Abstract
Large proteomic data sets identifying hundreds or thousands of modified peptides are becoming increasingly common in the literature. Several methods for assessing the reliability of peptide identifications both at the individual peptide or data set level have become established. However, tools for measuring the confidence of modification site assignments are sparse and are not often employed. A few tools for estimating phosphorylation site assignment reliabilities have been developed, but these are not integral to a search engine, so require a particular search engine output for a second step of processing. They may also require use of a particular fragmentation method and are mostly only applicable for phosphorylation analysis, rather than post-translational modifications analysis in general. In this study, we present the performance of site assignment scoring that is directly integrated into the search engine Protein Prospector, which allows site assignment reliability to be automatically reported for all modifications present in an identified peptide. It clearly indicates when a site assignment is ambiguous (and if so, between which residues), and reports an assignment score that can be translated into a reliability measure for individual site assignments.
Proteomic research is increasingly moving from simply cataloging proteins to trying to understand which components are most important for regulation and function (1, 2). Protein activity can be controlled over the long term by changes in expression levels, but for rapid and precise changes, the cell employs a range of post-translational modifications (PTMs)1. Mass spectrometry is the enabling tool for PTM characterization, as it is the only approach that can study thousands of modification sites in a single experiment (3). Modern mass spectrometers are able to produce large amounts of data in relatively short periods of time, such that the bottleneck in most proteomic research is the data analysis (4).
There is a broad spectrum of mass spectrometry search engines that can be employed for data analysis (5), and from most of these programs a measure of reliability for individual peptide identifications is reported, commonly in the form of a probability or expectation value. These calculations determine how much better than random a particular assignment is. As a modified peptide with an incorrect modification site assignment is highly homologous to the correct answer, assignments to the correct peptide sequence but with incorrect site assignment will generally give a confident identification score. Hence, although these tools can be applied for analyzing peptides bearing chemical or biological modifications they will report some incorrect modification site assignments and no search engine currently reports a measure of reliability for the assignment of a site of modification within a peptide.
To address this issue, a range of tools has been written to try to assess phosphorylation site assignments from search engine results (6–10). Phosphorylation is an obvious modification on which to focus on developing tools, as in addition to it being arguably the most important biological regulatory modification, it is also a modification that can occur on a broad spectrum of amino acid residues, although it is most commonly found on serines, threonines, and tyrosines. Typical peptides produced from proteolytic digests of proteins will contain multiple potential sites of phosphorylation, so there is a clear need for software that can determine how reliably a particular assignment is pinpointed. Most of these tools take outputs from a particular search engine, then look for diagnostic fragment ions that would distinguish between potential sites of modification. Probably the best known of these is Ascore (7), which calculates a probability for a site assignment by working out how many b or y ions could be observed to distinguish between potential sites, and what percentage of these were observed in the particular spectrum. However, this software was designed only for low mass accuracy ion trap collision induced dissociation (CID) data and requires an output from a particular version of the search engine Sequest (11).
Determining site assignment reliabilities directly from a search engine result has a number of advantages. It allows site assignment scoring for all data types that can be analyzed by the software. It also permits determining scores for all modifications; not just phosphorylation. Two groups have used results from the search engine Mascot (12) to report confidences for site assignments based on the difference in score between a particular site assignment and the next highest scoring modified version of the same peptide (10, 13). This information is not displayed directly in the search engine output, but software has been written to parse information out from links contained in the search engine results. In the study by Savitski et al., the authors created a range of fragmentation data from analyzing synthetic phosphopeptides where the modification sites were known, which allowed determination of a phosphorylation site false localization rate (FLR).
In this manuscript we present Site Localization In Peptide (SLIP) scoring that is automatically calculated for all modifications identified in peptides identified by the search engine Batch-Tag in the Protein Prospector suite of tools (14). To characterize its performance, we compare it to the results using alternative tools AScore and Mascot Delta Score and we also test it using a larger phosphopeptide data set, determining FLRs associated with a given site score.
MATERIALS AND METHODS
Samples Used for Creating Data Sets for Analysis
The quadrupole fragmentation data was acquired using a quadrupole time-of-flight (QTOF) Micro (Waters, Milford, MA) mass spectrometer and the details describing the sample creation and data acquisition are described in the publication where the data set was created (10). Briefly, 180 phosphopeptides were synthesized bearing a mixture of phosphorylated serine, threonine, and tyrosine residues. The majority of them were singly phosphorylated, but some doubly phosphorylated peptides were also present. Each peptide was analyzed separately by liquid chromatography (LC)-tandem MS (MSMS), then all the data from the 180 peptides were combined for data analysis. The raw data and peak lists created from this data are freely available for download from Tranche (https://proteomecommons.org/tranche/).
