Abstract
Water-forming NADH oxidases (encoded by noxE, nox2, or nox) are flavoproteins generally implicated in the aerobic survival of microaerophilic bacteria, such as lactic acid bacteria. However, some natural Lactococcus lactis strains produce an inactive NoxE. We examined the role of NoxE in the oxygen tolerance of L. lactis in the rich synthetic medium GM17. Inactivation of noxE suppressed 95% of NADH oxidase activity but only slightly affected aerobic growth, oxidative stress resistance, and NAD regeneration. However, noxE inactivation strongly impaired oxygen consumption and mixed-acid fermentation. We found that the A303T mutation is responsible for the loss of activity of a naturally occurring variant of NoxE. Replacement of A303 with T or G or of G307 with S or A by site-directed mutagenesis led to NoxE aggregation and the total loss of activity. We demonstrated that L299 is involved in NoxE activity, probably contributing to positioning flavin adenine dinucleotide (FAD) in the active site. These residues are part of the strongly conserved sequence LA(T)XXAXXXG included in an alpha helix that is present in other flavoprotein disulfide reductase (FDR) family flavoproteins that display very similar three-dimensional structures.
INTRODUCTION
Water-forming NADH oxidases (Nox, NoxE, or Nox2) are flavoproteins involved in the aerobic growth and survival of lactic acid bacteria (LAB) under fermentation conditions (7). Their inactivation in several streptococci strongly reduces the aerobic growth. Inactivation of nox2 in Streptococcus pneumoniae and Streptococcus agalactiae reduces growth in aerated media by 80% (37, 38). The growth defect of the S. agalactiae mutant has been attributed to a defect in fatty acid production resulting from nonproduction of the fatty acid precursor acetyl-coenzyme A (CoA) due to strictly homolactic fermentation (37). Nox2 inactivation in Streptococcus mutans also leads to strict homolactic fermentation on glucose; however, it does not affect growth on glucose but greatly reduces growth on mannitol (11). In Streptococcus pyogenes, Nox2 inactivation completely abolishes aerobic growth under carbon limitation conditions and increases sensitivity to the superoxide-generating agent paraquat (methyl viologen). This growth defect has been attributed to hydrogen peroxide (H2O2) accumulation (10). NoxE is the principal enzyme displaying NADH oxidase (NOX) activity in Lactococcus lactis (95% of the NADH oxidase activity of L. lactis TIL46), a LAB widely used in industrial milk fermentations and generally considered a model LAB. However, NoxE inactivation in L. lactis reduces the growth rate in milk by only 20%, although it strongly impairs oxygen consumption (34). Moreover, we isolated two natural L. lactis strains without detectable NADH oxidase activity from matured cheese (34), suggesting that NoxE is not important for L. lactis survival in dairy media. In one natural NADH oxidase-negative strain, the noxE gene was complete but the enzyme appeared to be inactive. Several water-forming NADH oxidases from LAB, including NoxE of L. lactis (18, 21), have been purified and characterized. They are homodimers with a flavin adenine dinucleotide (FAD) molecule bound to each 50-kDa subunit. NoxE from L. lactis shares at least 33% sequence identity with other NADH oxidases from LAB, and several regions are particularly well conserved, especially the NADH and FAD binding domains and the nonflavin redox center Cys42. The crystal structure of the Lactobacillus sanfranciscensis Nox2 has been described (22). Structural and sequence analyses suggest that the mechanisms of action of the NADH oxidases of L. sanfranciscensis and L. lactis are similar (22). By comparison with Enterococcus faecalis Nox, several amino acid residues, in addition to Cys42, have been implicated in NoxE activity. His10, which stabilizes Cys42, may stabilize the peroxy-flavin intermediate, formed during the enzymatic reaction, through hydrogen bonding (25). Ala300 of chain A and Phe427 of chain B in L. lactis NoxE are predicted to be involved in FAD binding through hydrogen bonding with the cofactor (22). Other important residues form part of the specific motifs of the NADH and FAD binding domains that are conserved in the flavoprotein disulfide reductase (FDR) family, to which NADH oxidases belong (1). This flavoprotein family, represented by the enzyme glutathione reductase, has been thoroughly studied. The FAD binding domain consists mostly of two associated regions, one in the N-terminal region of NADH oxidase and the other near the C terminus (8), although other partially conserved motifs also appear to be involved. In particular, the D(X)6GXXP motif (residues 237 to 247 in NoxE), located at the interface between the FAD and NADH binding domains, may interact with the isoalloxazine ring of FAD (8). Moreover, Wierenga et al. (36) reported the presence of a long α-helix in the two FDR proteins p-hydroxybenzoate hydroxylase (residues 298 to 319) and glutathione reductase (residues 339 to 354): its N terminus points toward the O-2α region of the isoalloxazine of FAD, where the negative charge of the isoalloxazine radical and anion is localized. The helix dipole can stabilize a negative charge around O-2α, so the helix may be important for catalysis. Another helix, corresponding to the C-terminal sequence of human glutathione reductase (residues 436 to 459), is involved in dimerization, essential for enzyme activity (27, 28).
