Abstract
Yeast cells lacking Ctf18, the major subunit of an alternative Replication Factor C complex, have multiple problems with genome stability. To understand the in vivo function of the Ctf18 complex, we analyzed chromatin composition in a ctf18Δ mutant using the quantitative proteomic technique of stable isotope labeling by amino acids in cell culture. Three hundred and seven of the 491 reported chromosomal proteins were quantitated. The most marked abnormalities occurred when cells were challenged with the replication inhibitor hydroxyurea. Compared with wild type, hydroxyurea-treated ctf18Δ cells exhibited increased chromatin association of replisome progression complex components including Cdc45, Ctf4, and GINS complex subunits, the polymerase processivity clamp PCNA and the single-stranded DNA-binding complex RPA. Chromatin composition abnormalities observed in ctf18Δ cells were very similar to those of an mrc1Δ mutant, which is defective in the activating the Rad53 checkpoint kinase in response to DNA replication stress. We found that ctf18Δ cells are also defective in Rad53 activation, revealing that the Ctf18 complex is required for engagement of the DNA replication checkpoint. Inappropriate initiation of replication at late origins, because of loss of the checkpoint, probably causes the elevated level of chromatin-bound replisome proteins in the ctf18Δ mutant. The role of Ctf18 in checkpoint activation is not shared by all Replication Factor C-like complexes, because proteomic analysis revealed that cells lacking Elg1 (the major subunit of a different Replication Factor C-like complex) display a different spectrum of chromatin abnormalities. Identification of Ctf18 as a checkpoint protein highlights the usefulness of chromatin proteomic analysis for understanding the in vivo function of proteins that mediate chromatin transactions.
Cells deploy multiple interconnected pathways to ensure accurate chromosome maintenance, especially during DNA replication when the unwound DNA helix is vulnerable to DNA damage. An essential component of the replication machinery is Replication Factor C (RFC)1, a “clamp-loading” complex that loads the polymerase processivity clamp PCNA (proliferating cell nuclear antigen) at replication forks (1, 2). PCNA is central to the replication machinery and is a multifunctional complex, acting as a platform that interacts with many proteins including DNA polymerases, DNA helicases, nucleases, DNA ligases, histone chaperones, DNA repair proteins, and sister chromatid cohesion factors (3). RFC is a pentamer consisting of a large subunit Rfc1 associated with four smaller proteins Rfc2 through Rfc5 (Rfc2–5).
Interestingly, all eukaryotic cells have a series of RFC-like complexes. These “RLC” complexes share the Rfc2–5 subunits with RFC, but Rfc1 is replaced by one of a series of “alternative” subunits: Rad24 (called Rad17 in human), Elg1, or Ctf18 (4). Rad24-RLC is the best understood, and acts to load the PCNA-like complex Rad17-Mec3-Ddc1 (the equivalent of the human 9–1-1 complex) at DNA damage sites. Elg1-RLC and Ctf18-RLC are more mysterious. Elg1-RLC binds PCNA, but has not been reported to load or unload it on DNA. elg1Δ yeast cells exhibit elevated rates of chromosome rearrangement, and are defective in sister chromatid cohesion (5–8). Ctf18-RLC is unique in the RLC family in forming a heptamer which contains two additional subunits, called Dcc1 and Ctf8. In vitro, the Ctf18-RLC can load PCNA onto DNA and also unload it from DNA (9–11). Yeast cells lacking Ctf18 have multiple problems with genome stability: a ctf18Δ mutant is viable but shows defective sister chromatid cohesion (12), fails to position telomeres at nuclear periphery or maintain telomere length (13, 14), and is hypersensitive to the DNA damaging agent MMS and the replication inhibitor hydroxyurea (HU) (15). Ctf18-RLC appears to act at DNA replication forks (16, 17), however, the in vivo function of Ctf18-RLC is unknown.
Because its pleiotropic effects suggest that various chromosome maintenance pathways could be affected in the ctf18Δ mutant, we took a proteomic strategy to investigate chromatin abnormalities. Possible targets of regulation by Ctf18-RLC include proteins involved in the processes of DNA replication, the replication stress response, or establishment of sister chromatid cohesion. Central to all these processes is the replisome, the multiprotein complex that replicates the DNA, as described below. Replisome assembly begins during G1 phase, with the formation of prereplication complexes by loading of heterohexameric Mcm2–7 complexes at origin sites (18). Replication initiation then involves recruitment by Mcm2–7 hexamers of Cdc45 and the GINS complex (containing the four subunits Sld5, Psf1, Psf2, and Psf3), leading to assembly of the Cdc45-MCM-GINS complex. Cdc45-MCM-GINS is central to replisome function, probably forming the activated replicative helicase that unwinds the DNA, enabling its replication on the leading strand by DNA polymerase ε and on the lagging strand by DNA polymerase δ (along with its processivity factor PCNA and the DNA polymerase α priming complex) (18–20). During the replication process, exposed single-stranded DNA is coated by the single-stranded binding protein RPA (containing subunits Rfa1, Rfa2, and Rfa3) to stabilize and protect it.
A recent biochemical study revealed that the Cdc45-MCM-GINS is the central component of the so-called “replisome progression complex” (RPC), which contains additional factors including Ctf4 (21). Ctf4 appears to link Cdc45-MCM-GINS to Polymerase α (22, 23), and is required for the establishment of cohesion (12), the mechanism through which duplicated sister chromatids are held together until anaphase. Sister chromatids are held together by the ring-shaped cohesin complex, which contains subunits Smc1, Smc3, Scc1, and Scc3 (24). Establishment of cohesion appears to be closely coupled to replication through RPC-mediated events, and yeast cells lacking RPC proteins such as Ctf4 display cohesion defects (25).
A further component of the RPC is the checkpoint mediator protein Mrc1, essential for cells to respond correctly to replication stress, such as fork blockage events induced by the DNA replication inhibitor HU (21, 26, 27). In response to replication stress, eukaryotic cells activate a DNA replication checkpoint pathway that suppresses new replication initiation events at unfired origins, stabilizes replication forks, induces transcription of DNA stress response genes, and delays cell cycle progression (28). The current model suggests that a checkpoint kinase Mec1, the budding yeast homolog of human ATR, is recruited to replication block sites and phosphorylates target proteins. One of the targets is Mrc1, which provides an activation platform for the checkpoint kinase Rad53 to initiate the checkpoint response (27). mrc1Δ cells demonstrate a significant delay in Rad53 activation in response to a replication block (26). In wild-type cells HU treatment causes suppression of further replication initiation events, but HU-treated mrc1Δ cells inappropriately initiate replication at late origins, because of failure of DNA replication checkpoint activation (26, 29).
To investigate the in vivo function of Ctf18-RLC, we analyzed the differences in chromatin composition between wild-type and ctf18Δ cells. We took advantage of recent advances in quantitative protein mass spectrometry, which provide the tools to enable the analysis of entire chromatin composition in an unbiased way. Here, we have applied stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics (30) for comparison of chromatin components from wild type and ctf18Δ Saccharomyces cerevisiae. Budding yeast provides an ideal organism for developing this work because of its small, relatively well-characterized proteome. This novel approach has provided us with a large-scale view of changes in chromatin composition in a ctf18Δ mutant, and revealed that the Ctf18-RLC complex acts in the same pathway as Mrc1 to mediate the DNA replication checkpoint.
EXPERIMENTAL PROCEDURES
Yeast Strains
S. cerevisiae strains used are listed in supplemental Table S1. SHY201 was generated by sporulation of the diploid strain BY4743 and selection of a MATa lysine auxotroph, followed by disruption of ARG4. To construct TKY1, a ctf18Δ::kanMX4 construct was PCR-amplified from the relevant EUROSCARF gene deletion strain and transformed into SHY201. TKY18, TYK130, and TKY131 were constructed in the same way, using elg1Δ::kanMX4 and rad9Δ::kanMX4 fragments. A strain carrying a deletion of the entire MRC1 gene was created by PCR-based one-step gene replacement using pFA6a-kanMX6 as a template (31). Myc-, FLAG-, and HA-tagging was carried out using standard PCR-based gene insertion methods (31). Disrupted and tagged alleles were confirmed by PCR. Primer sequences are available on request.
SILAC Labeling
For lysine and arginine double labeling, lys2Δ arg4Δ strains TKY1, TKY18, or TKY111 were grown for at least ten generations in “heavy” medium, which is synthetic yeast medium containing: 6.9 g/l yeast nitrogen base without amino acids (FORMEDIUM), 1.85 g/l amino acid dropout mixture without arginine and lysine (Kaiser formulation, FORMEDIUM), 2% glucose, 15 mg/l [13C6]l-arginine and 30 mg/l [13C6] or [13C6,15N2]l-lysine. SHY201 cells were grown in “light” medium, containing 15 mg/l l-arginine and 30 mg/l l-lysine. For lysine single labeling, TKY1 or SHY201 cells were grown for ten generations in the same synthetic medium but containing 15 mg/l l-arginine and either 30 mg/l [13C6]l-lysine or 30 mg/l l-lysine.
