Abstract
The collagenase gene was cloned from Grimontia (Vibrio) hollisae 1706B, and its complete nucleotide sequence was determined. Nucleotide sequencing showed that the open reading frame was 2,301 bp in length and encoded an 84-kDa protein of 767 amino acid residues. The deduced amino acid sequence contains a putative signal sequence and a zinc metalloprotease consensus sequence, the HEXXH motif. G. hollisae collagenase showed 60 and 59% amino acid sequence identities to Vibrio parahaemolyticus and Vibrio alginolyticus collagenase, respectively. In contrast, this enzyme showed <20% sequence identity with Clostridium histolyticum collagenase. When the recombinant mature collagenase, which consisted of 680 amino acids with a calculated molecular mass of 74 kDa, was produced by the Brevibacillus expression system, a major gelatinolytic protein band of ∼60 kDa was determined by zymographic analysis. This result suggested that cloned collagenase might undergo processing after secretion. Moreover, the purified recombinant enzyme was shown to possess a specific activity of 5,314 U/mg, an ∼4-fold greater activity than that of C. histolyticum collagenase.
INTRODUCTION
Bacterial collagenases are metalloproteases containing a consensus motif for zinc proteases, the HEXXH sequence and are capable of digesting both native and denatured collagen. They make multiple cleavages at the Y-Gly bond in repeating X-Y-Gly sequences within triple helical regions, where proline and hydroxyproline residues are most common in the X and Y positions, respectively (17). Because of their characteristics, bacterial collagenases have been widely used in biological experiment as tissue-dispersing enzymes, as well as in medical procedures such as the isolation of pancreatic islet cells for transplantation (14) and the treatment for Dupuytren's disease (6).
Much of our knowledge of bacterial collagenases has come from studies of the enzymes produced by Clostridium histolyticum (13, 15–17, 34). Analysis of the primary structure of the gene product from C. histolyticum has revealed that clostridial collagenases consist of three domains (catalytic domain, polycystic kidney disease [PKD] domain, and collagen-binding domain [CBD]) in their molecules. Moreover, CBD has utilized for anchoring molecule that growth factors fused to CBD can be functional to bind to collagen fibrils and maintain biological activities (21). On the other hand, one of the other well-investigated bacterial collagenases is Vibrio alginolyticus collagenase (7, 10, 11, 28). The collagenase activity of V. alginolyticus collagenase is higher than that of any other bacterial collagenase, and it was found highly efficient in debridement of necrotic burns, ulcers and decubitus. To date, bacterial collagenases have been purified from various species, and their genes have been cloned and sequenced (8, 12, 18, 24, 35). However, many collagenases have not yet been both enzymatically and structurally characterized.
Vibrio hollisae is a Gram-negative bacterium first described in 1982 (4) and recently reclassified as the novel genus Grimontia (29). Grimontia (Vibrio) hollisae has been reported as a toxic bacterium, whose toxin was clarified as thermostable direct hemolysin (22), and is primarily known to cause moderate to severe cases of gastroenteritis in healthy people (5). G. hollisae strain 1706B was isolated from seashore sand collected from the Shin-Kiba coast in Tokyo (27). This organism produces a collagenase with a very high specific activity in the presence of gelatin, and this enzyme even degrades the tanned leather (26). The characteristics of this organism and purified collagenase have been described in a series of papers (25–27). The properties of this collagenase are as follows: (i) it has a molecular mass of 60 kDa; (ii) it degrades insoluble collagen, soluble collagen, Z-GPLGP peptide, and Pz-PLGPR peptide, but not casein; (iii) it has an optimum pH of 7.0 to 8.0 for insoluble collagen hydrolysis; and (iv) it is stable in the range between pH 4.5 and 11 (25). In order to clarify its enzymatic characteristics and to utilize it for biological applications, the primary structure of the collagenase needs to be elucidate.
In the present study, we cloned and sequenced a novel collagenase gene from G. hollisae 1706B to elucidate its primary structure and demonstrated the expression and characterization of recombinant mature collagenase using the Brevibacillus expression system. Moreover, we discussed the characteristics of the corresponding amino acid sequence of this enzyme and its similarity to those of other bacterial collagenases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
G. hollisae strain 1706B was obtained around the shore of Shin-Kiba, Tokyo, Japan, and used throughout the present study (27). The plasmid pGEM-T Easy and the Escherichia coli competent cells JM109 (Promega, Madison, WI) were used as subcloning vector and host, respectively. The expression plasmid vector pCC1BAC and the host TransforMaxEPI300 E. coli (Epicentre Biotechnologies, Madison, WI) were used to make a bacterial artificial chromosome (BAC) clone library of G. hollisae. Bacillus brevis expression vector pNY326 and Brevibacillus choshinensis S5 (Takara Bio, Shiga, Japan) were used for expression of recombinant proteins.
Construction of a genomic library from G. hollisae 1706B.