We describe the mutation responsible for the loss of activity of a naturally occurring variant of NoxE and its involvement in an alpha-helix conserved in proteins of the FDR family. Various single mutations in this alpha-helix abolished normal NoxE conformation and activity. We also demonstrate that NoxE is the principal enzyme involved in oxygen consumption but plays a minor role in the aerobic growth and short-term survival of L. lactis in GM17 broth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The laboratory strains and plasmids used during this study are listed in Table 1. NZ9000noxE was constructed by single crossover using exactly the same strategy described for TIL46noxE (34). Wild-type (WT) L. lactis strains were provided by CSK Food Enrichment (Leeuwarden, Netherlands). Escherichia coli strains were grown at 37°C and 200 rpm in Luria-Bertani broth (LB) (Difco, Detroit, MI). When required, erythromycin, kanamycin, or chloramphenicol was added to final concentrations of 150 μg ml−1, 50 μg ml−1, or 50 μg ml−1, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source |
|---|---|---|
| Strains | ||
| L. lactis | ||
| TIL46 | WT strain derived from L. lactis NCDO763 cured of its 2-kb plasmid | National Collection of Food Bacteria (Shinfield, Reading, United Kingdom) |
| TIL46noxE | TIL46 noxE-negative mutant obtained by SCOa integration of pORInoxE; Eryr | 34 |
| TIL46noxE-pJIM::noxE | TIL46 noxE-negative mutant complemented with pJIM::noxE; Eryr Cmr | 34 |
| NZ9000 | MG1363; pepN::nisRK | 19a |
| NZ9000noxE | NZ9000 noxE-negative mutant obtained by SCO integration of pORInoxE; Eryr | This study |
| E. coli | ||
| TG1 repA+ | TG1 derivative with the repA gene integrated into the chromosome, allowing replication of L. lactis plasmids | P. Renault (INRA, Génétique Microbienne, Jouy-en-Josas, France) |
| Rosetta | F−ompT hsdSB (rB− mB−) gal dcm pRARE; Cmr | Novagen |
| Plasmids | ||
| pET28a | 5.4-kb, expression vector for C-terminal 6-His-tag fusion; Kanr | Novagen |
| pET28a::noxE_TIL46 | pET28a with noxE gene from TIL46 + C-terminal 6-His tag under the control of the T7 promoter | This study |
| pET28a::noxE_A144T | pET28a::noxE_TIL46 containing the A144T mutation in noxE | This study |
| pET28a::noxE_A303T | pET28a::noxE_TIL46 containing the A303T mutation in noxE | This study |
| pET28a::noxE_K384N | pET28a::noxE_TIL46 containing the K384N mutation in noxE | This study |
| pET28a::noxE_A303G | pET28a::noxE_TIL46 containing theA303G mutation in noxE | This study |
| pET28a::noxE_N302S | pET28a::noxE_TIL46 containing the N302S mutation in noxE | This study |
| pET28a::noxE_L299T | pET28a::noxE_TIL46 containing the L299T mutation in noxE | This study |
| pET28a::noxE_A300T | pET28a::noxE_TIL46 containing the A300T mutation in noxE | This study |
| pET28a::noxE_G307A | pET28a::noxE_TIL46 containing the G307A mutation in noxE | This study |
| pET28a::noxE_G307S | pET28a::noxE_TIL46 containing the G307S mutation in noxE | This study |
| pET28a::noxE_CSK1382 | pET28a with noxE gene from CSK1382 + C-terminal 6-His tag under the control of the T7 promoter | This study |
| pET28a::noxE_T303A | pET28a::noxE_CSK1382 containing the T303A mutation in noxE | This study |
| pNZ8048 | nisAp + NcoI + MCS + terminator | 19a |
| pNZ8048::noxE_TIL46 | pNZ8048 with noxE gene of TIL46 under nisAp control | This study |
| pNZ8048::noxE_CSK1382 | pNZ8048 with noxE gene of CSK1382 under nisAp control | This study |
| pNZ8048::noxE_6His | pNZ8048 with noxE gene of TIL46 + C-terminal 6-His tag under nisAp control | This study |
| pNZ8048::noxE_A303T_6His | pNZ8048::noxE_6His containing the A303T mutation in noxE | This study |
SCO, single-crossover.
L. lactis strains were grown at 30°C in M17 broth or agar (Difco, Detroit, MI) supplemented with 0.5% (wt/vol) glucose (GM17). When required, erythromycin or chloramphenicol was added to a final concentration of 5 μg ml−1. Cultures were grown in either standing filled flasks (static conditions) or under aeration (aerated conditions) in Erlenmeyer flasks filled to 1/10 volume and shaken at 200 rpm. All growth experiments were performed at least three times independently. Growth was estimated by measuring the optical density at 480 nm (OD480) or OD600. Growth data were modeled according to the Gompertz equation (three parameters) with Sigma Plot 10.0 (Jandel Scientific), and growth parameters were calculated as described previously (40). Survival in late stationary phase was determined in aerobic GM17 cultures (with agitation at 200 rpm) by counting CFU. The survival rate was calculated by dividing the total number of CFU at 24 h by the total number of CFU at 8 h.
Oxygen free-radical resistance assay.
Sensitivity to paraquat and H2O2 was determined as described previously (32) with a few modifications. To assess sensitivity to paraquat, cultures were grown in GM17 broth containing 10 mM paraquat, and growth was monitored by measuring the OD480 hourly for 7 h. To determine H2O2 sensitivity, GM17 cultures in mid-exponential growth phase (OD480 = 2) were exposed to 5 mM H2O2 for 30, 60, and 120 min. Appropriate dilutions were plated on GM17 agar, and colonies were counted after 24 h at 30°C. Survival is expressed as a percentage of the original CFU. All experiments were performed at least three times.
Spontaneous mutation rates.
The emergence of rifampin resistance was determined for the wild-type strain and the noxE mutant in aerated GM17 cultures after 4 h and 8 h of growth, as described previously (32).
Monitoring of acidification, oxygen consumption and glucose metabolism.
Acidification and oxygen consumption were monitored during L. lactis growth in GM17 at 30°C in 1-liter bioreactors (BioStat Q plus; Sartorius) as previously described (34). The oxygen sensors were calibrated in air-sparged water (partial O2 pressure [pO2] = 100%) and nitrogen-sparged water (pO2 = 0%). A pO2 of 100% corresponded to a saturated oxygen concentration of 2.4 × 104 M (in pure water at 30°C) (17). The cultures (300 ml) were stirred vigorously (250 rpm), and the bioreactor was closed. Under these conditions, there was equilibrium between the O2 concentrations in the headspace air and in the dissolved air. It was therefore possible to estimate that a pO2 of 100% corresponded to a total of 6,634 μmol of oxygen (6,562 μmol in the headspace and 72 μmol in the medium). The biomass was monitored by measuring the optical density at 480 nm and correlating the optical density with cell dry weight measurements obtained as described previously (17). One optical density unit at 480 nm was equivalent to 0.31 g of cells liter−1.
Fermentation end products and glucose were assayed in samples filtered through a 0.22-μm-pore-size filter. Glucose, lactate, formate, acetate, ethanol, and acetoin were separated by high-pressure liquid chromatography (Aminex HPX87H column [Bio-Rad, Hercules, CA]) at 40°C using 5 mM H2SO4 at a flow rate of 0.5 ml min−1 as the mobile phase. They were quantified with a Waters 2414 refractive-index detector (Millipore, Milford, MA).
DNA manipulations.
