Abstract
The opportunistic pathogen Pseudomonas aeruginosa forms biofilms, which render it more resistant to antimicrobial agents. Levels of iron in excess of what is required for planktonic growth have been shown to promote biofilm formation, and therapies that interfere with ferric iron [Fe(III)] uptake combined with antibiotics may help treat P. aeruginosa infections. However, use of these therapies presumes that iron is in the Fe(III) state in the context of infection. Here we report the ability of phenazine-1-carboxylic acid (PCA), a common phenazine made by all phenazine-producing pseudomonads, to help P. aeruginosa alleviate Fe(III) limitation by reducing Fe(III) to ferrous iron [Fe(II)]. In the presence of PCA, a P. aeruginosa mutant lacking the ability to produce the siderophores pyoverdine and pyochelin can still develop into a biofilm. As has been previously reported (P. K. Singh, M. R. Parsek, E. P. Greenberg, and M. J. Welsh, Nature 417:552-555, 2002), biofilm formation by the wild type is blocked by subinhibitory concentrations of the Fe(III)-binding innate-immunity protein conalbumin, but here we show that this blockage can be rescued by PCA. FeoB, an Fe(II) uptake protein, is required for PCA to enable this rescue. Unlike PCA, the phenazine pyocyanin (PYO) can facilitate biofilm formation via an iron-independent pathway. While siderophore-mediated Fe(III) uptake is undoubtedly important at early stages of infection, these results suggest that at later stages of infection, PCA present in infected tissues may shift the redox equilibrium between Fe(III) and Fe(II), thereby making iron more bioavailable.
INTRODUCTION
Iron is an essential component of many metabolically important proteins in living cells. Because we live in an aerobic world, it is often assumed that biologically available iron is present in its ferric [Fe(III)] form and thus extremely limiting; this is thought to be the case in diverse environments, ranging from seawater to soils to the human body (47, 58). This limitation arises from the fact that under aerobic, circumneutral pH conditions, Fe(III) is present as highly insoluble Fe(III) minerals or is tightly bound to mammalian host Fe(III)-sequestering proteins (1, 58). In the context of bacterial infections, much attention has been placed on controlling bacterial growth by limiting Fe(III) acquisition (4, 30, 68). This is especially relevant for Pseudomonas aeruginosa, an opportunistic pathogen that infects the lungs of people with cystic fibrosis (CF) (25). Not only is iron essential for its growth, but iron at levels in excess of what is required for growth promotes biofilm formation by signaling a transition from a motile to a sessile state (5, 7, 51, 62, 68). As a biofilm, P. aeruginosa becomes significantly more resistant to antimicrobial agents and much more difficult to eradicate (13), which leads to an onslaught of problems that compromise proper lung functioning, commonly resulting in the morbidity and mortality of CF patients (13, 45).
Traditionally, studies of iron acquisition by P. aeruginosa have focused on the Fe(III) uptake pathway mediated by heterogeneous siderophores, i.e., small molecules with high affinity for binding Fe(III) (53, 64). Although in laboratory experiments lactoferrin, an innate-immunity protein, was shown to inhibit P. aeruginosa biofilm development by sequestering Fe(III) from siderophores (5, 68), it is unclear whether this occurs in vivo, because such inhibition may be compromised due to cleavage of lactoferrin by cathepsins (62). Intriguingly, P. aeruginosa strains present in advanced CF infections can lose the ability to make the siderophore pyoverdine (16, 69), and the iron content of CF sputum is often highly elevated compared to that in the sputum of healthy controls (60). Nonetheless, the actual chemistry of iron in CF sputum is poorly defined and likely varies temporally for any given patient, from patient-to-patient, and within any given microenvironment. This is due to a complex set of interactions between the host and microbial cells, some of which are just beginning to be elucidated (23), that can affect the chemistry of iron and its release (59). That said, it is reasonable to assume that if lactoferrin is present in sufficient amounts to bind Fe(III) on mucosal surfaces, it would block biofilm formation at early stages of infection (5, 68). Building on this finding, a novel coculture system to permit the study of biofilm formation on CF lung epithelial cells showed that cells harboring the ΔF508 mutation (the most common mutation of the cystic fibrosis transmembrane conductance regulator, CFTR) release significantly more iron than wild-type CFTR cells, stimulating biofilm formation by P. aeruginosa. In this system, chelation of Fe(III) by conalbumin (a lactoferrin surrogate protein) dramatically reduced biofilm formation (45). A recent follow-up study using this cell culture system showed that the combined use of tobramycin and FDA-approved iron chelators reduced biofilm biomass by approximately 90% (46).
While there is good reason to be optimistic regarding the efficacy of therapeutics that interfere with Fe(III) acquisition at early stages of infection (4, 30, 46), an understudied and potentially important aspect of the problem is whether iron always exists in the ferric state as infections progress, especially as inflammation is stimulated, cell densities increase, and oxygen tensions in CF sputum decline (78). Although the total amount of extracellular iron can reach high values in CF sputum (59), the chemical nature of this iron (e.g., free, chelated to siderophores or some other microbial ligand, or heme) and what fraction is oxidized or reduced is unknown. What is known, however, is that many P. aeruginosa strains isolated from CF sputum make redox-active phenazine compounds (21, 69), that phenazines are present in CF sputum at concentrations in the 1 to 100 μM range (77), and that their concentration increases as lung function declines across a broad patient cohort (R. C. Hunter and D. K. Newman, unpublished data). Phenazines are capable of reducing a variety of high-reduction-potential oxidants, including Fe(III), through extracellular electron transfer (14, 27, 28, 54, 74). In vitro experiments have shown that phenazines can liberate Fe(III) from transferrin by reducing it to ferrous iron [Fe(II)] (14), as well as reducing highly insoluble Fe(III) (hydr)oxides to Fe(II) (27, 74). Thus, there is reason to speculate that at least some fraction of the total iron in CF sputum may be in the ferrous form and that efforts to iron limit P. aeruginosa through Fe(III) sequestration may be compromised by phenazine-promoted reduction of Fe(III) to Fe(II).
Although it has long been appreciated that P. aeruginosa may acquire iron via siderophore-independent pathways (48), whether phenazines might play a role in iron acquisition in the context of biofilm formation has not been studied. Here, we sought to test the hypothesis that phenazine-mediated Fe(II) acquisition through the ferrous iron uptake system (Feo [11, 40]) could alleviate Fe(III) limitation and stimulate biofilm formation.
MATERIALS AND METHODS
Chemicals.
Phenazine-1-carboxylate (PCA) was purified from aerobic stationary-phase cultures of Pseudomonas fluorescens strain 2-79 (NRRL B-15132) (70) grown in King's A medium (32) at 30°C, as previously described (74). Pyocyanin (PYO) was purified from aerobic stationary-phase cultures of P. aeruginosa strain UCBPP-PA14 (56) grown in Luria-Bertani (LB) broth at 37°C, as previously described (74). Pyoverdine, purified according to the method described in Albrecht-Gary et al. (2), and pyochelin, characterized and synthesized according to published procedures (80, 81), were provided by Schalk's group (Institut de Recherche de l'Ecole de Biotechnologie de Strasbourg, IREBS FRE3211 CNRS/Université de Strasbourg, France). Fe(OH)3(s), referred to as the Fe(III) mineral ferrihydrite, was synthesized according to the method by Schwertmann and Cornell (66), as previously described (74). Substantially iron-free conalbumin, 1,10-phenanthroline, hydroxylamine hydrochloride, ammonium acetate, ferrous ammonium sulfate, and carrier DNA for yeast transformation were purchased from Sigma-Aldrich. All enzymes used for DNA manipulation were purchased from New England BioLabs.
Strains, plasmids, primers, and growth conditions.
