Abstract
While telomeres must provide mechanisms to prevent DNA repair and DNA damage checkpoint factors from fusing chromosome ends and causing permanent cell cycle arrest, these factors associate with functional telomeres and play critical roles in the maintenance of telomeres. Previous studies have established that Tel1 (ATM) and Rad3 (ATR) kinases play redundant but essential roles for telomere maintenance in fission yeast. In addition, the Rad9-Rad1-Hus1 (911) and Rad17-RFC complexes work downstream of Rad3 (ATR) in fission yeast telomere maintenance. Here, we investigated how 911, Rad17-RFC and another RFC-like complex Ctf18-RFC contribute to telomere maintenance in fission yeast cells lacking Tel1 and carrying a novel hypomorphic allele of rad3 (DBD-rad3), generated by the fusion between the DNA binding domain (DBD) of the fission yeast telomere capping protein Pot1 and Rad3. Our investigations have uncovered a surprising redundancy for Rad9 and Hus1 in allowing Rad1 to contribute to telomere maintenance in DBD-rad3 tel1Δ cells. In addition, we found that Rad17-RFC and Ctf18-RFC carry out redundant telomere maintenance functions in DBD-rad3 tel1Δ cells. Since checkpoint sensor proteins are highly conserved, genetic redundancies uncovered here may be relevant to telomere maintenance and detection of DNA damage in other eukaryotes.
Keywords: telomere, checkpoint, Rad9-Rad1-Hus1, Rad17, Ctf18
Introduction
Telomeres, the ends of eukaryotic chromosomes, are essential for protection of chromosome ends against rearrangements and fusions. In most eukaryotes, telomeres also allow telomerase to counteract the gradual erosion of chromosome ends, caused by the inability of replicative DNA polymerases to fully replicate ends of linear DNA, by extending the telomeric GT-rich repeat sequences.1 GT-rich telomeric repeats consist of both double-stranded regions and 3′ single-stranded overhangs, known as G-tails. G-tails are essential for both telomeric repeat extension by telomerase and protection of telomeres by the telomere-capping complex.2
In mammalian cells, telomeres are capped and protected by the “shelterin” complex, composed of TRF1, TRF2, RAP1, TIN2, TPP1 and POT1.2 A corresponding shelterin-like complex, composed of Taz1, Rap1, Poz1, Ccq1, Tpz1 and Pot1, was recently identified in fission yeast.3 TRF1 and TRF2 specifically bind to the double-stranded telomeric repeats in mammalian cells, while Taz1 specifically binds the double-stranded telomeric repeats in fission yeast cells.2 In both mammals and fission yeast, Pot1, in collaboration with TPP1/Tpz1, is thought to protect telomeres by binding to the G-tails,3-5 and to contribute to the recruitment and/or activation of telomerase.3, 6, 7 In contrast, budding yeast cells lack TRF1/TRF2/Taz1 and Pot1 orthologs, and instead utilize Rap1 and Cdc13 to recognize the double-stranded DNA (dsDNA) and G-tail portions of telomeric repeats, respectively.2
The catalytic subunit of telomerase, known as telomerase reverse transcriptase (TERT), stably associates with the telomerase RNA subunit, which functions both as a platform for the assembly of telomerase subunits and as a template for telomeric repeat extension by telomerase.8, 9 In fission yeast, the trt1+ gene encodes the TERT subunit and the ter1+ gene encodes the telomerase RNA.10-12 Recent studies have shown that the fission yeast shelterin subunit Ccq1 is required to promote the interaction between Tpz1 and telomerase as well as the recruitment of telomerase to telomeres.3, 13, 14 Ccq1 is also required to protect telomeres against checkpoint activation and recombination,3, 13 and regulates heterochromatin formation at telomeres by interacting with the Snf2/Hdac-containing repressor complex (SHREC).15 However, it is currently unclear if the mammalian shelterin complex also associates with a Ccq1-like molecule to regulate telomere protection and/or telomerase recruitment.
Unlike other types of DNA double-strand breaks (DSBs), telomeres should not be repaired (i.e. fused) and should not fully activate DNA damage checkpoint responses. Thus, one might expect that functional telomeres would exclude DNA repair and DNA damage checkpoint proteins. However, studies have found that proteins involved in repair of DSBs, such as the Ku and Mre11-Rad50-Nbs1 (MRN) complexes, are recruited to telomeres, and they are in fact required for proper maintenance of telomeres.16, 17 Furthermore, proteins responsible for the detection of DSBs in DNA damage checkpoint responses, such as the PIKK (phosphatidyl inositol 3-kinase-like kinase) family kinases ATM and ATR, are required for stable maintenance of telomeres in a wide variety of organisms,18-23 and they are recruited to functional telomeres.24-26 In fission yeast, we have recently shown that Tel1 (ATM) and Rad3 (ATR) contribute redundantly to telomere protection and telomerase recruitment by promoting the efficient accumulation of Ccq1 at telomeres.14 In addition, other checkpoint “sensor” complexes, such as the proliferating cell nuclear antigen (PCNA)-like ring shaped Rad9-Rad1-Hus1 (911) complex as well as the alternative replication factor C (RFC)-like complexes Rad17-RFC and Ctf18-RFC, have been shown to contribute to telomere maintenance.19, 20, 22, 23, 27, 28 However it is still not understood how the 911, Rad17-RFC and Ctf18-RFC complexes contribute to the stable maintenance of telomeres.
In this study, we investigated how 911, Rad17-RFC and Ctf18-RFC complexes contribute to fission yeast telomere maintenance in the context of a novel hypomorphic allele of rad3, DBD-rad3, generated by the fusion between the Pot1 DNA binding domain and Rad3. While we expected to find that rad1Δ, rad9Δ, hus1Δ and rad17Δ cells would exhibit identical telomere phenotypes, our investigations uncovered a surprising redundancy for the 911-complex subunits Rad9 and Hus1 in allowing Rad1 to contribute to telomere maintenance in DBD-rad3 tel1Δ cells. Furthermore, we show that the Rad17-RFC and Ctf18-RFC complexes contribute redundantly to telomere maintenance in DBD-rad3 tel1Δ cells.
Results
Creation of fusion proteins between the DNA-binding domain (DBD) of Pot1 and checkpoint proteins
In budding yeast, studies utilizing a series of fusion proteins between the DNA binding domain of Cdc13 and various telomerase and telomere binding proteins have yielded valuable insights on how telomere maintenance is regulated.29-31 Motivated by such studies, we explored the possibility of creating fusion proteins between the DNA binding domain of Pot1 and various factors that might play a role in telomere function, in order to test if the DNA binding domain of Pot1 could be utilized to ectopically target the resulting fused proteins to the G-tails of fission yeast telomeres.
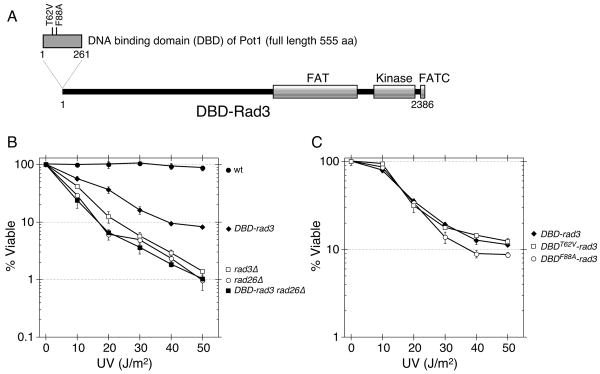
The DNA binding domain of Pot1 has been shown to reside within the N-terminal 185 amino acids and to form an oligonucleotide/oligosaccharide-binding (OB) fold,32 much like the DNA binding domain of budding yeast Cdc13.33 We chose to clone a cDNA version of the Pot1 DNA N-terminal fragment corresponding to amino acids 1-261 of Pot1 (this particular fragment will be hereafter referred to as DBD) into a plasmid, and utilized it further in our fusion protein studies. In addition to the wild-type Pot1 DBD, we also created mutant versions of DBD, which carry either Threonine 62 to Valine (T62V) or Phenylalanine 88 to Alanine (F88A) mutations that have previously been shown to abolish the G-tail binding activity of Pot1 DBD32 (Fig. 1A).