Standard recombinant DNA techniques were used for nucleic acid preparation and analysis (33), and plasmid DNA was prepared as described by O'Sullivan and Klaenhammer (29). Taq DNA polymerase (MP Biomedicals, Illkirch, France) or DNA polymerase Phusion (Finnzymes, Finland) was used for PCR, as advised by the manufacturers. The amplification products were sequenced with an ABI310 automated DNA sequencer, using the ABI Prism dye terminator cycle-sequencing kit (Applied Biosystems, Courtaboeuf, France). L. lactis electrocompetent cells were prepared and transformed as described previously (12). Table S1 in the supplemental material lists the oligonucleotides used in this study.
Site-directed mutagenesis and overproduction of recombinant NoxE and variants. (i) In E. coli
The noxE gene of L. lactis TIL46 or CSK1382 was PCR amplified using primers noxE_NcoI_F and noxE_XhoI_R and ligated between the NcoI and XhoI sites in pET28a (Novagen) (Table 1). In this construction (pET28a::noxE) (Table 1), the noxE gene is under the control of the T7 promoter, and its expression is induced by IPTG (isopropyl-β-d-thiogalactopyranoside). The encoded NoxE is fused to a 6-histidine tag at the carboxy terminus. Mutations were introduced into the NoxE protein of L. lactis TIL46 or CSK1382 using the QuikChange Site-Directed Mutagenesis protocol from Stratagene (La Jolla, CA). pET28a::noxE plasmids were PCR amplified with primer pairs (see Table S1 in the supplemental material) to replace Ala144, Leu299, Ala300, Asn302, Ala303, Gly307, or Lys384 in NoxE_TIL46 and T303 in NoxE_CSK1382 with other residues. The pET28a::noxE plasmids and derivatives were then introduced into E. coli strain Rosetta (Table 1). The plasmid constructions were verified by sequencing them using the primers Nox2F, Nox2R, T7terRev, and T7promRev (see Table S1 in the supplemental material). Expression of the genes inserted into pET28a under the T7 promoter was induced by addition of 0.1 mM IPTG to cultures of E. coli Rosetta containing pET28a derivatives in LB medium at an OD600 of 0.6. The cultures were further incubated at 20°C overnight.
(ii) In L. lactis.
The nisin-controlled expression (NICE) system (26) was used for noxE overexpression in L. lactis NZ9000. Briefly, the noxE genes of L. lactis TIL46 and CSK1382 were amplified with the primers noxE_NcoI_F and noxE_XbaI_R (see Table S1 in the supplemental material). The PCR products were then independently inserted into pNZ8048 under the control of the nisin-inducible promoter nisAp (Table 1) at the NcoI and XbaI sites. The resulting plasmids, pNZ8048::noxE_TIL46 and pNZ8048::noxE_CSK1382 (Table 1), were used to transform NZ9000noxE. A 6-histidine tag was added to the carboxy termini of TIL46 NoxE and the A303T variant as described above, using pET28a::noxE and pET28a::noxE_A303T (Table 1) as the templates and the noxE_XbaI_6H_R primer instead of noxE_XbaI_R (see Table S1 in the supplemental material). The resulting plasmids, pNZ8048::noxE_6His and pNZ8048::noxE_A303T_6His (Table 1), were introduced into L. lactis NZ9000noxE (Table 1) and verified by sequencing them using the Nox2F, Nox2R, pNZ8048_F, and pNZ8048_R primers (see Table S1 in the supplemental material). Expression of the genes inserted in pNZ8048 under the nisin-inducible promoter was induced by addition of 0.4 ng ml−1 nisin (Sigma, Saint-Quentin Fallavier, France) to cultures of L. lactis NZ9000noxE containing pNZ8048 derivatives (Table 1) in GM17 medium at an OD600 of 0.2 to 0.6. The cultures were further incubated at 30°C until they reached an OD600 of approximately 2.
Purification of recombinant 6-His-tagged NoxE and variants.
Recombinant His-tagged proteins were purified by affinity chromatography on Ni-nitrilotriacetic acid (NTA) agarose columns (Qiagen). The purification steps were as recommended by the manufacturer with the following modifications: all steps were performed at 4°C, 2 mM dithiothreitol (DTT) and 10% glycerol were added to the equilibration buffer, and the elution fractions were not ultrafiltered. For circular dichroism (CD) spectra and dynamic light scattering (DLS) analyses, the recombinant proteins were desalted on a PD SpinTrap G-25 (GE Healthcare, Buckinghamshire, United Kingdom) equilibrated with 20 mM MOPS (morpholinepropanesulfonic acid), pH 7.0. Protein concentrations were determined according to the Bradford method (5) using bovine serum albumin for calibration.
Analysis of FAD ligand.
To determine the amount of FAD associated with the recombinant enzymes, an aliquot of each enzyme was denatured for 40 min at 80°C, and FAD was separated from the enzyme by centrifugation (17,400 × g; 5 min). The amount of FAD in the supernatant was calculated from the absorbance at 450 nm using a calibration curve established with standard solutions of FAD (Sigma, St. Louis, MO; 5 to 100 μM) in the same buffer as the samples.
Measurement of NADH oxidase activity.
Whole-cell extracts or purified recombinant enzymes were used to measure NOX activity as previously described (34). Purified enzymes were reactivated as reported previously (13). Briefly, the purified enzyme was incubated for 5 min at 37°C in 50 mM phosphate buffer at pH 7.0 with 0.1 mM FAD and 125 mM cysteine. One unit of enzyme was defined as the amount that catalyzed the oxidation of 1 μmol NADH to NAD per minute at 25°C. Control experiments were performed without the enzyme to verify the absence of spontaneous NADH oxidation.
Absorption and CD spectra.
The absorption spectra of the recombinant enzyme solutions were recorded from 250 nm to 580 nm (Uvikon XL; Biotek Instruments, Colmar, France).
Circular dichroism spectra were recorded from 185 to 260 nm on a Jasco J-810 spectropolarimeter in 0.1-mm-path-length cuvettes. Measurements were made at 20°C using 10 μM protein in 20 mM MOPS buffer, pH 7.0. Each spectrum was an average of eight consecutive scans.
DLS.
Particle size was measured with a Zetasizer Nano-ZS (Malvern Instruments, United Kingdom) based on dynamic light scattering. This involves measuring fluctuations of scattered-light intensity as a function of the Brownian movement of the particles. The measurements were performed at 20°C in triplicate with recombinant proteins in 20 mM MOPS, pH 7.0, at a final concentration of 0.5 mg ml−1.
Protein electrophoresis.
