Abstract
Krüppel-like factor 4 (KLF4), a transcription factor that regulates cell fate in a context-dependent fashion, is normally induced upon growth arrest or differentiation. In many cancer cells there is dysregulation, with increased expression in proliferating cells. To identify sequence elements that mediate KLF4 suppression in normal epithelial cells, we utilized a luciferase reporter and RK3E cells, which undergo a proliferation-differentiation switch to form an epithelial sheet. A translational control element (TCE) within the KLF4 3′-untranslated region interacted with microRNAs (miRs) 206 and 344-1 to promote or inhibit KLF4 expression, respectively, in proliferating epithelial cells. Overall, the TCE suppressed expression in proliferating primary human mammary epithelial cells, but this suppressive effect was attenuated in immortalized mammary epithelial MCF10A cells, in which Dicer1 and miR-206 promoted KLF4 expression and TCE reporter activity. In contrast to MCF10A cells, in breast cancer cells the activity of miR-206 was switched, and it repressed KLF4 expression and TCE reporter activity. As miR-206 levels were KLF4 dependent, the results identify a KLF4–miR-206 feedback pathway that oppositely affects protein translation in normal cells and cancer cells. In addition, the results indicate that two distinct miRs can have opposite and competing effects on translation in proliferating cells.
INTRODUCTION
The zinc finger protein Krüppel-like factor 4 (KLF4) regulates gene transcription and cell fate in a context-dependent fashion, promoting cell differentiation, tumor suppression, stem cell properties, and malignant transformation (2, 21, 40, 58). Although Klf4 is dispensable for early development, analysis of postnatal, Klf4-deficient mice revealed roles in formation of the cutaneous water permeability barrier, in formation of mucosecreting goblet cells in the gut or conjunctiva, and in late fetal or early postnatal cardiac development (23, 24, 30, 42, 46, 61). In addition to its developmental roles, KLF4 regulates the phenotype of cancer cells and stem cells. While KLF4 appears to suppress tumor formation in tissues such as the gut (5, 12, 65), it can promote malignant properties in other tissues, such as the breast and skin (8, 10, 31, 37, 39, 45, 62). When expressed in adult somatic cells with other Yamanaka factors, KLF4 can promote the formation of induced pluripotent stem (IPS) cells (38, 47, 48, 58).
How KLF4 mediates its pleiotropic effects is an area of current study. KLF4 typically reduces cell proliferation rates, possibly through regulation of p21Waf1/Cip1 or other factors (39, 64). Even though KLF4 slows cell proliferation, human carcinomas are often slow growing, and KLF4 may promote malignant properties in this context through suppression of p53 or by upregulation of Notch1 and confer stem cell properties in embryonic stem (ES) cells through induction of factors such as Nanog (16, 31, 39, 63).
A seminal observation by Yang and colleagues was the induction of endogenous Klf4 transcripts and protein following in vitro growth suppression (43, 64). A variety of growth-suppressive signals lead to upregulation of KLF4, including contact inhibition, serum starvation, DNA damage, and differentiation signals, such as retinoids or cyclic AMP (3, 43, 54, 59, 64). These in vitro results suggest an inverse relationship between KLF4 levels and cell proliferation rates and are supported by extensive analyses in vivo that revealed that KLF4 mRNA and protein are selectively expressed in the postmitotic, differentiating cell layers of epithelia such as the skin, gut, and oral mucosa (10, 11, 42, 43).
Mechanisms accounting for induction of KLF4 upon growth arrest or differentiation potentially involve the gain of positive factors as well as the loss of suppressive influences on transcription, translation, or protein stability. In rapidly dividing colorectal cancer cells, ubiquitin-mediated proteolysis destabilizes KLF4, and protein stabilization therefore contributes to the induction of KLF4 upon serum starvation (4). Since KLF4 can induce its own transcription, stabilization of the protein in growth-arrested cells can potentially lead to positive feedback (6, 33).
Given its role as a stem cell factor that can promote malignant transformation, regulatory mechanisms that suppress KLF4 in proliferating cells may be important to restrict cancer progression and/or the acquisition of stem cell phenotypes. Support for this notion includes the observation that KLF4 is upregulated in the basal epithelial cells of dysplastic or malignant lesions in the skin and oropharynx (10, 14, 18) and of the activity of KLF4 as an oncogene when induced in the basal layer of mouse skin (10).
MicroRNAs (miRs), processed from pre-miR hairpin structures by DICER1 (DCR1), associate with Argonaute family members and other components to generate micro ribonucleoproteins (miRNPs) that can suppress or promote protein translation through regulatory elements within mRNAs (1, 13, 20, 27, 36, 44, 51–53). In the current study we observed cell-type-specific effects of DCR1 knockdown on cellular levels of KLF4. We identified a TCE coregulated by translation-stimulatory miRs (i.e., miR-206 in human and rodent cells) and translation-inhibitory miRs (i.e., miR-344 in rodent cells). The TCE suppressed the activity of a luciferase reporter in proliferating epithelial cells, where endogenous KLF4 was low, but promoted reporter activity in other contexts where the endogenous protein was increased. These effects were attributed to induction of miR-206 by KLF4, creating a positive feedback loop for translational control in primary human mammary epithelial cells (HMECs), MCF10A, and other epithelial cells. Unlike in normal epithelial cells, miR-206 inhibited translation in malignant cells, indicating a switch from a positive to a negative regulator during tumor progression. The results identify miR regulation of protein translation as a mechanism by which epithelial KLF4 is suppressed in normal proliferating cells. Altered TCE activity contributed to the upregulation of KLF4 in both immortalized epithelial cells and cancer cells. TCE regulation is intricate and involves competition between miRs that have opposing effects. Finally, the distinct effects of miR-206 in normal cells and cancer cells may account for some of the distinct and context-dependent effects of KLF4 on tumor progression (5, 8, 10, 12, 31, 37, 39, 45, 62, 65).
MATERIALS AND METHODS
Cell lines and cell culture.
Mammary epithelial MCF10A and breast cancer cell lines were obtained from ATCC. Primary HMECs (passage 6) were purchased from Invitrogen. MCF10A and 184A1 cells were cultured as previously reported (7, 8, 34). HMECs were cultured in mammary epithelial basal medium (Lonza) supplemented with bovine pituitary extract, hydrocortisone, epidermal growth factor, insulin, gentamicin, amphotericin B, transferrin, and isoproterenol. T47D and BT20 cells were cultured in RPMI 1640 supplemented with penicillin, streptomycin, and 10% fetal bovine serum. MCF7 and BT474 cells were cultured similarly except that insulin was added. MDA-MB-231 and RK3E cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with penicillin, streptomycin, and 10% fetal bovine serum. 4-Hydroxytamoxifen (4OHT) was dissolved in ethanol and, where indicated, was added to cultures at a final concentration of 0.3 μM.
Plasmid construction.
The pMIR-REPORT luciferase vector was purchased from Ambion/Invitrogen. pRL-TK was obtained from Promega (Madison, WI). KLF4 (GenBank accession number AF105036) fragments were excised from pCTV3K-SCC7-1 (10) and cloned into the 3′-untranslated region (UTR) of pMIR-REPORT luciferase. For pMIR-REPORT-K1S (K1S), a 446-bp SalI-SacI fragment was cloned into PmeI and SacI sites of the vector. SacI fragments of 465 bp (K2S), 495 bp (K3S), and 380 bp (K4S) were cloned into the SacI sites of the vector. For K5S, a 300-bp EcoRI fragment was cloned into the EcoRI site of the vector. For K6S, a 558-bp EcoRI-SalI fragment was treated with Klenow polymerase and doexynucleoside triphosphates (dNTPs) to generate blunt ends and cloned into the PmeI site of the vector. To insert a nearly full-length (FL) cDNA, a SalI-Asp718 fragment that included the KLF4 sequence in AF105036 was treated with Klenow polymerase and dNTPs and then cloned into the PmeI site of the vector.
Subfragments of K5S were generated by PCR using K5S reporter as template. The PCR products were digested with HindIII and XhoI and cloned into the same sites of pMIR-REPORT luciferase. Primers were as follows (with the KLF4 sequence shown in uppercase letters): K5S-1781Sense, 5′-gggagctcAATTCAGTATTTTTTACTTTTCAC-3′; Vector Antisense, 5′-aggcgattaagttgggtaacgc-3′; K5S- 1Antisense, 5′-cccaagcttAAGATGACTCAGTTGGGAACTTG-3′; K5S-2Sense, 5′-gggagctcGTGAGTGGATAATCAGGAAAAATG-3′; K5S-2Antisense, 5′-cccaagcttCGGATTTAGAATTGGAATGATAG-3′; K5S-3Sense, 5′-gggagctcACTTGAATATTCCTGGACTTAC-3′; K5S-4Sense, 5′-gggagctcCCAGCCAGAAAGCACTACAATC-3′; K5S-4Antisense, 5′-cccaagcttCTTTGATTTTTGTCTTTTGGATTC-3′; K5S-5Sense, 5′-gggagctcAACAGATGGGGTCTGTGACTGG-3′; K5S-5Antisense, 5′-cccaagcttCAACTTCCAGTCACCCCCTTGG-3′; K5S-6Antisense, 5′-cccaagcttCGGATTTAGAATTGGAATGATAG-3′; K5S-7Sense, 5′-gggagctcGTGAGTGGATAATCAGGAAAAATG-3′. Cloned PCR products were confirmed to be wild type by sequence analysis.
