Summary
Curli are extracellular amyloid fibers produced by Escherichia coli that are critical for biofilm formation and adhesion to biotic and abiotic surfaces. CsgA and CsgB are the major and minor curli subunits, respectively, while CsgE, CsgF, and CsgG direct the extracellular localization and assembly of curli subunits into fibers. The secretion and stability of CsgA and CsgB are dependent on the outer membrane lipoprotein CsgG. Here, we identified functional interactions between CsgG and CsgE during curli secretion. We discovered that CsgG overexpression restored curli production to a csgE strain under curli-inducing conditions. In antibiotic sensitivity and protein secretion assays, CsgG expression alone allowed translocation of erythromycin and small periplasmic proteins across the outer membrane. Co-expression of CsgE with CsgG blocked non-specific protein and antibiotic passage across the outer membrane. However, CsgE did not block secretion of proteins containing a 22 amino acid putative outer membrane secretion signal of CsgA (A22). Finally, using purified proteins, we found that CsgE prohibited the self-assembly of CsgA into amyloid fibers. Collectively, these data indicate that CsgE provides substrate specificity to the curli secretion pore CsgG, and acts directly on the secretion substrate CsgA to prevent premature subunit assembly.
Keywords: E. coli, curli, protein secretion, outer membrane, amyloid
Introduction
Curli represent a novel class of bacterial proteinaceous surface fibers that are produced by enteric bacteria such as Escherichia, Salmonella, and Citrobacter (Zogaj et al., 2003). Curli promote biofilm formation (Vidal et al., 1998), as well as mediate binding to a variety of eukaryotic proteins including fibronectin and MHC class I molecules (Olsen et al., 1989, Olsen et al., 1998). Curli share biochemical and structural properties with disease-associated fibers called amyloids (Chapman et al., 2002). Amyloids are characterized as unusually stable β-sheet rich fibrous protein aggregates that bind to the dyes Congo red and thioflavin T (Nilsson, 2004). Unlike disease-associated amyloid formation, which is the product of protein misfolding, curli assembly is regulated by a dedicated biosynthesis pathway (Romling et al., 1998, Barnhart & Chapman, 2006, Chapman et al., 2002). Therefore, the curli biogenesis pathway provides unique insight into how cells control amyloidogenesis.
The curli specific genes (csg) are present on the divergently transcribed csgBAC and csgDEFG operons (Hammar et al., 1995). CsgA and CsgB constitute the major and minor curli subunits, respectively. The major fiber subunit CsgA interacts with the CsgB nucleator protein at the cell surface and assembles into a highly insoluble and aggregative amyloid fiber (Hammar et al., 1996). The transcriptional activator CsgD is required for expression of the csgBAC operon, while CsgC, CsgE, CsgF, and CsgG comprise the curli subunit assembly machinery (Loferer et al., 1997, Gibson et al., 2007, Salgado et al., Nenninger et al., 2009, Chapman et al., 2002, Hammar et al., 1995, Robinson et al., 2006). The curli assembly machinery functions to restrict curli amyloid formation to the extracellular space, thus minimizing potential cytotoxic effects of intracellular amyloidogenesis. However, the mechanism(s) by which the cell produces and secretes amyloidogenic proteins while preventing their internal assembly has remained elusive.
The 30-kDa lipoprotein CsgG localizes to the outer membrane and in vivo is absolutely required for curli subunit stability and fiber synthesis (Robinson et al., 2006, Loferer et al., 1997). CsgG has periplasmic and extracellular domains, and is spatially clustered in cells at sites of curli assembly (Epstein et al., 2009). Consistent with its proposed role as the secretion channel for curli subunits, CsgG forms an SDS-resistant pore-like multimer at the outer membrane and interacts with the mature N-terminus of the fiber subunit CsgA (Loferer et al., 1997, Robinson et al., 2006). The small chaperone-like proteins CsgE and CsgF modulate the levels of CsgA (Chapman et al., 2002). CsgF is surface-exposed and requires CsgG for its stability and surface localization (Nenninger et al., 2009). Functionally, CsgF is critical for CsgB-mediated nucleation at the cell surface and thus a csgF mutant secretes CsgA away from cells (Chapman et al., 2002, Robinson et al., 2006, Hammer et al., 2007, Nenninger et al., 2009).
The role of CsgE in curli assembly is unclear, although cells lacking csgE display phenotypes similar to a csgG mutant strain. Like the csgG phenotype, deletion of csgE results in white colonies on Congo red indicator agar, and nearly undetectable levels of the CsgG secreted proteins, CsgA and CsgF (assessed previously) and CsgB (assessed in this study) (Chapman et al., 2002, Robinson et al., 2006, Loferer et al., 1997, Nenninger et al., 2009). Unlike the csgG mutant, csgE bacteria can assemble a small amount of CsgA-reactive fibers, with a distinct curved morphology compared to the straight and rigid morphology of wild type curli fibers (Chapman et al., 2002). Importantly, CsgG stability and localization to the outer membrane are unaffected in csgE cells (Epstein et al., 2009).
Since the csgG and csgE phenotypes closely resemble each other, we investigated the role of CsgE in CsgG-mediated secretion. In this work we show that CsgE and CsgG function in the same pathway to support the stability and secretion of CsgA, CsgB and CsgF. Our data indicate that CsgE modulates the pore-like properties of CsgG by acting as a specificity factor in CsgG-mediated translocation. Furthermore, we found that CsgE can inhibit the polymerization of purified CsgA into fibers, suggesting a model for how E. coli can prohibit intracellular amyloid formation.
Results
csgE phenotypes are suppressed by CsgG overexpression
CsgG is required for the localization and stability of CsgB, CsgA and CsgF. Thus, csgG mutants do not assemble curli (Loferer et al., 1997, Nenninger et al., 2009, Robinson et al., 2006, Hammar et al., 1995). csgE mutant phenotypes resemble csgG mutant phenotypes in that they produce little to no detectable CsgA, CsgF or curli fibers (Chapman et al., 2002, Robinson et al., 2006, Nenninger et al., 2009). Therefore, we examined the possibility that increased CsgG expression alleviates the requirement for CsgE in fiber subunit secretion and assembly. To test this hypothesis, we expressed CsgG in the csgE mutant strain at low levels using a medium-copy plasmid containing the csgBA promoter (pLR1, see Table 1), and at high levels using a high-copy overexpression plasmid (pTrc99a). Strains were plated on YESCA agar supplemented with the amyloid-binding dye Congo red (CR), which provides a measure of curli production. As shown in Figure 1A, csgE mutant bacteria without vector or with the empty vectors pLR1 or pTrc99a (csgE, csgE/v and csgE/v+, respectively) were white on a CR-indicator plate as compared to the wild type strain (WT). CR binding was restored to near wild type levels in the csgE mutant strain by low-level expression of CsgE (Fig. 1A, csgE/pE). Similarly, overexpression of CsgG restored wild type levels of CR binding to the csgE mutant (Fig. 1A, csgE/pG+). When CsgG was expressed at low levels, however, little CR binding was observed, suggesting high levels of CsgG expression were required to overcome the csgE mutant phenotype (Fig. 1A, csgE/pG). While low level CsgE expression resulted in approximately wild type levels of CR binding, overexpression of CsgE in the csgE strain resulted in reduced CR binding relative to both the wild type and csgE/pE strains (Fig. 1A, compare WT, csgE/pE and csgE/pE+). CsgF was unable to suppress the csgE mutant phenotype when expressed either at low or high levels (Fig. 1A, csgE/pF and csgE/pF+, respectively).
Table 1.