The phosphopeptide data for assessing ion trap fragmentation spectra was derived from a tryptic digestion of mouse synaptosomes. Mouse synaptosomes were resuspended in 1 ml buffer containing 50 mm ammonium bicarbonate, 6 m guanidine hydrochloride 6× Roche Phosphatase Inhibitor Cocktails I and II, and 6× PUGNAc inhibitor. The mixture was reduced with 2 mm Tris(2-carboxyethyl)phosphine hydrochloride and alkylated 4.2 mm iodoacetamide. The mixture was diluted to 1 m guanidine with ammonium bicarbonate and digested for 12 h at 37 °C with 1:50 (w/w) trypsin.
Phosphorylated peptides were enriched over an analytical guard column packed with 5 μm titanium dioxide beads (GL Sciences, Tokyo, Japan). Peptides were loaded and washed in 20% acetonitrile/1% trifluoroacetic acid then eluted with saturated KH2PO4 followed by 5% phosphoric acid. High pH reverse phase chromatography was performed using an ÄKTA Purifier (GE Healthcare, Piscataway, NJ) equipped with a 1 × 100 mm Gemini 3μ C18 column (Phenomenex, Torrance, CA). Buffer A consisted of (20 mm ammonium formate, pH 10). Buffer B consisted of buffer A with 50% acetonitrile. The gradient went from 1% B 100% B over 6.5 mls. Twenty fractions were collected and dried down using a SpeedVac concentrator.
Phosphopeptides were separated by low pH reverse phase chromatography using a NanoAcquity (Waters) interfaced to an LTQ-Orbitrap Velos (Thermo) mass spectrometer. Precursor masses were measured in the Orbitrap. All MS/MS data was acquired using CID in the linear ion trap and all fragment masses were measured using the linear ion trap.
Search Parameters
Data was searched using Protein Prospector version 5.8.0 (14). QTOF Micro data was searched against a concatenated database of SwissProt downloaded on August 10th 2010 and randomized versions of these entries. Only human entries were considered, meaning a total of 40574 entries were queried. Fully tryptic cleavage specificity was assumed, all cysteines were assumed to be carbamidomethylated and possible modifications considered included oxidation (M); Gln->pyro-Glu (peptide N-term); Met-loss, acetylation and the combination thereof (protein N-term); and phosphorylation (S, T, or Y). Precursor masses were considered with ± 200 ppm mass tolerance and fragments with ± 0.4 Da tolerance. The results are presented in supplemental Table S1.
Peak lists for LTQ-Velos data were created using in-house software “PAVA.” LTQ-Velos data was searched against a list of 3794 rodent proteins and randomized versions thereof that are all found in UniprotKB downloaded on August 10th 2010. Fully tryptic cleavage specificity was assumed and all cysteines were considered to be carbamidomethylated. A precursor mass tolerance of 15 ppm and fragment mass tolerance of 0.6 Da were permitted. Two searches were performed. In each case the same variable modifications were considered as for the QTOF data set described above. However, in one search phosphorylation of proline was also considered; in the other phosphorylation of glutamic acid was considered. Batch-Tag identifies both phosphorylated fragment ions and those for which phosphoric acid has been eliminated. Therefore, to fully simulate other potential sites of phosphorylation the code was adapted to consider phosphate loss from glutamates and prolines that were assigned as phosphorylated. Results were filtered to a 0.1% spectrum false discovery rate (FDR) according to target-decoy database searching.
Description of SLIP Scoring
SLIP scores are determined by comparing probability and expectation values for the same peptide with different site assignments. As a default, Protein Prospector was only saving the top five matches to each spectrum, and in some cases the match to the same peptide with the next best modification site assignment was not in these results (e.g. if the other potential modification site is at the other end of the peptide). Hence, the manner in which the results were saved was altered so that the score for the next best site assignment is always stored for all of the top five peptide matches to a particular spectrum. The SLIP score is derived by comparing the probability or expectation value for each peptide to the next best match of the same peptide but with different site assignment (the difference between probabilities and expectation values will be identical as the expectation value is the probability multiplied by a constant (number of precursors in the database within the precursor mass tolerance)). This difference is converted into a simple integer score by converting into Log10 scale and then multiplying by −10. This transformation is analogous to that used by the Mascot search engine for converting its probability scores into Mascot scores (12). Under this conversion, a SLIP score of 10 means an order of magnitude difference in probability scores for the different site assignments whereas a SLIP score of 5 corresponds to about a sevenfold higher confidence for a peptide assignment.
