Abstract
SIV infection of macaques is the most widely employed model for preclinical AIDS vaccine and pathogenesis research. In macaques, high-titer virus-specific antibodies are induced by infection, and antibody responses can drive evolution of viral escape variants. However, neutralizing antibodies (Nabs) induced in response to SIVmac239 and SIVmac251 infection or immunization are generally undetectable or of low titer, and the identification and cloning of potent Nabs from SIVmac-infected macaques remains elusive. Based on recent advances in labeling HIV-specific B lymphocytes [1-3], we have generated recombinant, secreted, soluble SIVmac envelope (Env) proteins (gp120 and gp140) for detection and quantification of SIVmac Env-specific B lymphocytes. In contrast to HIV-1, we found that soluble SIVmac239 gp140 retains the ability to form stable oligomers without the necessity for introducing additional, stabilizing modifications. Soluble oligomeric gp140 reacted with rhesus anti-SIV Env-specific monoclonal antibodies (MAbs), and was used to deplete Env-specific antibodies with SIV neutralization capability from plasma taken from a rhesus macaque immunized with live attenuated SIVmac239Δnef. Soluble gp120 and gp140 bound to SIV-specific immortalized B cells, and to SIV Env-specific B lymphocytes in peripheral blood of immunized animals. These reagents will be useful for analyzing develiopment of Env-specific B cell responses in preclinical studies using SIV-infected or vaccinated rhesus macaques.
Keywords: SIV, HIV, Env, gp140, rhesus macaque, B cells, neutralizing antibodies
1. Introduction
The development of an effective HIV vaccine has so far remained elusive, a situation that is exacerbated by the limitations of current animal models. Despite over two decades of research, there is still no convenient, small animal model that faithfully recapitulates the persistent infection and pathogenesis of HIV-1 infection in humans [4-8]. Consequently, the SIV/macaque model is critical for preclinical evaluation of AIDS vaccine candidates and for pathogenesis studies.
Numerous assays are currently available for investigation of the role of different T cell subsets in the immune response to HIV/SIV infection or vaccination. In contrast, the evaluation of humoral immune responses has largely been restricted to titration of virus antigen-specific binding antibodies or neutralizing antibodies from serum of infected individuals. In SIV-infected macaques, initial reports were limited to the description of total B cell populations in the periphery [9-11]. The phenotypic identification of different B (naïve and memory) cell subsets during the course of SIV infection is more recent [12]. To our knowledge, specific phenotypic identification of SIV envelope (Env)-binding B cells has not been reported. For HIV-1-infected individuals, the use of engineered trimeric HIV gp140 bearing a biotinylation target sequence (Avitag) has permitted the identification of HIV Env-binding B cells by flow cytometry [1-3]. This reagent also made it possible to clone HIV-Env-specific immunoglobulin genes directly from sorted B cells [2]. Because of the central role of the SIV-infected macaque as a model for studying AIDS pathogenesis and exploring vaccine strategies, we developed similar recombinant, soluble SIVmac239 gp120 and gp140 proteins for flow cytometry based identification of SIV Env-specific B Lymphocytes [1-3].
2. Materials and methods
2.1 Plasmids
SIVmac239 Env sequences have previously been optimized for expression in mammalian cells [13]. In order to generate soluble gp120 and gp140 proteins for efficient biotinylation and detection by flow cytometry, both proteins were modified to contain a protein tag commonly known as Avitag (LNDIFEAQKIEWHE) that can be biotinylated with high efficiency using biotin ligase; once biotinylated, the Avitag can serve as a high affinity substrate for streptavidin-based reagents. [3]. To generate the tagged proteins, gene sequences corresponding to SIVmac239 Env gp120 and gp140 were amplified by PCR and the Avitag was introduced as part of the reverse primer. The same Forward primer EcoRI-SIVmac239envF-BF (5’-AGCGAAT
TCATGGGATGTCTTGGGAATCAGCTGCTTATCGCCATC-3’) containing an EcoRI restriction site was used for both gp120 and gp140. Reverse primers gp120_avitag_XhoI-R-BF (5’-GGCCTCGAGTCACTCGTGCCA
CTCGATCTTCTGGGCCTCGAAGATGTCGTTCAGCCGCTTGTTCCGCGACGTCCCCCCGGTCGT-3’) and gp140_avitag_XhoI-R-BF (5’-GGCCTC GAGTCACTCGTGCCACTCGATCTTCTGGGCCTCGAAGATGTCGTTCAGCGACGCCAGGTCGAACCAGTTGCCGAACAC-3’) containing a sequence corresponding to the Avitag signal and an XhoI restriction site were used for gp120 and gp140, respectively. The PCR amplification product was clone into pCDNA3.1(+) via EcoRI and XhoI restriction sites.
A non-modified gp120 protein was produced as a control for immunoblotting experiments. For that purpose, a stop codon was created after the amino-acid position 525 by quick change mutation (Strategene, La Jolla, CA) using primers oli082 (5’-CGTCGCGGAACAAGCGGTGATCATTCGTCCTGGGGTTCC-3’) and oli083 (5’-GGAACCCCAGGACGAATGATCACCGCTTGTTCCGCGACG-3’).
2.2 Protein production and biotinylation
Protein expression plasmids were used to transfect 293T cells using the Transfectin reagent, according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). Culture supernatants (serum-free medium) were harvested twice, at 2 and 5 days post-transfection, and pooled. Pooled supernatants were centrifuged 2095 RCF for 5 min to remove cell debris. The supernatants were further clarified using a 0.45 μm syringe filter (Millipore, Billerica, MA). Protein was purified using Galanthus nivalis Lectin-Agarose (Sigma, St-Louis, MO). Biotinylation was performed with biotin ligase according to the manufacturer's suggestions (Avidity, Denver, Co). Biotinylation efficiency was evaluated by reference to a commercial, fully-biotinylated protein, BIS-300 (Avidity, Denver, Co). Proteins were quantified by determining adsorbance at 280 nm using a spectrophotometer (Nanodrop Technologies Inc, Wilmington, DE).
2.3 Glutaraldehyde (GA) cross-linking
Culture supernatant from transfected cells was harvested 48h post-transfection and centrifuged 2095 rcf for 5 min to pellet cell debris. Supernatant was further cleared using a 0.45 μm syringe filter (Millipore, Billerica, MA). 200 μl of filtered supernatant were mixed with gluteraldehyde (GA) at different concentrations (0, 1, 2.5, 5 and 10 mM). The mixture was then incubated for 5 minutes at room temperature and the reaction was quenched with 0.1 M Tris-HCl pH 7.4. Treated protein was then analyzed by denaturing SDS-PAGE on an 8% gel (BioRad, Hercules, CA).