The genomic DNA of G. hollisae 1706B was purified with a QIA genomic DNA extraction kit (Qiagen, Hilden, Germany). The purified DNA was digested with the restriction enzyme EcoRI and separated on a 0.6% (wt/vol) agarose gel. Only DNA fragments larger than 2 kb were ligated into the E. coli expression vector, pCC1BAC (Epicentre Biotechnologies). Then, pCC1BAC was transformed into TransforMaxEPI300 E. coli by electroporation, and the transformants were plated onto LB agar plates containing 50 μg of ampicillin/ml.
DNA probe preparation.
Degenerate primers were designed based on the internal peptide sequence of original collagenase (see Fig. 2, box 6) for F1 and the consensus sequence of catalytic site from V. alginolyticus and V. parahaemolyticus collagenase (see Fig. 2, underlined) for R1. The primer sets F1 (5′-GAGGCNATCTTTAGCTCCAATCATATGTAYAAY-3′) and R1 (5′-ATCTAAGTAATGCACGTATTCATGYTCNAGRTT-3′) were used for amplification of probe sequence. Y, R, and N represent C/T, A/G, and A/C/T/G, respectively. Amplification was performed in 50 cycles of 0.5 min at 95°C, 0.5 min at 45°C, and 1 min at 72°C. The amplified 1,080-bp PCR product was electro-eluted from 1% agarose gel, ligated into pGEM-T Easy vector (Promega), and then transformed into E. coli strain JM109. The purified plasmid was used as a template to create digoxigenin (DIG)-labeled DNA probes using DIG-high prime labeling reagent (Roche, Basel, Switzerland) according to the manufacturer's instructions.
Fig. 2.
DNA sequence and deduced amino acid sequence of Grimontia hollisae collagenase gene. The N-terminal amino acid sequences of the 74- and 60-kDa collagenases are indicated by box 1. Numbered boxes indicate biochemically identified peptides as follows: boxes 2, 3, and 4, lysyl endoprotease-digested fragments; box 5, trypsin-digested fragment; boxes 6 and 7, V8 protease-digested fragments. The zinc metalloprotease HEXXH consensus motif is underlined. The SD site is indicated by dotted line. The putative transcriptional terminator sequence is indicated by arrows.
Screening of the genomic library with DNA probe.
The genomic library was screened by colony hybridization according to the manufacturer's instructions (Roche). Briefly, the ampicillin-resistant transformants were blotted onto a nylon membrane (Roche) and lysed with 0.5 M NaOH. The denatured DNA was then immobilized, followed by protease K treatment. The DIG-labeled DNA probe was used for hybridization. Positive clones were picked from cultured LB agar plates and subcultured in LB liquid medium with ampicillin at 37°C. BAC DNA of collagenase-positive colonies was prepared by using the QIA genomic DNA extraction kit (Qiagen).
DNA sequencing and alignment of deduced amino acid sequence.
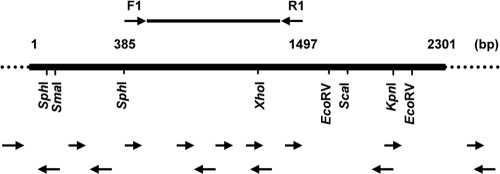
Purified BAC DNA was amplified with cycle sequencing using a thermal cycler (Takara Bio) and sequenced with a DNA autosequencer (ABI Prism 310). Appropriate oligonucleotide primers (Sigma-Aldrich, St. Louis, MO) and Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA) were used to walk along the sequence. The sequencing strategy for the pCC1BAC-2 insert was outlined in Fig. 1. Computer analysis of the DNA sequence data was performed using GenBank database and BLAST search programs. The deduced amino acid sequence alignment and homology data were generated by using the CLUSTAL W2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Fig. 1.
Sequencing strategy for the collagenase gene inserted into pCC1BAC-2. The thick line represents the collagenase gene inserted into pCC1BAC-2 plasmid. The thin line indicates probe region used for cloning. The lower arrows indicate the direction of sequence determinations, starting from specific primers.
Recombinant collagenase preparation.
pCC1BAC-2 was used as the DNA template. To add an NcoI site to the 5′ region and a HindIII site to the 3′ region of the mature collagenase gene, primers were designed as follows: forward, 5′-AAACCATGGCTTTCGCTGCGGTTGAACAGTGTGATCT-3′; and reverse, 5′-AAAAAGCTTTTACTGACGACACTGGTTAC-3′ (the restriction sites are underlined). The mature domain of 2.1-kb collagenase gene was amplified by using the Expand High Fidelity PCR system (Roche). After treatment with NcoI and HindIII, the double-digested fragment was ligated into the multiple cloning site of the Brevibacillus expression vector pNY326, which was located downstream of Brevibacillus signal sequence (pNY326-Col2). Plasmid pNY326-Col2, harboring the complete mature collagenase gene, was transformed into Brevibacillus choshinensis S5 to express the recombinant enzyme. The Brevibacillus transformant was aerobically cultured in 2SLN medium containing neomycin (50 μg/ml). After centrifugation, the supernatant was purified with a DEAE-Sepharose column (26 by 100 mm) with a fast-protein liquid chromatography (FPLC) system under a gradation of sodium concentrations (0.2 to 1.1 M NaCl). The column was eluted isocratically with 0.2 M NaCl–50 mM Tris-HCl (pH 7.5) for 10 min at 5 ml/min, followed by a linear gradient to 1.1 M NaCl for 50 min. The purified recombinant protein was concentrated by ultrafiltration with a 30-kDa cutoff (Pall, Port Washington, NY) and dialyzed against 50 mM Tris-HCl buffer (pH 7.5) at 4°C.