To generate K5S reporters with mutant miR seed sites, mFold was used to identify seed sequence alterations that retained the secondary structure predicted for the wild-type reporter (66). PCR-mediated mutagenesis was performed, and constructs were confirmed by sequence analysis. To test for complementation of mutant reporters, the following small interfering RNAs (siRNAs) were purchased from Dharmacon (seed mutations are indicated in lowercase letters): MtmiR344B, 5′-UGAaCgAGCCAAAGCCUGACUGU-3′; MtmiR206B, 5′-UGGAuaGUAAGGAAGUGUGUGG-3′.
To generate retroviral vectors encoding HA-KLF4-FL and HA-KLF4-FLΔK5S, a BspEI-SalI fragment containing the 3′-UTR of KLF4 was inserted into the BspEI and XhoI sites of pcDNA3.1-HA-KLF4 (37) to generate pcDNA3.1-HA-KLF4-FL. pcDNA3.1-HA-KLF4-FLΔK5S was generated by digestion with EcoRI followed by religation so as to delete the internal EcoRI fragment. BamHI-XbaI fragments were excised from these constructs and inserted into the same sites of the retroviral vector pB-puro. For rescue of KLF4 knockdown cells, the retroviral vector pLJD-KLF4-ERT, conferring Geneticin resistance, was constructed by transferring the insert from pBpuro-KLF4-ERT (9).
KLF4 knockdown (shKLF4) utilized pGIPZ lentiviral shRNAmir plasmid (Open Biosystems V2LHS_28277; mature sense, 5′-GCCAGAAAGCACTACAATC-3′; mature antisense, 5′-GATTGTAGTGCTTTCTGGC-3′), which targets 3′-UTR sequence within the K5S fragment, and nonsilencing-GIPZ lentiviral shRNAmir control (shCtrl; RHS4346). psPAX2 and pCMV-VSVg packaging plasmids were obtained from Addgene.
Transient-transfection, luciferase reporter, and translation efficiency assays.
The data shown below in Fig. 2 to 4 were generated using “forward transfection,” indicating that cells were first plated at 5 × 104 cells per well in 12-well plates 24 h prior to transfection. Forward transfections were performed when cells were at approximately 30% confluence. Transfection mixtures included 50 ng of pMIR-Report vector, with or without insert, and 20 ng of pRL-TK. TransIT-LT1 transfection reagent (3 μl; Mirus Bio LLC, Madison, WI) was used according to the manufacturer's instructions, except that keratinocyte growth medium (Invitrogen) was substituted for Opti-MEM.
Fig. 2.
The KLF4 transcript can suppress translation of a linked reporter prior to the epithelial proliferation-differentiation switch. (A) Full-length human KLF4 cDNA (KLF4-FL) was inserted in the sense orientation into the 3′-UTR of FLuc. (B) Following transfection of KLF4-FL or vector in parallel, the normalized FLuc activity ratio was determined (i.e., FLucKLF4-FL/FLucvector). Proliferating, RK3E cells were harvested at 50% confluence; PC24 h, RK3E cells were harvested 24 h after reaching confluence. (C) Wells transfected in parallel (see panel B) were used for preparation of RNA. Luc mRNAs were analyzed using qRT-PCR with normalization to the Gapdh gene. (D, left) Calculation of relative translation efficiency (RTE) of construct X relative to construct V. (Right) The RTE of KLF4-FL was calculated from the data shown in panels B and C (three independent experiments; bars show standard errors).
Fig. 4.
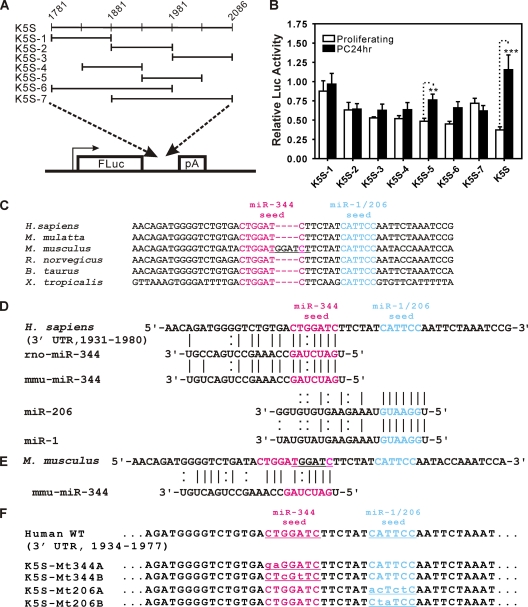
A 100-nt subfragment of the TCE directs growth-state-dependent expression and identifies evolutionarily conserved binding sites for candidate TCE-regulating miRs. (A) To localize potential functional elements, 100- to 200-nt fragments were analyzed in the reporter assay. (B) K5S-5, corresponding to bases 1931 to 2030 of GenBank accession number AF105036, showed growth-state-dependent activity (six independent experiments; bars show standard errors). (C) Alignment of sequences from the proximal one-half of K5S-5 (bases 1931 to 1980). A recent 4-nt insertion (GGAT) in the mouse created a potential new seed for miR-344 (underlined). (D) Alignments of candidate interacting miRs with the K5S-5 subfragment (rno, Rattus norvegicus; mmu, Mus musculus). (E) Conservation in mice of the miR-344 seed, 3′ pairing, and spacing relative to the seed for miR-1/206 despite the 4-base duplication (GGAT). (F) Seed mutations were introduced into the FLuc reporter K5S, corresponding to bases 1781 to 2086 of GenBank accession number AF105036.
For analysis of miR activities, subsequent experiments (summarized below in Fig. 5 to 9) utilized “reverse transfection,” in which cells were added to wells that contained preformed lipid-nucleic acid complexes. Anti-miR inhibitors (AMs; single-stranded, chemically modified nucleic acids) or miR mimics (PMs; mature miR/miR* duplexes that are loaded onto RISC without additional processing) were obtained from Ambion/Invitrogen, including miR-344-1 (IDAM10439, PM10439), miR-1 (AM10617, PM10617), and miR-206 (AM10409, PM10409), as well as AM negative control 1 (AM17010), and PM negative control 1 (AM17110). Inhibitors and mimics were diluted to 20 μM in water and used for cell transfections at a final concentration of 25 nM in a 0.50-ml total volume in 3.8-cm2 wells (12-well plates). Briefly, lipid-RNA complexes were allowed to form by incubation for 20 min in the wells by addition of 100 μl Opti-MEM I medium (Invitrogen), 2.0 μl of Lipofectamine RNAiMAX transfection reagent (Invitrogen), and nucleic acids. In a separate tube, lipid-DNA complexes were allowed to form for 20 min following addition of 100 μl serum-free medium, 3.0 μl TransIT-LT1, and plasmid DNA (quantities were as described for forward transfections). Lipid-DNA complexes were then added to the wells containing lipid-RNA complexes. Following trypsinization and washing, cells were resuspended in keratinocyte-SFM (Invitrogen; supplemented with epidermal growth factor and bovine pituitary extract) at 3.3 × 105 cells/ml, and 0.30 ml was added to each well. Plates were incubated for 2 h at 37°C in 5% CO2, and then 1.0 ml of complete growth medium was added to each well.
Fig. 5.
Regulation of epithelial Klf4 by miRs. (A) Endogenous miR levels were analyzed by SL-RTQ. miR levels (PC24hr) are presented as a fraction of that observed in proliferating cells (Table 1). (B) TCE regulation by exogenous (left panel) or endogenous (right panel) miRs was analyzed in proliferating RK3E cells. (C) Effects of miRs on the rate of KLF4 nascent protein synthesis were determined by metabolic labeling of proliferating cells for one protein half-life (48 min), followed by IP of N-terminally tagged HA-KLF4. RK3E cells transduced with empty vector served as a control. The species indicated by the asterisk was undetected in vector control cells and may therefore represent a cleavage product or else an alternate isoform of HA-KLF4. Equal cpm of radiolabeled protein were used as input for each IP (lower panels). (D) Endogenous Klf4 protein (upper panels) and mRNA (lower panels) were analyzed in proliferating RK3E cells treated in parallel with miR or anti-miR (three independent experiments where protein and mRNA was isolated; bars show standard errors). (E) Abilities of exogenous (left panel) or endogenous (right panel) miRs to compete for regulation of the TCE were assessed in proliferating RK3E cells. (F) WT or mutant versions of the TCE (K5S) reporter were analyzed in proliferating RK3E cells (left panel). miR mimics, containing base changes to complement the mutant TCE reporters, were assessed for regulation of reporter activity (center and right panels).
Fig. 9.
KLF4 autorepression through induction of miR-206 in breast cancer cells. (A) MDA-MB-231 cells were transduced with a lentivirus vector that expresses shRNA to KLF4 or a control shRNA. Transduced cells were selected in puromycin, and KLF4 protein levels were determined by immunoblot analysis. KLF4 shRNA targets the 3′-UTR within the TCE but outside K5S-5. (B) Transwell migration or invasion assay of KLF4 knockdown and control cells. (C) Endogenous miR-206 levels were analyzed by qRT-PCR. (D) KLF4 knockdown cells were transduced with KLF4-ERT vector or control. Cells were selected in Geneticin and puromycin, and resistant cells were induced with 4OHT as indicated. KLF4-ERT vector does not contain the 3′-UTR that is targeted by the shRNA. (E) Conditional rescue of migration by the KLF4-ERT vector. Cells were treated with 4OHT for 24 h prior to plating the cells in Transwell chambers. (F) K5S-5 reporter activity was measured in proliferating cells. (G) miR-206 was modulated by transfection of miR into KLF4 knockdown cells, where endogenous miR-206 was low (see panel C) or by transfection of anti-miR into control cells, in which endogenous miR-206 is high. Cells were analyzed 48 h after transfection. For panels A through G, cells were analyzed at approximately 70% confluence. Panels B through G show results for three independent experiments; bars indicate the standard errors.
