Abstract
Mouse incisors grow continuously throughout life. This growth is supported by the division of dental epithelial stem cells that reside in the cervical loop region. Little is known about the maintenance and regulatory mechanisms of dental epithelial stem cells. In the present study, we investigated how transforming growth factor β (TGF-β) signaling-mediated mesenchymal-epithelial cell interactions control dental epithelial stem cells. We designed two approaches using incisor organ culture and bromodeoxyuridine (BrdU) pulse-chase experiments to identify and evaluate stem cell functions. We show that the loss of the TGF-β type I receptor (Alk5) in the cranial neural crest-derived dental mesenchyme severely affects the proliferation of TA (transit-amplifying) cells and the maintenance of dental epithelial stem cells. Incisors of Wnt1-Cre; Alk5fl/fl mice lost their ability to continue to grow in vitro. The number of BrdU label-retaining cells (LRCs) was dramatically reduced in Alk5 mutant mice. Fgf10, Fgf3, and Fgf9 signals in the dental mesenchyme were downregulated in Wnt1-Cre; Alk5fl/fl incisors. Strikingly, the addition of exogenous fibroblast growth factor 10 (FGF10) into cultured incisors rescued dental epithelial stem cells in Wnt1-Cre; Alk5fl/fl mice. Therefore, we propose that Alk5 functions upstream of Fgf10 to regulate TA cell proliferation and stem cell maintenance and that this signaling mechanism is crucial for stem cell-mediated tooth regeneration.
INTRODUCTION
The growth of mouse incisors continues throughout life and is supported by dental epithelial stem cells localized within the cervical loop of the incisor tooth organ. The dental epithelium gives rise to the inner enamel epithelium (IEE), stratum intermedium, stellate reticulum (SR), and outer enamel epithelium (OEE). The IEE was previously proposed to represent transit-amplifying (TA) cells and will differentiate into ameloblasts (9). Investigations of adult stem cells have been difficult because of their low abundance and the lack of specific markers to differentiate them from surrounding cells. A bromodeoxyuridine (BrdU) pulse-chase experiment, in which stem cells are identified by taking advantage of their relative quiescence within the surrounding population of cells, is a widely accepted approach for identifying stem cells (8). This method allows the characterization of stem cells with few or no identified stem cell markers. Label-retaining cells (LRCs) have been found in various epithelial tissues, including the cornea (3), hair follicle (4), intestinal crypts (20), mammary gland (31), and skin epithelium (27). Pulse-chase experiments conducted on mouse incisors indicated that most LRCs are in the cervical loop region, localized among the stellate reticulum cells (24).
Adult stem cells typically reside in specialized niches where they receive microenvironmental cues to maintain homeostasis and function. Previous studies demonstrated that the inductive signals for dental epithelial stem cells originate from the dental mesenchyme (10). Different pathways have been proposed to regulate the dental epithelial stem cell population, including the Fgf, Spry, Follistatin, and Bmp pathways (10, 13, 23, 25, 28). Fgf10 appears to play a crucial role, because the knockout of Fgfr2b or Fgf10 in mice results in the absence of the cervical loop structure (10, 19). Nevertheless, the molecular mechanisms involved in the regulation of fibroblast growth factor (FGF) signaling and the maintenance of dental epithelial stem cells through mesenchymal-epithelial cell interactions remain largely unknown.
Transforming growth factor β (TGF-β) family members signal through a heteromeric complex composed of type I and type II receptors. Upon ligand binding, type II receptors recruit and phosphorylate type I receptors, which then propagate the signal via phosphorylated Smad proteins. Both type I and type II receptors play important roles in tooth development (18, 32). In our previous studies, we have shown that interactions between TGF-β and FGF signaling pathways in cranial neural crest (CNC)-derived mesenchymal tissues play crucial roles in the development of the tongue and calvaria (11, 22). In this study, we designed experiments to test the hypothesis that TGF-β-mediated FGF signaling is well conserved in other areas of embryonic development and may function to regulate dental epithelial stem cells. We used Wnt1-Cre; Alk5fl/fl mutant mice as a model to study how the TGF-β family may regulate dental epithelial stem cells through mesenchymal-epithelial cell interactions. We found that the loss of TGF-β type I receptor (Alk5) expression in the dental mesenchyme severely affected the maintenance of dental epithelial stem cells. Wnt1-Cre; Alk5fl/fl incisors no longer had the ability to support continuous growth in vitro, and the number of LRCs was significantly reduced. We also detected a reduced expression level of Fgf10 in Wnt1-Cre; Alk5fl/fl incisors. Strikingly, we found that the addition of exogenous FGF10 into organ cultures was able to rescue the dental epithelial cell defects of Wnt1-Cre; Alk5fl/fl incisors. Therefore, we propose that Alk5-mediated TGF-β signaling functions upstream of Fgf10 to regulate dental epithelial stem cells during tooth development.
MATERIALS AND METHODS
Mouse maintenance and genotyping.
The Wnt1-Cre transgenic line and Alk5 floxed alleles were described previously (5, 6, 14). Wnt1-Cre; Alk5fl/fl embryos were generated by mating Wnt1-Cre; Alk5fl/+ male mice with Alk5fl/fl female mice. Genotyping was carried out by using PCR on tail tip or yolk sac DNA. Mutant embryos were identified by PCR genotyping for the presence of the Cre transgene (Cre1 [TGATGAGGTTCGCAAGAACC] and Cre2 [CCATGAGTGAACGAACCTGG]) and the Alk5fl/fl allele (lnl5′ [ATGAGTTATTAGAAGTTGTTT], lnl3′ [ACCCTCTCACTCTTCCTGAGT], and llox3′ [GGAACTGGGAAAGGAGATAAC]). Noon on the plugging day was designated embryonic day 0.5 (E0.5). All mouse embryos used in this study were maintained in a C57BL/6J background.
Histology and in situ hybridizations.
For routine histological analyses, embryos were fixed in 4% paraformaldehyde, embedded, and sectioned at 7 μm. Paraffin sections of embryos were stained with hematoxylin and eosin (H&E) according to standard procedures.
For in situ hybridization, embryos were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 8 μm. RNA probes were labeled with digoxigenin (Roche). In situ hybridization was performed as previously described (30).
BrdU incorporation analysis.
Cell proliferation was assayed with BrdU incorporation experiments. Pregnant mice were injected with 1 ml 10 mM BrdU in water per 100 g of body weight, and embryos were collected 2 h later. For samples in organ cultures, the explants were incubated with BrdU (10 μM) for 2 h. Samples or embryos were then fixed overnight in 4% paraformaldehyde and embedded in paraffin. Histology sections (7 μm) were prepared according to standard techniques. BrdU was detected by using a BrdU staining kit (Invitrogen) according to the instructions provided by the manufacturer.