SDS-PAGE.
SDS-PAGE was carried out on 7.5 or 10% polyacrylamide gel according to the method of Laemmli (9) unless otherwise stated. After electrophoresis, the gel was stained with 0.25% Coomassie brilliant blue R-250 in 50% methanol and 10% acetic acid and then destained with 5% methanol and 7.5% acetic acid.
Determination of collagenolytic activity.
The collagenolytic activity of recombinant collagenase was measured by using fluorescein isothiocyanate (FITC)-labeled type I collagen as previously described (20). Briefly, the enzyme solution was mixed with 50 mM Tris-HCl (pH 7.5) containing 0.05% FITC-labeled type I collagen, 5 mM CaCl2, and 200 mM NaCl and incubated at 30°C for 30 min. After adding EDTA to stop the enzymatic reaction, the degraded FITC-labeled collagen fragment was extracted with 50 mM Tris-HCl (pH 9.5) containing 70% ethanol. The fluorescence intensity of the supernatant was measured by fluorescence spectrophotometry (530 nm [emission], 485 nm [excitation]). One unit of collagenolytic activity was defined as the amount degrading 1 μg of FITC-labeled collagen at 30°C per min. Protein concentrations were determined by using the Coomassie Plus–The Better Bradford Assay Reagent (Thermo Scientific, Rockford, IL). Collagenase from Clostridium histolyticum (Amano Enzyme, Nagoya, Japan) was used as a reaction standard. All assays were carried out in triplicate.
For the measurement of enzyme kinetics, 0.5 μg of enzyme was incubated with various amounts of FITC-labeled type I collagen (10 to 50 μg) at 30°C for 5 min, and the fluorescence intensity of the supernatant was measured. Specific collagenase substrate FALGPA [N-(3-[2-furyl]acryloyl)-Leu-Gly-Pro-Ala; Bachem AG, Bubendorf, Switzerland] was also chosen to determine the enzyme activity. An assay with FALGPA was performed according to the modified method of a previous report (31). Briefly, the enzyme and FALGPA were mixed in 50 mM Tricine buffer (pH 7.5) containing 0.4 M NaCl and 40 mM CaCl2 and incubated at 30°C for 5 min. After incubation, the absorbance change at 345 nm was measured by using a Corona SH-9000 microplate reader (Corona Electric, Ibaraki, Japan). The FALGPA concentrations were varied from 0.5 to 3.0 mM. In the FALGPA assay, one unit of activity was defined as the amount degrading 1 μmol of FALGPA peptide at 30°C per min. The Vmax and Km values for hydrolysis of native collagen and FALGPA were estimated from the Lineweaver-Burk plot by using the reaction rates at different substrate concentrations.
Real-time zymography.
Real-time zymography was performed as previously described (3). Briefly, recombinant collagenase was subjected to nonreducing SDS-PAGE using a 10% gel containing 0.05% FITC-labeled gelatin. After electrophoresis at 4°C, the gel was washed in 50 mM Tris-HCl (pH 7.5) containing 2.5% TritonX-100 for 30 min and then incubated in 50 mM Tris-HCl (pH 7.5) containing 5 mM CaCl2 and 200 mM NaCl at 37°C for 5 h. The collagenase-digested FITC-labeled gelatin was visualized by using a transilluminator.
The effect of protease inhibitors on collagenase activity was determined by adding the inhibitors to the incubation buffer. In inhibition studies, protease inhibitors such as EDTA, o-phenanthroline, N-ethylmaleimide (NEM), or phenylmethylsulfonyl fluoride (PMSF) were used at final concentrations of 20, 2.0, 5.0, and 1.0 mM, respectively.
Amino acid sequence.
Amino acid sequence analysis of N-terminal or internal peptide fragments of original collagenase was performed as described previously (19). Briefly, internal sequences were determined by lysyl endoprotease, trypsin, and V8 protease digestion. The enzyme-cleaved fragments and purified collagenase were separated by SDS–10% PAGE and electrophoretically transferred to Immobilon-P (Millipore, Billerica, MA). The membrane was stained with Coomassie brilliant blue R-250, and the protein band was excised from the membrane and then washed extensively with deionized distilled water. The N-terminal sequence was analyzed by using a Procise 491 protein sequencer (Applied Biosystems, Carlsbad, CA). The detected fragments are shown in Fig. 2.
Nucleotide sequence accession number.