A minigel system (Novex minicell; Invitrogen) with 4 to 12% N,N-methylenebisacrylamide (BIS)-Tris gel was used for sodium dodecyl sulfate-polyacrylamide gel electrophoreses (SDS-PAGE). Ten micrograms of total protein from cell extracts and 1 μg of purified proteins were loaded. Before being loaded, the purified proteins were heated with 5% β-mercaptoethanol and 2% SDS for 5 min at 95°C in the sample buffer. Proteins were visualized by Coomassie brilliant blue staining. NoxE purity was estimated by image analysis with SameSpot v2.0 software (Nonlinear Dynamics, Newcastle, United Kingdom).
Bioinformatics analyses.
NoxE orthologs were retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/). This supplies the results of pairwise genome comparisons for all protein-coding genes in the KEGG GENES database of all completely sequenced genomes. DNA and amino acid sequences were compared using the free software BioEdit (T. Hall, Ibis Biosciences), which includes CLUSTAL W. The @TOME v2 metaserver (http://abcis.cbs.cnrs.fr/AT2/; J.-L. Pons and G. Labesse, CBS, CNRS, France) was used to search in the Protein Data Bank (PDB) for proteins structurally similar to NoxE (31). The structure models were visualized using the molecular visualization and analysis program Visual Molecular Dynamics (VMD) (http://www.ks.uiuc.edu/Research/vmd/).
RESULTS
Disruption of the noxE gene slightly impairs aerobic growth in GM17 and does not affect oxidative stress resistance.
Under aerobic conditions, both the growth rate and final density of L. lactis TIL46 cultures were significantly decreased by noxE inactivation (Table 2). However, optimal growth was not restored when a noxE expression vector (pJIM::noxE) was introduced into the noxE mutant, although the complemented mutant displayed NADH oxidase activity 8-fold higher than that of TIL46 (34). This result suggests that the growth defect of the noxE mutant was not due to the absence of NADH oxidase activity. Under static conditions, the differences between strains were not significant. The growth rates of both the WT strain and TIL46noxE were significantly reduced by aeration. Surprisingly, the survival of the noxE mutant after 24 h of aerated culture (91% ± 22%) tended to be higher than that of the WT (78% ± 15%), although the difference was not significant.
Table 2.
Growth of wild-type strain TIL46, the noxE-negative mutant (TIL46noxE), and the complemented noxE-negative mutant (TIL46noxE+pJIM::noxE) in GM17 under static and aerated conditionsa
| Strain | Growth rate (μ, h−1) |
Maximal OD480 |
||
|---|---|---|---|---|
| Static | Aerated | Static | Aerated | |
| WT | 1.73 ± 0.15 | 1.31 ± 0.05 | 3.67 ± 0.07 | 3.50 ± 0.12 |
| TIL46noxE mutant | 1.39 ± 0.30 | 0.88 ± 0.07b | 3.29 ± 0.32 | 3.00 ± 0.06b |
| TIL46noxE+pJIM::noxE mutant | 1.32 ± 0.20 | 0.70 ± 0.15b | 3.40 ± 0.21 | 2.90 ± 0.15b |
Aerated conditions, shaking at 200 rpm. The values are means ± standard deviations of at least three independent measurements.
Significant difference (P < 0.01; Student's t test) compared to WT.
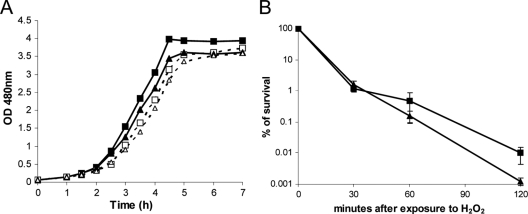
The sensitivities of the noxE mutant and WT strains to paraquat, which generates intracellular O2− radicals, and to H2O2 were tested. The growth rates of both strains were slightly and similarly delayed by 10 mM paraquat (Fig. 1A). After exposure to 5 mM H2O2 for 30 min, the survival of both strains similarly dropped to 1%, and after 2 h of exposure, the survival of the noxE mutant was only 1 log unit lower than that of the WT (Fig. 1B). Oxidative DNA damage is a major source of mutagenesis, so we examined the frequency of spontaneous mutations at the end of exponential growth in aerated GM17 broth. The noxE mutant exhibited the same rate of spontaneous mutation (1.41 × 10−7 ± 0.47 × 10−7) as the WT (1.76 × 10−7 ± 0.98 × 10−7). These findings indicate that NoxE is not required for resistance to oxidative stress.
Fig. 1.
Sensitivity of L. lactis TIL46 and the noxE mutant to oxidative stress. (A) Growth curves in GM17 broth at 30°C in the static condition. Shown are the wild-type strain in the absence (▪) and presence (□) of 10 mM paraquat and the noxE mutant in the absence (▴) and presence (▵) of 10 mM paraquat. (B) Survival of exponentially growing cells of the wild-type strain (▪) and the noxE mutant (▴) grown in GM17 broth after exposure to 5 mM H2O2 for 30, 60, or 120 min at 30°C. The error bars represent the standard deviations of three independent experiments.
Disruption of the noxE gene strongly impairs oxygen consumption and mixed-acid fermentation but does not affect NAD regeneration.
The biomass of noxE mutant cultures after fermentation for 6 h in aerated GM17 broth was about 15% lower than that of the WT (Table 3); the pO2 in the medium fell from 100% to approximately 74% in WT cultures but only to 95% in noxE mutant cultures. This allowed estimation of the specific oxygen uptake of the two strains (Table 3). The noxE mutant consumed only 26% of the amount of oxygen consumed by the WT, indicating that NoxE was responsible for 74% of the total oxygen consumption. In the noxE mutant, glucose fermentation was strictly homolactic, whereas the WT produced some acetic acid in addition to lactic acid (Table 3). Therefore, the amount of NADH reoxidized by NoxE in the WT (6.56 mmol per g of biomass) accounted for only 16% of that reoxidized by lactate dehydrogenase (LDH) (40.6 mmol per g of biomass). Moreover, the noxE mutant produced more lactic acid, regenerating more NAD (52.2 mmol per g of biomass) via this pathway, than the WT. Consequently, the noxE mutation led to increased acidification of the medium (Table 3).
Table 3.
