Abstract
Nucleosomes containing histone variant H2A.Z (Htz1) serve to poise quiescent genes for activation and transcriptional initiation. However, little is known about their role in transcription elongation. Here we show that dominant mutations in the elongation genes SPT5 and SPT16 suppress the hypersensitivity of htz1Δ strains to drugs that inhibit elongation, indicating that Htz1 functions at the level of transcription elongation. Direct kinetic measurements of RNA polymerase II (Pol II) movement across the 9.5-kb GAL10p-VPS13 gene revealed that the elongation rate of polymerase is 24% slower in the absence of Htz1. We provide evidence for two nonexclusive mechanisms. First, we observed that both the phospho-Ser2 levels in the elongating isoform of Pol II and the loading of Spt5 and Elongator over the GAL1 open reading frame (ORF) depend on Htz1. Second, in the absence of Htz1, the density of nucleosome occupancy is increased over the GAL10p-VPS13 ORF and the chromatin is refractory to remodeling during active transcription. These results establish a mechanistic role for Htz1 in transcription elongation and suggest that Htz1-containing nucleosomes facilitate Pol II passage by affecting the correct assembly and modification status of Pol II elongation complexes and by favoring efficient nucleosome remodeling over the gene.
INTRODUCTION
Transcription by RNA polymerase II (Pol II) takes place on a compositionally and topologically complex chromatin template at multiple steps, including promoter activation, transcription initiation, elongation, and termination. A number of mechanisms facilitate the initiation of gene transcription on chromatin templates. Two of the most prominent of these are the remodeling of nucleosomes by DNA-dependent ATPase complexes and the covalent posttranslational modification of histone proteins (20, 90). At a minimum, these pathways are thought to act by establishing a permissive chromatin structure at gene promoters, allowing access of site-specific transcription factors and the subsequent recruitment of polymerase and the general factors (25, 69).
Nucleosomes also pose a barrier to transcriptional elongation, and a variety of conserved elongation factors have been found to facilitate transcription on chromatin templates (6, 54, 57). Among these are the DSIF, Spt6, FACT, Elongator, PAF, and COMPASS complexes. While the mechanisms by which these factors carry out their elongation functions are less well understood, they include the ability to influence either transcription-coupled nucleosome assembly and disassembly, as in the case of Spt6 and FACT (9, 22, 42, 74, 85), or covalent protein modification, as in the case of Elongator, Set2, and COMPASS (44, 50, 70, 102, 104).
Histone variants provide a fundamentally different mechanism for modulating chromatin through altered composition (7, 37, 73). Histone variant H2A.Z is conserved from yeast to humans, comprising roughly 10% of the total H2A in the cell. It is an essential histone in many organisms and is conditionally essential in the budding yeast Saccharomyces cerevisiae, where it is encoded by the HTZ1 (YOL012C) gene (30, 78, 92). Htz1 has roles in a variety of distinct functions, including gene expression, chromosome segregation, silencing, and blocking the spread of heterochromatin (30, 63, 65, 78, 98). The X-ray crystal structure of nucleosomes containing H2A.Z has been determined and, together with the results of biochemical experiments, argues that H2A.Z alters the physical properties of the chromatin template (7). Histone H2A.Z is itself subject to posttranslational modifications, including acetylation, ubiquitylation, and sumoylation, and these may alter its chromatin association, exchange dynamics, structure, and function (8, 32, 39, 41, 43, 66, 81, 84, 95). H2A.Z is broadly but nonuniformly distributed throughout the chromosomes and is deposited by specific remodeling complexes, such as the yeast SWR1 complex (SWR1) (3, 45, 51, 60, 67).
H2A.Z has important functions during transcription initiation (19, 30, 32, 34, 92, 99). Early experiments in budding yeast revealed that Htz1 is partially redundant with the Swi/Snf nucleosome remodeling and SAGA histone modification complexes, providing genetic evidence for a role in transcription activation. Furthermore, at several individual loci Htz1 was preferentially enriched over the promoters of inactive genes and became relatively depleted when those genes were induced (83). Subsequent genome-wide mapping studies have shown that this principle of Htz1 occupancy is generally applicable to a large fraction of regulated genes, leading to the hypothesis that H2A.Z poises genes for transcriptional initiation (2, 29, 58, 77, 99, 108). Several mechanisms have been proposed for this activity, including recruitment of transcription factors, conditioning of promoter nucleosome architecture, and facilitating nucleosome eviction (1, 30, 99, 108).
Less well understood is the relationship between H2A.Z function and transcription elongation. Recent genome mapping experiments in Drosophila melanogaster tissue culture cells have highlighted differences in H2A.Z nucleosome compositions associated with polymerase pausing, suggesting potential roles in elongation (100). Genetic screens in budding yeast have revealed that many of the strongest genetic interactions involving htz1Δ strains are with genes encoding transcription elongation factors, including dst1, rpb9, set2, spt4, and spt16, and with multiple genes encoding subunits of the PAF complex (11, 15, 49, 51, 53, 62, 98, 101). At present, however, the interpretations of these data are unclear. It is possible that the observed genetic interactions stem from the combined insults of an Htz1 initiation defect and a separate defect in elongation. That is, for genes already defective for initiation, further impeding elongation could produce synthetic lethality. Alternatively, Htz1 could have more direct roles in transcription elongation that are currently unrecognized. Here we report the results of experiments designed to explore these alternative models. In these studies we employed a combination of mutational approaches to characterize new positive interactions with genes for elongation, together with molecular approaches to measure the rates of elongation, the composition of the elongation complexes, and the nucleosome occupancy of transcribed open reading frames (ORFs). The results of these experiments indicate that Htz1 plays a positive role in the process of transcription elongation by influencing the composition of elongation complexes and facilitating nucleosome remodeling over the gene.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The yeast strains used in this work are summarized in Table 1. Yeast extract-peptone-dextrose (YPD), synthetic complete (SC), 5-fluoroorotic acid (5-FOA), and presporulation and sporulation media were made as described previously (4). 6-Azauracil (6-AU) was added to SC medium lacking uracil (SC-Ura medium) at a final concentration of 50 μg/ml to make 6-AU plates. Gene disruptions were performed by one-step gene replacement selecting for URA3 or G418 resistance. The chromosomal ELP3-HA, SPT5-HA, and RPB3-HA alleles were created by a PCR-based method that fuses the sequence derived from plasmid pFA6a-3HA-kanMX6, encoding three copies of the hemagglutinin (HA) epitope, to the 3′ ends of the ELP3, SPT5, and RPB3 open reading frames (ORFs). SPT4 deletion strains were constructed by replacing the SPT4 ORF with the natMX4 cassette, which confers resistance to the antibiotic nourseothricin.
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| MSY590 | MATα (HHF2-HHT2)Δ leu2-3,112lys2Δ201 ura3-52 |
| CY397 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| CY398 | MATα ade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| JSY677 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 HTZ1 GAL10p-VPS13::TRP1 |
| MSY1262 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 |
| MSY1499 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ |
| MSY1786 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ pMSS104[HTZ1 LEU2 ADE3] |
| MSY1793 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 |
| MSY1795 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 dst1::KanR |
| MSY1798 | MATα ade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 |
| MSY1803 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ spt5-24::KlURA3 pMSS104[HTZ1 LEU2 ADE3] |
| MSY1804 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ spt5-28::KlURA3 pMSS104[HTZ1 LEU2 ADE3] |
| MSY2157 | MATα ade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1::URA3 HTZ1 dst1::KanR |
| MSY2158 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1::URA3 |
| MSY2165 | MATα ade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 elp3::KanR |
| MSY2175 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 |
| MSY2176 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 spt4::NatR |
| MSY2178 | MATα ade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1::URA3 |
| MSY2180 | MATα ade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1::URA3 spt4::NatR |
| MSY2268 | MATa/MATα ade2-1/ade2-1 ade3::hisG/ADE3 can1-100/can1-100 his3-11/his3-11 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 htz1Δ/HTZ1 DST1/dst1::KanRSPT5-18::KlURA3/SPT5 |
| MSY2278 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 SPT5-18::KlURA3 |
| MSY2279 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 SPT5-18::KlURA3 dst1::KanR |
| MSY2280 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT5-18::KlURA3 |
| MSY2281 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT5-18::KlURA3 dst1::KanR |
| MSY2287 | MATa/MATα (HHF2 HHT2)Δ/(HHF2 HHT2)Δ leu2-3,112/leu2-3,112 lys2Δ201/lys2Δ201 ura3-52/ura3-52 htz1::URA3/HTZ1elp3::KanR/ELP3 |
| MSY2301 | MATα (HHF2-HHT2)Δ leu2-3,112 lys2Δ201 ura3-52 ELP3-HA |
| MSY2302 | MATα (HHF2-HHT2)Δ leu2-3,112 lysΔ201 ura3-52 htz1::URA3 ELP3-HA |
| MSY2304 | MATα (HHF2-HHT2)Δ leu2-3,112 lys2Δ201 ura3-52 SPT5-HA |
| MSY2305 | MATα (HHF2-HHT2)Δ leu2-3,112 lys2Δ201 ura3-52 htz1::URA3 SPT5-HA |
| MSY2464 | MATa/MATα ade2-1/ade2-1 ade3::hisG/ade3::hisG can1-100/can1-100 his3-11/his3-11 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 htz1Δ/htz1Δ SPT16-18htz1::KlURA3/SPT16 |
| MSY3183 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 HTZ1 GAL10p-VPS13::TRP1 RPB3-HA |
| MSY3188 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1::URA3 GAL10p-VPS13::TRP1 RPB3-HA |
| MSY3973 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT16-61::KlURA3 pRS314 |
| MSY3974 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT16-61::KlURA3 pMSS132[SPT16 TRP1] |
| MSY3979 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT16-183::KlURA3 pRS314 |
| MSY3980 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT16-183::KlURA3 pMSS132[SPT16 TRP1] |
| MSY3993 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ spt5-15::KlURA3 pRS314 |
| MSY3994 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ spt5-15::KlURA3 pMSS107[SPT5 TRP1] |
| MSY3995 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT5-18::KlURA3 pRS314 |
| MSY3996 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1Δ SPT5-18::KlURA3 pMSS107[SPT5 TRP1] |
| MSY3999 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11/his3-11 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 htz1::URA3/htz1Δ SPT5/SPT5-18::KlURA3 |
| MSY4002 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1::URA3 pRS314 |
| MSY4003 | MATaade2-1 ade3::hisG can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 htz1::URA3 pMSS132[SPT16 TRP1] |
| MSY4447 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 pRS314 |
| MSY4448 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 htz1::URA3 pMSS107[SPT5 TRP1] |
| MSY4465 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 GAL10p-VPS13::TRP1 HA-HTZ1 |
| MSY4100 | MATα can1::Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 htz1::NatR |
| MSY4506 | MATαcan1::Prom-STE2-Sphis5 lyp1Δ cyh2 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 htz1::NatRswr1::HphR |
Plasmids.