Figure 1.
Construction and initial characterization of DBD-rad3 related strains. (A) A schematic diagram representing the DBD-rad3 construct, integrated at the rad3+ locus. The conserved functional domain of Rad3 and the two point mutations (T62V and F88A) in the Pot1 DNA binding domain used in this study are also indicated. (B, C) UV survival experiments involving DBD-rad3 strains. Experiments were repeated at least three times, and error bars correspond to standard deviations.
We were interested in understanding how checkpoint signaling is modulated at telomeres, and wondered if the forced targeting of DNA damage checkpoint proteins to telomeres might result in permanent cell cycle arrest. Thus, we decided to N-terminally fuse Pot1 DBD to either the checkpoint kinase Rad3ATR or the checkpoint adaptor protein Crb2/Rhp9. These two checkpoint proteins were chosen due to the contrasting telomere phenotypes exhibited by their deletions. Previous studies have established that the Rad3ATR-Rad26ATRIP complex is very important for maintenance of wild-type length telomeres, and it is recruited to functional telomeres during telomere replication.22, 24, 34 By contrast, crb2Δ cells were found to carry essentially wild-type length telomeres,22 and thus Crb2 might be normally excluded from telomeres to prevent permanent activation of the checkpoint by telomeric DNA ends. For Rad3ATR fusion, the wild-type rad3+ gene was replaced with the DBD-rad3 allele. For Crb2 fusion, the DBD-crb2 allele was integrated at the leu1+ locus in a crb2Δ strain. Therefore, the engineered Pot1 DBD fusion proteins were the only copies of Rad3ATR or Crb2 expressed in both cases (see Materials and Methods for details).
We first examined if the cells showed any sign of elongation or slow growth, possibly indicating an ectopic activation of checkpoint signaling at telomeres. We found that both DBD-rad3 and DBD-crb2 cells grew normally and did not generate highly elongated cells, expected of cells with activated DNA damage checkpoint response (data not shown). Thus, we concluded that these fusion proteins did not cause ectopic activation of DNA damage checkpoint responses.
Since we did not have the means to directly examine if the fusion proteins were expressed properly or indeed localized to telomeres, it was possible that these particular fusion constructs simply generated inactive Rad3ATR or Crb2 proteins. On the other hand, we examined DNA damage sensitivities of DBD-rad3 and DBD-crb2 strains, and found that they were more sensitive to both hydroxyurea (HU) and UV than wild-type cells, but less sensitive than rad3Δ or crb2Δ cells, respectively (Fig. 1B and data not shown). Thus, it is likely that both DBD-Rad3ATR and DBD-Crb2 are expressed, and that these fusion proteins retain at least some function. In addition, DBD-rad3 cells were found to carry intermediate telomere length that is shorter than rad3+ but longer than rad3Δ cells (Fig. 2), suggesting that the DBD-Rad3ATR protein retains at least partial functions of Rad3ATR in telomere maintenance. As for DBD-crb2 cells, they showed normal telomere length, much like crb2Δ cells (data not shown). Since we were more interested in the telomere length regulation mechanism and DBD-Crb2 was unable to induce DNA damage checkpoint activation, we decided to focus our attention solely on characterizing the DBD-rad3 mutant allele.
Figure 2.
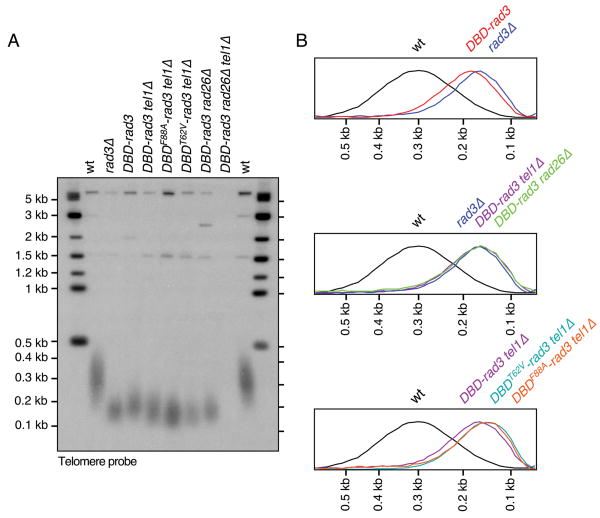
Telomeric repeat length analysis of DBD-rad3 strains. (A) Southern blot hybridization analysis of ApaI digested genomic DNA, probed with telomeric repeat sequences. All strains were extensively streaked on agar plates prior to preparation of genomic DNA to ensure terminal telomere phenotype, and all lanes contained comparable genomic DNA based on ethidium bromide (EtBr) staining (data not shown). (B) Quantification of hybridization signal shown in (A) by ImageQuant software (Molecular Dynamics). Peaks of telomeric repeat hybridization signal were normalized and plotted against DNA size.
Characterization of the DBD-Rad3 hypomorphic allele
We wished to test if the UV hypersensitivity of DBD-rad3 cells (Fig. 1B) might be caused by the ability of Pot1 DBD to bind to the telomeric G-tail and potentially sequester Rad3ATR to telomeres and therefore prevent it from recognizing DNA damage occurring elsewhere in the genome. Thus, we examined if we might be able to rescue the UV hypersensitivity of DBD-rad3 cells by introducing mutations in Pot1 DBD to abolish its G-tail binding activity.35 Since we observed similar UV hypersensitivities for DBD-rad3, DBDT62V-rad3 and DBDF88A-rad3 cells (Fig. 1C), we concluded that the UV hypersensitivity of DBD-rad3 cells is not necessarily caused by the G-tail binding activity of Pot1 DBD. Instead, it is more likely that addition of Pot1 DBD to the N-terminus of Rad3ATR is hindering its proper function and causing various hypomorphic phenotypes in DBD-rad3 cells.
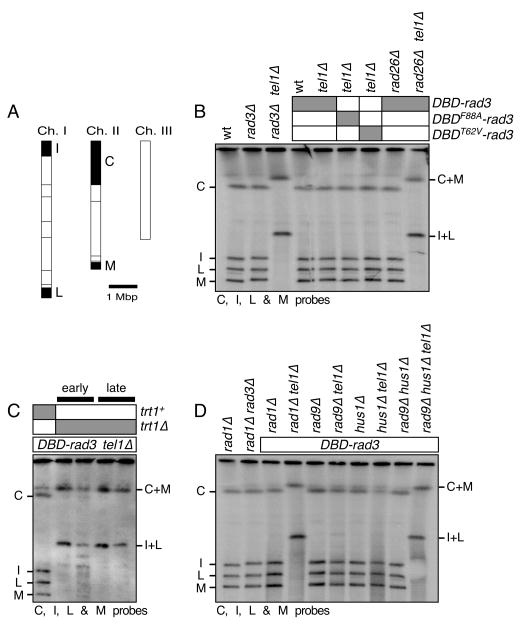
It has been previously established that Rad3ATR-Rad26ATRIP and Tel1ATM-MRN represent two redundant pathways essential for telomere maintenance in fission yeast.22, 36 Thus, cells simultaneously lacking these two pathways, such as rad3Δ tel1Δ and rad26Δ tel1Δ, lose their telomeres and circularize all three chromosomes.22, 34, 36 Since we observed that DBD-rad3 cells already exhibit telomere shortening (Fig. 2), we next examined if DBD-rad3 tel1Δ cells could still maintain telomeres or circularize their chromosomes. When average telomere-repeat length was analyzed by Southern blot hybridized to a telomeric repeat DNA probe (Fig. 2), we found that DBD-rad3 tel1Δ and rad3Δ cells carry identical telomere length. Chromosome circularization in fission yeast cells can be conveniently monitored by separating NotI-digested chromosomal DNA by pulsed-field gel electrophoresis (PFGE), and then performing Southern blot analysis with telomeric NotI fragment-specific probes C, I, L and M (Fig. 3A). In this assay, cells carrying circular chromosomes lose individual telomeric NotI fragments and show I+L and C+M bands, corresponding to the fused telomeric fragments from chromosomes I and II. As shown in Fig. 3B, we found that DBD-rad3 tel1Δ cells can stably maintain linear chromosomes. Thus, it appeared that the DBD-rad3 allele retained critical telomere function of Rad3ATR, essential for telomere maintenance in cells lacking the Tel1ATM kinase.