For BrdU pulse-chase experiments, samples were incubated with BrdU (10 μM) for 24 h. BrdU-containing medium was then replaced with regular medium and refreshed every day. Samples were continuously cultured for 14 days and then fixed with 4% paraformaldehyde and embedded in paraffin. Histology sections (7 μm) were prepared according to standard techniques. BrdU was detected by using a BrdU staining kit (Invitrogen).
Apoptosis assay.
Apoptosis was detected by a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay using the In Situ Cell Death Detection kit (fluorescein) (Roche) according to the manufacturer's instructions. The paraffin sections were rehydrated and treated with proteinase K according to the manufacturer's instructions. After the substrate reaction, slides were mounted with Vectashield mounting medium (Vector) and observed under a fluorescent microscope.
Organ culture experiments.
The incisors were dissected carefully from the lower jaws of newborn mice. The calcified tip was removed for better nutrient penetration. The incisors were then cultured in Trowell-type organ culture dishes (Corning) on filter paper supported by metal grids in a humidified atmosphere of 5% CO2 in air at 37°C. The culture medium consisted of BGJb medium (Invitrogen) supplemented with penicillin-streptomycin and vitamin C (Gibco BRL). The date when the incisors were plated onto the culture dish was day 0, and the length of the dental epithelium was measured from days 2 to 16. In some experiments, incisors were cultured in medium supplemented with FGF3 at 10 ng/ml (R&D systems), FGF9 at 10 ng/ml (R&D systems), or FGF10 at 10 ng/ml (R&D systems), and the medium was replaced every other day.
Quantitative real-time PCR.
For real-time PCR, lower incisors from E16.5 mouse embryos were dissected carefully. RNA was extracted with the RNeasy minikit (Qiagen) according to the manufacturer's instructions. RNA was reverse transcribed by using a Superscript III kit (Invitrogen) according to the manufacturer's protocols. Quantitative PCR was carried out by using 2× SYBR green PCR master mix (Bio-Rad) with an iCycleriQ instrument (Bio-Rad). Amplifications were performed with the following primers: Fgf3 forward primer 5′-AGCGGAGGCAGAAGAAG-3′, Fgf3 reverse primer 5′-AGCCAGTCCACCTGTATG-3′, Fgf10 forward primer 5′-ACCACCCACAACCAAA-3′, Fgf10 reverse primer 5′-TCCAACGTCTGCACTA-3′, Fgf9 forward primer 5′-CAACACCTACTCTTCCAACCTCTA-3′, and Fgf9 reverse primer 5′-GGAGTCCCGTCCTTATTTAATGC-3′. PCR products were subjected to gel electrophoresis to verify the absence of nonspecific reaction products and primer dimers. Data were analyzed by using Bio-Rad iCycler software and were normalized against β-actin.
RESULTS
Compromised TGF-β signaling in the dental mesenchyme causes defects in the dental epithelial stem cell compartment.
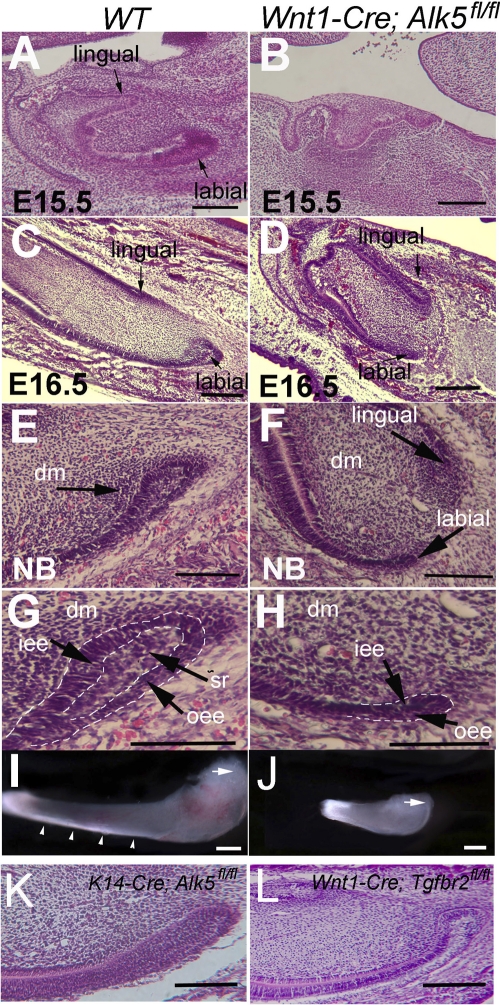
The development of the mouse incisor initiates at E11.0, when the thickening of the epithelium can first be seen (26). The incisor tooth germ reaches the bell stage at E15.5 (Fig. 1A). Enlarged cervical loops were detectable on both the labial and lingual sides of the incisor (Fig. 1A). However, the growth of the labial and lingual cervical loops was asymmetrical, and their difference in size was already detectable at E16.5 (25) (Fig. 1C). The labial side cervical loop is much larger than the lingual side cervical loop. At the newborn stage, the labial side cervical loop is composed of the inner enamel epithelium (IEE), the outer enamel epithelium (OEE), and stellate reticulum (SR) cells (25) (Fig. 1E and G). Enamel was detectable only on the labial side (Fig. 1I and 2A).
Fig. 1.
Development of the lower incisor and the labial side cervical loop. (A to H) H&E staining of sagittal sections of wild-type (WT) (A, C, E, and G) and Wnt1-Cre; Alk5fl/fl (B, D, F, and H) lower incisors at E15.5 (A and B), E16.5 (C and D), and the newborn (NB) stage (E to H). Cervical loops in E and F are shown enlarged in G and H, respectively. Arrows indicate cervical loops. (I and J) Whole-mount views of lower incisors dissected from newborn wild-type (I) and Wnt1-Cre; Alk5fl/fl (J) mice. Note that the wild-type lower incisor is much larger than the Wnt1-Cre; Alk5fl/fl mutant lower incisor. (K and L) H&E staining of the lower incisors of K14-Cre; Alk5fl/f (K) and Wnt1-Cre; Tgfbr2fl/fl (L) mice. Dashed lines outline the dental epithelia. Arrows indicate cervical loops, and arrowheads indicate enamel. dm, dental mesenchyme; iee, inner enamel epithelium; oee, outer enamel epithelium; sr, stellate reticulum. Scale bars, 200 μm.
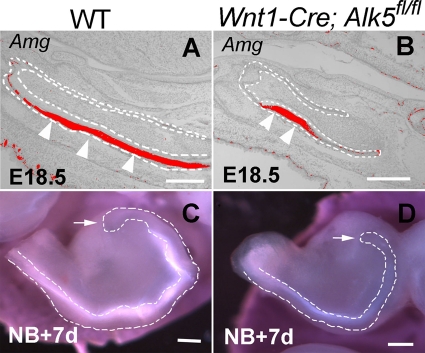
Fig. 2.
