Abstract
Genotyping of Francisella tularensis (A1a, A1b, A2, and type B) and Francisella novicida has identified multiple differences between species and among F. tularensis subspecies and subpopulations. Variations in virulence, geographic distribution, and ecology are also known to exist among this group of bacteria, despite the >95% nucleotide identity in their genomes. This study expands the description of phenotypic differences by evaluating the ability of F. tularensis and F. novicida to degrade chitin analogs and produce active chitinases. Endochitinase activities were observed to vary among F. tularensis and F. novicida strains. The activity observed for F. tularensis strains was predominantly associated with whole-cell lysates, while the chitinase activity of F. novicida localized to the culture supernatant. In addition, the overall level of chitinase activity differed among the subpopulations of F. tularensis and between the species. Bioinformatic analyses identified two new putative chitinase genes (chiC and chiD), as well as the previously described chiA and chiB. However, the presence of these four open reading frames as intact genes or pseudogenes was found to differ between Francisella species and F. tularensis subspecies and subpopulations. Recombinant production of the putative chitinases and enzymatic evaluations revealed ChiA, ChiB, ChiC, and ChiD possessed dissimilar chitinase activities. These biochemical studies coupled with bioinformatic analyses and the evaluation of chiA and chiC knockouts in F. tularensis A1 and A2 strains, respectively, provided a molecular basis to explain the differential chitinase activities observed among the species and subpopulations of Francisella.
INTRODUCTION
The second most abundant polysaccharide found in nature is chitin, a biopolymer composed of repeating units of β-1,4 linked N-acetyl-d-glucosamine (35). Chitin is synthesized by many eukaryotes and is an essential component of arthropod cuticles, insect peritrophic membranes, and fungal cell walls. The abundance of this biopolymer allows it to serve as a major environmental reservoir of carbon and nitrogen, and chitin is especially concentrated in marine environments. Several bacteria produce chitinases that depolymerize chitin into metabolically accessible subunits. However, chitinases are not unique to prokaryotes and can be found in vertebrates, arthropods, plants, protozoa, and fungi (22, 41). All chitinases possess one or more glycosyl hydrolase (GH) superfamily domains of the 18, 19, 20, or 48 superfamilies (6, 11). Nonenzymatic domains commonly found in chitinases such as N-acetylglucosamine-binding protein A, carbohydrate binding, and fibronectin type 3 domains facilitate depolymerization of or adherence to chitin (2, 21, 53). Individual chitinases are also differentiated based on whether they act as endo- or exochitinases (6). Thus, the form of hydrolyzing activity, organization of functional domains, primary amino acid sequence, and protein folds are all used to distinguish and subgroup individual chitinases (5, 6).
Chitinases are especially abundant in the gammaproteobacteria. This is likely due to the association of this bacterial class with marine environments (4). The gammaproteobacteria subdivision includes both saprophytic and pathogenic species (7, 9, 13, 28, 32, 47). However, the involvement of chitinases in bacterial infections or vector transmission has received limited evaluation. A study by Kirn et al. (21) found that a chitin-binding protein of Vibrio cholerae allowed for enhanced binding to glycan moieties on the surface of human epithelial cells. In Legionella pneumophila, a chitinase was shown to enhance bacterial survival in the lungs of mice, but the specific mechanism remains unknown (7). Adding to the potential importance of chitinases in human disease is the observation that chitinases enable vector based transmission of the protist Plasmodium falciparum (the etiological agent of malaria) by degrading the peritrophic membrane that surrounds the mosquito blood meal (51).
Tularemia is a potentially severe zoonotic infection that occurs in the northern hemisphere and is caused by the gammaproteobacterium Francisella tularensis (43). Infection with this bacterium occurs via contact with infected animal tissues or ingestion of contaminated water, inhalation of infectious aerosols, or an arthropod bite (ticks, flies, and mosquitoes) (38). Three species of Francisella are recognized—F. tularensis, Francisella novicida, and Francisella philomiragia—that share >91% nucleotide identity between all species and >95% between F. tularensis and F. novicida (3, 42). F. tularensis contains three subspecies, tularensis, holarctica, and mediasiatica. Of these subspecies, tularensis (also known as type A) and holarctica (also known as type B) are clinically relevant human pathogens. Epidemiological, molecular, and virulence data demonstrate that F. tularensis type A and type B strains have divergent ecologies, biochemical properties, and virulence characteristics (3, 19, 34, 38, 46). F. tularensis type A strains cause disease exclusively in North America, are maintained zoonotically in lagomorph/tick cycles, and cause the most severe form of tularemia (38). Molecular data have identified four subpopulations within F. tularensis type A: A1a, A1b, A2a, and A2b (10, 24). Epidemiological analyses and virulence studies further support the separation of F. tularensis type A (24, 34). Specifically, F. tularensis A1a and A1b infections occur primarily in the Eastern United States and are associated with higher mortality rates and severity of infection (highest in A1b infections) compared to A2 infections. A2 infections occur primarily in the Western United States, and differences in geographic distribution or virulence among the two subpopulations (A2a and A2b) have not been found (24, 34). Conversely, F. tularensis type B strains occur throughout the Northern Hemisphere, have close associations to water and rodents, are transmitted by mosquitoes, and cause a milder usually self-limiting form of tularemia (38). F. tularensis type B subpopulations exist based on molecular typing (8, 16, 27, 37, 52) and geographic distribution (23, 39, 40, 52); however, virulence differences among subpopulations have not been shown (39). F. novicida and F. philomiragia, in contrast to F. tularensis, are most commonly associated with marine environments but are not associated with arthropod vectors, have larger coding capacities, and are rarely implicated in human disease (3).
We previously identified genomic regions of difference between the F. tularensis A1 and A2 subpopulations (33). One of these regions of genomic variance, denoted RD12, included a putative chitinase gene chiA that was present in F. tularensis A1, but not A2 genomes. In a separate study, ChiA was described as highly upregulated (>20-fold) in vivo during a murine infection with F. tularensis A1a (50). In addition, two F. novicida chitinases (ChiA and ChiB) were shown to be secreted by a type II secretion system, to aid in biofilm formation on chitin surfaces, and to facilitate nutrient acquisition (13, 31). In this present study, chitinase production and activity were evaluated across the F. tularensis subspecies and subpopulations (A1a, A1b, A2, and type B) and compared to that of F. novicida. This included two previously undefined chitinase gene products (ChiC and ChiD), as well as the previously studied ChiA and ChiB (13, 31). Despite the similarity among Francisella genomes, variability was observed between chitinase genes and gene products of the Francisella species and F. tularensis subspecies and subpopulations. These differences correlated to the chitinase activity of individual species and subpopulations, and individual chitinases presented unambiguous variability in activity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Francisella strains (Table 1) were grown from frozen stocks on cysteine heart agar supplemented with 9% chocolatized sheep red blood cells (CHAB) at 35°C for 48 h, followed by subculture onto CHAB for 24 h at 35°C. Francisella liquid cultures were grown using a 24 h CHAB subculture to inoculate modified Mueller-Hinton (MMH) broth (1, 33). Liquid cultures of Francisella were incubated at 35°C overnight with shaking at 160 rpm. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth, with shaking at 160 rpm or on LB agar. When needed, appropriate antibiotics were added to the media.