Luciferase assays utilized the dual luciferase reporter assay system (DLR assay; Promega). Extracts were prepared in 0.15 ml of 1× passive lysis buffer (PLB), and luciferase activities were determined at 24 h posttransfection (for proliferating cells, at approximately 50 to 60% confluence) or at 72 h posttransfection (postconfluence for 24 h [PC24 h]). Briefly, 20 μl of lysate was added to 50 μl of luciferase substrate, and luciferase activity was measured using a Modulus microplate multimode reader (Promega). Experiments, performed in duplicate, were repeated for a total of three or more independent experiments.
To determine translation efficiency, culture wells were transfected in parallel to isolate either RNA or protein. To adjust for well-to-well variations in transfection efficiencies, the firefly luciferase (FLuc)/Renilla luciferase (RLuc) ratio (RNA or Luc activity) was first determined for each well, and the mean ratio was determined for a set of replicates. FLuc and RLuc RNA ratios were determined using the 2−ΔCT method with glyceraldehyde dehydrogenase (GAPDH) as the control. Translation efficiency for each construct was calculated as follows: (FLuc/RLuc)RLU/(FLuc/RLuc)RNA. A detailed example calculation is available upon request from the authors.
Cell transfection and virus transduction.
Human DCR1 (NM_030621) knockdown experiments utilized ON-TARGETplus RNA interference (RNAi) duplexes and, as a control, the siGENOME nontargeting siRNA 2 (Dharmacon D-001210-02). The sequence for catJ-003483-09 (si-DCR1 number 09) is 5′-UAAAGUAGCUGGAAUGAUGUU-3′ (sense) and 5′-P-CAUCAUUCCAGCUACUUUAUU-3′ (antisense); the sequence for catJ-003483-11 (si-DCR1 number 11) is 5′-ACACAGCAGUUGUCUUAAAUU-3′ (sense) and 5′-P-UUUAAGACAACUGCUGUGUUU-3′ (antisense).
Lentiviral particles were packaged in HEK293T cells following calcium phosphate-mediated cotransfection of shRNA vector, psPAX2, and pCMV-VSVg. Retroviral transduction of pBpuro constructs into RK3E cells was performed using supernatants of BOSC23 cells as described previously (10). Retroviral transduction of human cells utilized the supernatant of AM12 amphotropic packaging cells (a gift from L. T. Chow, University of Alabama at Birmingham). Following incubation with lentiviral or retroviral particles, cells were selected in puromycin (0.5 μg/ml) or Geneticin (200 μg/ml).
Cell growth rate and cell cycle analysis.
Cells were maintained at subconfluence. Following trypsinization, 1 × 103 cells were transferred into 96-well plates and cultured for the indicated intervals. Cell number was determined using the CellTiter-Glo luminescent cell viability assay (Promega). Fluorescence-activated cell sorting analysis of cell cycle distribution was performed following staining of cells with propidium iodide using the Cycle Test Plus DNA reagent kit (BD Biosciences).
Metabolic labeling, immunoprecipitation, and protein half-life determination.
To assess the rate of KLF4 protein synthesis, 2.2 × 106 cells were seeded in a 10-cm dish. Where miRs were analyzed the dishes contained preformed lipid-RNA complexes. After culture for 48 h the cells were washed twice in phosphate-buffered saline (PBS) and then cultured for an additional 2 h in l-methionine/l-cysteine (Met/Cys)-free DMEM supplemented with 10% FBS, 20 mM HEPES-NaOH (pH 7.2) and l-Gln. 35S[Met/Cys] (Perkin Elmer) was added directly to the culture medium at 200 μCi/ml. Labeled cells were rinsed twice in cold PBS and then scraped in 200 μl of boiling denaturation buffer (BDB; 0.5% SDS, 50 mM Tris-HCl [pH 8.0], 10 mM dithiothreitol). Extracts were passed through a 26-gauge needle 10 times to reduce the viscosity, heated to 100°C for 5 min, and then diluted with 4 volumes of radioimmunoprecipitation assay (RIPA) buffer without SDS (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5.0 mM EDTA, 1.0 mM NaF, 1% sodium deoxycholate, 1% NP-40, and 0.25 mM phenylmethylsulfonyl fluoride). Incorporated counts per minute were determined by trichloroacetic acid precipitation, and 6.0 × 107 cpm were subjected to immunoprecipitation (IP) using monoclonal anti-hemagglutinin (HA)–agarose (clone HA-7; Sigma-Aldrich). IPs were washed in RIPA containing 0.1% SDS, eluted in SDS sample buffer, and separated by SDS-PAGE (10%). The gel was processed for autoradiography using EN3HANCE as recommended by the supplier (PerkinElmer), and the dried gel was exposed to film at −80°C. The yields were analyzed by digitized phosphorimaging (Typhoon imager; GE Healthcare).
The HA-KLF4 protein half-life was estimated by immunoblot analysis of cycloheximide (CHX)-treated cells using anti-HA antibody. HA-KLF4 protein levels were normalized to a stable protein, β-actin, and protein half-life was determined from the first-order rate constant during the first 1 h of CHX treatment. The estimated half-life was further confirmed by comparison of the IP yield of radiolabeled protein when cells were labeled for 1 half-life (i.e., to 50% of steady state) versus 5 half-lives (i.e., to approximately the steady state).
In vitro Transwell migration and invasion assays.
For migration assays 1.0 × 104 cells were plated in the top chamber (24-well insert; pore size, 8.0 μm; BD Biosciences). For invasion assays, 1.0 × 104 cells were plated in the top chamber over a Matrigel-coated filter (24-well insert; pore size, 8.0 μm). In both assays, cells were plated in growth medium containing 0.5% FBS, and growth medium containing 10% FBS was used as a chemoattractant in the lower chamber. After 24 h, cells that did not migrate or invade through the pores were removed by scraping, and cells on the lower surface of the membrane were stained using the Diff-Quik stain set (Siemens) and counted.
Immunoblot analysis.
Cells were washed twice in PBS and then incubated for 30 min in ice-cold lysis buffer: 150 mM sodium chloride, 1.0% (wt/vol) sodium deoxycholate, 1.0% (vol/vol) Triton X-100, 5.0 mM EDTA, 50 mM Tris-HCl (pH 7.5), 0.25 mM phenylmethylsulfonyl fluoride, 1.0 mM benzamidine, 1.0 mM pepstatin, 1.0 μg/ml leupeptin, 1.0 μg/ml aprotinin, 0.4 mM sodium orthovanadate, and 1.0 mM sodium fluoride. Extracts were centrifuged at 15,000 × g for 15 min at 4°C. Protein concentrations were quantified by using the Bradford assay (Bio-Rad).
Following electrophoresis, proteins were transferred to nitrocellulose. Antibodies to DCR1 (H-212; Santa Cruz Biotechnology), HA (12CA5; Roche) and cyclin D1 (H-295; Santa Cruz Biotechnology) were used at 0.40 μg/ml. Antibodies to KLF4 (H-180; Santa Cruz Biotechnology) and p21 (C-19; Santa Cruz Biotechnology) were used at 0.25 μg/ml. p27 antibody (C-19; Santa Cruz antibody) was used at 0.10 μg/ml. Antibody to β-actin (C-4; Santa Cruz Biotechnology) or β-tubulin (clone 2-28-33; Santa Cruz Biotechnology) was used at 0.20 μg/ml. Bound antibodies were detected using the Pierce enhanced chemiluminescence Western blotting substrate (Thermo Scientific). Scanned images were quantitated using ImageJ software, with normalization to the loading control.
Reverse transcription and real-time PCR detection of miRNAs.
To profile miR levels in proliferating versus confluent RK3E cells, total RNA was extracted using the mirVana miRNA isolation kit (Ambion/Invitrogen) according to the manufacturer's instructions. A total of 381 individual miRs were analyzed in triplicate using TaqMan rodent microRNA array cards as recommended by the manufacturer (Applied Biosystems). Briefly, miRNA was converted to cDNA by using a pool of stem-loop hairpin reverse transcription (RT) primers, and this product was delivered to single wells preloaded with miR-specific primers and probes. Real-time PCR was performed on a 7900HT ABI Prism sequence detector system. miRNA fold changes were determined using the ΔΔCT method with 18S rRNA as the internal control (32).
Individual miRs were analyzed by stem-loop reverse transcription followed by quantitative, real-time PCR (SL-RTQ) using TaqMan microRNA assays (Applied Biosystems) according to the manufacturer's instructions; the individual miRs were miR-206 (4373092), miR-1 (4395333), miR-344-1 (4373340), and U6 snRNA (4395470). For PCRs we utilized an Mx3005P real-time PCR system (Stratagene). miRNA fold changes were determined using the ΔΔCT method with U6 snRNA as the internal control. Three independent experiments were performed, with analysis of each RNA sample in duplicate fashion for each experiment.
Quantitative, real-time RT-PCR (qRT-PCR) analysis of mRNA.
Total RNA was isolated using an RNeasy minikit (Qiagen) and reverse transcribed using SuperScript II reverse transcriptase (Invitrogen). PCR primers and TaqMan probes were from Assays-on-Demand (Applied Biosystems): human and rat KLF4 (ID Hs00358836_m1 and Rn00821506_g1) or human and rat GAPDH (Hs99999905_m1 and Rn99999916_s1). Firefly and Renilla luciferase transcripts were analyzed using SYBR GreenER qPCR SuperMix (Invitrogen), with normalization to GAPDH and the following primers: Firefly-Luc-forward, 5′-GCCCGCGAACGACATTTA-3′; Firefly-Luc-Reverse, 5′-TTTGCAACCCCTTTTTGGAA-3′; Renilla-Luc-forward, 5′-GCAGCATATCTTGAACCATTCAAA-3′; Renilla-Luc-Reverse, 5′-CATCACTTGCACGTAGATAAGCATTATA-3′; GAPDH-forward, 5′-TCACCACCATGGAGAAGGC-3′; GAPDH-Reverse, 5′-GCTAAGCAGTTGGTGGTGCA-3′. These GAPDH primers recognize mouse, rat, and human forms.