Enamel formation in Wnt1-Cre; Alk5fl/fl mutant lower incisors. (A and B) In situ hybridization of Amelogenin (Amg) expression in the labial side dental epithelium of E18.5 wild-type (WT) (A) and Wnt1-Cre; Alk5fl/fl (B) lower incisors. Arrowheads indicate expression. Dashed lines outline the dental epithelia. (C and D) After 7 days of organ culture, enamel formation can be visualized in the labial side of incisor explants from both newborn wild-type (C) and Wnt1-Cre; Alk5fl/fl (D) mice. Dashed lines outline the labial side dental epithelia, and arrows indicate the positions of the cervical loops. Scale bars, 200 μm.
The initiation and development of Wnt1-Cre; Alk5fl/fl lower incisors were delayed compared with those of the wild-type control (32). Wnt1-Cre; Alk5fl/fl lower incisor tooth germs also reached the cap stage at E15.5 (Fig. 1B) and the bell stage at E16.5 (Fig. 1D). However, the enlarged cervical loop structure appeared abnormal in Alk5 mutant lower incisors (Fig. 1D). At the newborn stage, the labial side cervical loop of the Wnt1-Cre; Alk5fl/fl lower incisor was much smaller than that of the control (Fig. 1E and F). There were few stellate reticulum cells in the mutant incisor cervical loop (Fig. 1G and H). At the newborn stage, the length of the Wnt1-Cre; Alk5fl/fl mutant lower incisor was only about one-third of that of the wild-type lower incisor (Fig. 1I and J). Interestingly, Wnt1-Cre; Alk5fl/fl upper incisors had different phenotypes than the lower incisors, and we will describe these results in a separate study. Here we focus on the phenotypes of the lower incisors.
We examined cell differentiation in Wnt1-Cre; Alk5fl/fl incisors. Mineralized dentin formed in Wnt1-Cre; Alk5fl/fl mutant incisors at the newborn stage (32; data not shown). Although we did not detect calcified enamel formation in Wnt1-Cre; Alk5fl/fl mutant incisors, we did detect Amelogenin expression on the labial sides of both wild-type and Wnt1-Cre; Alk5fl/fl lower incisors at E18.5 (Fig. 2A and B). Because Wnt1-Cre; Alk5fl/fl embryos died at birth, we dissected the incisor tooth germs from newborn mice and cultured them in Trowell-type organ culture dishes in order to investigate the development of incisors. After 7 days of in vitro culture, we detected enamel on the labial sides of both wild-type and mutant incisors (Fig. 2C and D). Thus, we conclude that ameloblast differentiation is still maintained in Wnt1-Cre; Alk5fl/fl mutant incisors.
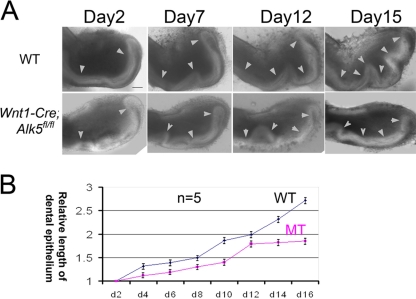
To evaluate the ability of the incisors to maintain continuous growth, we dissected the incisor tooth germs from newborn mice and cultured them in vitro. Pictures were taken every other day, and the length of the dental epithelium was measured by using NIH ImageJ software. The dental epithelia of both wild-type and Wnt1-Cre; Alk5fl/fl incisor tooth germs were elongated in vitro (Fig. 3A). After 16 days of culture, the dental epithelium of wild-type incisors increased in length by over 2.5-fold (Fig. 3B). The growth rate of the epithelium from mutant incisors was lower than that of the wild type (Fig. 3B). After 12 days the mutant epithelial growth rate began to decrease and reached a plateau stage, suggesting a growth arrest (Fig. 3B). Therefore, the ability of the dental epithelium to support the continuous growth of incisors was compromised in Wnt1-Cre; Alk5fl/fl mice.
Fig. 3.
The ability for continuous elongation is compromised in Wnt1-Cre; Alk5fl/fl mutant incisor dental epithelia. (A) Light microscope views of incisors dissected from wild-type (WT) and Wnt1-Cre; Alk5fl/fl (mutant [MT]) newborn mice after culture for 16 days. The date when the incisors were plated onto the culture dish was day 1, and length measurements started from day 2. Arrowheads indicate the dental epithelia. (B) Quantitation of the lengths of the visible dental epithelia in wild-type (WT) and Wnt1-Cre; Alk5fl/fl (MT) incisors on the indicated days using NIH ImageJ software. The measurements were compared to the lengths on day 2 to obtain the relative length values. The graph charts the average relative lengths from 5 samples. Scale bar, 200 μm.
The loss of the type I (Alk5) or type II TGF-β receptor in the neural crest-derived tissue has differential impacts on craniofacial bones and tooth development (12, 32). In order to evaluate the functional significance of Tgfbr2 in regulating the continued growth of mouse incisors, we investigated the cervical loop in Wnt1-Cre; Tgfbr2fl/fl mouse incisors. In parallel, to examine the functional significance of Alk5-mediated TGF-β signaling in the dental epithelium, we also investigated incisor cervical loops in mice lacking Alk5 in the epithelium (K14-Cre;Alk5fl/fl). The initiation of incisor development was normal for both mutants. Moreover, the cervical loops of both mutants were indistinguishable morphologically from that of the wild type (Fig. 1K and L and data not shown).
Loss of Alk5 in the dental mesenchyme results in reduced cell proliferation and fewer LRCs in the dental epithelium.