Strains and plasmids used in this study.
| Strains/ plasmids |
Description | Source/reference |
|---|---|---|
| Strains | ||
| MC4100 | F- araD139 Δ(argF-lac)U169 rspL150(Strr) relA1flbB5301 deoC1 ptsF25 rbsR | (Casadaban, 1976) |
| MHR480 | MC4100 ΔcsgE | (Hammar et al., 1996) |
| LSR11 | MC4100 Δcsg | This study |
| LSR35 | MC4100 csgEF::kanR | This study |
| NEB3016 | E. coli protein expression strain | New England Biolabs |
| Plasmids | ||
| pBAD33 | arabinose-inducible expression vector | (Guzman et al., 1995) |
| pET11d | IPTG-inducible expression vector | Novagen |
| pTrc99a | IPTG-inducible expression vector | Pharmacia Biotech |
| pAN65 | an ORF encoding the CsgA signal sequence fused to mature CpxP with a C-terminal 6-his tag in pBAD33 (CsgAss-CpxP-his) |
This study |
| pAN66 | an ORF encoding the CsgA signal sequence fused to mature CpxP with a C-terminal 6-his tag in pTrc99a (CsgAss-CpxP-his) |
This study |
| pAN69 | an ORF encoding the PapD signal sequence with a Gly-Thr linker (encoded by a ggtacc KpnI site) fused to mature CpxP with a C-teriminal 6-his tag, inserted into the NcoI-PstI sites of pLR92 (PapDss-[GT]-CpxP-his) |
This study |
| pAN70 | an ORF encoding the PapD signal sequence fused to the mature N-terminus of CsgA (A22) with a Gly-Thr linker (encoded by a ggtacc KpnI site) fused to mature CpxP with a C-terminal 6-his tag in pLR92 (PapDss-A22-[GT]-CpxP-his) |
This study |
| pAN87 | papD2-his cloned into the KpnI-PstI sites of pAN69 (PapDss-[GT]-PapD2-his) | This study |
| pAN88 | papD2-his cloned into the KpnI-PstI sites of pAN70 (PapDss-A22-[GT]-PapD2-his) | This study |
| pAN93 | the NcoI-PstI fragment of pAN87 cloned into pTrc99a (PapDss-[GT]-PapD2-his) | This study |
| pAN94 | the NcoI-PstI fragment of pAN88 cloned into pTrc99a (PapDss-A22-[GT]-PapD2- his) |
This study |
| pLR1 | csgBA promoter cloned into the BamHI-PstI sites of pACYC177 | (Robinson et al., 2006) |
| pLR42 | csgE inserted into the KpnI-PstI sites of pBAD33 | This study |
| pLR50 | an ORF encoding the CsgA signal sequence and mature N-terminus (A22) fused to mature CpxP with a C-terminal 6-his tag inserted into the KpnI-PstI sites of pBAD33 (CsgAss-A22-CpxP-his) |
This study |
| pLR51 | the CsgAss-A22-CpxP-his NcoI-PstI fragment of pLR50 inserted into pLR1 | This study |
| pLR58 | csgF-HA in pBAD33 | (Robinson et al., 2006) |
| pLR70 | The csgE NcoI-PstI fragment of pLR42 inserted into pLR1 | This study |
| pLR71 | csgE inserted into the NcoI-BamHI sites of pTrc99a | This study |
| pLR73 | csgF in pLR1 | (Nenninger et al., 2009) |
| pLR74 | csgF inserted into the NcoI-PstI sites of pTrc99a | This study |
| pLR75 | csgF in pBAD33 | (Nenninger et al., 2009) |
| pLR92 | a modified pBAD33 vector (modification: cmR gene does not contain an NcoI site) containing csgG-HA flanked by SacI-NcoI (5′) and KpnI-BglII-Pst1 (3′) sites |
(Robinson et al., 2006) |
| pLR93 | csgG in pLR1 | (Robinson et al., 2006) |
| pLR116 | the CsgAss-A22-CpxP-his NcoI-PstI fragment of pLR50 cloned into pTrc99a | This study |
| pMC1 | csgG in pTrc99a | (Chapman et al., 2002) |
| pNH27 | gene encoding cytoplasmic CsgE inserted into the NcoI-BamHI sites of pET11d | This study |
Figure 1.
Overexpression of CsgG restores curli fiber formation to the csgE mutant.
(A) The following strains were grown on YESCA-Congo red agar for 48 hrs at 26°C: wild type (WT, MC4100); csgE (MHR480); csgE/v (MHR480/pLR1); csgE/pE (MHR480/pLR70); csgE/pF (MHR480/pLR73); csgE/pG (MHR480/pLR93); csgE/v+ (MHR480/pTrc99a); csgE/pE+ (MHR480/pLR71); csgE/pF+ (MHR480/pLR74); csgE/pG+ (MHR480/pMC1). + indicates an overexpression vector.
(B) Negative stain electron micrographs of strains csgE/v (MHR480/pLR1), csgE/pE (MHR480/pLR70), csgE/pE+ (MHR480/pLR71) and csgE/pG+ (MHR480/pMC1) after 48 hrs of growth on YESCA agar at 26°C. Scale bars = 0.5 μm.
(C) Western blot analysis of the same strains as in panel A. Whole cells were collected from YESCA agar after 48 hrs of growth at 26°C, treated with formic acid and analyzed for the presence of CsgA, CsgB, CsgF and CsgG.
To investigate whether the CR binding phenotypes were due to curli production, we performed negative stain electron microscopy (EM) and Western blot analysis on each of the strains in Figure 1A. By negative stain EM, the csgE mutant harboring an empty vector displayed few extracellular fibers, most of which were characterized by a curved morphology (Fig. 1B, csgE/v), similar to what has been previously shown for the csgE mutant alone (Chapman et al., 2002). In contrast, the csgE mutant complemented with either low-level CsgE expression (pE) or high-level CsgG expression (pG+) exhibited extracellular fibers of the same abundance and morphology as wild type curli fibers, suggesting both plasmids were able to rescue the csgE mutant phenotype (Fig. 1B, csgE/pE and csgE/pG+).
By Western blot analysis of whole cell lysates, the csgE mutant contained dramatically decreased levels of CsgA, CsgF and CsgB (Fig. 1C, lane 2). Whole cell levels of CsgA, CsgB and CsgF were restored with low-level expression of CsgE, as well as overexpression of CsgG (Fig. 1C, lanes 4 and 10). Overexpression of CsgE resulted in a phenotype similar to the csgE/pE strain: CsgA, CsgB and CsgF protein levels were largely restored to wild type levels (Fig. 1C, lane 8), fibers were observed by EM (Fig. 1B, csgE/pE+) and very little CsgA was SDS-soluble, indicating that most or all of the CsgA produced by these strains was in an aggregated form (Fig. S1B). However, the csgE/pE+ strain did not exhibit full complementation as measured by CR binding (Fig. 1A). It is possible that fibers produced in the presence of excess CsgE may have subtle biochemical differences such as CR binding capacity. When CsgF was overexpressed in the csgE mutant, no extracellular fibers of any kind could be detected by EM (data not shown) and no CsgA or CsgB protein could be detected by Western analysis, even after prolonged exposure of the blot (Fig. 1C, lane 9), which is consistent previous findings indicating that CsgF and CsgA levels are inversely correlated (Chapman et al., 2002, Nenninger et al., 2009). Similar protein profiles of all ten strains in Figure 1 were obtained when samples containing the bacterial lawn and the underlying agar were analyzed by Western blot, indicating that the absence of CsgA, CsgB and CsgF is the result of protein instability and not due to secretion into the agar media (Fig. S1B and data not shown).
To confirm that CsgA, CsgB and CsgF were surface exposed in the complemented csgE strains (csgE/pE and csgE/pG+) as they are in the wild type strain (Nenninger et al., 2009, Hammar et al., 1996, Bian & Normark, 1997), we performed whole cell proteinase K assays. In both of the complemented csgE strains, CsgA, CsgB and CsgF were susceptible to digestion by proteinase K, while the periplasmic protein DsbA was not, indicating all three curli proteins were surface exposed (Fig. S1A, lanes 3-6). Even the small amounts of CsgA, CsgB and CsgF protein detected in the csgE mutant containing an empty vector were found to be surface exposed (Fig. S1A, lanes 1-2), suggesting that CsgE was not absolutely required for the surface localization of any of the secreted curli proteins. Collectively these data indicate that CsgE is dispensable for curli fiber formation when CsgG is overexpressed, and suggest that CsgE may play the role of an efficiency factor in the CsgG-mediated secretion of curli proteins.