Tables 1 and 2 list the strains, plasmids, and primers used in this study. For planktonic and biofilm experiments with P. aeruginosa PA14 strains, 0.3 g/liter of Bacto tryptic soy broth (TSB; 1% TSB, where 1% is relative to the usual concentration of TSB medium [30 g/liter]; Becton Dickinson) was used as the medium. Where indicated, for the PA14 siderophore-null strain (ΔpvdA ΔpchE), 1.0 μM Fe(OH)3(s), 10 μM PCA or PYO, or 1.0 μM Fe(OH)3(s) together with 10 μM PCA or PYO was added to 1% TSB. Also as indicated, for the PA14 wild type and the feoB::MAR2xT7 mutant (from the nonredundant PA14 mutant library [38]), 10 μM PCA or PYO alone or 40 μg/ml of iron-free conalbumin alone or together with 10 μM PCA or PYO was added to 1% TSB. Flow cell biofilm experiments with the PA14 wild type and phenazine deletion (Δphz) strains were also performed in 1% TSB supplemented with 10 μM PCA. To confirm that in contrast to the PA14 wild type, the ΔpvdA ΔpchE strain was unable to produce pyoverdine and pyochelin during planktonic growth, an iron-deficient morpholinepropanesulfonic acid (MOPS)-based medium (100 mM MOPS at pH 7.2, 20 mM succinate, 93 mM NH4Cl, 43 mM NaCl, 2.2 mM KH2PO4, and 1 mM MgSO4 [modified from reference 50]) was used. These planktonic growth experiments were performed in acid-washed iron-free sterile glass culture tubes or polypropylene flasks at 37°C with vigorous shaking at 250 rpm to generate aerobic conditions. To confirm that the PA14 feoB::MAR2xT7 mutant was disrupted in ferrous iron transport, cells were incubated, with anaerobic shaking, in Amberlite-treated 1% TSB containing 100 mM KNO3, 50 mM glutamate, 1% glycerol, and 100 μM iron source [either (NH4)2Fe(II)(SO4)2 or Fe(III)Cl3] at 37°C for 22 h. Culture densities were monitored at an optical density at 500 nm (OD500) in a Thermo Scientific Spectronic 20D+ or Shimadzu UV-2450 spectrophotometer.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Propertiesa | Reference or sourceb |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PA14 | Clinical isolate UCBPP-PA14; wild-type strain | 56 |
| PA14 Δphz | PA14 with deletions of operons phzA1-G1 and phzA2-G2 | 17 |
| PA14 ΔpvdA ΔpchE | PA14 with deletions of pvdA and pchE | This study |
| PA14 Δphz ΔpvdA ΔpchE | PA14 with deletions of pvdA, pchE, and operons phzA1-G1 and phzA2-G2 | This study |
| PA14-YFP | PA14 constitutively expressing YFP from a Tn7 insertion created by introducing plasmid pAKN69 | L. E. P. Dietrich, MIT |
| PA14 Δphz-YFP | PA14 Δphz constitutively expressing YFP; analogous to PA14-YFP | L. E. P. Dietrich, MIT |
| PA14 ΔpvdA ΔpchE-YFP | PA14 ΔpvdA ΔpchE constitutively expressing YFP plasmid pAKN69 | This study |
| PA14 Δphz ΔpvdA ΔpchE-YFP | PA14 Δphz ΔpvdA ΔpchE constitutively expressing YFP plasmid pAKN69 | This study |
| PA14 feoB::MAR2×T7 | PA14 mutant with an insertion of the MAR2×T7 transposon in the PA14_56680 ORF, which is the homolog of the PAO1 ORF PA4358 | 38 |
| E. coli | ||
| UQ950 | DH5α λ pir host for cloning | D. P. Lies, Caltech |
| BW29427 | Donor strain for conjugation | W. M. Metcalf, UIUC |
| β-2155 | Donor strain for conjugation | 15 |
| S. cerevisiae | Ura− for gap repair cloning | Invitrogen |
| InvSC1 | ||
| Plasmids | ||
| pMQ30 | Yeast-based allelic exchange vector, sacB, CEN6/ARSH4, URA3+, Gmr | 67 |
| pUX-BF13 | R6K replicon-based helper plasmid providing the Tn7 transposition functions in trans; can replicate only when the pir gene is supplied in trans; Ampr | 6 |
| pAKN69 | Transposon delivery plasmid containing the mini-Tn7(Gm)PA1/04/03::eyfp fusion | 35 |
| pYW01 | pvdA deletion fragments cloned into pMQ30 | This study |
| pYW02 | pchE deletion fragments cloned into pMQ30 | This study |
ORF, open reading frame.
UIUC, University of Illinois at Chicago.
Table 2.
PCR primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| pvdAKO1 | CCA GGC AAA TTC TGT TTT ATC AGA CCG CTT CTG CGT TCT GAT AGC GCT GGA ACT CGC CAC |
| pvdAKO2 | GCT TCA GGT GCT GGT ACA GTG CCT GAG TCA TTT CCA GTT CC |
| pvdAKO3 | GGA ACT GGA AAT GAC TCA GGC ACT GTA CCA GCA CCT GAA GC |
| pvdAKO4 | GGA ATT GTG AGC GGA TAA CAA TTT CAC ACA GGA AAC AGC TCT GAA GCC GAT GTT GAC CAC |
| pchEKO1 | CCA GGC AAA TTC TGT TTT ATC AGA CCG CTT CTG CGT TCT GAT CTG ATC CTC GTG CAG AGC |
| pchEKO2 | GGT CTG CAC CTG CAA GTG CAG GGC GGT ACG GGA ATC |
| pchEKO3 | GAT TCC CGT ACC GCC CTG CAC TTG CAG GTG CAG ACC |
| pchEKO4 | GGA ATT GTG AGC GGA TAA CAA TTT CAC ACA GGA AAC AGC TCG TCA GGT TGA GAC AGA ACG |
P. aeruginosa PA14 strains and Escherichia coli strains used for mutant construction were cultured in LB broth (Fisher Scientific) at 37°C. E. coli BW29427 and β-2155 were supplemented with 0.3 mM diaminopimelic acid. The yeast Saccharomyces cerevisiae uracil-auxotrophic strain InvSc1 (Invitrogen), used for gap repair cloning (10, 19), was grown with yeast extract-peptone-dextrose medium (YPD medium; 1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose) at 30°C, and selections were performed with synthetic defined agar (SDA) medium lacking uracil (URA; Qbiogene 4813-065). For selection and maintenance of plasmids pMQ30 and its derivatives, as well as pAKN69, gentamicin was used at 15 μg/ml for E. coli and 75 to 100 μg/ml for P. aeruginosa, respectively. Selection and maintenance of E. coli containing pUX-BF13 was carried out on 100 μg/ml of ampicillin.
Strain construction.
P. aeruginosa synthesizes two known siderophores, the stronger Fe(III)-binding pyoverdine and the weaker Fe(III)-binding pyochelin (53). We constructed the siderophore-null strain by generating unmarked deletions of the pyoverdine and pyochelin biosynthetic genes pvdA (71) and pchE (61), respectively, in PA14 wild type. Analogously, we constructed the phenazine-siderophore-null strain in the PA14 phenazine-null strain (Δphz). We first deleted pvdA and then pchE. Here we describe the protocol for in-frame deletion of pvdA using yeast gap repair cloning based on previously developed methods (10, 19, 49, 67). The 5′ and 3′ regions (both ∼1 kb in length) of the sequence flanking pvdA were amplified using primer pairs pvdAKO1/pvdAKO2 and pvdAKO3/pvdAKO4, respectively (Table 2). These 5′- and 3′-flanking DNA fragments and the gapped plasmid vector pMQ30 were simultaneously introduced into the yeast S. cerevisiae uracil auxotrophic strain InvSc1 (Invitrogen) for in vivo recombination (10, 19). The plasmid pMQ30 is an allelic exchange vector for Gram-negative bacteria unable to support replication of the ColE1 origin and contains CEN6/ARSH4 DNA sequences to support replication in S. cerevisiae, a URA3 yeast-selectable marker, a multicloning site in a lacZα allele for blue-white screening, an oriT for conjugation-mediated plasmid transfer, a gentamicin-resistance gene (aacC1), and the counterselectable marker sacB (67). Recombinant yeast cells were selected for on medium deficient in uracil. Plasmids were liberated from recombinant yeast and electroporated into E. coli UQ950, which was used as a host strain for plasmid replication (3). Transformants containing recombination products of pMQ30 with the PCR products were isolated by blue-white screening and gentamicin resistance (3), yielding the construct for deleting the PA14 pvdA gene. This plasmid, which we call pYW01 (Table 1), was purified from E. coli UQ950, transformed by heat shock into E. coli BW29497 (a diaminopimelic acid auxotroph), and then mobilized into PA14 (the wild type or the Δphz mutant) via biparental conjugation (76). PA14 (the wild type and the Δphz mutant) single recombinants (merodiploid containing the intact and the deleted pvdA gene) were isolated by selecting for gentamicin resistance. Resolution of the merodiploid was performed by selection on 10% sucrose, followed by PCR-based screening for loss of the wild-type gene to isolate the pvdA deletion mutants (referred to as ΔpvdA and Δphz ΔpvdA). The deletion of pchE from the ΔpvdA and Δphz ΔpvdA strains was performed the same way, using primer pairs pchEKO1/pchEKO2 and pchEKO3/pchEKO4 (Table 2). Besides using PCR-based diagnosis, we confirmed the ΔpvdA ΔpchE and Δphz ΔpvdA ΔpchE mutants by their inability to produce pyoverdine or pyochelin when grown in iron-deficient MOPS-based medium, in contrast to the wild-type and Δphz strains, respectively (see “Analyses of phenazines and siderophores” for pyoverdine and pyochelin detection methods).
To follow P. aeruginosa biofilm development using confocal microscopy, we generated YFP-labeled PA14 strains by introducing a chromosomally encoded constitutive enhanced yellow fluorescent protein (EYFP) into the wild type, Δphz, ΔpvdA ΔpchE, and Δphz ΔpvdA ΔpchE strains. Plasmid pAKN69, containing the mini-Tn7(Gm)PA1/04/03-eyfp fusion, was used for this purpose (35). This plasmid was cloned into E. coli BW29427 and then mobilized into each PA14 strain via triparental mating with the helper plasmid pUX-BF13 (carrying the transposase genes) in E. coli β-2155, as described previously (6, 33). PA14 transformants with YFP constructs were selected with gentamicin and confirmed by YFP fluorescence.