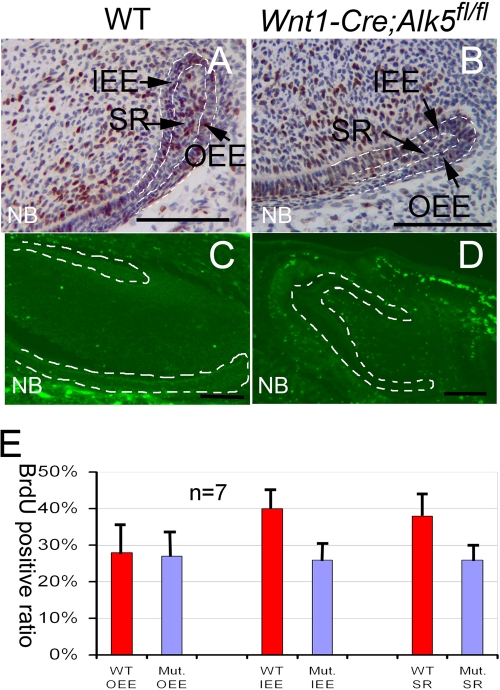
The reduced cervical loop size in Wnt1-Cre; Alk5fl/fl mutant incisors may be caused directly by reduced cell proliferation or increased apoptosis. To investigate these possibilities, we performed BrdU incorporation experiments with wild-type and Wnt1-Cre; Alk5fl/fl mice. In the lower incisors of wild-type newborn mice, we detected cells with BrdU incorporation in all three components of the cervical loops, including the IEE, OEE, and SR, although cell proliferation was higher in the IEE than in the OEE (Fig. 4A and E). Proliferation in the IEE and SR of Wnt1-Cre; Alk5fl/fl lower incisors was significantly decreased compared to that for the wild type (Fig. 4B and E). We detected a similar decrease in cell proliferation activity in E16.5 Wnt1-Cre; Alk5fl/fl incisors (data not shown). Next, we conducted TUNEL assays to investigate the apoptosis activity. We detected no positive apoptotic signals in the dental epithelium of either wild-type or Wnt1-Cre; Alk5fl/fl lower incisors at the newborn stage (Fig. 4C and D). We conclude that the loss of Alk5 in the dental mesenchyme results in reduced proliferation in the dental epithelium.
Fig. 4.
Proliferation is reduced in the cervical loop of Wnt1-Cre; Alk5fl/fl mutant incisors. Shown are BrdU incorporation (A, B, and E) and TUNEL (C and D) assay results for newborn wild-type (WT) (A and C) and Wnt1-Cre; Alk5fl/fl (B and D) lower incisors. (E) BrdU results were quantitated by cell type. At least three samples were analyzed for each experiment. Dotted lines (C and D) outline the dental epithelia. Mut, Wnt1-Cre; Alk5fl/fl mutant; OEE, outer enamel epithelium; IEE, inner enamel epithelium; SR, stellate reticulum. Error bars represent standard deviations. Scale bars, 200 μm.
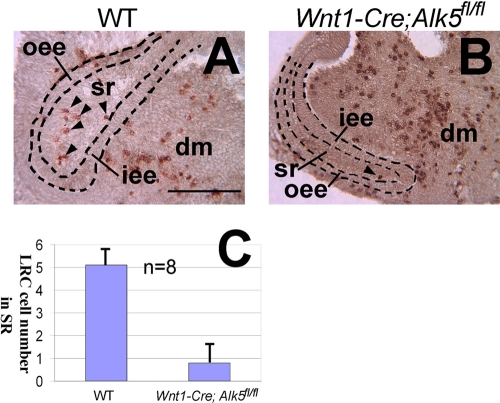
At present, no specific biochemical marker has been established that can be used to distinguish dental epithelial stem cells from surrounding cells. To test whether there is a dental epithelial stem cell defect in Wnt1-Cre; Alk5fl/fl mutant incisors, we used BrdU pulse-chase experiments, which have been established as a widely accepted approach to locate epidermal stem cells (2, 8, 27). To perform these experiments, we dissected wild-type and Wnt1-Cre; Alk5fl/fl tooth germs at the newborn stage and incubated them with medium containing BrdU for 24 h so that all the cells in the cervical loop would have incorporated BrdU (data not shown). The samples were then incubated in regular medium without BrdU for 2 weeks. We detected label-retaining cells (LRCs) in wild-type lower incisors throughout the stellate reticulum cells. On average, approximately 5 LRCs were detectable in each section (Fig. 5A and C). In Wnt1-Cre; Alk5fl/fl mutant mice, in contrast, less than 1 cell was detected on average in each section of the cervical loop (Fig. 5B and C). Therefore, we conclude that the cervical loop in Wnt1-Cre; Alk5fl/fl mice had defects in both TA cell and stem cell populations.
Fig. 5.
Label-retaining cell (LRC) numbers are decreased in Wnt1-Cre; Alk5fl/fl mutant incisors. (A and B) BrdU staining of lower incisors from newborn wild-type (WT) (A) and Wnt1-Cre; Alk5fl/fl (B) mice following pulse-labeling with BrdU for 24 h and 14 days of chase. Arrowheads indicate the BrdU LRCs within the cervical loops. Dashed lines outline the dental epithelia. (C) Quantitation of the average number of LRCs per section. At least eight samples were analyzed to obtain the mean values. dm, dental mesenchyme; iee, inner enamel epithelium; oee, outer enamel epithelium; sr, stellate reticulum. Error bars represent standard deviations. Scale bar, 200 μm.
Loss of Alk5 leads to reduced expression of FGF.
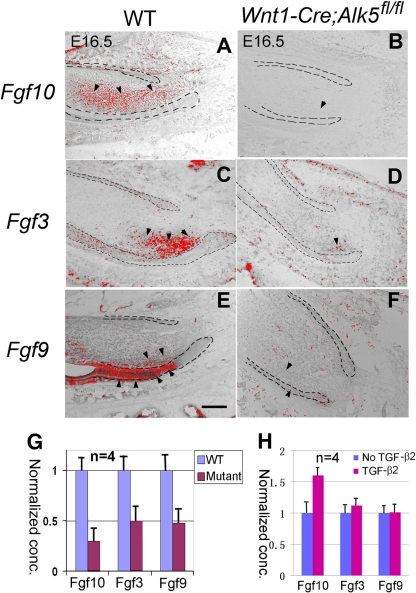
FGF3, FGF9, and FGF10 play important roles in regulating dental epithelial stem cells (29). In wild-type mice at E16.5, Fgf10 expression is detectable throughout the dental mesenchyme, with a stronger intensity at the labial side (Fig. 6A). We detected Fgf3 expression almost exclusively in the labial side dental mesenchyme near the cervical loop (Fig. 6C), whereas we detected Fgf9 expression in both the dental epithelium and mesenchyme. The strongest Fgf9 signal was localized to a small region of the dental epithelium just anterior to the labial side cervical loop (Fig. 6E). In E16.5 Wnt1-Cre; Alk5fl/fl lower incisors, the levels of expression of Fgf3, Fgf9, and Fgf10 were all significantly reduced (Fig. 6B, D, and F). We also detected similar expression level changes for Fgf3, Fgf9, and Fgf10 in Wnt1-Cre; Alk5fl/fl incisors at the newborn stage (data not shown), suggesting that the changes in Fgf expression are not due to a developmental delay in incisor development in Alk5 mutant mice.
Fig. 6.
Expression of Fgf3, Fgf10, and Fgf9 in Wnt1-Cre; Alk5fl/fl lower incisors and induction of Fgf10 by TGF-β2 in wild-type lower incisors. (A to F) In situ hybridization of Fgf3, Fgf9, and Fgf10 in E16.5 wild-type (WT) and Wnt1-Cre; Alk5fl/fl (mutant) lower incisors. Black dashed lines outline the dental epithelia. Arrowheads indicate the locations of positive signals. (G) Quantitation of real-time PCR analysis data using four samples for each genotype. (H) Real-time PCR analysis of Fgf10, Fgf3, and Fgf9 expression levels in wild-type incisors in the presence of 10 ng/ml TGF-β2. Error bars represent standard deviations. Scale bar, 200 μm.