Statistical analyses.
Data were analyzed using the unpaired t test or else a one-way analysis of variance (ANOVA) with Tukey's multiple comparison test, both using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Differences were considered significant when the two-tailed analysis yielded a P value of <0.05.
RESULTS
Identification of posttranscriptional control elements in the KLF4 transcript.
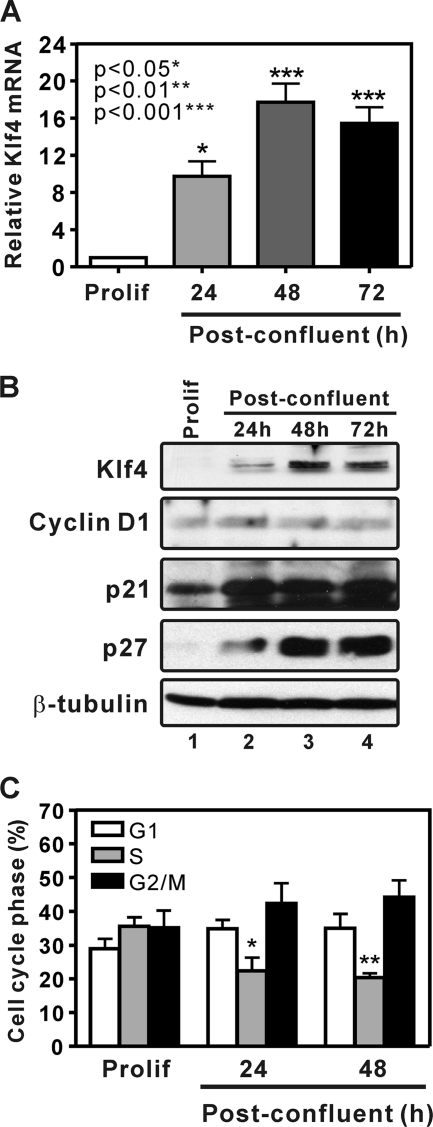
A signature feature of KLF4 is its induction upon growth arrest or differentiation (43, 64). To determine whether sequence elements within the KLF4 mRNA can function in posttranscriptional regulation, we utilized subconfluent and postconfluent cultures of adenovirus E1A-immortalized rat kidney cells (RK3E) (41). These diploid epithelial cells proliferate rapidly at subconfluence, with a doubling time of approximately 12 h. Under these conditions the levels of Klf4 mRNA (Fig. 1 A) and protein (Fig. 1B, lane 1) were low. At confluence the cells form an epithelial sheet of 1 to 3 cell layers that persists intact for several weeks, with apical tight junctions, adherens junctions, and frequent desmosomes (10). Typical of epithelial cells in vivo, these cells show peripheral staining of E-cadherin and β-catenin, with very low expression of mesenchymal markers, such as vimentin (10, 29). Similar to differentiated epithelial cells in vivo, postconfluent RK3E had increased amounts of Klf4 mRNA and protein (Fig. 1A and B, lanes 2 to 4) and increased amounts of the cyclin-dependent kinase inhibitors p21Waf1/Cip1 and p27Kip1 (Fig. 1B). Cell cycle analysis revealed a decrease in S phase from 36.0% in proliferating cells to 20.8% in postconfluent cultures (Fig. 1C). These studies identified a simple in vitro model of the proliferation/differentiation switch that epithelial cells undergo in vivo.
Fig. 1.
KLF4 and KLF4-regulated cell cycle inhibitors are induced during formation of a confluent epithelial sheet in vitro. (A) Real-time qRT-PCR analysis of endogenous Klf4. Total RNA was isolated from subconfluent (proliferating) or postconfluent RK3E epithelial cells (three independent experiments; bars show standard errors). (B) Klf4 and the cell cycle regulatory proteins cyclin D1, p21Waf1/Cip1, and p27Kip1 were analyzed by immunoblotting. β-Tubulin served as a loading control. (C) Cell cycle distribution of RK3E cells (three independent experiments performed in duplicate; bars show standard errors).
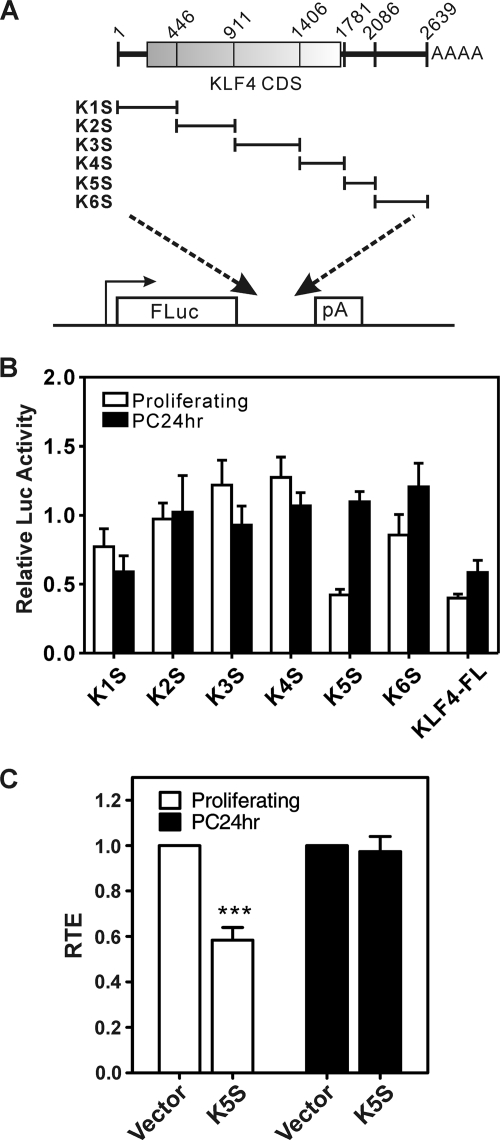
To screen for elements that could confer differential expression in proliferating versus postconfluent cells, we inserted the KLF4 cDNA (i.e., KLF4-FL, including the protein-coding region and 3′-UTR) into the 3′-UTR of a FLuc expression vector (Fig. 2 A). KLF4-FL or the empty vector was cotransfected with an RLuc expression vector into RK3E cells. Following normalization to RLuc, the effect of KLF4-FL sequence on FLuc activity was determined by comparison to the vector control (Fig. 2B). In proliferating cells, normalized FLuc activity derived from FLuc-KLF4-FL was about 60% lower than vector. In postconfluent cells there was likewise a suppression of FLuc derived from KLF4-FL, but the effect was attenuated compared to proliferating cells (Fig. 2B, PC24 h).
To distinguish effects on mRNA from effects on translation efficiency, we analyzed FLuc and RLuc mRNA levels by qRT-PCR (Fig. 2C). Relative translation efficiency (RTE) was calculated using Luc activity and RNA levels (Fig. 2D). As shown in Fig. 2C, insertion of the ∼2.6-kb KLF4-FL sequence did not decrease the abundance of FLuc transcripts relative to vector in proliferating cells. Furthermore, in postconfluent cells there was only a modest suppression of 27%. Rather, reduced FLuc activity in proliferating cells (Fig. 2B) was entirely attributed to inefficient translation (Fig. 2D). In these experiments the RTE was reduced by 72% (P < 0.001). After adjusting for mRNA abundance, the RTE in postconfluent cells was similar for the vector and KLF4-FL (Fig. 2D, PC24 h). These results suggested that KLF4 transcripts are inefficiently translated into protein when cells are proliferating, a condition where only low levels of endogenous Klf4 are observed (Fig. 1B). These mapping studies identified translational control as a mechanism leading to differential expression of KLF4 in proliferating and differentiating epithelial cells.
Fragment K5S represents a 3′-TCE that suppresses translation in proliferating cells.
We tested subfragments of the KLF4 cDNA for regulation of translation in proliferating cells (Fig. 3 A). K5S, a 3′-UTR-derived fragment of 306 nucleotides (nt), was sufficient to suppress Luc activity by 58% in proliferating cells (Fig. 3B) and suppressed RTE by 42% (Fig. 3C). As KLF4-FL had a larger effect on RTE (Fig. 2D), other regulatory elements may remain to be identified, or else the TCE may be context sensitive. As observed for KLF4-FL, the effect of K5S on RTE was specific, because there was no suppression in postconfluent cells (Fig. 3C).
Fig. 3.
A cis-acting TCE derived from the KLF4 3′-UTR inhibits translation of a linked reporter in proliferating epithelial cells. (A) Fragments of KLF4 were inserted into the 3′-UTR region of FLuc. (B) Following transfection of RK3E cells, FLuc activity relative to the vector was determined using the DLR assay as shown in Fig. 2B (five independent experiments; bars show standard errors). (C) RTE of the TCE construct (K5S, corresponding to bases 1781 to 2086 of GenBank accession number AF105036) in proliferating and PC24 h cells (three independent experiments; bars show standard errors).
A 100-nt subregion of the TCE contains translation regulatory activity.