Biofilm experiments.
A flow cell system was constructed for biofilm experiments. The size of each flow channel was 1.5 by 4 by 34 mm; continuous flow of 1% TSB (with or without the additives detailed in the Results section) at the rate of 3 ml/h was supplied with a Watson-Marlow peristaltic pump. The temperature for biofilm growth was 22°C. An early-stationary-phase culture grown in 10% TSB was diluted to an OD500 of 0.1 in biofilm control medium (1% TSB). Each flow cell was then inoculated with 300 μl of the diluted culture by injection with a 1-ml syringe. In order to allow cells to attach to the glass surface, the flow was arrested for 1.5 h and then resumed throughout the length of each experiment (up to 6 days).
To image biofilms, confocal laser scanning microscopy (CLSM) with a Leica TCS SPE inverted microscope was used. For PA14 strains constitutively expressing EYFP (the wild type, Δphz, ΔpvdA DpchE, and Δphz ΔpvdA ΔpchE strains), three-dimensional fluorescence images were acquired using an excitation wavelength of 488 nm with constant intensity and collecting emissions in the range of 510 to 618 nm. For the PA14 feoB::MAR2xT7 mutant, images were obtained using differential interference contrast (DIC) mode. To ensure images used for comparisons of biofilm formation were representative and reproducible, multiple fields of view were acquired over time with a 10× dry lens objective in each flow cell within a single experimental set, and at least four independent experimental sets were performed. Fluorescence and DIC images were processed using Bitplane Imaris and NIH ImageJ software. In most cases, fluorescence-based multiple biofilm image stacks (spaced 1 to 2 μm apart) were analyzed using AutoCOMSTAT software, a modified version of the COMSTAT biofilm evaluation package by Heydorn et al. (29, 44). For each image, a global threshold was calculated using the robust automated threshold selection algorithm with a critical-size setting of 20 μm, and connected volume filtering was performed with a connectivity setting of 18 to remove free-floating biomasses. Substratum coverage calculations were based on the first 3 μm above the substratum. The area of each analyzed image was 3.03 × 105 μm2. The results from measurements of one to six images for each strain and treatment were averaged, and the standard error of the mean was calculated. The biofilm parameters reported here are biovolume per image area (referred to as biomass), substratum coverage, maximum height, and average height of the biomass, which excludes any area not covered by cells.
Iron analysis.
The total concentrations of iron in TSB-based media and suspensions of the mineral ferrihydrite were analyzed by the phenanthroline assay according to published protocols (34). In summary, complete reduction of Fe(III) (soluble and/or mineral forms) to soluble Fe(II) was achieved by adding the reductant hydroxylamine hydrochloride to acidified samples. 1,10-Phenanthroline and the pH buffer ammonium acetate were then added and allowed enough time to fully develop an orange-red Fe(II)-phenanthroline complex at pH 3.5. The total iron concentrations reflected by the colored Fe(II) complex were calculated based on the absorbance readings at 510 nm in a Shimadzu UV-2450 spectrophotometer. Iron in sterile, aerobic, TSB-based media should be present as the oxidation state +3, even though the specific Fe(III) forms are unknown.
Analyses of phenazines and siderophores.
Filtrates (passed through a 0.2-μm-pore-size filter) were prepared from biofilm effluents or planktonic cultures. For characterizing and quantifying phenazines and the siderophore pyochelin, filtrates were directly loaded onto a Beckman System Gold reverse-phase high-pressure liquid chromatography (HPLC) instrument with a diode array UV-visible light (Vis) detector and a Waters Symmetry C18 analytical column (5-μm-particle size; 4.6 by 250 mm). Analysis was performed in a gradient of water-0.1% trifluoroacetic acid (TFA; solvent A) to acetonitrile-0.1% TFA (solvent B) at a flow rate of 1.0 ml/min using the following method. For 0 to 1 min, chromatography was performed in a linear gradient from 100% solvent A to 15% solvent B, for 1 to 12 min in a linear gradient to 58% solvent B, for 12 to 13 min in a linear gradient to 70% solvent B, for 13 to 25 min in a linear gradient to 85% solvent B, for 25 to 26 min in a linear gradient to 100% solvent A, and for 26 to 29 min in 100% solvent A. Phenazines (e.g., PCA, PYO, phenazine-1-carboxamide [PCN], and 1-hydroxyphenazine [1-OHPHZ]) and pyochelin, which are known to be potentially produced by P. aeruginosa, can all be detected based on their characteristic absorption wavelengths and retention times as long as their concentrations are higher than 0.05 to 0.1 μM (17, 43, 65, 74).
The same HPLC instrument was used for analyzing the siderophore pyoverdine, with gradient profiling and sample preparation as described in detail by Bultreys et al. (9). In summary, filtrates with Fe(III)-chelated pyoverdine(s) at pH 5.0 were prepared for HPLC by adding FeCl3 into the samples, followed by filtration (0.2-μm-pore-size filter) and pH adjustment. Analysis was carried out in a gradient profiling with solvent A as water-17 mM NaOH-acetic acid at pH 5.0 and solvent B as acetonitrile (solvent B) (9). For samples with pyoverdine being released at concentrations higher than 0.1 μM, a single Fe(III)-pyoverdine peak was detected at 403 nm. In addition, iron-free pyoverdine in filtered (0.2-μm-pore-size filter) biofilm effluents was analyzed using a fast fluorescence-based method by a BioTek Synergy 4 fluorescence plate reader with a xenon flash light source at the specific excitation/emission wavelength set of 405 nm/455 nm (20). Control experiments and independent HPLC analyses confirmed that the measured fluorescence signal was contributed predominantly by pyoverdine and hence can be used for its detection and quantification.
RESULTS
PCA can work together with the siderophore pyoverdine in promoting biofilm development.
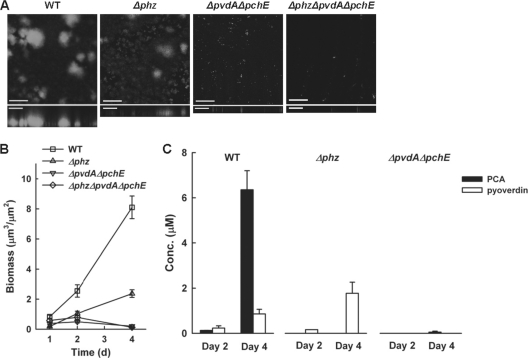
We first followed biofilm development in the P. aeruginosa PA14 wild type, a phenazine-null strain (Δphz), a siderophore-null strain (ΔpvdA ΔpchE), and a phenazine-siderophore-null strain (Δphz ΔpvdA ΔpchE) under a flow of 1% TSB medium over 4 days. In addition, we used AutoCOMSTAT software to analyze biofilm images for quantitative comparisons. As illustrated in Fig. 1, wild-type biofilm formation proceeded in the typical stages such that bacteria initially attached to the abiotic glass surface had by day 1 clustered into microcolonies, which enlarged over time and eventually matured by day 4. The Δphz mutant, despite initially attaching equally well, formed fewer and smaller microcolonies over time than the wild type, as was seen before in LB-based medium (57). The ΔpvdA ΔpchE and Δphz ΔpvdA ΔpchE mutants showed equally severe biofilm defects and failed to develop microcolonies even 4 days after initial attachment (Fig. 1A). More quantitatively, the wild-type biomass was only marginally higher than those of three other strains at day 1, but by day 4 was about 3 to 4 times greater than that of Δphz and 40 to 60 times greater than those of the ΔpvdA ΔpchE and Δphz ΔpvdA ΔpchE strains (Fig. 1B; Table 3). By day 4, the difference in total biomass between the wild-type and Δphz mutant biofilms reflected variation mainly in thickness. On the other hand, the biofilm thickness observed for the ΔpvdA ΔpchE and Δphz ΔpvdA ΔpchE strains was about the same as that of the Δphz strain; their extremely low total biomass results primarily from a lack of surface coverage (Table 3).
Fig. 1.
The phenazine PCA can work together with the siderophore pyoverdine in promoting P. aeruginosa biofilm formation. (A) Confocal microscopic images of biofilms at day 4 (A), total biomass over time as inferred by Auto-COMSTAT analysis (B), and the corresponding release of phenazine(s) and/or siderophore(s) into the biofilm effluents for the YFP-labeled PA14 wild type (WT), the phenazine-null strain (Δphz), the siderophore-null strain (ΔpvdA ΔpchE), and the phenazine-siderophore-null strain (Δphz ΔpvdA ΔpchE) under a flow of 1% TSB medium at 22°C (C) are shown. Confocal images consist of top-down views (x-y plane, top images) and side views (x-z plane, bottom images, enlarged and truncated to emphasize differences in the z dimension). Results are representative of six experiments. Data reported in panels B and C represent means ± standard errors of the means (SEMs). Related quantitative data can be found in Table 3. Scale bars, 100 μm for images with a top-down view and 50 μm for side-view images.
Table 3.