Cell Synchronization and Release
Cells were grown in heavy or light media at 30 °C to early log phase (∼3 × 106 cells/ml), and synchronized by treating with 5 μg/ml α-factor for 2.5 h at 30 °C. For release into HU, cells were spun down, washed once in heavy or light medium, re-suspended in HU-containing heavy or light medium, and incubated with shaking for 1.5 h at 30 °C. To release cells from α-factor arrest into normal S phase, cells were synchronized in the same way and resuspended in heavy or light media lacking HU. The cultures were then incubated with shaking at 30 °C and harvested at a mid-S phase time point, as determined by flow cytometry analysis. Flow cytometry was carried out using SYTOX Green nucleic acid stain (Invitrogen Molecular Probes) as previously described (32).
Preparation of Chromatin-enriched Fraction
Chromatin enriched-fractions were prepared according to Sheu et al. (33), modified to incorporate a nuclear isolation procedure (34). Approximately 4 × 109 cells (∼1 × 107 cells/ml) were harvested and resuspended in 10 ml of prespheroplasting buffer (100 mm PIPES/KOH, pH 9.4, 10 mm dithiotreitol (DTT), 0.1% sodium azide) then incubated for 10 min at room temperature, followed by incubation in 10 ml of spheroplasting buffer (50 mm KH2PO4/K2HPO4, pH 7.4, 0.6 m Sorbitol, 10 mm DTT) containing 200 μg/ml Zymolyase-100T and 5% Glusulase at 37 °C for 30 min with occasional mixing. Spheroplasts were washed with 5 ml of ice-cold wash buffer (20 mm KH2PO4/K2HPO4, pH 6.5, 0.6 m Sorbitol, 1 mm MgCl2, 1 mm DTT, 20 mm β-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride (PMSF), Protease inhibitor tablets (EDTA free, Roche)) and resuspended in 5 ml of ice-cold wash buffer. The suspension was overlaid onto 5 ml of 7.5% Ficoll-Sorbitol cushion buffer (7.5% Ficoll, 20 mm KH2PO4/K2HPO4, pH 6.5, 0.6 m Sorbitol, 1 mm MgCl2, 1 mm DTT, 20 mm β-glycerophosphate, 1 mm PMSF, Protease inhibitor tablets) and the spheroplasts were spun through the cushion buffer at 5000 rpm for 5 min to remove proteases derived from Zymolyase-100T. The pelleted spheroplasts were resuspended in 200 μl of ice-cold wash buffer and dropped into 14 ml of 18% Ficoll buffer (18% Ficoll, 20 mm KH2PO4/K2HPO4, pH 6.5, 1 mm MgCl2, 1 mm DTT, 20 mm β-glycerophosphate, 1 mm PMSF, Protease inhibitor tablets, 0.01% Nonidet P-40) with stirring. At this stage, it was confirmed microscopically that the cytoplasmic membranes were lysed, but that nuclei and vacuoles (often attached together) were intact. The suspension was subjected to 10 strokes with a loose-fitting pestle in a Potter-Elvehjem homogenizer (which releases nuclei from vacuoles and improves recovery of nuclei). Unbroken cells were removed by two low-speed spins (5000 × g for 5 min at 4 °C). Nuclei were then pelleted by a high-speed spin (16,100 × g for 20 min) and the cytoplasmic fraction removed. After washing nuclei in ice-cold wash buffer, the nuclei were resuspended in 200 μl of Extraction Buffer EB (50 mm HEPES/KOH, pH 7.5, 100 mm KCl, 2.5 mm MgCl2, 0.1 mm ZnSO4, 2 mm NaF, 0.5 mm spermidine, 1 mm DTT, 20 mm β-glycerophosphate, 1 mm PMSF, Protease inhibitor tablets) and lysed by addition of Triton X-100 to 0.25%, followed by incubation on ice for 10 min. The lysate was overlaid on 500 μl of EBX-S buffer (EB buffer, 30% sucrose, 0.25% Triton X-100), and spun at 12,000 rpm for 10 min at 4 °C. The top layer (nucleoplasmic fraction) was removed and the chromatin pellet was washed in 1 ml of EBX buffer (EB buffer, 0.25% Triton X-100) and spun at 10,000 rpm for 2 min at 4 °C. The chromatin pellet was resuspended in 40 μl of 1.5× Tris-Glycine SDS Sample Buffer and incubated for 2 min at 85 °C, followed by spinning at 10,000 rpm for 30 s before loading on a Novex 8–16% Tris-Glycine Gel (Invitrogen). Whole-cell extract was prepared by lysing the washed spheroplasts in EBX buffer. Protein concentration of whole cell extract was measured using Qubit Fluorometer and Quant-iT Protein Assay Kit (Invitrogen). Protein concentration of chromatin fraction was calculated by comparing intensities of protein bands of chromatin fraction with those of whole cell extract on SYPRO Ruby (Bio-Rad) stained gel.
S phase chromatin was prepared in the same way (see above) with the following adjustments: sodium azide was added directly to the culture to a final concentration of 0.1%, and the culture was immediately chilled on ice for 10 min. After harvesting, the cells were resuspended and incubated in prespheroplasting buffer for 10 min on ice (instead of at room temperature). 0.1% sodium azide was included in the spheroplasting buffer.
Wild-type and ctf18Δ chromatin were prepared separately and then mixed, to avoid complications arising from differences of these strains in cell size and susceptibility to spheroplasting enzymes.
Mass Spectrometry and Data Analysis
Equal amounts (60 μg each) of proteins from chromatin-enriched fractions (differentially-labeled with isotopes) were mixed and size-fractionated by one-dimensional SDS-PAGE (Novex 8–16% Tris-Glycine Gel, Invitrogen). Proteins were visualized by colloidal Coomassie staining (Colloidal Blue Staining Kit, Invitrogen), and the entire protein gel lane was excised and cut into 12 slices. The gel slices were destained in dH2O and 20 mm NH4HCO3. Each gel slice was subjected to in-gel digestion with trypsin (Trypsin Gold, Promega) (for lysine- and arginine-labeled proteins) or Lys-C (Lysyl Endopeptidase, Wako) (for lysine-labeled proteins) (35). The resulting peptides were extracted and analyzed in automated LC-MS/MS as described previously (36). Mass spectrometry analysis was performed using a nanoflow high performance liquid chromatography system connected to a linear ion trap-orbitrap hybrid mass spectrometer (linear trap quadrupole-Orbitrap XL or Velos, Thermo Fisher Scientific Inc) via a nanoelectrospray ion source (Proxeon Biosystems) (36).
Quantitation was performed using the program MaxQuant (version 1.0.12.31. or 1.0.13.13) (37). The derived peak list generated by Quant.exe (in the first part of MaxQuant) was searched using the Mascot search engine (Matrix Sciences, version 2.2.2) for peptide identifications against the yeast GenBank database (released May 2006), containing 11,168 S. cerevisiae protein sequences with the addition of 175 commonly observed contaminants and all the reversed sequences. The initial mass tolerance was set to 7 ppm, and tandem MS (MS/MS) mass tolerance was 0.5 Da. The enzyme was specified as trypsin or Lys-C, with a maximum of two missed cleavages. Carbamidomethylation of cysteine was searched as a fixed modification, whereas N-acetyl protein and oxidation of methionine were searched as variable modifications. As discussed in Cox et al. (37), peptides and proteins were accepted in reverse order of their PEP (probability of false hit) scores whereas the number of forward database identifications remained 100-fold higher than the number of reverse database identifications (i.e. until reverse identifications exceeded 1% of those accepted), thus resulting in a false discovery rate of 1%. Because the use of MaxQuant software has significantly improved reliability and accuracy of peptide quantitation and assignment to proteins (37), proteins were considered to be identified if represented by at least one unique peptide, and were considered quantified if they had at least one quantified SILAC pair. The data quality (in particular, the number of unique peptides and number of quantification events for each protein) was however an important additional parameter when considering the confidence to be given to specific results. Taking account of the data quality, we indicate the names of a selected number of proteins that were quantitated by at least two peptides in Figs. 3A, 4B, 6D, 7A, and 7C.
Fig. 3.