To aid in the identification of regulatory miRs, we tested 100- to 200-nt subfragments of K5S for differential reporter activity (Fig. 4 A). Of these, only K5S-5 was differentially active (Fig. 4B). For this construct, the RTE was 0.56 ± 0.04 (proliferating cells) and 0.84 ± 0.08 (PC24 h). While several other fragments inhibited translation, these failed to show differential activity and were not further investigated. These studies supported context dependence, as several larger fragments that included the K5S-5 region showed either no differential activity (K5S-7) or else an attenuated difference (K5S-6). Therefore, in addition to the nucleotide sequence, the secondary structure appears likely to be important for activity. Indeed the predicted structure of K5S indicates a highly structured stem with a more open region in the vicinity of K5S-5 (mfold data not shown).
Based upon target site prediction, at least eight different miRs, including miR-1, -26a, -26b, -145, -206, -344, -465, and -613 have the potential to regulate through K5S-5 (Table 1 ). In different contexts, KLF4 translation was previously found to be suppressed by miR-10b, miR-200c, miR-145, and miR-1 (50, 55–57). Profiling indicated that three of these known inhibitory miRs (miR-200c, -145, and -10b) are expressed at increased levels in PC24 h RK3E cells compared with proliferating cells (Table 1), even though translation of reporters is upregulated (Fig. 2D and 3C) and endogenous Klf4 is induced (Fig. 1A and B). miR-1 was unchanged (Table 1). Unlike these miRs, miR-344-1 (miR-344) was suppressed by 67-fold in PC24 h cells, potentially contributing to the induction of Klf4.
Table 1.
Potential TCE-regulating miRs and growth-state-specific expression in RK3E cells
| miRa | Mature miR sequenceb (5′–3′) | Start–stopc | Expressiond (proliferating) | Fold change (PC24 h/proliferating) |
|---|---|---|---|---|
| mmu-miR-429 | UAAUACUGUCUGGUAAAACCGU | 1770–1791 | Not detected | |
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | 1771–1791 | Not detected | |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | 1772–1791 | 0.188 ± 0.058 | 22.6 ± 0.2 |
| rno-miR-7 | UGGAAGACUAGUGAUUUUGUUGU | 1793–1815 | 0.667 ± 0.389 | 0.921 ± 0.357 |
| hsa-miR-7 | UGGAAGACUAGUGAUUUUGUUG | 1793–1815 | 0.498 ± 0.125 | 0.920 ± 0.355 |
| hsa-miR-425 | AUCGGGAAUGUCGUGUCCGCC | 1798–1818 | 0.262 ± 0.063 | 0.0440 ± 0.0010 |
| hsa-miR-149 | UCUGGCUCCGUGUCUUCACUCCC | 1818–1839 | 7.65 ± 1.80 | 1.06 ± 0.03 |
| hsa-miR-512-3p | AAGUGCUGUCAUAGCUGAGGUC | 1826–1848 | Not tested | |
| hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA | 1826–1849 | Not detected | |
| hsa-miR-520f | AAGUGCUUCCUUUUAGAGGGUU | 1828–1847 | Not tested | |
| has-miR-508 | UGAUUGUAGCCUUUUGGAGUAGA | 1829–1853 | Not tested | |
| hsa-miR-219 | UGAUUGUCCAAACGCAAUUCU | 1836–1854 | Not detected | |
| hsa-miR-591 | AGACCAUGGGUUUUCAUUGU | 1841–1859 | Not tested | |
| mmu-miR-201 | UACUCAGUAAGGCAUUGUUCU | 1855–1875 | Not detected | |
| hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU | 1877–1899 | 0.212 ± 0.078 | 0.995 ± 0.025 |
| hsa-miR-511 | GUGUCUUUUGCUCUGCAGUCA | 1899–1921 | Not tested | |
| mmu-miR-344 | UGAUCUAGCCAAAGCCUGACUGU | 1932–1956 | 0.740 ± 0.005 | 0.0150 ± 0.0010 |
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | 1948–1968 | Not tested | |
| hsa-miR-206 | UGGAAUGUAAGGAAGUGUGUGG | 1948–1968 | 0.114 ± 0.027 | 1.68 ± 0.61 |
| hsa-miR-1 | UGGAAUGUAAAGAAGUGUGUAU | 1949–1968 | 1.89 ± 0.43 | 1.06 ± 0.29 |
| mmu-miR-465 | UAUUUAGAAUGGCACUGAUGUGA | 1956–1978 | Not detected | |
| hsa-miR-26b | UUCAAGUAAUUCAGGAUAGGUU | 1966–1987 | 23.4 ± 0.6 | 2.648 ± 0.686 |
| hsa-miR-26a | UUCAAGUAAUCCAGGAUAGGC | 1966–1987 | 31.2 ± 7.1 | 2.08 ± 0.55 |
| hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU | 2003–2026 | 0.423 ± 0.154 | 3.98 ± 0.10 |
| mmu-miR-10b | UACCCUGUAGAACCGAAUUUGU | 2023–2045 | 0.266 ± 0.064 | 12.5 ± 0.3 |
| hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG | 2023–2045 | 7.68 ± 1.78 | 4.27 ± 1.13 |
| hsa-miR-642 | GUCCCUCUCCAAAUGUGUCUUG | 2053–2074 | Not tested |
Rodent and/or human miRs predicted to bind within or immediately adjacent to the TCE, represented by fragment K5S (Fig. 3). Web-based tools included MiRanda (19), TargetScan 3.1 (28), and PicTar (26).
Predicted seed sequences are indicated by underlining.
Nucleotide numbers correspond to GenBank accession number AF105036.
Expression relative to 18S RNA (2−ΔCT × 105). Not detected, no signal after 40 cycles, where the CT for 18S RNA was approximately 17.
We therefore used gain- and loss-of-function methods to investigate the functions of miR-344 and two miRs with overlapping, evolutionarily conserved binding sites, miR-1 and miR-206 (Fig. 4C and D). Unlike other vertebrates, mice have an insertion of 4 bases in the center of the miR-344 seed (Fig. 4C) (20). Interestingly, rather than disrupting the Klf4–miR-344 base pairing, this GGAT insertion duplicates the adjacent seed sequence and recreates an miR-344 seed with extensive 3′ pairing for both miR-344 and miR-1/206 (Fig. 4E and data not shown). As vertebrates therefore appear to strictly conserve the binding site for miR-344, the site for miR-1/206, and the 6-nt spacing between the two seeds (Fig. 4C and E), we constructed K5S FLuc reporters with mutations at the seed regions (Fig. 4F).
miR-344 and miR-206 mediate proliferation-dependent translation of the TCE.
To confirm the results obtained by miR profiling (Table 1), we used gene-specific SL-RTQ assays to analyze miRs in proliferating and PC24 h cells (Fig. 5 A). miR-344 was suppressed by 42-fold as cells became confluent. In contrast, miR-206 was increased by 2.8-fold at confluence, and miR-1 was unchanged. Cotransfection of FLuc and RLuc reporters with either miR mimic or anti-miR inhibitor into proliferating RK3E cells revealed opposing effects of miR-206 (activating) and miR-344 (inhibitory) on TCE activity (Fig. 5B). Interestingly, miR-1 reagents gave similar results as the nontargeting controls, indicating that this potentially inhibitory miR does not play a major role in these cells. miR-206 upregulated the TCE reporter by 1.4-fold compared with the vector, suggesting that it does not act solely by competing with the inhibitory miR-344 but rather that it has TCE-promoting activity of its own (Fig. 5B, left panel). Also, anti-miR-344 could induce TCE activity by 1.2-fold relative to the vector, supporting a role for unopposed, endogenous miR-206 as a promoter of TCE activity (Fig. 5B, right panel).
To further analyze these miRs we engineered a retroviral vector containing HA epitope-tagged, KLF4-FL cDNA with an intact 3′-UTR (pBpuro-HA-KLF4-FL). We used metabolic labeling and IP to determine the effect of miRs on the rate of synthesis of transgene-derived HA-KLF4. Using the two assays described in Materials and Methods, the HA-KLF4 protein half-life in proliferating RK3E cells was determined to be 48 min (data not shown). HA-KLF4 or vector control cells were then labeled with [35S]Met/Cys for 1 half-life (Fig. 5C). As observed above for the TCE reporter (Fig. 5B), exogenous miR-206 and anti-miR-344 each promoted the synthesis of HA-KLF4 in proliferating RK3E cells (miR-206, 2.6-fold induction; anti-miR-344, 2.1-fold induction) (Fig. 5C). This increase was attributed to an increased efficiency of translation, because miR reagents did not alter HA-KLF4 mRNA levels (data not shown). Furthermore, any increase in the HA-KLF4 protein half-life would have a smaller effect (≤1.3-fold), as only about 25% of the labeled protein will turn over during a labeling interval of 1 half-life (i.e., 48 min for control cells) (data not shown).
To determine whether endogenous Klf4 is regulated in a similar fashion as the TCE reporter and the HA-KLF4 transgene, we treated proliferating RK3E cells with miR-206, anti-miR-344, or the relevant controls (Fig. 5D). Each reagent induced the endogenous Klf4 protein (Fig. 5D, upper panels; miR-206 at 2.1-fold and anti-miR-344 at 3.7-fold). However, neither reagent significantly altered the Klf4 mRNA levels, consistent with a role for these miRs in regulating the translation of endogenous Klf4 (Fig. 5D, lower panels).