RESULTS
Comparison to AScore and Mascot Delta Score for Quadrupole CID Data
A previous study compared the performance of the Mascot Delta Score to the alternative phosphorylation site scoring software AScore (10). Generously, they made the raw data produced for this comparison freely available. Hence, this allows new tools for phosphorylation site assignment to be benchmarked against these results.
The data set given the most focus in the previous study was acquired on a QTOF Micro mass spectrometer, so we decided to employ Batch-Tag for analyzing this same data set, then assessed the SLIP scoring integrated into the Search Compare output to determine site assignment reliability. The results are summarized in the first column of Table I and the full results are supplemental Table S1. Search Compare reported 2334 correct peptide identifications, containing 2437 phosphorylation site assignments. Of these, 164 peptides contained only one possible site of modification, so site assignment scoring was not relevant. A further 220 sites were reported as completely ambiguous; i.e. multiple sites achieved exactly the same score. In the remaining 2053 cases, one possible site assignment scored better than the others, so a SLIP score was reported. Of these, 130 of the site assignments were incorrect, corresponding to a 6.3% FLR. The previously reported result for Ascore when analyzing this data set was 1584 IDs with 138 incorrect (9% FLR) and 1840 with 201 incorrect (11% FLR) for Mascot Delta Score.
Table I. Comparison of different peak list filtering approaches. Three different peak list filtering approaches are compared for their ability to identify phosphopeptides and pinpoint modification sites.
| 20 + 20 | 4 per 100 | 5 per 100 | |
|---|---|---|---|
| Spectra | 2334 | 2378 | 2282 |
| Phosphosites | 2437 | 2488 | 2397 |
| Correct | 1924 | 1910 | 1883 |
| Incorrect | 130 | 136 | 161 |
| Ambiguous | 220 | 255 | 211 |
| One Site | 164 | 187 | 142 |
| FLR (%) | 6.3 | 6.6 | 7.9 |
| Ambiguous (%) | 9.0 | 10.2 | 8.8 |
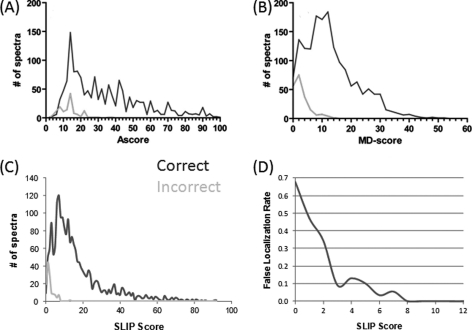
Results were broken down by SLIP score to try to determine a reliability estimate for a given SLIP score. Fig. 1 presents score histograms for correct and incorrect site assignments and the corresponding plots for Ascore and Mascot Delta Score are also presented. In Fig. 1D a plot of the FLR against score is shown. This plot is quite noisy, because of the fact that there are only 130 incorrect results (and only 32 with a score of three or greater), but it suggests that a SLIP score of six corresponds to a local FLR of about 5%; i.e. 5% of results with a SLIP score of exactly six are incorrect.
Fig. 1.
Distribution of correct and incorrect SLIP scores for the QTOF Micro data set. SLIP scores for analysis of the same data set of quadrupole CID data are plotted for Ascore (A); Mascot Delta Score (B), and PP Site Score (C). Correct site assignments are in blue; incorrect are plotted in red. Panel D plots a histogram of the local FLR for a given SLIP score. Panels A and B are reproduced from Savitski et al. (10).
Comparing Different Peak List Filtering Approaches
When raw data is converted into a peak list for analysis by a database search engine there is typically no peak detection step during the process; i.e. the peak list contains a mixture of real and noise peaks. Hence, most search engines employ a filtering step to try to maximize the number of real peaks while minimizing the number of noise peaks considered. Batch-Tag in Protein Prospector normally splits the observed mass range into two, then uses the 20 most intense peaks in each half of the mass range (a total of forty peaks if there are at least 20 peaks in each half of the mass range) for database searching (15). Ascore uses a more extensive binning approach where it considers a fixed number of peaks per 100 m/z, and varies this value to try to optimize site assignment discrimination (7).
