Abstract
Chlorite dismutase (Cld) is a unique heme enzyme catalyzing the conversion of ClO2− to Cl− and O2. Cld is usually found in perchlorate- or chlorate-reducing bacteria but was also recently identified in a nitrite-oxidizing bacterium of the genus Nitrospira. Here we characterized a novel Cld-like protein from the chemolithoautotrophic nitrite oxidizer Nitrobacter winogradskyi which is significantly smaller than all previously known chlorite dismutases. Its three-dimensional (3D) crystal structure revealed a dimer of two identical subunits, which sharply contrasts with the penta- or hexameric structures of other chlorite dismutases. Despite a truncated N-terminal domain in each subunit, this novel enzyme turned out to be a highly efficient chlorite dismutase (Km = 90 μM; kcat = 190 s−1; kcat/Km = 2.1 × 106 M−1 s−1), demonstrating a greater structural and phylogenetic diversity of these enzymes than was previously known. Based on comparative analyses of Cld sequences and 3D structures, signature amino acid residues that can be employed to assess whether uncharacterized Cld-like proteins may have a high chlorite-dismutating activity were identified. Interestingly, proteins that contain all these signatures and are phylogenetically closely related to the novel-type Cld of N. winogradskyi exist in a large number of other microbes, including other nitrite oxidizers.
INTRODUCTION
Perchlorate (ClO4−), chlorate (ClO3−), and chlorite (ClO2−) are a serious environmental concern since rising concentrations of these harmful compounds have been detected in groundwater, surface waters, and soils (7). While significant natural sources of perchlorate are restricted to mineral deposits in Chile, environmental contamination with this compound results from its extensive use as an oxidizer in pyrotechnics and rocket fuel and its presence in certain fertilizers (9). Intake of perchlorate by humans occurs mainly via drinking water, milk, and certain consumed plants and should be minimized since this chemical affects hormone production by the thyroid gland (39). Chlorate and chlorite are used as bleaching agents in the textile, pulp, and paper industries, as disinfectants and components of cleaning solutions, in pesticides, and in some other applications, such as chemical oxygen generators. Due to its oxidative nature, chlorite reacts with organic material and thus has toxic effects on living cells (44). Interestingly, some microorganisms are able to use perchlorate or chlorate [referred to as (per)chlorate] as terminal electron acceptors for anaerobic respiration, leading to the reduction of perchlorate to chlorate and subsequently to chlorite (reviewed by Coates and Achenbach [7]). These (per)chlorate-reducing bacteria (PCRB) also possess the unique enzyme chlorite dismutase (Cld), which detoxifies chlorite by converting ClO2− to Cl− and O2 (46). In combination, (per)chlorate reduction to chlorite and the activity of Cld enable the microbially mediated removal of these compounds, making bioremediation the primary approach for the treatment of (per)chlorate contaminations (7).
Until 2008, all isolated PCRB were facultatively anaerobic or microaerophilic bacteria from different subclasses of the Proteobacteria. However, the recent description of a novel member of the genus Moorella (phylum Firmicutes) capable of (per)chlorate reduction demonstrated phylum-level diversity of PCRB (2). The detection of Cld activity in cell extracts from this organism indicated that this enzyme also is not confined to the Proteobacteria. In the same year, environmental genomics and heterologous gene expression led to the identification and validation of a catalytically efficient Cld in the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii” (phylum Nitrospirae) (33). This enzyme (referred to as NdCld) is the first characterized Cld from a nonproteobacterial organism. The discovery of NdCld was unexpected, because nitrite-oxidizing bacteria (NOB) have not been considered relevant for (per)chlorate removal. Phylogenetic analyses made in the same study (33) showed that many of the sequenced bacterial and archaeal genomes, including those from several pathogenic bacteria, encode Cld-like proteins, although most of the respective organisms have never been observed to use (per)chlorate or convert chlorite. Most of these Cld-like proteins have not been characterized yet, but the enzyme from Thermus thermophilus was found to have only a very weak chlorite-degrading activity (12). Consistently, this Cld-like protein is phylogenetically not closely related to the catalytically efficient Clds from PCRB and “Ca. Nitrospira defluvii” in phylogenetic trees (33).
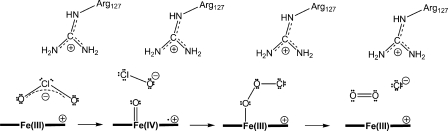
Recently we determined the crystal structure of NdCld and found a high degree of structural conservation compared to Clds from the Proteobacteria (22). Cld is a heme enzyme that, based on the published structural information, consists of five or six identical subunits (11, 12, 16, 22). Besides its importance for bioengineering, Cld is extremely interesting from a biochemical perspective. Besides photosystem II and a yet-uncharacterized enzyme of an anaerobic methane-oxidizing bacterium (14), Cld is the only known enzyme which efficiently catalyzes the formation of a covalent O-O bond as its principal function. A recently proposed reaction mechanism for Cld (Fig. 1) starts with the cleavage of the Cl-O bond at the heme cofactor to form a ferryl porphyrin radical species (compound I) and hypochlorite (28). Subsequently, the hypochlorite recombines with compound I to form Cl− and O2.
Fig. 1.
Proposed reaction mechanism of chlorite dismutase starting with the attack of anionic chlorite at ferric heme b. After formation of the Fe(III)-chlorite complex (not shown), heterolytic cleavage of the O-Cl bond leads to the formation of hypochlorite and the redox intermediate compound I [oxoiron(IV) porphyrin cation radical]. Finally, upon nucleophilic attack of anionic hypochlorite at the ferryl oxygen, compound I is reduced to the resting state and dioxygen and chloride are released. In addition, the scheme shows the putative role of conserved Arg127 in the orientation and stabilization of the substrate and the postulated intermediate(s).
In this study, we have addressed the question of whether yet-uncharacterized Cld-like proteins found in microorganisms other than the known PCRB and Nitrospira might efficiently degrade chlorite. For this purpose, we chose to determine the crystal structure and to analyze biochemical features of a Cld-like protein from the nitrite oxidizer Nitrobacter winogradskyi, which, like “Ca. Nitrospira defluvii,” does not belong to the functional group of heterotrophic PCRB. This protein (NwCld) was detected in the sequenced genome of this microbe (40) and is considerably smaller (183 amino acid residues) than all functionally validated (i.e., canonical) Clds (251 to 285 residues). Interestingly, NwCld represents a separate phylogenetic lineage of Cld-like proteins that are all similar in size and are encoded in the genomes of various Alpha-, Beta-, Gamma-, and Deltaproteobacteria as well as Cyanobacteria. For none of the proteins affiliated with this lineage has a chlorite transforming capability been shown yet. Due to these features, NwCld is a superb candidate for experimental evaluation of whether the phylogenetic and structural diversity of active Clds might be greater than previously anticipated. Our results demonstrate that N. winogradskyi encodes a structurally unusual and highly active Cld, and thus, they have implications for future research on the biochemistry of chlorite dismutases and possibly also for future studies of PCRB in the environment.
MATERIALS AND METHODS
Cloning of NwCld.
A DNA fragment containing the full-length coding region of chlorite dismutase from Nitrobacter winogradskyi (accession no. YP_319047) was amplified by PCR using the newly designed primers NwCldF (5′-CGA GCG CAT ATG ACG TTC ACA GTC TTC ACC-3′) and NwCldR (5′-GCG CGA GGA TCC CCT ATC GCG CGC GCC AAT CG-3′) and N. winogradskyi genomic DNA as a template. The amplicon was cloned into the expression vector pET-21b(+) (Merck/Novagen, Darmstadt, Germany) for the subsequent production of a C-terminally His-tagged fusion protein.
