Abstract
The glycolipid transfer protein (GLTP) superfamily is defined by the human GLTP-fold which represents a novel motif for lipid binding/transfer and for reversible interaction with membranes, i.e. peripheral amphitropic proteins. Despite limited sequence homology with human GLTP, we recently showed that HET-C2 GLTP of Podospora anserina is organized conformationally as a GLTP-fold. Currently, insights into the folding stability and conformational states that regulate GLTP-fold activity are almost nonexistent. To gain such insights into the disulfide-less GLTP-fold, we investigated the effect of changing pH on the fungal HET-C2 GLTP-fold by taking advantage of its two tryptophans and four tyrosines (compared to three tryptophans and ten tyrosines in human GLTP). pH-Induced conformational alterations were determined by changes in: i) intrinsic tryptophan fluorescence (intensity, emission wavelength maximum, anisotropy); ii) circular dichroism over the near-UV and far-UV ranges including thermal stability profiles of the derivatized molar ellipticity at 222 nm; iii) fluorescence properties of 1-anilinonaphthalene-8-sulfonic acid; iv) glycolipid inter-membrane transfer activity monitored by Förster resonance energy transfer. Analyses of our recently solved crystallographic structure of HET-C2 (1.9 Å) enabled identification of side chain electrostatic interactions that contribute to HET-C2 GLTP-fold stability and can be altered by pH change. Side-chain interactions include numerous salt bridges and interchain cation-pi interactions, but not intramolecular disulfide bridges. Histidine residues are especially important for stabilizing the local positioning of the two tryptophan residues and the conformation of adjacent chains. Induction of a low pH-induced, molten globule-like state inhibited glycolipid intermembrane transfer by the HET-C2 GLTP-fold.
Keywords: cation-π and π-stacking interactions involving histidine and tryptophan, networked salt bridges, tryptophan and ANS fluorescence, far-UV and near-UV circular dichroism, fungal cell incompatibility
Characterization of protein stability and folding intermediates can provide insights into normal folding mechanisms as well as pathological conditions involving protein aggregation (e.g. Alzheimer’s disease). A partially unfolded state of special interest is the molten globule (MG) state, which maintains a compact organization with pronounced secondary structure, but lacks tertiary conformation (1–4). MG-like states have been linked to a number of biological processes such as protein-protein interactions and protein insertion/translocation into membranes including the action of lipid binding/transfer proteins (5–12).
In filamentous fungi, het genes play a major role in vegetative incompatibility processes involving cell-cell recognition (13–16). One het gene product, HET-C2, is known to act as lipid binding/transfer protein that is specific for simple glycosphingo-lipids (17,18). Recently, we showed that HET-C2 is organized as a two-layer ‘sandwich’ dominated by alpha-helices (18) and is folded very similarly to human glycolipid transfer protein (GLTP), which serves as the prototype and founding member of the GLTP superfamily (19–21). Like human GLTP (22), HET-C2 acquires and delivers glycolipids by interacting transiently and reversibly with membranes (17,18). Thus HET-C2 displays the defining features of a peripheral amphitropic membrane protein, which typically has affinity for both aqueous and nonpolar environments but requires neither post-translational modifications nor anchor proteins for reversible interaction with membranes (23,24).
Presently, very limited information exists about the stability of the GLTP-fold and the physical conditions that can affect folding status. Moreover, it is not known whether partially-unfolded (e.g. MG-like states) play any role in GLTP-fold functionality. What is known is that human GLTP has a highly cooperative unfolding transition that occurs near 54°C at neutral pH (25) consistent with 55°C inactivation reported by Nylund and Mattjus (26). However, HET-C2 exhibits a 4–5°C lower unfolding transition temperature midpoint (~49°C) at neutral pH despite being a GLTP-fold (18). Further characterization under various physical conditions is likely to provide better understanding of the key parameters controlling GLTP-fold stability and folding. Our strategy was to take advantage of the simpler Trp and Tyr composition of HET-C2 (two Trp & four Tyr) compared to GLTP (three Trp & ten Tyr) and to use fluorescence and far-UV/near-UV circular dichroism (CD) spectroscopy to further elucidate the relationship between GLTP-fold conformation and functionality. Herein, we have characterized the pH dependence of HET-C2 stability and activity as well as investigated whether partially-unfolded, pH-induced, MG-like states regulate the glycolipid transfer activity of HET-C2.
