 2004. Cleavage-dependent activation of a chlamydia-secreted protease. Mol. Microbiol. 52:1487–1494 [DOI] [PubMed] [Google Scholar]
- 11. Dong F., Su H., Huang Y., Zhong Y., Zhong G. 2004. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect. Immun. 72:3863–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engel J. 2004. Tarp and Arp: how Chlamydia induces its own entry. Proc. Natl. Acad. Sci. U. S. A. 101:9947–9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan T., et al. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fields K. A., Mead D. J., Dooley C. A., Hackstadt T. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671–683 [DOI] [PubMed] [Google Scholar]
- 15. Hackstadt T., Fischer E. R., Scidmore M. A., Rockey D. D., Heinzen R. A. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5:288–293 [DOI] [PubMed] [Google Scholar]
- 16. Hackstadt T., Scidmore M. A., Rockey D. D. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 92:4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hackstadt T., Scidmore-Carlson M. A., Shaw E. I., Fischer E. R. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1:119–130 [DOI] [PubMed] [Google Scholar]
- 18. Hobolt-Pedersen A. S., Christiansen G., Timmerman E., Gevaert K., Birkelund S. 2009. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol. Med. Microbiol. 57:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hower S., Wolf K., Fields K. A. 2009. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol. Microbiol. 72:1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang Z., et al. 2008. Structural basis for activation and inhibition of the secreted chlamydia protease CPAF. Cell Host Microbe 4:529–542 [DOI] [PubMed] [Google Scholar]
- 21. Karunakaran K. P., et al. 2003. Molecular analysis of the multiple GroEL proteins of chlamydiae. J. Bacteriol. 185:1958–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar Y., Valdivia R. H. 2008. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 4:159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Z., et al. 2008. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect. Immun. 76:2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z., Chen D., Zhong Y., Wang S., Zhong G. 2008. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 76:3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z., et al. 2008. Antibodies from women urogenitally infected with C. trachomatis predominantly recognized the plasmid protein pgp3 in a conformation-dependent manner. BMC Microbiol. 8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marrichi M., Camacho L., Russell D. G., DeLisa M. P. 2008. Genetic toggling of alkaline phosphatase folding reveals signal peptides for all major modes of transport across the inner membrane of bacteria. J. Biol. Chem. 283:35223–35235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McClarty G. 1994. Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 2:157–164 [DOI] [PubMed] [Google Scholar]
- 28. Misaghi S., et al. 2006. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol. Microbiol. 61:142–150 [DOI] [PubMed] [Google Scholar]
- 29. Peterman T. A., et al. 2006. High incidence of new sexually transmitted infections in the year following a sexually transmitted infection: a case for rescreening. Ann. Intern. Med. 145:564–572 [DOI] [PubMed] [Google Scholar]
- 30. Pirbhai M., Dong F., Zhong Y., Pan K. Z., Zhong G. 2006. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J. Biol. Chem. 281:31495–31501 [DOI] [PubMed] [Google Scholar]
- 31. Rockey D. D., Heinzen R. A., Hackstadt T. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15:617–626 [DOI] [PubMed] [Google Scholar]
- 32. Rockey D. D., Scidmore M. A., Bannantine J. P., Brown W. J. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333–340 [DOI] [PubMed] [Google Scholar]
- 33. Scidmore M. 2008. Chlamydia weave a protective cloak spun of actin and intermediate filaments. Cell Host Microbe 4:93–95 [DOI] [PubMed] [Google Scholar]
- 34. Scidmore M. A., Fischer E. R., Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma J., Bosnic A. M., Piper J. M., Zhong G. 2004. Human antibody responses to a Chlamydia-secreted protease factor. Infect. Immun. 72:7164–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sharma J., et al. 2006. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect. Immun. 74:1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sherman K. J., et al. 1990. Sexually transmitted diseases and tubal pregnancy. Sex. Transm. Dis. 17:115–121 [DOI] [PubMed] [Google Scholar]
- 38. Spaargaren J. 2005. Slow epidemic of lymphogranuloma venereum l2b strain. Emerg. Infect. Dis. 11:1787–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Su H., et al. 2004. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 279:9409–9416 [DOI] [PubMed] [Google Scholar]
- 40. Subtil A., et al. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 56:1636–1647 [DOI] [PubMed] [Google Scholar]
- 41. Valdivia R. H. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11:53–59 [DOI] [PubMed] [Google Scholar]
- 42. Vandahl B. B., Stensballe A., Roepstorff P., Christiansen G., Birkelund S. 2005. Secretion of Cpn0796 from Chlamydia pneumoniae into the host cell cytoplasm by an autotransporter mechanism. Cell. Microbiol. 7:825–836 [DOI] [PubMed] [Google Scholar]
- 43. Wang J., et al. 2010. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 185:1670–1680 [DOI] [PubMed] [Google Scholar]
- 44. Wolf K., et al. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wright H. R., Turner A., Taylor H. R. 2008. Trachoma. Lancet 371:1945–1954 [DOI] [PubMed] [Google Scholar]
- 46. Zhong G. 2009. Killing me softly: chlamydial use of proteolysis for evading host defenses. Trends Microbiol. 17:467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhong G., Fan P., Ji H., Dong F., Huang Y. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhong G., Fan T., Liu L. 1999. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 189:1931–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhong G., Liu L., Fan T., Fan P., Ji H. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J. Exp. Med. 191:1525–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhong G., Reis e Sousa C., Germain R. N. 1997. Production, specificity, and functionality of monoclonal antibodies to specific peptide-major histocompatibility complex class II complexes formed by processing of exogenous protein. Proc. Natl. Acad. Sci. U. S. A. 94:13856–13861 [DOI] [PMC free article] [PubMed] [Google Scholar]