Heterologous expression and purification of NwCld.
Recombinant NwCld was expressed in Escherichia coli Tuner (DE3) cells (Merck/Novagen) grown in heme-enriched Luria-Bertani (LB) medium. Briefly, LB medium supplemented with carbenicillin (100 μg/ml) and hemin (50 μg/ml) was inoculated with a freshly prepared overnight culture (at a dilution ratio of 1:100). The culture was grown at 37°C under agitation (220 rpm) until the early stationary phase was reached (optical density at 600 nm [OD600] = 1.8). For NwCld expression, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, and the temperature as well as the shaker speed were reduced to 20°C and 180 rpm, respectively. After 12 h, the culture was centrifuged and the resulting cell pellet was either processed immediately or frozen in liquid nitrogen and stored at −80°C. When needed, cell pellets were resuspended in 50 mM Tris, 300 mM NaCl, 2% glycerol, and 20 mM imidazole (pH 8.5) supplemented with 1 mM phenylmethylsulfonylfluoride (PMSF) and 100 μM hemin. The resulting cell suspension was lysed by sonication and clarified by centrifugation. Subsequently, the supernatant was loaded onto 20 ml HisTrap FF crude (GE Healthcare, Vienna, Austria) columns. The eluted proteins were screened by SDS-PAGE, and fractions containing NwCld were pooled and applied on a HiLoad 26/60 Superdex 200 pg column (GE Healthcare) equilibrated with buffer (50 mM Tris, 150 mM NaCl, pH 8.5). Aliquots of purified protein were concentrated to 11 mg/ml, frozen in liquid nitrogen, and stored at −80°C until further use.
Crystallization of NwCld.
Initial screening for suitable crystallization conditions for NwCld was performed at 22°C with commercially available crystallization screens, using the sitting-drop vapor diffusion technique and a nanodrop-dispensing robot (Phoenix RE; Rigaku Europe, Kent, United Kingdom). The first crystals of NwCld appeared within 1 week in 1.0 M Na2HPO4-NaH2PO4, pH 8.2 (condition no. 55 of the SaltRX crystallization screen; Hampton Research, Aliso Viejo, CA). After several rounds of optimization trials using the same aforementioned setup, single, well-diffracting crystals of NwCld were obtained from 53.6 mM Na2HPO4 and 846.4 mM NaH2PO4. Prior to data collection, crystals were transferred into crystallization solution supplemented with 30% glycerol and were flash-frozen in liquid nitrogen.
Data collection, processing, and phasing.
Diffraction data were collected from a cube-shaped crystal with approximate dimensions 300 by 200 by 120 μm3 at ESRF beamline ID14-4 using a 1.278-Å wavelength to a maximum resolution of 2.1 Å. Four diffraction data sets collected on distinct positions on the same crystal were integrated and scaled using the software program XDS (20). Data collection statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics for NwCld
| Parameter | Value for NwCld or identifiera |
|---|---|
| Data collection | |
| Beamline | ID14-4 (ESRF) |
| Wavelength (Å) | 1.278 |
| Resolution (Å) | 45.8–2.1 (2.15–2.10) |
| Space group | P41 |
| Unit cell (Å) | a = b = 102.62; c = 49.12 |
| No. of molecules/AUb | 2 |
| No. of unique reflections | 54,986 (3,612) |
| No. of total reflections observed | 2,080,683 (132,332) |
| Completeness (%) | 94.0 (83.1) |
| Rmeasc | 0.081 (0.769) |
| Rmerged | 0.043 (0.007) |
| Multiplicity | 37.8 (36.6) |
| I/σ(I) | 39.38 (7.43) |
| Phasing | |
| No. of sites | 2 |
| Figure of merit acentric (centric) | 0.308 (0.176) |
| Refinement | |
| Rcryste/Rfreef | 0.191/0.239 |
| RMSD, bonds (Å) | 0.012 |
| RMSD, angles (°) | 1.314 |
Except where otherwise noted, values in parentheses are for the highest-resolution shell.
AU, asymmetric units.
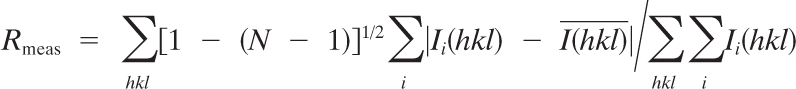 where Ii(hkl) and
where Ii(hkl) and 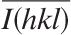 are the ith and mean measurements of the intensity of reflection hkl, and N is the redundancy.
are the ith and mean measurements of the intensity of reflection hkl, and N is the redundancy.
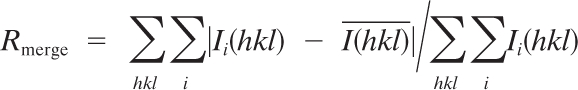 See footnote c for definitions of symbols.
See footnote c for definitions of symbols.
Rcryst = Σ|F0 − Fc|/F0
Rfree is the cross-validation Rfactor computed for the test set of reflections (5%) which are omitted in the refinement process.
The phase problem was solved by single-wavelength anomalous dispersion (SAD) methodology, exploiting the anomalous signal of iron. Heavy atom search, SAD phasing, solvent flattening, and autobuilding were carried out using AutoSol from the PHENIX software suite (1).
Model building, refinement and validation of the structure.
The structure of NwCld was refined using the software programs phenix.refine, of the PHENIX software package, and TLSMD (1, 35). Manual model building was performed in the COOT program (13), while validation of the model was done using the MOLPROBITY program (10). Final refinement statistics are summarized in Table 1.
Structural analysis and superposition.
Structure comparisons and superpositions were performed by using the SSM server (Protein Structure Comparison service SSM at the European Bioinformatics Institute [EBI]), the software program SUPERPOSE of the CCP4 package, and the PyMOL program (http://www.pymol.org) (8, 24). The subunit interface of structures was analyzed using the Protein Interfaces, Surfaces and Assemblies service (PISA) at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (23).
Steady-state kinetics.
The chlorite-dismutating activity of NwCld was measured continuously using a Clark-type oxygen electrode (Oxygraph Plus; Hansatech Instruments, Norfolk, United Kingdom) inserted into a stirred water bath kept at 30°C. The electrode was equilibrated to 100% air saturation by bubbling air to the reaction mixture for at least 15 min and for 0% air saturation by bubbling with N2 for at least 15 min to derive an offset and calibration factor. Reactions were carried out in O2-free 100 mM phosphate buffer (pH 7.0) with 50 μM to 100 mM NaClO2 added from a stock made in the same buffer. Reactions were started by addition of 20 nM solutions of NwCld. With increasing chlorite concentrations, irreversible inactivation of NwCld occurred, as was evident with inspection of individual time traces. Thus, it was important to use only the initial linear phase for rate calculation and to deduce Michaelis-Menten parameters from a set of chlorite concentrations below 1 mM. Molecular oxygen production rates (μM O2 s−1) were obtained from initial linear time traces (<10% substrate consumed) and plotted against chlorite concentrations.
Spectral analysis and heme determination.