EXPERIMENTAL PROCEDURES
Expression and Purification of HET-C2
HET-C2 was expressed and purified similarly to GLTP (18,27) using pET-30 Xa/LIC (Novagen) containing het-c2 ORF of P. anserina (NCBI GenBank # U05236). Transformed BL21 cells (E. coli) grown at 37°C overnight in Luria-Bertani medium were induced with IPTG (0.1 mM) and then grown for 16–20 h at 15°C. Purification of rHET-C2 from soluble lysate protein was accomplished by Ni-NTA affinity chromatography. The N-terminal His-S-tag was removed using factor Xa, yielding protein identical in sequence to native HET-C2. HET-C2 was repurified by FPLC size exclusion chromatography using a HiLoad 16/60 Superdex-75 prep grade column (Amersham). HET-C2 fractions were pooled, concentrated centrifugally (10 kDa cut-off membrane), and checked for purity by SDS-PAGE.
Fluorescence Measurements
Trp fluorescence of HET-C2 was measured at 25°C from 310 to 420 nm with a SPEX Fluoromax spectrofluorimeter (Horiba Scientific, Edison NJ) using excitation and emission bandpasses of 5 nm. Excitation at 295 nm minimized contributions from Tyr and protein concentration was kept at OD295<0.1 to avoid inner filter effects (25,28,29). Fluorescence polarization measurements of Trp were performed at room temperature (25 °C) using a Hitachi polarization accessory. Anisotropy values were calculated using equation 1:
| (1) |
where IVV and IVH are the background-subtracted, measured fluorescence intensities obtained with the excitation polarizer vertically oriented and the emission polarizer vertically and horizontally oriented, respectively. The grating correction factor (G) is the efficiency ratio of the detection system for vertically and horizontally polarized light and equals IVH/IHH. For measurements of 1-anilinonaphthalene-8-sulfonic acid (ANS), excitation occurred at 375 nm and emission intensity was recorded from 400 to 560 nm.
Circular Dichroism Spectroscopy
CD spectra were collected using a J-810 spectro-polarimeter (JASCO, Japan) equipped with a CTC-345 temperature-control system at 50 μM HET-C2 and 10°C in 10 mM Na bicarbonate (pH 10), carbonate-bicarbonate (pH 8 or 9), phosphate (pH 6 or 7.4), citrate-phosphate (pH 3, 4, or 5) or glycine-HCl (pH 2) while purging continuously with N2. Spectral and temperature dependence measurements in the far-UV (185–250 nm) and near-UV range (250–320 nm) were performed as detailed previously (18, 25, & references therein). Also detailed are calculations of HET-C2 secondary structure from far-UV CD spectra using the CDPro program and of tertiary structure class using the CDPro CLUSTER program.
Protein Concentration
The concentration of HET-C2 (molar absorptivity at 280 nm = 17.12 mM−1cm−1) was measured using a Beckman DU 640 spectrophotometer at 1.8 nm bandwidth and averaging results from four calculation methods (30–33). Spectra were corrected for turbidity by plotting the log dependence of solution absorbance versus the log of the wavelength and extrapolating their linear dependence in the 340–440 nm range to the 240–300 nm absorption range using the DU-640 scatter correction routine. Extrapolated absorbance values were subtracted from measured values, decreasing the apparent protein absorbance at ~280 nm by ~15%.
HET-C2 Transfer Activity of Glycolipid
Continuous real-time monitoring of glycolipid intermembrane transfer by HET-C2 and determination of initial lipid transfer rates were obtained by Förster resonance energy transfer (FRET) using a continuously stirred sample (~100 rpm) maintained at 25 ±0.1°C (Neslab, RTE-111) while measuring emission intensity at 425 nm (17). Both fluorescent lipids [1 mole% AV-glycolipid containing the anthyrlvinyl fluorophore the omega acyl position [(11E)-12-(9-anthryl)-11-dodecenoyl acyl chain] and 1.5 mole% 1-acyl- 2-[9-(3-perylenoyl)-nonanoyl]-sn-glycero-3-phosphocholine] (Per-PC) were initially localized in the donor vesicles comprised of POPC matrix and formed by rapid ethanol injection. Minimal emission by AV-glycolipid occurs upon excitation (370 nm) because of resonance energy transfer to nearby Per-PC. Minimal emission change also is elicited by the addition of pure sonicated POPC acceptor vesicles (10-fold excess) because of the slow time course of spontaneous lipid transfer. Addition of catalytic HET-C2 amounts causes a sudden, exponential increase in AV emission intensity (425 nm) as the protein extracts AV-labeled glycolipid from donor vesicles and delivers to the acceptors (creating separation from the ‘nontransferable’ perylenoyl lipid). Addition of Tween 20 detergent at the end of the kinetic time course provides a measure of maximum AV intensity achievable by ‘infinite’ separation from perylenoyl fluorophore. The initial lipid transfer rate νo is obtained by nonlinear regression analysis and fitting to first-order exponential behavior using ORIGIN 7.0 software (Origin Lab, Northhampton, MA). ΔF represents the AV emission intensity difference after extended incubation with HET-C2 compared to time zero with no HET-C2. The standard deviations in fitting are calculated at 95% confidence intervals and R2 values are >0.96 for all estimates.