The determined nucleotide sequence was deposited in the DDBJ database under accession number AB600550.
RESULTS
Cloning of collagenase gene from G. hollisae 1706B.
In order to amplify a fragment of the G. hollisae collagenase coding gene, degenerate primers were designed. A primer set yielded a single amplification product, and nucleotide sequencing revealed that the amino acid sequence deduced from this PCR product contained four partial peptide sequences (Fig. 2, boxes 2, 3, 4, and 7). Therefore, the plasmid containing this PCR product was used as a template to create DIG-labeled DNA probes as a hybridization probe for genomic library screening.
A partial genomic library was constructed with EcoRI-digested genomic DNA fragments from G. hollisae using a pCC1BAC vector system. The plasmids were transformed into TransforMaxEPI300 E. coli using electroporation, yielding about 5,000 colonies on LB-ampicillin plates. By using the colony hybridization technique, 30 positive clones were picked up from the library, and all of the purified BAC contained 2.3-kb inserts encoding the collagenase gene. One of the former clones, designated pCC1BAC-2, was chosen for further study. Restriction analysis revealed that pCC1BAC-2 contained a >50-kb EcoRI insert (data not shown).
Nucleotide sequence of G. hollisae collagenase gene.
The nucleotide sequence of the pCC1BAC-2 insert was sequenced using the strategy outlined in Fig. 1. The obtained sequences were aligned by their overlaps to form a single contiguous sequence, and the 2.3-kb sequence of G. hollisae collagenase was determined (Fig. 2). The entire open reading frame (ORF) of G. hollisae collagenase was sequenced on both strands. Analysis of the sequence revealed a complete ORF extending from an ATG codon at nucleotide +1 to a TAA stop codon at position 2301, which encodes a protein of 767 amino acids (aa). A Shine-Dalgarno sequence (AGAAGAA) is observed 5 to 11 bp upstream from the ATG codon. A stem-loop sequence (nucleotide positions 2328 to 2347) with a short run of T's is present downstream of the termination codon.
The N-terminal amino acid sequence (Ala-Val-Glu-Gln-Cys-Asp-Lys-Ser-Gln) of the purified original enzyme corresponded perfectly to the deduced amino acid sequences of our determined sequence in position 262 to 288 downstream of the ATG codon. Moreover, the deduced amino acid sequence of ORF includes a zinc metalloprotease HEXXH consensus motif, which was detected as HEYVH in positions 1474 to 1488. It is known that the amino acid sequence HEXXH is important in facilitating the electron transfer with zinc in enzyme catalysis (30). Moreover, the internal peptide sequences were found to agree completely with the protein sequence deduced from DNA sequencing.
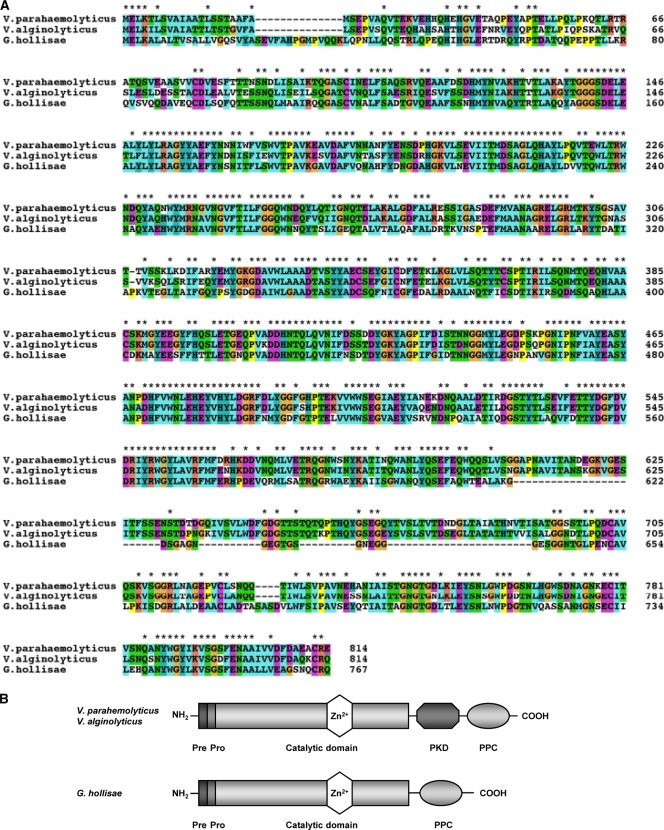
We next compared the deduced primary sequence of G. hollisae collagenase gene with other known protein sequences using the BLASTP programs of the National Center for Biotechnology Information. The predicted amino acid sequence of G. hollisae collagenase showed 59 and 60% identities with collagenase from V. alginolyticus (28) and V. parahaemolyticus (8), respectively (Fig. 3 A). Moreover, the alignment predicted that G. hollisae collagenase consists of a pre-pro region (aa 1 to 87), a catalytic domain (aa 88 to 615), and a bacterial pre-peptidase C-terminal (PPC) domain (aa 688 to 749) (33) (Fig. 3B). Furthermore, a database search revealed that G. hollisae collagenase showed <35% similarity to any other reported Vibrio metalloproteases (data not shown). On the other hand, G. hollisae collagenase showed 12 and 11% identities to ColG and ColH, respectively, from Clostridium histolyticum (data not shown).