CsgE confers erythromycin resistance to cells overexpressing CsgG
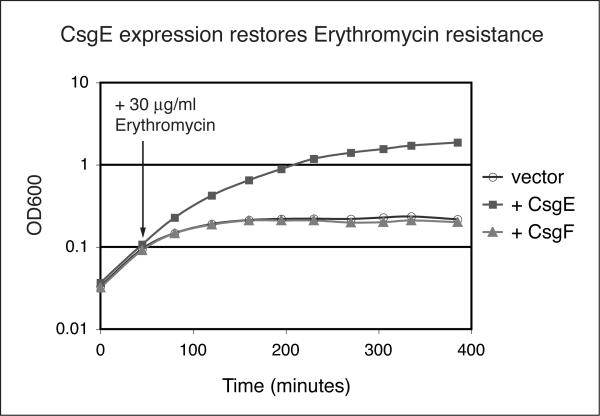
Our initial observations suggested that CsgE might modulate the activity of the CsgG pore. The pore-like properties of CsgG were previously demonstrated, in part, using antibiotic sensitivity assays. In these assays, E. coli containing an empty vector are erythromycin resistant while E. coli overexpressing CsgG are susceptible to erythromycin (Robinson et al., 2006). To determine whether CsgE affects CsgG-mediated erythromycin sensitivity, CsgG was coexpressed with CsgE in the presence of erythromycin. As seen in Figure 2, CsgE expression significantly increased the growth rate of bacteria overexpressing CsgG (“+ CsgE”, filled squares) relative to cells expressing CsgG alone (“vector”, open circles), demonstrating that coexpression of CsgE prevented CsgG-dependent erythromycin sensitivity. Since CsgG levels were similar in all strains as measured by Western blot (data not shown), and we’ve shown previously that CsgG membrane localization is not affected by CsgE (Epstein et al., 2009), this data suggests the CsgG translocon was blocked in the presence of CsgE. Coexpression of CsgF with CsgG had no effect on erythromycin sensitivity (Fig. 2, “+ CsgF”, filled triangles), suggesting the CsgG pore was in an open state in these cells and demonstrating that restoration of erythromycin resistance to CsgG-expressing cells was an activity specific to CsgE.
Figure 2.
Effect of CsgE on erythromycin sensitivity conferred by the overexpression of CsgG. LSR11 (MC4100Δcsg) overexpressing CsgG from pMC1 along with an empty vector (pBAD33, open circles), CsgE (pLR42, filled squares), or CsgF (pLR58, filled triangles) were grown in LB shaking cultures at 37°C with 30 μg/ml erthromycin added 30 min after plasmid induction, and then measured by OD600 at ~40 min intervals.
CsgE blocks non-specific CsgG-mediated secretion
Elucidating the concerted roles of CsgG and CsgE in curli protein secretion is complicated since all three secreted proteins (CsgA, CsgB and CsgF) are undetectable, or nearly so, in strains lacking CsgG or CsgE. Therefore, we developed a CsgG protein secretion assay using small periplasmic proteins not directly related to curli biogenesis that remain detectable in the absence of CsgG or CsgE. In this way we were able to separate the issue of protein stability from protein secretion, allowing us to track CsgG-mediated secretion in the presence and absence of CsgE. In order to find proteins that were suitable CsgG secretion substrates, we screened a number of periplasmic proteins for their ability to be translocated to the supernatant in the presence of CsgG overexpression. Many proteins tested were not amenable to this assay (e.g., PhoA, DsbA, PpiA, PapD, FimC) perhaps because they are too large or tightly folded to pass through the CsgG secretion channel, which is predicted to be approximately 2 nm in diameter (Stathopoulos et al., 1996, Robinson et al., 2006). However, we discovered that domain 2 of the P pilus chaperone, PapD (Bann & Frieden, 2004)(referred to as “PapD2”), as well as the cpx stress response adaptor protein, CpxP (Isaac et al., 2005), were amenable for CsgG-mediated secretion. Both the PapD2 domain (93 amino acids) and CpxP (145 amino acids) are similar in size to the three secreted curli proteins (118-131 amino acids). Overexpression of CsgG from pTrc99a and PapD2 from pBAD33 in liquid cultures of the LSR11 strain (MC4100Δcsg) resulted in secretion of the PapD2 protein to the supernatant (Fig. 3B, lane 2). No PapD2 was detected in the supernatant in the absence of CsgG, even after prolonged exposure of the blot, demonstrating that localization of PapD2 to the supernatant was CsgG-dependent (Fig. 3B, lane 1). Similar results were obtained for the CpxP secretion construct (Fig. S2B). This data suggests that the CsgG translocon is an ungated pore under these experimental conditions, allowing secretion of certain periplasmic proteins that are not associated with curli biogenesis. The idea that overexpressed CsgG forms an ungated pore is consistent with the previous observation that CsgG overexpression renders cells sensitive to the antibiotic erythromycin (Robinson et al., 2006).
Figure 3.
Effects of CsgE on the CsgG-dependent secretion of PapD2 and A22-PapD2.
(A) A schematic diagram of the PapD2 and A22-PapD2 constructs used in the secretion assays. PapD2 consists of the 21 amino acid sec-dependent signal sequence of PapD (ssD) fused to domain 2 of PapD with a C-terminal 6-his tag; domain 2 is the C-terminal 93 amino acids of PapD encompassing the second domain of this 2-domain chaperone. A22-PapD2 consists of the PapD signal sequence (ssD), the N-terminal 22 amino acids of mature CsgA (A22), domain 2 of PapD and a C-terminal 6-his tag. The carat indicates the sec cleavage site.
(B-C) CsgG-dependent protein secretion assay: cells were grown in LB shaking cultures at 37°C to OD600~0.8, protein expression induced for 1 hr, then whole cell and supernatant (or concentrated supernatant) fractions were subjected to Western blot analysis using PapD-specific antisera. < corresponds to the predicted size of ssD-A22-PapD2 (i.e., prior to sec signal cleavage); * indicates a non-specific background band.
(B) The bacterial strain LSR11 (MC4100Δcsg) expressing PapD2 (lanes 1-2; pAN87) or A22-PapD2 (lanes 3-4; pAN88) and an empty vector (lanes 1 and 3; pTrc99A) or CsgG (lanes 2 and 4; pMC1).
(C) The bacterial strain LSR35 (MC4100ΔcsgEF::kanR) expressing CsgG from the chromosome contained the following plasmids: PapD2 (lanes 5-7; pAN93) or A22-PapD2 (lanes 8-10; pAN94); and, empty vector (lanes 5 and 8; pBAD33), CsgE (lanes 6 and 9; pLR42) or CsgF (lanes 7 and 10; pLR75).
(D) Whole cell samples of the strains in panel B and C were subjected to Western blot analysis using CsgG-specific antiseria. Lane numbers correlate to those of panels B and C.
We next determined whether the addition of CsgE altered the secretion profile described above. For these experiments, the strain LSR35 (MC4100ΔcsgEF::kanR) was used such that CsgG was expressed off the chromosome, while the secretion constructs (PapD2 or CpxP) and CsgE could be expressed individually from separate plasmids. Chromosomal CsgG expression in LSR35 was less than plasmid CsgG expression in LSR11 (Fig. 3D, compare lanes 2 and 4 to 5-10), but was sufficient to allow secretion of PapD2 (Fig. 3C, lane 5). When CsgE was coexpressed with PapD2, PapD2 secretion to the supernatant was suppressed (Fig. 3C, compare lanes 5 and 6), indicating that CsgE can inhibit the secretion of non-curli related proteins through the curli translocon. Similar results were obtained for the CpxP construct (Fig. S2C, lane 5). Together with the erythromycin sensitivity assay, these data suggest that CsgE provides a gating mechanism to an otherwise permissive outer membrane pore.