UV-visible (UV-Vis) spectra were recorded on a Nanodrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA) at 22°C. Heme type and NwCld:heme b stoichiometry were determined by the pyridine hemochrome assay (37). In detail, oxidized protein samples in 50 mM potassium phosphate (pH 7.0) were mixed with pyridine and NaOH (final concentrations, 20% [vol/vol] pyridine and 0.1 M NaOH). After the oxidized spectrum was recorded, sodium dithionite was added to a final concentration of 10 mM, the spectrum of the reduced pyridine hemochrome was recorded, and the difference spectrum was calculated. The heme content was calculated using the molar extinction coefficient: ε418 = 191,500 M−1 cm−1 (37).
Analytical size exclusion chromatography (SEC).
To determine the stability of the NwCld dimer, recombinant protein (4 mg/ml) was diluted to a final concentration of 0.4 mg/ml in 50 mM Tris (pH 8.5) supplemented with 0.1 M, 0.25 M, 1 M, or 2 M NaCl. After incubation for at least 3 h at 4°C, individual protein samples were applied on an analytical Superdex 75 10/300 column equilibrated with 50 mM Tris (pH 8.5) containing respective amounts of NaCl. For calculating the expected elution volume of the NwCld monomer, calibration of the column was performed for each particular salt concentration by using the LMW gel filtration calibration kit (GE Healthcare).
Phylogenetic analyses.
All phylogenetic analyses were performed by using the ARB software package (32) and an already existing database of aligned amino acid sequences of Clds and Cld-like proteins (33). The alignment was manually refined based on the crystal structures of NwCld (this study), NdCld (22) (PDB: 3NN1, 3NN2, 3NN3, and 3NN4), Cld of Azospira oryzae (AoCld) (PDB: 2VXH), Cld of Dechloromonas aromatica (DaCld) (PDB: 3DQ08 and 3DQ09), a Cld-like protein from Geobacillus stearothermophilus (GsCld) (PDB: 1T0T), a Cld-like protein from Thermoplasma acidophilum (TaCld) (PDB: 3DTZ), and a Cld-like protein from Thermus thermophilus (TtCld) (PDB: 1VDH). Highly conserved secondary and tertiary structure motifs were used to identify homologous residues. Phylogenetic trees were calculated by applying protein maximum-likelihood (PhyML [17]) and maximum-parsimony (PHYLIP version 3.66 with 100 bootstrap iterations) methods, both with the Jones, Taylor, and Thornton (JTT) substitution model. In total, 243 alignment columns were used for phylogenetic analysis. To determine the coverage of cld genes by cld-targeted PCR primer sets, an ARB PT_SERVER database was established from the nucleotide gene sequences of the Clds and Cld-like proteins in the database. Primer sequences were then matched to this PT_SERVER database by using the ARB probe match tool. All possible sequence permutations of degenerate primers were resolved and matched by using the respective tools of ARB.
Miscellaneous methods.
Protein concentrations were determined by UV280 absorption. The extinction coefficient was calculated using the ProtParam software program (15) and experimentally confirmed by comparison of protein concentrations determined by this extinction coefficient with those measured by the Lowry assay using bovine serum albumin as a standard. The oligomeric state of NwCld in solution was determined with analytical size exclusion chromatography using a Superdex 75 10/300 (GE Healthcare) column equilibrated with 100 mM Tris buffer at pH 8.5 and 200 mM NaCl. All molecular graphics figures were prepared using PyMOL. Signal peptides for protein translocation via the Sec and Tat pathways were detected by using the SignalP (4), PrediSi (www.predisi.de), and TatP (5) online tools.
Protein structure accession number.
The structure for NwCld has been deposited at the RCSB Protein Data Bank (accession no. 3QPI).
RESULTS AND DISCUSSION
The heterologous overexpression of NwCld in E. coli and subsequent purification yielded sufficient amounts of pure protein for successful crystallization, structure analysis, and kinetic characterization. The structure of NwCld was determined to a resolution of 2.1 Å. The protein crystallizes in the P41 space group with the following unit cell dimensions: a = b = 102.62 Å; c = 49.12 Å. The structural analysis revealed several interesting differences at the tertiary and quaternary structure levels from previously determined structures of Clds and Cld-like proteins. Most remarkably, NwCld was found to be a homodimer. This is consistent with analytical size exclusion chromatography, where NwCld eluted in a single peak with an apparent molecular mass of 43 kDa, which corresponds to an NwCld dimer (see Fig. S1 in the supplemental material). In contrast to NwCld, all canonical Clds and all Cld-like proteins whose structures have been determined are homopentamers or homohexamers, respectively (11, 12, 16, 22).
Subunit structure of NwCld and its impact on oligomeric state.
The NwCld primary sequence is about 30% shorter than those of the canonical Clds, with a significant deletion in the N-terminal region. These unique structural properties of NwCld are also reflected by phylogenetic analysis, which revealed that NwCld does not cluster with any other structurally characterized member of the Cld-like protein superfamily (Fig. 2). Due to weak electron density, we could not model residues 41 to 47 in subunit A and residues 41 to 48 in subunit B, suggesting that these regions are flexible and were disordered in the crystal lattice (Fig. 3 a).
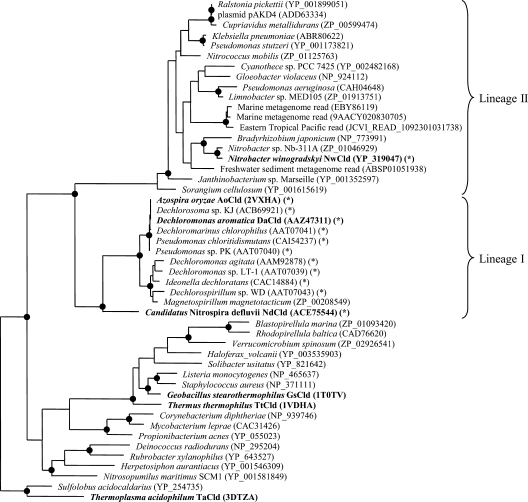
Fig. 2.
Maximum-likelihood phylogenetic tree based on the amino acid sequences of selected Clds and Cld-like proteins. Names printed in boldface represent proteins with known crystal structures. Validated and catalytically efficient Clds are marked with an asterisk. The proposed phylogenetic lineages I (canonical Clds, mainly from PCRB) and II (NwCld and related proteins) are delimited by curly brackets. Black circles on tree nodes symbolize high-parsimony bootstrap support (≥90%) based on 100 iterations. Accession numbers are indicated for all sequences.
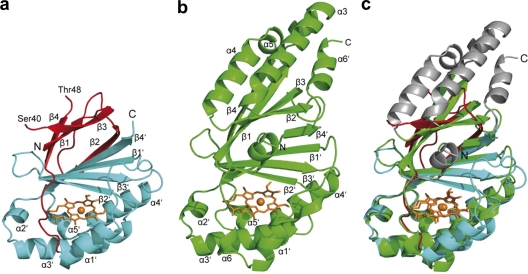
Fig. 3.
NwCld subunit and structural comparison with canonical NdCld. (a) Ribbon representation of one NwCld subunit. The N-terminal domain is shown in red, whereas the C-terminal domain is shown in cyan. The N and C termini are labeled, and the heme b is presented as orange sticks, with iron shown as an orange sphere. The labeling of secondary structure elements follows the labeling used for NdCld as shown in panel b. (b) Ribbon representation of one subunit of NdCld from “Ca. Nitrospira defluvii.” Secondary structure elements are labeled according to the method of Kostan et al. (22). Heme group and iron are presented as in panel a. (c) A subunit of NdCld (shown in green and gray) superimposed on a subunit of NwCld (shown in red and cyan). The orientation of the subunits is the same as in panels a and b. Structural elements missing in the subunit structure of NwCld compared to the NdCld subunit structure are depicted in gray.