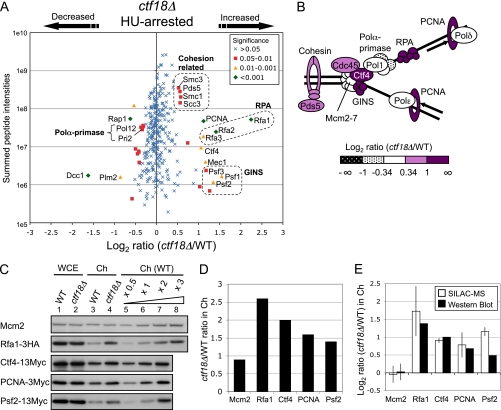
Increased chromatin binding of DNA replication proteins, the checkpoint kinase Mec1, and cohesin proteins in HU-treated ctf18Δ cells. A, Plot shows log2 ratios of all chromosome proteins identified and their summed peptide intensities. In this and subsequent plots, the marker symbols indicate significance scores for the changes observed, with green diamonds indicating the highly significant abnormalities and blue crosses changes less likely to be significant. ctf18Δ cells were labeled with [13C6]-Lys. Strains used are SHY201 and TKY1. B, Schematic representation of replisome proteins, colored according to their altered chromatin loading. C, Western blot analysis confirms changed chromatin binding levels. Western blots show whole cell extract (WCE; lanes 1 and 2) and chromatin-enriched (Ch; lanes 3 and 4) fractions from strains with epitope-tagged proteins Rfa1–3HA, Ctf4–13Myc, PCNA-3Myc, or Psf2–13Myc. Loading of Ch fractions was adjusted to be appropriate for each protein analyzed. A dilution series of wild-type chromatin (lanes 5–8) allows the assembly of a standard plot for quantification. Strains used are TKY27, TKY33, TKY25, TKY31, Y1109, SHY164, TKY22, and TKY23. Top panel (Mcm2) shows TKY27 and TKY33. D, Histogram shows ctf18Δ/WT ratios in Ch fraction for each protein, as measured by Western blots. Ratios were calculated based on signal intensities normalized against levels of histone H3 (see also Supplemental Fig. S3). E, Histogram shows ctf18Δ/WT ratios in Ch fraction on log scale, as measured in SILAC analysis (open bars) and by Western blotting (filled bars). For Western blot analysis ratios, mean value and standard deviation (error bar) of Mcm2 from four experiments is shown. For the SILAC analysis ratios, mean values and standard deviations (error bars) are derived from two independent experiments.
Fig. 4.
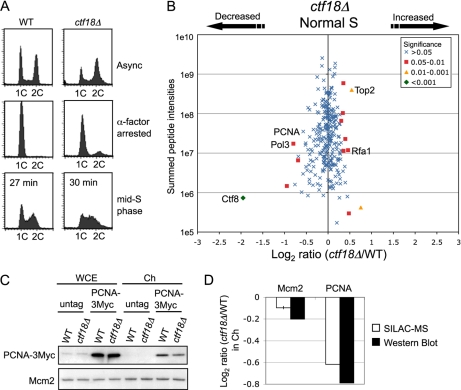
Chromatin binding levels of DNA replication proteins did not increase in ctf18Δ cells during normal S phase. A, Cell cycle progression on release from α-factor analyzed by flow cytometry. Wild-type and ctf18Δ cells were harvested when cells were in mid-S phase (27 min and 30 min respectively after release at 30 °C). Positions of cells with 1C and 2C DNA contents are indicated. Strains used are SHY201 and TKY1. B, Plot shows log2 ratios of all chromosome proteins identified and their summed peptide intensities. ctf18Δ cells were labeled with [13C6]-Arg and [13C6,15N2]-Lys. C, Western blot analysis confirms that levels of chromatin-bound PCNA are slightly decreased in ctf18Δ cells undergoing normal S phase. Strains used are SHY201, TKY1, Y1109, and SHY164. D, Histogram shows ctf18Δ/WT ratios in Ch fraction for Mcm2 and PCNA, as measured in SILAC analysis and by Western blots. For the SILAC analysis ratios, mean values and standard deviations (error bars) are derived from two independent experiments. Levels of Mcm2 in contrast changed only marginally, as measured by SILAC or Western blotting.
Fig. 6.
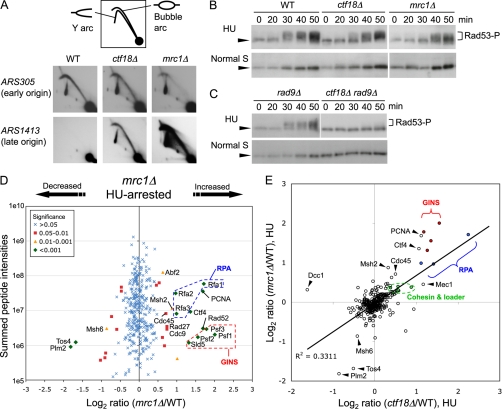
Ctf18 acts in the DNA replication checkpoint response to HU. A, Two-dimensional gel analysis of EcoRI fragments containing ARS305 (early origin) and ARS1413 (late origin) in WT, ctf18Δ and mrc1Δ cells released into S phase in the presence of 200 mm HU for 90 min at 30 °C. Strains used are SHY201, TKY1, and TKY111. B, ctf18Δ cells are defective in activating Rad53 in response to HU. Cells were arrested in G1 phase then released into S phase in the presence (upper panels) or absence (lower panels) of 200 mm HU at 25 °C. Cells were collected at the indicated time points and protein extracts prepared, followed by Western blotting to detect Rad53. Arrowhead indicates unmodified Rad53 and the bracket, phosphorylated forms of Rad53. Strains used are SHY201, TKY1, and TKY111. C, An experiment similar to that in B shows that Rad53 activation in ctf18Δ depends on the DNA damage (Rad9-mediated) checkpoint. Strains are TKY130 and TKY131. D, Chromatin abnormalities in cells lacking the checkpoint mediator Mrc1 treated with HU. Plot shows log2 ratios of all chromosome proteins identified and their summed peptide intensities. mrc1Δ cells were labeled with [13C6]-Arg and [13C6,15N2]-Lys. Strains used are SHY201 and TKY111. E, Abnormalities in chromatin composition in HU-treated ctf18Δ cells resemble those of HU-treated mrc1Δ cells. Log2 ratios of chromatin association for mrc1Δ/WT in HU, plotted against log2 ratios of chromatin association for ctf18Δ/WT in HU.
Fig. 7.
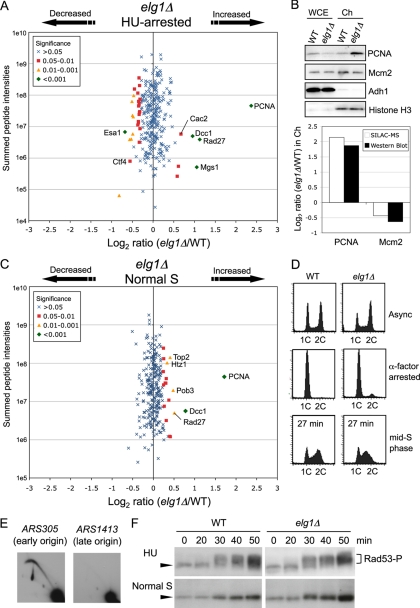
Chromatin association of PCNA and the flap endonuclease Rad27, but not Cdc45, GINS, or Ctf4, was increased in cells lacking Elg1. A, Changes in chromatin composition in HU-treated elg1Δ cells, compared with HU-treated wild-type. elg1Δ cells were labeled with [13C6]-Arg and [13C6,15N2]-Lys. Strains used are SHY201 and TKY18. B, Increase in chromatin-bound PCNA in HU-treated elg1Δ cells confirmed by Western blot analysis. Strains used are SHY201 and TKY18. C, Changes in chromatin composition in elg1Δ cells during normal S phase. elg1Δ cells were labeled with [13C6]-Arg and [13C6,15N2]-Lys. Strains used are SHY201 and TKY18. D, Cell cycle progression was analyzed by flow cytometry. Wild-type and elg1Δ cells were synchronized by blocking with α-factor in G1 phase and release into medium without HU. Both cultures were harvested in mid-S phase, 27 min after release. Positions of cells with 1C and 2C DNA contents are indicated. E, Two-dimensional gel analysis of EcoRI fragments containing ARS305 and ARS1413 in elg1Δ cells released into S phase in the presence of 200 mm HU for 90 min at 30 °C. Strain used is TKY18. F, Kinetics of Rad53 phosphorylation in elg1Δ cells. Rad53 was detected as in Fig. 6B.