Biomass concentration, specific uptake of glucose and oxygen, and specific production of lactate and acetate by the wild-type strain TIL46 and the noxE-negative mutant (noxE) after fermentation for 6 h in GM17 medium under vigorously aerated conditions
| Strain | Biomass concn (g [dry wt] liter−1) | Specific uptake (mmol·g biomass−1) |
Specific production (mmol·g biomass−1) |
Specific acidification (ΔpH·g biomass−1) | ||
|---|---|---|---|---|---|---|
| Glucose | Oxygen | Lactate | Acetate | |||
| WT | 1.23 ± 0.01 | 23.9 ± 0.2 | 4.43 ± 0.29 | 40.6 ± 0.6 | 3.37 ± 0.45 | 1.21 ± 0.04 |
| noxE mutant | 1.03 ± 0.03b | 27.0 ± 1.3a | 1.15 ± 0.07b | 52.2 ± 0.6b | NDb | 1.45 ± 0.09b |
Significant difference (P < 0.05; Student's t test) compared to WT.
Significant difference (P < 0.001; Student's t test) compared to WT. ND, not detected.
NoxE inactivation in the natural L. lactis strain CSK1382 is due to point mutations.
We previously found one natural L. lactis subsp. cremoris strain (CSK1382) without any detectable NADH oxidase activity (34). To check that this NADH oxidase activity defect was associated with the production of an inactive NoxE, the noxE genes of this strain (noxE_CSK1382) and TIL46 were overexpressed in a noxE mutant of L. lactis. The noxE_TIL46-overexpressing strain showed a NADH oxidase activity of 19 ± 4 U mg−1, while the noxE_CSK1382-overexpressing strain did not display any detectable activity, although a band corresponding to the molecular mass of NoxE (49 kDa) was detected in the cell extract by SDS-PAGE (data not shown), confirming that NoxE of CSK1382 was produced but inactive. The noxE sequence of TIL46 was identical to that of MG1363 (NC_ 009004), whereas 17 nucleotides differed in the noxE sequence of CSK1382. However, there were only three differences between the deduced amino acid sequences of NoxE from CSK1382 and that from TIL46: A144T, A303T, and K384N.
Mutations in the 299-to-307 region affect NoxE conformation.
To identify which of the three differences was responsible for the inactivation of CSK1382 NoxE, TIL46 NoxE residues 144, 303, and 384 were individually replaced by those of CSK1382 NoxE. Most of the recombinant NoxE from TIL46 and the A144T and K384N variants produced in E. coli were in the soluble cell fractions (see Fig. S1 in the supplemental material). On the other hand, most of the A303T variant was in the insoluble fractions of E. coli (see Fig. S1 in the supplemental material) and L. lactis (not shown) cell extracts. The Ala303 residue was replaced with a Gly residue, which, like Ala, is small and nonpolar (3). Moreover, four other residues flanking A303 and conserved in NADH oxidases were replaced (Table 4). The amount of the A300T variant in the soluble cell fraction was similar to that of the WT, and that of L299T was slightly greater. In contrast, the amounts of the A303G, N302S, G307A, and G307S variants in the soluble cell fraction were much smaller, as for A303T. After purification on an Ni-NTA resin, the amount of protein retrieved in the elution fractions was 5- to 8-fold smaller for N302S, A303, and G307 variants than for the NoxE of TIL46 (Table 4). SDS-PAGE analysis of the elution fractions from the Ni-NTA resin revealed one major band (>95% of the total intensity) corresponding to the calculated mass of the His-tagged NoxE (49.5 kDa) for the NoxE of TIL46 and the A144T, K384N, A300T, and L299T variants, whereas the elution fractions of the other recombinant proteins were less pure (Table 4; see Fig. S2 in the supplemental material). To estimate the aggregation status of the purified recombinant proteins, the particle size distribution in the elution fractions was determined by DLS (Table 4; see Fig. S3 in the supplemental material). The proteins that were produced in large quantities in the soluble cell fraction were essentially mono dispersed in solutions, and the hydrodynamic diameter of the proteins was estimated to be 8.7 nm, corresponding to an apparent molecular mass of 100 kDa for a globular protein. This is consistent with the 49.56-kDa NoxE protein forming dimers. DLS analysis did not detect particles of the expected size for NoxE in the elution fractions of the N302S, A303, and G307 variants, despite the fractions containing 6 to 38% NoxE protein: all particles present in these fractions were much larger than expected. Therefore, the N302, A303, and G307 variants were presumably not in the active dimeric form or a compact globular form. A more purified preparation of the A303T variant (70% purity estimated from gel electrophoresis), overproduced in L. lactis, was used for CD analysis. The CD spectra did not reveal any apparent difference between the secondary structures of the NoxE from TIL46 and the A303T variant (see Fig. S4 in the supplemental material). The shape of the spectra in the far-UV region was typical of a mixture of α-helix and β-sheet elements and did not indicate an unfolded secondary structure. All of the various observations strongly suggest that N302, A303, and G307 mutations in NoxE affect its tertiary structure or prevent protein dimerization, leading to protein aggregation and the formation of inclusion bodies in E. coli.
Table 4.
Characteristics of recombinant His-tagged NoxE of L. lactisTIL46 and the different variants produced in E. coli after purification on Ni-NTA agarose resin
| Protein | Amta (mg) | % Purityb | Particles with expected sizec | Sp actd (U/mg) | FAD/NoxE ratioe |
|---|---|---|---|---|---|
| NoxE | 2.5 | 100 | + | 130 ± 9 | 1 |
| NoxE_A144T | 2.6 | 100 | + | 137 ± 10 | − |
| NoxE_K384N | 3.5 | 100 | + | 139 ± 11 | − |
| NoxE_A303T | 0.3 | 9 | − | 0.13 ± 0.1 | ND |
| NoxE_A303G | 0.3 | 20 | − | 9 ± 8 | ND |
| NoxE_L299T | 6 | 100 | + | 35 ± 3 | 0.9 |
| NoxE_A300T | 2 | 100 | + | 123 ± 12 | 1 |
| NoxE_N302S | 0.4 | 38 | − | 53 ± 10 | ND |
| NoxE_G307S | 0.25 | 9 | − | 1 ± 1 | ND |
| NoxE_G307A | 0.35 | 6 | − | 2 ± 1 | ND |
Amount of protein retrieved in the elution fraction from the Ni-NTA column.
Estimated by SDS-PAGE (see Fig. S2 in the supplemental material).
Presence (+) or absence (−) of particles with a hydrodynamic diameter of 8.7 nm (corresponding to an apparent molecular mass of 100 kDa) as determined by dynamic light scattering (see Fig. S3 in the supplemental material).