To confirm the changes in Fgf expression levels, we collected lower incisor tooth germs from E16.5 wild-type and Wnt1-Cre; Alk5fl/fl embryos and extracted RNA for real-time PCR experiments to quantify the expression levels of Fgf3, Fgf9, and Fgf10. We found that the levels of expression of Fgf3, Fgf9, and Fgf10 were reduced 2- to 3-fold in Wnt1-Cre; Alk5fl/fl samples compared with those of the wild type (Fig. 6G).
To confirm the regulatory effects of TGF-β signals on Fgf genes, we collected lower incisors from newborn wild-type embryos and treated them with 10 ng/ml TGF-β2 in the organ culture system. Samples were collected 3 h after treatment, and RNA was extracted to quantify the expression levels of Fgf3, Fgf9, and Fgf10. We found that the expression level of Fgf10 was elevated by over 1.5-fold in the presence of 10 ng/ml TGF-β2 compared with untreated samples. In contrast, there was no significant change of Fgf3 and Fgf9 expression levels in the presence of TGF-β2 (Fig. 6H).
Analysis of gene expression known to be critical for regulating dental epithelial stem cells in Wnt1-Cre; Alk5fl/fl mice.
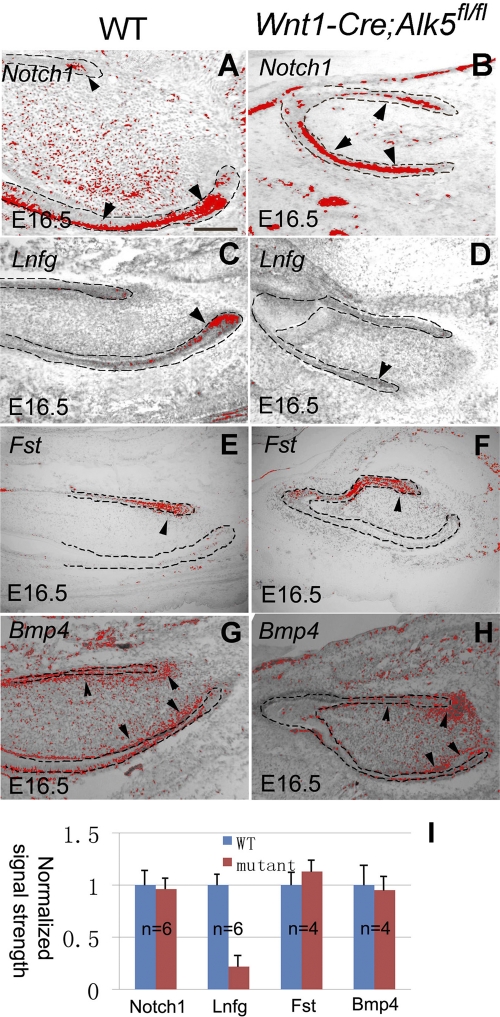
To determine the status of genes that play a role in dental epithelial stem cells, we assayed their expressions using in situ hybridization. Notch receptors and their modulators have been shown to be important for tooth morphogenesis and ameloblast differentiation. Notch1 and lunatic fringe (Lnfg) were previously proposed to regulate the fate of dental epithelial stem cells (9). At E16.5, Notch1 expression in the stellate reticulum cells of labial and lingual side dental epithelia was indistinguishable in intensity and pattern for wild-type and Wnt1-Cre; Alk5fl/fl mice (Fig. 7A and B). This result suggests that Notch1 expression in the dental epithelium is independent of Alk5 in the mesenchyme. In contrast, the level of Lnfg expression, which was restricted to a small region of IEE cells in wild-type mice (Fig. 7C), was significantly reduced in Wnt1-Cre; Alk5fl/fl incisors (Fig. 7D). Follistatin (Fst) plays a critical role in regulating the dental epithelial stem cell niche by interacting with Fgf and inhibiting stem cell proliferation (29). At E16.5, Fst expression was detectable mainly in the lingual side dental epithelium (Fig. 7E). Its expression pattern and intensity in Wnt1-Cre; Alk5fl/fl mutant incisors showed no significant difference compared with those of the control sample (Fig. 7F and I). Bmp4 is critical for regulating the differentiation of dental epithelial stem cells into ameloblasts (29). At E16.5, Bmp4 expression was detectable mainly in the dental mesenchyme (Fig. 7G). Its expression pattern and intensity in Wnt1-Cre; Alk5fl/fl mutant incisors showed no significant difference compared to those of the control (Fig. 7H and I).
Fig. 7.
Expression of genes related to dental epithelial stem cell regulation in Wnt1-Cre; Alk5fl/fl mutant incisors. In situ hybridizations of Notch1 (A and B), Lnfg (C and D), Follistatin (Fst) (E and F), and Bmp4 (G and H) in E16.5 wild-type (WT) or Wnt1-Cre; Alk5fl/fl mutant embryos are shown. Dashed lines outline dental epithelia. Arrowheads indicate signals. (E) Quantitation of signal strength measured with ImageJ software. Error bars represent standard deviations. Scale bar, 200 μm.
We also investigated other genes that may play a role in incisor development, including p63, Activin βA, Bmp7, Lef1, and Msx1, but found no evidence of altered expression in Wnt1-Cre; Alk5fl/fl lower incisors (data not shown).
Exogenous FGF10 rescues the dental epithelial stem cell defects in Wnt1-Cre; Alk5fl/fl lower incisors.