Figure 3.
PFGE analysis of NotI digested chromosomal DNA for the indicated DBD-rad3 related strains. (A) NotI restriction enzyme map of fission yeast chromosomes (horizontal lines). The telomeric probes C, I, L and M, used in Southern blot hybridization are filled black and marked. Chromosome circularization is indicated by the appearance of C+M and I+L fusion bands. Chromosome III was not monitored since it lacks NotI site. (B-D) Chromosomal DNA was prepared in agarose plugs, digested with NotI, and then used in PFGE for the indicated strains, which were extensively restreaked on agar plates to ensure terminal telomere phenotype. DNA was then transferred to a Nylon membrane and hybridized to C, I, L and M specific probes. For (C), “early” samples are prepared from confirmed trt1Δ strains immediately after transformation with the trt1Δ construct, while “late” samples are prepared from strains restreaked four times on agar plates after transformation with the trt1Δ construct.
We next tested if the potential G-tail binding activity provided by Pot1 DBD might be critical for telomere maintenance in DBD-rad3 tel1Δ cells. Mutations in Pot1 DBD (T62V and F88A) of DBD-Rad3 did cause a slight but reproducible telomere shortening in DBDT62V-rad3 tel1Δ and DBDF88A-rad3 tel1Δ cells when compared to DBD-rad3 tel1Δ cells (Fig. 2). However, both DBDT62V-rad3 tel1Δ and DBDF88A-rad3 tel1Δ cells stably maintained linear chromosomes (Fig. 2 and 3B). Thus, while the G-tail binding activity of Pot1 DBD might provide a slight advantage for the DBD-Rad3 fusion protein to function more effectively at telomeres, the G-tail binding activity of Pot1 DBD is not essential for telomere maintenance in DBD-rad3 tel1Δ cells.
Rad26ATRIP is still necessary for telomere function(s) provided by DBD-Rad3ATR
The Rad3ATR kinase forms a complex with its regulatory subunit Rad26ATRIP, and previous studies have shown that the ATRIP subunit plays a critical role in the recruitment of ATR kinase to sites of DNA damage and telomeres.37-39 Thus, we hypothesized that we might be able to bypass the requirement for Rad26ATRIP in telomere maintenance if Pot1 DBD was indeed able to target the DBD-Rad3ATR to telomeres, independently of Rad26ATRIP. We have previously established that rad3Δ, rad26Δ and rad3Δ rad26Δ cells show identical telomere length by Southern blot analysis, leading to the conclusion that Rad3ATR and Rad26ATRIP work in the same epistasis group for telomere maintenance.22 Moreover, we have shown that rad26Δ tel1Δ cells carry circular chromosomes, much like rad3Δ tel1Δ cells.22
Thus, if DBD-Rad3ATR is able to bypass the requirement for Rad26ATRIP for Rad3ATR function, we expect DBD-rad3 rad26Δ cells to show longer telomere length than rad3Δ or rad26Δ cells, and DBD-rad3 rad26Δ tel1Δ cells to still maintain telomeres. However, we found that DBD-rad3 rad26Δ cells carry the same telomere length as rad3Δ cells (Fig. 2), and DBD-rad3 rad26Δ tel1Δ cells lose telomeres and circularize their chromosomes (Fig. 2A and 3B). Recent studies have provided evidence that the ATRIP subunit is not only important for the recruitment of ATR to sites of DNA damage, but also for the regulation of ATR kinase activity.40, 41 Therefore, a failure of DBD-Rad3ATR to bypass the telomere function of Rad26ATRIP might simply reflect the fact that Rad3ATR kinase cannot function without Rad26ATRIP. In any case, based on our current data, we were unable to establish whether the Pot1 DBD module could indeed target Rad3ATR to telomeres in the absence of Rad26ATRIP.
Telomere maintenance in DBD-rad3 tel1Δ cells depends on the telomerase catalytic subunit Trt1TERT
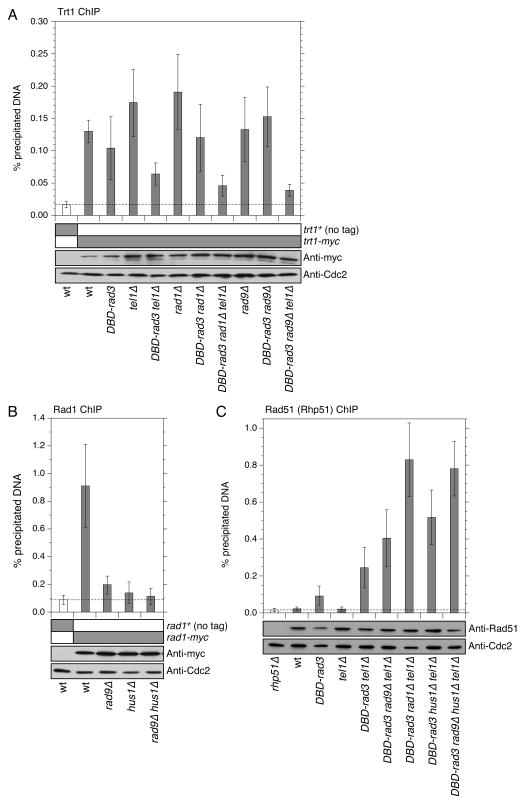
We have recently shown that rad3Δ tel1Δ cells are defective in both telomerase recruitment to telomeres and telomere protection.14 Thus, we next examined if DBD-rad3 tel1Δ cells could still recruit telomerase to telomeres, and if telomere maintenance in DBD-rad3 tel1Δ is dependent on telomerase. When the telomerase catalytic subunit Trt1TERT was eliminated, we found that the resulting DBD-rad3 tel1Δ trt1Δ cells were no longer able to maintain linear chromosomes (Fig. 3C). Therefore, telomere maintenance in DBD-rad3 tel1Δ cells depends on telomerase. Based on quantitative chromatin immunoprecipitation (ChIP) assays, we found that Trt1TERT recruitment to telomeres was reduced in DBD-rad3 tel1Δ cells compared to rad3+, DBD-rad3 or tel1Δ cells (Fig. 4A); however, Trt1TERT was still detected at telomeres significantly above the untagged control in DBD-rad3 tel1Δ cells. We thus concluded that the residual telomere function provided by DBD-Rad3ATR was sufficient to allow the recruitment of telomerase to telomeres and maintain telomeres in DBD-rad3 tel1Δ cells.
Figure 4.