We decided to investigate the effect of using a 100 m/z binning approach, comparing it to the standard Batch-Tag 20 + 20 approach, assessing it both for peptide identification and site assignment reliability. Values from three to nine peaks per 100 m/z were investigated. The results for 20 + 20, four per 100 and five per 100 are presented in Table I.
Using four peaks per 100 identified marginally more spectra than the other two analyses, but had the highest number of ambiguous modification site assignments. Considering five peaks per 100 identified the fewest number of both spectra and sites and made the most mistakes, partly because it reported fewer sites as being ambiguous. The 20 + 20 results correctly identified the most sites and gave the most reliable site assignments.
Testing SLIP Scoring on a Larger Scale with Ion Trap CID Data
Determining the accuracy of site assignments can be difficult, as it requires knowledge of the correct answer, which is practically never the case when analyzing large, biologically derived data sets. The results presented so far were created from synthetic peptides with known sites of modification, but there were only 130 incorrect results (using the 20 + 20 peak filtering approach) from which to measure the FLR. Therefore, to create a larger data set for testing, and also to test a different fragmentation data type, we employed an alternative strategy for assessing site assignment scoring. We temporarily changed the settings in Batch-Tag such that it would consider phosphorylation of proline and glutamic acid residues. We also allowed it to consider loss of phosphoric acid from these amino acids. Hence, we were able to perform searches considering phosphorylation of two amino acids that if assigned as the site of modification, would always be incorrect.
We analyzed a large phosphopeptide data set acquired using ion trap CID fragmentation on an LTQ-Orbitrap Velos mass spectrometer. Two searches were performed: in the first we considered phosphorylation of S, T, Y, and P; in the second search we considered phosphorylation of S, T, Y, and E. In each search, over 90,000 spectra were identified at an estimated 0.1% false discovery rate according to searching against a concatenated normal/random database (16). Of these spectra, roughly 60,000 were of phosphopeptides.
For testing site assignment scoring we wanted spectra where the correct site assignment is known. Hence, we restricted our subsequent analysis to those spectra matched to phosphopeptides where there was only one serine, threonine, or tyrosine in the peptide sequence. For the “phosphoproline” search results there were 5433 of these peptide identifications, and for the “phosphoglutamate” results there were 5415. Of these, 361 and 245 respectively reported phosphorylations on the relevant decoy amino acid, corresponding to 6.6% and 4.5% global FLRs.
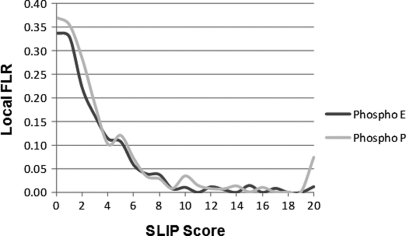
Fig. 2 plots false localization rates against SLIP score for the two different search results. As can be seen, very similar results were produced for the two decoy amino acid searches, and as in the QTOF Micro data analysis results, a SLIP score of six corresponded to a 5% local FLR.
Fig. 2.
FLR as a function of SLIP score for an ion trap CID data set. Histograms of FLR for a given SLIP score are plotted for the searches considering phosphoglutamate and phosphoproline modification.
SLIP Scoring of Electron Transfer Dissociation (ETD) Data of O-GlcNAcylated Peptides
To test the performance of the SLIP scoring for a third type of fragmentation and also for a different regulatory PTM we re-analyzed data used in a previous publication studying O-GlcNAc modified peptides in mouse brain (17). In this study 58 sites of O-GlcNAc modification were identified from roughly eighty modified peptide spectra. Clearly, with this limited number of modified peptide spectra it is not possible to accurately calculate a FLR for a particular SLIP score. However, it is possible to examine spectra with assignments of a particular SLIP score and make general comments about the amount of evidence supporting the site assignment.