Categorization of Proteins
Protein categorization annotations are generally according to the Gene Ontology Cellular Component in the Saccharomyces Genome Database. Where Gene Ontology Cellular Component makes multiple assignments for a particular gene product, it was assigned to the category appearing first in the following list: Chromosome, Nucleolus, Nucleoplasm, Cytoplasm, Other. For example, a protein with a Gene Ontology Cellular Component annotation “chromosomal, nuclear, and cytoplasmic” was assigned to the category “chromosome” (and not to “nucleoplasm” or “cytoplasm”). Potential proteins encoded by dubious open reading frames were included in the category “Other.” Some probable chromatin proteins (e.g. transcription, chromatin remodelling and repair proteins, such as RNA polymerase components, SWI/SNF, SAGA, and RSC subunits, “Rad” proteins, Mec1, and Tel1) were manually re-allocated from “nucleoplasm” to “chromosome” categories.
Quantification of Proteins in Chromatin-enriched Fractions by Western Blotting
Whole cell extracts and chromatin-enriched fractions were prepared from HU-arrested cultures of epitope-tagged strains as described above. Samples were electrophoresed on NuPAGE 4–12% Bis-Tris gels (Invitrogen) gels and blotted on PVDF membrane (Hybond-P, GE Healthcare). Antibodies used in detection of epitope-tagged proteins were mouse anti-HA (HA.11, Covance Research Products, Princeton, NJ) and mouse anti-Myc (ab32, Abcam, Cambridge, MA). Antibodies used in detection of Mcm2, Adh1, and histone H3 were goat anti-Mcm2 (sc-6680, Santa Cruz, Santa Cruz, CA), rabbit anti-Adh1 (ab34680, Abcam), and rabbit anti-histone H3 (ab46765, Abcam). Secondary antibodies were alkaline phosphatase (AP)-conjugated anti-mouse IgG (sc-2008, Santa Cruz), AP-conjugated anti-rabbit IgG (S3731, Promega, Madison, WI), and AP-conjugated anti-goat IgG (sc-2022, Santa Cruz). The detection substrate was ECF Western blotting reagent (GE Healthcare). Chemifluorescent signals were scanned by FLA-3000 (excitation, 473 nm; filter, O580, FUJIFILM) and quantified using ImageGauge V4.21 software. To compare the amounts of proteins in chromatin fractions from wild type and mutant (see Figs. 3, 4, and 7), a standard plot was drawn based on analysis of a dilution series of chromatin from wild type (WT) (e.g. supplemental Fig. S3), and values for experimental samples were measured by placement on that plot. Values were adjusted for variations in loading based on histone H3 signal. To compare chromatin-bound PCNA in wild type and elg1Δ cells by Western blot, chromatin fractions were prepared as described above and PCNA was detected using mouse monoclonal anti-PCNA antibody (ab70472, Abcam).
Immunoprecipitation
Approximately 2 × 108 spheroplasts were resuspended in 600 μl of lysis buffer (50 mm HEPES/KOH, pH 7.5, 150 mm NaCl, 3 mm MgCl2, 10% glycerol, 0.2% Nonidet P-40, 10 mm NaF, 20 mm β-glycerophosphate) containing protease inhibitors (1× Complete (Roche), 1% Protease Inhibitor Mixture (Sigma P8215)). Lysates were treated with 200 units of DNase I (Roche) at 4 °C for 30 min and then centrifuged to produce cleared whole cell extract. This extract was incubated with 2 μg of anti-FLAG M2 antibody (Sigma) conjugated with 20 μl of Dynabeads Protein G (Dynal) at 4 °C for 2 h. Samples were washed four times with lysis buffer, and then resuspended in SDS sample buffer. To examine Mcm4 binding for Cdc45 and Pol1, EBX buffer (as used for chromatin fractionation) or low salt buffer (50 mm HEPES/KOH, pH 7.5, 50 mm potassium acetate, 10 mm magnesium acetate, 1 mm DTT, 10% glycerol, 0.1% Nonidet P-40, 2 mm NaF, 2 mm β-glycerophosphate, 1× Complete (Roche), 1% Protease Inhibitor Mixture (Sigma P8215)) was used instead of lysis buffer. Cdc45 protein was detected using sheep polyclonal anti-Cdc45 antibody (kindly gifted from Dr. Karim Labib).
Two-dimensional Gel
Genomic DNA was prepared as described (38, 39). DNA fragments digested using EcoRI were separated by neutral/neutral two-dimensional agarose gel electrophoresis (40) and transferred to Neutral membrane (Qbiogene, Illkirch, France) by capillary blotting. EcoRI fragments containing ARS305 and ARS1413 were detected using suitable 32P-labeled probes.
Rad53 Phosphorylation
SHY201, TKY1, TKY18, TKY111, TKY130, or TKY131 cells were arrested in G1 phase using α-factor, and released into S phase in the presence or absence of 200 mm HU at 25 °C. Cells were sampled at indicated time points, washed twice with water and incubated in 0.1 m NaOH for 5 min at room temperature. The cells were spun down and resuspended in SDS sample buffer before SDS-PAGE electrophoresis and Western blotting as described above. Rad53 protein was detected using a goat polyclonal anti-Rad53 antibody (sc-6749, Santa Cruz Biotechnology).
RESULTS
Workflow for Quantitative Analysis of Chromatin Components in Wild-type and ctf18Δ Cells
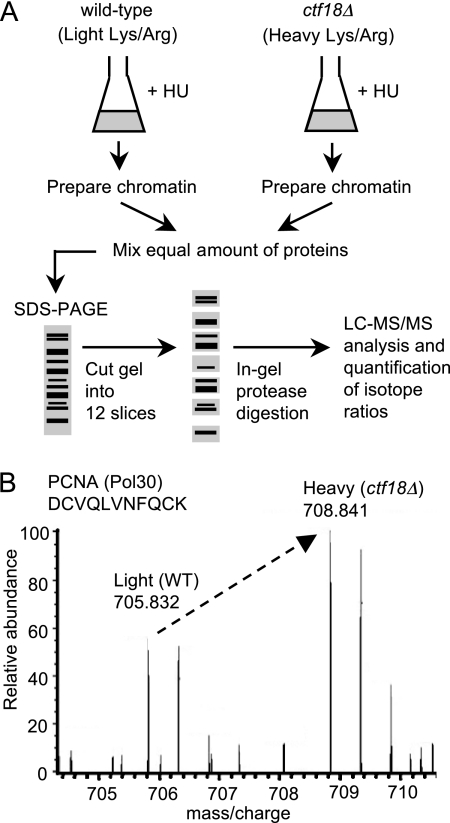
To understand the in vivo function of Ctf18-RLC, we used SILAC-based comparative proteomics (30) to compare chromatin composition in wild-type and ctf18Δ cells (Fig. 1A). ctf18Δ cells were grown in “heavy” media, i.e. containing 13C/15N-substituted arginine and/or lysine; complete labeling of cellular proteins was facilitated by the use of lysine and arginine auxotrophic mutants (lys2Δ arg4Δ). Wild-type cells were grown in “light”, i.e. unlabeled (12C/14N) media. Both wild-type and ctf18Δ cultures were synchronized during DNA replication by release from G1 phase arrest into medium containing the replication inhibitor HU. Chromatin proteins were prepared from wild-type and ctf18Δ cells separately (see Fig. 2A), mixed and then size-fractionated using SDS-PAGE, followed by trypsin or Lys-C digestion (as appropriate for the labeling regime used in each experiment). The resulting peptides were analyzed by high sensitivity mass spectrometry linear trap quadrupole-Orbitrap.
Fig. 1.
Outline of procedure for SILAC-based quantitative proteomic analysis of chromatin. A, ctf18Δ cells were metabolically labeled by culturing in “heavy” media containing 13C/15N-isotopes of arginine and lysine, whereas wild-type cells were grown in “light” media containing the 12C/14N-arginine and lysine isotopes. After 10 generations, both cultures were synchronized by blocking in G1 phase with α-factor then releasing into HU-containing heavy or light medium. After 90 min, chromatin-enriched fractions were prepared (see Fig. 2) and mixed. Proteins were separated by 1D SDS-PAGE, digested, and subjected to LC-MS/MS analysis, allowing calculation of heavy:light ratios for the peptides and proteins identified. Strains used are TKY1 and SHY201. B, Specimen MS spectrum for a PCNA peptide, showing increased loading of PCNA on chromatin in the ctf18Δ strain.
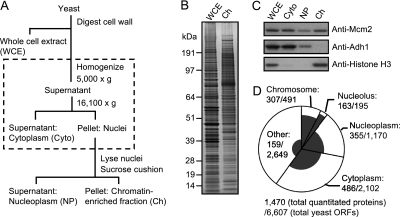
Fig. 2.