Quantitative ChIP assays to monitor the recruitment of Trt1 (A), Rad1 (B) and Rad51 (Rhp51) (C) to telomeres for the indicated strains. Error bars represent standard deviation among at least three independent experiments. Expression levels for the indicated proteins were monitored by Western blot analysis, and Western blots with anti-Cdc2 served as loading controls. For (A), Trt1-myc showed statistically significant enrichment of telomeric DNA over no tag control for all strains tested (P < 0.015), while the difference in % precipitated (ppt.) telomeric DNA values between DBD-rad3 rad1Δ tel1Δ and DBD-rad3 rad9Δ tel1Δ was not statistically significant (P = 0.47). For (B), Rad1-myc showed statistically significant enrichment of telomeric DNA over no tag control for wild-type (wt) (P = 0.0010) and rad9Δ (P = 0.020) but not for hus1Δ (P = 0.30) and rad9Δ hus1Δ (P = 0.51). On the other hand, Rad1-myc ChIP analyses found no statistically significant differences among rad9Δ, hus1Δ and rad9Δ hus1Δ strains (P = 0.067 ∼ 0.60). For (C), Rhp51 showed statistically significant enrichment of telomeric DNA over rhp51Δ control for all strains tested (P < 0.0089), except wt (P = 0.54) and tel1Δ (P = 0.50) strains. In addition, % ppt. telomeric DNA values for DBD-rad3 tel1Δ rad1Δ cells showed statistically significant difference against DBD-rad3 tel1Δ (P = 0.00075), DBD-rad3 tel1Δ rad9Δ (P = 0.0044), DBD-rad3 tel1Δ hus1Δ (P = 0.013), but not against DBD-rad3 tel1Δ rad9Δ hus1Δ cells (P = 0.57) for Rhp51 ChIP.
Rad1, but not Rad9 or Hus1, is essential for telomere maintenance in DBD-rad3 tel1Δ cells
Even though we could not establish if the Pot1 DBD can target Rad3ATR to the telomeric G-tail, we were still intrigued by the telomere phenotypes exhibited by DBD-rad3 mutant cells. We also reasoned that the ability to stably maintain telomeres in DBD-rad3 tel1Δ cells might provide us with a convenient functional assay to characterize factors that collaborate with the Rad3ATR-Rad26ATRIP complex in telomere maintenance. Therefore, we next investigated what additional factors besides Trt1TERT and Rad26ATRIP might be required for telomere maintenance in DBD-rad3 tel1Δ cells.
We focused our attention on the 911 checkpoint sensor complex, which forms a stable ring shaped complex resembling PCNA.42, 43 Deletions of individual components of this complex (rad1Δ, rad9Δ or hus1Δ) result in identical telomere shortening phenotypes,22, 44 and previous epistasis analyses have allowed us to establish that the 911-complex represents one of multiple redundant pathways that are regulated by the Rad3ATR-Rad26ATRIP complex in telomere maintenance.22 The fission yeast 911-complex has also been shown to associate with telomeres by ChIP assays.22
Based on our previous characterization of the 911-complex deletion mutants, we expected identical telomere phenotypes for DBD-rad3 tel1Δ rad1Δ, DBD-rad3 tel1Δ rad9Δ, and DBD-rad3 tel1Δ hus1Δ cells. Surprisingly, we found that while DBD-rad3 tel1Δ rad1Δ cells are unable to maintain telomeres and circularize their chromosomes, DBD-rad3 tel1Δ rad9Δ and DBD-rad3 tel1Δ hus1Δ cells stably maintain telomeres (Fig. 3D). Additionally, we found that DBD-rad3 tel1Δ rad9Δ hus1Δ cells are unable to maintain telomeres (Fig. 3D). Thus, Rad1 appears to provide the most critical telomere maintenance function, while Rad9 and Hus1 play redundant roles that allow Rad1 to carry out its essential telomere function in DBD-rad3 tel1Δ cells.
To better understand why Rad9 and Hus1 are redundantly required for telomere maintenance in DBD-rad3 tel1Δ cells, we monitored how telomere recruitment of Rad1 is affected in rad9Δ, hus1Δ or rad9Δ hus1Δ cells, compared to wild-type cells. Our working hypothesis was that Rad9 and Hus1 would provide redundant functions for the recruitment of Rad1 to telomeres, and thus cells would completely lose the ability to maintain telomeres in the rad9Δ hus1Δ background, much like in rad1Δ cells. Accordingly, we hoped to observe a significant association of Rad1 with telomeres in rad9Δ or hus1Δ cells, but not in rad9Δ hus1Δ cells. However, we found that rad9Δ and hus1Δ cells already show reduced levels of telomeric DNA pulled down by Rad1, and we could not detect any differences among rad9Δ, hus1Δ and rad9Δ hus1Δ cells in terms of Rad1 recruitment efficiency to telomeres (Fig. 4B). Thus, while we still presume that Rad1 would probably need to associate with telomeres to perform its telomere function, we were unable to correlate the efficiency of Rad1 association with the observed differences in telomere maintenance phenotypes among DBD-rad3 tel1Δ rad9Δ, DBD-rad3 tel1Δ hus1Δ and DBD-rad3 tel1Δ rad9Δ hus1Δ cells.
Studies in mammalian cells have shown that the 911-complex interacts with telomerase and positively regulates telomerase activity.19 In addition, studies in Caenorhabditis elegans have found that the 911-complex is essential for telomerase-dependent telomere maintenance.20, 45 Since we observed that DBD-rad3 tel1Δ cells show residual Trt1TERT recruitment to telomeres (Fig. 4A) while rad3Δ tel1Δ cells fail to recruit telomerase to telomeres,14 we next examined if the deletion of rad1 or rad9 differentially affects the ability of DBD-rad3 tel1Δ cells to recruit Trt1TERT to telomeres by ChIP assay. However, we did not observe significant differences in Trt1TERT recruitment between DBD-rad3 tel1Δ rad1Δ and DBD-rad3 tel1Δ rad9Δ cells (Fig. 4A). Furthermore, we have thus far been unsuccessful in detecting an interaction between Rad1 and Trt1TERT or between Rad1 and TER1 telomerase RNA by co-immunoprecipitation assays (data not shown). Taken together, it thus appears that the 911-complex does not significantly contribute to telomerase recruitment to telomeres in DBD-rad3 tel1Δ cells. Furthermore, it is unlikely that differences in telomerase recruitment efficiency could account for the difference in the ability of DBD-rad3 tel1Δ rad1Δ and DBD-rad3 tel1Δ rad9Δ cells to maintain telomeres.
Next, we tested if Rad1 might contribute to telomere protection against chromosome fusion in DBD-rad3 tel1Δ cells. Since we have previously observed that rad3Δ tel1Δ cells are defective in preventing the association of the homologous recombination (HR) repair protein Rhp51Rad51 with telomeres,14 we next monitored the recruitment of Rhp51Rad51 in various mutant backgrounds. As shown in Fig. 4C, deletion of rad1 as well as the simultaneous deletion of rad9 and hus1 in DBD-rad3 tel1Δ cells led to a significant increase in Rhp51Rad51 recruitment to telomeres, compared to DBD-rad3 tel1Δ, DBD-rad3 tel1Δ rad9Δ or DBD-rad3 tel1Δ hus1Δ cells. Thus, reduced protection against telomere fusions appears to account for the chromosome circularization phenotype observed in DBD-rad3 tel1Δ rad1Δ and DBD-rad3 tel1Δ rad9Δ hus1Δ cells.