Superposing NwCld and all other available structures of Clds and Cld-like proteins revealed a high structural similarity at the subunit level, with a root mean square deviation (RMSD) of 1.40 Å over 119 superposed Cα atoms, although the amino acid sequence identities between NwCld and these proteins are as low as 13 to 27%. Consistent with the phylogeny of Cld and Cld-like proteins (Fig. 2), NwCld is structurally most similar to DaCld from D. aromatica (structure 3Q08, RMSD of 1.30 Å; structure 3Q09, RMSD of 1.30 Å, both over 144 Cα atoms) and to NdCld (RMSD of 1.68 Å over 146 Cα atoms) and least similar to TaCld from T. acidophilum (RMSD of 1.78 Å over 128 Cα atoms).
All previously determined subunit structures of Clds and Cld-like proteins (11, 12, 16, 22) consist of two similar domains with a ferredoxin-like fold. Both domains are characterized by a four-stranded antiparallel β-sheet flanked by six α-helices. The two β-sheets from each domain pack together at an angle of about 65°, forming a central flattened β-barrel surrounded on both sides by α-helices (Fig. 3b). NwCld, however, lacks the major part of the N-terminal domain (Fig. 3a and c). In particular, all N-terminal α-helices are missing, and only the β-sheet has been conserved. Furthermore, this β-sheet is also different: the β2 strand, which in canonical Clds interacts with the β3 strand, is no longer part of the first β-sheet but rather forms the fifth strand of the second β-sheet interacting with the β4′ strand of the C-terminal ferredoxin-like domain (Fig. 3a). Therefore, the central β-barrel no longer consists of two similar four-stranded β-sheets but of one three-stranded and one five-stranded β-sheet. The C-terminal domain is structurally highly similar to the C-terminal domains of previously described Clds (Fig. 3b), although it lacks the last α-helix α6′ that is present in the canonical Clds (Fig. 3a and c).
Subunit interface: molecular determinants of NwCld dimerization.
Until now, all structurally characterized Clds and Cld-like proteins are pentamers (12, 16, 22) or, in the case of the Cld from A. oryzae, a hexamer (11). The interface between the subunits in all these structures is very similar. The N- and C-terminal domains of each subunit interact with the corresponding halves of the neighboring subunit. The interface consists mainly of residues from the N-terminal helix α4 and strand β4, which interact with residues in the loop between strands β2 and β3 from the N-terminal domain of the neighboring subunit. The second half of the interface consists of the corresponding parts of the C-terminal domain. In contrast, NwCld is a dimer (Fig. 4 a), and the interface between monomers is entirely different from that observed in the pentameric or hexameric Clds (Fig. 4b). First, the NwCld subunit lacks all helices in the N-terminal domain, whereas the loop between strands β2 and β3 is much longer and adopts a completely different conformation. The β2 strand is now part of the second β-sheet interacting with β4′, and therefore NwCld lacks half of the interacting surface, which would be needed for pentamerization. Second, the β2 strand in this position along strand β4′ would block the interaction between the C-terminal domains. Instead, the dimer interface of NwCld is formed by residues from the loop between β4 and α1′ from one subunit which interacts with the loop between helix α2′ and helix α3′ plus the loop between strand β2′ and strand β3′ (Fig. 4c). The surface buried in this interface is 980 Å2, representing 11% of the total subunit surface. For comparison, the surface buried between two subunits in the pentameric NdCld is 1,400 Å2, corresponding to 11.5% of the subunit surface (22).
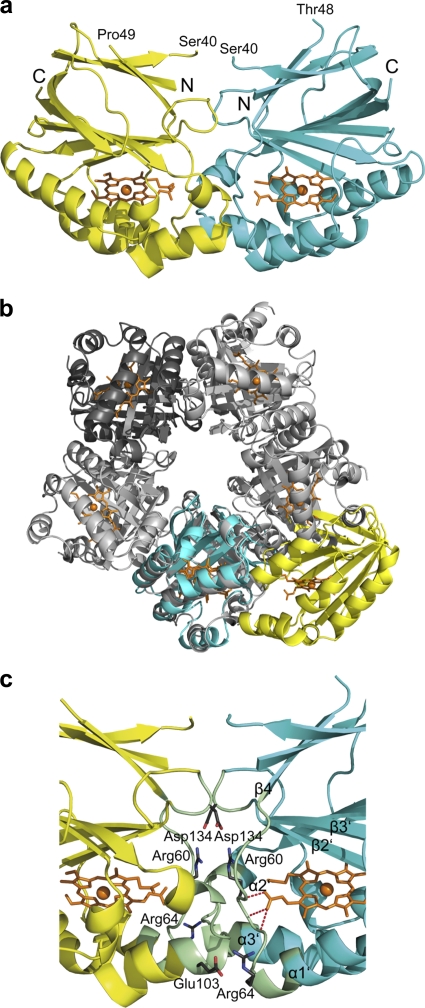
Fig. 4.
NwCld dimer structure and interface. (a) Ribbon representation of the NwCld dimer viewed perpendicular to the vertical 2-fold symmetry axis. Subunits are shown in different colors. The heme group is shown as an orange stick model in either subunit. The iron is displayed as an orange sphere. The N and C termini are labeled. Parts of the structure between Ser40 and Thr48 in one subunit and Ser40 and Pro49 in the other subunit could not be modeled in the structure due to weak electron density in these regions. (b) Superposition of an NwCld dimer with an NdCld pentamer (shown in gray), with one subunit of NwCld (cyan) spatially aligned to one NdCld monomer. Note the different location of the second NwCld subunit (yellow) compared to those of the subunits of NdCld, illustrating differences in the interfaces between the subunits of dimeric and pentameric Clds. To more easily follow the packing of subunits in the NdCld pentamer, one of its subunits is depicted in dark gray. (c) Detailed view of the NwCld dimer interface. Residues involved in interactions between the two subunits of the NwCld holoenzyme are shown in pale green. Side chains of amino acids involved in the formation of salt bridges are shown as sticks, with carbon, oxygen, and nitrogen atoms depicted in gray, red, and blue, respectively. Hemes are presented as orange stick models, with heme irons shown as orange spheres. Interactions of heme propionate with the protein backbone are visualized by red dashes.