Chromatin enrichment procedure and summary of proteins quantitated. A, Outline of chromatin enrichment procedure. The nuclear isolation step incorporated into the published chromatin enrichment method is shown within the dashed box. B, Whole cell extract (WCE) and chromatin-enriched fraction (Ch) were analyzed by SDS-PAGE followed by SYPRO ruby staining. 100× cell equivalents were loaded for Ch. C, Distribution of proteins in cytoplasmic (Cyto), nucleoplasm (NP), and chromatin (Ch) fractions during the enrichment procedure, analyzed by Western blotting. 10× cell equivalents loaded for NP and Ch fractions. D, Pie chart summarizing S. cerevisiae protein categories and the distribution of proteins quantitated by SILAC. The circle represents all 6607 S. cerevisiae ORFs, with pie slices representing the number of ORFs with gene products in each category. Within each pie slice, the shaded region represents those gene products quantitated in the SILAC experiment. Protein annotations generally according to the Saccharomyces Genome Database (see Experimental Procedures).
Peptides originating from ctf18Δ cells mirror those from wild-type cells, but show a shift in mass because of their content of heavy arginine and/or lysine. For proteins whose loading on chromatin is altered in ctf18Δ, the change is reflected in the abundance of heavy-labeled peptides. A specimen MS spectrum for a PCNA peptide is shown in Fig. 1B. This heavy and light peptide pair (SILAC peptide pair) indicates an ∼2-fold increase in PCNA in the ctf18Δ chromatin preparation, compared with wild-type. Using MaxQuant software (37), protein ratios are calculated as the median of all SILAC peptide pair ratios for each protein identified, including a normalization step adjusting for any inequality in protein loading in the two samples.
Validation of Chromatin Preparation
When we used a published chromatin enrichment protocol (33), we found that only 44% of proteins quantitated were nuclear or chromosomal and the remainder were cytoplasmic or other contaminants of the chromatin preparation (data not shown). To refine the analysis, we incorporated a nuclear isolation procedure (adapted from Young et al. (34)) into the protocol (Fig. 2A and supplemental Fig. S1). A chromatin-enriched fraction prepared by this modified procedure was analyzed by SDS-PAGE and SYPRO Ruby staining (Fig. 2B). Western blotting demonstrated that chromatin proteins (the MCM complex subunit Mcm2 and histone H3) were efficiently recovered, whereas the cytoplasmic protein Adh1 was undetectable in the chromatin-enriched fraction (Fig. 2C). With this modified procedure, 56% of proteins identified by mass spectrometry were nuclear or chromosomal, covering 63% (307 out of 491) of chromosomal proteins (Fig. 2D). As a category, chromosomal and nucleolar proteins were recovered and identified in the highest proportions. Many contaminating cytoplasmic proteins were nevertheless quantitated, as expected because our chromatin enrichment procedure does not constitute a full chromatin purification. Because our purpose was to measure, rather than to identify, chromatin proteins, the contaminating cytoplasmic proteins do not interfere with quantification of chromatin components. Thus, we had established a suitable methodology for analyzing chromatin composition using SILAC quantitative proteomics.
Chromatin Binding of DNA Replication Proteins (PCNA, RPA, GINS, and Ctf4), the Checkpoint Kinase Mec1, and Cohesin are Increased in HU-treated ctf18Δ Cells
To emphasize consistent changes within related groups of proteins, we plot graphs showing both heavy/light ratios and relative peptide abundance (i.e. MS peptide intensity) (37, 41). Log2 ratios of the 307 identified chromosomal proteins are plotted against summed MS peptide intensities for each protein (Fig. 3A). Validating the approach, “heavy” peptides derived from Ctf18 protein were not identified, and the protein showing the largest decrease on chromatin in the ctf18Δ mutant was the Dcc1 subunit of Ctf18-RLC (Fig. 3A). The Ctf8 subunit of Ctf18-RLC was not identified in this experiment. The list of chromosomal proteins quantitated is shown in supplemental Table S2. One striking abnormality observed in HU-treated ctf18Δ cells was a two- to fourfold increase in many replisome factors-including PCNA, single-strand DNA-binding proteins Rfa1, Rfa2, and Rfa3, GINS complex proteins Psf1, Psf2, and Psf3, the Ctf4 connector protein that binds GINS and Pol α, and the checkpoint kinase Mec1. We also observed a modest, but reproducible, increase in chromatin loading of the cohesin complex components Smc1, Smc3, and Scc3, and the cohesion establishment factor Pds5 (Fig. 3A). We observed a slight decrease in the telomeric chromatin component Rap1, and Pol α-primase subunits Pol12 and Pri2 (Fig. 3A). Because Rap1 binds telomere repeat sequences, a reduction in chromatin-bound Rap1 is consistent with the shortened telomeres observed in ctf18Δ cells (14). Fig. 3B shows a color-coded schematic representation of the changes we observed. We repeated this experiment with minor modifications and observed similar chromatin abnormalities (Supplemental Fig. S2 and Table S3).
To confirm the changes observed, we epitope-tagged specimen proteins and performed Western blotting to examine their representation in chromatin preparations from HU-treated wild-type and ctf18Δ cells. Overall expression levels of Rfa1–3HA, Ctf4–13Myc, PCNA-3Myc, and Psf2–13Myc were similar in wild-type and ctf18Δ, as assessed by analysis of whole cell extracts (Fig. 3C). However, these proteins were increased, when compared with wild-type, in chromatin-enriched fractions prepared from HU-treated ctf18Δ (Fig. 3C and 3D and Supplemental Fig. S3). The levels of increase closely reflect the changes observed by SILAC mass spectrometry (Fig. 3E). In summary, we successfully used SILAC quantitative proteomics to detect abnormalities in ctf18Δ chromatin, observing the most significant change to be increased loading of various replisome components.
Chromatin Loading of Replisome Components did not Increase in the ctf18Δ Mutant in Normal S Phase
ctf18Δ cells show cohesion and telomere defects in normal growth (12, 13), so we tested for chromatin abnormalities in ctf18Δ cells in an unchallenged S phase (“normal” S phase). We used SILAC to compare chromatin isolated from wild-type and ctf18Δ cultures progressing synchronously through normal DNA replication. For these experiments, unlabeled wild-type and labeled ctf18Δ cultures were released from G1 arrest and sampled at a mid-S phase time point (Fig. 4A). Chromatin was then prepared for SILAC-based chromatin profiling as described above. In general, we observed only slight abnormalities in ctf18Δ chromatin composition, and the changes observed on HU treatment were not apparent in normal S phase (Fig. 4B and supplemental Table S4). The levels of cohesin loading appeared very similar to wild-type (supplemental Fig. S4). Chromatin loading of DNA replication proteins (such as RPA components Rfa2 and Rfa3, GINS, and Ctf4) was in general not substantially changed in ctf18Δ when compared with wild type. We did observe a slight increase in loading of Rfa1, and a slight decrease in chromatin loading of the DNA polymerase δ catalytic subunit Pol3 (Fig. 4B). Loading on chromatin of PCNA was also slightly decreased in ctf18Δ during normal S phase (Fig. 4B, 4C and 4D), in contrast to its behavior in HU-treated ctf18Δ cells where PCNA loading was increased. We repeated this experiment and observed similar changes (supplemental Fig. S5 and Table S5). Taken the results in Figs. 3 and 4 together, we conclude that the increased loading of replisome components onto chromatin in ctf18Δ cells occurs in response to HU, and is not observed in ctf18Δ cells undergoing normal S phase.
Excess Active Helicase Complex is Present in HU-Treated ctf18Δ Cells
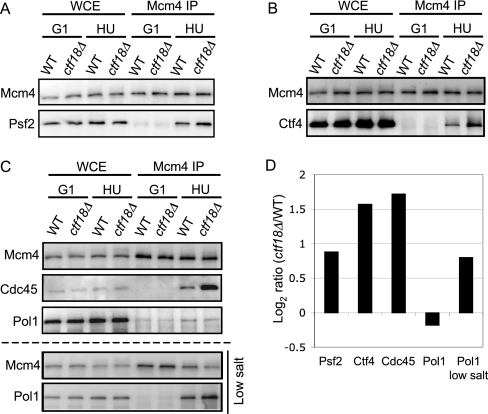
GINS and Ctf4 are subunits recruited to the MCM helicase during RPC formation (21). The increased GINS and Ctf4 observed in the chromatin fraction of HU-treated ctf18Δ cells (Fig. 3) suggested an increase in the proportion of GINS and Ctf4 complexed with MCM (suggesting a larger number of RPCs). To test this possibility, we immunoprecipitated Mcm4–3FLAG and compared the amount of Ctf4–13Myc or the GINS subunit Psf2–13Myc that co-immunoprecipitated in wild-type and ctf18Δ cells (Figs. 5A and 5B). We found that the amounts of Ctf4 and Psf2 complexed with Mcm4 were noticeably increased in HU-treated ctf18Δ cells (Figs. 5A, 5B, and 5D).