Table 1.
F. tularensis and F. novicida strains used in this study
| Francisella straina | Species | Subspecies | Subpopulationb | Source | Geographic origin |
|---|---|---|---|---|---|
| OK01-2528 | tularensis | A | A1a | Human | Oklahoma |
| MO02-4195 | tularensis | A | A1a | Human | Missouri |
| SCHU S4 | tularensis | A | A1a | Human | Ohio |
| MA00-2987 | tularensis | A | A1b | Human | Massachusetts |
| MD00-2970 | tularensis | A | A1b | Human | Maryland |
| WY96-3418 | tularensis | A | A2a | Human | Wyoming |
| NM99-1823 | tularensis | A | A2b | Human | New Mexico |
| MI00-1730 | tularensis | B | UK | Human | Michigan |
| KY99-3387 | tularensis | B | B.II | Human | Kentucky |
| LVS | tularensis | B | B.Br.LVS | Rat | Russia |
| GA99-3550 (U112) | novicida | Water | Utah | ||
| GA99-3548 | novicida | Human | Louisiana |
PCR and cloning of chitinase genes.
PCR amplification of chitinase genes was performed using 30 to 75 ng of genomic DNA, 2.5 U of PrimeSTAR HS DNA polymerase (Takara Bio, Inc., Otsu, Shiga, Japan), 1× PrimeSTAR buffer (Mg2+ plus), 200 μM concentrations of each deoxynucleoside triphosphate, and 1 μM concentration of each primer in a final reaction volume of 50 μl. Primers (see Table S1 in the supplemental material) were designed by using VectorNTI advance 11.0 software (Invitrogen, Carlsbad, CA) and included specific restriction enzyme sites for use in downstream cloning. PCR conditions were 98°C for 2 min, followed by 30 cycles of 98°C for 10 s, 45°C to 65°C for 5 s, and 72°C for 2.5 min. Amplicons were cloned by using a Zero Blunt TOPO PCR cloning kit (Invitrogen) according to the manufacturer's instructions. Engineered restriction enzyme sites were used to isolate the cloned chitinase gene from the TOPO vector and for ligation into the pET23b-(+) expression vector (Novagen, San Diego, CA). The expression plasmids are listed in Table S2 in the supplemental material. All clones were sequence verified by the Proteomics and Metabolomics Facility at Colorado State University using the primers listed in Tables S1 and S3 in the supplemental material.
Construction of chiA and chiC knockouts and complemented strains.
Constructs for in-frame deletions of chiA (FTMG_00598) in F. tularensis strain MA00-2987 (A1b) and chiC (FTW_0313) in F. tularensis strain WY96-3418 (A2) were made using the sacB-based allelic exchange vector pMP590 (29). Specifically, constructs pMP590-ΔchiA and pMP590-ΔchiC (see Table S2 in the supplemental material) were created by PCR amplification of 477- to 941-bp DNA fragments that encompassed both flanking regions and 65 to 120 bp of the 5′ and 3′ regions of chiA and chiC, followed by ligation of the corresponding amplicons into the multiple cloning site of pMP590. The primers used are listed in Table S4 in the supplemental material. Electroporation was used to transform competent F. tularensis cells with the pMP590-chiA or pMP590-chiC constructs (29). Plasmid DNA (0.1 to 1.0 μg) was added to 100 μl of competent cells, and electroporation was performed by using a Bio-Rad Gene Pulsar (Bio-Rad, Hercules, CA) with the following parameters: 2.0 kV, 25 μF, and 200 Ω. Electroporated cells were incubated in 1 ml of MMH broth for 3 h at 35°C with shaking (160 rpm). Transformants were selected by plating on MMH agar containing 5 μg of kanamycin/ml, followed by incubation at 35°C for 2 to 4 days. Individual clones were transferred to MMH plates containing 8% sucrose to select for homologous recombination events. Genomic DNA was isolated from the knockout strains, and the genomic fragment representing the regions of homologous recombination was amplified by PCR and sequence verified. The knockouts were termed ΔchiA and ΔchiC.
Deletion mutants were complemented by cloning the full-length target genes, chiA (FTMG_00598) and chiC (FTW_0313), into the shuttle vector pMP529 to create pMP529-chiA and pMP529-chiC, respectively (see Tables S2 and S4 in the supplemental material). Electroporation of the deletion mutants with the complementing plasmids was performed as described for wild-type (WT) F. tularensis. To select for transformants, electroporated cells were plated onto MMH plates containing 200 μg of hygromycin/ml and incubated for 2 to 4 days at 35°C. Complementation was confirmed by PCR amplification, sequencing, and Western blotting.
Recombinant chitinase production and purification.
To produce recombinant chitinases, expression vectors possessing individual chitinase genes (see Table S1 in the supplemental material) were transformed into E. coli BL21(DE3)/pLysS cells (Invitrogen). Recombinant clones were grown in 2 liters of LB broth with 100 μg of ampicillin/ml and 34 μg of chloramphenicol/ml at 37°C for 3 h while shaking at 160 rpm prior to the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cultures were grown for an additional 4 h, and the cells were harvested by centrifugation at 3,500 × g. Cells were suspended in 10 ml of breaking buffer (phosphate-buffered saline [PBS; pH 7.4], 1.2 μg of DNase I/ml, 1.2 μg of RNase A/ml, 1 μg of lysozyme/ml, and one Complete EDTA-free protease inhibitor cocktail tablet [Roche Applied Sciences, Mannheim, Germany] per 50 ml of buffer), placed on ice, and lysed by probe sonication using a Vibra Cell VCX750 sonicator (Sonics and Materials, Inc., Newton, CT). Sonication was performed at an amplitude setting of 30 with seven 60-s pulses and a 60-s pause between the pulse cycles. Unbroken cells and debris were removed by centrifugation at 12,000 × g, and the clarified lysate was applied to a 0.8-by-0.4-mm Poly-Prep (Bio-Rad) column prepacked with 1.5 ml of His-Resin (EMD Chemicals, Gibbstown, NJ) pre-equilibrated in binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]). Unbound proteins were eluted with 15 column volumes (CV) of binding buffer followed sequentially with 10 CV of wash buffer A (20 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]), 23 CV of wash buffer B (40 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]), and 20 CV of 10 mM Tris-HCl (pH 8.0). The bound recombinant protein was eluted with 5 ml of elution buffer (0.5 M imidazole, 10 mM Tris-HCl [pH 8.0]). All purification steps were performed at 4°C. The eluent of purified protein was dialyzed at 4°C against 10 mM ammonium bicarbonate using a 3,500-Da molecular mass cutoff dialysis membrane and concentrated by using a 10,000-Da molecular mass cutoff Amicon Ultra centrifugal filter unit (Millipore, Billerica, MA). Protein concentrations were determined by using the bicinchoninic assay (44). Samples were divided into aliquots and frozen at −80°C until further use.