Rad17-RFC and Ctf18-RFC are redundantly required for telomere maintenance in DBD-rad3 tel1Δ cells
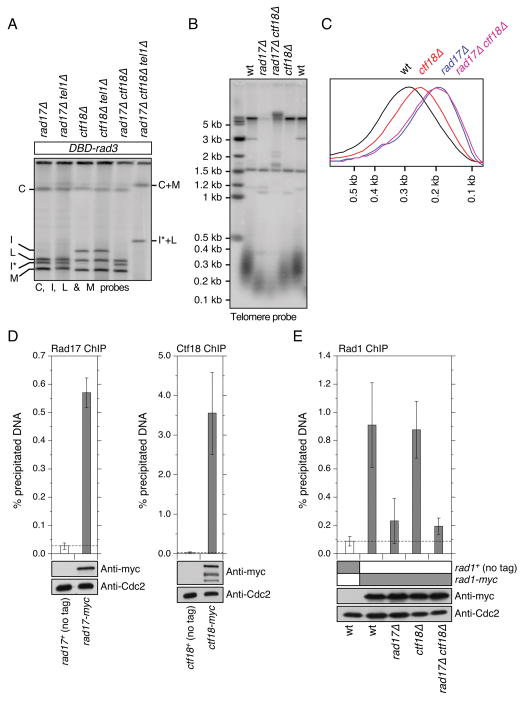
Previous studies have established that the replication factor C (RFC)-like checkpoint complex Rad17 (Rad17-RFC) is important for the loading of the 911-complex to sites of DNA damage.46-48 Additionally, Rad17 is essential for telomere maintenance in the same pathway as the 911-complex in C. elegans,23 and Rad17 and the 911-complex work in the same pathway for telomere length regulation in fission yeast.22 Thus, we investigated if Rad17 is also required for telomere maintenance in DBD-rad3 tel1Δ cells. Since we believed Rad17 might be essential for recruitment of Rad1 to telomeres, we expected DBD-rad3 tel1Δ rad17Δ cells to circularize their chromosomes, much like DBD-rad3 tel1Δ rad1Δ cells. However, contrary to our prediction, we found that DBD-rad3 tel1Δ rad17Δ cells can stably maintain telomeres (Fig. 5A). Therefore, it appears that DBD-rad3 tel1Δ cells have the ability to allow Rad1 to function at telomeres even in the absence of the Rad17-RFC complex.
Figure 5.
Characterization of telomere phenotypes involving Rad17 and Ctf18. (A) PFGE analysis of NotI digested chromosomal DNA for the indicated strains. DNA was then transferred to a Nylon membrane and hybridized to C, I, L and M specific probes. Since rad17+ gene resides on the “I” fragment and rad17Δ introduced an additional NotI site, the “I” fragment migrates faster (marked “I*”) in rad17Δ background. (B) Southern blot hybridization analysis of ApaI digested genomic DNA, probed with telomeric repeat sequences. All strains were extensively streaked on agar plates prior to preparation of genomic DNA to ensure terminal telomere phenotype, and all lanes contained comparable genomic DNA based on EtBr staining (data not shown). (C) Quantification of hybridization signal shown in (B) by ImageQuant software (Molecular Dynamics). Peaks of telomeric repeat hybridization signal were normalized and plotted against DNA size. (D, E) Quantitative ChIP assays to monitor the recruitment of Rad17, Ctf18 and Rad1 to telomeres for the indicated strains. Error bars represent standard deviation among at least three independent experiments. Expression levels for the indicated proteins were monitored by Western blot analysis, and Western blots with anti-Cdc2 served as loading controls. For (D), both Rad17-myc (P = 0.000067) and Ctf18-myc (P = 0.0044) showed statistically significant enrichment of telomeric DNA over no tag control. For (E), Rad1-myc ChIP showed statistically significant changes against wild-type (wt) cells for rad17Δ (P = 0.0021) and rad17Δ ctf18Δ (P = 0.00075), but not for ctf18Δ (P = 0.83) cells.
We wondered if another RFC-like complex containing Ctf18 (Ctf18-RFC) might have redundant telomere functions with Rad17-RFC and allow Rad1 to provide telomere maintenance functions in DBD-rad3 tel1Δ cells. We suspected the involvement of the fission yeast Ctf18-RFC complex in telomere maintenance since the budding yeast Ctf18-RFC complex functions redundantly with the Rad24-RFC complex (ortholog of fission yeast Rad17-RFC) for the regulation of the 911-complex in DNA replication checkpoint.49 When we monitored telomere status by PFGE, we found that DBD-rad3 tel1Δ rad17Δ ctf18Δ cells circularized their chromosomes while DBD-rad3 tel1Δ ctf18Δ cells still maintained telomeres (Fig. 5A). Thus, Rad17-RFC and Ctf18-RFC are indeed redundantly required for telomere maintenance in DBD-rad3 tel1Δ cells. Southern blot analysis revealed that ctf18Δ cells carry short telomeres, although the extent of telomere shortening was less than rad17Δ cells (Fig. 5B, C). On the other hand, since rad17Δ and rad17Δ ctf18Δ cells showed identical telomere length distribution, it appears that Rad17-RFC and Ctf18-RFC contribute to telomere length maintenance through a single pathway.
We have previously established that Rad1 and Rad17 work in the same pathway to regulate telomere length, based on the observation that rad1Δ, rad17Δ and rad1Δ rad17Δ cells show identical telomere length distribution.22 Additionally, we monitored the association of Ctf18 with telomeres by ChIP assay, and found that Ctf18 can efficiently pull down telomeric DNA, even better than Rad17 (Fig. 5D). Taken together, we concluded that both Rad17-RFC and Ctf18-RFC are recruited to normal telomeres, and that they are likely to play redundant roles in allowing the 911-complex to contribute positively to the maintenance of telomeres in fission yeast.
Based on these observations, we hypothesized that Rad17-RFC and Ctf18-RFC might contribute redundantly to telomere recruitment of Rad1. Therefore, we next examined how loss of Rad17 and/or Ctf18 would affect the recruitment of Rad1 to telomeres by ChIP assay. We observed that Rad1 recruitment to telomeres was greatly reduced in rad17Δ cells compared to wild-type cells, and that the level of Rad1 recruitment in rad17Δ cells became essentially background (Fig. 5E). Moreover, the level of Rad1 recruitment in rad17Δ ctf18Δ cells was indistinguishable from rad17Δ cells. On the other hand, ctf18Δ cells showed Rad1 recruitment level similar to wild-type cells (Fig. 5E). Thus, our data did not enable us to provide support for the notion that Ctf18-RFC contributes to telomere length regulation by promoting the recruitment of the 911-complex to telomeres.
Discussion
Can Pot1 DBD be used to target proteins to telomere G-tails?
In this study, we described our initial attempts to ectopically target proteins to the G-tail of fission yeast telomeres by creating fusion proteins between the target protein and the G-tail binding domain of Pot1. We chose to fuse amino acids 1-261 of Pot1 to either Rad3ATR or Crb2, with the hope that such fusion constructs will be targeted to the G-tails of fission yeast telomeres. Our choice to use the N-terminal 261 amino acids was based on previous studies that have mapped the G-tail binding activity of Pot1 to the region within the N-terminal 185 amino acid fragment of Pot1.5, 32 Unfortunately, based on our characterization of these fusion constructs, we were unable to obtain evidence that our constructs were indeed targeted to the G-tail by the Pot1 DBD.
It's possible that we would need to use longer N-terminal fragments of Pot1 to target fusion proteins to the G-tail, since recent studies have found that the N-terminal 389 amino acid fragment of Pot1 binds much tighter to single-stranded G-rich telomere primers than the Pot1 187 amino acid N-terminal fragment.50, 51 Of course, it is also possible that the addition of the Pot1 DBD at the N-terminus might have interfered with the normal functions of Rad3ATR and Crb2, and thus our 261 amino acid Pot1 DBD construct might be more successful in targeting other fused proteins to the G-tails of fission yeast telomeres. In any case, further optimization is clearly necessary to ensure successful targeting of the fused proteins to the telomeric G-tails by the G-tail binding domain of Pot1.