Plasmid pRS314::KlURA3, used in recombination-mediated PCR-directed mutagenesis, contains the Kluyveromyces lactis URA3 gene cloned into the EcoRI site of pRS314 (kindly provided by M. Christman). Plasmid pMSS102 contains the ADE3 gene cloned into pRS425. Plasmid pMSS104, used to provide HTZ1 in the mutagenesis of the SPT genes, has a 1.5-kb SalI-HindIII (blunted) genomic fragment containing HTZ1 cloned into the SalI-SmaI sites of pMSS102. Plasmid pMSS107 was constructed by cloning an NsiI 4.3-kb fragment from ATCC clone 70978 containing SPT5 into the PstI site of pRS314. Plasmid pMSS110 is the BseRI gapped version of pMSS107 used to rescue the mutation in spt5-28. A 4,667-bp BamHI fragment from plasmid pCC48 (kindly provided by Fred Winston) containing SPT16 was cloned into pRS314 to construct pMSS132. A 5-kb PCR fragment amplified from spt16-18 genomic DNA and digested with AatII-BglII was cloned into pMSS132 to generate pMSS137. Plasmid pMSS115, the disruption construct for ELP3, was constructed by cloning a HincII kanamycin resistance (KanR) cassette between the HincII sites of ELP3. The disruption plasmid for DST1, pMSS86, was constructed by cloning a BglII-XhoI (blunted) fragment from a KanR cassette into the BglII-BsmI (blunted) sites of DST1.
Synthetic lethal screens.
Recombination-mediated PCR mutagenesis was carried out as described previously (80). The yeast strain MSY1786 was created to screen for mutations that are synthetic lethal with htz1Δ by colony color sectoring (10, 38, 47). This strain has a deletion of HTZ1 and carries ade2 and ade3 mutant alleles as well as a plasmid containing HTZ1, ADE3, and LEU2. Cells that carry ade2 are blocked in the purine biosynthetic pathway and accumulate a red imidazole intermediate. However, the ade3 mutation is epistatic to ade2, blocking the purine biosynthetic pathway at an upstream step, and therefore ade2 ade3 double mutants form white colonies (38, 47). In the absence of selection for the plasmid the host yeast strain gives red/white sectored colonies. Mutations that confer synthetic lethality together with htz1Δ make cells dependent on the plasmid and thus give nonsectoring red colonies.
Analysis of SPT16-18htz1.
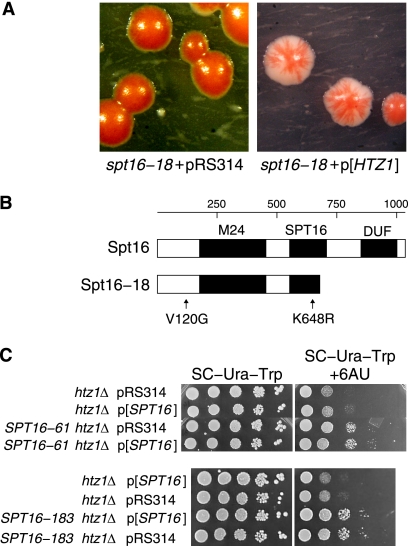
Sequence analysis of spt16-18 revealed several sites at which a mixture of wild-type and mutant nucleotides were recovered, suggesting heterozygosity. Flow cytometry of its DNA content confirmed that the spt16-18 strain contained diploid DNA, suggesting that the mutated spt16-18 DNA fragment integrated into a cell that had undergone diploidization either prior to or during the transformation to become MATa/MATa SPT16/spt16-18. The MATa/MATa diploid was crossed to a MATα/MATα strain, and the resulting tetraploid strain was dissected down to haploid spores through two rounds of sporulation. Dissection of tetrads from the intermediate MATa/MATα SPT16/spt16-18 diploid produced a 2:2 ratio of live to dead spores in which the viable segregants carried wild-type SPT16. These results show that spt16-18 is nonviable on its own and recessive to wild-type SPT16 but causes either haploinsufficiency or dominant synthetic lethality in the absence of HTZ1. Haploinsufficiency was ruled out by the observation that introduction of SPT16 on a plasmid into the SPT16/spt16-18 diploid failed to restore growth and red/white sectoring in the absence of HTZ1. We PCR amplified a DNA fragment spanning the spt16-18 KlURA3 locus and integrated it by one-step gene replacement into a homozygous htz1Δ/htz1Δ SPT16/SPT16 diploid carrying wild-type HTZ1 on a plasmid. The resulting heterozygous SPT16/SPT16-18htz1 strain required the HTZ1 plasmid for viability, confirming the dominant synthetic lethality of SPT16-18htz1.
Synthetic genetic array analysis.
Yeast strains for the specialized miniarrays were manually selected from the systematic gene knockout collection created in the MATa BY4741 background (26) (EUROSCARF, Institute for Molecular Biosciences, Frankfurt, Germany). Deletions having negative genetic interactions with htz1Δ and set3Δ were chosen based on annotations in the Saccharomyces Genome Database (http://www.yeastgenome.org). Synthetic genetic array analysis was carried out as described previously (96), crossing htz1Δ (MSY4100), and htz1Δ swr1Δ (MSY4506) query strains against the miniarray deletion collections. The growth of the final haploid pinned colonies was recorded by digital photography, and colony sizes were determined using custom software based on the Python Imaging Library (Pythonware). Colony sizes were normalized across different plates using a set of control strains arrayed with the test sets.
Antibodies and immunodetection.
Yeast whole-cell extracts and nuclei were prepared as described elsewhere (104). Approximately 30 μg of protein was used in SDS-PAGE gels. Anti-HA antibody 12CA5 (University of Virginia Hybridoma Center) was used at 1:2,000 (0.5 μg/ml) dilution. Antibodies 8WG16, H14, and H5 (Covance) were used at dilutions of 1:500, 1:10,000, and 1:1,000, respectively. The 8WG16 and 12CA5 antibodies were detected using horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG at 1:10,000 (Amersham Pharmacia Biotech). The H14 and H5 antibodies were detected using conjugated HRP-donkey anti-mouse IgM at 1:10,000 (Jackson Immunoresearch Laboratories). Rabbit anti-glucose-6-phosphate dehydrogenase (Sigma) was used at 1:5,000. This antibody was detected with HRP-conjugated donkey anti-rabbit IgG (Amersham Pharmacia Biotech) at 1:5,000 dilution.
ChIP.
Strains were grown to a density of 1.5 × 107 to 1.6 × 107 cells/ml in 2% raffinose media, and then galactose was added to a final concentration of 2% and cells were incubated for 1 h. Cells were fixed with 1% formaldehyde for 1 h (Elp3-HA, Spt5-HA, C-terminal domain [CTD], phospho-Ser5 [Ser5-P]) or 20 min (Ser2-P). Subsequent steps in the Elp3-HA and Spt5-HA chromatin immunoprecipitation (ChIP) experiments were carried out as described previously (83). For Pol II-CTD, Ser5-P, and Ser2-P, after quenching the formaldehyde with 250 mM glycine for 5 min, cells were lysed with glass beads in FA lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate [DOC]) for Pol II and Ser5-P or FA lysis buffer containing 0.1% SDS for Ser2-P. Chromatin was sheared by sonication to an average size of 300 bp. One milliliter of chromatin solution containing approximately 1 mg/ml of protein was incubated with 8WG16 antibody or H14 antibody (Covance) overnight at 4°C. For Ser2-P, 10 μl of H5 antibody (Covance) was preincubated overnight with 40 μl of magnetic beads coupled to a goat anti-IgM (Dynal), washed with phosphate-buffered saline (PBS), and then added to the chromatin solution overnight. For Ser5-P, chromatin solution containing H14 antibody was incubated with polyclonal goat anti-mouse IgM (Caltag) for 2 h at 4°C. Protein A-Sepharose beads (Pharmacia) were added to Pol II or Ser5-P samples, and samples were incubated for 2 h at room temperature. Samples were washed once for 5 min at room temperature with 1 ml of FA lysis buffer, i.e., FA buffer containing 500 mM NaCl, LiCl wash (250 mM LiCl, 1% NP-40, 1% DOC, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1), and twice with Tris-EDTA (TE). After reversal of cross-linking, DNA was precipitated and analyzed by PCR under conditions ensuring that the products were linearly proportional to the amount of input template. The sequences of the primers used for GAL1 are available on request. The sequences for primers in the intergenic region of chromosome V are as in reference 46.