Quantitative analyses of flow cell biofilms formed by P. aeruginosa PA14 strainsa
| Day | PA14 strain | No. of images | Total biomass (μm3/μm2) | Substratum coverage (%) | Avg thickness of biomass (μm)b | Maximum biofilm thickness (μm) |
|---|---|---|---|---|---|---|
| 1 | WT | 6 | 0.83 ± 0.11 | 7.1 ± 0.8 | 13.2 ± 0.4 | 40 |
| Δphz | 6 | 0.18 ± 0.03 | 1.9 ± 0.3 | 11.3 ± 0.8 | 35 | |
| ΔpvdA ΔpchE | 6 | 0.38 ± 0.08 | 4.5 ± 0.9 | 10.8 ± 0.4 | 28 | |
| Δphz ΔpvdA ΔpchE | 6 | 0.59 ± 0.08 | 5.7 ± 0.9 | 13.0 ± 0.6 | 37 | |
| 2 | WT | 5 | 2.54 ± 0.41 | 14.0 ± 1.0 | 19.9 ± 3.4 | 89 |
| Δphz | 6 | 1.04 ± 0.15 | 10.0 ± 0.9 | 13.0 ± 1.9 | 60 | |
| ΔpvdA ΔpchE | 6 | 0.50 ± 0.08 | 4.5 ± 0.7 | 12.2 ± 0.2 | 33 | |
| Δphz ΔpvdA ΔpchE | 6 | 0.79 ± 0.13 | 6.3 ± 0.8 | 13.4 ± 0.5 | 31 | |
| 4 | WT | 3 | 8.10 ± 0.75 | 28.1 ± 0.6 | 28.9 ± 3.0 | 95 |
| Δphz | 4 | 2.37 ± 0.26 | 23.1 ± 3.3 | 13.8 ± 0.7 | 44 | |
| ΔpvdA ΔpchE | 2 | 0.20 ± 0.09 | 1.5 ± 0.1 | 13.8 ± 4.3 | 47 | |
| Δphz ΔpvdA ΔpchE | 4 | 0.13 ± 0.05 | 1.0 ± 0.3 | 14.0 ± 0.9 | 36 |
Biofilms were formed under a flow of 1% TSB medium at 22°C for 4 days. Representative images from this data set are shown in Fig. 1. All values are means of the results of n images ± the standard error of the mean. Each image covers an area of 3.03 × 105 μm2. WT, wild type.
Calculated for the area covered by the biomass.
To further correlate phenazine and/or siderophore production with biofilm development, we simultaneously analyzed the compounds being released into the biofilm effluents for the wild type and the Δphz and ΔpvdA ΔpchE strains (Fig. 1C). The wild type started to release measurable levels of PCA (the precursor phenazine produced by all phenazine-making pseudomonads [43, 54]) and the siderophore pyoverdine beginning on day 2. Concomitant with biofilm development, both PCA and pyoverdine concentrations increased with time, with a dramatic rise in the PCA concentration by day 4. Surprisingly, no other phenazines, including the blue-colored PYO, ever reached concentrations above their respective HPLC detection limits (0.05 to 0.1 μM) under our biofilm growth conditions. Given that PYO is a highly diffusible zwitterion at circumneutral pH, we interpret the absence of PYO in the biofilm effluents to reflect little or no production rather than its retention in the biofilm matrix. The Δphz mutant started to release a measurable level of pyoverdine on day 2, and the pyoverdine concentration increased along with biofilm development. Despite releasing more pyoverdine than the wild type, the Δphz mutant generated 3 to 4 times less biofilm biomass by day 4. Adding 10 μM PCA (comparable to the levels detected in wild-type day 4 biofilm effluents) to the medium of the Δphz mutant throughout the experimental timescale increased the biofilm biomass 4 times by day 6, resulting in a biomass similar to that of the wild type (Fig. S1). The ΔpvdA ΔpchE mutant that failed to form biofilms did not release phenazines until day 4, and even then only at trace levels. Because phenazines are typically released only at a high cell density due to quorum-sensing regulation (17, 54), the lack of phenazine production by this strain in light of its sparse growth is not surprising. The lack of phenazine production by the ΔpvdA ΔpchE strain matches our observation that the ΔpvdA ΔpchE and Δphz ΔpvdA ΔpchE strains show equally severe biofilm defects. Collectively, these data indicate that both siderophores and phenazines (particularly PCA under these conditions) promote biofilm formation.
PCA can promote biofilm formation by generating Fe(II) in the absence of siderophores.
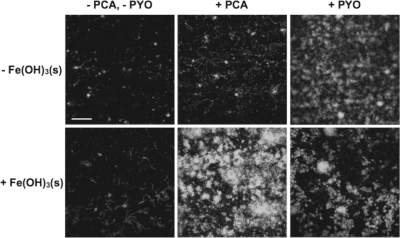
It is well established that pyoverdine promotes P. aeruginosa biofilm formation by facilitating iron acquisition (5). Effective iron acquisition is essential for biofilm development because more iron is required than for planktonic growth (68). This is due to the fact that iron acts as a signal to trigger the transition from a motile to a sessile state (68). From the phenanthroline assay, we calculated the total iron concentration to be 0.2 μM in 1% TSB. Because the medium was prepared aerobically at pH 7, the oxidation state of iron originally present in the medium should be +3, even though the specific Fe(III) forms are unknown. It is commonly believed that bioavailable iron in the micromolar concentration range is required for optimal bacterial growth (53, 58), and hence the iron content in 1% TSB verges on the lower threshold. If the observed PCA-promoted biofilm formation (Fig. 1) were due to a stimulation in iron acquisition as a result of phenazine-facilitated Fe(III) reduction to Fe(II), we would expect that PCA would be able to circumvent the pyoverdine pathway if enough Fe(III) were provided. To test whether PCA could rescue the biofilm defect in the PA14 ΔpvdA ΔpchE mutant in the presence of sparingly soluble Fe(III), we investigated the effect of adding 1.0 μM ferrihydrite mineral suspension [Fe(OH)3(s); Ksp [the solubility product constant] = 10−38.8 M (47)] alone, 10 μM PCA alone, or 1.0 μM Fe(OH)3(s) together with 10 μM PCA to the base medium (1% TSB). Adding exogenous PCA was necessary because, unlike the wild type, the ΔpvdA ΔpchE strain produced very little PCA on its own (Fig. 1C). We found that adding Fe(OH)3(s) or PCA alone could not rescue biofilm formation by the ΔpvdA ΔpchE mutant strain with statistical significance (Fig. 2; Table 4). By contrast, when Fe(OH)3(s) and PCA were added together, dramatic rescue was observed (Fig. 2; Table 4). This is consistent with our prior abiotic experiments which showed that some phenazines, including reduced PCA, can promote Fe(OH)3(s) reduction over a broad pH range and liberate bioavailable Fe(II) (27, 74). The fact that adding PCA to 1% TSB can promote biofilm formation by the Δphz mutant (which makes pyoverdine but not phenazines) but not the ΔpvdA ΔpchE mutant (which cannot make pyoverdine and is severely delayed in producing phenazines; Fig. 1C) further supports the conclusion that PCA and pyoverdine both contribute to biofilm formation. However, even in the absence of pyoverdine, PCA-promoted rescue can be achieved by adding just 1 μM Fe(III) to the base medium with 0.2 μM total Fe (Fig. 2; Table 4). This rescue occurred even with the total iron concentration (1.2 μM) being more than 10 times lower than that reported in CF sputum (generally greater than 10 μM, ranging from 17 to 200 μM) (60). Therefore, PCA can stimulate P. aeruginosa biofilm formation in the presence of otherwise biologically unavailable Fe(III), even in the absence of pyoverdine if sufficient iron levels can be attained.
Fig. 2.
PCA and PYO can circumvent the siderophore pathway for promoting P. aeruginosa biofilm development via Fe(II) uptake-dependent and -independent mechanisms, respectively. The effectively insoluble Fe(III) mineral ferrihydrite [Fe(OH)3(s)] was the Fe(III) source. Confocal microscopic images of the YFP-labeled P. aeruginosa PA14 siderophore-null strain (ΔpvdA ΔpchE) incubated in biofilm flow cells at 22°C for 6 days with no additions or with the addition of 1.0 μM Fe(OH)3(s), 10 μM phenazine (PCA or PYO), or 1.0 μM Fe(OH)3(s) together with 10 μM phenazine (PCA or PYO) to 1% TSB medium. Images are top-down views (x-y plane). Results are representative of four experiments. Related quantitative data can be found in Table 4. Scale bar, 100 μm.
Table 4.