When the TCE reporter activity was maximized by exogenous miR-206 (Fig. 5E, left panel) or by suppression of endogenous miR-344 (Fig. 5E, right panel), modulation of the other miR could revert TCE activity to approximately 1.0, supporting a competitive model rather than a model in which either miR plays a dominant role. Further support for a competitive model was obtained by mutation of either miR seed sequence (Fig. 4F). Reporters with mutation of the miR-344 seed were hyperactive in proliferating cells, consistent with regulation by miR-206, while reporters with mutation of the miR-206 seed showed similar or reduced activity compared to WT, consistent with repression by miR-344 (Fig. 5F, left panel). For the activated mutant Mt344B, transfection of an miR complementary to the mutated transcript repressed the activity of the reporter, restoring levels similar to WT (Fig. 5F, middle panel). For Mt206B, transfection of a miR complementary to the mutated transcript stimulated reporter activity, but not as efficiently as when exogenous WT miR-206 was used (compare Fig. 5F, right panel, and B, left panel). This result suggests that the suppressive activity of miR-344 is independent of its primary structure, similar to other miRs that repress translation. In contrast, for miR-206 the primary structure may be important for its ability to promote translation. These results indicate that miR-344 and miR-206 have opposing roles in TCE regulation and that the induction of TCE activity at confluence may be due to the combined effects of loss of TCE-inhibitory miR-344 and gain of TCE-activating miR-206.
KLF4 promotes its own translation by signaling through miR-206 to alter TCE activity.
To determine whether KLF4 can regulate TCE activity, we transduced RK3E cells with a retrovirus encoding a conditional KLF4-estrogen receptor (KLF4-ERT) fusion protein or with empty vector. With little or no background activity in the absence of 4OHT, activated KLF4-ERT induced KLF4 target genes, such as Notch1 and p21Waf1/Cip1, within 1 to 3 h and the outgrowth of foci of transformed cells by 2 to 3 weeks (9, 17, 31). In RK3E/KLF4-ERT cells, 4OHT promoted TCE-dependent reporter activity in proliferating cells (Fig. 6A). A time course showed that this effect occurred within 12 h of exposure to 4OHT (Fig. 6B). These results suggested that KLF4 can alter the inhibitory effect of the TCE and promote its own translation. To further investigate this possibility we analyzed the function of the TCE in the context of a full-length (FL) KLF4 transgene. We compared protein levels derived from an HA-KLF4-FL transgene and a construct with deletion of the K5S/TCE (pBpuro-HA-KLF4-FLΔK5S). Transduced cell populations were selected in puromycin and then analyzed at subconfluence and postconfluence (Fig. 6C). Regardless of confluence, immunoblotting of transduced cells revealed that the TCE appeared to promote expression of the KLF4 transgene, yielding 2.5-fold more KLF4 protein (Fig. 6C, proliferating cells, normalized to β-tubulin). Because TCE reporter activity approached 1.0 in the KLF4 active setting (Fig. 6A and B) but did not exceed 1.0, the TCE appears to function in a context-dependent fashion to promote KLF4 expression in cells with active KLF4 (i.e., positive feedback) (Fig. 6C).
Fig. 6.
KLF4 promotes TCE reporter activity and miR-206 expression in proliferating RK3E epithelial cells. (A) Cells were transduced with pBpuro-KLF4-ERT or vector retrovirus and then selected for puromycin resistance. TCE reporter activity was determined following treatment with 4OHT for 24 h. (B) TCE reporter activity was determined following treatment with 4OHT for the indicated interval. (C) RK3E cells transduced with the indicated vector were analyzed for expression of transgene-derived, HA-KLF4 protein. (D) Cell proliferation assay results for stably transduced populations. (E) miRs were analyzed in RK3E/KLF4-ERT cells or vector control cells following treatment with 4OHT for 24 h. (F) TCE reporter activity was analyzed in proliferating RK3E/KLF4-ERT cells following treatment with 4OHT and/or the indicated anti-miR (three independent experiments; bars show standard errors).
In RK3E cells KLF4-FL, which is sufficient to transform these cells (10), induced a slower growth phenotype at subconfluence (Fig. 6D). In contrast, the lower expression resulting from the ΔK5S allele actually promoted cell proliferation compared with the vector (Fig. 6D). Despite these differences, KLF4-FL and KLF4-ΔK5S induced foci of transformed cells with similar efficiencies (data not shown). In these studies, transgene-derived mRNA levels were similar (data not shown), indicating that differential expression is due to altered translation. Consistent with the FLuc reporter assay results shown above (Fig. 5B), these results indicated that the TCE can promote translation of KLF4 and influence a cellular slow growth phenotype. The results also suggest that malignant transformation occurs at a lower threshold than does suppression of cell proliferation.
As miR-206 (Fig. 5B), KLF4 (Fig. 6A and B), and the TCE (Fig. 6C) each appeared to promote translation of KLF4, we examined whether KLF4 could regulate miR-206. When RK3E/KLF4-ERT cells or vector control cells were treated with 4OHT for 24 h, miR-206 was increased by 2.2-fold in KLF4-ERT cells (Fig. 6E). No difference was observed for miR-344. Furthermore, induction of TCE reporter activity by KLF4-ERT was prevented by anti-miR-206 (Fig. 6F). These results suggest that KLF4 promotes its own translation through the TCE-regulating function of miR-206, identifying a positive regulatory feedback loop.
Although miR-344 appeared to play a critical role in rodent cells, it has not been reported to be expressed in human cells. Given the unusual role of miR-206 as a promoter of translation in proliferating cells, subsequent studies were focused upon this miR.
DCR1 differentially regulates KLF4 expression in mammary epithelial cells and cancer cells.
To examine the regulation of KLF4 in human cells, we first analyzed KLF4 mRNA and protein in proliferating cells, including normal, primary HMECs, immortalized mammary epithelial cells (184A1, MCF10A), and malignant, breast tumor-derived cell lines (Fig. 7A and B). Compared with primary HMECs, KLF4 mRNA levels were increased in the immortalized cell lines and in tumor cells (Fig. 7A, left panel; range, 1.9- to 8.4-fold). Similar results were obtained using a SYBR Green assay with distinct primers (data not shown).
Fig. 7.
Differential regulation of TCE reporter activity and KLF4 by DCR1 in mammary epithelial cells and cancer cells. (A) KLF4 mRNA and protein levels and the levels of miR-206 were analyzed for a panel of normal, primary cells (HMECs), immortalized cells (184A1 and MCF10A), and human tumor cell lines (proliferating cells). HMECs were analyzed at passage 7. (B) For the indicated cell lines, protein levels were assessed in triplicate. For each cell line, three cryovials were thawed, expanded, and analyzed by immunoblotting in a parallel fashion (left panel). Images were analyzed by densitometry to determine mean KLF4/β-actin ratios (right panel; bars show standard deviations). (C) TCE reporter activity was analyzed in proliferating cells. (D) Endogenous DCR1 and KLF4 proteins were analyzed following transfection of DCR1 siRNAs or a control. Extracts were prepared at 48 h posttransfection when cells were subconfluent (proliferating). Three independent experiments produced similar results. (E) TCE reporter activity was analyzed in proliferating cells following transfection of si-DCR1 11 or si-Ctrl. Histograms in panels A, C, and E represent three independent experiments (bars show standard errors).
Immunoblot analysis of these lines indicated that the basal type cell lines MCF10A and MDA-MB-231 expressed higher levels of both mRNA and protein (Fig. 7A, middle panel). However, KLF4 expression did not correlate with miR-206 levels (Fig. 7A, right panel). To further quantitate KLF4, we analyzed a subset of lines in triplicate—HMEC, MCF10A, MCF7, and MDA-MB-231—representing normal, immortalized, estrogen receptor (ER)-positive, and ER-negative cells (Fig. 7B). Relative to HMECs, KLF4 was increased in MCF10A (6.9-fold), MCF7 (4.3-fold), and MDA-MB-231 (12-fold) cells. In the current study three different sources of MDA-MB-231 cells showed similarly high levels of KLF4 protein by Western blotting (data not shown): early passage cells from the ATCC, the highly metastatic isolate MDA-MB-231-luc-D3H2LN (Caliper), and metastatic MDA-MB-231 cells from the lab of Theresa A. Guise (22). These results suggest that KLF4 protein expression is consistently elevated in MDA-MB-231 cells.
Similarly to RK3E epithelial cells, TCE reporter activity was low in primary HMECs (0.56-fold) (Fig. 7C). As observed for endogenous KLF4 (Fig. 7A and B), TCE reporter activity was increased relative to HMECs in established cell lines (range, 0.95 to 1.04-fold) (Fig. 7C).
The results presented in Fig. 5 and 6 identified specific miRs as promoters or inhibitors of KLF4 expression. Consistent with these data, siRNA-mediated suppression of DCR1 had distinct effects on KLF4 expression in human mammary cell lines (Fig. 7D). Whereas DCR1 and, by inference, miRs could promote KLF4 protein expression in MCF10A cells, this enzyme suppressed expression in mammary tumor-derived cells such as MDA-MB-231 and MCF7.
Opposite effects of miR-206 on KLF4 expression and TCE reporter activity in normal and cancer cells.
Consistent with its effect on endogenous KLF4 in MCF10A cells (Fig. 7D), DCR1 knockdown suppressed TCE reporter activity (Fig. 7E). In contrast, DCR1 knockdown in tumor lines either resulted in increased reporter activity (MCF7) or else had no significant effect (MDA-MB-231). These results indicate that DCR1 regulates endogenous KLF4 expression in MCF10A and MCF7 cells, at least in part through regulation of the TCE. In MDA-MB-231 cells the suppression of endogenous KLF4 by DCR1 (Fig. 7D) was discordant with its regulation of the TCE (Fig. 7E), suggesting a potential role for KLF4-repressive miRs that bind outside the TCE (Table 1).