Fig. 3.
Amino acid sequence comparison of Grimontia hollisae collagenase with homologous collagenase. (A) The amino acid sequences from G. hollisae (the present study), Vibrio parahaemolyticus (NP_797719), and Vibrio alginolyticus (CAA44501) were aligned by using the CLUSTAL W2 program. Identical residues among the three sequences are indicated by asterisks. (B) Schematic representation of the domain architecture of G. hollisae (the present study), V. parahaemolyticus (NP_797719) and V. alginolyticus (CAA44501). Pre, signal peptide; pro, putative pro-domain; PKD, polycystic kidney disease-like domain; PPC, pre-peptidase C-terminal domain.
Expression and characterization of recombinant collagenase in Brevibacillus.
To examine whether the product of the G. hollisae collagenase gene possesses similar proteolytic activity as the original enzyme, we first made a construct for expressing the recombinant mature collagenase (aa 88 to 767) and produced the recombinant enzyme using the Brevibacillus expression system. When the collagenolytic activity of the culture supernatant of transformants (carrying pNY326-Col2) was measured using FITC-labeled collagen, the activity was found to be higher than that of mock transformants (data not shown). This result indicated that recombinant collagenase was successively produced.
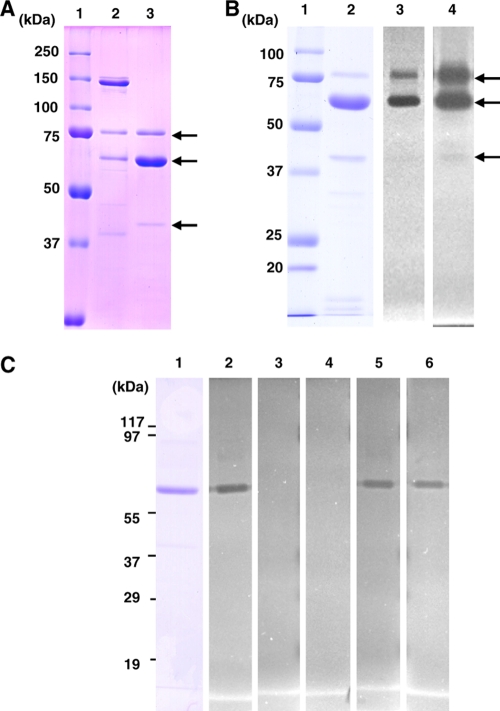
To characterize certain biochemical properties, recombinant G. hollisae collagenase was purified from a Brevibacillus culture medium using a DEAE-Sepharose column with an FPLC system. The expression typically yielded 0.2 g of pure collagenase from 1 liter of culture. When the Brevibacillus culture medium and purified recombinant enzyme were confirmed by SDS-PAGE, three bands (74, 60, and 40 kDa) derived from this enzyme were detected (Fig. 4 A, lane 3). The deduced molecular mass of mature protein, without the possible pre-pro peptide of 87 aa, was 74 kDa, and matched with the molecular mass determined by SDS-PAGE. The major 60-kDa form was the same molecular mass as the purified form from the original bacterial collagenase. Zymography showed that all three forms of collagenase possessed gelatinase activity; however, the activity of the 40-kDa form was weaker than that of the other forms (Fig. 4B). Moreover, the gelatinolytic band disappeared in the presence of the metal ion chelators, EDTA and o-phenanthroline, but not cysteine and serine protease inhibitors, such as NEM and PMSF (Fig. 4C). We also found that the three forms of collagenase have the same N-terminal amino acid sequence (data not shown), suggesting that the C-terminal region of the mature enzyme would be autodegraded and then become 60- and 40-kDa enzymes. Furthermore, the 60-kDa enzyme seems to be the most stable form. The purified recombinant enzyme could digest insoluble and soluble type I collagen, and a collagenolytic activity assay showed a specific activity of 5,841 U/mg using FITC-collagen, which was elevated by ∼5.8-fold compared to the culture medium (data not shown). In contrast, this recombinant enzyme could not degrade casein (data not shown).
Fig. 4.
Analysis of recombinant collagenase purified from Brevibacillus culture media. (A) Purified recombinant collagenase was analyzed by SDS-PAGE using a reducing 7.5% gel. Lane 1, molecular weight marker; lane 2, culture medium from Brevibacillus (carrying pNY326-Col2); lane 3, purified collagenase. (B) Real-time gelatin zymography using a nonreducing 10% gel. Lane 1, molecular weight marker; lane 2, SDS-PAGE of purified collagenase; lanes 3 and 4, gelatin zymogram of purified collagenase after 2 h (lane 3) or 19 h (lane 4) of incubation. (C) Inhibition assay using real-time gelatin zymography. Lane 1, SDS-PAGE of purified collagenase; lanes 2 to 6, gelatin zymogram of purified collagenase in the absence (lane 2) or presence of the inhibitors EDTA (lane 3), o-phenanthroline (lane 4), NEM (lane 5), and PMSF (lane 6).