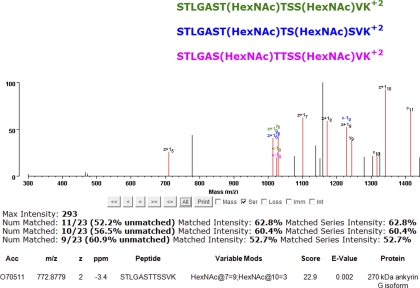
Fig. 3 compares site assignments in a peptide from Ankyrin G that contains two O-GlcNAc (HexNAc) modifications. One modification is assigned to the seventh residue in the peptide (middle threonine) with a SLIP score of nine; the second to the tenth residue (final serine) with a SLIP score of three. The next best assignment alternative to residue 10 is residue nine. Assignments that are unique to serine 9 are shown in blue, whereas assignments unique to serine 10 are in green. A peak at m/z 1024.97 matches a c-19 ion containing no modification, which supports the assignment of serine 10 as the modification site. Conversely, the peak at 1228.18 would correspond to a modified c-19 ion. However, this peak can also be explained as a z+19 ion from either site assignment interpretation. Hence, the assignment of serine 10 explains more peaks, but there is some level of ambiguity, which is consistent with the assignment only getting a SLIP score of three. The assignment of modification to threonine 7 is supported by the mass difference between z+15 and z+16 ions corresponding to a modified threonine. Hence, this interpretation explains an extra z+1 ion in comparison to site assignments on neighboring residues. z+1 ions are very common in ETD spectra of doubly charged precursors (18) and Protein Prospector scoring takes this into consideration (19). Therefore, this extra peak assignment gives a SLIP score of 9.
Fig. 3.
Comparison of three different site assignment combinations for a doubly O-GlcNAc modified peptide from Ankyrin G. Assignments in common among the different assignments are in black, those that differentiate between different assignments are labeled in the color of the peptide.
A second example is shown in supplemental Fig. S1 for a spectrum with a SLIP score of five. In this example there is some evidence to suggest that the spectrum may be a mixture of two different site assignments, but the mass difference between z+19 and z+110 strongly suggests modification of the second most N-terminal serine in the sequence.
DISCUSSION
Mass spectrometry instrumentation has improved dramatically in the last 15 years, such that it can now produce a vast amount of high quality data. This puts tremendous pressure on database search engines to be able to analyze these large data sets and produce reliable, unsupervised peptide and protein identification summaries. Initially, there was a period in which results of uncertain reliability were being produced and published. However, through pressure from the community and proteomic journals (20), software has now “caught up” such that reliability metrics are now associated with most published peptide sequence identifications.
PTM analysis is progressing through a similar cycle where the ability to reliably identify modified peptides is ahead of the software's ability to measure the confidence in the modification site assignment (21). However, again spearheaded by pressure from journals, programmers are busily developing tools to report reliabilities for site modifications reported. Most of these are stand-alone tools that re-analyze spectra for site assignment reliability using peptide identification results from a particular search engine as the reference. However, there are now attempts to use the search engine results directly to report site assignment reliabilities. The first of these have used score differences between different site assignments from the search engine Mascot (10, 13). Researchers have written software to extract results from the Mascot output and report score differences. In this manuscript we present an analogous approach using the Batch-Tag search engine in Protein Prospector. The difference here is that the site assignment scoring is directly incorporated into the output from the search engine. It reports SLIP scores that are based on the difference in expectation value for identifications with different site assignments, and these are calculated for all modifications in reported peptides.
We used two data sets to benchmark the FLR for a given SLIP score. The first was a data set that has previously been used to characterize the performance of two other site assignment scoring methods (10). This data set was acquired using quadrupole-type fragmentation, so will be representative of data acquired on QTOF instruments and also high-energy collision-induced dissociation data. For this data set, we identified a higher number of phosphopeptide spectra compared with the previously published results, due to Batch-Tag correctly interpreting more spectra than Mascot. However, the salient information for this study is the reliability of site assignment scoring. The SLIP scoring returned results with a global 6.3% FLR, whereas the Ascore and MD-score result had 9 and 11% FLRs. This suggests that Batch-Tag is making fewer mistakes in site assignments compared with the competing tools.