The N-terminus of CsgA (A22) is a curli-specific outer membrane secretion signal
Our results thus far have identified CsgE as a potential specificity factor for CsgG-mediated protein secretion, as it blocks secretion of proteins unrelated to curli biogenesis. We reasoned that the secreted curli proteins might contain a curli-specific outer membrane secretion signal that would allow secretion of curli proteins, similar to the sec signal sequence that directs proteins across the inner membrane through the Sec translocon. We previously demonstrated that a fusion protein containing the N-terminal 22 amino acid residues of mature CsgA (A22) specifically interacts with CsgG (Robinson et al., 2006). Further, A22 is not thought to participate in the core structure of curli fibers (Collinson et al., 1999), making it a prime candidate for a curli-specific secretion signal. Therefore we created an A22 fusion protein with PapD2 (Fig. 3A) to test the hypothesis that A22 is sufficient to allow secretion past the CsgE gate and through the CsgG pore. Like PapD2, A22-PapD2 was secreted to the supernatant in a CsgG-dependent manner in LSR11 cells expressing CsgG from a plasmid (Fig. 3B, lane 4). No A22-PapD2 could be detected in the supernatant in the absence of CsgG (Fig. 3B, lane 3). A22-PapD2 was also secreted to the supernatant in LSR35 cells expressing CsgG from the chromosome (Fig. 3C, lane 8). However, in contrast to the PapD2 construct whose secretion was suppressed in the presence of CsgE, secretion of A22-PapD2 was not changed with CsgE expression. The pore selectivity function is specific to CsgE, as coexpression of CsgF decreased secretion of both PapD2 and A22-PapD2 relative to the vector control (Fig. 3C, compare lanes 7 to 5 and 10 to 8). This result is not unexpected, as CsgF itself is known to be secreted through CsgG and would thus compete with other potential secretion substrates (such as PapD2 and A22-PapD2) for engagement with the CsgG secretion complex (Chapman et al., 2002, Nenninger et al., 2009, Hammer et al., 2007). As before, similar results were obtained with an A22-CpxP fusion protein (Fig. S2B-C). These data suggest that the A22 domain of CsgA is a sufficient specificity signal to allow protein secretion through a CsgE-gated CsgG pore.
CsgE inhibits self-assembly of CsgA into amyloid fibers
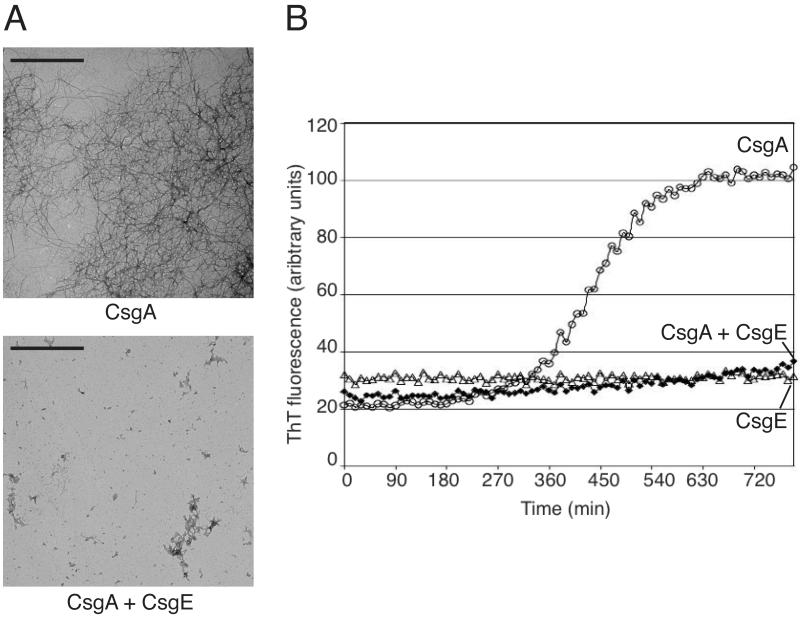
One outstanding question in the biosynthesis of curli fibers is how cells prevent amyloid formation on the periplasmic side of the curli assembly apparatus. The observation that CsgE provided substrate specificity to CsgG secretion raised the possibility that CsgE may also act on the secretion substrates themselves. Furthermore, CsgE appears to modulate the ultrastructural or biochemical properties of curli, as the absence of CsgE (csgE/v) produced fewer fibers of aberrant morphology relative to wild type (Fig. 1B), and the overexpression of CsgE (csgE/pE+) results in reduced CR binding despite having similar levels of polymerized CsgA to the csgE/pE strain (Fig. 1A and S1B). This may be due to interactions with, or modifications of, pre-assembled CsgA. We were unable to detect a direct protein-protein interaction between CsgA and CsgE by immunoprecipitation from cell lysates or by far-Western analysis (data not shown). However, we went on to examine the effect of CsgE on in vitro polymerization of CsgA. CsgA is soluble and unstructured immediately following purification, but after incubation for several hours CsgA begins to self-assemble into β-sheet rich amyloid fiber aggregates (Chapman et al., 2002). In vitro polymerization of CsgA can be monitored over time by the appearance of fibers by EM, and by characteristic reactivity with thioflavin T (ThT). Freshly purified CsgA was incubated with or without purified CsgE, as described in Experimental Procedures. As previously shown, CsgA incubated alone polymerized into large collections of 4- to 6-nm wide amyloid fibers (Fig. 4A, top)(Chapman et al., 2002, Wang et al., 2007). When incubated in a near 1:1 molar ratio with CsgE, however, CsgA failed to assemble into fibers after 24 hours; only a few, small, amorphous aggregates could be detected in this reaction (Fig. 4A, bottom). CsgA polymerization with or without CsgE was also monitored by a real-time ThT fluorescence assay. A reaction containing 25 μM CsgA alone exhibited exponential growth after 270 minutes of incubation at room temperature (Fig. 4B, open circles), while a reaction containing 25μM CsgA and 22.5 μM of CsgE did not exhibit substantial ThT reactivity after 800 minutes of incubation (Fig. 4B, filled diamonds). Reactions containing CsgE alone showed no increase in ThT fluorescence in this assay (Fig. 4B, open triangles). As a control, strains expressing an empty vector instead of a vector containing CsgE were subjected to the same CsgE purification protocol, and the fractions corresponding to the CsgE fractions (which contain similar contaminating bands by Coommassie stain as that of a CsgE purification) were added to 25 μM CsgA. This resulted in a similar ThT profile to that of CsgA alone, suggesting that inhibition of CsgA polymerization was CsgE specific (data not shown). Purified CsgE added to pre-formed CsgA fibers did not depolymerize CsgA into monomers, as no change in ThT fluorescence was observed for CsgA fibrils incubated with purified CsgE over a 24 hr time period (data not shown). These results suggest that, in vitro, CsgE can prevent the self-assembly of soluble CsgA into amyloid fibers.
Figure 4.
CsgE prevents self-assembly of CsgA into amyloid fibers.
(A) Freshly purified CsgA was incubated alone (top) or in the presence of CsgE in a near 1:1 molar ratio (bottom) for 24 hours before visualization with transmission electron microscopy (TEM). Scale bars equal 1 μm.
(B) Real-time monitoring of CsgA self-assembly into amyloid fibers by ThT fluorescence. 25 μM CsgA was incubated in reaction buffer containing ThT alone (open circles) or with 22.5 μM CsgE (filled diamonds). 22.5 μM CsgE was also incubated alone in ThT reaction buffer (open triangles).
Discussion
While protein secretion across the inner membrane is largely carried out by the Sec channel, gram-negative bacteria employ numerous secretion systems to translocate proteins across the outer membrane (Gerlach & Hensel, 2007, Saier, 2006, Filloux et al., 2008). During curli biogenesis, at least three curli-related proteins are translocated across the outer membrane in a CsgG-dependent fashion: CsgB, CsgA and CsgF. In the absence of CsgG, these proteins are not secreted and no curli are assembled (Hammar et al., 1995, Loferer et al., 1997, Robinson et al., 2006, Nenninger et al., 2009). Likewise, csgE mutants assemble fewer curli than WT cells and secrete less CsgB, CsgA and CsgF (Fig. 1)(Chapman et al., 2002, Nenninger et al., 2009). Here, we found evidence that CsgE promotes efficient subunit secretion, gates the outer membrane pore, and prevents aggregation of the major fiber subunit protein, CsgA.