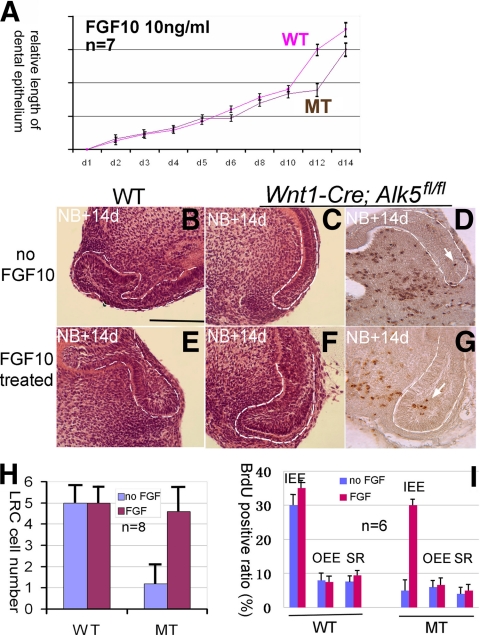
Because of the central role that Fgf10 plays in regulating dental epithelial stem cells, we examined whether the addition of exogenous FGF10 could rescue the stem cell defects in Wnt1-Cre; Alk5fl/fl mutant lower incisors. We added 10 ng/ml FGF10 into the culture medium of the organ culture system and measured the length of the labial side dental epithelium in each sample every other day for 2 weeks to generate a growth curve. In the presence of exogenous FGF10, the Wnt1-Cre; Alk5fl/fl lower incisor dental epithelium elongated at the same rate as that of the wild type in the first 10 days, and their growth curves overlapped beginning on the second day (Fig. 8A). After 10 days, the growth curve of Wnt1-Cre; Alk5fl/fl lower incisors treated with FGF10 progressed slowly, but no plateau stage was observed during the 14 days of culture (Fig. 8A). The incisor samples were embedded and examined by using microscopy after 14 days of culture. In the absence of FGF10, the size of the Wnt1-Cre; Alk5fl/fl cervical loop was much smaller than that of the wild type (Fig. 8B and C). The addition of FGF10 had no significant effect on the size of wild-type cervical loops (Fig. 8B and E). In contrast, we detected a significant increase in the size of the cervical loop in Wnt1-Cre; Alk5fl/fl samples following FGF10 treatment (Fig. 8C and F).
Fig. 8.
Rescue of dental epithelial stem cell defects by exogenous FGF10. (A) Growth curves of the dental epithelium from wild-type (WT) or Wnt1-Cre; Alk5fl/fl (MT) incisors cultured in the presence of FGF10 (10 ng/ml). (B to G) H&E staining shows the cervical loop morphology of wild-type (B and E) or Wnt1-Cre; Alk5fl/fl (C and F) samples and BrdU pulse-chase of Wnt1-Cre; Alk5fl/fl samples (D and G) after 14 days of culture in the absence or presence of FGF10. Dashed lines outline dental epithelia. Arrows indicate BrdU signals. (H) Quantification of the number of LRCs in wild-type (WT) and Wnt1-Cre; Alk5fl/fl (MT) samples to evaluate the effect of FGF10 on dental epithelial stem cell numbers. Eight samples were included for each treatment. (I) Quantification of BrdU-positive cells in wild-type (WT) and Wnt1-Cre; Alk5fl/fl (MT) samples to evaluate the effect of FGF10 on cervical loop proliferation. Six samples were included for each treatment. OEE, outer enamel epithelium; IEE, inner enamel epithelium; SR, stellate reticulum. Error bars represent standard deviations. Scale bar, 200 μm.
Next, we examined the effect of exogenous FGF10 on the number of LRCs in the cervical loop. In the absence of FGF10, we detected five LRCs on average in each section of the wild-type lower incisor and less than 1 cell in the Wnt1-Cre; Alk5fl/fl incisor sections (Fig. 8D and H). The addition of FGF10 (10 ng/ml) did not change the LRC numbers of the wild-type incisors significantly (Fig. 8H); however, the LRC numbers of the Wnt1-Cre; Alk5fl/fl incisors were restored to nearly 5 per section in the presence of FGF10 (Fig. 8G and H). Therefore, we conclude that FGF10 is able to rescue the stem cell maintenance defects in Wnt1-Cre; Alk5fl/fl incisors.
To examine the effects of FGF10 on different cell populations of the cervical loop, we cultured incisors in vitro for 24 h with or without FGF10 and analyzed proliferation using BrdU incorporation experiments. We found that the addition of 10 ng/ml FGF10 did not significantly increase the BrdU incorporation ratio in any of the three cell types of the wild-type cervical loop (Fig. 8I). In contrast, the addition of FGF10 increased the BrdU incorporation ratio of IEE cells in Wnt1-Cre; Alk5fl/fl samples by 3-fold (Fig. 8I), to a level comparable to that of wild-type IEE cells, but had no significant effects on SR and OEE cells in the same mutant samples. Thus, we conclude that FGF10 is able to rescue the proliferation defects of TA cells in Wnt1-Cre; Alk5fl/fl incisors.
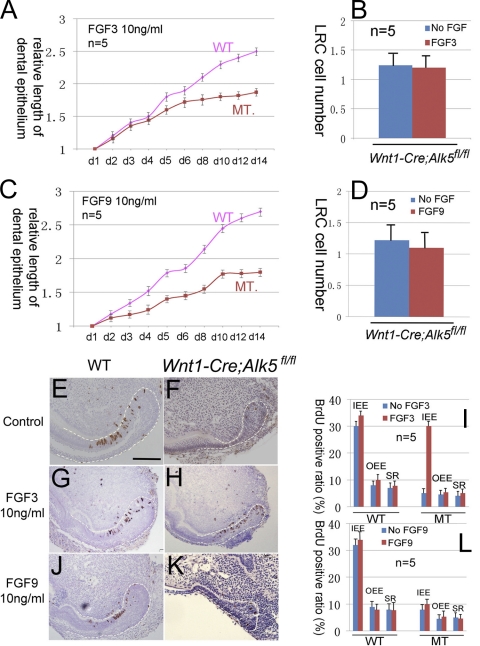
We also added FGF3 (10 ng/ml) or FGF9 (10 ng/ml) into the culture medium of organ cultures to examine their effects on cervical loop growth. The growth curve of Wnt1-Cre; Alk5fl/fl incisors was restored to the wild-type level in the first 3 days following the addition of FGF3. After 4 days, the growth curve of mutant samples progressed slowly and reached a plateau stage (Fig. 9). LRC cell numbers were not restored following the addition of FGF3 (Fig. 9B). BrdU incorporation experiments indicated that the addition of 10 ng/ml FGF3 significantly restored the proliferation of IEE cells but not OEE or SR cells in Wnt1-Cre; Alk5fl/fl incisor samples (Fig. 9G, H, and I). The addition of 10 ng/ml FGF9 had no effect on the growth curve of Wnt1-Cre; Alk5fl/fl incisor cervical loops (Fig. 9C). FGF9 also had no effect on the BrdU incorporation ratios of any of the three cell types of the cervical loop in Wnt1-Cre; Alk5fl/fl incisor samples (Fig. 9J, K, and L).
Fig. 9.