The interactions involved in NwCld dimer formation are mainly of electrostatic and to a lesser extent hydrophobic nature. Namely, only about 17% of the area buried in the interface involves hydrophobic amino acids, while the PISA server (23) identified 16 interdomain hydrogen bonds and four salt bridges. One salt bridge is formed between Arg60, located in the loop between β4 and α1′, and Asp134, found in the loop between strand β2′ and strand β3′. A second bridge connects Arg64, located in the beginning of helix α1′, to Glu103, found in the beginning of helix α3′. These two pairs of salt bridges are repeated two times due to the symmetric dimer interface (Fig. 4c). In order to address the importance of salt bridges for dimer stability, we incubated purified NwCld in the presence of increasing salt concentrations (0.1 to 2 M NaCl) and subsequently subjected it to analytical SEC (for details, see Materials and Methods). Interestingly, in none of the tested conditions was NwCld observed in an elution peak corresponding to a monomer (Fig. S1), indicating that the NwCld dimer is stable at high ionic strengths and that the overall stability of the dimer is due to the constructive interplay of both hydrophobic and polar interactions. The residues involved in these salt bridges (Arg60-Asp134 and Arg64-Glu103) are conserved in most Clds of lineage II (see Fig. S5 in the supplemental material), indicating that these proteins could form a dimer with an interface similar to that of NwCld. In addition, Arg60 interacts with the heme b via a hydrogen bond between its backbone amide hydrogen and a propionate group of the heme b (Fig. 4c). Thus, Arg60 is a direct link of interactions from the heme to the dimer interface, which might indicate that formation of the NwCld dimer influences the binding of the heme. This is in contrast to the canonical (lineage I) Clds, which lack any obvious link between the subunit interface and heme binding. The interface of the pentameric NdCld has a similar number of interdomain hydrogen bonds and salt bridges, but a larger part of the surface has a hydrophobic character (about 30%).
Active site structure of NwCld.
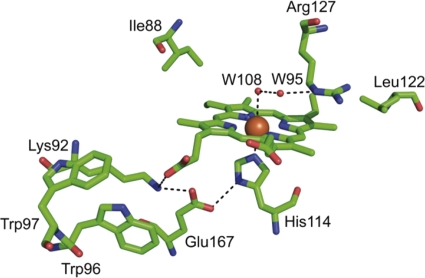
Similar to other known Cld structures, the active site of NwCld with the bound heme b is located in the C-terminal domain of the molecule, in a cavity formed between the second β-sheet (β1′ to β4′) and helices α2′, α3′, and α4′ (Fig. 3a). Most residues within 4 Å from heme b have a hydrophobic nature and interact with the prosthetic group via van der Waals contacts. Furthermore, several residues are involved in specific interactions that are important for heme binding and enzyme function. On the proximal side of the heme b, His114 from the α3′ helix coordinates the heme iron at a distance of 2.16 Å (Fig. 5) and forms a hydrogen bond with Glu167. The latter residue is hydrogen bonded to Lys92, which is also within hydrogen bonding distance of one propionate group of the heme b (Fig. 5). This intricate hydrogen bonding network increases the imidazolate character of the proximal histidine residue, thereby shifting the reduction potential of the heme iron to more negative values. This might be important for the stabilization of the higher heme oxidation state(s) involved in chlorite dismutation (28) (Fig. 1). The second propionate of heme b is hydrogen bonded to the backbone of two residues located in the loop between β4 and α1′: Arg60 and Tyr61, of which Arg60 is also important for the stability of the dimer interface (Fig. 4c). On the proximal heme side at a distance of 3.8 Å from one of the propionate groups, there is a pair of tryptophan residues (Trp96 and Trp97) whose aromatic ring systems are oriented perpendicularly to each other (Fig. 5). Trp96 may play a role in the catalytic mechanism, acting as the electron donor for the reduction of compound I to compound II (28) (Fig. 1). The second tryptophan (Trp97) contributes together with Lys92, Met99, Arg104, and Ile107 to the stabilization of the Trp96 side chain orientation that is suitable for interaction with the heme b propionate group. In addition, Trp97 influences the electronic structure of Trp96 through interaction of its partially negative π-electron cloud and the partially positive dipole of the indole ring of Trp96. At the inner part of the active site cavity, the two vinyl groups of the heme ligand interact with Ile88 and Leu122, respectively. These two amino acid residues define the depth of the ligand cavity and thus determine size and type of the ligand that can be accommodated here (Fig. 5).
Fig. 5.
Active site of NwCld. The illustration shows selected residues involved in heme b binding and in the catalytic mechanism of chlorite dismutation. Carbon, oxygen, and nitrogen atoms are depicted in green, red, and blue, respectively. The heme iron and water molecules (W) are shown as orange spheres. The hydrogen bonding network spanning from Arg127 to Glu167 is visualized by dashed lines.
At the distal side, the heme iron is coordinated by a water molecule (Wat108) at a distance of 2.9 Å. This water molecule is linked via a hydrogen bonding network to a second water molecule (Wat95) and Arg127 (Fig. 5). The Arg127 side chain, which points away from the heme, is further stabilized by hydrogen bonds to four additional water molecules (Wat1, Wat80, Wat174, and Wat193) and to the side chain of Gln74 (see Fig. S2a in the supplemental material). In our previous study on NdCld, an arginine residue at this position was experimentally shown to be important for catalytic activity, most likely by regulating substrate uptake and by stabilizing a reaction intermediate (22) (Fig. 1). Consistent with its functional importance, this arginine residue occurs in all catalytically efficient Clds (see below), and its side chain conformation is stabilized by a similar pattern of interactions in these enzymes. In NwCld, this involves the two water molecules Wat1 and Wat193. In the NdCld structure, these water molecules are replaced by a molecule of ethylene glycol (originating with the cryosolution) with its oxygen atoms in the position of the waters. In addition, while Arg127 of NwCld interacts with the side chain of Gln74, Arg173 of NdCld forms two hydrogen bonds to the backbone (see Fig. S2b).
The water molecule Wat108, which in NwCld coordinates the iron atom, is replaced by one of the nitrogen atoms of imidazole in the structure of NdCld. The same is true for the structures of AoCld (see Fig. S2c in the supplemental material) and DaCld, which were determined in complex with thiocyanate and nitrite, respectively: both of these inhibitors replace Wat108. In these two structures, the arginine side chain is no longer stabilized by hydrogen bonds to the glutamine side chain or to the backbone (as in NdCld) but is found in an orientation closer to the iron, with one of its nitrogen atoms replacing Wat95 (see Fig. S2c).
Similar to all Clds and Cld-like proteins whose structures are known, NwCld has a positive electrostatic potential on the surface around the entrance to the active site (see Fig. S3 in the supplemental material). For chlorite dismutases, this is clearly important for attracting the anionic substrate chlorite. In the case of the Cld-like proteins, this indicates that their substrate, if it is not chlorite, might also be negatively charged.
Heme analysis and UV-Vis spectral characterization.
Absorption spectra of reduced pyridine hemochromes of NwCld showed maxima at 418 nm (Soret), 526 nm (β), and 557 (α), typical for protoporphyrin IX (heme b) (37), as was seen in other so far characterized Clds (11, 22). The heme content was determined to be 0.58 heme b per NwCld subunit (with the Reinheitszahl A407/A280 equal to 1.9). This ratio is lower than expected, since the crystal structure is fully loaded with heme. Incomplete heme occupancy in recombinant proteins has been reported for other Clds (34) and might be related to bottlenecks in heme supply by the host due to high protein overexpression and/or suboptimal cultivation conditions. Native NwCld showed a Soret maximum at 411 nm, which shifted to 435 nm upon reduction by dithionite (data not shown). Similar Soret maxima of ferric and ferrous proteins have been reported for other Clds (33, 41, 46).
Steady-state kinetics of NwCld.