CsgG is a limiting factor in the stability of CsgA, CsgB and CsgF, as CsgG overexpression increases the steady-state levels of these proteins (Loferer et al., 1997, Nenninger et al., 2009, Robinson et al., 2006). We found that CsgG overexpression also suppresses the csgE mutant defect and restores the stability of all three CsgG-secreted proteins and the assembly of curli fibers (Fig. 1). Thus CsgE is required for curli fiber biogenesis when CsgG is expressed at physiological levels, but not when CsgG is artificially overexpressed. This suggests that CsgE might act as an efficiency factor for CsgG function. CsgG is functionally similar to both ushers and secretin-like proteins, which form outer membrane channels that support protein secretion to the extracellular milieu (Thanassi, 2002). Secretin-like pores are often dependent on small, periplasmic pilot proteins for their localization to the outer membrane and stability (Hardie et al., 1996a, Shevchik & Condemine, 1998, Crago & Koronakis, 1998, Hardie et al., 1996b). CsgE does not change CsgG steady state levels or localization to the outer membrane (Fig. 1)(Epstein et al., 2009). Because CsgE does not modulate CsgG stability or localization to the outer membrane, its role in supporting subunit secretion is apparently different than that of secretin pilot proteins but may play a role in gating and ungating CsgG.
To better define the role of CsgE in curli protein secretion, we developed a CsgG-dependent secretion assay using non-curli proteins as secretion cargo – PapD2 and CpxP. Unlike the native curli secretion substrates (CsgA, CsgB and CsgF), PapD2 and CpxP are stable in the periplasm in the absence of CsgG or CsgE; like the curli secreted proteins, however, both are secreted to the culture supernatant in a CsgG-dependent manner. In this way we were able to separate protein stability and protein secretion, which are inseparable for the native substrates. The secretion assays show that CsgE is able to restrict CsgG-dependent secretion of non-curli substrates while not affecting the secretion of substrates tagged with a putative curli secretion signal of CsgA, A22, suggesting that CsgE confers substrate selectivity to the CsgG pore (Fig. 3 and S2). The A22 domain of CsgA was sufficient to provide a curli-specific signal and allow passage past the CsgE gate. However, this does not exclude the involvement of other domains of CsgA during secretion. Overexpression of A22-fusion proteins did not exhibit dominant negative activity in wild type MC4100 cells (Fig. S3), suggesting an abundance of the A22 domain did not interfere with the secretion of wild type curli proteins. CsgB and CsgF are also secreted to the cell-surface in a CsgG-dependent manner, but we have not yet identified domains in CsgB or CsgF that function in a similar manner to the A22 domain of CsgA. Further work is necessary to determine if all three known extracellular curli proteins are secreted by the same or different mechanisms.
Although overexpression of CsgG mostly suppresses the csgE phenotype, curli formation does not appear as efficient when compared to WT. When CsgG is overexpressed in a csgE strain, an increased proportion of unpolymerized CsgA can be detected in the underlying agar compared to WT cells (Fig. S1B). Further, we observed less protease-resistant CsgB in the csgE/pG+ strain compared to csgE/pE (see Fig. S1A, compare lanes 4 and 6). This may be an indication that less CsgB is in a protease-resistant amyloid-like conformation, and also supports the idea that a portion of curli subunits fail to polymerize in the csgE/pG+ strain, resulting in inefficient curli formation.
The suggestion that CsgE participates in gating the outer membrane pore is supported by our finding that CsgE restores erythromycin resistance to cells rendered erythromycin sensitive by CsgG overexpression (Fig. 2). Thus, CsgE prevents non-specific translocation of small proteins and large molecules through the CsgG pore, providing a gate to an otherwise permissive outer membrane channel. Other outer membrane pores also require gating mechanisms in order to maintain membrane integrity and pore specificity, such as FepA and PapC (Liu et al., 1993, Remaut et al., 2008, Buchanan et al., 1999). The recently solved crystal structure of the PapC usher protein showed that PapC contains a folded plug domain that occludes the central channel to prevent non-specific translocation (Remaut et al., 2008). Conformational changes of the usher during pilus biogenesis results in a reorganization of the plug, thus ungating the pore, allowing subunit translocation across the usher (Phan et al., in press). In a similar manner, several members of the secretin family are thought to contain plug domains that fold back into and occlude the channel and thus provide the gating mechanism (PulD, XcpQ, OutD and possibly TcpC) (Bose & Taylor, 2005, Nouwen et al., 2000, Brok et al., 1999, Shevchik et al., 1997, Nouwen et al., 1999). Thus, the CsgG-CsgE pore gating mechanism represents a variation of the theme for most ushers and secretins that are self-gating. It remains possible that CsgG contains a plug or gating domain itself, but requires CsgE to fully engage this domain such that it properly functions as a selectivity barrier. An interesting comparison can be made between the CsgGE curli secretion complex and outer membrane components of the type IV bundle forming pilus (bfp) system of EPEC. Like CsgG, the BfpB secretin is a lipoprotein that does not require a pilot protein for stability or outer membrane targeting, but does have a small interacting protein, BfpG, that is not a lipoprotein. It has been proposed that BfpG functions as a gate to the BfpB pore, but this has not been directly tested (Daniel et al., 2006, Schmidt et al., 2001).
In addition to providing a gating mechanism, CsgE may possess a second role in curli biogenesis. We found that purified CsgE was able to prevent the polymerization of purified CsgA in an in vitro reaction. This suggests a potential chaperone-like role for CsgE with respect to CsgA. If biologically relevant, this activity in vivo could ensure that secretion of soluble CsgA precedes fiber formation, thereby preventing formation of periplasmic amyloid oligomers or fibers and abrogating intracellular amyloid toxicity. This activity would also promote secretion efficiency, as the CsgG pore could more easily accommodate a CsgA monomer rather than an aggregate. This is analogous to the role of the periplasmic chaperones of the chaperone-usher pathway (CUP) of pilus biogenesis. Pilus chaperones cap the interactive surfaces of the subunits, to prevent nonproductive interactions of subunits in the periplasm (Barnhart et al., 2000, Sauer et al., 1999, Sauer et al., 2000). Specific targeting of chaperone/subunit complexes to the usher promotes conformational changes resulting in the opening of the usher gate and translocation across the usher and assembly of subunits (Kuehn et al., 1991, Jones et al., 1997, Bullitt et al., 1996, Dodson et al., 1993). Amyloid inhibition has been described for a number of small heat shock proteins (e.g., HSP-16 and α-Crystallin) and an abundant cerebrospinal fluid protein, lipocalin-type prostaglandin D synthase, all of which inhibit aggregation of the clinically important Aβ peptide involved in the pathogenesis of Alzheimer’s disease (Fonte et al., 2002, Fonte et al., 2008, Hatters et al., 2001, Tanaka et al., 2008, Kanekiyo et al., 2007). While an exciting possibility, whether the activity that we observed for CsgE in vitro also occurs in vivo remains to be determined. Certainly, there are a number of bacterial periplasmic chaperones (FkpA, Skp and chaperones of CUP systems), that prevent the aggregation and facilitate the folding of periplasmic proteins (Walton & Sousa, 2004, Sauer et al., 2004, Ramm & Pluckthun, 2000). It would not be surprising, then, that the curli assembly apparatus includes a protein that prevents premature interactions of a highly aggregative protein, CsgA, until it is successfully delivered to and exported through its outer membrane secretion pore, CsgG. Given the rapid and toxic nature of amyloid polymerization reactions, there are likely additional control mechanisms in place in order to prevent periplasmic curli subunit aggregation. These mechanisms could be at the transcriptional, translational or post-translational levels. Results from reporter fusion assays show only a small decrease in csgBA translation in the absence of CsgG, suggesting the majority of this control occurs at the post-translational level (Loferer et al., 1997). Indeed, we have never observed periplasmic intermediates of either curli subunit, including in the csgE mutant, nor have we observed intracellular aggregates or fibers, suggesting that in the absence of secretion, curli subunits are very efficiently degraded.