Assays for chitinase activity.
To assess secreted chitinase activities of Francisella strains, the cells were grown in MMH broth (10 ml) overnight. Immediately prior to harvesting of the culture supernatant (CS), an aliquot (10 μl) of each culture was serially diluted and plated on CHAB agar in duplicate. The agar plates were incubated for 48 h at 35°C, and the colonies were enumerated. The CS was separated from the bacterial cells by centrifugation at 5,000 × g for 15 min, concentrated using a 10-kDa molecular mass cutoff Amicon Ultra centrifugal filter unit, and brought to a volume of 500 μl with PBS (pH 7.4). An aliquot of the processed CS corresponding to approximately 6.0 × 108 CFU of the original 10-ml culture was assayed against three chitin analogs (p-nitrophenyl N-acetyl-β-d-glucosaminide, p-nitrophenyl N,N′-diacetyl-β-d-chitobioside, and p-nitrophenyl β-d-N,N′,N″-triacetylchitotriose) provided in the colorimetric chitinase assay (Sigma) according to the manufacturer's instructions, except the activity was assessed after incubation of the enzyme reaction for 16 h at 35°C. The release of p-nitrophenol was measured at an absorbance of 405 nm using a Multiskan EX spectrophotometer with Ascent v2.6 software (Thermo-Fisher, Waltham, MA).
To evaluate cell-associated chitinase activity, Francisella strains were grown on CHAB agar for 24 h at 35°C. The cells were scraped from the agar and suspended in PBS to a turbidity of 0.6 to 0.7 measured by using a Microscan turbidity meter (Siemens Healthcare Diagnostics, Deerfield, IL). To ensure consistency and to normalize the enzymatic activity between each culture, the number of CFU in the cell suspension was determined as described for the CS. An aliquot (300 μl) of the cell suspension was subjected to seven freeze-thaw cycles using a dry ice-ethanol bath and a 37°C heat block. This process allowed for the lysis of F. tularensis without generating hazardous aerosols. Unbroken cells and the whole-cell lysates (WCL) were separated by centrifugation at 17,000 × g for 15 min. An aliquot of the WCL corresponding to approximately 1.6 × 108 CFU of the original cell suspension was applied to the colorimetric chitinase assay (Sigma), and the chitinase activity was measured as described for the CS. Chitinase activity was reported as the average of six assays (two technical replicates from three independent experiments). The background activity from the substrate incubated in the absence of CS or WCL was subtracted from all the activity values reported.
The enzymatic activity of individual recombinant chitinases (ChiA, ChiB, ChiC, or ChiD) produced in E. coli was determined by using a variation of the colorimetric chitinase assay. An aliquot (10 μl) of the recombinant chitinase at 1 μg/μl in 0.1 M sodium phosphate buffer (pH 5.9) was added to 90 μl of the appropriate chitin analog at a concentration of 267 μM in 0.1 M sodium phosphate (pH 5.9). Separate assays were conducted at ambient temperature and 37°C. The release of the p-nitrophenol was measured at 0, 15, 30, 45, 60, 90, 120, 150, 180, 210, 240, 300, 360, 420, 480, and 540 s during the enzymatic reaction. Three technical replicates were used for each time point and activity was reported as the average of these replicates. The enzymatic activity of the individual recombinant chitinases was confirmed using a glycol chitin substrate as previously described (7).
Antibody production, SDS-PAGE, and Western blotting.
Antisera were generated in rabbits to histidine tagged products of F. tularensis A1a ChiA (FTT_0715), F. tularensis A1a ChiB (FTT_1768c), and F. tularensis A2 ChiC (FTW_0313). All antisera were produced by SDIX (Windham, ME).
Recombinant proteins (0.1 μg) and WCL (15 μg) of F. tularensis or F. novicida strains were separated by SDS-PAGE using NuPAGE 4 to 12% Bis-Tris SDS-polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes by electroblotting (45). The nitrocellulose membranes were incubated in PBS (pH 7.4) containing 5% nonfat milk for 1 h and washed in PBS. Incubation of the primary antibody (anti-chitinase serum diluted 1:500 in PBS with 0.1% Tween 20 plus 5% nonfat dry milk or anti-histidine antibody diluted 1:5,000 [Qiagen, Valencia, CA]) with the nitrocellulose membranes was performed at ambient temperature with gentle shaking for 3 h, followed by washing with PBS (pH 7.4). The secondary antibody, alkaline phosphatase-conjugated goat anti-rabbit IgG (1:5,000; Calbiochem, Darmstadt, Germany), was applied for 1 h at room temperature with gentle shaking. A final series of PBS washes were performed, and antibody reactive proteins were detected using a solution of BCIP/NBT made with SigmaFAST tablets (Sigma). Western blot and SDS-PAGE images were digitized by using a ScanJet 4850 photo scanner (Hewlett-Packard, Palo Alto, CA).
Bioinformatic analyses of Francisella chitinases.
DNA and protein similarity searches were performed using BLASTN and BLASTP, respectively, against the nonredundant and whole-genome shotgun reads databases of the National Center for Biotechnology Information. To predict similarity to other known products, the largest product from each chitinase class (ChiA, ChiB, ChiC, and ChiD) of F. tularensis or F. novicida was used for BLAST similarity searches. When multiple gene products of the same chitinase class were of the same size, the F. novicida gene product was used in the BLAST search. Francisella genomes that were available in GenBank and used in the present study were: F. tularensis subsp. tularensis A1b strain MA00-2987 (accession number ABRI00000000), F. tularensis subsp. tularensis A1a strain SCHU S4 (accession number AJ749949), F. tularensis subsp. tularensis A2 strain WY96-3418 (accession number CP000608), F. tularensis subsp. holarctica OSU18 (accession number CP000437), F. tularensis subsp. holarctica LVS (accession number AM233362), and F. novicida strain GA99-3550/U112 (accession number CP000439). DNA and protein sequence alignments were performed using LALIGN (www.ch.embnet.org/software/LALIGN_form.html) and CLUSTAL W (www.ebi.ac.uk/clustalw/#). Conserved domains within the individual chitinase sequences of F. tularensis were identified using the NCBI Conserved Domain search software (concise display) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (30). The presence of GH18, GH19, GH20, and GH48 domains were identified in Francisella genomes using the UniProt database (www.uniprot.org). Signal peptide predictions were accomplished using the SignalP 3.0 software (http://www.cbs.dtu.dk/services/SignalP).
RESULTS
In vitro chitinase activity of F. tularensis (A1a, A1b, A2, and type B) and F. novicida.