FGF3 and FGF9 are unable to rescue the dental epithelial stem cell defects in Wnt1-Cre; Alk5fl/fl mutant incisors. (A and C) Growth curves of the dental epithelium from wild-type (WT) or Wnt1-Cre; Alk5fl/fl (MT) incisors cultured in the presence of FGF3 (10 ng/ml) (A) or FGF9 (10 ng/ml) (C). (B and D) Quantification of the numbers of LRCs in wild-type (WT) and Wnt1-Cre; Alk5fl/fl (MT) samples in the presence of 10 ng/ml FGF3 (B) or FGF9 (D) to evaluate the effect of FGF3 on dental epithelial stem cells. (E to H, J, and K) BrdU incorporation in newborn wild-type (WT) (E, G, and J) and Wnt1-Cre; Alk5fl/fl (F, H, and K) lower incisors in the presence of 10 ng/ml FGF3 (G and H) or 10 ng/ml FGF9 (J and K). (I and L) BrdU results were quantitated by cell type. At least five samples were analyzed for each experiment. Dotted lines outline the dental epithelia. OEE, outer enamel epithelium; IEE, inner enamel epithelium; SR, stellate reticulum. Error bars represent standard deviations. Scale bar, 200 μm.
DISCUSSION
As the mouse incisor continues to grow throughout life, cells at various differentiation stages, from the initial quiescent stem cells to transit-amplifying cells, preameloblasts, and the terminally differentiated ameloblasts, align along the dental epithelium (9). The slowly dividing stem cells within the SR give rise to TA cells that reside within the IEE. TA cells in the IEE undergo active division, move toward the incisal direction, and differentiate into ameloblasts, forming enamel (9). These features make the incisor an excellent model for studying the regulatory mechanisms of stem cell maintenance, proliferation, and differentiation. In our previous study, we reported that the loss of Tgf-β type I receptor (Alk5) expression in neural crest cells leads to delayed tooth initiation (32). In this study, we focus on the functional significance of Alk5 in regulating the development of the lower incisors, especially the cervical loops. We show that an Alk5-Fgf signaling cascade plays a crucial role in regulating mesenchymal-epithelial cell interactions to control the proliferation of TA cells and the maintenance of epithelial stem cells during incisor growth.
Alk5 is essential for the proliferation of TA cells and maintenance of dental epithelial stem cells in the cervical loop.
In this study we found that the size of the cervical loop in Wnt1-Cre; Alk5fl/fl mutant lower incisors is dramatically reduced compared to that of the wild type. Consequently, mutant incisors are smaller than those of the wild type, and their ability to support continuous growth is compromised. Based on our experiments, we conclude that the defects in the Alk5 mutant incisors are due to a reduced proliferation of TA cells residing in the IEE and a reduced number of dental epithelial stem cells residing in the stellate reticulum.
Our model of a TA cell defect is directly supported by data from BrdU incorporation experiments that indicated a significant reduction in the proliferation of the IEE populations in Alk5 mutant samples (Fig. 4E). In our in vitro organ culture experiments, the elongation of Alk5 mutant incisors occurred at a lower rate than that of the wild type during the first couple of days. This provides further evidence supporting a TA cell defect, because the division of TA cells is the immediate driving force of incisor elongation. Lnfg expression is restricted to the IEE area and is considered to be related to the regulation of TA cells in the cervical loop (9). The reduced Lnfg expression levels in Wnt1-Cre; Alk5fl/fl lower incisors are also consistent with a TA cell defect.
Evidence for the dental epithelial stem cell defect comes from morphological examinations and BrdU pulse-chase experiments. In vitro organ cultures of the dental epithelium have been used for assaying the function of dental epithelial stem cells (10). In our experiments, the elongation of the Wnt1-Cre; Alk5fl/fl incisor dental epithelium was arrested after 10 days. This result suggests that the cervical loops in Wnt1-Cre; Alk5fl/fl incisors have lost the ability to support continuous growth after the TA cells supporting initial growth have been depleted due to their limited ability for self-renewal. FGF rescue experiments are also consistent with a stem cell defect. FGF3 stimulates IEE cell proliferation in Wnt1-Cre; Alk5fl/fl incisors to a level comparable to that of the wild type but is not able to restore the LRC numbers. Consequently, the growth curve of Wnt1-Cre; Alk5fl/fl incisors in the presence of FGF3 was restored only in the first 3 days but then slowed down significantly due to stem cell depletion.
Alk5 is upstream of Fgf10 in regulating dental epithelial stem cells.
FGF family members play critical roles in regulating dental epithelial stem cells. The cervical loop is absent in the incisors of Fgf10 null mice, and enamel formation is defective in the incisors of Fgf3 null mice (10, 19). A complicated interaction network involving Fgf3, Fgf10, Fst, and Activin βA was proposed previously to regulate dental epithelial stem cells and to establish the asymmetrical labial and lingual cervical loop structures (29). In our study, the levels of expression of Fgf10, Fgf3, and Fgf9 were significantly reduced, whereas Fst, Bmp4, and Activin βA expression levels were unchanged, in the lower incisors of Wnt1-Cre; Alk5fl/fl mice. The addition of FGF10 to the tooth organ culture medium restored the cervical loop size, restored the self-renewal ability of dental epithelial stem cells in Wnt1-Cre; Alk5fl/fl incisors, and restored LRC numbers in Wnt1-Cre; Alk5fl/fl cervical loops to a level comparable to that of the wild type. Moreover, in the in vitro organ culture system, TGF-β2 was able to induce the expression of Fgf10. Thus, we conclude that Alk5 is upstream of Fgf10 in regulating the proliferation and maintenance of dental epithelial stem cells. In our previous studies, we have shown that interactions between TGF-β and FGF signaling pathways are crucial for tongue and calvaria development (11, 22). Collectively, our studies suggest that TGF-β-mediated FGF signaling is a well-conserved signaling cascade that mediates tissue-tissue interactions during craniofacial development.
Our experiments indicate that FGF10 and FGF3 may have differential impacts on the dental epithelial progenitors in the incisor cervical loop. In the Wnt1-Cre; Alk5fl/fl explant culture, FGF10 was able to stimulate TA cell proliferation and maintain the stem cell population, whereas FGF3 acted only on TA cells to stimulate their proliferation and had no effect on the stem cell population. Such findings are intriguing because both FGF3 and FGF10 bind to FGFR2IIIb to mediate their functions. In some studies, FGF3 and FGF10 were previously considered to be redundant to each other in incisor development (10). The incisor phenotype differences between Fgf3−/− and Fgf10−/− mice were explained as their different expression patterns in vivo (10). More experiments need to be conducted to investigate the functional difference between FGF3 and FGF10 under physiological conditions.