For the GAL10p-VPS13 ChIPs, strain JSY677, generously provided by J. Svejstrup, was crossed to MSY1262 and one of the RPB3 copies of the resulting diploid strain was tagged with the HA epitope. Strains MSY3183 and MSY3188, used in the ChIP experiments, were spore segregants from dissection of that diploid strain. Cells were grown to a density of 0.8 × 107 to 1 × 107 cells/ml in rich media containing 2% raffinose as the carbon source, and then galactose was added to a 2% final concentration for 2 h (time [t] = 0), followed by addition of glucose to a 4% final concentration. At 1-min time intervals following glucose addition, cells were fixed in 1% formaldehyde for 20 min and quenched with glycine at a final concentration of 125 mM. Cells were processed for ChIP as described above. Antibody 12CA5 against HA-tagged RPB3 was preincubated overnight with protein-Dynal magnetic beads, washed, and then added to chromatin extracts for an overnight incubation at 4°C. PCRs were set up using SYBR green master mix (Applied Biosystems) and carried out in a 7300 ABI system.
Estimation of Pol II elongation rates.
The rates for Pol II runoff elongation were estimated assuming a constant rate specific for each genotype. The sets of Pol II ChIP occupancy data for each strain were fit to the function S(t) = A/ [1.0 + e−(t − t1/2)/σ] + b, where A is the amplitude of the ChIP signal, t is the time after glucose addition, t1/2 is the time when the initial Pol II occupancy at a probe has decreased by one-half, σ is the decay constant defining the sharpness of Pol II passage, and b is an offset for the background ChIP signal. The inflection points of the curves at the set of ChIP probes, t1/2, are related by the function t1/2 = (p/r) + δ, where r is the elongation rate in kilobases per minute, p is the position of the ChIP probe in kilobases, and δ is a time constant for the phenotypic lag between the time of glucose addition and the start of elongation runoff. Twenty-eight data points were fit to five parameters (A, b, σ, r, and δ) separately for the HTZ1 and htz1 data using the Levenberg-Marquardt least-squares algorithm, as implemented in ScientificPython (http://dirac.cnrs-orleans.fr/plone/software/scientificpython/). The statistical variance in the fit was estimated by bootstrap analysis in which each of the fitted data points was perturbed by a randomly selected residual from the set of residuals to generate derived data sets. The least-squares fit was then calculated for each of 10,000 such data sets.
RESULTS
We first sought to determine whether transcription elongation was in fact impaired in the htz1Δ mutant. Previous systematic pharmacogenetic screens had revealed that htz1Δ single mutants have decreased resistance to 6-azauracil and mycophenolic acid (MPA) (17, 82). By inhibiting inosine 5′-monophosphate (IMP) dehydrogenase, both of these drugs decrease the pool of ribonucleotides in vivo, which is not well tolerated in mutants with known defects in transcription elongation (21, 35, 61, 64, 76). Thus, the hypersensitivity of the single htz1Δ mutant to these agents is potentially strong evidence for a role in elongation. However, both drugs also have other physiological targets that do not involve elongation (17, 82). Only 6-AU-sensitive cells that fail to induce the PUR5 gene, encoding IMP dehydrogenase, are actually defective in transcription elongation (88). Under permissive growth conditions PUR5 induction is only modestly affected by htz1. However, in the presence of hydroxyurea (HU), which blocks DNA replication, htz1Δ cells fail to induce PUR5 transcription, suggesting that Htz1 becomes important for PUR5 induction under conditions that make transcription-dependent chromatin remodeling necessary (55). We reasoned that if the 6-AU-sensitive phenotype of htz1Δ cells is at the level of Pol II elongation, then mutant alleles of dedicated elongation genes might either exacerbate that phenotype or, more tellingly, suppress the hypersensitivity.
Mutations in SPT5 both enhance and suppress htz1Δ phenotypes.
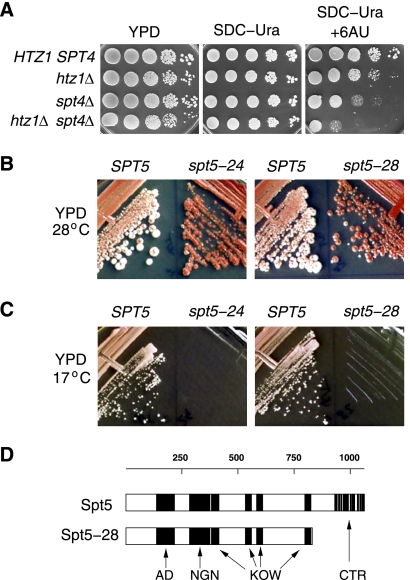
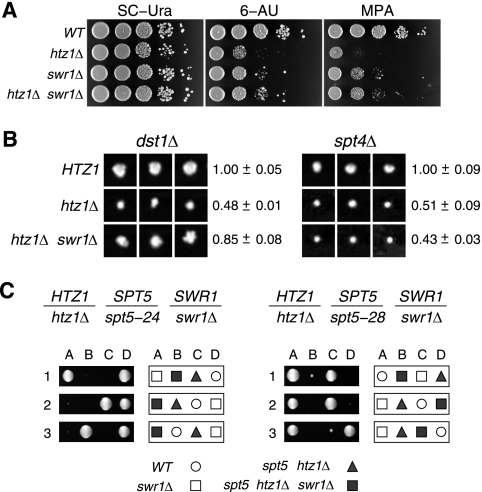
SPT4 and SPT5 encode subunits of a transcription elongation factor that is conserved from yeast to humans, where it is termed DSIF, and the complex is known to play both positive and negative roles in regulating elongation (103, 107). SPT4 is not an essential gene, and a previous deletion mutant screen identified spt4 as enhancing the slow-growth phenotype of htz1Δ (15). In agreement with that result, we find that htz1Δ spt4Δ double mutants in a W303 strain background exhibit synthetic hypersensitivity to 6-AU (Fig. 1A).
Fig. 1.
Synthetic interactions of htz1Δ with mutations in SPT4 and SPT5. (A) Serial dilutions of the cell cultures were plated on YPD and semidefined medium lacking uracil (SDC-Ura), with or without 6-AU (50 μg/ml). Wild-type and spt4 single mutants carried an integrated copy of the URA3 plasmid pRS306 to confer the uracil prototrophy required for growth in 6-AU plates. HTZ1 SPT4, MSY2178; htz1Δ, MSY2175; spt4Δ, MSY2180; htz1Δ spt4Δ, MSY2176. (B) The strains indicated were streaked on YPD plates and incubated at either 28°C or 17°C. Mutant alleles spt5-24 (MSY1803) and spt5-28 (MSY1804) are synthetic lethal with htz1Δ, and strains form solid red colonies. Transformation with an SPT5 plasmid complements the synthetic lethality, giving red/white sectored colonies. (C) Both spt5-24 and spt5-28 mutants are cold sensitive. Introducing wild-type SPT5 on a plasmid complements the cold sensitivity. (D) Schematic of the functional domains of the yeast Spt5 and the predicted Spt5-28 mutant protein showing the deletion of the CTR domain. AD, acidic domain; NGN, NusG, N terminus (InterPro IPR006645); KOW, Kyprides-Ouzounis-Woese motif (InterPro IPR005824); CTR, C-terminal repeat domain.
SPT5 is an essential gene, and therefore we targeted recombination-mediated PCR mutagenesis directly to the chro mosomal copy of SPT5 (80). Mutagenized cells were then screened for synthetic lethality, as revealed by a nonsectoring red colony morphology (see Materials and Methods). This strategy identified two alleles, spt5-24 and spt5-28, that conferred a severe slow-growth phenotype in combination with htz1Δ and a strong nonsectoring red phenotype (Fig. 1B). Both of the single mutants also displayed a severe cold-sensitive growth defect (Fig. 1C), a phenotype shared with other spt5 alleles characterized previously (35). Using a series of SPT5 plasmids, linearized to create gaps across the gene, we localized the mutation in spt5-28 to a 409-bp BseRI restriction fragment toward the 3′ end of the open reading frame (ORF) (see Materials and Methods). DNA sequence analysis of this segment revealed a single nucleotide substitution that changed the codon for residue 861 from leucine to an amber stop codon. This allele truncates the final 200-amino-acid C-terminal regulatory domain (Fig. 1D), which contains 15 copies of a 6-amino-acid repeat sequence (93).