Fig. 5.
Increased association of GINS, Ctf4 and Cdc45 with MCM in HU-treated ctf18Δ, when compared with HU-treated wild-type cells. Cells were synchronized in G1 phase (G1) using α-factor, and then released in the presence of 200 mm HU for 90 min (HU). Mcm4–3Flag was immunoprecipitated (Mcm4 IP), followed by analysis of co-precipitated Psf2–13Myc (A), Ctf4–13Myc (B), or Cdc45 and Pol1–6HA (C: upper panels). Immunoblotting detection used anti-Flag, anti-Myc, anti-Cdc45 or anti-HA antibodies. Immunoprecipitation was also performed under low salt conditions (50 mm potassium acetate) (C: lower panels). Strains used are TKY52, TKY78, TKY59, and TKY85. D, Histogram shows ctf18Δ/WT ratios of Mcm4 binding for Psf2–13Myc, Ctf4–13Myc, Cdc45, and Pol1–6HA. Values were normalized based on the amount of Mcm4–3Flag precipitated in each experiment.
Next, we examined whether Cdc45 showed increased association with Mcm4. Cdc45 forms another central subunit of the RPC, and showed slightly increased chromatin association in HU-treated ctf18Δ cells in SILAC experiments (Fig. 3B and supplemental Fig. S4). Using co-IP analysis, we observed a substantial increase in Cdc45-Mcm4 complex formation in HU-treated ctf18Δ cells (Fig. 5C and 5D), of similar magnitude to that observed for Psf2 and Ctf4. These co-immunoprecipitation experiments show increased binding to Mcm4 of Cdc45, GINS subunit Psf2, and Ctf4 and together suggest that excess active replicative helicase is present in HU-treated ctf18Δ cells.
Ctf4 binds to GINS and DNA Pol α and is thought to form a physical connector between the replicative helicase complex Cdc45-MCM-GINS and the DNA Pol α-primase complex (22, 23). Although Ctf4 association with chromatin increased in HU-treated ctf18Δ cells, the SILAC analysis suggested that chromatin association of Pol α-primase subunits, such as Pol1, Pol12 and Pri2, actually decreased (Fig. 3A and supplemental Fig. S4), potentially suggesting an imbalance in replisome components. We used co-IP to examine whether the amount of Pol1 binding to the helicase complex in HU-treated ctf18Δ cells is really reduced. Consistent with the SILAC data, slightly less Pol1 appeared to associate with Mcm4 in HU-treated ctf18Δ when pull-down was carried out under the same buffer conditions as for SILAC chromatin fractionation (Fig. 5C upper three panels). However, the signal from co-immunoprecipitated Pol1 was weak and close to background level. To improve Pol1 signal strength, we performed immunoprecipitation under lowered salt conditions (50 mm potassium acetate) as described previously (22). With this adjustment, Pol1 signal strength was increased and it was clear that in HU-treated ctf18Δ cells, the amount of Pol1 associated with Mcm4 is elevated (Fig. 5C bottom and 5D). This observation suggests that in HU-treated ctf18Δ cells the amount of active, replisome-associated Pol1 (and Pol α-primase) is in fact increased, like other RPC components. It appears however that particular characteristics of the Pol α-primase complex preclude identification of this increase by SILAC (see Discussion).
Ctf18 is Required for Full Activation of Rad53 in Response to HU
The hundreds of replication origins in the S. cerevisiae genome initiate replication sequentially according to a temporal program (42). In wild-type cells exposed to HU, initiation at late origins is inhibited by the DNA replication checkpoint pathway. In mutant cells deficient in the DNA replication checkpoint (e.g. mrc1Δ), late origins are activated (26, 29). One possibility is that the excess active helicase complex present in HU-treated ctf18Δ cells is caused by inappropriate initiation and RPC formation at late replication origins, because of a defective DNA replication checkpoint. To test this idea, we examined whether late origin ARS1413 fires in HU-treated ctf18Δ cells using two-dimensional gel analysis. No replication intermediates were detected at ARS1413 in HU-treated wild-type cells, but a clear bubble arc was detected in HU-treated ctf18Δ (Fig. 6A), indicative of initiation at ARS1413 and suggestive of a defect in the replication checkpoint. An even more intense bubble arc at ARS1413 in mrc1Δ cells may reflect differing kinetics in ctf18Δ and mrc1Δ strains of activating the DNA damage checkpoint, which also affects late origins (see below).
HU exposure causes hyperphosphorylation of the checkpoint kinase Rad53 and stimulation of its kinase activity (43). We compared the phosphorylation kinetics of Rad53 in HU-treated wild-type and ctf18Δ cells (Fig. 6B). We found that Rad53 phosphorylation was delayed in HU-treated ctf18Δ cells, similar to the delay in mrc1Δ cells (compare 30 min time points). We conclude that Ctf18 is required for timely activation of Rad53 and efficient DNA replication checkpoint engagement in response to HU. In the absence of the Mrc1-mediated replication checkpoint, HU treatment causes accumulating DNA damage, with the result that Rad53 becomes phosphorylated through the Rad9-dependent DNA damage checkpoint pathway (26). To test whether the somewhat delayed Rad53 activation in HU-treated ctf18Δ cells depends on the Rad9 pathway, we constructed a ctf18Δ rad9Δ double mutant and tested phosphorylation kinetics of Rad53. On HU treatment, phosphorylated forms of Rad53 accumulate in rad9Δ cells, but are hardly detected in the ctf18Δ rad9Δ double mutant in HU (Fig. 6C). These results suggest that Ctf18, like Mrc1, is required to activate the DNA replication checkpoint pathway. In the absence of Ctf18, Rad53 activation occurs only through the Rad9-dependent checkpoint pathway, and probably reflects accumulating DNA damage.
We conclude that the excess active helicase present in HU-treated ctf18Δ cells results from inappropriate initiation at late replication origins because of a defect in the DNA replication checkpoint.
Alteration of Chromatin Composition in HU-treated ctf18Δ Cells is Similar to that in HU-treated Cells Lacking a Checkpoint Mediator Mrc1
We used SILAC quantitative proteomics to examine the chromatin of HU-treated mrc1Δ cells, to test whether the chromatin abnormalities observed in ctf18Δ cells are similar to those of another checkpoint-deficient mutant. We found increased chromatin association of replication proteins including PCNA, Cdc45, GINS components, Ctf4, and RPA subunits in HU-treated mrc1Δ cells (Fig. 6D and Supplemental Table S6), very similar to the changes observed for the ctf18Δ mutant exposed to HU. The similarity in chromatin profile between HU-treated ctf18Δ and mrc1Δ included a slight increase in chromatin association of the cohesin complex (Fig. 6E). The close resemblance of chromatin profiles of HU-treated mrc1Δ and ctf18Δ mutants supports the suggestion that Ctf18 acts in the same pathway as Mrc1 in the cellular response to HU, affecting chromatin through the DNA replication checkpoint.
HU Treatment of Cells Lacking the Elg1-RLC Reveals a Spectrum of Chromatin Abnormalities that Differs from ctf18Δ
The function of Elg1-RLC, another alternative RFC complex, is not well understood. To investigate the function of Elg1-RLC, we used SILAC to compare chromatin fractions from HU-treated wild-type and elg1Δ cells. There was no significant increase in chromatin association of Cdc45, GINS, Ctf4, RPA, Mec1 and the cohesin complex subunits Scc1 and Scc3 in HU-treated elg1Δ cells (Fig. 7A and supplemental Table S7). This chromatin profile differs sharply from the ctf18Δ mutant and suggests that Elg1 is not essential for activation of DNA replication checkpoint. Interestingly, we observed that chromatin association of PCNA and the flap endonuclease Rad27 (a yeast FEN1 homolog) was substantially increased in HU-treated elg1Δ cells (Figs. 7A and 7B). Rad27 is believed to act in Okazaki fragment processing and maturation. Chromatin association of PCNA and Rad27 was also abnormally high during normal S phase in elg1Δ cells (Figs. 7C and 7D and supplemental Table S8). Increased chromatin loading of PCNA in elg1Δ is consistent with recently published data (44). Finally, we confirmed that the DNA replication checkpoint is intact in elg1Δ cells, by demonstrating that late origin ARS1413 is repressed and Rad53 phosphorylation occurs normally in HU-treated elg1Δ cells (Fig. 7E and 7F). Elg1 is therefore not required for the DNA replication checkpoint, and that the chromatin composition abnormalities seen in elg1Δ presumably result from a different defect. Despite certain phenotypic similarities (7, 8), Ctf18-RLC and Elg1-RLC therefore appear to have distinct in vivo functions in DNA metabolism.