Kinetic parameters for the hydrolysis of type I collagen by collagenase.
The kinetic parameters of collagenase were determined using native type I collagen and the synthetic peptide substrate, FALGPA (Table 1). G. hollisae collagenase showed a 4.2-fold lower Km value and a slightly higher Vmax value against FITC-collagen than C. histolyticum collagenase, resulting in a higher specific constant (∼6.7-fold higher). On the other hand, the two enzymes have comparable substrate affinity to FALGPA, whereas the Vmax values increased 25-fold in G. hollisae compared to C. histolyticum collagenase. As a result, G. hollisae collagenase showed a high specific constant also against FALGPA (∼24-fold higher). These results help to explain the specific activity of the two enzymes. Previously, V. parahaemolyticus collagenase has been reported to have a Km value of 1.06 mM toward FALGPA, at pH 8.0 and 25°C (35), and this Km value was comparable to that of the present collagenase. Based on the specific constant, G. hollisae collagenase had a higher specificity for type I collagen and FALGPA than did C. histolyticum collagenase.
Table 1.
Kinetic constants of Grimontia hollisae and Clostridium histolyticum collagenasesa
| Substrate | Enzyme strain | Sp act (U/mg) | Mean ± SD |
||
|---|---|---|---|---|---|
| Km (mM) | Vmax (mM/min) | Specific constant (Vmax/Km) | |||
| FITC-collagen | G. hollisae | 5,314 | (2.83 ± 0.76) × 10−3* | 8,889 ± 924† | (3.18 ± 0.27) × 106* |
| C. histolyticum | 1,289 | (11.8 ± 1.95) × 10−3 | 5,556 ± 962 | (0.48 ± 0.04) × 106 | |
| FALGPA | G. hollisae | 7.40 | 2.41 ± 0.19 | 33.2 ± 1.11* | 13.8 ± 0.81* |
| C. histolyticum | 0.39 | 2.40 ± 0.52 | 1.35 ± 0.12 | 0.57 ± 0.07 | |
The activities of G. hollisae and C. hystolyticum collagenase were determined by using FITC-labeled collagen (FITC-collagen) or the synthetic peptide substrate FALGPA. Assays were carried out in 50 mM Tris-HCl–0.3 M NaCl–10 mM CaCl2 (pH 7.5) at 30°C for the FITC-labeled collagen or 50 mM Tricine–0.4 M NaCl–40 mM CaCl2 (pH 7.5) at 30°C for FALGPA. Each collagenase was used at the amount of 0.5 μg for FITC-collagen. When used for FALGPA, the amounts of G. hollisae and C. hystolyticum collagenase were 1.0 and 20 μg, respectively. The data represent the means three separate experiments. *, P < 0.01; †, P < 0.05.
DISCUSSION
In the present study, we isolated the G. hollisae collagenase gene by a cloning and sequencing method. The isolated gene consisted of 2,301 nucleotides, and the 767-amino-acid protein deduced from the ORF revealed high homology to several Vibrio collagenases. Moreover, we succeeded in the effective expression of its recombinant enzyme in Brevibacillus. It hydrolyzed type I collagen, gelatin, and FALGPA peptide more efficiently than C. histolyticum collagenase, and the inhibition study showed that the enzyme was inhibited by metal ion chelators, such as EDTA, indicating that the cloned collagenase was a metalloprotease.
A BLASTP search indicated that the deduced amino acid sequences of G. hollisae collagenase show extensive homology with V. alginolyticus and V. parahaemolyticus metalloproteases. On the other hand, G. hollisae collagenase shared <20% identity with ColG and ColH from C. histolyticum and ColA from C. perfringens (data not shown). This result indicates that G. hollisae collagenase can be classified into the M9A subfamily in the MEROPS database (23). In the previous study, Vibrio metalloproteases were classified into three subgroups (class I, II, and III) based on the HEXXH sequence and substrate specificity (8). According to this classification, G. hollisae collagenase belongs to the class III group that includes V. alginolyticus and V. parahaemolyticus. However, G. hollisae collagenase has little caseinase activity (data not shown), while V. alginolyticus and V. parahaemolyticus collagenase reportedly possess apparent caseinase activity. One of the possible reasons for this difference in substrate specificity may be the difference in domain structures among these enzymes. Given that the PKD domain was absent from G. hollisae collagenase (Fig. 3), this domain might participate in caseinase activity when bound to substrate. These results indicate that G. hollisae collagenase should be classified into a new group among the Vibrio collagenases and suggest that the three enzymes, which differ in function and origin, are evolutionary related.