Next, we screened for SPT5 alleles that suppress htz1Δ strain 6-AU hypersensitivity. This screen identified two alleles, SPT5-15 and SPT5-18 (Fig. 2). The SPT5-15 allele has mutations changing Asn920 to Asp and Ser925 to Ala, while SPT5-18 carries mutations changing Gln342 to Arg, Tyr725 to Cys, and Asp767 to Glu (Fig. 2D). In addition to suppressing the 6-AU sensitivity of htz1Δ cells, these alleles were also able to suppress the 6-AU sensitivity of the dst1 deletion mutant and partially suppress the severe defect of the htz1Δ dst1Δ double mutant (shown for SPT5-18 in Fig. 2A). This phenotype is not a bypass of 6-AU sensitivity since both SPT5-15 and SPT5-18 improve the growth of HTZ1 DST1 wild-type cells on drug plates, evident at the limit dilutions shown in Fig. 2A. The suppression phenotype is dominant, as demonstrated by the continued suppression of the 6-AU sensitivity in the htz1Δ/htz1Δ SPT5/SPT5-18 diploid strain (Fig. 2B). Identical results were obtained for SPT5/SPT5-15 cells (data not shown). Furthermore, introducing an episomal wild-type copy of SPT5 on a plasmid into either htz1Δ SPT5-15 or htz1Δ SPT5-18 haploid strains does not revert the suppression phenotype (Fig. 2C).
Fig. 2.
Suppression of htz1 by SPT5 mutations. (A) SPT5-18 suppresses the 6-AU sensitivity of htz1Δ, dst1Δ, and dst1Δ htz1Δ strains. Serial dilutions of the indicated strains are shown after 6 days of growth. HTZ1 DST1 SPT5, MSY2158; htz1Δ, MSY1798; dst1Δ, MSY2157; dst1Δ htz1Δ, MSY1795; SPT5-18, MSY2278; dst1Δ SPT5-18, MSY2279; SPT5-18 htz1Δ, MSY2280; SPT5-18 dst1Δ htz1Δ, MSY2281. (B) SPT5-18 is a dominant suppressor of htz1Δ 6-AU sensitivity. (Top) Strains MSY1793 (SPT5 htz1Δ) and MSY2280 (SPT5-18 htz1Δ) were crossed to obtain the SPT5/SPT5-18 htz1Δ/htz1Δ diploid (MSY3999). In this diploid, SPT5 does not revert suppression of the htz1Δ 6-AU sensitivity of SPT5-18. Growth is shown after 4 days of incubation. (Bottom) Four spore segregants of a tetrad from dissection of the diploid SPT5/SPT5-18 htz1Δ/htz1Δ strain are shown. Sensitivity to 6-AU segregates 2:2 as expected. (C) Transformation with wild-type SPT5 does not revert suppression of the htz1Δ 6-AU sensitivity of SPT5-15 and SPT5-18 strains. (Top) The htz1Δ strain (MSY1793) was transformed with either plasmid pRS314-SPT5 (MSY4448) or the pRS314 vector alone (MSY4447). Growth is shown on SDC-Trp-Ura plates with or without 6-AU after 4 days. (Bottom) Growth of SPT5-15 htz1Δ and SPT5-18 htz1Δ strains in the presence of 6-AU after 4 days. The strains were transformed either with plasmid pRS314-SPT5 (p[SPT5]) (MSY3994 and MSY3996) or with the pRS314 vector alone (MSY3993 and MSY3995). (D) The domain structure and locations of mutations in Spt5 proteins are shown. Labels are as in Fig. 1D.
Mutations in SPT16 both enhance and suppress htz1Δ phenotypes.
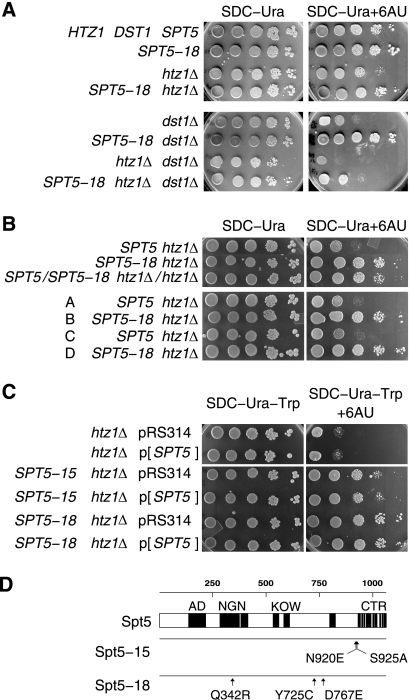
The FACT complex promotes transcription elongation on chromatin templates (23). In budding yeast, subunits of the FACT complex are encoded by SPT16, POB3, and NHP6A (13, 24). Previous work identified spt16-11 as synthetic lethal with htz1Δ (11), making SPT16 a good candidate for further analysis. We carried out a mutational screen of SPT16, similar to that for SPT5, and identified one synthetic lethal mutant, spt16-18, with unusual properties (Fig. 3A). Analysis revealed that this mutant strain had undergone spontaneous diploidization to MATa/MATa SPT16/spt16-18 (see Materials and Methods). Interestingly, spt16-18 alone is recessive, but in combination with htz1Δ it becomes dominant synthetic lethal. We will refer to the spt16-18 allele in this context as SPT16-18htz1. Using a primer pair in which the reverse primer was specific for the KlURA3 sequence, we amplified the spt16-18 locus and determined the DNA sequence of the mutant allele. From this analysis, spt16-18 is predicted to code for a truncated Spt16 protein due to the introduction of a stop codon in place of the Arg 670 codon. This truncation deletes the DUF1747 domain, which has homology to Rtt106 and other histone chaperone proteins, and partially deletes the conserved SPT16 motif. In addition, Spt16-18 is predicted to have two conservative amino acid substitutions: V120G and K648R (Fig. 3B). We were also able to identify spt16 alleles, such as SPT16-61 and SPT16-183, which suppress the 6-AU sensitivity of htz1Δ cells. Both mutant alleles are also dominant suppressors, as transformation of the haploid strains with a wild-type SPT16 plasmid fails to revert the 6-AU resistance (Fig. 3C).
Fig. 3.
Alleles of SPT16 genetically interact with htz1Δ. (A) The SPT16-18htz1 allele is dominant synthetic lethal with htz1Δ. Colonies of the SPT16-18/SPT16htz1 htz1Δ/htz1Δ (MSY2464) double mutant are nonsectoring, indicating that the cells require wild-type HTZ1 contained in the ADE3 plasmid to grow (left). Introduction of HTZ1 on a CEN ARS TRP1 plasmid restores sectoring in the double mutant (right). (B) Schematic of the functional domains of the yeast Spt16 protein and the predicted Spt16-18 mutant protein showing the sites of mutation. M24, peptidase M24 motif (InterPro IPR000994); SPT16, Spt16 motif (InterPro IPR013953); DUF, domain of unknown function (InterPro IPR013719) shared with Rtt106 and other histone chaperones. (C) Alleles of SPT16 suppress the 6-AU sensitivity of htz1Δ. (Top) The poor growth of the htz1Δ strain on media containing 6-AU is suppressed in the SPT16-61 htz1Δ strain, and the introduction of wild-type SPT16 on a pRS314-SPT16 plasmid (p[SPT16]) does not reverse the suppression. Growth is shown after 4 days. htz1Δ pRS314, MSY4002; htz1Δ p[SPT16], MSY4003; SPT16-61 htz1Δ pRS314, MSY3973; SPT16-61 htz1Δ p[SPT16], MSY3974. (Bottom) Similar results are shown for SPT16-183. SPT16-183 htz1Δ pRS314, MSY3979; SPT16-183 htz1Δ p[SPT16], MSY3980; other strains are as above.
Htz1 and SWR1 phenotypes.
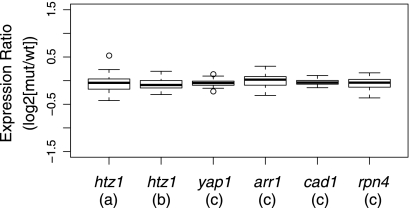
To evaluate whether Htz1 might function indirectly in elongation by activating the expression of elongation genes, we examined the 56 genes in budding yeast with the Gene Ontology annotation of “RNA elongation from RNA polymerase II promoter” (GO:0006368) and compared their expression profiles in two independent wild-type HTZ1 and htz1Δ data sets (65, 67). For comparison, the expression profiles of these same elongation genes were also examined in the data sets for deletions of four genes encoding specialized sequence-specific transcription factors that do not regulate elongation genes: YAP1, ARR1, CAD1, and RPN4 (36). As shown in Fig. 4, the range of gene expression ratios for htz1Δ cells is comparable to those for the control transcriptional activators.
Fig. 4.
Expression ratios of elongation genes. The 56 yeast genes with GO annotations for Pol II transcription elongation were compared for their expression ratios in two independent htz1Δ data sets and four site-specific DNA binding transcriptional activators. The distribution of normalized log2 expression ratios is shown in the box plots. Expression ratios within the second and third quartiles are boxed, and the whiskers extend to 1.5 times the interquartile range. The medians are shown as the solid bars in the boxes, and outliers are indicated by open circles. (a) Data for htz1Δ from reference 65; (b) data for htz1Δ from reference 67; (c) data for transcriptional activator genes YAP1, ARR1, CAD1, and RPN4 from reference 36 (Gene Expression Omnibus series GSE1798).