DBD-rad3 as an unusual but useful new hypomorphic allele of rad3 to dissect factors involved in telomere maintenance
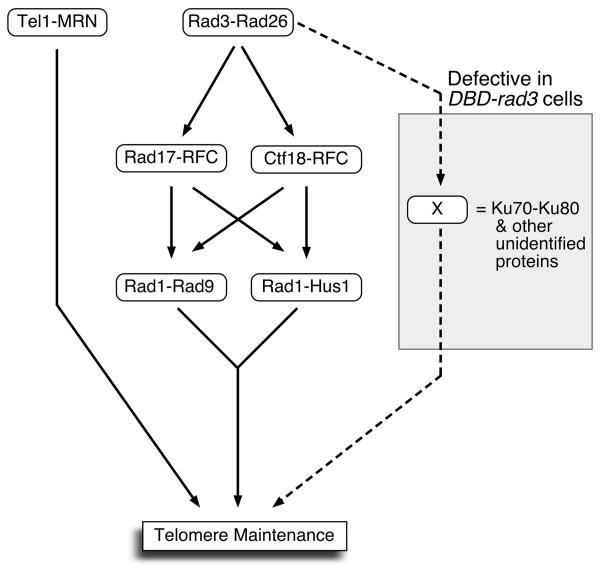
Despite the fact that we could not be sure if DBD-Rad3ATR is indeed targeted to G-tails by the Pot1 DBD more than wild-type Rad3ATR, we utilized the DBD-rad3 mutant allele as a new interesting hypomorphic allele in order to investigate how the 911, Rad17-RFC and Ctf18-RFC complexes contribute to telomere maintenance in fission yeast. Previously, epistasis analyses based on measurements of steady state average telomere lengths for single and multiple combinations of deletion mutants among DNA repair and checkpoint factors established that Rad3ATR-Rad26ATRIP and Tel1ATM-MRN represent two redundant pathways that are essential for telomere maintenance in fission yeast22, 36 (Fig. 6).
Figure 6.
A possible genetic interaction model for telomere maintenance in fission yeast. The model incorporates the observed redundant telomere maintenance functions of Rad1-Rad9 and Rad1-Hus1 as well as redundant telomere functions for Rad17-RFC and Ctf18-RFC in DBD-rad3 tel1Δ cells. It should be noted that the current study does not provide direct molecular evidence for the existence of Rad1-Rad9 and Rad1-Hus1 sub-complexes. In addition, observed redundancy for the Rad17-RFC and Ctf18-RFC complexes in telomere maintenance could involve downstream target(s) other than the 911-complex.
In addition, we have previously determined that the 911, Rad17-RFC, and Rad3ATR-Rad26ATRIP complexes contribute to telomere maintenance in a single pathway. This conclusion was reached because double mutants, that carried one mutation from Rad1, Rad9, Hus1 or Rad17 and another mutation from either Rad3ATR or Rad26ATRIP, behaved like single deletion mutants of the Rad3ATR-Rad26ATRIP complex. However, since only rad3Δ and rad26Δ mutants, but not rad1Δ, rad9Δ, hus1Δ, and rad17Δ mutants, showed synergistic chromosome circularization phenotypes when they were combined with deletion of Tel1ATM or MRN subunits, the Rad3ATR-Rad26ATRIP complex was hypothesized to have additional target(s) other than the 911 and Rad17-RFC complexes that are important for telomere maintenance. Further epistasis analyses revealed that the Ku70-Ku80 non-homologous end-joining (NHEJ) DNA repair complex represents an additional epistasis group which, in addition to the 911 and Rad17-RFC complexes, acts in the Rad3ATR-Rad26ATRIP pathway because telomere shortening phenotypes exhibited by rad3Δ and rad26Δ mutants were epistatic to that of pku70Δ mutation. However, since cells simultaneously lacking the Tel1ATM-MRN, 911/Rad17-RFC and Ku70-Ku80 pathways maintained short but stable telomeres, we concluded that the Rad3ATR-Rad26ATRIP complex must have an additional unidentified telomere maintenance pathway besides the 911/Rad17-RFC and Ku70-Ku80 pathways22 (Fig. 6).
We have not been successful in determining what factor(s) represent the unknown pathway(s) regulated by the Rad3ATR-Rad26ATRIP complex. However, DBD-rad3 mutant cells must be simultaneously defective in both the Ku-dependent mechanism of telomere maintenance/protection and at least one additional pathway that is regulated by the Rad3ATR-Rad26ATRIP complex, so that the Rad1-dependent pathway now represents the only remaining mechanism that allows telomere maintenance in DBD-rad3 tel1Δ cells (Fig. 6). Alternatively, it is possible that DBD-Rad3 (but not wild-type Rad3) strictly depends on Rad1 for the phosphorylation of critical telomere target(s), since the 911-complex, in collaboration with TopBP1/Dpb11 (ortholog of fission yeast Cut5/Rad4), is required to activate ATR/Mec1 kinase in mammals and budding yeast.40, 41, 52 We have previously generated triple mutant cells simultaneously carrying tel1Δ, rad1Δ and various hypomorphic alleles of rad3, hoping to see that such triple mutant cells would show chromosome circularization. However, to date, DBD-rad3 has been the only allele of rad3 found to cause chromosome circularization in a tel1Δ rad1Δ background. In any case, future screens for suppressors of chromosome circularization in DBD-rad3 tel1Δ rad1Δ might finally allow us to identify the factor(s) regulated by Rad3ATR-Rad26ATRIP that work redundantly with the 911/Rad17-RFC and Ku70-Ku80 for telomere maintenance in fission yeast.
The 911-complex subunits have non-equivalent roles in telomere maintenance
The 911-complex forms a stable heterotrimeric complex resembling PCNA,42, 43 and we and others have previously found that rad1Δ, rad9Δ and hus1Δ cells exhibit a similar extent of telomere shortening.22, 44 Thus, we were surprised to find that deletions of different subunits of the 911-complex resulted in completely different telomere phenotypes for rad1Δ versus rad9Δ or hus1Δ in DBD-rad3 tel1Δ cells (Fig. 3D). In fact, our ChIP data indicated that Rad1 recruitment to telomeres is greatly reduced in rad9Δ or hus1Δ cells (Fig. 4B), consistent with the earlier finding that all three proteins must exist for the 911-complex to maintain wild-type telomere length.22, 44
On the other hand, there has been a report that observed shorter telomere lengths for various alleles of rad1 mutant cells (rad1-1, rad1-S3, rad1-S4 and rad1Δ), but wild-type telomere lengths for rad9-192 and hus1Δ cells.27 Thus, unknown genetic variations, which somehow allow rad9 and hus1 mutant cells to retain wild-type length telomeres, might exist in fission yeast. Importantly, this earlier report is in agreement with the notion that Rad1 plays more critical roles in telomere maintenance than Rad9 or Hus1.27
Since previous studies have found direct interactions between Rad1 and Rad17 in fission yeast and humans,53-55 Rad1 alone, in the absence of both Rad9 and Hus1, could potentially collaborate with Rad17-RFC to provide telomere function. However, based on our observations that Rad9 and Hus1 provide redundant essential telomere maintenance functions in DBD-rad3 te1lΔ (Fig. 3D), we favor the notion that two redundant sub-complexes of the 911-complex, Rad1-Rad9 and Rad1-Hus1, represent minimal functional units that would allow telomere maintenance in DBD-rad3 tel1Δ cells (Fig. 6). Furthermore, previous biochemical studies of the 911-complex in budding yeast and humans support the notion that two subunits of the 911-complex can indeed form a stable complex in the absence of the third subunit.56, 57
There have been other reports where mutations in different subunits of the 911-complex caused different phenotypes. For example, a study in fission yeast has found that rad1Δ but not hus1Δ cells are hypersensitive to the microtubule-depolymerizing drug TBZ.58 Previous budding yeast studies also reported that rad17Δ (fission yeast rad1 ortholog) cells show telomere shortening59 but mec3Δ (fission yeast hus1 ortholog) cells carry either longer59, 60 or wild-type61 length telomeres. Thus, the budding yeast Rad1 ortholog also appears to play a more critical role in telomere maintenance than the other subunits of the 911-complex. Budding yeast Rad17Rad1 has been reported to show DNA damage-induced self-interaction.62 Thus, a Rad17Rad1 dimer could be loaded to telomeres by Rad24Rad17-RFC and provide telomere function in the absence of both Ddc1Rad9 and Mec3Hus1 subunits. Alternatively, analogous to our findings in fission yeast, Ddc1Rad9 and Mec3Hus1 subunits could function redundantly in assisting Rad17Rad1 to fulfill its telomere function in budding yeast.