2.4 Protein antigenicity
Prior to biotinylation, antigenicity of Avitag bearing gp120 and gp140 was determined by immunoblotting. Purified and cross-linked proteins were denatured by addition of equal volume of 2x concentrated sample buffer containing SDS and ß-mercaptoethanol (Sigma, St-Louis, MO) and boiling at 99°C for 5 min. Denatured proteins were separated by SDS-PAGE on an 8 or 12% gel (BioRad, Hercules, CA). Staining with Coomassie blue was performed to verify equal loading of purified proteins. The presence of Avitag sequence on the non-biotinylated proteins was tested using anti-Avitag monoclonal antibody (Mab) while reactivity with SIV Env-specific antibodies was tested using monoclonal antibody 3.11H [14] and pooled plasma from SIV-infected macaques.
The antigenicity of none or biotinylated gp120 and gp140 was analyzed by ELISA using plasma of SIV-infected macaque (Mm376-04) and SIV gp120-specific macaque MAbs obtained from Dr James Robinson (Tulane University Medical School, New Orleans, Lo). The binding sites of these MAbs have been more or less characterized and they have been assigned to different competition groups [14, 16, 17]. MAb 3.11H is V3-loop specific and belongs to the competition group IV while MAb 1.10A assigned to group VII has a less defined binding site but encompasses the V4 loop. MAb 3.4E and 1.9C respectively assigned to group V and VI have an undefined binding sites but seems to exclude the variable loops of gp120. ELISA was performed as previously described [15]. ELISA plates were coated at 50 ng of antigen per well. HRP-conjugated anti-human IgGs (Santa Cruz Biotechnology, Santa Cruz, CA) was used for detection with TMB solution (BioRad, Hercules, CA).
2.5 Binding to immortalized, SIV-specific rhesus B cell lines
Rhesus macaque B cell lines producing anti-SIV gp120 MAb 3.11H and MAb 3.4E were obtained from Dr James Robinson (Tulane University Medical School, New Orleans, Lo).
Rhesus macaque B cell lines were stained with biotinylated 10 μg/ml of gp120 or gp140 for 30 min at 4°C in 100 μl of staining buffer corresponding to phosphate buffer saline PBS (Invitrogen, Carlsbad, CA) containing 0.1% bovine serum albumin (Sigma, St-Louis, MO). Cells were then incubated with streptavidin-APC (BD Biosciences, San Jose, CA). Data were acquired on a FACS Calibur (BD Biosciences, San Jose, CA) and analyzed using FlowJo software (TreeStar, Cupertino, CA).
2.6 Plasma adsorption and elution of anti-gp120 and -gp140 antibodies
Adsorption and elution of plasma anti-HIV-1 gp120 or anti-membrane proximal external region (MPER) antibodies have previously been described [18, 19]. Purified gp120, gp140 or control BSA (BioRad, Hercules, CA) were coated to Dynabeads MyOne Tosylactivated at the ratio of 40 μg protein per mg of beads according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA).
To verify integrity of gp120 and gp140, protein-coated beads were first stained with MAb 3.11H or pooled plasmas from SIV-infected animals and then FITC-conjugated anti-human IgGs (BD Biosciences, San Jose, CA). Data were acquired on a FACS calibur (BD Biosciences, San Jose, CA) and analyzed using FlowJo software (TreeStar, Cupertino, CA).
For antibody depletion, 50 μl of plasma from a rhesus macaque (Mm 372-04) previously immunized with SIVmac239Δnef was diluted with 150 μl of RPMI (Invitrogen, Carlsbad, CA). Diluted plasma was separately added in 2ml Eppendorf tubes to 1 mg of beads previously coated with 40 μg of corresponding proteins. Tubes were incubated for 1 hour at room temperature with rotation and the beads where collected using a magnet. A second round of adsorption was performed with fresh beads (1 mg, 40 μg of proteins). The efficiency of the adsorption was tested by ELISA, using plates coated with gp120 or gp140.
gp120- and gp140-binding antibodies were eluted by addition of 200 μl of 0.1 M glycine to beads pellets. Beads were then retained with a magnet and supernatants (eluates) were collected and brought to neutral pH by addition of 7.5 μl of 1.5 M Tris-HCl pH 8.5. The antigenic specificity of eluted IgGs was evaluated by ELISA, using plates coated with gp120 or gp140. The IgG content of eluates was determined using an IgG quantification ELISA Kit following recommendations from the manufacturer (Columbia Bio LLC, Elmhurst, NY). A cut-off value was set at 2x mean background values.
2.7 Virus neutralization assay
To test interaction of the Env proteins with neutralizing antibodies, plasma was depleted by treatment with g120- and gp140- coated beads and tested for virus neutralization using the CEMx174-SEAP indicator cell line as previously described [17]. Untreated plasma, plasma treated with “beads-only”, and plasma treated wtih BSA-coated beads were all included as controls. Neutralization assays were performed using the SIVmac316 (neutralization sensitive) and SIVmac239 (neutralization resistant) isolates as test viruses.
2.8 Detection of gp120- and gp140-specific B cells in peripheral blood
Mononuclear cells were prepared from peripheral blood using Lymphocyte Separation Medium (MP Biomedicals LLC, Irvine, CA) following the manufacturer's instructions. CD3+ cells were depleted using anti-human CD3 clone SP34-2 (BD Biosciences, San Jose, CA) and Pan Mouse IgG Dynabeads (Invitrogen, Carlsbad, CA) in order to enrich for B cells. CD3+ cell depletion resulted in depletion of CD4+ T cells, which gp120 and gp140 could bind through the CD4 receptor. Enriched B cells fractions were stained as previously described [1]. At first, cells were stained with non-conjugated purified anti-CD4 antibody (NIH Nonhuman Primate Reagent Resource, Boston, MA) at the ratio of 10 μg/ml at 4°C in order to prevent gp120 and gp140 binding to potentially remaining CD4 positive cells. After 15 min of incubation, cells were stained with 10 μg/ml of biotinylated gp120 or gp140 for 30 min at 4°C. Cells were then incubated with streptavidin-V450 and anti-human antibodies IgG-PE-Cy5, CD3-V500 and CD20-APC-H7 (BD Biosciences, San Jose, CA). Data were acquired (800 000 events) using an LSRII Flow Cytometer (BD Biosciences, San Jose, CA) and analyzed using FlowJo software (TreeStar, Cupertino, CA).
2.9 Animals
Mm 376-04 is a rhesus macaque that was immunized with SIVmac239Δnef. The animal had an ongoing but limited replication of the vaccine strain SIVmac239Δnef with viral loads of ~1000 ceq/ml. Two months after immunization, SIVmac239Δnef viral load was equal to 1200 ceq/ml. Mm 376-04 was sterilely protected following intravenous challenge with wild type SIVmac239, 5 weeks after SIVmac239Δnef-immunization (wild type SIVmac239 viral load always below 60 ceq/ml).