Quantitative analyses of flow cell biofilms formed by the P. aeruginosa PA14 ΔpvdA ΔpchE-YFP straina
| Day | Additive(s) to biofilm control medium (1% TSB) | No. of images | Total biomass (μm3/μm2) | Substratum coverage (%) | Avg thickness of biomass (μm)b | Maximum biofilm thickness (μm) |
|---|---|---|---|---|---|---|
| 2 | None | ND | ||||
| 1.0 μM Fe(OH)3(s) | 4 | 0.03 ± 0.00 | 0.4 ± 0.1 | 10.7 ± 2.0 | 30 | |
| 10 μM PCA | 4 | 0.05 ± 0.01 | 0.6 ± 0.2 | 12.6 ± 1.4 | 32 | |
| 1.0 μM Fe(OH)3(s), 10 μM PCA | 4 | 0.45 ± 0.21 | 6.3 ± 3.3 | 11.1 ± 1.2 | 30 | |
| 10 μM PYO | 3 | 0.09 ± 0.03 | 0.8 ± 0.3 | 13.5 ± 0.4 | 34 | |
| 1.0 μM Fe(OH)3(s), 10 μM PYO | 4 | 0.57 ± 0.29 | 7.3 ± 3.7 | 10.3 ± 0.5 | 34 | |
| 4 | None | ND | ||||
| 1.0 μM Fe(OH)3(s) | 3 | 0.05 ± 0.01 | 0.5 ± 0.2 | 12.1 ± 2.3 | 30 | |
| 10 μM PCA | 4 | 0.19 ± 0.09 | 2.9 ± 1.4 | 10.8 ± 0.9 | 34 | |
| 1.0 μM Fe(OH)3(s), 10 μM PCA | 4 | 1.90 ± 1.08 | 12.5 ± 6.4 | 15.8 ± 1.8 | 44 | |
| 10 μM PYO | 4 | 0.83 ± 0.22 | 9.9 ± 2.4 | 10.1 ± 0.4 | 42 | |
| 1.0 μM Fe(OH)3(s), 10 μM PYO | 4 | 3.1 ± 1.76 | 19.2 ± 9.2 | 10.8 ± 2.4 | 48 | |
| 6 | None | 4 | 0.63 ± 0.16 | 6.0 ± 1.0 | 10.4 ± 0.8 | 40 |
| 1.0 μM Fe(OH)3(s) | 4 | 0.75 ± 0.33 | 3.3 ± 1.3 | 24.3 ± 1.6 | 82 | |
| 10 μM PCA | 3 | 0.21 ± 0.09 | 3.3 ± 2.1 | 16.1 ± 4.4 | 66 | |
| 1.0 μM Fe(OH)3(s), 10 μM PCA | 4 | 4.87 ± 1.32 | 29.3 ± 8.5 | 18.9 ± 1.8 | 80 | |
| 10 μM PYO | 3 | 9.54 ± 0.18 | 44.3 ± 5.1 | 24.1 ± 2.1 | 94 | |
| 1.0 μM Fe(OH)3(s), 10 μM PYO | 3 | 4.20 ± 0.17 | 31.8 ± 3.5 | 19.9 ± 2.6 | 56 |
Biofilms were formed at 22°C for 6 days with no addition or with the addition of 1.0 μM Fe(OH)3(s), 10 μM phenazine (PCA or PYO), or 1.0 μM Fe(OH)3(s) together with 10 μM phenazine (PCA or PYO) to 1% TSB medium. Representative day 6 images from this data set are shown in Fig. 2. All values are means of the results for n images ± the standard error of the mean. Each image covers an area of 3.03 × 105 μm2. ND, not determined.
Calculated for the area covered by the biomass.
PCA can promote biofilm formation in the presence of conalbumin by facilitating Fe(II) uptake.
The previous experiments used the effectively insoluble mineral ferrihydrite to limit cells for Fe(III). While this may be relevant to understanding Fe(III) acquisition by P. aeruginosa in soil environments, in a clinical context, Fe(III) limitation results from binding by host-produced proteins of the transferrin family (including lactoferrin, serotransferrin, and conalbumin; Kd [dissociation constant] ∼ 1020 to 23 M−1) (1, 58). We note, however, that recent studies have documented formation of nanoparticles of Fe(III) (hydr)oxides on human serum transferrin fibrils (22), so the two may not be as distinct as previously thought. Regardless, it has been suggested that the Fe(III)-sequestering function of lactoferrin presents an innate defense against P. aeruginosa; by competing against high-affinity siderophore(s) for binding Fe(III), a subinhibitory concentration of lactoferrin that does not impose a planktonic growth defect prevents P. aeruginosa from accessing the level of iron required to signal the transition to the biofilm mode of growth (5, 68). Furthermore, earlier experiments performed with whole-bacterial-cell suspensions showed that PYO could mediate Fe(II) uptake by iron-limited P. aeruginosa by reducing transferrin-bound Fe(III) with concomitant liberation of Fe(II), due to its much weaker affinity for Fe(II) (∼103 M−1) (58). This led us to hypothesize that PCA could stimulate biofilm development by facilitating Fe(II) uptake through reductive liberation of Fe(II) from host Fe(III)-binding proteins, a pathway that is independent of siderophore-mediated Fe(III) uptake.
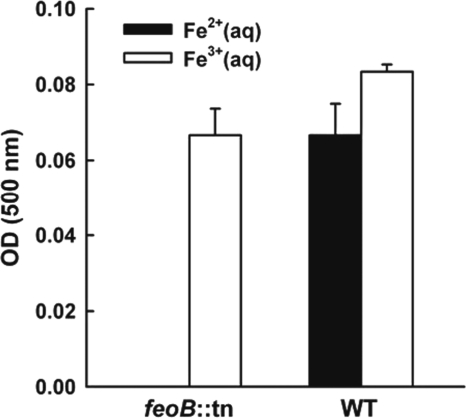
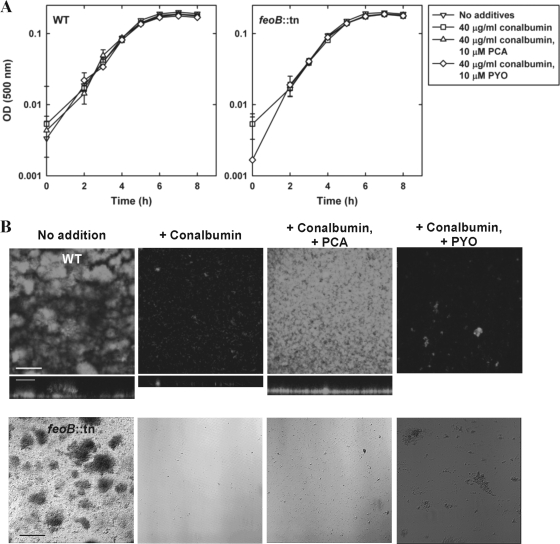
To test this hypothesis, we compared biofilm development under a flow of 1% TSB between the wild type and the feoB::MAR2xT7 mutant, a strain disrupted in gene PA14_56680, which encodes the cytoplasmic membrane protein FeoB (11, 40). We confirmed the identity of this mutant by sequencing and performing a diagnostic phenotypic test, as follows. Because FeoB was recently shown to be the transporter required for energy-dependent Fe(II) uptake across the cytoplasmic membrane of P. aeruginosa (11, 40), distinct from the more extensively studied TonB1-dependent ABC transport system for siderophore-mediated Fe(III) uptake (53, 64, 82), we predicted that the feoB::MAR2xT7 mutant would not grow on Fe(II). To test this prediction, we performed experiments in iron-free medium to which we added back either Fe(II) or Fe(III). As expected, the feoB::MAR2xT7 mutant could not grow anaerobically in planktonic batch cultures when given Fe(II) as its sole iron source, yet it could grow in the presence of Fe(III); under the same conditions, the wild type grew regardless of whether iron was in the ferric or ferrous form (Fig. 3). In 1% TSB medium, both strains developed into mature biofilms over time, as expected (Fig. 4B). However, when we reduced the amount of available iron by adding the iron chelator conalbumin (in its iron-free form), neither strain formed biofilms. Previous researchers have also used conalbumin as a lactoferrin surrogate in these types of experiments (45, 68) because they have similar structures, Fe(III)-binding capacities, and effects on biofilm formation. To confirm that the amounts of conalbumin and PCA used in our biofilm experiments specifically affected the amount of iron required to signal biofilm formation and not the amount of iron required for planktonic growth, we measured planktonic growth by the wild type and the feoB::MAR2xT7 mutant; the additive(s) had no effect on the growth of either strain (Fig. 4A). When we added both conalbumin and PCA, making the acquisition of sufficient iron to signal biofilm formation dependent upon PCA-mediated reduction of conalbumin-bound Fe(III) to Fe(II), the wild type could form biofilms but the feoB::MAR2xT7 mutant could not.
Fig. 3.
The P. aeruginosa PA14 feoB::MAR2xT7 mutant (feoB::tn) cannot grow when given Fe(II) as its sole iron source, yet it can grow in the presence of Fe(III); the wild type (WT) can grow regardless of whether iron is in the ferric or ferrous form. Cells were incubated, with anaerobic shaking, in Amberlite-treated 1% TSB medium containing100 mM KNO3, 50 mM glutamate, 1% glycerol, and 100 μM iron source [either (NH4)2Fe(II)(SO4)2 or Fe(III)Cl3] at 37°C for 22 h. Data reported are the means of triplicate experiments ± standard deviations (SDs).
Fig. 4.