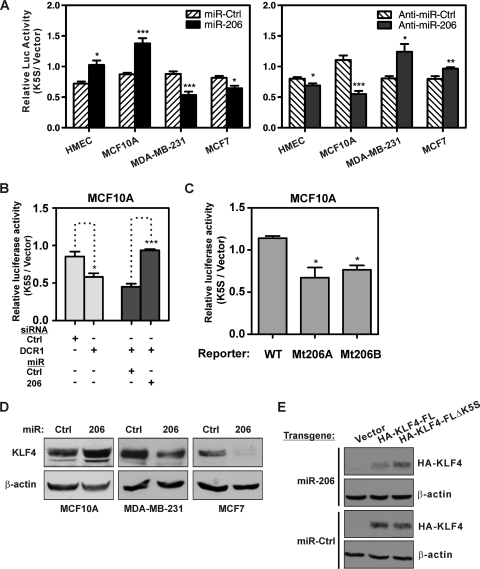
Regardless of overall TCE reporter activity (Fig. 7C), transfection of miRs or anti-miRs indicated that exogenous and endogenous miR-206 promotes TCE activity in nonmalignant cells (HMEC and MCF10A) but inhibits this activity in the breast cancer cells that we examined (Fig. 8A). In each of these lines miR-206 was detected by SL-RTQ, whereas the closely related miR-1 was not (data not shown). The ability of exogenous miR-206 to yield reporter activity greater than that of the vector (Fig. 8A, MCF10A; 1.4-fold) is most consistent with a mechanism in which miR-206 complexes have an intrinsic ability to positively regulate translation in these human cells. In support of this, loss of TCE reporter activity following suppression of DCR1 was rescued by exogenous miR-206 in MCF10A cells (Fig. 8B), and mutation of the miR-206 binding site gave reduced reporter activity (Fig. 8C).
Fig. 8.
miR-206 differentially regulates TCE reporter activity and KLF4 in mammary epithelial cells and cancer cells. (A) TCE reporter activity was analyzed in proliferating cells following transfection of miR-206 mimic, antagonist, or the relevant control. HMECs were at passage 10. (B) Exogenous miR-206 was analyzed for regulation of TCE reporter activity following suppression of DCR1 in MCF10A cells. (C) The activities of WT and mutant reporters were analyzed in MCF10A cells. (D) Endogenous KLF4 protein was analyzed following transfection of miR-206 mimic or a control. Extracts were prepared at 48 h posttransfection, when cells were subconfluent. β-Actin served as loading control. (E) MDA-MB-231 cells were transduced with the indicated retrovirus. Following transfection of miR-206 mimic or a control, transgene-derived KLF4 protein was analyzed by detection with HA antibody. Extracts were prepared at 48 h posttransfection, when cells were subconfluent.
Consistent with its effects on the reporter, miR-206 regulated the expression of endogenous KLF4 in human cells (Fig. 8D). miR-206 effects were consistent with its distinct activities on the TCE reporter (Fig. 8A and B). Exogenous miR-206 could either induce KLF4 in MCF10A (1.8-fold) or repress KLF4 in the tumor cell lines (MCF7, 83%; MDA-MB-231, 38%) (Fig. 8D). In addition, miR-206 repressed expression from a transgene containing the TCE in its normal context, as shown by analysis of MDA-MB-231 cells expressing exogenous KLF4-FL or -ΔK5S transgenes (Fig. 8E). In summary, miR-206 promotes TCE activity and KLF4 levels in normal (HMECs) or immortalized (RK3E or MCF10A) epithelial cells but represses these in cancer cells.
KLF4 represses its own translation in breast cancer cells through induction of miR-206.
KLF4 mRNA and protein and miR-206 are readily detected in the majority of primary human breast tumors and cell lines, particularly tumors with aggressive properties, such as low ER expression or elevated Notch1 (8, 15, 31, 37). In vitro, KFL4 and miR-206 are coinduced as RK3E cells attain confluence (Fig. 1A and B and 5A), and miR-206 is upregulated following activation of KLF4-ERT in RK3E cells (Fig. 6E), where it functions to promote TCE reporter activity and KLF4 expression.
To determine whether KLF4 regulates the expression of miR-206 in breast cancer cells, we generated KLF4 knockdown MDA-MB-231 cells (Fig. 9A). qRT-PCR analysis indicated 68% suppression of the mRNA (data not shown). These cells showed less migration in Transwell chambers (Fig. 9B) or wound healing assays (data not shown), and invasion of a Matrigel layer was similarly suppressed (Fig. 9B). Compared with control shRNA cells, miR-206 levels were reduced in KLF4 shRNA cells by 96% (Fig. 9C). Supporting a role for KLF4 in maintenance of miR-206, expression was rescued within 0.5 h following activation of the KLF4-ERT fusion and was increased by 14-fold at 24 h (Fig. 9D). Similarly, activation of KLF4-ERT was sufficient to rescue the migration defect (Fig. 9E). Invasion was not rescued within the time frame examined (24 h) (data not shown).
In KLF4 knockdown MDA-MB-231 cells, in which miR-206 was reduced, there was increased activity of the K5S-5 reporter (Fig. 9F). In KLF4 knockdown or control cells, miR-206 repressed the reporter, as indicated by transfection of miR-206 or anti-miR-206 (Fig. 9G). The KLF4 shRNA binds within the TCE (K5S) reporter but does not affect the K5S-5 reporter utilized in these studies. These results identified a KLF4–miR-206 feedback loop that operates in a distinct fashion in normal epithelial cells (positive feedback) and cancer cells (negative feedback).
The cell context-dependent activity of miR-206 upon TCE function and KLF4 levels are presented in Fig. 10. In certain contexts the TCE suppresses translation even though miR-206 is stimulatory (e.g., HMECs and RK3E), indicating that TCE activity is regulated by additional factors, such as proteins and/or miRNPs termed TCE regulatory factors (TCE-RFs). In MCF10A cells, overall TCE activity of ∼1.0 resulted from increased expression of miR-206 and its translation-promoting activity, while in breast cancer cells the reduced expression of miR-206, a translational repressor, permitted a similar overall TCE activity (similar to 1.0; see Fig. 7A [right panel] and C and 10).
Fig. 10.
Context-dependent effects of miR-206 on TCE activity. Approximate TCE reporter activity, as determined in the current study, is indicated in parentheses. Inductive (↑) or inhibitory (⊤) effects of miRNP/miR-206 are indicated. The roles of other potential TCE regulatory factors (TCE-RFs) were inferred from the overall activity of the TCE and from the stimulatory or inhibitory effect of miR-206. The observed relative expression levels of endogenous KLF4 and endogenous miR-206 are indicated by the font size.
DISCUSSION
Mechanisms that suppress KLF4 in dividing cells are potentially important for the restriction of stem cell and malignant properties (18, 40, 55, 58). As shown for other cellular transcripts, miRs that were previously shown to regulate KLF4 consistently repressed its expression. These studies analyzed the role of miR-145 in differentiation of human embryonic stem cells (57), the role of Zeb1 and miR-200c in the induction of KLF4 and other stem cell factors in pancreatic cancer cells (55), the role of miR-10b and KLF4 in the migration and invasion of esophageal tumor cells (50), and the role of miR-1 in retinoid-induced differentiation of ES cells to smooth muscle cells (56). We have shown that miR-344, which thus far has been reported in rodent cells but not in human cells, functions similarly to these other miRs, suppressing Klf4 translation in proliferating cells.
Consistent with the observation that miR-206 can promote translation, Steitz and colleagues previously demonstrated that specific miRs can repress or promote translation, depending upon the cell growth state (44, 51–53). For the miRs that were analyzed, repression of translation was a property of proliferating cells, whereas in growth-arrested cells miRs consistently promoted translation. In the current study we observed that DCR1, acting through miRs, has distinct effects on KLF4 in proliferating human MCF10A cells and tumor cells. Analysis of the TCE identified multiple aspects of its regulation, including regulation by both positively and negatively acting miRs, KLF4 autoregulation through miR-206, altered modes of miR and TCE function following exposure to exogenous KLF4, and altered activity of the TCE and its regulatory miRs in human tumor cell lines compared to normal or immortalized cells. In these studies the TCE could regulate activity of an artificial reporter over a range from 0.17-fold (proliferating pancreatic cancer cells [Panc 02.03]) to 4.4-fold (confluent NIH 3T3 cells [data not shown]). In the context of the KLF4 transcript, the TCE was required for high-level expression sufficient to prolong the cell doubling time.
Although other potentially inhibitory miRs were expressed in proliferating RK3E epithelial cells (Table 1), miR-344 was essential for repression of TCE translation (Fig. 5B), and miR-344 was greatly reduced at confluence, whereas translation was upregulated (Table 1; Fig. 5A). More surprising was the observation that miR-206 promoted translation in these cells (Fig. 5B to D). miR-206 could compete with miR-344 (Fig. 5E), consistent with the overlap of their binding sites (Fig. 4C and D). In addition to competition with a repressor, other observations support an independent role for the mRNPmiR-206 complex as a positive regulator of translation. For example, in several contexts the TCE reporter was more active than empty vector in response to exogenous or endogenous miR-206 (Fig. 5B and F and 8A; data not shown for NIH 3T3 cells).
The molecular basis by which miRNP complexes promote translation, for example, in growth-arrested cells, is not understood (51, 52). Steitz and colleagues used tethering studies and other approaches to show that association of soluble FXR1 with miRNPs led to upregulation of translation. This activity switch occurred in growth-arrested cells that expressed endogenous FXR1, or else following transfection of exogenous FXR1, even in proliferating cells. By inference, miR-206 may promote incorporation of FXR1 or an FXR1-like factor into miRNP in epithelial cells. Studies to investigate this possibility are under way. To regulate translation in the opposite fashion from other miRs, the sequence or structure, possibly including covalent modifications, of miR-206 may specify a distinct miRNP function. The data obtained to date would support an epithelium-specific factor or mechanism that is present in cells such as MCF10A cells.