Surprisingly, we found that G. hollisae collagenase possessed markedly greater activity compared to C. histolyticum collagenase (Table 1). Since G. hollisae collagenase showed high affinity to native collagen and high catalytic activity to FALGPA compared to C. histolyticum collagenase, these results suggested that the degradation mechanism of the two collagenases appear to be different against collagen or gelatin. However, the C. histolyticum collagenase used in the present study should be considered a commercially available enzyme, which is a purified native protein. Since we found that CBD and PKD domains were absent from this purchased enzyme by SDS-PAGE (data not shown), this result raises the possibility that the loss of CBD and PKD domains may lead to a decrease in activity of this purchased enzyme compared to the intact form.
Analysis of kinetic parameters led to our considerable interest in why and how G. hollisae collagenase degrades collagen effectively. Because collagen is highly resistant to most proteases, collagenase seems to possess an effective degradation mechanism against collagen. For example, a recent study indicated that mammalian collagenases locally unwind the triple-helical structure through the coordinated action of the catalytic domain and collagen-binding domain (called the hemopexin domain) and then hydrolyze the peptide bonds (1). Moreover, the PKD domain of deseasin MCP-01, which is a bacterial collagenolytic serine protease, is reported to bind collagen and to swell collagen fascicles, suggesting that the PKD domain may improve the collagenolytic efficiency of the catalytic domain (32, 36). However, G. hollisae collagenase possesses neither a CBD nor a PKD domain. Since CBD has been reported to be necessary for the collagenolytic activity of mammalian and bacterial collagenase (2, 16), G. hollisae collagenase may contain an unidentified CBD in the 60-kDa form and/or possess a novel degradation mechanism against collagen. The domain structure-function relationship remains to be clarified in order to elucidate the mechanism of action of this enzyme.
It is noteworthy that the recombinant collagenase of G. hollisae was produced with stable activity using the Brevibacillus expression system. Since Brevibacillus is a Gram-positive bacterium, this system leads to the expression of recombinant proteins with low endotoxin contamination, which has been known to enhance the immunological response of higher animals. In addition, this recombinant enzyme can be used for dispersion of human fibroblasts in collagen gel and appears to have no obvious cytotoxicity (data not shown). Therefore, it can be utilized for biological applications, specifically for medical applications.
In conclusion, we cloned a novel collagenase gene from G. hollisae 1706B and produced a high yield of recombinant enzyme by using the Brevibacillus expression system. Moreover, we provided evidence that this enzyme showed higher collagenolytic activity than C. histolyticum collagenase, indicating that G. hollisae collagenase is suitable for both basic and applied research.
ACKNOWLEDGMENTS
We thank Kiyoko Ogawa-Goto and Tomonori Ueno for their technical advice and support. We also thank Osamu Matsushita and Takehiko Mima (Department of Microbiology, Kitasato University School of Medicine) for their stimulating discussions.
Footnotes
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Chung L., et al. 2004. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 23:3020–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clark I. M., Cawston T. E. 1989. Fragments of human fibroblast collagenase: purification and characterization. Biochem. J. 263:201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hattori S., Fujisaki H., Kiriyama T., Yokoyama T., Irie S. 2002. Real-time zymography and reverse zymography: a method for detecting activities of matrix metalloproteinases and their inhibitors using FITC-labeled collagen and casein as substrates. Anal. Biochem. 301:27–34 [DOI] [PubMed] [Google Scholar]
- 4. Hickman F. W., et al. 1982. Identification of Vibrio hollisae sp. nov. from patients with diarrhea. J. Clin. Microbiol. 15:395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hinestrosa F., Madeira R. G., Bourbeau P. P. 2007. Severe gastroenteritis and hypovolemic shock caused by Grimontia (Vibrio) hollisae infection. J. Clin. Microbiol. 45:3462–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hurst L. C., et al. 2009. Injectable collagenase clostridium histolyticum for Dupuytren's contracture. N. Engl. J. Med. 361:968–979 [DOI] [PubMed] [Google Scholar]
- 7. Keil B. 1992. Vibrio alginolyticus (“Achromobacter”) collagenase: biosynthesis, function, and application. Matrix Suppl. 1:127–133 [PubMed] [Google Scholar]
- 8. Kim S. K., Yang J. Y., Cha J. 2002. Cloning and sequence analysis of a novel metalloprotease gene from Vibrio parahaemolyticus 04. Gene 283:277–286 [DOI] [PubMed] [Google Scholar]
- 9. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 10. Lecroisey A., Keil B. 1979. Differences in the degradation of native collagen by two microbial collagenases. Biochem. J. 179:53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lecroisey A., Keil-Dlouha V., Woods D. R., Perrin D., Keil B. 1975. Purification, stability, and inhibition of the collagenase from Achromobacter iophagus. FEBS Lett. 59:167–172 [DOI] [PubMed] [Google Scholar]