We also assessed the negative impact of the SWR1 complex on elongation phenotypes in the absence of its normal Htz1 substrates (32, 68). The deletion of SWR1 is partially able to suppress the sensitivity of htz1Δ cells to 6-AU and MPA (Fig. 5 A). The swr1Δ single mutant is hypersensitive to both drugs, consistent with a loss of Htz1 deposition, and the phenotype of the double htz1Δ swr1Δ mutant is similar to that of the swr1Δ mutant. Thus, the loss of Htz1 histone causes a major impairment of these drug-dependent pathways, and SWR1 contributes an additional partially negative impact. Deletion of SWR1 is also able to partially suppress the synthetic growth defect of htz1Δ dst1Δ cells, but, interestingly, swr1 has little to no effect on htz1Δ spt4Δ cells (Fig. 5B). Similarly, swr1 cannot suppress the synthetic lethality of htz1Δ spt5-24 and can only weakly suppress htz1Δ spt5-28 (Fig. 5C). These results indicate that pathways involving Spt4 and Spt5 are primarily sensitive to the loss of Htz1 itself and not the function of SWR1 in the absence of Htz1.
Fig. 5.
SWR1 partially contributes to elongation phenotypes in the absence of Htz1. (A) swr1Δ is sensitive to the elongation inhibitor drugs 6-azauracil (6-AU) and mycophenolic acid (MPA) and partially suppresses the hypersensitivity of htz1Δ. Tenfold serial dilutions of cultures of each of the indicated strains were spotted on permissive media (SC-Ura), 6-AU media, and MPA media. (B) swr1Δ partially suppresses the synthetic growth defect of htz1Δ dst1Δ double mutants but not htz1Δ spt4Δ double mutants. Synthetic genetic analysis was carried out on a miniarray of deletion strains that are synthetic lethal with htz1Δ (see Materials and Methods). The pinned spots from three independent biological replicates are shown for crosses of dst1Δ and spt4Δ strains against the strains indicated to the left. The averages and standard deviations of the colony growth sizes are shown to the right. (C) swr1Δ fails to suppress the synthetic lethality of htz1Δ spt5-24 and only weakly suppresses htz1Δ spt5-28. Diploid strains with the genotypes shown at the top were sporulated, and the resulting tetrads were dissected. The four spore colonies (A to D) from three representative tetrads (1 to 3) are shown for each cross. Keys to the relevant spore colony genotypes are shown at the bottom.
Htz1 is required for efficient phosphorylation of the CTD Ser2 in RNA Pol II.
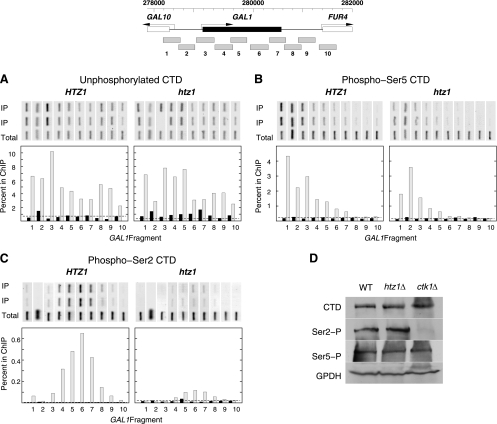
An attractive model for how promoter-localized Htz1 nucleosomes might function in downstream Pol II elongation is through the assembly and maturation of the productive elongation complex. To test this prediction, we assayed Pol II and other elongation factors at the inducible GAL1 gene by ChIP assays. We first examined the distribution of RNA Pol II at GAL1 using an antibody (8WG16) directed against the unmodified C-terminal domain (CTD) of Rpb1. As shown in Fig. 6A, the association of RNA Pol II with the GAL1 ORF recognized by this antibody depends on transcriptional activation by galactose. However, the levels and patterns of this association were similar in wild-type and htz1Δ cells.
Fig. 6.
Ser2 CTD phosphorylation over the GAL1 ORF is partially dependent on Htz1. (A to C) ChIP analysis of unphosphorylated CTD (antibody 8WG16) (A), phospho-Ser5 (antibody H14) (B), and phospho-Ser2 (antibody H5) (C) at the GAL1 locus in HTZ1 wild-type (MSY2301) and htz1Δ (MSY2302) strains. A diagram of the GAL1 locus, illustrating the locations of the 10 fragments assayed by PCR, is given at the top. For each panel, the PCRs are shown on top. Duplicate reactions were run for each immunoprecipitated sample (IP), and one reaction was performed for the total input chromatin sample (total). Quantification of the ChIP results is shown below as the percentages of input chromatin immunoprecipitated for the 10 fragments across the locus. Black bars, cells before galactose induction; gray bars, cells after 1 h of galactose induction. In each plot, the dashed line shows the ChIP results for a control nontranscribed region of chromosome V. The results shown are representative of three independent experiments for the unphosphorylated CTD, three for the Ser5-P CTD, and four for the Ser2-P CTD. (D) Global levels of Ser2, Ser5, and the unphosphorylated CTD are not affected by htz1Δ. A Western blot of whole-cell extracts of the indicated strains was probed with H5 (anti-Ser2), H14 (anti-Ser5), and 8WG16 (unphosphorylated CTD). Glucose-6-phosphate dehydrogenase was used as the loading control. The ctk1Δ strain is from the systematic deletion collection strain (EUROSCARF).
The phosphorylation of CTD Ser2 and Ser5 provides two well-characterized markers for different stages of Pol II elongation (14, 46). As shown in Fig. 6B, polymerase with Ser5-P was associated with the GAL1 gene only following galactose induction and it was preferentially localized to the 5′ end of the gene as expected. Furthermore, this pattern of association was found in both wild-type and htz1Δ strains. The results of Ser2-P assays were significantly different. In wild-type cells, CTD Ser2-P was preferentially cross-linked over the body of the GAL1 gene, as expected, and this association required galactose induction. However, in htz1Δ cells the relative level of CTD Ser2-P was severely reduced (Fig. 6C). The global levels of unphosphorylated CTD, Ser2-P, and Ser5-P in whole-cell extracts were not significantly affected by deletion of HTZ1, while deletion of CTK1 results in a global loss of Ser2-P as expected (Fig. 6D). These results indicate that Htz1 is important for the normal distribution of CTD Ser2-P in elongating Pol II at GAL1 and that this function acts at the level of the local chromatin environment.
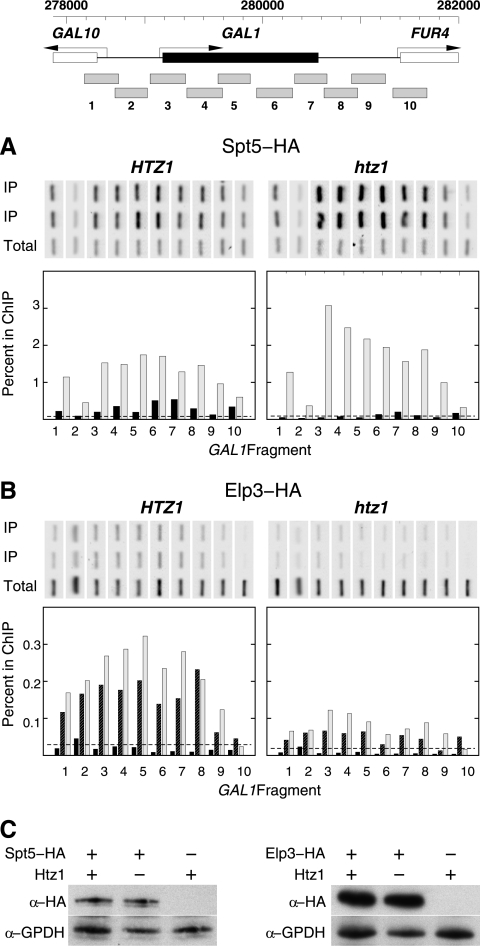
Htz1 affects the association of elongation factors with Pol II.
The htz1Δ-dependent defect in CTD Ser2-P predicted that there should be corresponding defects in associated elongation factors. To test this prediction, we assayed wild-type and htz1Δ cells for the loading of Spt5, which associates with Pol II independent of CTD phosphorylation (35, 52, 59, 75, 89). Spt5 protein was readily detected by ChIP assays over the body of the GAL1 gene following galactose induction, in agreement with previous studies (75). Surprisingly, in the absence of Htz1 we observed an increase in the ChIP signal for Spt5, particularly in the 5′-proximal region of the ORF (Fig. 7 A). Next, we tested the loading of Elongator, a multifunctional protein complex originally purified by its preferential association with the Ser2-P hypermodified form of elongating Pol II and known to physically associate with nascent RNA transcripts during elongation (27, 33, 72, 91). The Elp3 Elongator subunit was also detected at the GAL1 ORF following induction in wild-type cells (Fig. 7B). The ChIP signals for Elp3 were well above control levels in each of six independent experiments using different protocols and yeast strains, and Elp3 cross-linking for wild-type cells was both time and induction dependent. However, unlike that of Spt5, the association of Elp3 with GAL1 was severely impaired in cells with HTZ1 deleted (Fig. 7B). Western blot analysis showed that equivalent levels of Spt5 and Elp3 proteins were expressed in HTZ1 and htz1Δ cells (Fig. 7C). Taken together, these results suggest that the establishment or maintenance of a normal Pol II elongation complex, or both, are facilitated by chromatin containing histone variant Htz1.
Fig. 7.