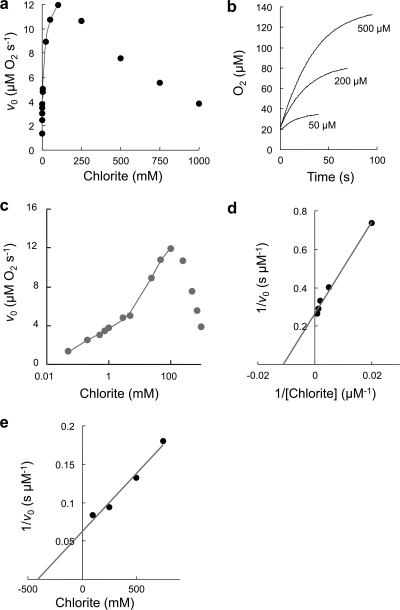
The stoichiometry of the Cld reaction has been reported to be 1 mol Cl− and 1 mol O2 out of 1 mol ClO2− (28). Figure 6 a depicts the plot of the initial rate of O2 release by NwCld versus chlorite concentration. It demonstrates saturation in the initial rate (v0) with increasing chlorite concentrations followed by a decrease in v0 at very high chlorite concentrations (>100 mM), which indicates uncompetitive substrate inhibition. At these high concentrations, where the inhibitory effect predominates, an inhibition constant (Ki = 310 mM) could be determined from the Dixon plot depicted in Fig. 6e. Inactivation of NwCld by chlorite was evident with inspection of the individual time traces and the total amount of released dioxygen at a defined [ClO2−]. Only at low chlorite concentrations (<50 μM) was the substrate completely consumed, whereas at higher concentrations residual substrate remained due to inactivation of the enzyme in the course of the reaction (Fig. 6b). At [ClO2−] of >1 mM, less than 10% of chlorite was converted to Cl− and O2. Thus, in order to minimize interference with inhibition kinetics, Michaelis-Menten parameters were calculated from a set of chlorite concentrations with [ClO2−] of ≤1 mM, which probably reflect physiological conditions better than higher concentrations (for better clarity, see also the semilogarithmic plot in Fig. 6c). Michaelis-Menten parameters were determined from the double-reciprocal plot (Fig. 6d and Table 2). The obtained values resemble the kinetic parameters of the canonical Clds of PCRB and “Ca. Nitrospira defluvii” (Table 2). For comparison, the kinetic parameters of the Cld-like protein TtCld from T. thermophilus indicate a much weaker chlorite-dismutating activity (Table 2). The temperature and pH optima of the chlorite-dismutating activity of NwCld (20°C and 5.5; see Fig. S4 in the supplemental material) are slightly lower than the respective optima of other Clds (Table 2). Thus, despite the pronounced structural differences from the canonical Clds (Fig. 3 and 4), NwCld is an efficient chlorite dismutase. This result is consistent with the phylogenetic analysis. The distinct and well-supported lineage that contains NwCld shares a common ancestor with another lineage containing all known and highly efficient canonical Clds to the exclusion of all other Cld-like proteins (Fig. 2). Here we refer to the canonical Clds as “lineage I,” whereas the cluster formed by NwCld and similar proteins is designated “lineage II” (Fig. 2). The monophyly of lineages I and II indicates that these groups share steps of their evolutionary history, which most likely were key to the development of efficient chlorite-dismutating enzymes and separated these lineages from the other Cld-like proteins. Since the small size of NwCld and the other lineage II proteins is an exception within the Cld-like protein superfamily (see Fig. S5), it probably does not represent the ancestral state but was rather caused by a partial loss of the N-terminal domain during the evolution of lineage II.
Fig. 6.
Kinetics of chlorite dismutation activity by NwCld. (a) Plot of the initial rate (v0) of molecular oxygen evolution as a function of chlorite concentration. Points represent averages of three measurements. Applied conditions were as follows: 50 mM phosphate buffer (pH 7.0), 20 nM NwCld, 30°C. (b) Selected time traces at different chlorite concentrations. Conditions were as for panel a. (c) Semilogarithmic plot of the data in panel a. Double-rectangular hyperbolar fits are shown in gray. (d) Double reciprocal plot of v0 versus chlorite concentration for the determination of kinetic parameters. (e) Dixon plot for the determination of Ki.
Table 2.
Kinetic parameters, temperature, and pH optima of validated chlorite dismutasesa
| Parameter | Value for enzyme |
||||||
|---|---|---|---|---|---|---|---|
| NwCld | NdCld | AoCld | IdCld | DaCld | PcCld | TtCld | |
| Km (μM) | 90 | 58 | 170 | 260 | 220 | 80 | 13,000 |
| kcat (s−1) | 190 | 35 | 1,200 | 1,800 | 1,880 | 230 | 0.77 |
| kcat/Km (M−1 s−1) | 2.1 × 106 | 6.0 × 105 | 7.1 × 106 | 6.9 × 106 | 35.4 × 106 | 2.7 × 106 | 59 |
| Temp optimum (°C) | 20 | 25 | 30 | NDb | ND | 25 | ND |
| pH optimum | 5.5 | 6 | 6 | ND | ND | 6 | ND |
From N. winogradskyi (NwCld; this study), “Ca. Nitrospira defluvii” (NdCld) (22), A. oryzae (AoCld) (46), Ideonella dechloratans (IdCld) (41), D. aromatica (DaCld) (42), and Pseudomonas chloritidismutans (PcCld) (34). Values for the Cld-like protein of T. thermophilus (TtCld) (12) are included for comparison.
ND, not determined.
Signature residues of catalytically efficient Clds.
The currently available structures and kinetic data for Clds and Cld-like proteins allowed us to identify potential signature amino acid residues that could define a catalytically efficient Cld. Some of the aforementioned residues in the vicinity of the active site of NwCld (Lys92, Trp96, and His114; NwCld numbering) are strictly conserved in all Clds and Cld-like proteins (see Fig. S5 in the supplemental material). However, the five residues Ile88, Trp97, Leu122, Arg127, and Glu167 are conserved only in NwCld, the canonical Clds (including NdCld), and in sequences that are closely related to these reference proteins (see Fig. S5). As outlined above, these residues are proposed to stabilize the heme cofactor or to play roles in the catalytic mechanism of Cld. For Arg127, an important function in substrate positioning and activation during chlorite degradation has been experimentally demonstrated (22) (Fig. 1). All Cld-like proteins whose structure has been determined and found to be without heme or which have been tested for Cld activity but found to be inactive (TtCld, GsCld, and TaCld) have different residues at these key positions (see Fig. S5). In these proteins Arg127 is replaced by a polar residue (glutamine or serine), while Glu167, which stabilizes the proximal His114 and connects it via a network of hydrogen bonds to Lys92 (see above), is replaced by a small hydrophobic residue (alanine or valine) that cannot form a similar interaction network. At the position corresponding to Ile88, these Cld-like proteins have a tyrosine residue. In silico replacement of Ile88 with tyrosine leads to a steric clash between the tyrosine side chain and the vinyl group of the heme moiety. Similarly, the second vinyl group in NwCld interacts with Leu122, while the Cld-like protein structures without heme (GsCld, TaCld, and TtCld) have an alanine, a proline, or a glutamine in the corresponding position, leading to less-favorable interactions with a putative heme b. Moreover, in these Cld-like proteins, the loop between strand β4 and helix α1′, whose backbone interacts with a heme propionate group in NwCld (see above), is in a different position (not shown). These subtle differences suggest that if these Cld-like proteins bind heme b, they might bind it more weakly and in a slightly different orientation than in the canonical Clds and NwCld. Assuming these proteins still bind heme, it would be expected that their Cld activity is low due to the differences in the key residues discussed above.