To determine the chitinase activity of Francisella grown in vitro, endochitinase and exochitinase (chitobiosidase and N-acetylglucosaminidase) activities were evaluated by using a panel of 12 Francisella strains (Table 1) representing F. tularensis (A1a, A1b, A2, and type B) and F. novicida (Fig. 1). Comparisons were normalized to the cell numbers used to generate the WCL and CS. Endochitinase activity differed between the Francisella species and subpopulations (Fig. 1A and B). The A2 strains showed the highest level of endochitinase activity in the WCL (average A405 of 0.27), but displayed minimal activity in the CS (Fig. 1A and B). Endochitinase activity was detectible in the WCL of F. tularensis A1a/A1b strains; however, this activity was minimal. The WCL of F. tularensis type B strains displayed approximately half of the endochitinase activity of the F. tularensis A2 strains (average A405 of 0.13). In contrast, the endochitinase activity of F. novicida strains was observed in the WCL and CS, with the CS possessing the greatest activity (average A405 of 0.57) and the WCL activity (average A405 of 0.11) similar to that of F. tularensis type B strains. Little to no chitobiosidase or N-acetylglucosaminidase activities were detected in the WCL of F. tularensis or F. novicida strains (data not shown). However, both types of exochitinase activities were detectable in the CS of F. novicida (Fig. 1C and D). These data provide strong evidence of differential production of chitinases among the Francisella species and subpopulations and suggest underlying differences in their chitinase genetic profiles.
Fig. 1.
In vitro chitinase activities of F. tularensis (A1a, A1b, A2, and type B) and F. novicida strains. A panel of 12 characterized strains (three A1a, two A1b, two A2, three type B, and two F. novicida) were evaluated for endochitinase, chitobiosidase, and N-acetylglucosaminidase activity. (A) endochitinase activities of WCL; (B) endochitinase activities of CS; (C) chitobiosidase activities of CS; (D) N-acetylglucosaminidase activities of CS. The average absorbance at 405 from three biological replicates (two technical replicates) are reported as a dot (●), with the average of all replicates indicated by a bar (—). Little to no chitobiosidase or N-acetylglucosaminidase activities was detected in WCL.
F. tularensis and F. novicida encode four putative chitinase genes (chiA, chiB, chiC, and chiD).
To address whether genetic differences exist between Francisella species and subpopulations with respect to chitinase genes, a comprehensive bioinformatics analysis was performed. Genes encoding for chiA and chiB were previously described in F. novicida and F. tularensis (13, 26). Domain searches of the chiA and chiB gene products found they possessed a GH18 domain (PfamID PF00704), a characteristic domain of bacterial chitinases. Thus, a search for chitinase proteins was performed by BLAST analyses using the previously described Francisella ChiA and ChiB and their GH18 domains. This identified a total of four chitinase genes—chiA, chiB, chiC, and chiD—in F. tularensis and F. novicida (Fig. 2). The chiA gene product shared the greatest similarity with a probable chitinase of Polysphondylium pallidum (E value 7 × 10−51). The Francisella ChiB most closely resembled a chitinase from Saccharophagus degradans (E value 3 × 10−11), and the ChiC displayed the greatest similarity to a hypothetical chitinase of Aureococcus anophagefferens (E value 4 × 10−56). F. tularensis and F. novicida ChiD showed limited similarity to other known chitinases but most closely resembled a chitinase from Lactococcus lactis (E value 9 × 10−8). The GH18 of the Francisella ChiD, however, is similar (E value 2.65 × 10−79) to a specific type of GH18 domain (cd02871) previously identified in Bacillus circulans.
Fig. 2.
Domain features of F. tularensis (A1a, A1b, A2, and type B) and F. novicida chitinases. The relative positions of conserved domains were identified in the chitinase gene products of F. tularensis and F. novicida. Solid black boxes indicate the predicted signal peptide cleavage site, striped boxes indicate the location and completeness of the GH18 domain (truncated GH18 domains appear as a pentagon), open boxes indicate the position of fibronectin type 3 domains, gray boxes indicate carbohydrate binding domains, and dark gray chevrons represent incomplete N-acetylglucosamine-binding protein A domains. (A) chiA gene products; (B) chiB gene products; (C) chiC gene products; (D) chiD gene products. A similar chiC gene product was identified in the genome of F. novicida GA99-3548 and GA99-3549 but not F. novicida GA99-3550.
The presence of these four chitinase genes differed among F. tularensis subpopulations (A1a, A1b, A2, and type B), and F. novicida (Fig. 2). F. tularensis A1a and A1b possessed genes for chiA, chiB, chiC, and chiD with an altered chiB and chiC. Specifically, the chiC of F. tularensis A1a possessed a point mutation causing a premature stop codon and two predicted open reading frames (FTT_1592c and FTT_1593c) encoding products of 387 and 207 amino acids, respectively. The F. tularensis A1a FTT_1592c open reading frame encodes for the C-terminal portion of ChiC that includes a complete GH18 domain. This was found to be identical in the F. tularensis A1b genome (MA00-2987); however, it is currently annotated as one reading frame (FTMG_01551). The ChiB of the F. tularensis A1a/A1b subpopulations contained a 124-amino-acid N-terminal truncation that removed a partial N-acetylglucosamine-binding protein A domain and the conserved signal peptide for translocation. F. tularensis A2 appeared to encode a functional ChiC and ChiD. F. tularensis type B was predicted to have all four chitinase genes, but the chiC product possessed a 58-amino-acid C-terminal truncation that appeared to impact the essential GH18 domain. F. novicida strain U112 was predicted to only lack chiC. Two additional F. novicida genomes became available during the course of the present study: the genome for strain GA99-3548 (accession number ABAH00000000) and strain GA99-3549 (accession number AAYF00000000). Both of these F. novicida strains possessed a chiC. In addition to the overall absence or presence of the four chitinases, the domain structure between ChiA, ChiB, ChiC, and ChiD differed (Fig. 2). Detailed bioinformatic analyses are described in the supplemental material.
These in silico analyses predicted that each Francisella species, subspecies, and subpopulation possessed two or more functional chitinases. When applied to the differential endochitinase activities observed with the WCL of Francisella species, subspecies and subpopulations (Fig. 1), this bioinformatics data led to several hypotheses. (i) Enzyme kinetics of the individual chitinases or different levels of chitinase production in the various Francisella strains significantly influence the overall endochitinase activity; (ii) the strong chitinase activity of the A2 subpopulation is attributable to ChiC or ChiC in combination with ChiD; and (iii) the moderate cell-associated endochitinase activity of F. tularensis type B and F. novicida is a result of activity from ChiA, ChiB, or ChiD or a combination of these proteins.
Differential endochitinase activities of ChiA, ChiB, ChiC, and ChiD.