In stark contrast to disease-associated amyloidogenesis that underlies such neurodegenerative aliments like Alzeihmer’s and Parkinson’s disease, assembly of curli fibers is accomplished in a directed manner, without associated cellular toxicity. The curli assembly machine functions to guide the secretion of its amyloidogenic subunits across the outer membrane such that fiber formation is promoted on the cell surface, and restricted in the periplasm (Chapman et al., 2002, Nenninger et al., 2009, Robinson et al., 2006, Loferer et al., 1997, Hammar et al., 1996, Hammer et al., 2007). These opposing tasks appear to be executed by the two small chaperone-like proteins of curli biogenesis, CsgE and CsgF. Future definition of the molecular mechanisms of CsgE and CsgF will increase our understanding of how amyloid formation is controlled, a biological problem that E. coli has solved in the form of this unique biogenesis pathway.
Experimental Procedures
Bacterial Strains and Plasmids
Bacterial strains and plasmids used in this study can be found in Table 1. Primers used to make genetic constructs can be found in Table 2.
Table 2.
Primers used in this study.
| Primer name | Primer sequence |
|---|---|
| csgE KO F no FRT | AGCGGTTTCCTGGGCAAACGATAACCTCAGGCGATAAAGCCACGCTGCCGCAAGCACT C |
| csgF KO R no FRT | ATAAGCGCTGCATGATTATTTTCCTTATGAAGCTGGGGAATAGGAACTTCAAGATCC |
| csgA F Kpn-RBS-Nco | GTTTGGTACCACACAGGAAACAGACCATGGCGAAACTTTTAAAAGTAGCAGCAATTGC AGC |
| csgA-cpxP R | CGCCTGAACCGACTTCAGCATTTGGGCCGCTATTATTACC |
| csgA-cpxP F | GGTAATAATAGCGGCCCAAATGCTGAAGTCGGTTCAGGCG |
| cpxP his R Pst (LR) | GTTTCTGCAGTTAGTGGTGGTGGTGGTGGTGCTGGGAACGTGAGTTGCTAC |
| csgE F Kpn-RBS-Nco | GTTTGGTACCACACAGGAAACAGACCATGGCGAAACGTTATTTACGCTGGATTGTGGC |
| csgE R 3Xs-Pst | GTTTCTGCAGTTATTAGATCCTTAGAATTCATCATGCGCCAAATCGCCC |
| csgE R BamHI | GTTTGGATCCTTAGAATTcATCATGCGCCAAATCGCC |
| csgF F Nco | GTTTCCATGGCACGTGTCAAACATGCAGTAG |
| csgF R Pst | GTTTCTGCAGTTAAAAATCGGTTGAGTTATTTTGTAAACC |
| a22cpxP F delA22 | CGGTAGCGCTCTGGCAGCTGAAGTCGGTTC |
| a22cpxP R delA22 | GAACCGACTTCAGCTGCCAGAGCGCTACCG |
| papDss(ala) KpnI mcpxP F NcoI |
CGAGCCATGGCGATTCGAAAAAAGATTCTGATGGCTGCCATCCCCCTGTTTGTTATATC CGGGGCAGACGCTGGTACCGCTGAAGTCGGTTCAGGCGATAAC |
| cpxP his R PstI (AN) | CTCGCTGCAGTTAGTGGTGGTGGTGGTGGTGCTGGGAACGTGAGTTGCTACTACT |
| papDss(ala) mcsgA F NcoI |
CGAGCCATGGCGATTCGAAAAAAGATTCTGATGGCTGCCATCCCCCTGTTTGTTATATC CGGGGCAGACGCTGGTGTTGTTCCTCAGTACGGCGG |
| csgA22 R KpnI | CGAGGGTACCATTTGGGCCGCTATTATTACCGCCACC |
| papD2 F KpnI | CGACGGTACCGAAGTATGGCAGGACCAGTTAATTCTG |
| papD2-his R PstI | CGACCTGCAGTTAGTGGTGGTGGTGGTGGTGTTTCTCTTTTTTCACAGAGCAACGGCTA C |
| Sec- CsgE For NcoI | GCGTTTCCATGGCCGTTGAGGTAGAAGTCCCGGGA |
| CsgE 6x His Rev BamHI |
GTTTAAAGCTTGGATCCTTAGTGATGGTGATGGTGATGGAATTCATCATGCGCCAAATC |
Strain construction
LSR11 (MC4100 Δcsg) was created by removing the kanR cassette from LSR5 (MC4100 Δcsg::kanR)(Chapman et al., 2002) using FLP recombinase according to standard techniques (Datsenko & Wanner, 2000). LSR35 was constructed by deleting the csgEF locus with a PCR product encoding the kanR cassette from pKD13 using the phage λ Red recombinase method (Datsenko & Wanner, 2000). The resistance cassette was amplified from pKD13 with the primers csgE KO F no FRT and csgF KO R no FRT.
Plasmid construction
Unless otherwise indicated, chromosomal DNA from MC4100 was used as a template in PCRs. pLR50 containing the CsgAss-A22-CpxP-his fusion was synthesized by overlap PCR. An N-terminal portion encoding residues 1-42 of premature CsgA and the first 20 residues of mature CpxP was amplified with the primers csgA F Kpn-RBS-Nco and csgA-cpxP R. A C-terminal region encoding residues 36-42 of CsgA and his-tagged mature CpxP was amplified with the primers csgA-cpxP F and cpxP his R Pst (LR). The two regions were fused together in a third PCR, and the product was cloned into the Kpn1-Pst1 sites of pBAD33. pLR116 and pLR51 were made by subcloning the NcoI-PstI fragment from pLR50 into pTrc99a and pLR1, respectively. pAN65 and pAN66 were made by deleting the region encoding the CsgA A22 domain using site directed mutagenesis (Stratagene QuickChange protocol) of pLR50 and pLR116, respectively, with the primers a22cpxP F delA22 and a22cpxP R delA22. pAN69 was made by ligating the PCR product of primers papDss(ala) KpnI mcpxP F NcoI and cpxP his R PstI (AN) and template pAN66 into the NcoI-PstI sites of pLR92. pAN70 was created by ligating the PCR product of primers papDss(ala) mcsgA F NcoI and csgA22 R KpnI and template pLR116 into the NcoI-KpnI sites of pAN69. pAN87 and pAN88 were made by ligating the PCR product of primers papD2 F KpnI and papD2-his R PstI into the KpnI-PstI sites of pAN69 and pAN70, respectively. pAN93 and pAN94 were made by subcloning the NcoI-PstI fragments of pAN87 and pAN88, respectively, into pTrc99a. pLR42 was made by ligating the PCR product of csgE F Kpn-RBS-Nco and csgE R 3XS-Pst into the KpnI-PstI sites of pBAD33. pLR70 was created by subcloning the NcoI-PstI fragment of pLR42 into pLR1. pLR71 was made by ligating the PCR product of csgE F Kpn-RBS-Nco and csgE R BamHI into the NcoI-BamHI sites of pTrc99a. pLR74 was made by ligating the PCR product of csgF F Nco and csgF R Pst into the NcoI-PstI sites of pTrc99a. pNH27 encodes cytoplasmic CsgE (lacking the sec signal sequence) with a C-terminal 6-his tag and was cloned into the NcoI-BamHI sites of pET11d using the primers Sec- CsgE For NcoI and CsgE 6x His Rev BamHI.