Two RFC-like clamp loaders, Rad17-RFC and Ctf18-RFC, play redundant roles in telomere maintenance
Our analyses of factors critical for telomere maintenance in DBD-rad3 tel1Δ cells also revealed that Rad17-RFC and Ctf18-RFC play redundant roles in telomere maintenance (Fig. 6). Since only rad17Δ, but not ctf18Δ, affects the efficiency of Rad1 recruitment to telomeres (Fig. 5D), it is currently unclear if both Rad17-RFC and Ctf18-RFC are involved in the regulation of the 911-complex in fission yeast telomere maintenance. In fact, since mammalian Ctf18-RFC has been reported to interact with PCNA but not with the 911-complex,63 it's possible that Ctf18-RFC could collaborate with PCNA, rather than the 911-complex to regulate telomere length. In that case, the observed redundant requirement for Rad17-RFC and Ctf18-RFC in the 911-complex-dependent telomere maintenance mechanism might be caused by the indirect role of the Ctf18-RFC in regulating DNA replication in general.64, 65
On the other hand, budding yeast studies have found that rad17Δ (fission yeast rad1 ortholog) and ctf18Δ cells show telomere shortening while rad24Δ (fission yeast rad17 ortholog) cells maintain wild type telomere length.59, 66 Moreover, it has been shown that budding yeast Rad24Rad17-RFC and Ctf118-RFC contribute redundantly to the Rad17Rad1-dependent activation of the DNA replication checkpoint.49 Thus, evidence from budding yeast support the notion that Rad24Rad17-RFC and Ctf118-RFC are redundantly required to recruit/regulate the 911-complex for its functions in the DNA replication checkpoint and telomere maintenance. In any case, further investigation is necessary to fully understand how Rad17-RFC and Ctf18-RFC redundantly provide essential telomere maintenance functions in DBD-rad3 tel1Δ cells.
Perspectives
By utilizing an unusual hypomorphic allele of rad3, which artificially joins the Pot1 DBD to the N-terminus of Rad3ATR, we have uncovered unsuspected non-equivalent roles for the subunits of the 911-complex and redundant roles of two RFC-like complexes in fission yeast telomere maintenance. While our current study has only focused on the regulation of telomere maintenance by these factors in fission yeast, similar redundancies appear to exist in budding yeast telomere length regulation as well. Furthermore, since studies in recent years have uncovered many similarities in the way DNA repair factors and DNA damage/replication checkpoint sensor proteins treat telomeres and accidental DNA double-strand breaks, our current findings might be more generally applicable to help us understand how these factors collaborate in DNA damage responses.
Materials and Methods
Fission yeast strains and plasmids
The fission yeast strains used in this study were constructed by standard techniques,67 and they are listed in supplementary Table S1. Original sources for trt1Δ, rad3Δ, rad26Δ, tel1Δ, rad1Δ, rad9Δ, hus1Δ, rad17Δ, ctf18Δ, rhp51Δ, rad1-myc, rad17-myc, ctf18-myc and trt1-myc were described previously.10, 11, 14, 22, 65 DNA primers used for construction of plasmids are listed in supplementary Table S2. Plasmids used in this study are listed in supplementary Table S3.
The cDNA version of the Pot1 DNA binding domain (DBD), corresponding to amino acids 1-261 of Pot1, was amplified by PCR using DNA primers pot1-DB1 and pot1-DB2, and cloned as a SacI-KpnI fragment into pBluescript II SK+, generating the pBS-pot1-DBD plasmid. The SacI-BamHI fragment from pBS-pot1-DBD was exchanged with the T62V mutant version, generated by PCR using pot1-DB1 and pot1-T62V-rev primers, to construct the pBS-pot1-DBD-T62V plasmid. The BamHI-ApaI fragment from pBS-pot1-DBD was exchanged with the F88A mutant version, generated by PCR using pot1-F88A-fwd and Pot1-DB2 primers, to construct the pBS-pot1-DBD-F88A plasmid.
These wild-type or mutant Pot1 DBD fragments were then cloned as a NdeI-NdeI fragment into the pBF150 plasmid, which carries genomic DNA corresponding to the promoter and the N-terminal ∼3.6 kb of the Rad3 ORF with an engineered NdeI site just prior to the start codon of the Rad3 ORF. The resulting DBD-rad3 constructs (wild-type, T62V or F88A) were then excised as PstI-SmaI fragments, and then used to transform fission yeast strains carrying the ura4+ marker inserted within the promoter region of the rad3+ gene (TN2101; see Table S1), and then selected for the loss of the ura4+ gene on 5-FOA-containing agar plates. Correct integration of the DBD-Rad3 constructs was then confirmed by sequencing.
For DBD-crb2, Pot1 DBD was excised from the pBS-pot1-DBD plasmid as a BglII-BglII fragment, and cloned into the BamHI site of pJK148-Crb2(BamHI), which carries the crb2+ genomic locus with an engineered BamHI site at the N-terminus of the Crb2 ORF and the fission yeast leu1+ gene. The resulting pJK148-DBD-Crb2 plasmid was then digested within the leu1+ ORF with NruI, and used to integrate the DBD-crb2 construct at the leu1-32 locus of the crb2Δ∷ura4+ strain (TN862; see Table S1). Correct integration of the DBD-Crb2 construct was then confirmed by sequencing.
UV viability
Exponentially growing cells were counted and plated in triplicates on YES plates. The plates were then irradiated in a UV cross-linker (Stratagene) at the indicated doses. Following three days of incubation at 32 °C, colonies were counted and the percent viabilities, compared to non-irradiated controls were calculated.
Pulsed-Field gel electrophoresis (PFGE)
Chromosomal DNA samples were prepared in agarose plugs from fission yeast strains as previously described.22 NotI-digested DNA samples were fractionated in a 1% agarose gel with 0.5X TAE buffer (20 mM Tris acetate and 0.5 mM EDTA) at 14 °C, using the CHEF-DR III system (Bio-Rad) at 6 V/cm (200 V) and a pulse time of 60 to 120 seconds for 24 hours. The telomeric repeat C, I, L, and M probes were prepared as previously described.68
Southern blot analysis
ApaI-digested DNA was prepared from the indicated fission yeast strains and separated in a 2% agarose gel at 100 V for 5 hours. DNA was then transferred to a Hybond XL membrane (GE Amersham Biosciences) overnight in transfer buffer (1.5M NaCl, 0.02M NaOH). The membrane was then hybridized with a telomeric repeat DNA probe.22
Chromatin immunoprecipitation (ChIP) analysis
Exponentially growing cells were processed for ChIP as previously described.24 For Trt1-myc, Rad1-myc, Rad17-myc and Ctf18-myc ChIP, monoclonal anti-myc antibody (9B11, Cell Signaling) was used. For Rad51 (Rhp51) ChIP, polyclonal anti-rad51 (A-92, Santa Cruz) was used. While Rad1-myc ChIP data were quantified using dot blot hybridization,69 Trt1-myc, Rad17-myc, Ctf18-myc and Rad51 ChIP data were analyzed by several independent triplicate SYBR Green-based real-time PCR (Bio-Rad) using TAS1 primers jk380 and jk381.24 Western blot analysis was performed to monitor the expression levels of the proteins tested by ChIP as previously described.69 Anti-Cdc2 (y100.4, Abcam) antibody was used as loading control. Two-tailed Student's t-tests were performed, and P values ≤0.05 were considered as statistically significant differences.