To establish the level of endochitinase enzyme activity associated with individual gene products, F. tularensis and F. novicida chitinases predicted to be functional (those with a complete or partially altered GH18 domain) were produced as recombinant proteins in E. coli and assayed for endochitinase, chitobiosidase, and N-acetylglucosaminidase function. The ChiA, ChiB, and ChiC proteins possessing a complete GH18 domain were positive for endochitinase activity (Fig. 3). The F. tularensis type B ChiC was found to be negative for all chitinase activity, a result that correlates with the 58-amino-acid C-terminal deletion that truncates the GH18 domain of this protein. The greatest endochitinase activity over a 9-min assay was observed for the ChiB product of F. tularensis A1a/A1b and type B and F. novicida. The full-length ChiC of F. tularensis A2 was slightly less active than the intact ChiB proteins. Interestingly, the recombinant F. tularensis A1a/A1b C-terminal ChiC fragment (FTT_1592c) was active but at decreased levels compared to the full-length A2 ChiC. The ChiA of F. tularensis A1a/A1b and type B and F. novicida yielded modest endochitinase activity (A405 values of 0.40, 0.38, and 0.40, respectively), and none of the ChiD proteins were found to possess chitinase activity. Only ChiA and ChiC chitinases displayed minimal measurable activity to the chitobiosidase analog, and N-acetylglucosaminidase activity was not observed for any chitinase enzyme (Fig. 3).
Fig. 3.
Chitinase activity and substrate specificities of F. tularensis (A1a/A1b, A2, and type B) and F. novicida recombinant chitinases. F. tularensis and F. novicida chitinases with complete or partial GH18 domains were produced as recombinants in a heterologous system and assayed for 9 min to determine their degree and specificity to analogs capable of distinguishing endochitinase (●), chitobiosidase (▵), and N-acetylglucosaminidase (♢) activities. The absorbance at 405 nm was determined for three technical replicates of each enzyme at 37°C. The average of the technical replicates is reported as a bar of the corresponding color and shading.
The enzyme kinetics of individual chitinases were evaluated to further define and differentiate these proteins (Fig. 4). ChiD was excluded from these analyses since no chitinase activity was associated with this protein. The kinetics of the ChiA proteins of F. tularensis A1a/A1b and type B and F. novicida were similar to one another (Fig. 4A), but these chitinases yielded a much slower rate of catalysis than ChiB or ChiC (Fig. 4D). The ChiB recombinant proteins of F. tularensis A1a/A1b and type B and F. novicida all presented similar kinetics, and these recombinant products provided the greatest activity over the 9-min endochitinase assay (Fig. 4B and D). Although the maximum release of p-nitrophenol from the endochitinase substrate by ChiC of F. tularensis A2 was slightly less than that observed with the most active ChiB protein, the ChiC protein had a dramatically higher rate of catalysis, with activity plateauing after 90 s. The F. tularensis A1a/A1b C-terminal ChiC fragment (FTT_1592c) containing the intact GH18 domain again presented significantly less activity than the full-length ChiC of F. tularensis A2, but its activity also plateaued at 90 s (Fig. 4C and D).
Fig. 4.
Comparative endochitinase kinetics of F. tularensis (A1a, A1b, A2, and type B) and F. novicida recombinant chitinases. Recombinants that tested positive for endochitinase activity (Fig. 3) were assayed with an endochitinase analog to determine the relative activity of each functional enzyme at 37°C. The average absorbance at 405 nm of three technical replicates is reported for time points at 0, 15, 30, 45, 60, 90, 120, 150, 180, 210, 240, 300, 360, 420, 480, and 540 s, except for ChiC, where activity plateaued at 90 s. The kinetics of ChiA recombinant proteins (A), ChiB recombinant proteins (B), and ChiC recombinant proteins (C) are presented, as well as a relative comparison of all functional chitinases (D).
To validate the observations made with the small chitin analogues and the colorimetric assay, a second assay with glycol chitin (a soluble polymeric chitin) as the substrate was performed (7). The presence or absence of chitinase activity for each of the recombinant chitinases was the same as that observed using the colorimetric assay (data not shown). To account for the neutral pI of the ChiD products, the glycol chitin assay was also performed at a neutral pH, and no activity was observed. In addition to test whether the form of the recombinant protein influenced activity, each chitinase was also produced as a recombinant product without a predicted signal peptide or without a histidine tag and then assayed for chitinase activity. The relative endochitinase activities between ChiA, ChiB, ChiC, and ChiD did not change with these other recombinant forms (data not shown).
Production of individual chitinases by F. tularensis (A1a, A1b, A2, and type B) and F. novicida.
Western blot analyses of WCL from in vitro grown F. tularensis and F. novicida were performed with antiserum generated against the recombinant proteins possessing endochitinase activity (Fig. 5). Antiserum specificity was confirmed using the individual recombinant chitinases (data not shown). A protein band of the correct molecular mass (83 kDa) reactive to anti-ChiA antiserum was detected in WCL of F. tularensis A1a/A1b and type B, but, as predicted by bioinformatics, not to F. tularensis A2. A ChiA product of ∼120 kDa was detected in F. novicida WCL. This was greater than the predicted mass of 95.5 kDa. The ChiB antiserum recognized products at a predicted mass of 79 kDa in the WCL of F. tularensis type B and F. novicida but did not identify a product in the F. tularensis A1a/A1b or A2. A protein band corresponding to ChiC was only detected in F. tularensis A2. Products corresponding to the F. tularensis A1a/A1b ChiC fragments or the inactive F. tularensis type B ChiC were not detected.
Fig. 5.
Chitinases produced in vitro by F. tularensis (A1a, A1b, A2, and type B) and F. novicida. Western blots of WCL (15 μg) of each Francisella strain with anti-ChiA antiserum, anti-ChiB antiserum, and anti-ChiC antiserum. Lanes 1 to 3, A1a strains OK01-2528, MO02-4195, and SCHU S4, respectively; lanes 4 and 5, A1b strains MA00-2987 and MD00-2970, respectively; lanes 6 and 7, A2 strains NM99-1823 and WY96-3418, respectively; lanes 8 to 10, type B strains KY99-3387, LVS, and MI00-1730, respectively; lanes 11 and 12, F. novicida strains GA99-3548 and GA99-3550, respectively.
It is noted that the ChiA of F. novicida migrated at a molecular mass of ∼120 kDa, although its predicted mass was ∼96 kDa. This aberrant migration is hypothesized to be a result of the chemical properties of the protein's amino acid sequence or an uncharacterized posttranslational modification. The recombinant F. novicida ChiA with a poly-His tag had a predicted molecular mass of 97 kDa but also migrated at ∼120 kDa as determined by SDS-PAGE (data not show). Thus, we are confident that the protein observed in Fig. 5 is the actual native ChiA and not an artifact. The ChiA gene of F. novicida also possesses several tandem repeat sequences that might have resulted in errors in the assembly of the gene sequence or annotation. However, we sequenced the PCR product used to generate the recombinant F. novicida ChiA and did not find differences with the reported gene sequence. This further suggests that the aberrant migration by SDS-PAGE is an intrinsic property of this protein.
Analysis of ChiA and ChiC knockouts of F. tularensis.