In the conventional signaling model, Alk5 functions concomitantly with the TGF-β type II receptor to mediate the TGF-β signaling pathway (7, 16). However, our previous study has shown that these two receptors may have independent roles to regulate tooth initiation. The initiation of both incisors and molar tooth germs is delayed in Wnt1-Cre; Alk5fl/fl mice but not in Wnt1-Cre; Tgfbr2fl/fl mice (32). In this study, we show that these two receptors also have independent roles in regulating dental epithelial stem cells. Cervical loops in Wnt1-Cre; Tgfbr2fl/fl incisors appear to be unaffected morphologically. One possible explanation for the phenotype discrepancy between Alk5 and Tgfbr2 mutant mice is that Alk5 may bind to TGF-β superfamily ligands other than TGF-β to regulate dental epithelial stem cells. Besides TGF-β, growth/differentiation factor 8 (GDF8), GDF9, and GDF11 are able to bind to Alk5 (1, 15, 21). Only GDF11 is expressed in the craniofacial region, and no tooth phenotype has been detected in Gdf11−/− mice (17). Another possibility is that TGF-β binds to the type II receptor or Alk5 separately and initiates different downstream signaling cascades. More studies will be needed to reveal the intracellular processes downstream of Alk5 that regulate dental epithelial stem cells.
Mesenchymal-epithelial cell interactions are crucial for maintaining the dental epithelial stem cell compartment.
A niche is required for stem cells to maintain their quiescent status. The stellate reticulum within the cervical loop was previously proposed to be the niche for dental epithelial stem cells (9). The cervical loop structure is not self-sustaining and requires signals from the dental mesenchyme, such as FGF10 and FGF3, for its maintenance (10, 19). The cervical loop undergoes apoptosis in the absence of the dental mesenchyme (10). The instructive role of the dental mesenchyme is further supported by our results showing that the loss of Alk5 in the dental mesenchyme leads to an epithelial cell defect in the cervical loop. Interestingly, the loss of Alk5 in the dental epithelium had no visible effect on the cervical loop. Based on our FGF10 rescue experiments, we propose that ALK5-mediated TGF-β signaling controls FGF10 expression and regulates mesenchymal-epithelial cell interactions to maintain dental epithelial stem cells and the continuous growth of mouse incisors. Our discovery may ultimately facilitate stem cell-mediated tissue regeneration for oral tissue.
ACKNOWLEDGMENTS
We thank Julie Mayo for critical reading of the manuscript; Pablo Bringas, Jr., for technical assistance; and Irma Thesleff and Ophir Klein for in situ probes.
This study was supported by grants from the National Institute of Dental and Craniofacial Research, NIH (grants R37 DE012711, R01 DE014078, and R01 DE017007) to Yang Chai.
Footnotes
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Andersson O., Reissmann E., Ibáñez C. F. 2006. Growth differentiation factor 11 signals through the transforming growth factor-beta receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep. 7:831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bickenbach J. R. 1981. Identification and behavior of label-retaining cells in oral mucosa and skin. J. Dent. Res. 60:1611–1620 [DOI] [PubMed] [Google Scholar]
- 3. Cotsarelis G., Cheng S. Z., Dong G., Sun T. T., Lavker R. M. 1989. Existence of slow-cycling limbal epithelial basal cells that can be referentially stimulated to proliferate: implications on epithelial stem cells. Cell 57:201–209 [DOI] [PubMed] [Google Scholar]
- 4. Cotsarelis G., Sun T. T., Lavker R. M. 1990. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61:1329–1337 [DOI] [PubMed] [Google Scholar]
- 5. Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P. 1998. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8:1323–1326 [DOI] [PubMed] [Google Scholar]
- 6. Dudas M., et al. 2006. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev. Biol. 296:298–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feng X. H., Derynck R. 2005. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 21:659–693 [DOI] [PubMed] [Google Scholar]
- 8. Fuchs E. 2009. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell 137:811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harada H., et al. 1999. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J. Cell Biol. 147:105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harada H., et al. 2002. FGF10 maintains stem cell compartment in developing mouse incisors. Development 129:1533–1541 [DOI] [PubMed] [Google Scholar]
- 11. Hosokawa R., et al. 2010. TGF-beta mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev. Biol. 341:186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ito Y., et al. 2003. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 130:5269–5280 [DOI] [PubMed] [Google Scholar]
- 13. Klein O. D., et al. 2008. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development 135:377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsson J., et al. 2001. Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J. 20:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazerbourg S., et al. 2004. Growth differentiation factor-9 signaling is mediated by the type I receptor, activin receptor-like kinase 5. Mol. Endocrinol. 18:653–665 [DOI] [PubMed] [Google Scholar]
- 16. Moustakas A., Heldin C. H. 2009. The regulation of TGF-beta signal transduction. Development 136:3699–3714 [DOI] [PubMed] [Google Scholar]
- 17. Nakashima M., Toyono T., Akamine A., Joyner A. 1999. Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech. Dev. 80:185–189 [DOI] [PubMed] [Google Scholar]
- 18. Oka S., et al. 2007. Cell autonomous requirement for TGF-beta signaling during odontoblast differentiation and dentin matrix formation. Mech. Dev. 124:409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parsa S., et al. 2010. Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development 137:3743–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Potten C. S., Owen G., Booth D. 2002. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 115:2381–2388 [DOI] [PubMed] [Google Scholar]
- 21. Rebbapragada A., Benchabane H., Wrana J. L., Celeste A. J., Attisano L. 2003. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol. Cell. Biol. 23:7230–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sasaki T., et al. 2006. TGFbeta-mediated FGF signaling is crucial for regulating cranial neural crest cell proliferation during frontal bone development. Development 133:371–381 [DOI] [PubMed] [Google Scholar]
- 23. Seidel K., et al. 2010. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development 137:3753–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith C. E. 1980. Cell turnover in the odontogenic organ of the rat incisor as visualized by graphic reconstructions following a single injection of 3H-thymidine. Am. J. Anat. 158:321–343 [DOI] [PubMed] [Google Scholar]
- 25. Thesleff I., Wang X. P., Suomalainen M. 2007. Regulation of epithelial stem cells in tooth regeneration. C. R. Biol. 330:561–564 [DOI] [PubMed] [Google Scholar]
- 26. Tucker A. S., Sharpe P. T. 2004. The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 5:499–508 [DOI] [PubMed] [Google Scholar]
- 27. Tumbar T., et al. 2004. Defining the epithelial stem cell niche in skin. Science 303:359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X. P., et al. 2004. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev. Cell 7:719–730 [DOI] [PubMed] [Google Scholar]
- 29. Wang X. P., et al. 2007. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 5:1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilkinson D. G. 1992. Whole mount in situ hybridization of vertebrate embryos, p. 75–83 In Wilkinson D. G. (ed.), In situ hybridization. IRL, Oxford, United Kingdom [Google Scholar]
- 31. Zeps N., Bentel J. M., Papadimitriou J. M., D'Antuono M. F., Dawkins H. J. 1998. Estrogen receptor-negative epithelial cells in mouse mammary gland development and growth. Differentiation 62:221–226 [DOI] [PubMed] [Google Scholar]
- 32. Zhao H., Oka K., Bringas P., Kaartinen V., Chai Y. 2008. TGF-beta type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev. Biol. 320:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]