Growth Conditions
Chromosomal curli expression was induced by growing bacteria on YESCA agar (per liter: 10 g Casamino acids, 1 g yeast extract, 20 g agar; for CR-YESCA agar, supplement with 50 μg/ml Congo red) for 48 hours at 26°C (Fig. 1). Plasmid gene expression in liquid cultures (Figs. 2, 3, S2) was induced with IPTG (0.1-0.5 mM) for pTrc99a-based vectors and arabinose (0.05-0.2% (w/v)) for pBAD33-based vectors in LB broth, shaking at 37°C. Inducer concentrations were the same within individual experiments, except for Fig. S2B, where pLR50 strains were induced with 0.001% ara and pAN65 strains induced with 0.025% ara; this was to equalize expression between the A22-CpxP-his and CpxP-his constructs, since CpxP-his expression levels were typically lower than A22-CpxP-his. For maintenance of plasmids in broth cultures, bacteria were grown with 100 μg/ml ampicillin, and 34 μg/ml chloramphenicol. LSR35 cultures were further supplemented with 10 μg/ml kanamycin.
Electron microscopy
Electron microscopy was performed with a JEOL 1200 EX II transmission electron microscope. For whole bacteria, cells were grown on YESCA for 48 hours and then resuspended in PBS and fixed with 1% glutaraldehyde. Fixed cells or purified protein were applied to Formvar-coated copper grids for 1-2 minutes. Specimens were then stained with 0.2% uranyl acetate for 1-2 minutes prior to visualization.
Western blot analysis
Sample preparation
For whole cell Western blot in Figure 1, bacteria were scraped from YESCA plates, resuspended in PBS, and normalized by OD600. A cell suspension volume corresponding to 1 optical density unit (ODU = 1 ml of OD600=1.0) was collected for each sample (i.e., whole cells + PBS, so that no protein was lost by pelleting of the bacteria and aspiration of PBS). For samples not pre-treated with formic acid (FA), cell suspensions were brought to 200 μl using SDS loading buffer. For FA treated samples, FA was added to cell suspensions (final = 70% or greater FA), the acid evaporated in a vacuum centrifuge, the pellet resuspended in 200 μl SDS sample buffer and pH adjusted with 1N NaOH, if necessary. For plug samples (Fig. S1B), a circular plug (d=8mm) was cut from the agar and collected. +/− FA treatment of plugs was as above for whole cells, except that FA treated plugs were solubilized in 100μl of 96% FA. All other samples for Western blot were prepared as indicated.
Western blotting
All samples were boiled for 5 min prior to SDS-PAGE in 15% acrylamide gels; resolved proteins were transferred to nitrocellulose membrane at 4°C in 25mM CAPS, pH 11.2 (50 V for 3 hrs or 12 V overnight). Blocking: RT 2h with rocking in 1X TBST, 1.5% milk, 1.5% BSA. Primary antibodies and their respective dilutions can be found in Table 3. Horseradish peroxidase conjugated secondary antibody (Pierce): RT 1h at 1:5000-1:10,000 dilution in blocking buffer. Detection: Supersignal West Femto chemiluminescent substrate (Pierce). The PapD antibody was raised in rabbits against purified wild type PapD protein (SigmaGenosys custom antisera production).
Table 3.
Antibodies used in this study.
| Antibodies | Description | Dilution used in Western blot |
Source/Reference |
|---|---|---|---|
| α-CsgA | Polyclonal rabbit antiserum to CsgA | 1:5000-1:10,000 | (Barnhart et al., 2006) |
| α CsgB | Polyclonal rabbit antiserum to the peptide (EGSSNRAKIDQTGDY) of CsgB |
1:2000 | (Robinson et al., 2006) |
| α-CsgF | Polyclonal rabbit antiserum to CsgF | 1:5000-1:10,000 | (Nenninger et al., 2009) |
| α-CsgG | Polyclonal rabbit antiserum to CsgG | 1:5000 | (Robinson et al., 2006) |
| α-DsbA | Polyclonal rabbit antiserum to DsbA | 1:20,000 | Kind gift of the James Bardwell Laboratory, University of Michigan |
| α-PapD | Polyclonal rabbit antiserum to PapD | 1:5000 | This study |
| α-CT-His | Mouse anti- C-terminal his tag | 1:5000 | Invitrogen |
| α CpxP | Polyclonal rabbit antiserum to CpxP | 1:10,000 | Kind gift of the Tracy Raivio Laboratory, University of Alberta |
Whole Cell Proteinase K (PK) Treatment
Intact cells were scraped from YESCA agar, normalized by OD600 and 2 ODUs of each strain collected, as described in Western blot analysis. 2 ODUs of cells were brought to 270μl with 1X PBS. 30μl of water or a 10X protease stock (1 mg/ml PK) in water was then added and vortexed to begin the reaction. After incubation for 2 hr at 37°C, the reaction was quenched with 2mM PMSF. Cells were pelleted; cell pellets were treated with 100 μl 96% formic acid and prepared for immunoblotting as described in Western blot analysis.
Antibiotic sensitivity assay
E. coli strain LSR11 containing pMC1 (csgG in pTrc99a) and pBAD33 (vector), pLR42 (csgE in pBAD33) or pLR58 (csgF in pBAD33) was diluted 1:500 from overnight cultures and grown to an OD600 of 0.05 in LB broth at 37°C shaking. Plasmid gene expression was induced with 0.1 mM IPTG and 0.04% arabinose. 30 min later, 30 μg/ml of erythromycin was added to the media. OD600 measurements were then recorded at 40 min intervals for 6 hrs.
CsgG-dependent Secretion Assays
After growth and induction (described in Growth Conditions) of the indicated strains, cultures were normalized by OD600 and whole cell pellet and supernatant samples taken for Western blot analysis. For the secretion assay presented in Figure 3C and S2C, supernatants were concentrated as follows: supernatants were obtained by pelleting cells (3600 rpm 10 min), then filter-sterilizing (0.22 μm) the spent media; a metal affinity resin (Clontech, Talon®) was added to the filter-sterilized supernatant and allowed to incubate overnight at 4°C with rocking; the resin was then collected, washed 3x in 1X PBS buffer, resuspended in SDS sample buffer and boiled for 5 min prior to gel loading. Filtered supernatant volumes used for the pull-down were also normalized by OD600 of the corresponding induced culture.
Purification of CsgE
CsgE-his was expressed from pNH27 in strain NEB 3016 with 100 μg/ml ampicillin. Cells were grown to late log phase and induced for 1 hr with 0.5mM IPTG. Cells were harvested by centrifugation at 5,000×g for 20 minutes and resuspended in lysis buffer (50 mM KPi pH 7.3, 100 mM NaCl, 10 mM 2-mercaptoethanol, 0.1% Tween-20, 1U/mL DNase, 1mM PMSF). Cells were lysed using a French Press. The lysate was centrifuged at 10,000×g for 20 minutes. The supernatant was incubated with HIS-Select HF Nickel Affinity Gel (Sigma, H0527) rotating at 4°C for 1 hour and applied to a 5mL column (Thermo Scientific, 29922). The column was washed with lysis buffer, followed by 12.5 mM Imidazole in 50mM KPi pH 7.3. CsgE-his was eluted with 250mM imidazole in 50mM KPi pH 7.3 and then dialyzed overnight at 4°C against 50mM KPi pH 7.3, 100mM NaCl, 1mM PMSF. Precipitated proteins were removed by centrifugation. The same expression and purification strategy was carried out for NEB 3016 containing an empty expression vector (pET11d), and the dialyzed eluate from this purification was used as a negative control for the CsgA polymerization assay (see Real time CsgA polymerization assay).
Real time CsgA polymerization assay
Experiments were carried out as previously described (Wang et al., 2007, Hammer et al., 2007), except that purified CsgE-his or a negative control (see Purification of CsgE) were added to freshly purified, monomeric CsgA prior to monitoring of thioflavin T fluorescence.
Supplementary Material
Acknowledgements
We are extremely grateful to Julia Wong and Tracy Raivio (University of Alberta) for providing purified CpxP antisera, to the Bardwell laboratory for providing the DsbA antisera and to members of the Hultgren and Chapman labs for the helpful discussions during the writing of this manuscript. This work was supported by NIH AI48689 (S.J.H.) and NIH AI073847 (M.R.C.).