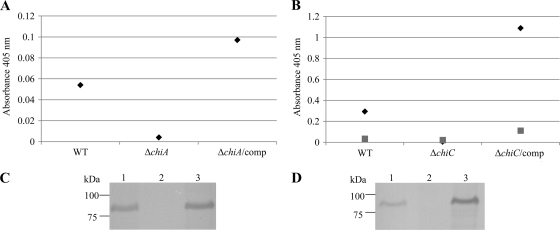
To demonstrate that the observed endochitinase activity of F. tularensis A2 was attributable solely to ChiC, a ΔchiC mutant and the corresponding complement ΔchiC/comp were generated in F. tularensis A2 strain WY96-3418 and evaluated for chitinase activity (Fig. 6B). The ΔchiC strain displayed minimal to no endochitinase activity (A405 of 0.005). When this mutant was complemented with chiC, the endochitinase activity was restored (A405 of 1.088) to a level greater than that of the WT F. tularensis A2 strain (A405 of 0.239). Western blot analysis with anti-ChiC antiserum verified the production of ChiC in WCL of the WT F. tularensis A2 and the ΔchiC/comp strains and confirmed its absence in WCL of the ΔchiC mutant (Fig. 6D). In addition, to determine whether the low endochitinase activity observed for F. tularensis A1a and A1b strains was provided by ChiA and not the fragmented ChiC, a ΔchiA mutation in A1b strain MA00-2987 and the corresponding complement ΔchiA/comp were generated (Fig. 6A). The endochitinase activity of the WT strain was low (A405 of 0.054) but was reduced further to an A405 of 0.004 in the ΔchiA mutant. The ΔchiA strain complemented with chiA yielded a level of chitinase activity moderately higher (A405 of 0.097) than the WT F. tularensis A1b strain (Fig. 6A). Western blot analyses with anti-ChiA serum verified the production of ChiA in WCL of the WT F. tularensis A1b and the complemented ΔchiA strains and demonstrated its absence in the ΔchiA mutant (Fig. 6C). These results indicate that the ChiC fragment of F. tularensis A1a/A1b is not active under the conditions tested and that the low endochitinase activity observed is due to ChiA, a protein that yields modest endochitinase activity as a purified recombinant product.
Fig. 6.
Analyses of ChiA and ChiC chitinase knockouts in F. tularensis A1b strain MA00-2987 and F. tularensis A2 strain WY96-3418, respectively. (A) Endochitinase (◆) activity of WCL from WT strain MA00-2987, ΔchiA MA00-2987, and ΔchiA/comp MA00-2987; (B) endochitinase (◆) and chitobiosidase (■) activity of WCL from WT strain WY96-3418, ΔchiC WY96-3418, and ΔchiC/comp WY96-3418. (C) Western blot with anti-ChiA against WCL from WT MA00-2987 (lane 1), ΔchiA MA00-2987 (lane 2), and ΔchiA/comp MA00-2987 (lane 3) strains. (D) Western blot with anti-ChiC against WCL from WT WY96-3418 (lane 1), ΔchiC WY96-3418 (lane 2), and ΔchiC/comp WY96-3418 (lane 3) strains.
DISCUSSION
The genomes of F. tularensis and F. novicida present >95% identity (3), and the genomes of F. tularensis strains are considered monomorphic with a pairwise average nucleotide identity of >99.2% (25). Despite this, genetic differences do exist among the Francisella species, and F. tularensis subspecies and subpopulations, and are reflected in the documented phenotypic variations within members of this genus (43). The present study now expands this phenotypic variability to include endochitinase activity. A detailed comparison of F. tularensis A1a, A1b, A2, and type B strains and F. novicida, coupled with evaluation of two previously identified chitinase genes (chiA and chiB) and two newly identified putative chitinase genes (chiC and chiD), provided a molecular basis for the observed chitinase phenotypes. Table 2 provides a summary of the chitinase gene products for each of the Francisella species, subspecies, and subpopulations. Among the four predicted chitinases, ChiD was not a factor in defining the variable chitinase activity. The chiD gene was ubiquitous in all strains evaluated, but the purified recombinant ChiD protein was the only putative chitinase that failed to display activity. Examination of the other predicted chitinases (ChiA, ChiB, and ChiC) revealed that ChiC was the only active chitinase in F. tularensis A2, accounting for the relatively robust endochitinase activity of the F. tularensis A2 strains. Both F. tularensis A2a and A2b strains were tested, indicating that this phenotype is conserved among the A2 subpopulations. In contrast, F. tularensis A1 strains had two predicted functional chitinases (ChiA and ChiB) and a fragment of ChiC with a functional GH18 family domain but barely detectible enzymatic activity. This weak activity was a result of ChiA, since neither ChiB nor the ChiC fragment were detected in the WCL of F. tularensis A1 strains grown in vitro. Compared to the F. tularensis type A1 and A2 subpopulations, F. tularensis type B had a dissimilar chitinase profile comprised of a predicted functional ChiA and ChiB. The differences between the chitinase profiles of F. tularensis type A and type B were not surprising given the number of phenotypic variations that exist among these two subspecies (3, 19, 34, 38). The low endochitinase activity of purified F. tularensis ChiA proteins and the dominant activity of the ChiB protein support the conclusion that the robust endochitinase activity observed for F. tularensis type B is attributable to ChiB. A subset of the type B subpopulations identified by Vogler et al. (52) were tested in the present study, suggesting that that ChiB activity is conserved among at least 2 of the 10 type B subpopulations. Unlike F. tularensis, F. novicida displayed high chitinase activity in the CS with less activity detected in the WCL. However, the WCL activity of F. novicida was similar to that of F. tularensis type B, and there were similar patterns of reactivity to the chitinase-specific antibodies. These data, along with the enzymatic assays of the recombinant products, suggest that the WCL endochitinase activity F. novicida is attributable to ChiB, just as in F. tularensis type B.
Table 2.
Summary of Francisella chitinase bioinformatics and activities
| Francisella group | Chitinase class | Predicted pseudogenea | Recombinant activity | In vitro production |
|---|---|---|---|---|
| F. tularensis | ||||
| A1a/A1b | ChiA | No | + | + |
| ChiB | No* | + | – | |
| ChiC | Yes/Yes† | –/+ | – | |
| ChiD | No | – | NTb | |
| A2 | ChiA | Yes‡ | NT | – |
| ChiB | Yes§ | NT | – | |
| ChiC | No | + | + | |
| ChiD | No | – | NT | |
| Type B | ChiA | No | + | + |
| ChiB | No | + | + | |
| ChiC | No¶ | – | – | |
| ChiD | No | – | NT | |
| F. novicida | ChiA | No | + | + |
| ChiB | No | + | + | |
| ChiC | No# | NT | – | |
| ChiD | No | – | NT |
The pseudogene designation is based on existing notations made to the annotated genome sequences. *, F. tularensis A1a/A1b ChiB has an N-terminal truncation resulting in deletion of the signal peptide and the N-acetylglucosamine-binding protein A domain but does not impact the GH18 domain. †, F. tularensis A1a/A1b chiC contains a point mutation that causes a premature stop codon and results in two predicted open reading frames. One reading frame (FTT_1592c) encodes an unaltered GH18 domain. ‡, F. tularensis A2 chiA is truncated due to a mutation resulting in a premature stop codon; the GH18 domain is missing. §, F. tularensis A2 chiB is truncated due to a mutation resulting in a premature stop codon; the GH18 domain is missing. ¶, F. tularensis type B ChiC contains a C-terminal truncation of that impacts the GH18 domain, but its gene is not annotated as a pseudogene. #, F. novicida strain GA99-3550 did not encode chiC. However, F. novicida GA99-3548 encoded a chiC whose product was predicted functional.