DISCUSSION
To investigate the role of Ctf18-RLC in S phase, we used SILAC-based proteomics to analyze chromatin composition. About 63% of known chromatin proteins were identified and quantified (Fig. 2D). We found significant and reproducible changes in chromatin composition in the ctf18Δ mutant, which were confirmed by Western blot analysis (Fig. 3). The results demonstrate that our quantitative proteomic approach is useful to obtain a large-scale view of changes in chromatin composition.
Ctf18 is Required for DNA Replication Checkpoint Activation
SILAC proteomic analysis revealed increases in chromatin-bound RPC components such as GINS, Cdc45, and Ctf4 in HU-treated ctf18Δ cells, suggesting the presence of excess active DNA helicase complex (Fig. 3). Immunoprecipitation assays confirmed that the amount of Mcm4-bound GINS, Cdc45, and Ctf4 (Fig. 5) was increased, suggesting increased RPC formation and the presence of a larger number of active replisomes. Because this effect was observed only when replication was challenged by HU and not during normal S phase (Figs. 3 and 4), we suspected that the apparent increase in active replisomes might reflect inappropriate late origin initiation because of defective DNA replication checkpoint activation. We found that a late origin is derepressed in HU-treated ctf18Δ cells (Fig. 6A) and phosphorylation of the checkpoint kinase Rad53 is delayed (Fig. 6B), showing that Ctf18 is required for normal activation of the replication checkpoint. The close resemblance of chromatin composition profiles from HU-treated ctf18Δ cells and mrc1Δ cells (Fig. 6E) supports the idea that most of the abnormalities observed in HU-treated ctf18Δ chromatin are caused by defective DNA replication checkpoint activation. Using a different approach Crabbé et al. (45) also recently showed that Ctf18-RLC is required for the DNA replication checkpoint. Our results are moreover consistent with previous data suggesting that the Ctf18-RLC subunits Dcc1 and Ctf8 are required for the DNA replication checkpoint pathway (46), and with the suggestion that lack of human Ctf18-RLC alters the dynamics of replication (47).
The Rad53 phosphorylation that does still occur in the ctf18Δ mutant on HU treatment depends on the Rad9-mediated DNA damage checkpoint. The delay to Rad53 activation in the absence of Ctf18 probably provides a time window allowing initiation at late origins. The fairly slight retardation of Rad53 phosphorylation probably explains why earlier studies of the ctf18Δ mutant (which used less rigorous sampling protocols) did not detect a checkpoint defect (5, 48).
Chromatin Profile Abnormalities of HU-treated ctf18Δ Cells Could Result From a Defective DNA Replication Checkpoint
ctf18Δ cells show defects in sister chromatid cohesion (12). Cohesin loading on chromatin appeared normal in ctf18Δ cells in an unchallenged S phase. Unexpectedly however, we observed slightly increased loading of the cohesin complex on chromatin in HU-treated ctf18Δ cells (Fig. 3 and supplemental Fig. S4). Extra cohesin is therefore loaded onto chromatin when replication is blocked in ctf18Δ cells, when compared with wild-type. In addition to the normal quota of cohesin loaded in late G1 phase, extra cohesin can be loaded onto chromatin during G2/M phase to reinforce cohesion at sites of DNA damage (49). We suspect that the additional cohesin observed on chromatin in HU-challenged ctf18Δ cells reflects a cellular response to abnormal levels of DNA damage, which may result from the formation of extended tracts of single-stranded DNA in HU-treated ctf18Δ cells because of the replication checkpoint activation defect. Regions of single-stranded DNA have been observed at HU-blocked replication forks in checkpoint-deficient mutants (50). Increased chromatin loading of the repair protein Rad52 in HU-treated ctf18Δ and mrc1Δ cells (supplemental Fig. S2 and Fig. 6D) supports the idea that these mutants accumulate DNA damage when challenged with HU, as does their accumulation of H2A(X) phosphorylation (45). HU-treated mrc1Δ cells display an increase of chromatin-bound cohesin complex similar to that of ctf18Δ (Fig. 6E), consistent with its resulting from the DNA replication checkpoint defect.
The checkpoint kinase Mec1-Ddc2, homolog of human ATR-ATRIP, is recruited to RPA-coated single-stranded DNA in response to HU or DNA damage (28). We therefore suspect that increased chromatin loading of Mec1 is probably because of increased RPA loading in HU-arrested ctf18Δ (Fig. 3), caused by single-stranded DNA accumulating at HU-blocked replication forks in this checkpoint-deficient mutant. The reduction in chromatin-bound Plm2 we observed when either ctf18Δ or mrc1Δ is HU-treated probably also reflects the checkpoint defect. Plm2 is a putative transcription factor that is phosphorylated in a Rad53-dependent way (51, 52), and so altered Plm2 behavior is consistent with defective Rad53 activation in HU-treated ctf18Δ and mrc1Δ cells. In general, most of the chromatin abnormalities we observe in the HU-blocked ctf18Δ mutant can be interpreted as resulting from failure to activate the DNA replication checkpoint.
What is the Molecular Activity of the Ctf18-RLC?
In vitro, the Ctf18-RLC can load PCNA onto DNA and also unload it from DNA (9–11). Increased PCNA on chromatin in HU-treated ctf18Δ cells (Fig. 3) might be taken to suggest that Ctf18-RLC unloads PCNA from DNA in vivo. However, because chromatin-associated PCNA also increased in HU-treated mrc1Δ cells (Fig. 6), it is likely that the increase in chromatin-bound PCNA in HU-treated ctf18Δ cells results mainly from loading of PCNA at late origins, possibly mediated by RFC. In contrast to our SILAC results, ChIP-microarray and ChIP-qPCR studies reported decreased PCNA in HU-treated ctf18Δ cells (16). This apparent inconsistency may reflect different quantification methods. In ChIP-microarray and ChIP-qPCR datasets it is difficult to distinguish between PCNA destabilization at replication forks, and replisomes that are themselves abnormally spread out (perhaps as a result of defective checkpoint activation).
During normal S phase in ctf18Δ cells, we did observe a decrease in PCNA loading on chromatin (Fig. 4). This result suggests that Ctf18 may load PCNA during normal S phase, possibly at specific genomic sites. Further experiments will however be needed to test this idea, because reduced PCNA loading could equally be an indirect effect of the ctf18Δ mutation.
It is unclear how the Ctf18-RLC mediates DNA replication checkpoint activation. Because it binds DNA polymerase ε (53, 54), one possibility is that Ctf18-RLC coordinates the replisome components to allow checkpoint activation by RPA and Mec1. Alternatively, the Ctf18-RLC might load or unload a modified form of PCNA required for checkpoint activation.
Distinct Functions of Ctf18-RLC and Elg1-RLC
In contrast to the ctf18Δ mutant, yeast cells lacking Elg1 showed no increase in GINS, Cdc45 and Ctf4 on chromatin, and displayed normal Rad53 activation on HU treatment (Fig. 7). Ctf18-RLC and Elg1-RLC therefore appear to have distinct roles in maintaining genome stability.
Association of PCNA with chromatin was increased in elg1Δ cells both during normal S phase and when replication forks are challenged by HU addition (Fig. 7). This increased chromatin association of PCNA is unlikely to result from inappropriate late origin initiation, because there was no matching increase in Cdc45-MCM-GINS complex formation. The accumulation of PCNA in elg1Δ could potentially result from failure of PCNA unloading by Elg1-RLC (although so far there has been no in vitro demonstration of PCNA unloading by Elg1-RLC). Increased loading of the flap endonuclease Rad27 in elg1Δ cells may suggest involvement of Elg1 in PCNA transactions during Okazaki fragment maturation, a possibility requiring further investigation.