Elp3 association with the GAL1 ORF is partially dependent on Htz1. Results of ChIP assays are shown as in Fig. 4. (A) ChIP analysis of Spt5-HA at the GAL1 locus in wild-type HTZ1 (MSY2304) and htz1Δ (MSY2305) strains. Black bars, cells before galactose induction; gray bars, cells after 1 h of galactose induction. (B) ChIP analysis of Elp3-HA at the GAL1 locus in HTZ1 wild-type (MSY2301) and htz1Δ (MSY2302) strains. The results shown are representative of five independent experiments. Black bars, cells before galactose induction; cross-hatched bars, cells 30 min after galactose induction; gray bars, cells after 1 h of galactose induction. (C) Elp3 and Spt5 levels as assayed by Western blotting are comparable in htz1Δ and wild-type (WT) strains. Nuclear extracts were prepared from the same strains used in the ChIP analyses. The untagged strain is MSY590. Blots were first probed with 12CA5 antibody against the HA epitope, then stripped and probed with antibody against glucose-6-phosphate dehydrogenase as the loading control. α, anti.
Pol II elongation is impaired in htz1Δ cells.
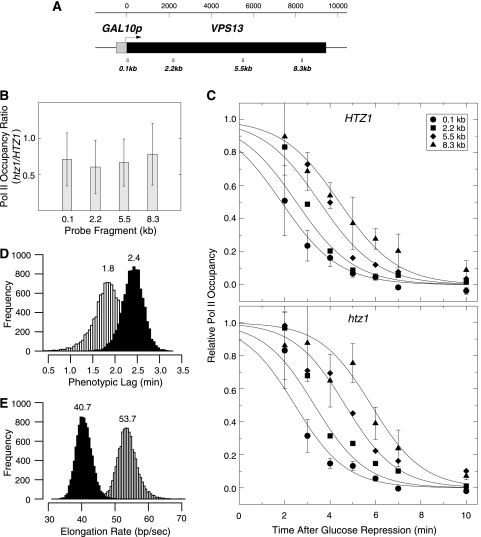
To directly measure the dependence of Pol II elongation rates on HTZ1, we assayed Pol II by kinetic quantitative chromatin immunoprecipitation (ChIP) (64) across the 9.5-kb VPS13 ORF driven by the inducible GAL10 promoter (GAL10p-VPS13) (Fig. 8) (48). Cultures of HTZ1 and htz1Δ cells were grown in raffinose and then induced with galactose for 2 h. Under conditions of active transcription on galactose, the relative distribution patterns of polymerase occupancy from 5′ to 3′ across the ORF were similar for both strains, suggesting similar processivities of Pol II; however, the ChIP signal for Pol II in htz1Δ cells was approximately 70% that in HTZ1 wild-type cells throughout (Fig. 8B), which may indicate decreased polymerase loading on the gene in the absence of Htz1 (1).
Fig. 8.
The rate of Pol II elongation on the GAL10p-VPS13 gene is decreased in the htz1Δ strain. (A) The positions of the four PCR fragments used in the ChIP analysis of Rpb3-HA cross-linking across the GAL10p-VPS13 open reading frame are shown. (B) The ratios of Pol II occupancy are shown for the htz1Δ mutant relative to the HTZ1 wild-type strain grown at steady state on galactose. Polymerase cross-linking is decreased in the htz1Δ strain, but the relative ratio is maintained across the ORF. (C) The normalized background-subtracted occupancy values of Pol II are shown for the four ChIP probe locations following repression of transcription by the addition of glucose at 0 min. The upper panel shows the results for the HTZ1 wild-type strain (MSY3183). The bottom panel shows the results for the htz1Δ mutant strain (MSY3188). The points shown are the averages of five independent experiments. The error bars are the standard deviations of the data, shown for the 0.1-kb and 8.3-kb locations for clarity. The data sets were fit to a sigmoid function of Pol II elongation through the probe locations to estimate the apparent elongation rates and the phenotypic lag between the addition of glucose and the start of Pol II runoff (see Materials and Methods). (D) The variance of the fit to the model shown in panel C for phenotypic lag was estimated by bootstrap analysis of 10,000 derived data sets perturbed by random permutation of the residuals to the fit (see Materials and Methods). A histogram of the resulting phenotypic lag estimates is shown. Open bars, HTZ1 wild type; filled bars, htz1Δ mutant. The apparent last wave of Pol II elongation began at 1.8 ± 0.3 min following glucose addition for the wild-type strain and at 2.4 ± 0.2 min for the htz1Δ mutant (P < 8 × 10−6). (E) The variance of the fit to the model shown in panel C for Pol II elongation rate is shown as the histogram of the fit to 10,000 derivative data sets. Open bars, HTZ1 wild type; filled bars, htz1Δ mutant. The apparent elongation rates are 53.7 ± 3.0 bp/s for the wild type and 40.7 ± 2.4 bp/s for the htz1Δ mutant (P < 3 × 10−15).
The GAL10 promoter was then repressed by the addition of glucose to the media, and Pol II occupancy was assayed by ChIP of its HA-Rpb3 subunit at 1-min intervals following repression using four probe locations across the ORF. These assays were repeated for five separate experiments, starting with independent cultures, and the ChIP signals were determined by quantitative PCR from the average of two technical replicates for each experiment. When transcription was repressed in HTZ1 wild-type cells, we observed a well-behaved Pol II depletion that progressed sequentially across the ORF as the last wave of elongating polymerase completed transcription of the gene (Fig. 8C). To estimate the rate of elongation, we modeled the Pol II runoff as a shock wave with a phenotypic lag, defined by a sigmoidal function, and determined the midpoint of the inflection at each location along the ORF (see Materials and Methods). The runoff of Pol II began at the 5′ ORF probe within approximately 1.8 min of glucose addition and then propagated across the ORF at 54 ± 3 bp/s (Fig. 8D and E). This estimate of elongation rate is consistent with previous estimates of the in vivo rate, which range from 20 to 100 bp/s (5, 16, 64, 71, 87, 97).
When transcription of GAL10p-VPS13 was repressed in htz1Δ mutant cells, we also observed a pattern of elongation runoff similar to that of the HTZ1 wild type. The runoff of elongating Pol II began at the 5′ ORF probe approximately 2.4 min after glucose addition. However, it took longer for the last wave of polymerase to clear the downstream 8.3-kb probe in htz1Δ cells than it did in HTZ1 wild-type cells. The elongation rate in htz1Δ cells is approximately 41 ± 2 bp/s. Thus, the average elongation rate for Pol II on GAL10p-VPS13 is approximately 24% slower in the htz1Δ mutant than in wild-type cells.
Htz1 decreases nucleosome occupancy during transcription.
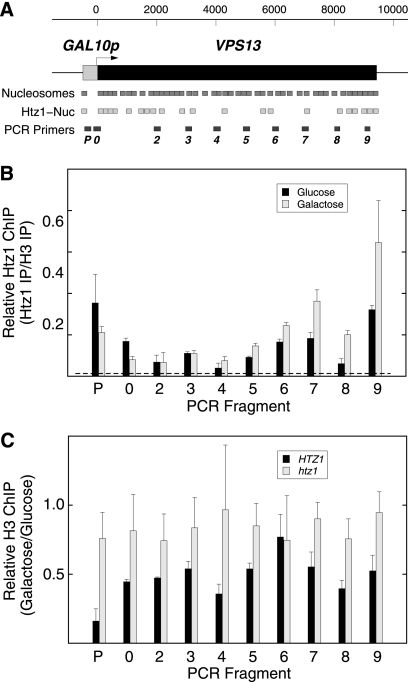
Pol II transcription from the GAL10 promoter normally displaces nucleosomes across the open reading frame, causing a decrease in bulk nucleosome occupancy as measured by ChIP (48, 86). We reasoned that in htz1Δ cells the replacement of Htz1-containing nucleosomes by canonical H2A nucleosomes might impede Pol II elongation because of their more stable occupancy (18, 108). Indeed, Htz1-containing nucleosomes have been detected over the normal VPS13 coding sequence in previous assays involving genome-wide ChIP followed by high-throughput sequencing (ChIP-seq) (2) (Fig. 9 A). Therefore, we first measured Htz1 occupancy across the GAL10p-VPS13 open reading frame by ChIP, using the occupancy of histone H3 to normalize for nucleosome density. The maps of total and Htz1-containing nucleosomes at the normal GAL10 promoter and over the wild-type VPS13 gene are diagrammed in Fig. 9A (2). Consistent with these maps and previous reports (29, 58, 83, 108), we found that the fraction of nucleosomes containing Htz1 is enriched in the promoter region relative to the open reading frame under glucose repression (Fig. 9B, black bars). Total nucleosome density decreases over the promoter when transcription is activated by galactose (Fig. 9C), and the fraction of the remaining nucleosomes that contain Htz1 decreases as well (Fig. 9B). Nevertheless, Htz1-containing nucleosomes were also readily detected above background (Fig. 9B, dashed line) across the VPS13 coding sequence under both activating and repressing conditions. Surprisingly, the fraction of nucleosomes containing Htz1 actually increased over the 3′ third of the gene in the presence of active transcription.
Fig. 9.
Htz1-containing nucleosomes decorate the GAL10p-VPS13 locus under inducing and repressing conditions. (A) The positions for the Htz1-containing nucleosomes and canonical nucleosomes are shown (2). At the bottom, the positions for the 10 PCR fragments used in the ChIP analysis of HA-Htz1 and H3 across the GAL10p-VPS13 locus are shown. (B) The ratio of Htz1 abundance versus H3 is shown for cells growing at steady state under repressing conditions on glucose and under inducing conditions on galactose. The data shown are the averages of two independent experiments. (C) The relative H3 occupancy across the GAL10p-VPS13 locus on galactose versus glucose is shown for HTZ1 and htz1 strains.