(A) The presence of subinhibitory levels of conalbumin alone or together with phenazine (PCA or PYO) does not inhibit planktonic growth of P. aeruginosa PA14 strains. Experiments were performed in batch cultures in 1% TSB-based biofilm medium at 37°C. Data reported are the means of triplicate experiments ± SDs. (B) PCA but not PYO can rescue the conalbumin-induced P. aeruginosa biofilm defect by reducing protein-sequestered Fe(III) with concomitant release of Fe(II). Shown are confocal microscopic images of YFP-labeled P. aeruginosa PA14 wild type (WT) and DIC microscopic images of the P. aeruginosa PA14 feoB::MAR2xT7 mutant (feoB::tn) disrupted in Fe2+ transport into the cytoplasm and incubated in biofilm flow cells at 22°C for 6 days, with no addition or with the addition of 40 μg/ml of iron-free conalbumin alone or together with 10 μM phenazine (PCA or PYO) to 1% TSB medium. Confocal images consist of top-down views (x-y plane, top images) and side views (x-z plane, bottom images, enlarged and truncated to emphasize differences in the z dimension). DIC images are top-down views (x-y plane). Results are representative of four experiments. Related quantitative data can be found in Table 5. Scale bars, 100 μm for images with top-down views and 50 μm for side-view images.
For both the wild type and the feoB::MAR2xT7 mutant, in contrast to the mature biofilms formed in medium without additives, in medium with added conalbumin, attached bacteria mostly remained as separated individual cells and failed to form clusters even after 6 days (Fig. 4B). This is similar to what was observed by other investigators studying P. aeruginosa strain PAO1 in the presence of lactoferrin (5, 30, 68). In accord with the conalbumin-induced severe biofilm defect, little PCA was released into the biofilm effluents by either strain (∼ 0.05 to 0.1 μM), as expected for density-dependent phenazine production (17, 54). For the wild type, the addition of PCA to conalbumin-treated medium rescued the defect by promoting biofilm growth into a uniformly distributed lawn type structure, distinct from the mushroom-like structures observed in control medium (Fig. 4B), within 6 days. With PCA-induced rescue, by day 6, the biofilm biomass increased by a factor of 13 compared to biofilms treated with conalbumin alone; the total biofilm biomass was comparable to that in medium without additives (Table 5). The wild-type biofilm developed similarly with respect to structure and biomass regardless of whether PCA was added to the control medium (data not shown), indicating that PCA-induced rescue was specific to the biofilm defect caused by the conalbumin treatment. On the other hand, for the feoB::MAR2xT7 mutant with a disrupted Fe(II) transporter, PCA addition failed to rescue the conalbumin-induced biofilm defect (Fig. 4B). Together, these results indicate that PCA's ability to rescue biofilm formation in the presence of the Fe(III)-binding protein conalbumin is due to the fact that it makes Fe(II) bioavailable by reducing protein-sequestered Fe(III) through extracellular electron transfer.
Table 5.
Quantitative analysis of flow cell biofilms formed by WT P. aeruginosa PA14 with different additives by day 6a
| Additive(s) to biofilm control medium (1% TSB) | No. of images | Total biomass (μm3/μm2) | Substratum coverage (%) | Avg thickness of biomass (μm)b | Maximum biofilm thickness (μm) |
|---|---|---|---|---|---|
| None | 1 | 7.20 | 46.7 | 16.4 | 62 |
| 40 μg/ml conalbumin | 6 | 0.80 ± 0.08 | 11.8 ± 1.3 | 9.0 ± 0.7 | 32 |
| 40 μg/ml conalbumin, 10 μM PCA | 4 | 10.70 ± 0.49 | 80.0 ± 4.3 | 13.2 ± 0.8 | 36 |
| 40 μg/ml conalbumin, 10 μM PYO | 1 | 0.34 | 4.2 | 8.7 | 32 |
Biofilms were formed at 22°C for 6 days with no addition or with the addition of 40 μg/ml conalbumin alone or together with 10 μM phenazine (PCA or PYO) to 1% TSB medium. Representative day 6 images from this data set are shown in Fig. 4B. All values are means of the results for n images ± the standard error of the mean. Each image covers an area of 3.03 × 105 μm2.
Calculated for the area covered by the biomass.
The phenazine PYO can affect biofilm formation independently of facilitating Fe(II) uptake.
PYO, a well-studied phenazine produced by P. aeruginosa and also detected in CF sputum (77), has been previously shown to affect biofilm formation in LB-based medium (57). Even though PYO was not released under our 1% TSB-based biofilm medium conditions, we performed analogous experiments to examine whether PYO might promote biofilm development in the same manner as PCA. We first tested whether PYO could rescue the biofilm defect in the PA14 ΔpvdA ΔpchE mutant by comparing the effect of adding 10 μM PYO alone to the effect of adding 1.0 μM Fe(OH)3(s) and 10 μM PYO together to the base medium (1% TSB). In contrast to PCA's rescue occurring only together with the addition of 1.0 μM Fe(OH)3(s), PYO rescued the biofilm defect regardless of whether Fe(OH)3(s) was present; the rescue without Fe(OH)3(s) was 2 times greater than that with the addition of Fe(OH)3(s) (Fig. 2; Table 4).
We next tested whether the addition of PYO to conalbumin-treated 1% TSB medium could rescue the PA14 wild-type biofilm defect. Unlike PCA, PYO did not rescue the conalbumin-induced biofilm defect when added to the medium (Fig. 4B; Table 5). This implies that PYO cannot efficiently reduce conalbumin-bound Fe(III) under these conditions, which is consistent with PYO being a thermodynamically less favorable reductant than PCA (74). However, this result was somewhat unexpected, given an earlier study by Cox that reported that P. aeruginosa could reduce transferrin-bound Fe(III) (14). We speculate that the reason PYO did not stimulate conalbumin-bound Fe(III) reduction for us, but did stimulate transferrin-bound Fe(II) reduction for Cox, is that one or more of the following caveats apply: (i) even though proteins within the transferrin family (such as conalbumin and transferrin) have similarly strong Fe(III)-binding capacities, the Kd values can be different by 2 to 3 orders of magnitude (1); (ii) a given phenazine's Fe(III) reduction activity depends on factors such as concentration ratios between reactants [e.g., phenazine versus chelated Fe(III)] and/or the presence of other oxidants that could react more readily with reduced phenazines. For example, reduced PYO has been shown to be much more reactive toward oxygen than reduced PCA, making PYO more subject to oxygen competing with Fe(III) as an electron acceptor (74). Consistent with this, in the whole bacterial cell suspension system studied by Cox, PYO was demonstrated to be efficient at reducing transferring-bound Fe(III) only under a strict anaerobic condition (14). Considering that our flow cell biofilm system is very different from the one studied by Cox both with respect to oxygen content and other factors, it makes sense that phenazine reactivity would be different. Interestingly, PYO did not stimulate iron-independent rescue under these conditions either, in contrast to what we observed for the ΔpvdA ΔpchE mutant under different iron-limited conditions (Fig. 2).
DISCUSSION
Phenazines can affect cells in multiple ways, ranging from deleterious (generating toxic superoxide species; see references 26, 36, 37, and 39) to beneficial (serving as signaling molecules that foster biofilm development [18] and promote redox homeostasis [55] and survival when oxidants are limited [73]). To this list we now add another function: the potential for phenazines to enhance biofilm formation by making iron more available. While it is well known that all organisms need iron to grow, it has become clear that an amount of iron in excess of this need is required for biofilm formation (5, 7, 51, 62, 68). In this context, more attention has been placed on ferric iron acquisition by P. aeruginosa than on ferrous iron acquisition (59); however, our results suggest that the latter might be important in environments where phenazines are present, ranging from the rhizosphere (24) to the CF lung (13). Here we have shown that in the presence of Fe(III) and the phenazine PCA, biofilm formation can proceed in the absence of siderophores and in the presence of host iron-binding proteins such as conalbumin. While we have focused on the electron-shuttling properties of the phenazine PCA in this study, we note that similar iron acquisition mechanisms have been proposed for phenazine-like riboflavin produced by Shewanella species (41) and Helicobacter pylori (79) and for humic-like pyomelanin produced by Legionella pneumophila (12), although these have not been tested in biofilm systems.
By highlighting the importance of phenazines in changing the redox state of iron, we do not mean to imply that other means of iron acquisition are somehow less essential for P. aeruginosa. On the contrary, it is well established that siderophore production is crucial for Fe(III) acquisition by P. aeruginosa (as well as many other bacteria) and that this is necessary for their growth in many contexts. Moreover, we hasten to point out that phenazine production is relevant for P. aeruginosa only once it has achieved a sufficiently high density for phenazine biosynthesis to be initiated. What we suggest is that the relative importance of different types of iron acquisition strategies likely change over time in response to changes in the chemistry of the microenvironment that P. aeruginosa and other bacteria inhabit. In environments where iron is primarily in the ferric form, growth of P. aeruginosa necessitates siderophores. However, as cultures grow in cell density, inflammation progresses, and phenazine production begins, siderophores would be expected to decline in importance for iron acquisition as phenazines mediate the reduction of Fe(III) to Fe(II) and/or heme becomes more available. Consistent with this, we have observed that one of the major effects of phenazine production at the transcriptional level is the repression of genes involved in Fe(III) acquisition (e.g., downregulation of siderophore biosynthesis and transport) (18). While for the sake of clarity we genetically separated siderophore from phenazine production in this study, in the wild type, both siderophores and phenazines contribute to iron acquisition: siderophores play a role early in biofilm development, but likely become less important once phenazine production begins.