Mm 376-02 is a rhesus macaque that was immunized with SIVmac239Δnef and had persistent low-level replication of the vaccine strain of 60-300 ceq/ml. Mm 376-02 was sterilely protected following vaginal challenge with SIVmac251 at 20 weeks after SIVmac239Δnef-vaccination (SIVmac251 viral load always below 10 ceq/ml).
Naïve animals Mm 400-08 and 9-010 are SIV-negative.
All 4 animals were utilized in experiments aiming to detect gp120- and gp140-binding B cells. Only Mm 376-04 was involved in plasma antibody adsorption experiments.
3. Results
3.1 Protein antigenicity / recognition
To identify SIV Env-specific B cells in the peripheral blood of SIV-infected animals, we engineered recombinant, soluble SIVmac239 Env proteins bearing a high affinity biotinylation target sequence, or Avitag (Avidity, Denver, Co) at the C-terminus. Soluble gp120 encompasses the entire coding region of the SIVmac239 surface subunit (SU or gp120), whereas soluble gp140 includes all of gp120 and the extracellular portion of the transmembrane subunit (TM or gp41). The Avitag sequence was placed at the C-terminus of gp120 and gp140; thus, in the case of gp140, processing of the protein into the SU and TM subunits results in only the TM subunit (the truncated gp41) bearing the Avitag. Proteins were generated by transient transfection of 293T cells and purified using Galanthus nivalis Lectin-Agarose (Sigma, St-Louis, MO).
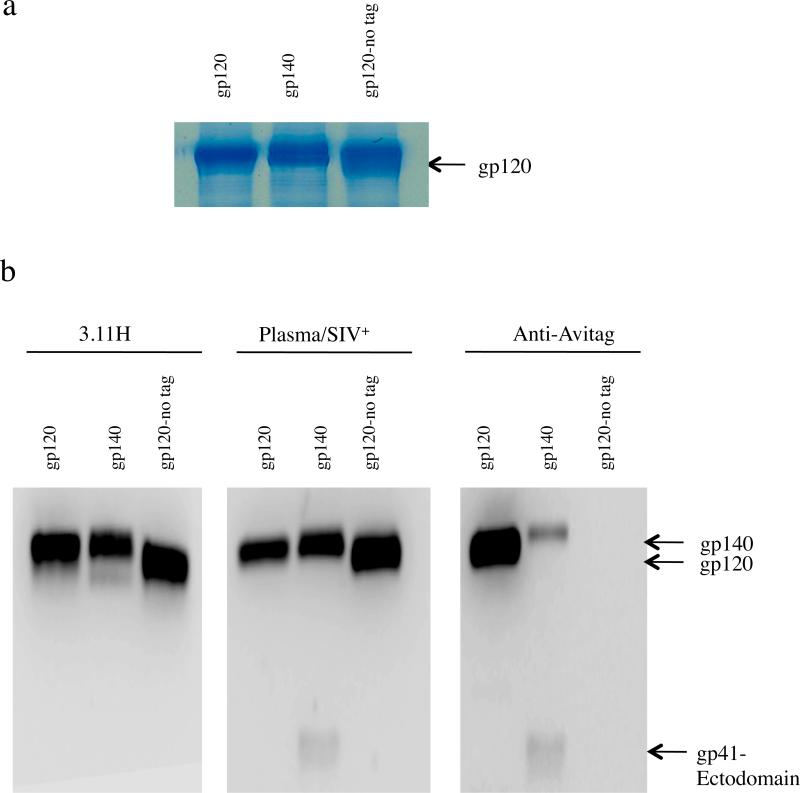
The purified non-biotinylated, soluble gp120 and gp140 proteins were denatured (99°C, 5 min) and separated on a 12% SDS-PAGE gel in reducing conditions, along with untagged soluble gp120 as a control (Fig. 1a). We confirmed expression of the proteins by coomassie staining and by immunoblotting using Env-specific and Avitag specific antibodies. As expected, all three proteins were detected with the SIV V3 loop-specific MAb 3.11H (Fig. 1b, left panel) and pooled plasma from SIV-infected rhesus macaques (Fig. 1b, center panel). In contrast, only the Avitag-bearing proteins were detected with an Avitag-specific MAb (Fig. 1b, right panel). With both SIV+ plasma and the anti-Avitag MAB a band of smaller size (~ 25 Kda) corresponding to the truncated, gp41-derived subunit of gp140 was detected; importantly, the intensity of the 25kD band is similar whether probing with SIV+ plasma or the anti-Avitag MAb (Fig. 1b, center and right panels). The combined results confirmed that purified soluble SIV gp140 is comprised of two subunits, gp120 and the ectodomain region of gp41. The 2 subunits were separated under denaturing/reducing conditions as shown by the detection of gp41 ectodomain (Fig. 1, center and right panels). In the same lanes, the higher molecular weight fragment is a mixture of gp140 and gp120. Probing with SIV+ plasma detected both gp140 and gp120 (Fig. 1, center panel) generating a strong band. In contrast, because the Avitag sequence is at the C-terminus of the TM subunit, the anti-Avitag antibody detected only gp140 but not gp120 (Fig. 1, right panel); this result also reveals that the gp140 preparation contains both cleaved and uncleaved gp140 proteins.
Fig. 1.
Recognition of Avitag-bearing gp120 and gp140 by immunoblotting. SIV gp120 and gp140 specificities were detected with monoclonal antibody (MAb) 3.11H (V3 loop-specific) and pooled plasma from SIV-infected macaques. Avitag specificity was determined with anti-Avitag MAb. Purified proteins were separated by SDS-PAGE . (a) Coomassie stained gel showing biotinylated gp120 and gp140 and untagged control gp120. (b) immunoblotting with MAb 3.11H (left panel), pooled plasma from SIV-infected macaques (center panel) and anti-Avitag MAb (right panel).