Finally, we measured the transcription-dependent decrease in nucleosome density in HTZ1 and htz1Δ strains (Fig. 9C). As expected from previous studies (48, 86), in HTZ1 cells H3 nucleosome density is reduced by approximately 50% over the VPS13 ORF and by 90% over the GAL10 promoter when transcription is activated with galactose (Fig. 9C, black bars). Remarkably, this transcription-dependent decrease in nucleosome occupancy is largely absent in htz1Δ cells, over both the promoter and open reading frame (Fig. 9C, gray bars). Taken together, these data are consistent with models in which Htz1-containing nucleosomes serve to facilitate the remodeling of chromatin during Pol II transcription elongation.
DISCUSSION
Histone H2A.Z is strongly implicated in the activation of transcription initiation (19, 30, 32, 34, 92, 99). The results of the experiments reported here argue that H2A.Z also influences one or more steps in transcription elongation. First, htz1Δ produces strong synthetic growth defects in combination with mutations in genes for the transcription elongation factors Spt5 and Spt16. Second, htz1Δ single mutants are hypersensitive to elongation inhibitors, and our genetic results suggest that the mechanism of this sensitivity is at the level of transcription elongation. The mutations in SPT5 and SPT16 that suppress 6-AU sensitivity create dominant alleles, consistent with active suppression of an htz1Δ-dependent elongation defect. Third, direct biochemical measurements at GAL10p-VPS13 show that htz1Δ reduces the rate of RNA Pol II elongation runoff by approximately 24% compared to that for the wild-type HTZ1 strain. Fourth, transcription-dependent nucleosome remodeling over the GAL10p-VPS13 ORF requires Htz1. Together, these results indicate that budding yeast cells experience actual defects in Pol II transcription elongation in the absence of histone H2A.Z nucleosomes.
The specific mutations in SPT5 and SPT16 reported here fit well with their known functions and domains. The recessive synthetic lethal allele, spt5-28, expresses a truncation of Spt5 that is missing the C-terminal repeat (CTR) domain. In human cells this region is phosphorylated by P-TEFb and is critical for the elongation activation role of DSIF (106). In yeast, the BUR kinase is important for the phosphorylation of Spt5 in vivo and can also phosphorylate the Spt5 CTR in vitro (109). It has been suggested that the CTR may serve as a platform to recruit other elongation factors such as the PAF complex (106, 109). The dominant suppressor SPT5-15 encodes a sequence with two mutations immediately upstream of the CTR, one of which introduces an acidic charge that theoretically could serve as a phosphomimetic. The second dominant suppressor, SPT5-18, also encodes a sequence with two amino acid substitutions. One of these, Q342R, falls in the NGN domain, which serves as the binding interface between Spt5 and Spt4 (31). We suggest that the Q342R substitution might further stabilize the NGN domain and activate the positive elongation function of the Spt4-Spt5 complex.
The SPT16-18htz1 allele is particularly intriguing. Aside from encoding two conservative amino acid substitutions, its main feature is that it encodes a truncation that deletes 357 residues from the C-terminal end of the protein. The truncation in Spt16-18 deletes a conserved acidic C-terminal domain that is shared with histone chaperones such as Rtt106. Deletion of this domain from Spt16 destroys the ability of reconstituted FACT to bind mononucleosomes and stimulate Pol II transcription on chromatin templates in vitro (9).
While the phenotypic properties of SPT5-15 and SPT5-18 suggest functional interactions with htz1Δ during Pol II elongation, it is important to note that SPT genes have been linked to transcription initiation and that, in particular, FACT has been shown to have roles in establishing transcription initiation complexes as well as modulating promoter accessibility (11, 12, 79, 94, 105).
There are at least two models for how Htz1 might positively regulate Pol II transcription elongation. First, we suggest that Htz1 may facilitate Pol II elongation by helping to establish the complete assembly, or correct modification state, of the elongation complex. Acting within promoter-proximal chromatin, the positional cues established by Htz1-containing nucleosomes (2, 29, 77) or direct factor recruitment (1, 28, 56) could provide space, time, or cofactors needed to assemble a fully functional elongation complex. In support of this model we have detected physical alterations in the elongation complexes formed in htz1Δ mutants, including increased association of Spt5 and decreases in both the phosphorylation of CTD Ser2 and the loading of Elongator. This model also fits well with the preferential occupancy of Htz1 within promoter chromatin.
A second model for the function of Htz1 in elongation is that it serves to modulate the properties of the nucleosomes encountered by Pol II as it traverses the open reading frame. Although Htz1 is generally concentrated at promoters genome wide, it is not entirely absent from ORFs and can actually be enriched within the ORFs of individual genes (2, 32, 39, 100). Acetylated Htz1-K14ac has also been observed over the ORF and positively correlates with the level of transcription (66). The altered stability of H2A.Z nucleosomes could then facilitate Pol II elongation just as it is thought to help remodel promoter chromatin for initiation (39, 40, 60, 100, 108). Consistent with this model, we found that the transcription-dependent decrease in nucleosome occupancy is strongly attenuated in htz1Δ cells. These results are in agreement with genome-wide studies which found that the global turnover of histone H3 is decreased in htz1Δ cells, especially in regions with the highest turnover (18). This dynamic cycling of nucleosomes could be driven by the exchange of Htz1 into canonical nucleosomes through the action of the SWR1 complex (60), and this activity provides an attractive explanation for the partial role for SWR1 in specific elongation phenotypes of htz1Δ that we observed.
These models are not mutually exclusive and could be interrelated. For example, it is formally possible that the increased nucleosome density in htz1Δ cells causes the change in the composition of the Pol II elongation complex. Alternatively, the defective elongation complex could be less efficient in remodeling nucleosomes for eviction, resulting in a higher nucleosome occupancy in ChIP assays. Further studies focused on the physical and temporal relationships between Htz1 and elongation cofactors and modifications will help to discriminate among alternate models. Nevertheless, the results of the genetic and molecular experiments reported here support the hypothesis that Htz1-containing chromatin has an important direct mechanistic role in facilitating Pol II transcription elongation.
ACKNOWLEDGMENTS
We thank Jesper Svejstrup for reagents and Carl Wu, Stefan Bekiranov, David Auble, and members of the Smith lab for helpful discussions.
This work was supported by an ASCB MAC Visiting Professorship Award to M.S.S., a Robert R. Wagner Fellowship to M.H., and grants GM28920 and GM60444 from the National Institutes of Health to M.M.S.
Footnotes
Published ahead of print on 28 February 2011.