Regardless of how Fe(III) is bound—be it in the form of insoluble ferric (hydr)oxides or chelated to host proteins—it is susceptible to reductive liberation to Fe(II) by reacting with phenazines, rendering it more bioavailable to P. aeruginosa (Fig. 5). Our results suggest that reduced PCA can efficiently reduce host protein-bound Fe(III) even though, paradoxically, this is predicted to be thermodynamically unfavorable at pH 7: the redox potentials for Fe(III)/Fe(II) and PCA (oxidized versus reduced) redox pairs are reported as −500 mV (52) and −116 mV (74), respectively. Note that these numbers correspond to equilibrium conditions, which likely are quite different from those within a biofilm. For example, biofilms are known to experience oxygen limitation (28), which would shift the redox equilibrium of PCA toward its reduced form. In addition, Fe(II) librated by reducing Fe(III) can be taken up by cells. Both of these effects would drive redox reactions between host protein-bound Fe(III) with reduced PCA in a more thermodynamically favorable direction.
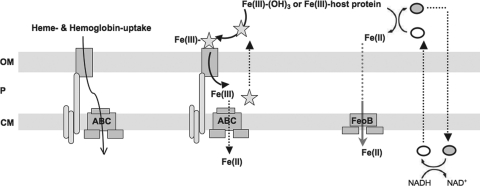
Fig. 5.
A highly simplified schematic view of P. aeruginosa iron transport via phenazine-facilitated Fe(II) uptake (right), siderophore-mediated Fe(III) uptake (middle), and heme uptake (left). In Fe(III) uptake, a siderophore (e.g., pyoverdine, indicated by the star) binds extracellular Fe(III) and crosses the outer membrane (OM) via a TonB-dependent transporter (53, 64). In the periplasm, Fe(III) is released from the siderophore, which can then be recycled; Fe(III) is reduced by an unknown mechanism to Fe(II) in the periplasm (P) and transported across the cytoplasmic membrane (CM), presumably by an ABC transport system (53, 64). By contrast, phenazines can reduce extracellular Fe(III) to Fe(II). After entering the periplasm, presumably via an OM porin, Fe(II) is transported across the CM via FeoB (11, 40). Phenazines themselves are recycled (27, 73) and enter and leave the cell through various transporters (not drawn, for simplicity). Intracellularly, phenazine reduction is coupled to NADH oxidation to NAD+ (55), although whether this reduction is enzyme mediated is unknown. Reduced phenazine is indicated by the open oval and oxidized phenazine by the filled oval. Additionally, P. aeruginosa can acquire iron from heme and heme-containing proteins (e.g., hemoglobin), which are transported through a specific outer membrane receptor channel in a TonB-dependent manner (31, 48, 72). In P. aeruginosa, two distinct heme uptake systems have been found, although the mechanistic details of heme capture and delivery and the fate of heme after entering the cytoplasm are not fully understood (31, 48).
Given their high redox potentials, ferric (hydr)oxides can be easily reduced by PCA even under equilibrium conditions. This has broad relevance, given that PCA is the precursor phenazine produced by all phenazine-making pseudomonads (43, 54). Other natural phenazines, such as PCN and 1-OHPHZ, can react with Fe(III) at rates comparable to that of PCA (74). Interestingly, however, PYO is the one phenazine whose ability to reduce Fe(III) is poor (74). We therefore think it is unlikely that the observed PYO-enhanced biofilm rescue of our siderophore mutant in the presence of Fe(OH)3(s) was due to more efficient iron reduction under these conditions than that achieved by PCA. In fact, PYO is known to be a signaling molecule that helps coordinate P. aeruginosa biofilm development (18, 57). Thus, PYO's enhancement of biofilm formation under these conditions could be due to an iron-independent mechanism. Curiously, the enhancement of biofilm formation observed in the presence of PCA plus Fe(OH)3 or PYO plus Fe(OH)3 was less than that achieved in the presence of PYO alone (Table 4). This suggests that PCA- or PYO-mediated iron reduction is a double-edged sword: while it can promote biofilm formation relative to that achieved in the presence of Fe(OH)3 alone, it can also exact a toxic price. The detailed mechanisms by which different phenazines affect biofilm formation remain to be elucidated, but what is clear is that different phenazines can affect biofilm formation in different ways under different environmental conditions.
Although it has long been recognized that the microbial community that forms in CF sputum is complex (25), what controls the structure and dynamics of this population is unclear. While many factors can be anticipated to impact the ecological success of different species in the lung, one important parameter is competition for iron (59). In coculture experiments pitting Staphylococcus aureus against P. aeruginosa, it was found that in the presence of S. aureus, P. aeruginosa decreased transcription of genes known to be regulated by iron availability (42). It was concluded that this effect was most likely due to lysis of S. aureus by P. aeruginosa, leading to iron release. While the iron source was not identified in this study, one reasonable possibility is heme, which is thought to be an important nutrient for Pseudomonas and other bacteria in CF sputum, particularly as inflammation progresses (31, 48, 72). In contrast, P. aeruginosa can lose out in the Fe(III) battle to other organisms such as Burkholderia spp., which produce competing siderophores, such as ornibactin (75), as well as phenazines. Such positive and negative interactions involving iron may help explain the dynamic takeovers between different pathogens in the lungs of CF patients over time. Yet given that phenazines can alleviate the dependence of Pseudomonas on Fe(III) by generating Fe(II), competition for Fe(III) may be relevant only when phenazines are not present. Indeed, it seems likely that phenazine-enabled Fe(II) generation could benefit not only phenazine producers like P. aeruginosa but also other organisms, much in the same way that siderophores can be used by different species (53, 64).
The full complexity of the microbial community in the CF lung and the interactions among its members are only beginning to be appreciated (8, 25, 63). Systematic studies are needed to understand how iron availability—impacted by both host cells and the microbiome—affects this ecosystem. As recently pointed out by Reid et al. (59), we do not have a clear understanding of iron chemistry in CF sputum, and this must be determined before we can properly understand how iron might be acquired by P. aeruginosa cells or any other cell at different stages in lung function decline. In the future, it will be interesting to determine whether phenazines—possibly by way of promoting Fe(II) acquisition, among other effects (17, 18, 27, 28, 54, 55, 57, 73, 74)—influence the composition of the microbial community present in infections and its stability.
Supplementary Material
Acknowledgements
We thank N. C. Caiazza and L. E. P. Dietrich for assistance with mutagenesis; I. J. Schalk, G. L. A. Mislin, and F. Hoegy for the gifts of pyoverdine and pyochelin; and L. E. P. Dietrich, Alexa Price-Whelan, and the anonymous reviewers for constructive feedback.