The data are means of 2 determinations ± standard deviations and take into account the percent purity of protein preparations estimated by SDS-PAGE. Activity was determined after reactivation with cysteine and FAD.
−, not determined; ND, not detected.
Mutations in the 299-to-307 region affect NoxE activity.
The optimum activity of the purified recombinant His-tagged NoxE from TIL46 determined using the fresh preparation and after reactivation with cysteine and FAD was 130 ± 9 U mg−1, with a Km value for NADH of 4 μM and a Vmax value of 68 μM min−1. The calculation of the specific activity took into account the percentage purity estimated by image analysis of SDS-PAGE (Table 4). The recombinant NoxE from TIL46 and the A144T, K384N, and A300T variants displayed similar high levels of NADH oxidase activity. The specific activities of the L299T and N302S variants were 3.7 and 2.6 times lower, respectively, than that of the WT, and the activities of the A303 and G307 variants were close to zero. Therefore, A303, G307, and, to a lesser extent, L299 and N302 are required for NoxE activity.
Mutations in the 302-to-307 region of NoxE affect FAD binding.
Cell extracts of L. lactis strains overproducing the NoxE of TIL46 were yellow, while those of strains overproducing the NoxE of CSK1382 or the A303T variant were colorless. Similarly, solutions of the purified recombinant NoxE of TIL46 and the A300T and L299T variants produced in E. coli were yellow, while those of the other variants were colorless, suggesting that no FAD was bound to the inactive enzymes (Table 4). However, the absence of color in certain solutions (T303, N302S, and G307 variants) may be due to the very low concentration of NoxE protein, which was 50- to 100-fold lower than in the colored solutions. The presence of FAD in the yellow proteins was confirmed by the two absorption peaks specific to FAD at 370 and 446 nm in their spectra (not shown). The FAD released after heat denaturation of the recombinant proteins was assayed; the FAD/NoxE monomer ratio was close to 1 for the NoxE from TIL46 and the A300T and L299T variants, consistent with the equimolar ratio found previously for the native NoxE enzyme (21). FAD was not detected for the other variants, indicating that they did not accommodate a FAD molecule. This was probably due to inappropriate conformation of these variant proteins.
The T303A mutation in NoxE of CSK1382 restored the activity of the enzyme.
The reverse mutation T303A was introduced into noxE of CSK1382. The modified gene was overexpressed in E. coli. After induction with IPTG, the noxE_CSK1382-T303A-overexpressing strain displayed cellular activity similar to that of the noxE_TIL46-overexpressing strain (10 ± 2 U mg−1), while the strain overexpressing noxE_CSK1382 displayed very low activity (0.1 U mg−1). This confirms that the natural mutation A303T was responsible for the loss of activity of CSK1382 NoxE.
The 299-to-307 sequence of NoxE is conserved among bacterial NADH oxidases and other flavoproteins.
Sequence identity between L. lactis subsp. cremoris NoxE and other NADH oxidases of LAB and more distant bacteria is high (see Fig. S5 in the supplemental material). In addition to the known conserved motifs, we found that the 299-to-307 sequence of NoxE is highly conserved. However, in the NoxE of L. lactis subsp. cremoris SK11 (gene ID 4433101), the consensus N302 is replaced by a serine residue.
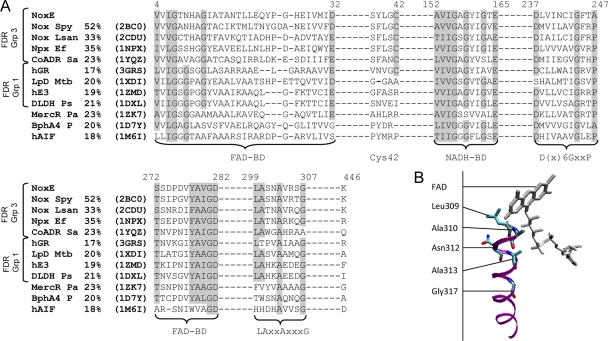
Structural comparisons between NoxE and proteins in the PDB revealed the highest similarity with S. pyogenes Nox (@tome relative score, 98.70) and significant structural similarities with other flavoproteins. The structure-based amino acid alignment of L. lactis NoxE with other flavoproteins (Fig. 2A) showed conserved sequences, including the known FAD and NADH binding domain motifs and the LAXXAXXXG sequence (residues 299 to 307 of NoxE), which appears to be highly conserved in flavoproteins of FDR groups 1 and 3. In other flavoproteins, only the Ala residue in position 303 and the Gly in position 307 in NoxE are conserved.
Fig. 2.
(A) Structure-based amino acid alignment of flavoproteins sharing strong structural similarities with L. lactis NoxE. Flavoproteins of the group (Grp) 3 FDR family were as follows: NoxE, NADH oxidase NoxE of L. lactis; Nox Spy, NADH oxidase of S. pyogenes; Nox Lsan, NAD(P)H oxidase of L. sanfranciscensis; Npx Ef, NADH peroxidase of E. faecalis; CoADR Sa, coenzyme A-disulfide reductase of S. aureus. Flavoproteins of the group 1 FDR family were as follows: hGR, Homo sapiens glutathione reductase; LpD Mtb, lipoamide dehydrogenase of Mycobacterium tuberculosis; hE3, human dihydrolipoamide dehydrogenase; DLDH Ps, dihydrolipoamide dehydrogenase of Pisum sativum. The flavoprotein of the group 2 FDR family was MercR Pa, mercuric reductase of P. aeruginosa. Other flavoproteins were as follows: BphA4 P, NADH-dependent ferredoxin reductase component in biphenyl dioxygenase of Pseudomonas sp.; hAIF, human apoptosis induction factor. PDB accession codes are in parentheses. Percentage identities with NoxE are shown in the second column. The positions of amino acid residues of the NoxE sequence are indicated above the sequences. Conserved sequence motifs are shaded. BD, binding domain. (B) Visualization of the alpha-helix formed by residues 309 to 317 and FAD in the structural model of S. pyogenes NoxE (PDB code 2BC0).
Visualization of NoxE structural models (PDB codes 2BC0 and 2CDU) revealed that residues 299 to 307 are in an alpha-helix at the si-face of FAD (Fig. 2B), at the interface of the second monomer of the enzyme and the N-terminal FAD binding alpha-helix formed by His10 and its surrounding residues (see Fig. S6 in the supplemental material).