REFERENCES
- 1. Adam M., Robert F., Larochelle M., Gaudreau L. 2001. H2A. Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albert I., et al. 2007. Translational and rotational settings of H2A. Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446:572–576 [DOI] [PubMed] [Google Scholar]
- 3. Altaf M., Auger A., Covic M., Cote J. 2009. Connection between histone H2A variants and chromatin remodeling complexes. Biochem. Cell Biol. 87:35–50 [DOI] [PubMed] [Google Scholar]
- 4. Amberg D., Burke D., Strathern J. 2005. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 5. Ardehali M. B., et al. 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28:1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armstrong J. A. 2007. Negotiating the nucleosome: factors that allow RNA polymerase II to elongate through chromatin. Biochem. Cell Biol. 85:426–434 [DOI] [PubMed] [Google Scholar]
- 7. Ausio J. 2006. Histone variants—the structure behind the function. Brief. Funct. Genomics Proteomics 5:228–243 [DOI] [PubMed] [Google Scholar]
- 8. Babiarz J., Halley J., Rine J. 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A. Z in Saccharomyces cerevisiae. Genes Dev. 20:700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belotserkovskaya R., et al. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090–1093 [DOI] [PubMed] [Google Scholar]
- 10. Bender A., Pringle J. R. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biswas D., et al. 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 25:4479–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biswas D., Yu Y., Prall M., Formosa T., Stillman D. J. 2005. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 25:5812–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brewster N. K., Johnston G. C., Singer R. A. 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21:3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cho E. J., Kobor M. S., Kim M., Greenblatt J., Buratowski S. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collins S. R., et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446:806–810 [DOI] [PubMed] [Google Scholar]
- 16. Darzacq X., et al. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14:796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desmoucelles C., Pinson B., Saint-Marc C., Daignan-Fornier B. 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 277:27036–27044 [DOI] [PubMed] [Google Scholar]
- 18. Dion M. F., et al. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405–1408 [DOI] [PubMed] [Google Scholar]
- 19. Draker R., Cheung P. 2009. Transcriptional and epigenetic functions of histone variant H2A.Z. Biochem. Cell Biol. 87:19–25 [DOI] [PubMed] [Google Scholar]
- 20. Feng Q., Zhang Y. 2003. The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 274:269–290 [DOI] [PubMed] [Google Scholar]
- 21. Fish R. N., Kane C. M. 2002. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577:287–307 [DOI] [PubMed] [Google Scholar]
- 22. Fleming A. B., Kao C. F., Hillyer C., Pikaart M., Osley M. A. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31:57–66 [DOI] [PubMed] [Google Scholar]
- 23. Formosa T. 2008. FACT and the reorganized nucleosome. Mol. Biosyst. 4:1085–1093 [DOI] [PubMed] [Google Scholar]
- 24. Formosa T., et al. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fry C. J., Peterson C. L. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185–R197 [DOI] [PubMed] [Google Scholar]
- 26. Giaever G., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 27. Gilbert C., Kristjuhan A., Winkler G. S., Svejstrup J. Q. 2004. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14:457–464 [DOI] [PubMed] [Google Scholar]
- 28. Gligoris T., Thireos G., Tzamarias D. 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol. Cell. Biol. 27:4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guillemette B., et al. 2005. Variant histone H2A. Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guillemette B., Gaudreau L. 2006. Reuniting the contrasting functions of H2A.Z. Biochem. Cell Biol. 84:528–535 [DOI] [PubMed] [Google Scholar]
- 31. Guo M., et al. 2008. Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation. Structure 16:1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halley J. E., Kaplan T., Wang A. Y., Kobor M. S., Rine J. 2010. Roles for H2A. Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 8:e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han Q., et al. 2008. Gcn5- and Elp3-induced histone H3 acetylation regulates hsp70 gene transcription in yeast. Biochem. J. 409:779–788 [DOI] [PubMed] [Google Scholar]
- 34. Hartley P. D., Madhani H. D. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hartzog G. A., Wada T., Handa H., Winston F. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haugen A., et al. 2004. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 5:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Henikoff S. 2008. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9:15–26 [DOI] [PubMed] [Google Scholar]
- 38. Hieter P., Mann C., Snyder M., Davis R. W. 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40:381–392 [DOI] [PubMed] [Google Scholar]
- 39. Jin C., Felsenfeld G. 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21:1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jin C., et al. 2009. H3.3/H2A. Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat. Genet. 41:941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kalocsay M., Hiller N. J., Jentsch S. 2009. Chromosome-wide Rad51 spreading and SUMO-H2A. Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 33:335–343 [DOI] [PubMed] [Google Scholar]
- 42. Kaplan C. D., Laprade L., Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096–1099 [DOI] [PubMed] [Google Scholar]
- 43. Keogh M., et al. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20:660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim J. H., Lane W. S., Reinberg D. 2002. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. U. S. A. 99:1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kobor M. S., et al. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A. Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Komarnitsky P., Cho E. J., Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koshland D., Kent J. C., Hartwell L. H. 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell 40:393–403 [DOI] [PubMed] [Google Scholar]
- 48. Kristjuhan A., Svejstrup J. Q. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krogan N. J., et al. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. U. S. A. 101:13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krogan N. J., et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721–729 [DOI] [PubMed] [Google Scholar]
- 51. Krogan N. J., et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565–1576 [DOI] [PubMed] [Google Scholar]
- 52. Krogan N. J., et al. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krogan N. J., et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kulaeva O. I., Gaykalova D. A., Studitsky V. M. 2007. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat. Res. 618:116–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larochelle M., Gaudreau L. 2003. H2A. Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J. 22:4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lemieux K., Larochelle M., Gaudreau L. 2008. Variant histone H2A. Z, but not the HMG proteins Nhp6a/b, is essential for the recruitment of Swi/Snf, Mediator, and SAGA to the yeast GAL1 UAS(G). Biochem. Biophys. Res. Commun. 369:1103–1107 [DOI] [PubMed] [Google Scholar]
- 57. Li B., Carey M., Workman J. L. 2007. The role of chromatin during transcription. Cell 128:707–719 [DOI] [PubMed] [Google Scholar]
- 58. Li B., et al. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 102:18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lindstrom D. L., et al. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Luk E., et al. 2010. Stepwise histone replacement by SWR1 requires dual activation with histone H2A. Z and canonical nucleosome. Cell 143:725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Malagon F., et al. 2006. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics 172:2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Malagon F., Tong A. H., Shafer B. K., Strathern J. N. 2004. Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation-elongation transition. Genetics 166:1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Martins-Taylor K., Sharma U., Rozario T., Holmes S. G. 26 October 2010, posting date H2A.Z (Htz1) controls the cell-cycle dependent establishment of transcriptional silencing at Saccharomyces cerevisiae telomeres. Genetics doi:10.1534/genetics.110.123844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mason P. B., Struhl K. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831–840 [DOI] [PubMed] [Google Scholar]
- 65. Meneghini M. D., Wu M., Madhani H. D. 2003. Conserved histone variant H2A. Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725–736 [DOI] [PubMed] [Google Scholar]
- 66. Millar C., Xu F., Zhang K., Grunstein M. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20:711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mizuguchi G., et al. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343–348 [DOI] [PubMed] [Google Scholar]
- 68. Morillo-Huesca M., Clemente-Ruiz M., Andujar E., Prado F. 2010. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One 5:e12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Narlikar G. J., Fan H. Y., Kingston R. E. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475–487 [DOI] [PubMed] [Google Scholar]
- 70. Ng H. H., Robert F., Young R. A., Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709–719 [DOI] [PubMed] [Google Scholar]
- 71. O'Brien T., Lis J. T. 1993. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 13:3456–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Otero G., et al. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109–118 [DOI] [PubMed] [Google Scholar]
- 73. Panchenko T., Black B. E. 2009. The epigenetic basis for centromere identity. Prog. Mol. Subcell. Biol. 48:1–32 [DOI] [PubMed] [Google Scholar]
- 74. Pavri R., et al. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703–717 [DOI] [PubMed] [Google Scholar]
- 75. Pokholok D. K., Hannett N. M., Young R. A. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799–809 [DOI] [PubMed] [Google Scholar]
- 76. Powell W., Reines D. 1996. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem. 271:6866–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Raisner R. M., et al. 2005. Histone variant H2A. Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Raisner R. M., Madhani H. D. 2006. Patterning chromatin: form and function for H2A. Z variant nucleosomes. Curr. Opin. Genet. Dev. 16:119–124 [DOI] [PubMed] [Google Scholar]
- 79. Ransom M., et al. 2009. FACT and the proteasome promote promoter chromatin diassembly and transcriptional initiation. J. Biol. Chem. 284:23461–23471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reid R. J., Lisby M., Rothstein R. 2002. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350:258–277 [DOI] [PubMed] [Google Scholar]
- 81. Ren Q., Gorovsky M. A. 2003. The nonessential H2A N-terminal tail can function as an essential charge patch on the H2A. Z variant N-terminal tail. Mol. Cell. Biol. 23:2778–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Riles L., Shaw R. J., Johnston M., Reines D. 2004. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21:241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Santisteban M. S., Kalashnikova T., Smith M. M. 2000. Histone H2A. Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411–422 [DOI] [PubMed] [Google Scholar]
- 84. Sarcinella E., Zuzarte P. C., Lau P. N., Draker R., Cheung P. 2007. Monoubiquitylation of H2A. Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 27:6457–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Saunders A., et al. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301:1094–1096 [DOI] [PubMed] [Google Scholar]
- 86. Schwabish M. A., Struhl K. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sehgal P. B., Derman E., Molloy G. R., Tamm I., Darnell J. E. 1976. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science 194:431–433 [DOI] [PubMed] [Google Scholar]
- 88. Shaw R. J., Reines D. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Simic R., et al. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Strahl B. D., Allis C. D. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 91. Svejstrup J. Q. 2007. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19:331–336 [DOI] [PubMed] [Google Scholar]
- 92. Svotelis A., Gevry N., Gaudreau L. 2009. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem. Cell Biol. 87:179–188 [DOI] [PubMed] [Google Scholar]
- 93. Swanson M. S., Malone E. A., Winston F. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11:3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Takahata S., Yu Y., Stillman D. J. 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 34:405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Thambirajah A. A., Li A., Ishibashi T., Ausio J. 2009. New developments in post-translational modifications and functions of histone H2A variants. Biochem. Cell Biol. 87:7–17 [DOI] [PubMed] [Google Scholar]
- 96. Tong A. H., et al. 2004. Global mapping of the yeast genetic interaction network. Science 303:808–813 [DOI] [PubMed] [Google Scholar]
- 97. Ucker D. S., Yamamoto K. R. 1984. Early events in the stimulation of mammary tumor virus RNA synthesis by glucocorticoids. Novel assays of transcription rates. J. Biol. Chem. 259:7416–7420 [PubMed] [Google Scholar]
- 98. Venkatasubrahmanyam S., Hwang W. W., Meneghini M. D., Tong A. H., Madhani H. D. 2007. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. U. S. A. 104:16609–16614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wan Y., et al. 2009. Role of the histone variant H2A. Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell. Biol. 29:2346–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Weber C. M., Henikoff J. G., Henikoff S. 7 November 2010, posting date H2A.Z nucleosomes enriched over active genes are homotypic. Nat. Struct. Mol. Biol. doi:10.1038/nsmb.1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wilmes G. M., et al. 2008. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell 32:735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Winkler G. S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U. S. A. 99:3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Winston F. 2001. Control of eukaryotic transcription elongation. Genome Biol. 2:reviews1006–reviews1006.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739–34742 [DOI] [PubMed] [Google Scholar]
- 105. Xin H., et al. 2009. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35:365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yamada T., et al. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21:227–237 [DOI] [PubMed] [Google Scholar]
- 107. Yamaguchi Y., Narita T., Inukai N., Wada T., Handa H. 2001. SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J. Biochem. 129:185–191 [DOI] [PubMed] [Google Scholar]
- 108. Zhang H., Roberts D. N., Cairns B. R. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhou K., Kuo W. H., Fillingham J., Greenblatt J. F. 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U. S. A. 106:6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]