This work was supported by grants to D.K.N. from the Packard Foundation and the Howard Hughes Medical Institute and an NSF graduate research fellowship to J.C.W. D.K.N. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Aisen P. 1998. Transferrin, the transferrin receptor, and the uptake of iron by cells, p. 585–631In Sigel A., Sigel H.(ed.), Metal ions in biological systems, vol. 35 Marcel Dekker, Inc., New York, NY: [PubMed] [Google Scholar]
- 2. Albrecht-Gary A.-M., Blanc S., Rochel N., Ocaktan A. Z., Abdallah M. A. 1994. Bacterial iron transport: coordination properties of pyoverdin PaA, a peptidic siderophore of Pseudomonas aeruginosa. Inorg. Chem. 33:6391–6402 [Google Scholar]
- 3. Ausubel F. M., et al. 1992. Current protocols in molecular biology. John Wiley & Sons, New York, NY [Google Scholar]
- 4. Banin E., et al. 2008. The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. U .S. A. 105:16761–16766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banin E., Vasil M. L., Greenberg E. P. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U .S. A. 102:11076–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bao Y., Lies D. P., Fu H., Roberts G. P. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168 [DOI] [PubMed] [Google Scholar]
- 7. Berlutti F., et al. 2005. Iron availability influences aggregation, biofilm, adhesion and invasion of Pseudomonas aeruginosa and Burkholderia cenocepacia. Int. J. Immunopathol. Pharmacol. 18:661–670 [DOI] [PubMed] [Google Scholar]
- 8. Bouchara J. P., et al. 2009. Development of an oligonucleotide array for direct detection of fungi in sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 47:142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bultreys A., Gheysen I., Wathelet B., Maraite H., de Hoffmann E. 2003. High-performance liquid chromatography analyses of pyoverdin siderophores differentiate among phytopathogenic fluorescent Pseudomonas species. Appl. Environ. Microbiol. 69:1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burke D., Dawson D., Stearns T. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 11. Cartron M. L., Maddocks S., Gillingham P., Craven C. J., Andrews S. C. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157 [DOI] [PubMed] [Google Scholar]
- 12. Chatfield C. H., Cianciotto N. P. 2007. The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect. Immun. 75:4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costerton J. W., Stewart P. S., Greenberg E. P. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 14. Cox C. D. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dehio C., Meyer M. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179:538–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Vos D., et al. 2001. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch. Microbiol. 175:384–388 [DOI] [PubMed] [Google Scholar]
- 17. Dietrich L. E. P., Price-Whelan A., Petersen A., Whiteley M., Newman D. K. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of pseudomonas aeruginosa. Mol. Microbiol. 61:1308–1321 [DOI] [PubMed] [Google Scholar]
- 18. Dietrich L. E. P., Teal T. K., Price-Whelan A., Newman D. K. 2008. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321:1203–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elble R. 1992. A simple and efficient procedure for transformation of yeasts. Biotechniques 13:18–20 [PubMed] [Google Scholar]
- 20. Folschweiller N., et al. 2002. The interaction between pyoverdine and its outer membrane receptor in Pseudomonas aeruginosa leads to different conformers: a time-resolved fluorescence study. Biochemistry 41:14591–14601 [DOI] [PubMed] [Google Scholar]
- 21. Fothergill J. L., et al. 2007. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghosh S., Mukherjee R., Sadler P. J., Verma S. 2008. Periodic iron nanomineralization in human serum transferrin fibrils. Angew. Chem. Int. Ed. Engl. 47:2217–2221 [DOI] [PubMed] [Google Scholar]
- 23. Griffin A. S., West S. A., Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430:1024–1027 [DOI] [PubMed] [Google Scholar]
- 24. Haas D., Defago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307–319 [DOI] [PubMed] [Google Scholar]
- 25. Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923 [DOI] [PubMed] [Google Scholar]
- 26. Hassan H. M., Fridovich I. 1980. Mechanism of the antibiotic action of pyocyanine. J. Bacteriol. 141:156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hernandez M. E., Kappler A., Newman D. K. 2004. Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70:921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hernandez M. E., Newman D. K. 2001. Extracellular electron transfer. Cell. Mol. Life Sci. 58:1562–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heydorn A., et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 30. Kaneko Y., Thoendel M., Olakanmi O., Britigan B. E., Singh P. K. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117:877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaur A. P., Lansky I. B., Wilks A. 2009. The role of the cytoplasmic heme-binding protein (PhuS) of Pseudomonas aeruginosa in intracellular heme trafficking and iron homeostasis. J. Biol. Chem. 284:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. King E. O., Ward M. K., Raney D. E. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 33. Koch B., Jensen L. E., Nybroe O. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187–195 [DOI] [PubMed] [Google Scholar]
- 34. Komadel P., Stucki J. W. 1988. Quantitative assay of minerals for Fe2+ and Fe3+ using 1,10-phenanthroline. III. A rapid photochemical method. Clay Clay Miner. 36:379–381 [Google Scholar]
- 35. Lambertsen L., Sternberg C., Molin S. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6:726–732 [DOI] [PubMed] [Google Scholar]
- 36. Lau G. W., Hassett D. J., Ran H., Kong F. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599–606 [DOI] [PubMed] [Google Scholar]
- 37. Lau G. W., Ran H., Kong F., Hassett D. J., Mavrodi D. 2004. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 72:4275–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liberati N. T., et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U .S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahajan-Miklos S., Tan M.-W., Rahme L. G., Ausubel F. M. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47–56 [DOI] [PubMed] [Google Scholar]
- 40. Marshall B., Stintzi A., Gilmour C., Meyer J. M., Poole K. 2009. Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology 155:305–315 [DOI] [PubMed] [Google Scholar]
- 41. Marsili E., et al. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U .S. A. 105:3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mashburn L. M., Jett A. M., Akins D. R., Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187:554–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mavrodi D. V., Blankenfeldt W., Thomashow L. S. 2006. Phenazine compounds in fluorescent pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44:417–445 [DOI] [PubMed] [Google Scholar]
- 44. Merritt P. M., Danhorn T., Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189:8005–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moreau-Marquis S., et al. 2008. The ΔF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am. J. Physiol. Lung Cell. Mol. Physiol. 295:L25–L37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moreau-Marquis S., O'Toole G. A., Stanton B. A. 2009. Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am. J. Respir. Cell Mol. Biol. 41:305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morel F. M. M., Hering J. G. 1993. Principles and applications of aquatic chemistry. John Wiley & Sons, New York, NY [Google Scholar]
- 48. Ochsner U. A., Johnson Z., Vasil M. L. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185–198 [DOI] [PubMed] [Google Scholar]
- 49. Orr-Weaver T. L., Szostak J. W. 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. U. S. A. 80:4417–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Palmer K. L., Mashburn L. M., Singh P. K., Whiteley M. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187:5267–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patriquin G. M., et al. 2008. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 190:662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pierre J. L., Fontecave M., Crichton R. R. 2002. Chemistry for an essential biological process: the reduction of ferric iron. Biometals 15:341–346 [DOI] [PubMed] [Google Scholar]
- 53. Poole K., McKay G. A. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8:d661–d686 [DOI] [PubMed] [Google Scholar]
- 54. Price-Whelan A., Dietrich L. E. P., Newman D. K. 2006. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2:71–78 [DOI] [PubMed] [Google Scholar]
- 55. Price-Whelan A., Dietrich L. E. P., Newman D. K. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189:6372–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rahme L. G., et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 57. Ramos I., Dietrich L. E. P., Price-Whelan A., Newman D. K. 2010. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 161:187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ratledge C., Dover L. G. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881–941 [DOI] [PubMed] [Google Scholar]
- 59. Reid D. W., Anderson G. J., Lamont I. L. 2009. Role of lung iron in determining the bacterial and host struggle in cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 297:L795–L802 [DOI] [PubMed] [Google Scholar]
- 60. Reid D. W., Carroll V., O'May C., Champion A., Kirov S. M. 2007. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur. Respir. J. 30:286–292 [DOI] [PubMed] [Google Scholar]
- 61. Reimmann C., Serino L., Beyeler M., Haas D. 1998. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144:3135–3148 [DOI] [PubMed] [Google Scholar]
- 62. Rogan M. P., et al. 2004. Loss of microbicidal activity and increased formation of biofilm due to decreased lactoferrin activity in patients with cystic fibrosis. J. Infect. Dis. 190:1245–1253 [DOI] [PubMed] [Google Scholar]
- 63. Rogers G. B., Carroll M. P., Bruce K. D. 2009. Studying bacterial infections through culture-independent approaches. J. Med. Microbiol. 58:1401–1418 [DOI] [PubMed] [Google Scholar]
- 64. Schalk I. J. 2008. Metal trafficking via siderophores in gram-negative bacteria: specificities and characteristics of the pyoverdine pathway. J. Inorg. Biochem. 102:1159–1169 [DOI] [PubMed] [Google Scholar]
- 65. Schneider T. L., Walsh C. T. 2004. Portability of oxidase domains in nonribosomal peptide synthetase modules. Biochemistry 43:15946–15955 [DOI] [PubMed] [Google Scholar]
- 66. Schwertmann U., Cornell R. M. 2000. Iron oxides in the laboratory: preparation and characterization. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 67. Shanks R. M. Q., Caiazza N. C., Hinsa S. M., Toutain C. M., O'Toole G. A. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Singh P. K., Parsek M. R., Greenberg E. P., Welsh M. J. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555 [DOI] [PubMed] [Google Scholar]
- 69. Smith E. E., et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U .S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thomashow L. S., Weller D. M. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Visca P., Ciervo A., Orsi N. 1994. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdine biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J. Bacteriol. 176:1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wandersman C., Stojiljkovic I. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215–220 [DOI] [PubMed] [Google Scholar]
- 73. Wang Y., Kern S. E., Newman D. K. 2010. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 192:365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang Y., Newman D. K. 2008. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ. Sci. Technol. 42:2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Weaver V. B., Kolter R. 2004. Burkholderia spp. alter Pseudomonas aeruginosa physiology through iron sequestration. J. Bacteriol. 186:2376–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Whiteley M., Lee K. M., Greenberg E. P. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U .S. A. 96:13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wilson R., et al. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Worlitzsch D., et al. 2002. Effects of reduced mucus oxygen concentration in airway pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Worst D. J., Gerrits M. M., Vandenbroucke-Grauls C. M., Kusters J. G. 1998. Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J. Bacteriol. 180:1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Youard Z. A., Mislin G. L., Majcherczyk P. A., Schalk I. J., Reimmann C. 2007. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 282:35546–35553 [DOI] [PubMed] [Google Scholar]
- 81. Zamri A., Abdallah M. A. 2000. An improved stereocontrolled synthesis of pyochelin, siderophore of Pseudomonas aeruginosa and Burkholderia cepacia. Tetrahedron 56:249–256 [Google Scholar]
- 82. Zhao Q. X., Poole K. 2002. Differential effects of mutations in tonB1 on intrinsic multidrug resistance and iron acquisition in Pseudomonas aeruginosa. J. Bacteriol. 184:2045–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.