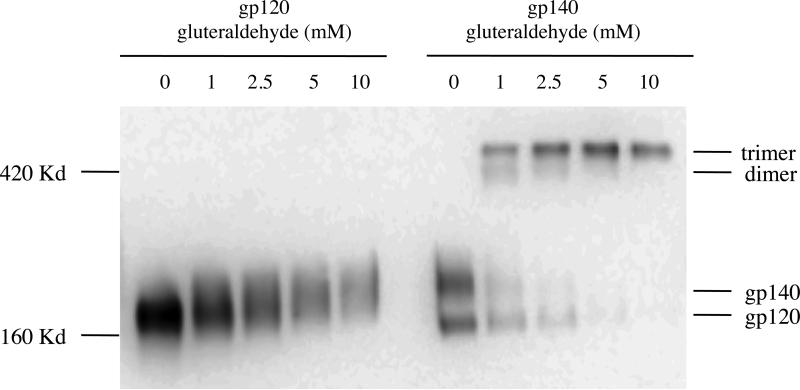
We next anaylzed the oligomeric structure of soluble SIV gp140 by glutaraldehyde cross-linking. In the absence of glutaraldehyde, gp140 migrates as two bands on an 8% SDS gel under denaturing/reducing conditions (Fig. 2). In the presence of increasing concentrations of glutaraldehyde, the gp140 bands shift into a slower migrating band corresponding to oligomeric gp140. In contrast, glutaraldehyde cross-linking did not result in oligomerization of gp120, even at the highest concentration tested. On the 8% SDS gel the Avi-tagged gp120 ran slightly above the 160 kD marker. The dramatic shift in size between gp140 in the absence and presence of glutaraldehyde suggests that the slowest migrating gp140 oligomer is most likely a trimer. An additional, faint band estimated to be a gp140 dimer was also observed, although this disappeared with increasing concentrations of glutaraldehyde (and was undetectable at the highest concentration tested).
Fig. 2.
Oligomerization of soluble SIV gp140. Gluteraldehyde (GA) cross-linking was used to determine the oligomeric structure of soluble gp140 on a 12% SDS gel and in reducing conditions following transfection of 293T cells with Avitag-bearing gp120 or gp140 expression vectors. Culture supernatant was harvested 48 h post-transfection. gp140 supernatant was cross-linked with different concentrations (mM) of GA. The reaction was stopped after 5 min by addition of 0.1 M Tris-HCl. Samples were loaded along with gp120 supernatant as control.
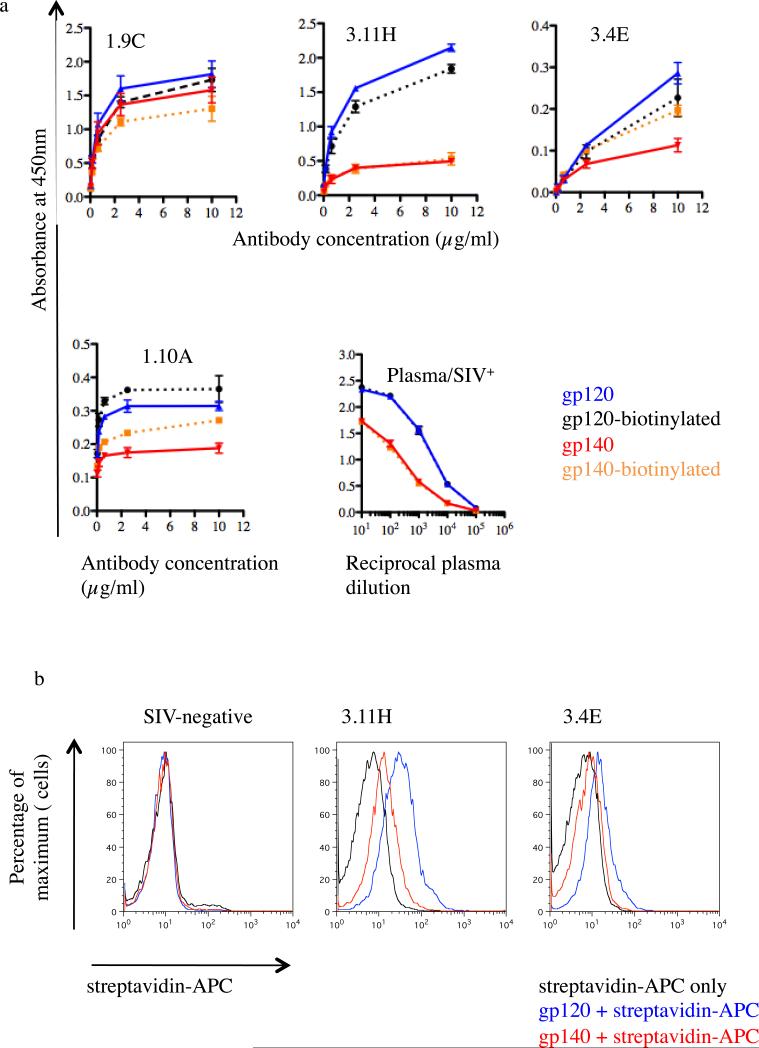
Following the analysis of the oligomeric structure of gp140, purified Avi-tagged gp120 and gp140 were biotinylated using biotin ligase. Efficacy of biotiylation was verified by ELISA by comparison to a reference protein BIS300 (Supplementary Figure 1). The integrity of biotinylated gp120 and gp140 proteins was analyzed by ELISA using plasma from an SIV+ macaque and SIV-gp120 specific rhesus macaque MAbs. For SIV+ plasma, binding to gp120 and gp140 was identical to corresponding biotinylated forms (Fig. 3). These data suggest that biotinylation did not grossly alter the antigenic structure of these proteins. Even though the results were not as striking as for polyclonal plasma, MAbs presented a similar trend (with the exception of MAbs 3.4E and 1.10A where biotinylation improved antibody binding to gp140). For all MAbs and SIV+ plasma, binding to gp120 was always superior to binding to gp140. These results are consistent with the assumption that oligomeric gp140 is structurally constrained and some surfaces are less accessible to binding antibodies.
Fig 3.
Recognition of biotinylated gp120 and gp140 by ELISA and binding to macaque B cell lines. (a) SIV gp120 and gp140 were detected with monoclonal antibodies (MAbs) 3.11H, 3.4E, 1.9C and 1.10A (competition group IV, V, VI and VII, respectively) and plasmas (Plasma/SIV+) from sterilely protected SIVmac239Δnef immunized macaque (Mm376-04) followed by HRP-conjugated anti-human IgGs. ELISA plates were coated with biotinylated gp120 and gp140 or non-biotinylated gp120 and gp140. OD, optical density. Background values were obtained using control wells (no antigen). Bars represent OD value obtained after background subtraction. Error bars are based on standard deviation of OD values for triplicate wells for each antigen. (b) Anti-SIV ENV gp120-directed monoclonal antibody (MAb) producing B cell lines (3.11H and 3.4E) were stained with biotinylated gp120 and gp140 or buffer only. Cells were washed and further stained with APC-conjugated streptavidin. The control B cell line was developed from a naïve (SIV-negative) macaque.
In order to demonstrate the ability of biotinylated gp120 and gp140 to bind SIV Env-specific antibody producing cells, we used two immortalized rhesus macaque B cell lines (BLCL) that produce MAb 3.11H and MAb 3.4E [14, 16, 17]. Both proteins readily stained these SIV-specific BLCLs, whereas neither of the biotinylated proteins (gp120 and gp140) bound to BLCL generated from naïve (SIV-negative) rhesus macaques (Figure 3, left panel). Binding of gp120 and gp140 to BLCL-3.11H was superior to BLCL-3.4E consistent with the ELISA results (Fig. 3). In addition, gp120 binding was also superior to gp140, similar to the result obtained by ELISA (Fig. 3). Because the Avitag is located at the C-terminus of the gp41 ectodomain, and binding to 3.11H and 3.4E occurs through the gp120 subunit, the gp120 subunit and the gp41 ectodomain must have remained associated throughout the assay.