The induction of miR-206 by KLF4, and upregulation of these factors in ER-negative breast cancers (8, 15, 35), indicates that miR-206 could mediate some phenotypic effects of KLF4 by regulating the translation or stability of other mRNAs (25). Like KLF4, enforced expression of miR-206 prolongs the cell doubling time of MDA-MB-231 cells (unpublished observations), suggesting that miR-206 could contribute to this effect. Our studies also suggest the interesting possibility that KLF4–miR-206 signaling could promote opposite phenotypic changes in normal epithelial cells versus tumor cells, as miR-206 appears to have opposite effects on translation in these contexts. This “switch” might help to explain how KLF4 activities that promote epithelial differentiation in normal cells (60) can be usurped for promotion of a malignant phenotype (9, 10, 39). Indeed, KLF4-transformed RK3E cells have a distinctive, spindled, mesenchymal morphology consistent with loss of epithelial features (10), and KLF4 translation appears to be promoted in human tumor cells through suppression of miR-200c by the EMT-inducing factor Zeb1 (55).
Overexpression of exogenous miR-206 or KLF4 has been reported to suppress the tumorigenicity of MDA-MB-231 cells (49, 60). However, the Ai group recently reported that endogenous KLF4 functions as an oncogene in these cells (62), consistent with our knockdown/rescue analysis (Fig. 9B and E). miR-206 knockdown studies have not yet been reported. It will be interesting to identify the signaling defects in breast cancer cells following KLF4 knockdown and to determine whether the miR-206 activity switch indeed contributes to the loss of epithelial features that occurs during tumor progression.
ACKNOWLEDGMENTS
We thank Katri S. Selander (University of Alabama at Birmingham) for providing MDA-MB-231 cells.
This work was supported by grant NCI RO1 CA127405, NIH/NCRR RR 016440, and the Jo and Ben Statler Chair in Breast Cancer Research.
Footnotes
Published ahead of print on 25 April 2011.
REFERENCES
- 1. Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bieker J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276: 34355–34358 [DOI] [PubMed] [Google Scholar]
- 3. Birsoy K., Chen Z., Friedman J. 2008. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 7: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Z. Y., Wang X., Zhou Y., Offner G., Tseng C. C. 2005. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 65: 10394–10400 [DOI] [PubMed] [Google Scholar]
- 5. Dang D. T., et al. 2003. Overexpression of Kruppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene 22: 3424–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dang D. T., Zhao W., Mahatan C. S., Geiman D. E., Yang V. W. 2002. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 30: 2736–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Debnath J., Muthuswamy S. K., Brugge J. S. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256–268 [DOI] [PubMed] [Google Scholar]
- 8. Foster K. W., et al. 2000. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 60: 6488–6495 [PubMed] [Google Scholar]
- 9. Foster K. W., et al. 2005. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 24: 1491–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foster K. W., et al. 1999. Oncogene expression cloning by retroviral transduction of adenovirus E1a-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 10: 423–434 [PubMed] [Google Scholar]
- 11. Garrett-Sinha L. A., Eberspaecher H., Seldin M. F., de Crombrugghe B. 1996. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J. Biol. Chem. 271: 31384–31390 [DOI] [PubMed] [Google Scholar]
- 12. Ghaleb A. M., et al. 2007. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 67: 7147–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henke J. I., et al. 2008. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27: 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang C. C., et al. 2005. KLF4 and PCNA identify stages of tumor initiation in a conditional model of cutaneous squamous epithelial neoplasia. Cancer Biol. Ther. 4: 1401–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iorio M. V., et al. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65: 7065–7070 [DOI] [PubMed] [Google Scholar]
- 16. Jiang J., et al. 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10: 353–360 [DOI] [PubMed] [Google Scholar]
- 17. Jiang W., et al. 2009. Prevention of KLF4-mediated tumor initiation and malignant transformation by UAB30 rexinoid. Cancer Biol. Ther. 8: 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang W., Lobo-Ruppert S. M., Ruppert J. M. 2008. Bad things happen in the basal layer: KLF4 and squamous cell carcinoma. Cancer Biol. Ther. 7: 783–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. John B., et al. 2004. Human microRNA targets. PLoS. Biol. 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joshua-Tor L., Hannon G. J. 1 September 2010, posting date. Ancestral roles of small RNAs: an ago-centric perspective. Cold Spring Harb. Perspect. Biol. doi: 10.1101/cshperspect.a003772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaczynski J., Cook T., Urrutia R. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kakonen S. M., et al. 2002. Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J. Biol. Chem. 277: 24571–24578 [DOI] [PubMed] [Google Scholar]
- 23. Katz J. P., et al. 2005. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology 128: 935–945 [DOI] [PubMed] [Google Scholar]
- 24. Katz J. P., et al. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129: 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H. K., Lee Y. S., Sivaprasad U., Malhotra A., Dutta A. 2006. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 174: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krek A., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37: 495–500 [DOI] [PubMed] [Google Scholar]
- 27. Lee Y. S., Dutta A. 2009. MicroRNAs in cancer. Annu. Rev. Pathol. 4: 199–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lewis B. P., Burge C. B., Bartel D. P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- 29. Li X., Deng W., Lobo-Ruppert S. M., Ruppert J. M. 2007. Gli1 acts through Snail and E-cadherin to promote nuclear signaling by β-catenin. Oncogene 26: 4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao X., et al. 2010. Kruppel-like factor 4 regulates pressure-induced cardiac hypertrophy. J. Mol. Cell Cardiol. 49: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Z., et al. 2009. Epithelial transformation by KLF4 requires Notch1 but not canonical Notch1 signaling. Cancer Biol. Ther. 8: 1840–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 33. Mahatan C. S., Kaestner K. H., Geiman D. E., Yang V. W. 1999. Characterization of the structure and regulation of the murine gene encoding gut-enriched Kruppel-like factor (Kruppel-like factor 4). Nucleic Acids Res. 27: 4562–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nijjar T., et al. 1999. p57KIP2 expression and loss of heterozygosity during immortal conversion of cultured human mammary epithelial cells. Cancer Res. 59: 5112–5118 [PubMed] [Google Scholar]
- 35. O'Day E., Lal A. 2010. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 12: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orom U. A., Nielsen F. C., Lund A. H. 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- 37. Pandya A. Y., et al. 2004. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin. Cancer Res. 10: 2709–2719 [DOI] [PubMed] [Google Scholar]
- 38. Park I. H., et al. 2008. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451: 141–146 [DOI] [PubMed] [Google Scholar]
- 39. Rowland B. D., Bernards R., Peeper D. S. 2005. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 7: 1074–1082 [DOI] [PubMed] [Google Scholar]
- 40. Rowland B. D., Peeper D. S. 2006. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 6: 11–23 [DOI] [PubMed] [Google Scholar]
- 41. Ruppert J. M., Vogelstein B., Kinzler K. W. 1991. The zinc finger protein GLI transforms rodent cells in cooperation with adenovirus E1A. Mol. Cell. Biol. 11: 1724–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Segre J. A., Bauer C., Fuchs E. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22: 356–360 [DOI] [PubMed] [Google Scholar]
- 43. Shields J. M., Christy R. J., Yang V. W. 1996. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J. Biol. Chem. 271: 20009–20017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steitz J. A., Vasudevan S. 2009. miRNPs: versatile regulators of gene expression in vertebrate cells. Biochem. Soc. Trans. 37: 931–935 [DOI] [PubMed] [Google Scholar]
- 45. Suzuki T., et al. 2002. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32: 166–174 [DOI] [PubMed] [Google Scholar]
- 46. Swamynathan S. K., et al. 2007. Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Mol. Cell. Biol. 27: 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takahashi K., et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- 48. Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- 49. Tavazoie S. F., et al. 2008. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tian Y., et al. 2010. MicroRNA-10B promotes human esophageal cancer cell migration and invasion through KLF4. J. Biol. Chem. 285: 7986–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vasudevan S., Steitz J. A. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vasudevan S., Tong Y., Steitz J. A. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934 [DOI] [PubMed] [Google Scholar]
- 53. Vasudevan S., Tong Y., Steitz J. A. 2008. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 7: 1545–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang C., Han M., Zhao X. M., Wen J. K. 2008. Kruppel-like factor 4 is required for the expression of vascular smooth muscle cell differentiation marker genes induced by all-trans retinoic acid. J. Biochem. 144: 313–321 [DOI] [PubMed] [Google Scholar]
- 55. Wellner U., et al. 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11: 1487–1495 [DOI] [PubMed] [Google Scholar]
- 56. Xie C., et al. 2011. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 20: 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu N., Papagiannakopoulos T., Pan G., Thomson J. A., Kosik K. S. 2009. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658 [DOI] [PubMed] [Google Scholar]
- 58. Yamanaka S. 2009. A fresh look at iPS cells. Cell 137: 13–17 [DOI] [PubMed] [Google Scholar]
- 59. Yoon H. S., Chen X., Yang V. W. 2003. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J. Biol. Chem. 278: 2101–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yori J. L., Johnson E., Zhou G., Jain M. K., Keri R. A. 2010. Kruppel-like Factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J. Biol. Chem. 285: 16854–16863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoshida T., et al. 2010. Smooth and cardiac muscle-selective knock-out of Kruppel-like factor 4 causes postnatal death and growth retardation. J. Biol. Chem. 285: 21175–21184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu F., et al. 17 January 2011, posting date. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene doi:10.1038/onc.2010.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang P., Andrianakos R., Yang Y., Liu C., Lu W. 2010. Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 285: 9180–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang W., et al. 2000. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 275: 18391–18398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao W., et al. 2004. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene 23: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]