References
- Bann JG, Frieden C. Folding and domain-domain interactions of the chaperone PapD measured by 19F NMR. Biochemistry. 2004;43:13775–13786. doi: 10.1021/bi048614u. [DOI] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Lynem J, Chapman MR. GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J Bacteriol. 2006;188:5212–5219. doi: 10.1128/JB.00234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Pinkner JS, Soto GE, Sauer FG, Langermann S, Waksman G, Frieden C, Hultgren SJ. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc Natl Acad Sci U S A. 2000;97:7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. Embo J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose N, Taylor RK. Identification of a TcpC-TcpQ outer membrane complex involved in the biogenesis of the toxin-coregulated pilus of Vibrio cholerae. J Bacteriol. 2005;187:2225–2232. doi: 10.1128/JB.187.7.2225-2232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brok R, Van Gelder P, Winterhalter M, Ziese U, Koster AJ, de Cock H, Koster M, Tommassen J, Bitter W. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J Mol Biol. 1999;294:1169–1179. doi: 10.1006/jmbi.1999.3340. [DOI] [PubMed] [Google Scholar]
- Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- Bullitt E, Jones CH, Striker R, Soto G, Jacob-Dubuisson F, Pinkner J, Wick MJ, Makowski L, Hultgren SJ. Development of pilus organelle subassemblies in vitro depends on chaperone uncapping of a beta zipper. Proc Natl Acad Sci U S A. 1996;93:12890–12895. doi: 10.1073/pnas.93.23.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J Mol Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- Crago AM, Koronakis V. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol. 1998;30:47–56. doi: 10.1046/j.1365-2958.1998.01036.x. [DOI] [PubMed] [Google Scholar]
- Daniel A, Singh A, Crowther LJ, Fernandes PJ, Schreiber W, Donnenberg MS. Interaction and localization studies of enteropathogenic Escherichia coli type IV bundle-forming pilus outer membrane components. Microbiology. 2006;152:2405–2420. doi: 10.1099/mic.0.28860-0. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson KW, Jacob-Dubuisson F, Striker RT, Hultgren SJ. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci U S A. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EA, Reizian MA, Chapman MR. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J Bacteriol. 2009;191:608–615. doi: 10.1128/JB.01244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- Fonte V, Kapulkin V, Taft A, Fluet A, Friedman D, Link CD. Interaction of intracellular beta amyloid peptide with chaperone proteins. Proc Natl Acad Sci U S A. 2002;99:9439–9444. doi: 10.1073/pnas.152313999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonte V, Kipp DR, Yerg J, 3rd, Merin D, Forrestal M, Wagner E, Roberts CM, Link CD. Suppression of in vivo beta-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J Biol Chem. 2008;283:784–791. doi: 10.1074/jbc.M703339200. [DOI] [PubMed] [Google Scholar]
- Gerlach RG, Hensel M. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297:401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Gibson DL, White AP, Rajotte CM, Kay WW. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology. 2007;153:1131–1140. doi: 10.1099/mic.0.2006/000935-0. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A. 2007;104 doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie KR, Lory S, Pugsley AP. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. Embo J. 1996a;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- Hardie KR, Seydel A, Guilvout I, Pugsley AP. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol Microbiol. 1996b;22:967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- Hatters DM, Lindner RA, Carver JA, Howlett GJ. The molecular chaperone, alpha-crystallin, inhibits amyloid formation by apolipoprotein C-II. J Biol Chem. 2001;276:33755–33761. doi: 10.1074/jbc.M105285200. [DOI] [PubMed] [Google Scholar]
- Isaac DD, Pinkner JS, Hultgren SJ, Silhavy TJ. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc Natl Acad Sci U S A. 2005;102:17775–17779. doi: 10.1073/pnas.0508936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Danese PN, Pinkner JS, Silhavy TJ, Hultgren SJ. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. Embo J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo T, Ban T, Aritake K, Huang ZL, Qu WM, Okazaki I, Mohri I, Murayama S, Ozono K, Taniike M, Goto Y, Urade Y. Lipocalin-type prostaglandin D synthase/beta-trace is a major amyloid beta-chaperone in human cerebrospinal fluid. Proc Natl Acad Sci U S A. 2007;104:6412–6417. doi: 10.1073/pnas.0701585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Normark S, Hultgren SJ. Immunoglobulin-like PapD chaperone caps and uncaps interactive surfaces of nascently translocated pilus subunits. Proc Natl Acad Sci U S A. 1991;88:10586–10590. doi: 10.1073/pnas.88.23.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rutz JM, Feix JB, Klebba PE. Permeability properties of a large gated channel within the ferric enterobactin receptor, FepA. Proc Natl Acad Sci U S A. 1993;90:10653–10657. doi: 10.1073/pnas.90.22.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley AP. Secretin PulD: association with pilot PulS, structure, and ion- conducting channel formation. Proc Natl Acad Sci U S A. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen N, Stahlberg H, Pugsley AP, Engel A. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. Embo J. 2000;19:2229–2236. doi: 10.1093/emboj/19.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Olsen A, Wick MJ, Morgelin M, Bjorck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect Immun. 1998;66:944–949. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan G, Remaut H, Wang T, Allen WJ, Lebedev A, Pirker KF, Henderson NS, Geibel S, Volkan E, Yan J, Kunzel M, Pinkner J, Ford BA, Kay CWM, Li H, Hultgren S, Thanassi DG, Waksman G. Crystal structure of the FimD usher bound to its cognate FimC:FimH substrate. Nature. doi: 10.1038/nature10109. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm K, Pluckthun A. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. II. Isomerase-independent chaperone activity in vitro. J Biol Chem. 2000;275:17106–17113. doi: 10.1074/jbc.M910234199. [DOI] [PubMed] [Google Scholar]
- Remaut H, Tang C, Henderson NS, Pinkner JS, Wang T, Hultgren SJ, Thanassi DG, Waksman G, Li H. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr. Protein secretion and membrane insertion systems in gram-negative bacteria. J Membr Biol. 2006;214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- Salgado PS, Taylor JD, Cota E, Matthews SJ. Extending the usability of the phasing power of diselenide bonds: SeCys SAD phasing of CsgC using a non-auxotrophic strain. Acta Crystallogr D Biol Crystallogr. 67:8–13. doi: 10.1107/S0907444910042022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer FG, Barnhart M, Choudhury D, Knight SD, Waksman G, Hultgren SJ. Chaperone-assisted pilus assembly and bacterial attachment. Curr Opin Struct Biol. 2000;10:548–556. doi: 10.1016/s0959-440x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Remaut H, Hultgren SJ, Waksman G. Fiber assembly by the chaperone-usher pathway. Biochim Biophys Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Schmidt SA, Bieber D, Ramer SW, Hwang J, Wu CY, Schoolnik G. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J Bacteriol. 2001;183:4848–4859. doi: 10.1128/JB.183.16.4848-4859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchik VE, Condemine G. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology. 1998;144(Pt 11):3219–3228. doi: 10.1099/00221287-144-11-3219. [DOI] [PubMed] [Google Scholar]
- Shevchik VE, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. Embo J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos C, Georgiou G, Earhart CF. Characterization of Escherichia coli expressing an Lpp’OmpA(46-159)-PhoA fusion protein localized in the outer membrane. Appl Microbiol Biotechnol. 1996;45:112–119. doi: 10.1007/s002530050657. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Tanaka R, Tokuhara M, Kunugi S, Lee YF, Hamada D. Amyloid fibril formation and chaperone-like activity of peptides from alphaA-crystallin. Biochemistry. 2008;47:2961–2967. doi: 10.1021/bi701823g. [DOI] [PubMed] [Google Scholar]
- Thanassi DG. Ushers and secretins: channels for the secretion of folded proteins across the bacterial outer membrane. J Mol Microbiol Biotechnol. 2002;4:11–20. [PubMed] [Google Scholar]
- Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Bokranz W, Nimtz M, Romling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.