DISCUSSION
In pathogenic streptococci that encounter oxygen upon host colonization, NADH oxidases contribute to oxygen tolerance by decreasing the intracellular NADH/NAD ratio and/or by direct elimination of oxygen (2, 10, 14, 37, 38). In L. lactis, which is particularly exposed to oxygen when it is used for milk fermentation in the dairy industry, the role of NADH oxidases in oxygen tolerance has not been investigated. Here, we characterized the lactococcal H2O-forming NADH oxidase NoxE and found it was not essential for either aerobic growth or oxidative stress resistance in a rich synthetic medium. LDH regenerated 6-fold more NAD than NoxE in the wild-type strain, indicating that NoxE plays only a minor role in NAD regeneration during growth in GM17 medium. Moreover, noxE inactivation increased the amount of NAD regenerated by LDH, which largely compensated for the loss of NAD regenerated by NoxE. We demonstrated that NoxE is the major enzyme consuming oxygen in growing L. lactis, but the noxE mutant was not more sensitive to aeration than the wild-type strain. This is surprising, because noxE is among the most strongly induced genes (15-fold) in response to aeration (30). Nevertheless, many other genes involved in stress responses, especially genes encoding detoxification enzymes, are also induced during aerobic fermentation (30) and may manage oxygen toxicity in the noxE mutant. Although NoxE is not by itself critical for resistance to oxidative stress, it might make a significant contribution to this resistance, in which many processes act jointly. Moreover, even if NoxE is not essential for the aerobic growth and survival of L. lactis in rich synthetic medium or milk (34), it may be needed in other environments, like Nox2 in S. pyogenes, S. mutans, and L. sanfranciscensis (10, 11, 16). In carbohydrate-limited media, for instance, carbon catabolite repression is reduced, leading to increased NoxE activity and decreased LDH activity (9, 24, 39). Under these conditions, NAD regeneration by NoxE may be much more important than in carbohydrate-rich media.
The identification of natural L. lactis strains that have no NADH oxidase activity is consistent with NoxE being nonessential for the growth and survival of L. lactis (34). One of these strains produced an inactive NoxE, allowing the identification of novel residues important for NADH oxidase activity.
We demonstrated that an Ala-to-Thr substitution at position 303 was responsible for total loss of activity, largely due to protein aggregation and probably defective FAD binding. So far, we do not know whether the mutation leads to an inappropriate conformation that prevents FAD binding or whether it prevents FAD binding, which in turn is likely to cause gross changes in the protein conformation. The crystal structures of NoxE homologues (PDB codes 2BC0 and 2CDU) showed that residue 303 is part of an alpha-helix close to FAD (see Fig. S6 in the supplemental material) and to the C-terminal part of the dimerization domain of the second subunit. This helix is in the same spatial position in other flavoproteins of the FDR family, implicating it in the catalytic processes (36). Therefore, it is not surprising that mutation of a residue in this tightly packed buried region close to the active site disrupts protein folding and cofactor binding. However, the importance of this helix in flavoprotein structure and activity had never been demonstrated. Here, we identified residues in this region of NoxE that were essential for protein folding and FAD binding and activity. Moreover, these residues constitute a sequence of amino acids conserved in flavoproteins that had not previously been reported (8). Mutation of A303 or G307, which are strictly conserved in the 1 and 3 family FDR proteins, was very disruptive: their replacement, even with residues with similar properties, affected protein folding and led to loss of FAD. L299 and A300 are also strongly conserved in flavoproteins, and their structural proximity to FAD implicates them in FAD binding (22). Replacement of L299 by a Thr did not affect protein dimerization or FAD binding but greatly decreased NoxE activity. L299 is close to the isoalloxazine ring of FAD in NoxE homologues (data not shown) and in other flavoproteins (1NPX and 3GRS), so our results suggest that L299 is involved in positioning FAD appropriately in the active site, possibly through hydrophobic interactions with the isoalloxazine ring. Although A300 is conserved in most enzymes, its replacement with Thr, as in human glutathione reductase (3GRS), did not affect protein folding, FAD binding, or NoxE activity. We do not know whether other mutations at this site would be detrimental. The polar residue N302 is replaced by residues with different sizes and charges (G, V, or K) in other flavoproteins and thus does not appear to be essential for flavoprotein activity. However, it is conserved in NADH oxidases, and its replacement with Ser, a small polar residue, as in NoxE of L. lactis SK11, affected the conformation and activity of NoxE. This explains the low NADH oxidase activity of L. lactis SK11 (34). However, after reactivation with cysteine and FAD, the variant recovered some activity, suggesting that the N302S variant may be in a partially folded and expanded dimeric state, like unfolding intermediates previously described for other flavoproteins (6, 23). This partially unfolded form may partly recover the compact dimer structure after reactivation. We therefore propose that LA(T)XXAXXXG constitutes a new typical sequence of flavoproteins and LA(T)XNAXXXG a typical sequence of NADH oxidases.
Contrary to the general view, we show that L. lactis does not need to totally eliminate dissolved oxygen from the medium to grow and acidify. Therefore, strains with active or inactive NoxE can be used to modulate dissolved oxygen levels during the manufacture of fermented products. Such modulation may allow diversification of the development of secondary flora and the production of aroma compounds that are dependent on the redox potential of the medium (4, 19). The new identification of amino acids required for NADH oxidase activity will make it possible to develop molecular tests based on their detection to identify strains with or without NADH oxidase activity. Such selection could also be of interest for other LAB. As this amino acid sequence is highly conserved in numerous flavoproteins, it would be useful to determine whether it is also essential for the activity of other flavoproteins, especially those of the FDR family, including enzymes with important physiological functions, such as thioredoxin reductase and glutathione reductase (15, 20, 35).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Eureka Research grant (Σ!3562-LABREDOX). We are grateful to CSK Food Enrichment for their financial support.