These results indicate that soluble SIVmac239 gp140 can form an oligomeric structure. The oligomeric structure of soluble SIVmac gp140 has been previously demonstrated with the Env protein of CP-MAC, a lab-adapted variant of SIVmacBK28 [20, 21]. These authors showed that introduction of a premature stop codon in the transmembrane (TM) domain of gp41 resulted in a highly stable gp120/gp41 association and the modification of the cleavage site between gp120 and gp41 was not necessary. Our results with soluble SIVmac239 gp140 are in accordance with these previous findings. In contrast, the association between HIV-1 gp120 and gp41 is apparently more labile, and retention of stable oligomeric structures typically requires experimental modifications, such as elimination of the gp120-gp41 cleavage site, introduction of cysteine residues into the gp41 N36 coiled coil, and inclusion of a heterologous trimerization motif [22-25]. The minimal alteration of SIVmac239 gp140 is desirable in order to retain the original antigenicity of the trimeric Env complex.
3.2 Plasma antibody adsorption and elution
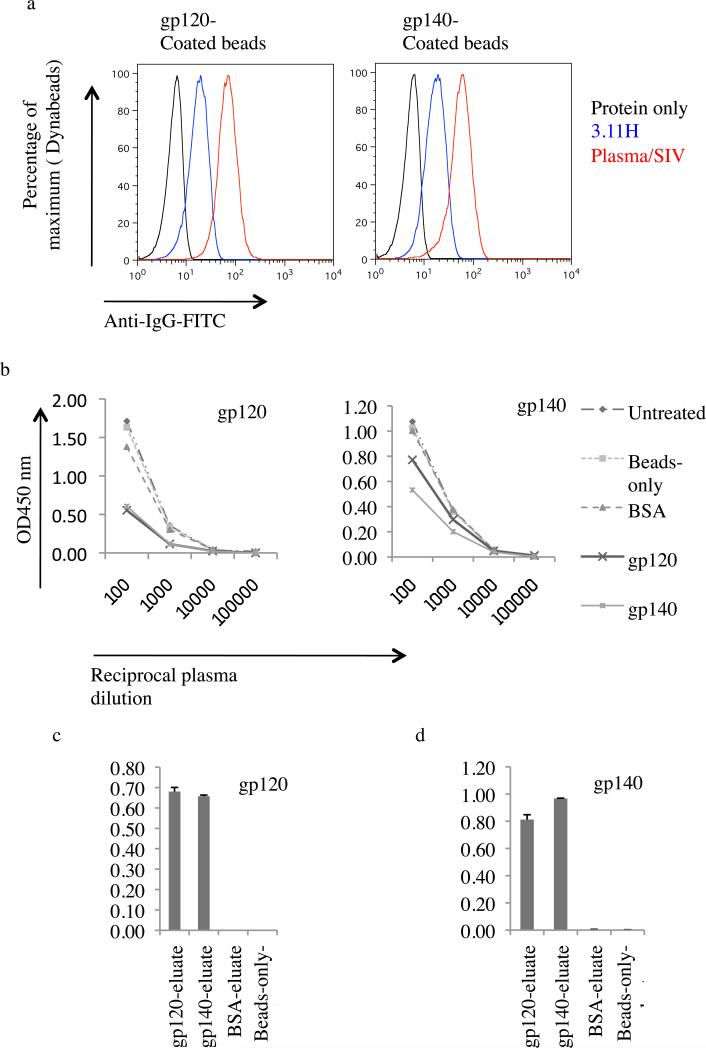
In order to further assess the structural integrity of soluble SIV gp120 and gp140, the proteins were tested for ability to adsorb SIV Env-specific antibodies in plasma from an animal immunized with SIVmac239Δnef (Mm 376-04). To do this, soluble gp120 and gp140 were coated to Dynabeads. Both gp120 and gp140 coated beads reacted with MAb 3.11H (V3 loop-specific) and pooled SIV+ plasmas (Fig. 4a).
Fig. 4. Plasma antibody adsorption and elution.
(a) Recognition of gp120 and gp140 coated Dynabeads. Coated beads were stained with monoclonal antibody (MAb) 3.11H (V3 loop-specific) and pooled plasmas from SIV-infected monkey (Plasma/SIV+) or buffer only. Antibody binding to gp120 and gp140 was revealed by flow cytometry using an anti-human FITC-conjugated IgG. (b) Anti-gp120 and anti-gp140 antibody adsorption from plasma from SIVmac239Δnef-immunized macaque (Mm 376-04). Antibodies were adsorbed with gp120-coated beads, gp140-coated beads, BSA-coated beads or beads-only or left untreated. Resulting plasmas were then assayed by ELISA for binding to gp120 (left panel) or gp140 (right panel). (c) and (d) Bead-eluate content in anti-SIV ENV-directed antibodies. gp120 and gp140 binding IgGs were eluted with glycine. Eluates were then brought to neutral with Tris-HCl and tested using ELISA plates coated with gp120 and gp140. OD, optical density.
gp120 and gp140 coated Dynabeads were then used to adsorb SIV Env-specific antibodies from the plasma. The efficiency of plasma adsorption was evaluated by ELISA (Fig. 4b). gp120 and gp140 adsorptions resulted in reduction of SIV Env-specific antibodies in plasma in comparison to controls (controls included untreated plasma, plasma treated with beads-only, and plasma treated with BSA-coated beads controls) (Fig. 4). Both gp120 and gp140 adsorption resulted in similar levels of depletion (Fig. 4b, left panel). BSA-coated beads, beads-only and untreated plasma resulted in little or no depletion. We concluded that both SIV gp120 and SIV gp140 bind anti-Env-IgGs. Oligomeric SIV gp140 most likely encompasses the same specificities as SIV gp120 with regard to the gp120 component.
Plasma anti-gp140 IgG adsorption was more efficient with gp140-coated beads than gp120-coated beads (Fig. 4b, right panel). BSA-coated beads and beads-only had a similar level of plasma anti-gp120 IgG content compared to untreated plasma (Fig. 4b, right panel). We conclude that oligomeric SIV gp140 possesses additional specificities to those of SIV gp120 subunit, possibly due to epitopes in the ectodomain of gp41 or epitopes unique to the oligomeric structure.