NT, not tested.
It was interesting that ChiB of F. tularensis A1 only differed from that of F. tularensis type B and F. novicida by a short truncation at its N terminus but was not observed by Western blotting. Likewise, the ChiC of F. novicida and F. tularensis type B were predicted to be nearly identical or only slightly truncated compared to the ChiC of F. tularensis A2, but only the ChiC of F. tularensis A2 was detected by Western blotting. It is possible that beyond the genetic lesions resulting in truncated chitinase proteins or pseudogenes, the regulatory mechanisms also may differ between species and subspecies of Francisella and result in differential chitinase gene expression or protein stability. These potential regulatory mechanisms were not a focus of these studies or would not be uncovered by the experimentation presented. However, they could possibly explain the unexpected Western blot data and should be a focus of future experiments.
The differential chitinase phenotypes of the Francisella species, subspecies, and subpopulations were not a simple reflection of the number of active chitinases produced but also resulted from variable activity among the predicted chitinases. Nevertheless, all of the putative intact F. tularensis and F. novicida chitinases contained GH18 family domains. The chiC fragments identified in F. tularensis A1a and A1b were annotated as pseudogenes, but the C-terminal encoding fragment contained an intact GH18 domain and was active as a recombinant product. On the contrary, chiC of F. tularensis type B was annotated as a functional gene; however, its GH18 domain was truncated and displayed no activity. The inactivity of ChiD is also likely a result of a nonfunctional GH18 domain. Functional GH18 domains contain a conserved catalytic motif of “DXDXE” where the glutamate residue acts as an acid critical for catalysis (14). Examination of this motif in the ChiD products (487-NFDLS-491) revealed the glutamic acid was replaced by a neutral serine residue. This, along with a nonconserved aspartic acid-to-asparagine substitution, likely inactivated this catalytic domain in ChiD.
Beyond GH18 domains, other regions of the individual F. tularensis chitinases are hypothesized to contribute to their variable activity. A fibronectin type 3 domain, was present only in the functional ChiA proteins and was positioned between the GH18 and carbohydrate binding domains, a common observation in other chitinases (48). This domain is thought to serve as a linker that adjusts the relative position of the chitinase catalytic and carbohydrate binding domains (48). Fibronectin type 3 domains of cellulases are also known to help disorganize polymers of cellulose (18, 53). Thus, it may play a similar role in chitin depolymerization. Tandem bacterial (type 3) carbohydrate binding domains were identified in ChiA and ChiC. These domains localize chitinases to their substrate and aid in chitin depolymerization (15, 49). The carbohydrate binding domains were not essential for the chitinolytic activity, as demonstrated with the C-terminal ChiC fragment of F. tularensis A1 strains, but a comparison of activity from this C-terminal fragment and the intact ChiC of F. tularensis A2 strains demonstrated that the presence of the nonenzymatic carbohydrate binding domains enhanced activity. A second binding domain (N-acetylglucosamine-binding protein A domain) was identified in ChiB and ChiD, though in a truncated form. This domain is present in the chitin-binding protein of V. cholerae and enhances bacterial attachment to chitin and human epithelial cells (21). The linkage of these bioinformatic analyses to the different activities observed for ChiA, ChiB, and ChiC further explains the dissimilar chitinase phenotypes of the various Francisella species and subpopulations. It also strongly suggests that each chitinases may act on different substrates or provide different biological functions.
Putative signal peptides were identified in all of the chitinases, except the ChiB and C-terminal fragment of ChiC in F. tularensis A1a/A1b. Despite this, F. tularensis chitinase activity was dominant in the WCL in contrast to F. novicida, where activity was focused in the CS. F. novicida chitinase secretion is dependent on a type II secretion system and in particular four “pilus” proteins (PilA [FTN_0415], PilB [FTN_1115], PilC [FTN_1116], and PilQ [FTN_1137]) (13). Homologues of these proteins are encoded by all Francisella strains analyzed in the present study, except F. tularensis LVS that lacks PilA (12). Thus, the absence of chitinase secretion in F. tularensis is hypothesized to result from differences in the expression and production of secretion machinery between F. novicida and F. tularensis. Alternatively, with the larger number of pseudogenes in F. tularensis compared to F. novicida, it is possible that one or more unidentified products essential for secretion are absent from F. tularensis (3, 42).
Our studies demonstrated that chitinase activity differed between Francisella species, subspecies, and subpopulations. Nevertheless, at least one chitinase was found to be functional within all Francisella strains examined, thus implying a need for this enzyme. F. tularensis type A strains are considered more virulent than type B strains, and virulence differences occur among the F. tularensis type A subpopulations (A1a, A1b, and A2) (24, 34). The chiA gene is highly upregulated (>20 times) in mice infected with an F. tularensis A1 strain FSC033 (50), but a knockout of chiA in a F. tularensis A1 strain revealed no difference in virulence compared to WT strains (17). These data suggest that F. tularensis A1 strains produce ChiA during in vivo growth for a function not directly linked to pathogenesis. No studies have been conducted to decipher the potential roles of ChiB and ChiC in F. tularensis virulence. However, virulence differences that exist between F. tularensis A1, A2, and type B strains (34) provide a justification to study ChiB and ChiC mutants in animal models. Ecological niche modeling also predicts that F. tularensis A1 and A2 subpopulations occupy distinct habitats (36). Thus, the differences in chitinase phenotypes between these two subpopulations should be studied with respect to pathogen maintenance and survival. Likewise, the more complete repertoire and higher in vitro secreted chitinase activities of F. novicida may reflect this bacterium's ecological niche as an environmental organism that only rarely causes infections within humans (20). The variability we have described in the structure and function of specific chitinases now provides a foundation to investigate the role of these proteins in the growth and survival of pathogenic F. tularensis subpopulations (A1a, A1b, A2, and type B) and the environmental F. novicida strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martin Pavelka for kindly supplying the genetic tools necessary for conducting this work and Matthew De Miranda for technical assistance.