- 12. Lee J. H., et al. 1998. Isolation and sequence analysis of metalloprotease gene from Vibrio mimicus. Biochim. Biophys. Acta 1384:1–6 [DOI] [PubMed] [Google Scholar]
- 13. Maclennan J. D., Mandl I., Howes E. L. 1953. Bacterial digestion of collagen. J. Clin. Invest. 32:1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsumoto S., et al. 2005. Insulin independence after living-donor distal pancreatectomy and islet allotransplantation. Lancet 365:1642–1644 [DOI] [PubMed] [Google Scholar]
- 15. Matsushita O., et al. 1999. Gene duplication and multiplicity of collagenases in Clostridium histolyticum. J. Bacteriol. 181:923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsushita O., et al. 1998. A study of the collagen-binding domain of a 116-kDa Clostridium histolyticum collagenase. J. Biol. Chem. 273:3643–3648 [DOI] [PubMed] [Google Scholar]
- 17. Matsushita O., Koide T., Kobayashi R., Nagata K., Okabe A. 2001. Substrate recognition by the collagen-binding domain of Clostridium histolyticum class I collagenase. J. Biol. Chem. 276:8761–8770 [DOI] [PubMed] [Google Scholar]
- 18. Matsushita O., Yoshihara K., Katayama S., Minami J., Okabe A. 1994. Purification and characterization of Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J. Bacteriol. 176:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miura-Yokota Y., et al. 2005. Cloning and nucleotide sequence of a novel 28-kDa protein from the mantle muscle of the squid Todarodes pacificus with homology to tropomyosin. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 141:3–12 [DOI] [PubMed] [Google Scholar]
- 20. Nagai Y., et al. 1984. A micro-assay method for collagenase activity and its application in the study of collagen metabolism in pathological tissues. Japan J. Inflamm. 4:121–130 [Google Scholar]
- 21. Nishi N., et al. 1998. Collagen-binding growth factors: production and characterization of functional fusion proteins having a collagen-binding domain. Proc. Natl. Acad. Sci. U. S. A. 95:7018–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishibuchi M., Janda J. M., Ezaki T. 1996. The thermostable direct hemolysin gene (tdh) of Vibrio hollisae is dissimilar in prevalence to and phylogenetically distant from the tdh genes of other vibrios: implications in the horizontal transfer of the tdh gene. Microbiol. Immunol. 40:59–65 [DOI] [PubMed] [Google Scholar]
- 23. Rawlings N. D., Barrett A. J., Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res. 38:D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakurai Y., et al. 2009. Purification and characterization of a major collagenase from Streptomyces parvulus. Biosci. Biotechnol. Biochem. 73:21–28 [DOI] [PubMed] [Google Scholar]
- 25. Suzuki K. 2000. Purification and properties of collagenase from Vibrio hollisae 1706B strain. Hikakukagaku 45:272–283 [Google Scholar]
- 26. Suzuki K. 2002. Studies on practical application of vibrio hollisae collagenase. Hikakukagaku 48:209–213 [Google Scholar]
- 27. Suzuki K., Matsubara Y. 1998. Determination of aerobic collagenolytic bacterium isolated from seashore sand. Hikakukagaku 44:64–71 [Google Scholar]
- 28. Takeuchi H., et al. 1992. Structural gene and complete amino acid sequence of Vibrio alginolyticus collagenase. Biochem. J. 281(Pt. 3):703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson F. L., Hoste B., Vandemeulebroecke K., Swings J. 2003. Reclassification of Vibrio hollisae as Grimontia hollisae gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 53:1615–1617 [DOI] [PubMed] [Google Scholar]
- 30. Vallee B. L., Auld D. S. 1990. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29:5647–5659 [DOI] [PubMed] [Google Scholar]
- 31. Van Wart H. E., Steinbrink D. R. 1981. A continuous spectrophotometric assay for Clostridium histolyticum collagenase. Anal. Biochem. 113:356–365 [DOI] [PubMed] [Google Scholar]
- 32. Wang Y. K., et al. 2010. Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. strain SM9913: binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix. J. Biol. Chem. 285:14285–14291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeats C., Bentley S., Bateman A. 2003. New knowledge from old: in silico discovery of novel protein domains in Streptomyces coelicolor. BMC Microbiol. 3:3–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoshihara K., Matsushita O., Minami J., Okabe A. 1994. Cloning and nucleotide sequence analysis of the colH gene from Clostridium histolyticum encoding a collagenase and a gelatinase. J. Bacteriol. 176:6489–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu M. S., Lee C. Y. 1999. Expression and characterization of the prtV gene encoding a collagenase from Vibrio parahaemolyticus in Escherichia coli. Microbiology 145(Pt. 1):143–150 [DOI] [PubMed] [Google Scholar]
- 36. Zhao G. Y., et al. 2008. Hydrolysis of insoluble collagen by deseasin MCP-01 from deep-sea Pseudoalteromonas sp. strain SM9913: collagenolytic characters, collagen-binding ability of C-terminal polycystic kidney disease domain, and implication for its novel role in deep-sea sedimentary particulate organic nitrogen degradation. J. Biol. Chem. 283:36100–36107 [DOI] [PMC free article] [PubMed] [Google Scholar]