In order to confirm that Env-specific IgGs in the plasma from an animal immunized with SIV239Δnef bound to gp120- and gp140-coated beads, the beads were treated with a low pH buffer to elute any bound IgG and the eluate was tested for the presence of Env-specific binding antibodies by ELISA. Similar antibody levels were obtained for the gp120-eluate (247 ng/ml) and the gp140-eluate (247 ng/ml). Eluates from control BSA-coated beads and the beads-only control were below the cut-off value. An equal volume of each eluate (50 μl) was assayed using ELISA plates coated with gp120 and gp140. Similar gp120-specific IgG content was detected in the eluates from gp120-coated beads and gp140-coated beads (Fig. 4c). A similar level of gp120- and gp140-specific IgG content was observed in the eluate from both gp120- or gp140-coated beads (Fig. 4d). Neither gp120- nor gp140-specific IgGs were detected in beads-only eluate or BSA eluate. These results demonstrate that purified soluble SIV gp120 and oligomeric SIV gp140 can bind SIV Env-specific antibodies from a macaque immunized with SIVmac239Δnef.
3.3 Depletion of Neutralizing Activity from SIV+ Plasma with gp120 and gp140
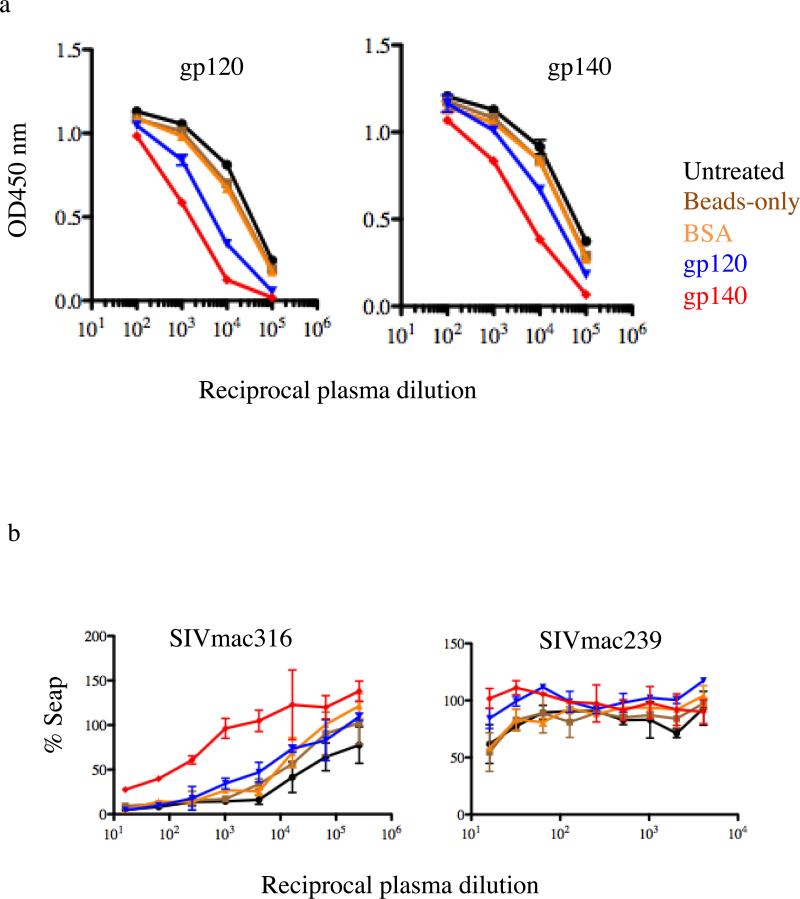
Finally, we sought to analyze the functionality of depleted anti-Env IgGs by performing virus neutralization assays. The SIV strain SIVmac316, like many laboratory strains of HIV-1, is sensitive to antibody-mediated neutralization when treated with SIVmac+ sera or anti-SIV Env antibodies [17]. Pre-depletion of plasma from macaque Mm376-04 with gp140 resulted in a significant decrease in 50% neutralizing titer (from 1:16,384 for the untreated control to 1:265 for the gp140 depleted control)(Figure 5b, left panel). SIVmac239 is generally resistant to neutralization by autologous plasma, and consequently we found that only the lowest plasma dilution (1:16) achieved close to 50% neutralization compared to controls. Nevertheless, depletion with gp120 and gp140 reduced neutralization at the 1:16 dilution from 50% to 80% (gp120) and 100% (gp140) (Figure 5b, right panel). No detectable difference was observed for neutralization of SIVmac239 using plasma from control samples (untreated, beads-only and BSA-coated beads) (Fig. 5b). Together, these results confirm that the SIV gp140 protein is recognized and bound by antibodies that also recognize native envelope spikes as they exist on the surface of infectious virions.
Fig. 5. Plasma antibody adsorption and virus neutralization assay.
(a) Anti-gp120 and anti-gp140 antibody adsorption from plasma from SIVmac239Δnef-immunized macaque (Mm 376-04). Antibodies were adsorbed with gp120-coated beads, gp140-coated beads, BSA-coated beads or beads-only or left untreated. Resulting plasmas were then assayed by ELISA for binding to gp120 (left panel) or gp140 (right panel). OD, optical density. (b) anti-gp120 and -gp140 IgG depleted plasma were assayed for neutralization of SIVmac239 and SICmac316.
3.4 Detection of SIV gp120- and gp140-specific B cells in peripheral blood
We next asked whether biotinylated gp120 and gp140 proteins could be used to identify SIV Env-specific B lymphocytes in peripheral blood using flow cytometry. The assay was performed using PBMC from SIVmac239Δnef-immunized macaques (Mm 376-02 and Mm 376-04) and a naïve (SIV-negative) control. To do this, CD3-positive cells were first depleted in order to enrich for B lymphocytes. Additionally, to prevent binding to residual CD4+ T-cells via the Env receptor binding domain, cells were treated with a CD4 blocking antibody prior to staining. While HIV gp140-binding CD19+IgG+ memory B cells have previously been reported [2, 3], the anti-human CD19 (Non-Human Primate Reagent Resources, Boston, MA) is less efficient for staining rhesus B cells even when used at high concentrations (data not presented). Preliminary studies using a number of surface markers specific for B-cells showed that in general CD19, CD21 and CD22 were detected on the surface of CD20+ cells, while a significant number of CD20+ cells were negative for CD19, CD21 and CD22 (data not presented). Therefore, in this study we chose CD20 as B-cell marker using an anti-human CD20 antibody (BD Biosciences, San Jose, CA). In human, one advantage of using anti-CD19 over anti-CD20 antibody can be the detection of plamablasts/early plasma cells. However, our preliminary studies in rhesus macaques failed to detect CD19+CD20- plamablasts/early plasma cells. Our lab and others are actively pursuing alternative protocols for phenotypic identification of plasmablasts/early plasma cells in macaques.