The use of different peak list filtering algorithms for peptide and modification site assignment was investigated, as multiple groups have proposed using binning of peaks per 100 Da for peak filtering and site assignment (6, 7). Our results suggest that as you increase the number of peaks considered by Batch-Tag you actually achieve fewer correct peptide identifications and make more mistakes in incorrectly assigning modification sites through the consideration of more noise peaks. When considering fewer peaks, more site assignments are deemed ambiguous. This is not necessarily a bad thing in that many would argue that it is better to report a result as ambiguous rather than reporting results with an elevated FLR. However, our results suggest that the 20 + 20 peak list approach normally employed by Batch-Tag is generally slightly better at reporting site assignments than the “per 100 Da” binning approach: it makes relatively few incorrect assignments while reporting fewer ambiguous site assignments than use of a similar number of peaks with 100 Da peak binning. Batch-Tag now has the option of using either of these peak filtering approaches, and as it is possible in Search Compare with combine or compare multiple search results of the same data set one could use both approaches (and vary the number of peaks considered), then take consensus site identifications. However, this would be extra work and we think for relatively small gain compared with just using the 20 + 20 results alone.
The second data set used for benchmarking the SLIP scoring was ion trap CID data. For this study we temporarily altered the Batch-Tag search engine to allow consideration of phosphorylation of glutamate and proline residues; two residues commonly found close to phosphorylation sites. Phosphorylation of proline does not exist in nature. Phosphorylated free glutamate (γ-glutamyl phosphate) is an intermediate in the biosynthesis of both glutamine and proline, but there are only a couple of reports suggesting it may occur in proteins (22). As it is chemically very unstable, it is highly unlikely it could ever survive to be found in a proteomic study. By knowing that all of these modification assignments were incorrect, this allowed analysis of a large phosphopeptide data set and by filtering the results to only those spectra of peptides that contain a single serine, threonine or tyrosine (such that we know the correct modification site) we were able to characterize incorrect site assignment scores based on phosphorylation matches to proline and glutamate residues.
Fig. 2 shows that a few phosphoproline and phosphoglutamate site assignments with SLIP scores all the way up to 20 (indeed there are a few with even greater scores but not plotted on this scale) were observed. However, these high scoring results are probably not examples of incorrect site assignments, but rather of incorrect peptide identifications. Although these results as a whole have an estimated 0.1% false discovery rate, we believe that the incorrect identifications are heavily enriched in the identifications with assignments of phosphorylated prolines and glutamates. Evidence for this is that, for example, in the phosphoproline search results as a whole 6465 out of the 93,554 reported results (7%) contained a phosphoproline site assignment. However, of the decoy database hits 33% (289/872) contained a phosphoproline. Incorrect peptide identifications are going to have randomly distributed SLIP scores. As one moves to higher SLIP scores there are fewer spectra with a given score. Hence, each incorrect identification creates a larger “spike” in the FLR plot (two phosphoproline site assignments with a score of 20 gave a large spike because of there being only 25 correct site assignments with a score of exactly 20).
The global false localization rate to prolines was similar to incorrect assignments in the quadrupole CID data set, whereas the incorrect phosphoglutamate assignment rate was lower. The difference is probably because of the frequency of these amino acids occurring next to real phosphorylation sites. The most common incorrect site assignments are when there are two potential residues next to each other that could be modified. Kinases have sequence recognition motifs, and a large number of biological phosphorylation sites are on sites surrounded by other serines and threonines (23). Similarly, there are many proline-directed kinases, so prolines are commonly found in proximity to phosphorylation sites. There are also kinases that contain acidic residues in their recognition motif, but these are less prevalent, so this may explain the reduced number of incorrect phosphoglutamate identifications.
Global FLRs are likely to be data set dependent: the higher the quality of the data (greater information content), the lower one would expect the FLR to be. However, for a given SLIP score, there should be a more consistent measure of reliability. In plots for the quadrupole and ion trap CID data a SLIP score of 6 corresponded to a local FLR of 5% and results analyzing ETD data of O-GlcNAc modified peptides were consistent with this score having a similar reliability.
A point of discussion is how site assignment results should be reported. Simply by reporting all site assignments that have any SLIP score would have returned 93–96% correct results in the described phosphopeptide analyses, which some would argue is an acceptable level of error. On the other hand, one could draw a site score threshold and only report site assignments that score above this. This is an option in the Search Compare output; it will report all results below this threshold as ambiguous. This will, of course, remove more correct site identifications than incorrect. The middle ground is to report all results but attempt to attach a measure of reliability to each SLIP score. The problem with this is that for real data sets, correct assignments are not known for enough spectra to calculate statistics unique to that set of data. Hence, one would have to extrapolate from results from a similar standard data set. In the two large studies described in this manuscript the 5% local FLR corresponded to the same score, but it may be a dangerous conclusion to assume that this is always the case.