Given the high diversity of Cld-like proteins (Fig. 2) (33) and the relatively low degree of raw sequence conservation in this superfamily, the aforementioned five signature residues are valuable indicators for a tentative functional classification of novel Cld-like sequences. For example, the presence of these signatures indicates that not only NwCld but also all known lineage II proteins from other bacteria (Fig. 2; see Fig. S5 in the supplemental material) could be efficient Clds. Interestingly, this group includes other nitrite oxidizers (Nitrobacter strain Nb-311A and Nitrococcus mobilis) and important pathogens of humans (Klebsiella pneumoniae and Pseudomonas aeruginosa) (Fig. 2). However, future experimental work is needed to validate the activity of additional lineage II Clds and to verify the proposed functional importance of all conserved signature residues (22).
Physiological function of NwCld and related proteins.
The observed high chlorite-dismutating activity of NwCld raises the question of which physiological function this enzyme has in Nitrobacter. The key enzyme for nitrite oxidation in Nitrobacter, nitrite oxidoreductase (Nxr) (43), is highly similar to the membrane-bound dissimilatory nitrate reductase (Nar) of E. coli and many other bacteria (21). Chlorate is transformed to chlorite by Nar due to the structural similarity between chlorate and nitrate (see Logan et al. [30] and references cited therein). Consistent with the similarity of Nxr to Nar, Nitrobacter winogradskyi and an environmental Nitrobacter isolate indeed are able to reduce chlorate and even can oxidize nitrite using chlorate instead of O2 as an electron acceptor during anoxic incubations (18, 29). An active NwCld should offer Nitrobacter protection from the chlorite produced under these conditions, but in a previous study, chlorate-driven nitrite oxidation by Nitrobacter diminished after a few hours, most likely due to inhibition by chlorite (29). However, this inhibition was observed after incubation of Nitrobacter in nitrite media containing relatively high chlorate concentrations (4.2 to 17 mM [29] or 10 mM [18]), which presumably led to the production of large amounts of chlorite. The same studies found that Nitrobacter was strongly inhibited by externally added chlorite above 1 mM, but inhibition was only weak with 0.3 mM (29) or less than 0.1 mM (18). In this context, it is noteworthy that toxic effects of chlorite on other bacteria have been observed at much lower concentrations of 0.01 to 0.02 mM (47). Based on these observations, Nitrobacter indeed has a limited chlorite resistance, which could enable it to persist and even oxidize nitrite in the presence of low concentrations of chlorate. However, this poses the next question of how chlorate or chlorite can get in contact with the active sites of Nxr or Cld, respectively. Nxr of Nitrobacter is attached to the cytoplasmic membrane, with its active site located on the cytoplasmic side of the membrane (38). Chlorate readily crosses the outer membrane of Gram-negative bacteria (25), but experiments using E. coli or Paracoccus denitrificans showed that without addition of detergents, the cytoplasmic membrane is a strong diffusion barrier for chlorate and that chlorate is not actively transported into the cells either (19, 25). Other studies, however, have indicated that passive diffusion of chlorate over the native cytoplasmic membrane of P. denitrificans occurs to a limited extent (26, 27). Assuming that the chlorate was reduced by Nxr (and not by an unidentified periplasmic enzyme), it must have entered Nitrobacter cells by either passive diffusion or active transport in the aforementioned experiments, which used intact Nitrobacter cells without adding detergents to the cultivation media. Since membrane permeability for chlorite is even lower than that for chlorate (26), any chlorite produced by Nxr would be entrapped in the cytoplasm and could be degraded only by cytoplasmic Cld. In silico analyses suggested that NwCld does not contain a signal peptide for protein translocation into the periplasm and that the cleaving of a signal peptide at the N terminus would severely disturb the structure of the N-terminal domain (data not shown). Thus, for NwCld we predict a cytoplasmic localization, which is compatible with the detoxification of chlorite resulting from chlorate reduction by Nxr. Interestingly, the localizations of Nxr and chlorite dismutases are also consistent in other NOB since they are always predicted to be located on the same side of the cytoplasmic membrane. In detail, Nitrococcus, which also has a cytoplasmically oriented Nxr (38), possesses a lineage II Cld that is predicted to be cytoplasmic (Fig. 2). In contrast, Nxr of “Ca. Nitrospira defluvii” faces the periplasmic side of the plasma membrane (31), and the lineage I Cld of this organism (NdCld) contains an N-terminal signal peptide for translocation into the periplasmic space via the Sec pathway (33). Thus, the chlorite dismutases of all analyzed nitrite oxidizers could be involved in chlorite protection. At present, however, we cannot exclude other and yet-undiscovered biological functions of these enzymes in NOB.
The lineage II Cld-like proteins of the other organisms (Fig. 2) are also predicted to be cytoplasmic based on in silico analyses (data not shown). In contrast to Nitrobacter, none of these bacteria is known to reduce chlorate. Therefore, we assume that their lineage II Cld-like enzymes have yet-unrecognized in vivo functions other than chlorite dismutation.
Two additional observations exemplify the need for further research on lineage II Clds. First, one lineage II cld gene was found on a highly promiscuous broad-host-range plasmid, pAKD4 (36) (Fig. 2). The presence of this gene on a mobile genetic element indicates that lineage II cld genes are subject to horizontal transfer and may be widespread among environmental bacteria. Existing PCR primers, which target the canonical lineage I cld genes (3, 33) and were used to detect these genes in the environment (3) and in pure cultures (6), have numerous base mismatches to all lineage II genes (data not shown). Thus, the environmental distribution of lineage II cld genes, for example at (per)chlorate-contaminated sites, is unknown at present. Second, the genome of the poplar tree Populus trichocarpa encodes a lineage II Cld-like protein (GenBank accession no. EEE70332), which is the only known Cld in the eukaryotic domain. Experiments using sterile plants and tissue cultures revealed perchlorate reduction by poplar to chlorate and chlorite and also the transformation to Cl− (45). It is thus very likely that the lineage II Cld-like protein of poplar, which contains all the above-described signature amino acids of functional chlorite dismutases, is involved in this activity, although this function remains to be experimentally verified.
Conclusions.
Recent progress in genome and metagenome sequencing has enormously increased the size and phylogenetic complexity of many groups of homologous proteins, including the Cld-like protein superfamily (33). However, for many of these groups it remains unclear whether all affiliated proteins share a specific function or whether modified functions occur in different species or evolutionary lineages. Tackling this question is often extremely difficult, in particular because most microorganisms cannot be grown under laboratory conditions and/or no genetic tools are available for their manipulation. For the Cld-like protein superfamily, only the canonical Clds of lineage I had been assigned a biological function in PCRB, whereas the substrates and activities of the numerous other Cld-like enzymes remained an enigma. In this study, by combining heterologous expression with structural and biochemical analyses, we have identified the N. winogradskyi Cld-like protein representing another lineage from this superfamily as a novel and structurally distinct chlorite dismutase. This result is interesting from a biochemical perspective and may also have implications for our understanding of the microbiology of (per)chlorate and chlorite bioremediation.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a University Research Focus project (Symbiosis research and molecular principles of recognition; project no. FS573001, Molecular interactions between intracellular bacteria and their eukaryotic host cells) of the University of Vienna.
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities, and we thank Andrew McCarthy for assistance in using beam line ID14-4.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 25 March 2011.