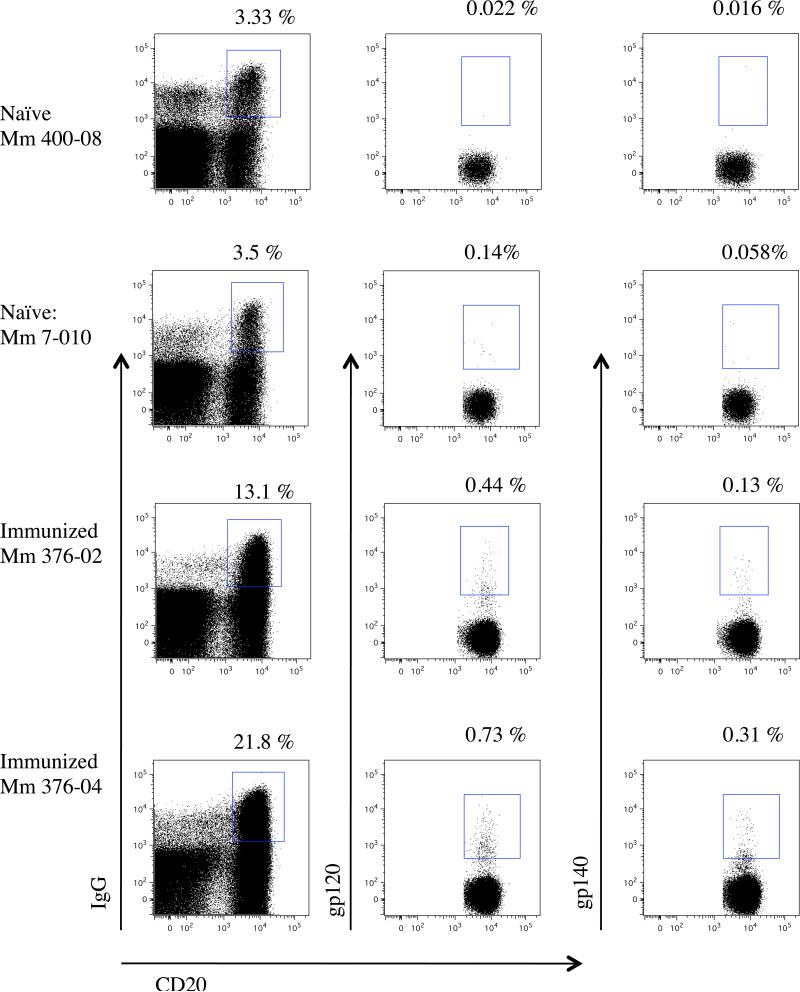
SIV Env-specific (gp120- and gp140-binding) CD20+ IgG+ B cells were detected in both animals (Mm 376-02 and Mm 376-04) that had been immunized with the live attenuated vaccine strain SIVmac239Δnef (Fig. 6). In contrast, detection of gp120-binding and gp140-binding B cells was about 10-fold lower in lymphocytes prepared in parallel from naïve uninfected (SIV-negative) macaques, which is likely to represent non-specific binding (Fig. 6). These results demonstrate the utility of biotinylated gp120 and gp140 for specific detection and phenotyping of SIV Env-specific CD20+IgG+ B cells. It is noteworthy that the number of gp120 or gp140 CD20+ IgG+ B detected cells was not particularly increased (less than double) with higher protein concentrations (up to 60 μg/ml). It is possible that Env-specific B cells in the peripheral blood reach an optimum number during the chronic phase for a sterilely protected SIVmac239Δnef immunized macaque with low titer of vaccine strain. A longitudinal study and comparison with other group of SIV infected macaque could provide more clarification on that matter.
Fig. 6.
Identification of gp120- and gp140-binding B cells. Gating was performed on CD3-negative lymphocytes from SIVmac239-immunized macaques (Mm 376-02 and Mm 376-04) and naïve (SIV-negative) control. The plots on the left show the gating on CD20+IgG+ B cells. The plots on the center show the gating on gp120-binding CD20+IgG+ B cells. The plots on the right show the gating on gp140-binding CD20+IgG+ B cells. Numbers indicate the frequency of CD20+IgG+ B cells (left) and the frequency of gp120-binding CD20+IgG+ B cells (center) and the frequency of gp140-binding CD20+IgG+ B cells (right).
4. Conclusion
Sterile protection induced by SIVmac239Δnef in the SIV/macaque model has been extensively studied, but the mechanism of protection remains poorly understood [26-30]. The exact contribution of the commonly studied parameters (such as anti-viral memory T cells, gag specific IFN-γ, binding and neutralizing antibodies) remain unclear. Importantly, characterization of the virus-specific antibody response to SIV infection or vaccination with SIV-based immunogens has been hampered by the lack of reagents for identifying SIV-specific B-lymphocytes. In the current study we demonstrated that soluble SIVmac239 gp140 forms oligomers without the need for additional modifications, and has characteristics similar to those of engineered soluble HIV gp140 [1-3]. This soluble SIV gp140 is capable of depleting Env-specific antibodies from the plasma from an animal immunized with SIVmac239Δnef as well as or better than soluble gp120. Importantly, both soluble SIV gp120 and SIV gp140 allowed the identification of SIV-specific B lymphocytes in animals vaccinated with the replication-competent, attenuated vaccine strain SIVmac239Δnef.
These novel tools will be helpful for identification, phenotyping and quantification of ENV-specific B cells (including memory and plasma cells) as previously demonstrated with HIV-1 gp140 [1-3]. As with HIV-1 gp140, SIV gp140 should permit the single-cell sorting and cloning of SIV-specific monoclonal antibodies. Importantly, SIV gp120 and SIV gp140 can be used to monitor the dynamics of ENV-specific B cell subsets during the course of SIV infection and in response to vaccination and challenge in SIV/macaque AIDS vaccine studies.
Supplementary Material
Supplementary Figure 1. Efficiency of gp120 and gp140 biotinylation. Avitag bearing gp120 and gp140 were biotyinlated using biotin ligase. Biotinylation was assessed by ELISA. a. Comparison of none and biotinylated proteins. Elisa plates were coated with Avitag bearing but not biotinylated (gp120 or gp140) proteins; and Avitag bearing and biotinylated (gp120-biotinylated or gp140-biotinylated) proteins at 1 ng/μl. All proteins were detected with plasma pool of SIV-infected monkey (Plasma/SIV+) followed by HRP-conjugated anti-human IgGs. Biotinylation specificity was verified with HRP-conjugated streptavidin. b. Assessment f biotinylation completion by comparison to a fully biotynlated standard. Elisa plates were coated with Avitag bearing biotinylated gp120 or gp140 and BIS-A a commercial fully biotinylated standard at 1; 0.6 and 0.2 ng/μl. Detection was performed with HRP-conjugated streptavidin. OD, opticlal density.