Unlike many of the site scoring tools developed thus far, the Batch-Tag scoring is applicable to all modifications. We used SLIP scoring to examine site assignments of O-GlcNAc modified peptides fragmented by ETD (17). This data set was not large enough to determine a FLR for a given score, but it is possible to manually assess site assignments with different SLIP scores to get an impression of the reliability of a given score. Example spectra are presented in Fig. 3 and supplemental Fig. S1. In these examples an assignment with a SLIP score of three was ambiguous, whereas scores of five and nine both seemed reliable. These scores are consistent with a given SLIP score having a similar meaning in O-GlcNAc ETD data to the two phosphopeptide CID data sets. Nevertheless, the distribution of scores for other modifications may differ from those for phosphorylation and O-GlcNAcylation. As these modifications can occur on multiple amino acid side-chains and serines and threonines are relatively common amino acids, there are regularly going to be multiple potential sites of modification in a typical peptide and often quite close together. In contrast, methionines are much less common, so peptides containing multiple potential sites of modification are going to be infrequent. Thus, in most cases a site assignment score is not required and if there are multiple sites they will often be many residues apart. As a result, it will typically be easier to confidently distinguish between candidate sites of methionine oxidation and few incorrect site assignments will be made. On the other hand, if oxidation of tryptophan and oxidized proline (hydroxyproline) are also considered, then site assignment scoring to differentiate between multiple potential residues would be more important. For large data sets, a similar approach to employed here; i.e. allowing the search engine to consider modification of a residue that cannot bear the particular modification, could be employed to evaluate SLIP scoring for other modification types.
One type of analysis that could benefit significantly from SLIP scoring is mass modification searching, where unpredicted or unknown modifications are being sought (14, 24). In these types of analyses residue specificity is not known, so the reliability of site assignments is particularly troublesome. SLIP scoring quickly assesses the reliability of these site assignments.
An important feature of any site assignment software is having the ability to admit that it does not know the correct answer. In many spectra there may be no evidence at all to distinguish between potential sites. In this circumstance Search Compare does not report a site assignment score; instead it indicates between which residues the ambiguity exists; e.g. ‘Phospho@4 |5′ indicates that either the fourth or fifth amino acid in the peptide sequence is phosphorylated. If there are multiple phosphorylated residues in the peptide then the site assignment can be more ambiguous; for example for a couple of spectra of the doubly phosphorylated peptide “TSSFAEPGGGGGGGGGGPGGSASGPGGTGGGK” the site assignment was reported as ‘Phospho@1&2 |1&3 |2&3′; i.e. two of the first three residues are modified, but there is no evidence to determine which two. In addition to reporting the residue/s within the peptide sequence that are modified and their SLIP scores, SearchCompare can also report this information at the protein residue number level. By sorting results using this information it is easier to determine how many unique phosphorylation sites within a protein there is evidence for, as opposed to instances where the same phosphorylation site is identified in two related peptides where one contains a missed cleavage.
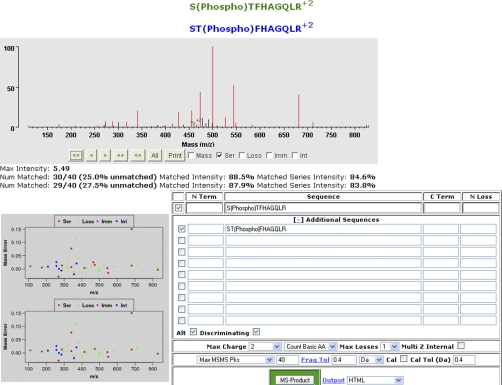
One advantage of site scoring being integrated into the Search Compare results is the ability to make use of linked programs to visually assess the results. Protein Prospector allows annotation of two or more different assignments to the same spectrum, so it is possible to display and compare the different site assignments (25). Fig. 4 shows an example of a spectrum with an incorrect site assignment that scored seven; i.e. one of the higher scoring incorrect site assignments. The incorrect assignment is labeled in green and the correct in blue. All explained fragment ions are in red; unexplained in black. By selecting the “discriminating” tickbox it is only labeling the peak assignments (in this example, a single peak) that differentiates between the two site assignments. A low intensity peak was matched to a doubly charged y8 ion, which presumably was a noise peak. From this interface it is also possible to vary the number of peaks considered (including switching to peaks per 100 m/z). If there are multiple site assignments that are reported as ambiguous, then when the user clicks on the peptide sequence from the Search Compare results report, Protein Prospector will display the alternative site assignments as in Fig. 3. This is primarily useful if a SLIP score threshold has been applied to the results, as this will allow the user to compare site assignments that are below the site assignment threshold applied, but there may be limited evidence to differentiate between potential sites.