This research was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant U54 AI065357). C.R.M. was funded for a portion of this study by CCID/ASM as a postdoctoral fellow.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Baker C. N., Hollis D. G., Thornsberry C. 1985. Antimicrobial susceptibility testing of Francisella tularensis with a modified Mueller-Hinton broth. J. Clin. Microbiol. 22:212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Champion M. D., et al. 2009. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 5:e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cottrell M. T., Wood D. N., Yu L., Kirchman D. L. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the alpha- and gamma-subclasses of the proteobacteria. Appl. Environ. Microbiol. 66:1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coutinho P. M., Deleury E., Davies G. J., Henrissat B. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307–317 [DOI] [PubMed] [Google Scholar]
- 6. Dahiya N., Tewari R., Hoondal G. S. 2006. Biotechnological aspects of chitinolytic enzymes: a review. Appl. Microbiol. Biotechnol. 71:773–782 [DOI] [PubMed] [Google Scholar]
- 7. DebRoy S., Dao J., Soderberg M., Rossier O., Cianciotto N. P. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U. S. A. 103:19146–19151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dempsey M. P., et al. 2007. Genomic deletion marking an emerging subclone of Francisella tularensis subsp. holarctica in France and the Iberian Peninsula. Appl. Environ. Microbiol. 73:7465–7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eppinger M., et al. 2007. The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 3:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farlow J., et al. 2005. Francisella tularensis in the United States. Emerg. Infect. Dis. 11:1835–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujita K., Shimomura K., Yamamoto K., Yamashita T., Suzuki K. 2006. A chitinase structurally related to the glycoside hydrolase family 48 is indispensable for the hormonally induced diapause termination in a beetle. Biochem. Biophys. Res. Commun. 345:502–507 [DOI] [PubMed] [Google Scholar]
- 12. Gil H., Benach J. L., Thanassi D. G. 2004. Presence of pili on the surface of Francisella tularensis. Infect. Immun. 72:3042–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hager A. J., et al. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227–237 [DOI] [PubMed] [Google Scholar]
- 14. Hsieh Y. C., et al. 2010. Crystal structures of Bacillus cereus NCTU2 chitinase complexes with chito-oligomers reveal novel substrate binding for catalysis: a chitinase without chitin binding and insertion domains. J. Biol. Chem. 285:31603–31615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ikegami T., et al. 2000. Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 275:13654–13661 [DOI] [PubMed] [Google Scholar]
- 16. Johansson A., et al. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 186:5808–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kadzhaev K., et al. 2009. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS One 4:e5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kataeva I. A., et al. 2002. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68:4292–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keim P., Johansson A., Wagner D. M. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105:30–66 [DOI] [PubMed] [Google Scholar]
- 20. Keim P. S., Wagner D. M. 2009. Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nat. Rev. Microbiol. 7:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirn T. J., Jude B. A., Taylor R. K. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866 [DOI] [PubMed] [Google Scholar]
- 22. Koga D., Mitsutomi M., Kono M., Matsumiya M. 1999. Biochemistry of chitinases. Exs 87:111–123 [DOI] [PubMed] [Google Scholar]
- 23. Kudelina R. I., Olsufiev N. G. 1980. Sensitivity to macrolide antibiotics and lincomycin in Francisella tularensis holarctica. J. Hyg. Epidemiol. Microbiol. Immunol. 24:84–91 [PubMed] [Google Scholar]
- 24. Kugeler K. J., et al. 2009. Molecular epidemiology of Francisella tularensis in the United States. Clin. Infect. Dis. 48:863–870 [DOI] [PubMed] [Google Scholar]
- 25. Larsson P., et al. 2009. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 5:e1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larsson P., et al. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153–159 [DOI] [PubMed] [Google Scholar]
- 27. Larsson P., et al. 2007. Canonical insertion-deletion markers for rapid DNA typing of Francisella tularensis. Emerg. Infect. Dis. 13:1725–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li X., Roseman S. 2004. The chitinolytic cascade in vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. U. S. A. 101:627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. LoVullo E. D., Sherrill L. A., Perez L. L., Pavelka M. S., Jr 2006. Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology 152:3425–3435 [DOI] [PubMed] [Google Scholar]
- 30. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Margolis J. J., et al. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl. Environ. Microbiol. 76:596–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McClelland M., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 33. Molins-Schneekloth C. R., Belisle J. T., Petersen J. M. 2008. Genomic markers for differentiation of Francisella tularensis subsp. tularensis A. I and A. II strains. Appl. Environ. Microbiol. 74:336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Molins C. R., et al. 2010. Virulence differences among Francisella tularensis subsp. tularensis clades in mice. PLoS One 5:e10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muzzarelli R. A. 1999. Native, industrial, and fossil chitins. EXS 87:1–6 [DOI] [PubMed] [Google Scholar]
- 36. Nakazawa Y., et al. 2010. Ecological niche modeling of Francisella tularensis subspecies and clades in the United States. Am. J. Trop. Med. Hyg. 82:912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pandya G. A., et al. 2009. Whole genome single nucleotide polymorphism based phylogeny of Francisella tularensis and its application to the development of a strain typing assay. BMC Microbiol. 9:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petersen J. M., Mead P. S., Schriefer M. E. 2009. Francisella tularensis: an arthropod-borne pathogen. Vet. Res. 40:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petersen J. M., Molins C. R. 2010. Subpopulations of Francisella tularensis subsp. tularensis and holarctica: identification and associated epidemiology. Future Microbiol. 5:649–661 [DOI] [PubMed] [Google Scholar]
- 40. Pilo P., Johansson A., Frey J. 2009. Identification of Francisella tularensis cluster in central and western Europe. Emerg. Infect. Dis. 15:2049–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robertus J. D., Monzingo A. F. 1999. The structure and action of chitinases. Exs 87:125–135 [DOI] [PubMed] [Google Scholar]
- 42. Rohmer L., et al. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sjostedt A. 2005. Family XVII. Francisellaceae genus I. Francisella, p. 200–210 In Brenner D. J., et al. (ed.), Bergey's manual of systematic bacteriology. Springer-Verlag, New York, NY [Google Scholar]
- 44. Smith P. K., et al. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76–85 [DOI] [PubMed] [Google Scholar]
- 45. Sonnenberg M. G., Belisle J. T. 1997. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infect. Immun. 65:4515–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Staples J. E., Kubota K. A., Chalcraft L. G., Mead P. S., Petersen J. M. 2006. Epidemiologic and molecular analysis of human tularemia, United States, 1964-2004. Emerg. Infect. Dis. 12:1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stover C. K., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 48. Toratani T., Kezuka Y., Nonaka T., Hiragi Y., Watanabe T. 2006. Structure of full-length bacterial chitinase containing two fibronectin type III domains revealed by small angle X-ray scattering. Biochem. Biophys. Res. Commun. 348:814–818 [DOI] [PubMed] [Google Scholar]
- 49. Tsujibo H., Kubota T., Yamamoto M., Miyamoto K., Inamori Y. 2003. Characterization of chitinase genes from an alkaliphilic actinomycete, Nocardiopsis prasina OPC-131. Appl. Environ. Microbiol. 69:894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Twine S. M., et al. 2006. In vivo proteomic analysis of the intracellular bacterial pathogen, Francisella tularensis, isolated from mouse spleen. Biochem. Biophys. Res. Commun. 345:1621–1633 [DOI] [PubMed] [Google Scholar]
- 51. Vinetz J. M., et al. 1999. The chitinase PfCHT1 from the human malaria parasite Plasmodium falciparum lacks proenzyme and chitin-binding domains and displays unique substrate preferences. Proc. Natl. Acad. Sci. U. S. A. 96:14061–14066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vogler A. J., et al. 2009. Phylogeography of Francisella tularensis: global expansion of a highly fit clone. J. Bacteriol. 191:2474–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Watanabe T., et al. 1994. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 176:4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.