We thank Hans Brandsma (CSK Food Enrichment), who provided the L. lactis subsp. cremoris strains; Lucy Henno (INRA) for technical assistance; Gilles Labesse (CNRS) for allowing us access to the IBiSA structural bioinformatics platform @tome; Human Rezaei (INRA) for CD and DLC measurements; Vincent Juillard (INRA) for the calculation of growth parameters and statistics; Philippe Gaudu (INRA) for fruitful discussion; and Véronique Monnet (INRA) for her critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Argyrou A., Blanchard J. S. 2004. Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 78:89–142 [DOI] [PubMed] [Google Scholar]
- 2. Auzat I., et al. 1999. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34:1018–1028 [DOI] [PubMed] [Google Scholar]
- 3. Betts M. J., Russell R. B. 2003. Amino acid properties and consequences of substitutions, p. 289–316 In Barnes M. R., Gray I. C. (ed.), Bioinformatics for geneticists. John Wiley and Sons Ltd., Chichester, United Kingdom [Google Scholar]
- 4. Boucher B., Brothersen C., Broadbent J. 2006. Influence of starter and nonstarter lactic acid bacteria on medium redox. Aust. J. Dairy Technol. 61:116–118 [Google Scholar]
- 5. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6. Caldinelli L., et al. 2004. Unfolding intermediate in the peroxisomal flavoprotein D-amino acid oxidase. J. Biol. Chem. 279:28426–28434 [DOI] [PubMed] [Google Scholar]
- 7. Condon S. 1987. Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Lett. 46:269–280 [Google Scholar]
- 8. Dym O., Eisenberg D. 2001. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10:1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaudu P., Lamberet G., Poncet S., Gruss A. 2003. CcpA regulation of aerobic and respiration growth in Lactococcus lactis. Mol. Microbiol. 50:183–192 [DOI] [PubMed] [Google Scholar]
- 10. Gibson C. M., Mallett T. C., Claiborne A., Caparon M. G. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182:448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Higuchi M., et al. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181:5940–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holo H., Nes I. F. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoskins D. D., Whiteley H. R., Mackler B. 1962. The reduced diphosphopyridine nucleotide oxidase of Streptococcus faecalis: purification and properties. J. Biol. Chem. 237:2647–2651 [PubMed] [Google Scholar]
- 14. Hwang I., Kaminogawa S., Yamauchi K. 1981. Purification and properties of a dipeptidase from Streptococcus cremoris. Agric. Biol. Chem. 45:159–165 [Google Scholar]
- 15. Imlay J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jänsch A., Freiding S., Behr J., Vogel R. F. 2011. Contribution of the NADH-oxidase (Nox) to the aerobic life of Lactobacillus sanfranciscensis DSM20451. Food Microbiol. 28:29–37 [DOI] [PubMed] [Google Scholar]
- 17. Jensen N. B., Melchiorsen C. R., Jokumsen K. V., Villadsen J. 2001. Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Appl. Environ. Microbiol. 67:2677–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang R., Riebel B. R., Bommarius A. S. 2005. Comparison of alkyl hydroperoxide reductase (AhpR) and water-forming NADH oxidase from Lactococcus lactis ATCC 19435. Adv. Synthesis Catalysis 347:1139–1146 [Google Scholar]
- 19. Kieronczyk A., Cachon R., Feron G., Yvon M. 2006. Addition of oxidizing or reducing agents to the reaction medium influences amino acid conversion to aroma compounds by Lactococcus lactis. J. Appl. Microbiol. 101:1114–1122 [DOI] [PubMed] [Google Scholar]
- 19a. Kuipers O. P., de Rayter P. G. G. A., Kleerebezem M., de Vos W. M. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 20. Li Y., Hugenholtz J., Abee T., Molenaar D. 2003. Glutathione protects Lactococcus lactis against oxidative stress. Appl. Environ. Microbiol. 69:5739–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez de Felipe F., Hugenholtz J. 2001. Purification and characterisation of the water forming NADH-oxidase from Lactococcus lactis. Int. Dairy J. 11:37–44 [Google Scholar]
- 22. Lountos G. T., et al. 2006. The crystal structure of NAD(P)H oxidase from Lactobacillus sanfranciscensis: insights into the conversion of O2 into two water molecules by the flavoenzyme. Biochemistry 45:9648–9659 [DOI] [PubMed] [Google Scholar]
- 23. Louzada P. R., Sebollela A., Scaramello M. E., Ferreira S. T. 2003. Predissociated dimers and molten globule monomers in the equilibrium unfolding of yeast glutathione reductase. Biophys. J. 85:3255–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luesink E. J., van Herpen R. E., Grossiord B. P., Kuipers O. P., de Vos W. M. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789–798 [DOI] [PubMed] [Google Scholar]
- 25. Mallett T. C., Claiborne A. 1998. Oxygen reactivity of an NADH oxidase C42S mutant: evidence for a C(4a)-peroxyflavin intermediate and a rate-limiting conformational change. Biochemistry 37:8790–8802 [DOI] [PubMed] [Google Scholar]
- 26. Mierau I., Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705–717 [DOI] [PubMed] [Google Scholar]
- 27. Nordhoff A., Bucheler U. S., Werner D., Schirmer R. H. 1993. Folding of the four domains and dimerization are impaired by the Gly446→Glu exchange in human glutathione reductase. Implications for the design of antiparasitic drugs. Biochemistry 32:4060–4066 [DOI] [PubMed] [Google Scholar]
- 28. Nordhoff A., et al. 1997. Denaturation and reactivation of dimeric human glutathione reductase—an assay for folding inhibitors. Eur. J. Biochem. 245:273–282 [DOI] [PubMed] [Google Scholar]
- 29. O'Sullivan D. J., Klaenhammer T. R. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pedersen M. B., et al. 2008. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J. Bacteriol. 190:4903–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pons J.-L., Labesse G. 2009. @TOME-2: a new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 37:W485–W491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poyart C., et al. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sambrook J., Russel D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Tachon S., Brandsma H., Yvon M. 2010. NADH oxidase NoxE and the electron transport chain are responsible for the ability of Lactococcus lactis to decrease the redox potential of milk. Appl. Environ. Microbiol. 76:1311–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vido K., et al. 2005. Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. J. Bacteriol. 187:601–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wierenga R. K., Drenth J., Schulz G. E. 1983. Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J. Mol. Biol. 167:725–739 [DOI] [PubMed] [Google Scholar]
- 37. Yamamoto Y., et al. 2006. The Group B Streptococcus NADH oxidase Nox-2 is involved in fatty acid biosynthesis during aerobic growth and contributes to virulence. Mol. Microbiol. 62:772–785 [DOI] [PubMed] [Google Scholar]
- 38. Yu J., et al. 2001. Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. Microbiology 147:431–438 [DOI] [PubMed] [Google Scholar]
- 39. Zomer A. L., Buist G., Larsen R., Kok J., Kuipers O. P. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zwietering M. H., Jongenburger I., Rombouts F. M., Riet K. van 't. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.