REFERENCES
- 1. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balk M., van Gelder T., Weelink S. A., Stams A. J. M. 2008. (Per)chlorate reduction by the thermophilic bacterium, Moorella perchloratireducens sp. nov., isolated from an underground gas storage. Appl. Environ. Microbiol. 74:403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bender K. S., Rice M. R., Fugate W. H., Coates J. D., Achenbach L. A. 2004. Metabolic primers for detection of (per)chlorate-reducing bacteria in the environment and phylogenetic analysis of cld gene sequences. Appl. Environ. Microbiol. 70:5651–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 5. Bendtsen J. D., Nielsen H., Widdick D., Palmer T., Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinform. 6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cladera A. M., Garcia-Valdes E., Lalucat J. 2006. Genotype versus phenotype in the circumscription of bacterial species: the case of Pseudomonas stutzeri and Pseudomonas chloritidismutans. Arch. Microbiol. 184:353–361 [DOI] [PubMed] [Google Scholar]
- 7. Coates J. D., Achenbach L. A. 2004. Microbial perchlorate reduction: rocket-fueled metabolism. Nat. Rev. Microbiol. 2:569–580 [DOI] [PubMed] [Google Scholar]
- 8. Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 9. Collette T. W., et al. 2003. Analysis of hydroponic fertilizer matrixes for perchlorate: comparison of analytical techniques. Analyst 128:88–97 [DOI] [PubMed] [Google Scholar]
- 10. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32:W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Geus D. C., et al. 2009. Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. J. Mol. Biol. 387:192–206 [DOI] [PubMed] [Google Scholar]
- 12. Ebihara A., et al. 2005. Structure-based functional identification of a novel heme-binding protein from Thermus thermophilus HB8. J. Struct. Funct. Genomics 6:21–32 [DOI] [PubMed] [Google Scholar]
- 13. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 14. Ettwig K. F., et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 15. Gasteiger E., et al. 2005. Protein identification and analysis tools on the ExPASy server, p. 571–607 In Walker J. M. (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ [Google Scholar]
- 16. Goblirsch B. R., Streit B. R., Dubois J. L., Wilmot C. M. 2010. Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. J. Biol. Inorg. Chem. 15:879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 18. Hynes R. K., Knowles R. 1983. Inhibition of chemoautotrophic nitrification by sodium chlorate and sodium chlorite: a reexamination. Appl. Environ. Microbiol. 45:1178–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. John P. 1977. Aerobic and anaerobic bacterial respiration monitored by electrodes. J. Gen. Microbiol. 98:231–238 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26:795–800 [Google Scholar]
- 21. Kirstein K., Bock E. 1993. Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Arch. Microbiol. 160:447–453 [DOI] [PubMed] [Google Scholar]
- 22. Kostan J., et al. 2010. Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: Identification of a catalytically important amino acid residue. J. Struct. Biol. 172:331–342 [DOI] [PubMed] [Google Scholar]
- 23. Krissinel E., Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 24. Krissinel E., Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256–2268 [DOI] [PubMed] [Google Scholar]
- 25. Kristjansson J. K., Hollocher T. C. 1979. Substrate binding site for nitrate reductase of Escherichia coli is on the inner aspect of the membrane. J. Bacteriol. 137:1227–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kucera I. 2006. Interference of chlorate and chlorite with nitrate reduction in resting cells of Paracoccus denitrificans. Microbiology 152:3529–3534 [DOI] [PubMed] [Google Scholar]
- 27. Kucera I. 2003. Passive penetration of nitrate through the plasma membrane of Paracoccus denitrificans and its potentiation by the lipophilic tetraphenylphosphonium cation. Biochim. Biophys. Acta 1557:119–124 [PubMed] [Google Scholar]
- 28. Lee A. Q., Streit B. R., Zdilla M. J., Abu-Omar M. M., DuBois J. L. 2008. Mechanism of and exquisite selectivity for O-O bond formation by the heme-dependent chlorite dismutase. Proc. Natl. Acad. Sci. U. S. A. 105:15654–15659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lees H., Simpson J. R. 1957. The biochemistry of the nitrifying organisms. V. Nitrite oxidation by Nitrobacter. Biochem. J. 65:297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Logan B. E., et al. 2001. Kinetics of perchlorate- and chlorate-respiring bacteria. Appl. Environ. Microbiol. 67:2499–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lücker S., et al. 2010. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:13479–13484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maixner F., et al. 2008. Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium ‘Candidatus Nitrospira defluvii.’ Environ. Microbiol. 10:3043–3056 [DOI] [PubMed] [Google Scholar]
- 34. Mehboob F., et al. 2009. Purification and characterization of a chlorite dismutase from Pseudomonas chloritidismutans. FEMS Microbiol. Lett. 293:115–121 [DOI] [PubMed] [Google Scholar]
- 35. Painter J., Merritt E. A. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450 [DOI] [PubMed] [Google Scholar]
- 36. Sen D., et al. 2010. Comparative genomics of pAKD4, the prototype IncP-1delta plasmid with a complete backbone. Plasmid 63:98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith K. M. 1975. Porphyrins and metalloporphyrins. Elsevier Scientific, Amsterdam, Netherlands [Google Scholar]
- 38. Spieck E., Bock E. 2005. The lithoautotrophic nitrite-oxidizing bacteria, p. 149–153 In Staley J. T., Boone D. R., Brenner D. J., de Vos P., Garrity G. M., Goodfellow M., Krieg N. R., Rainey F. A., Schleifer K. H. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer Science+Business Media, New York, NY [Google Scholar]
- 39. Stanbury J. B., Wyngaarden J. B. 1952. Effect of perchlorate on the human thyroid gland. Metabolism 1:533–539 [PubMed] [Google Scholar]
- 40. Starkenburg S. R., et al. 2006. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 72:2050–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stenklo K., Thorell H. D., Bergius H., Aasa R., Nilsson T. 2001. Chlorite dismutase from Ideonella dechloratans. J. Biol. Inorg. Chem. 6:601–607 [DOI] [PubMed] [Google Scholar]
- 42. Streit B. R., DuBois J. L. 2008. Chemical and steady-state kinetic analyses of a heterologously expressed heme dependent chlorite dismutase. Biochemistry 47:5271–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanaka Y., Fukumori Y., Yakamaka T. 1983. Purification of cytochrome a1c1 from Nitrobacter agilis and characterization of nitrite oxidation system of the bacterium. Arch. Microbiol. 135:265–271 [Google Scholar]
- 44. Ueno H., Oishi K., Sayato Y., Nakamuro K. 2000. Oxidative cell damage in Kat-sod assay of oxyhalides as inorganic disinfection by-products and their occurrence by ozonation. Arch. Environ. Contam. Toxicol. 38:1–6 [DOI] [PubMed] [Google Scholar]
- 45. van Aken B., Schnoor J. L. 2002. Evidence of perchlorate (ClO4−) reduction in plant tissues (poplar tree) using radio-labeled 36ClO4−. Environ. Sci. Technol. 36:2783–2788 [DOI] [PubMed] [Google Scholar]
- 46. van Ginkel C. G., Rikken G. B., Kroon A. G., Kengen S. W. 1996. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166:321–326 [DOI] [PubMed] [Google Scholar]
- 47. van Wijk D. J., Kroon S. G., Garttener-Arends I. C. 1998. Toxicity of chlorate and chlorite to selected species of algae, bacteria, and fungi. Ecotoxicol. Environ. Saf. 40:206–211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.