Fig. 4.
Visual comparison of different modification site assignments. Protein Prospector allows superimposition of multiple interpretations onto the same spectrum. In this example, the reported modification site assignment result is compared with the correct identification. All peaks labeled in red are explained by both peptide interpretations; only the single green y8 2+ ion differentiates between the two interpretations.
The proteomics community has encountered problems with determining the reliability of PTM site assignments, to the extent that some journals currently require annotated spectra for all site assignments (20). Hopefully with site scoring tools such as the one described here, more reliable results will populate the literature, so that biologists can start to unravel the function of different modifications on cellular regulation and signaling.
Acknowledgments
We thank Bernard Küster and his colleagues for making his synthetic phosphopeptide data sets available for others to use for software training.
* This work was supported by the National Center for Research Resources Grant P41 RR001614, the Howard Hughes Medical Institute, and the Vincent J. Coates Foundation.
 This article contains supplemental material and Fig. S1.
This article contains supplemental material and Fig. S1.
1 The abbreviations used are:
- PTM
- post-translational modification
- CID
- collision induced dissociation
- FLR
- false localization rate
- SLIP
- Site Localization In Peptide
- QTOF
- quadrupole time-of-flight.
REFERENCES
- 1. Zhao Y., Jensen O. N. (2009) Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9, 4632–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 3. Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 4. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 5. Nesvizhskii A. I. (2010) A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics 73, 2092–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 7. Beausoleil S. A., Villen J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 8. Bailey C. M., Sweet S. M., Cunningham D. L., Zeller M., Heath J. K., Cooper H. J. (2009) SLoMo: automated site localization of modifications from ETD/ECD mass spectra. J. Proteome Res. 8, 1965–1971 [DOI] [PubMed] [Google Scholar]
- 9. Ruttenberg B. E., Pisitkun T., Knepper M. A., Hoffert J. D. (2008) PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J. Proteome Res. 7, 3054–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. (2011) Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc.. Mass Spectrm. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 12. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 13. Mischerikow N., Altelaar A. F., Navarro J. D., Mohammed S., Heck A. J. (2010) Comparative assessment of site assignments in CID and electron transfer dissociation spectra of phosphopeptides discloses limited relocation of phosphate groups. Mol. Cell Proteomics 9, 2140–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., Burlingame A. L. (2008) In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell Proteomics 7, 2386–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chalkley R. J., Baker P. R., Huang L., Hansen K. C., Allen N. P., Rexach M., Burlingame A. L. (2005) Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer: II. New developments in Protein Prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol. Cell Proteomics 4, 1194–1204 [DOI] [PubMed] [Google Scholar]
- 16. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 17. Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U. S. A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chalkley R. J., Medzihradszky K. F., Lynn A. J., Baker P. R., Burlingame A. L. (2010) Statistical analysis of Peptide electron transfer dissociation fragmentation mass spectrometry. Anal. Chem. 82, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baker P. R., Medzihradszky K. F., Chalkley R. J. (2010) Improving software performance for peptide ETD data analysis by implementation of charge-state and sequence-dependent scoring. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bradshaw R. A., Burlingame A. L., Carr S., Aebersold R. (2006) Reporting protein identification data: the next generation of guidelines. Mol. Cell Proteomics 5, 787–788 [DOI] [PubMed] [Google Scholar]
- 21. Bradshaw R. A., Medzihradszky K. F., Chalkley R. J. (2010) Protein PTMs: post-translational modifications or pesky trouble makers? J. Mass Spectrom. 45, 1095–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Attwood P. V., Besant P. G., Piggott M. J. (2011) Focus on phosphoaspartate and phosphoglutamate. Amino Acids [DOI] [PubMed] [Google Scholar]
- 23. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 24. Tsur D., Tanner S., Zandi E., Bafna V., Pevzner P. A. (2005) Identification of post-translational modifications by blind search of mass spectra. Nat. Biotechnol. 23, 1562–1567 [DOI] [PubMed] [Google Scholar]
- 25. Chu F., Baker P. R., Burlingame A. L., Chalkley R. J. (2010) Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol. Cell Proteomics 9, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]