Supplementary Material
Acknowledgments
We thank A. M. Carr, T. Enoch, S. L. Forsburg, and M. Yanagida for fission yeast strains, and B. A. Moser and E. Noguchi for comments on the manuscript. Initial portions of this work were carried out by T. M. N. at The Scripps Research Institute, and they were supported in part by Damon Runyon Cancer Research Foundation postdoctoral fellowship DRG-1565 awarded to T. M. N. and the NIH grants CA77325 and GM59447 awarded to P. R. Our telomere research at the University of Illinois at Chicago has been supported by UIC start-up fund, Sidney Kimmel Scholar Program, and the NIH grant GM078253 awarded to T. M. N.
Abbreviations
- 911
Rad9-Rad1-Hus1
- ChIP
Chromatin immunoprecipitation
- DBD
DNA binding domain
- DSBs
double-strand breaks
- dsDNA
double-stranded DNA
- HU
hydroxyurea
- MRN
Mre11-Rad50-Nbs1
- NHEJ
non-homologous end-joining
- OB fold
oligonucleotide/oligosaccharide-binding fold
- PCNA
proliferating cell nuclear antigen
- PFGE
Pulsed-Field gel electrophoresis
- PIKK
phosphatidyl inositol 3-kinase-like kinase
- RFC
replication factor C
- SHREC
Snf2/Hdac-containing repressor complex
- TERT
telomerase reverse transcriptase
- UV
ultraviolet
References
- 1.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–31. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–4. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 4.Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, et al. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–61. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 5.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–5. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 6.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, et al. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–62. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 8.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 9.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–9. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–9. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 11.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 13.Tomita K, Cooper JP. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22:3461–74. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. Fission yeast Tel1ATM and Rad3ATR promote telomere protection and telomerase recruitment. PLoS Genet. 2009;5:e1000622. doi: 10.1371/journal.pgen.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22:125–40. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lydall D. Taming the tiger by the tail: modulation of DNA damage responses by telomeres. EMBO J. 2009;28:2174–87. doi: 10.1038/emboj.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vespa L, Couvillion M, Spangler E, Shippen DE. ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev. 2005;19:2111–5. doi: 10.1101/gad.1333805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francia S, Weiss R, Hande M, Freire R, d'Adda di Fagagna F. Telomere and Telomerase Modulation by the Mammalian Rad9/Rad1/Hus1 DNA-Damage-Checkpoint Complex. Curr Biol. 2006;16:1551–8. doi: 10.1016/j.cub.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–64. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6065–75. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura TM, Moser BA, Russell P. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics. 2002;161:1437–52. doi: 10.1093/genetics/161.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boerckel J, Walker D, Ahmed S. The Caenorhabditis elegans Rad17 homolog HPR-17 is required for telomere replication. Genetics. 2007;176:703–9. doi: 10.1534/genetics.106.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–20. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–61. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–20. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 27.Dahlén M, Olsson T, Kanter-Smoler G, Ramne A, Sunnerhagen P. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:611–21. doi: 10.1091/mbc.9.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–70. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 29.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–20. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 30.Grandin N, Damon C, Charbonneau M. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol. 2000;20:8397–408. doi: 10.1128/mcb.20.22.8397-8408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–96. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 32.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 33.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–7. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 34.Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20:203–6. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 35.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–8. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 36.Chahwan C, Nakamura TM, Sivakumar S, Russell P, Rhind N. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol Cell Biol. 2003;23:6564–73. doi: 10.1128/MCB.23.18.6564-6573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell. 2002;9:857–69. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- 38.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–11. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian L, Nakamura TM. A kinase-independent role for the Rad3ATR-Rad26ATRIP complex in recruitment of Tel1ATM to telomeres in fission yeast. PLoS Genet. 2010;6:e1000839. doi: 10.1371/journal.pgen.1000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A. 2008;105:18730–4. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–89. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sohn SY, Cho Y. Crystal structure of the human Rad9-Hus1-Rad1 clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 43.Doré AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the Rad9-Rad1-Hus1 DNA damage checkpoint complex—implications for clamp loading and regulation. Mol Cell. 2009;34:735–45. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 44.Kanoh J, Francesconi S, Collura A, Schramke V, Ishikawa F, Baldacci G, et al. The fission yeast spSet1p is a histone H3-K4 methyltransferase that functions in telomere maintenance and DNA repair in an ATM kinase Rad3-dependent pathway. J Mol Biol. 2003;326:1081–94. doi: 10.1016/s0022-2836(03)00030-5. [DOI] [PubMed] [Google Scholar]
- 45.Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12:1908–18. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 46.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:e33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, et al. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100:1633–8. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci U S A. 2003;100:2249–54. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naiki T, Kondo T, Nakada D, Matsumoto K, Sugimoto K. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol Cell Biol. 2001;21:5838–45. doi: 10.1128/MCB.21.17.5838-5845.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croy JE, Altschuler SE, Grimm NE, Wuttke DS. Nonadditivity in the recognition of single-stranded DNA by the Schizosaccharomyces pombe protection of telomeres 1 DNA-binding domain, Pot1-DBD. Biochemistry. 2009;48:6864–75. doi: 10.1021/bi900307x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J Mol Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 52.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, et al. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–62. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parker AE, Van de Weyer I, Laus MC, Verhasselt P, Luyten WH. Identification of a human homologue of the Schizosaccharomyces pombe rad17+ checkpoint gene. J Biol Chem. 1998;273:18340–6. doi: 10.1074/jbc.273.29.18340. [DOI] [PubMed] [Google Scholar]
- 55.Rauen M, Burtelow MA, Dufault VM, Karnitz LM. The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRad9. J Biol Chem. 2000;275:29767–71. doi: 10.1074/jbc.M005782200. [DOI] [PubMed] [Google Scholar]
- 56.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J Biol Chem. 2001;276:25903–9. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 57.Majka J, Burgers PM. Function of Rad17/Mec3/Ddc1 and its partial complexes in the DNA damage checkpoint. DNA Repair (Amst) 2005;4:1189–94. doi: 10.1016/j.dnarep.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 58.Wolkow TD, Enoch T. Fission yeast Rad26 responds to DNA damage independently of Rad3. BMC Genet. 2003;4:6. doi: 10.1186/1471-2156-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Longhese MP, Paciotti V, Neecke H, Lucchini G. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics. 2000;155:1577–91. doi: 10.1093/genetics/155.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corda Y, Schramke V, Longhese MP, Smokvina T, Paciotti V, Brevet V, et al. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–8. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 61.Grandin N, Damon C, Charbonneau M. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 2001;20:6127–39. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H, Zhu Z, Vidanes G, Mbangkollo D, Liu Y, Siede W. Characterization of DNA damage-stimulated self-interaction of Saccharomyces cerevisiae checkpoint protein Rad17p. J Biol Chem. 2001;276:26715–23. doi: 10.1074/jbc.M103682200. [DOI] [PubMed] [Google Scholar]
- 63.Merkle CJ, Karnitz LM, Henry-Sánchez JT, Chen J. Cloning and characterization of hCTF18, hCTF8, and hDCC1. Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J Biol Chem. 2003;278:30051–6. doi: 10.1074/jbc.M211591200. [DOI] [PubMed] [Google Scholar]
- 64.Kim J, Robertson K, Mylonas KJ, Gray FC, Charapitsa I, MacNeill SA. Contrasting effects of Elg1-RFC and Ctf18-RFC inactivation in the absence of fully functional RFC in fission yeast. Nucleic Acids Res. 2005;33:4078–89. doi: 10.1093/nar/gki728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ansbach AB, Noguchi C, Klansek IW, Heidlebaugh M, Nakamura TM, Noguchi E. RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell. 2008;19:595–607. doi: 10.1091/mbc.E07-06-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21:3144–58. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alfa C, Fantes P, Hyams J, McLoed M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 68.Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–6. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 69.Khair L, Subramanian L, Moser BA, Nakamura TM. Roles of heterochromatin and telomere proteins in regulation of fission yeast telomere recombination and telomerase recruitment. J Biol Chem. 2009;285:5327–37. doi: 10.1074/jbc.M109.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.