ACKNOWLEDGMENTS
We thank Mackenzie Bartlett for technical assistance. Purified non-conjugated anti-CD4 antibody and anti-CD19 antibody were kind gifts from the NIH Nonhuman Primate Reagent Resource (nhpreagents.bidmc.harvard.edu/NHP/default.aspx). This research was supported by NIH grants A1083118 (WEJ), AI07306 (RPJ) and RR00168 (NEPRC), a grant from the Japan Health Sciences Foundation (H.S.), and a grant from the International AIDS Vaccine Initiative (RPJ). RKR is supported by a CHAVI/HVTN Early Career Investigation award (U19 AI 067854-04).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Doria-Rose NA, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83(1):188–99. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–40. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 3.Scheid JF, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343(2):65–7. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato S, Johnson W. Antibody-mediated neutralization and simian immunodeficiency virus models of HIV/AIDS. Curr HIV Res. 2007;5(6):594–607. doi: 10.2174/157016207782418515. [DOI] [PubMed] [Google Scholar]
- 5.Okada H, et al. Synergistic effect of human CycT1 and CRM1 on HIV-1 propagation in rat T cells and macrophages. Retrovirology. 2009;6:43. doi: 10.1186/1742-4690-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keppler OT, et al. Susceptibility of rat-derived cells to replication by human immunodeficiency virus type 1. J Virol. 2001;75(17):8063–73. doi: 10.1128/JVI.75.17.8063-8073.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keppler OT, et al. Progress toward a human CD4/CCR5 transgenic rat model for de novo infection by human immunodeficiency virus type 1. J Exp Med. 2002;195(6):719–36. doi: 10.1084/jem.20011549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browning J, et al. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc Natl Acad Sci U S A. 1997;94(26):14637–41. doi: 10.1073/pnas.94.26.14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykhuizen M, et al. Determinants of disease in the simian immunodeficiency virus-infected rhesus macaque: characterizing animals with low antibody responses and rapid progression. J Gen Virol. 1998;79(Pt 10):2461–7. doi: 10.1099/0022-1317-79-10-2461. [DOI] [PubMed] [Google Scholar]
- 10.Mattapallil JJ, Letvin NL, Roederer M. T-cell dynamics during acute SIV infection. AIDS. 2004;18(1):13–23. doi: 10.1097/00002030-200401020-00002. [DOI] [PubMed] [Google Scholar]
- 11.Steger KK, et al. CD4+-T-cell and CD20+-B-cell changes predict rapid disease progression after simian-human immunodeficiency virus infection in macaques. J Virol. 1998;72(2):1600–5. doi: 10.1128/jvi.72.2.1600-1605.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhrt D, et al. Evidence of early B-cell dysregulation in simian immunodeficiency virus infection: rapid depletion of naive and memory B-cell subsets with delayed reconstitution of the naive B-cell population. J Virol. 84(5):2466–76. doi: 10.1128/JVI.01966-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosati M, et al. DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J Virol. 2005;79(13):8480–92. doi: 10.1128/JVI.79.13.8480-8492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson JE, et al. Production and characterization of SIV envelope-specific rhesus monoclonal antibodies from a macaque asymptomatically infected with a live SIV vaccine. AIDS Res Hum Retroviruses. 1998;14(14):1253–62. doi: 10.1089/aid.1998.14.1253. [DOI] [PubMed] [Google Scholar]
- 15.Hammonds J, et al. Induction of neutralizing antibodies against human immunodeficiency virus type 1 primary isolates by Gag-Env pseudovirion immunization. J Virol. 2005;79(23):14804–14. doi: 10.1128/JVI.79.23.14804-14814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole KS, et al. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology. 2001;290(1):59–73. doi: 10.1006/viro.2001.1144. [DOI] [PubMed] [Google Scholar]
- 17.Johnson WE, et al. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J Virol. 2003;77(18):9993–10003. doi: 10.1128/JVI.77.18.9993-10003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–4. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray ES, et al. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J Virol. 2009;83(21):11265–74. doi: 10.1128/JVI.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaBranche CC, et al. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol. 1995;69(9):5217–27. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edinger AL, et al. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J Virol. 2000;74(17):7922–35. doi: 10.1128/jvi.74.17.7922-7935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, et al. Modifications that stabilize human immunodeficiency viru senvelope glycoprotein trimers in solution. J Virol. 2000;74(10):4746–54. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, et al. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000;74(12):5716–25. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farzan M, et al. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J Virol. 1998;72(9):7620–5. doi: 10.1128/jvi.72.9.7620-7625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansfield K, et al. Vaccine protection by live, attenuated simian immunodeficiency virus in the absence of high-titer antibody responses and high-frequency cellular immune responses measurable in the periphery. J Virol. 2008;82(8):4135–48. doi: 10.1128/JVI.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauduin MC, et al. Induction of a virus-specific effector-memory CD4+ T cell response by attenuated SIV infection. J Exp Med. 2006;203(12):2661–72. doi: 10.1084/jem.20060134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gauduin MC, et al. Inhibition of simian immunodeficiency virus (SIV) replication by CD8(+) T lymphocytes from macaques immunized with live attenuated SIV. J Virol. 1998;72(8):6315–24. doi: 10.1128/jvi.72.8.6315-6324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauduin MC, et al. Characterization of SIV-specific CD4+ T-helper proliferative responses in macaques immunized with live-attenuated SIV. J Med Primatol. 1999;28(4-5):233–41. doi: 10.1111/j.1600-0684.1999.tb00274.x. [DOI] [PubMed] [Google Scholar]
- 30.Gauduin MC, et al. Immunization with live attenuated simian immunodeficiency virus induces strong type 1 T helper responses and beta-chemokine production. Proc Natl Acad Sci U S A. 1999;96(24):14031–6. doi: 10.1073/pnas.96.24.14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Efficiency of gp120 and gp140 biotinylation. Avitag bearing gp120 and gp140 were biotyinlated using biotin ligase. Biotinylation was assessed by ELISA. a. Comparison of none and biotinylated proteins. Elisa plates were coated with Avitag bearing but not biotinylated (gp120 or gp140) proteins; and Avitag bearing and biotinylated (gp120-biotinylated or gp140-biotinylated) proteins at 1 ng/μl. All proteins were detected with plasma pool of SIV-infected monkey (Plasma/SIV+) followed by HRP-conjugated anti-human IgGs. Biotinylation specificity was verified with HRP-conjugated streptavidin. b. Assessment f biotinylation completion by comparison to a fully biotynlated standard. Elisa plates were coated with Avitag bearing biotinylated gp120 or gp140 and BIS-A a commercial fully biotinylated standard at 1; 0.6 and 0.2 ng/μl. Detection was performed with HRP-conjugated streptavidin. OD, opticlal density.