Limitations of SILAC-based Chromatin Profiling
Using SILAC analysis of HU-treated ctf18Δ chromatin we observed changes consistent with increased Cdc45-MCM-GINS-Ctf4 complex formation; however, the amount of Pol α-primase in the chromatin fraction appeared slightly decreased (Fig. 3). Further investigation of the behavior of Pol α-primase using co-IP experiments, including adjustment of assay conditions, suggested that interaction between the helicase complex and the Pol α subunit is actually increased in HU-treated ctf18Δ cells (Fig. 5C), in a way that resembles the increased association of other RPC components with helicase. Increased Pol α-primase loading was not detected in our chromatin fractionation partly because of salt sensitivity of its interaction with the replisome (Fig. 5D), and partly because background levels of Pol α-primase binding to chromatin reduce the proportional increase observed on genuine replisome formation. Similarly, only small increases were observed in the chromatin association of certain other RPC subunits (e.g. Tof1, Spt16, Pob3, and Top1) in HU-treated ctf18Δ cells (supplemental Fig. S4), probably because the proportion of these proteins bound non-specifically to chromatin obscures genuine increases in their replisome association (i.e. where background association of a protein with chromatin is high, biologically meaningful changes can appear marginal). These cases highlight the importance of considering the limitations of the chromatin enrichment procedure, and the implications for interpreting results.
Conclusion and prospects
Chromatin profiling using SILAC-based proteomics represents the first method to obtain a large-scale view of changes in chromatin composition. This method is particularly useful in highlighting the significance of relatively small changes that nonetheless occur consistently among specific groups of proteins, and in this way has revealed that Ctf18 acts in the DNA replication checkpoint. Our chromatin profiling approach will be very useful to investigate change in chromatin composition that occur in other mutant cells and in response to drugs.
Acknowledgments
We thank Frederick van Deursen and Karim Labib for kindly gifting us anti-Cdc45 antibody and J. Julian Blow, Laura Trinkle-Mulcahy, David A. Stead, Sara T. Have, Motoharu Ono, Douglas J. Lamont, and Kenneth A. Beattie for advice and help with mass spectrometry and MaxQuant analysis.
* AIL is a Wellcome Trust Principal Research Fellow. This work was funded by Cancer Research UK grants C1445/A8791 and C1445/A11646.
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S8.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S8.
1 The abbreviations used are:
- RFC
- replication factor C
- SILAC
- stable isotope labeling by amino acids in cell culture
- RPC
- replisome progression complex
- RLC
- RFC-like complex
- RPA
- replication protein A
- HU
- hydroxyurea
- PCNA
- proliferating cell nuclear antigen
- DTT
- dithiotreitol
- PMSF
- phenylmethylsulfonyl fluoride.
REFERENCES
- 1. Gomes X. V., Burgers P. M. (2001) ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J. Biol. Chem. 276, 34768–34775 [DOI] [PubMed] [Google Scholar]
- 2. Bowman G. D., O'Donnell M., Kuriyan J. (2004) Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 [DOI] [PubMed] [Google Scholar]
- 3. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 4. Kim J., MacNeill S. A. (2003) Genome stability: A new member of the RFC family. Curr. Biol. 13, R873–5 [DOI] [PubMed] [Google Scholar]
- 5. Kanellis P., Agyei R., Durocher D. (2003) Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13, 1583–1595 [DOI] [PubMed] [Google Scholar]
- 6. Ben-Aroya S., Koren A., Liefshitz B., Steinlauf R., Kupiec M. (2003) ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. U.S.A. 100, 9906–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maradeo M. E., Skibbens R. V. (2009) The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS One 4, e4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parnas O., Zipin-Roitman A., Mazor Y., Liefshitz B., Ben-Aroya S., Kupiec M. (2009) The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS One 4, e5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bermudez V. P., Maniwa Y., Tappin I., Ozato K., Yokomori K., Hurwitz J. (2003) The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl. Acad. Sci. U.S.A. 100, 10237–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shiomi Y., Shinozaki A., Sugimoto K., Usukura J., Obuse C., Tsurimoto T. (2004) The reconstituted human Chl12-RFC complex functions as a second PCNA loader. Genes Cells 9, 279–290 [DOI] [PubMed] [Google Scholar]
- 11. Bylund G. O., Burgers P. M. (2005) Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol. Cell. Biol. 25, 5445–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanna J. S., Kroll E. S., Lundblad V., Spencer F. A. (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21, 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hiraga S., Robertson E. D., Donaldson A. D. (2006) The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J. 25, 1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hiraga S., Botsios S., Donaldson A. D. (2008) Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning. J. Cell Biol. 183, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bellaoui M., Chang M., Ou J., Xu H., Boone C., Brown G. W. (2003) Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22, 4304–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K. P., Shirahige K., Uhlmann F. (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell 23, 787–799 [DOI] [PubMed] [Google Scholar]
- 17. Ansbach A. B., Noguchi C., Klansek I. W., Heidlebaugh M., Nakamura T. M., Noguchi E. (2008) RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in schizosaccharomyces pombe. Mol. Biol. Cell 19, 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Labib K. (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aparicio T., Ibarra A., Méndez J. (2006) Cdc45-MCM-GINS, a new power player for DNA replication. Cell. Div. 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moyer S. E., Lewis P. W., Botchan M. R. (2006) Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 22. Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanaka H., Katou Y., Yagura M., Saitoh K., Itoh T., Araki H., Bando M., Shirahige K. (2009) Ctf4 coordinates the progression of helicase and DNA polymerase alpha. Genes Cells 14, 807–820 [DOI] [PubMed] [Google Scholar]
- 24. Uhlmann F. (2009) A matter of choice: The establishment of sister chromatid cohesion. EMBO Rep. 10, 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu H., Boone C., Brown G. W. (2007) Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 176, 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., Bousset K., Furuya K., Diffley J. F., Carr A. M., Elledge S. J. (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3, 958–965 [DOI] [PubMed] [Google Scholar]
- 27. Osborn A. J., Elledge S. J. (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17, 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Branzei D., Foiani M. (2009) The checkpoint response to replication stress. DNA Repair 8, 1038–1046 [DOI] [PubMed] [Google Scholar]
- 29. Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 30. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 31. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 32. Haase S. B., Reed S. I. (2002) Improved flow cytometric analysis of the budding yeast cell cycle. Cell. Cycle 1, 132–136 [PubMed] [Google Scholar]
- 33. Sheu Y. J., Stillman B. (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 24, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Young M. R., Tye B. K. (1997) Mcm2 and Mcm3 are constitutive nuclear proteins that exhibit distinct isoforms and bind chromatin during specific cell cycle stages of saccharomyces cerevisiae. Mol. Biol. Cell 8, 1587–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 36. Boulon S., Ahmad Y., Trinkle-Mulcahy L., Verheggen C., Cobley A., Gregor P., Bertrand E., Whitehorn M., Lamond A. I. (2010) Establishment of a protein frequency library and its application in the reliable identification of specific protein interaction partners. Mol. Cell. Proteomics 9, 861–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 38. Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. (1987) The in vivo replication origin of the yeast 2 microns plasmid. Cell 51, 473–481 [DOI] [PubMed] [Google Scholar]
- 39. Brewer B. J., Lockshon D., Fangman W. L. (1992) The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71, 267–276 [DOI] [PubMed] [Google Scholar]
- 40. Friedman K. L., Brewer B. J. (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262, 613–627 [DOI] [PubMed] [Google Scholar]
- 41. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raghuraman M. K., Brewer B. J. (2010) Molecular analysis of the replication program in unicellular model organisms. Chromosome Res. 18, 19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P. P., Foiani M. (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18, 6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parnas O., Zipin-Roitman A., Pfander B., Liefshitz B., Mazor Y., Ben-Aroya S., Jentsch S., Kupiec M. (2010) Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 29, 2611–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Crabbé L., Thomas A., Pantesco V., De Vos J., Pasero P., Lengronne A. (2010) Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat. Struct. Mol. Biol. 17, 1391–1397 [DOI] [PubMed] [Google Scholar]
- 46. Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., Boeke J. D. (2006) A DNA integrity network in the yeast saccharomyces cerevisiae. Cell 124, 1069–1081 [DOI] [PubMed] [Google Scholar]
- 47. Terret M. E., Sherwood R., Rahman S., Qin J., Jallepalli P. V. (2009) Cohesin acetylation speeds the replication fork. Nature 462, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naiki T., Kondo T., Nakada D., Matsumoto K., Sugimoto K. (2001) Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 21, 5838–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heidinger-Pauli J. M., Unal E., Koshland D. (2009) Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell 34, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sogo J. M., Lopes M., Foiani M. (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 [DOI] [PubMed] [Google Scholar]
- 51. Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., Gerstein M., Snyder M. (2002) Complex transcriptional circuitry at the G1/S transition in saccharomyces cerevisiae. Genes Dev. 16, 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H. (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 54. Murakami T., Takano R., Takeo S., Taniguchi R., Ogawa K., Ohashi E., Tsurimoto T. (2010) Stable interaction between the human PCNA loader complex Ctf18-RFC and DNA polymerase ε is mediated by the cohesion specific subunits, Ctf18, Dcc1 and Ctf8. J. Biol. Chem. 285, 34608–34615 [DOI] [PMC free article] [PubMed] [Google Scholar]