RESULTS
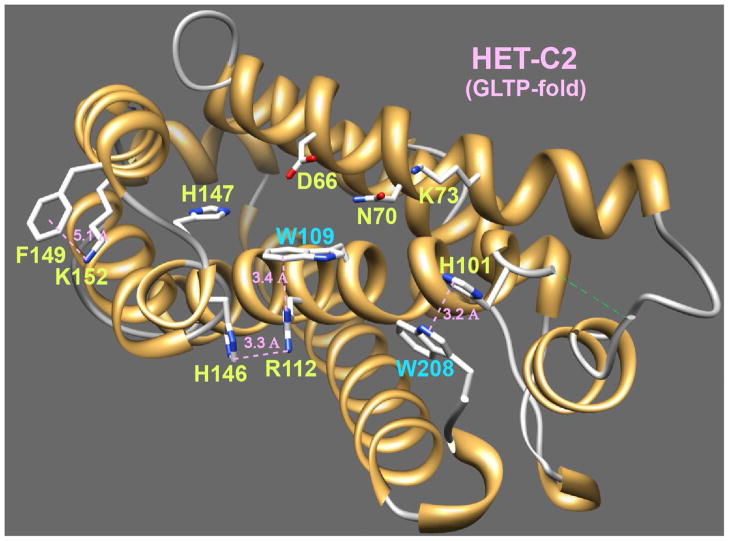
Our recent resolution of HET-C2 structure by X-ray diffraction (1.9 Å) shows the global architecture to be a GLTP-fold (18). However, unlike human GLTP, which has three Trp residues (19,28,34), HET-C2 contains only two Trp residues. As illustrated in Figures 1 and S1, Trp109 of α-helix 4 acts as a stacking plate in the glycolipid headgroup recognition center and helps orient the ceramide-linked sugar for hydrogen bonding with Asp66, Asn70, and Lys73, thus functioning analogously to Trp96 of human GLTP. Figure 1 also shows how Arg112 stabilizes the orientation of the Trp109 indole ring from beneath by cation-pi interaction (~3.4Ǻ) (35). The orientation of the planar guanidinium moiety of Arg112 also is influenced by stacking (~3.3Ǻ) against the imidazole ring of His146 (α5-α6 loop) (36). The other Trp residue (Trp208) forms the C-terminus of HET-C2, is surface accessible, and stacks (~3.2Ǻ) against the imidazole ring of His101 on the α3-α4 loop, thus distinguishing it from Trp85 and Trp142 of human GLTP. Because of the apparent involvement of the intrinsically fluorescent Trp109 and Trp208 residues in stabilizing HET-C2 architecture via their interactions with His146 and His101, pH-induced conformational changes in HET-C2 were initially investigated using fluorescence spectroscopy.
Figure 1.
HET-C2 Architecture. A GLTP-fold has been determined for HET-C2 (PDB 3kv0) by X-ray diffraction (1.9 Å) (18). Also depicted are the locations of Trp109 and Trp208 along with nearby residues (Arg112, His146, His101) that affect their topology.
pH Dependence of Trp Fluorescence in HET-C2
As shown in Figure 2A, the intrinsic Trp fluorescence of HET-C2 has an emission wavelength maximum (λmax) that is strongly red-shifted (~354 nm at neutral pH), consistent with a highly polar environment for the fluorophore and in agreement with X-ray data showing surface localizations for the indole rings of Trp109 and Trp208 (Fig. S2). The HET-C2 Trp emission intensity is relatively high at and above neutral pH with maximum intensity occurring near pH 10. Below neutral pH, Trp emission intensity decreases dramatically. This response contrasts that of λmax (Fig. 2B) which remains constant (~354 nm) over the pH 5.5 to 7 range, but displays discernible λmax blue shifts to ~353 nm at pH 5, 8, and 9 as well as to ~352 nm at pH 4 and pH 10. More pronounced λmax blue shifts to ~349 nm occur at pH values of 2 and 3. The blue-shifted λmax suggests pH-induced conformational change that results in decreased polarity of the Trp average environment. To evaluate further, the anisotropy of the Trp emission signal was measured (Fig. 2C). The overall pattern of pH-induced anisotropy change correlated remarkably well with that of the λmax blue-shifts. For instance, Trp anisotropy was maximal and unaltered at pH values between 5.5 and 7. Moderate decreases in anisotropy are observed at pH 5, 8, and 9, slightly larger decreases occur at pH 4 and pH 10, and the most pronounced decreases are observed at pH 3 and 2. Because diminished anisotropy indicates motional increase by the fluorophore, we concluded that low pH is particularly effective at inducing HET-C2 unfolding. Altogether, the fluorescence data suggest that HET-C2 is relatively stable over a wide pH range (e.g. pH 5 to 9) but becomes destabilized at more extreme pH conditions (pH ≤4 or ≥10).
Figure 2.
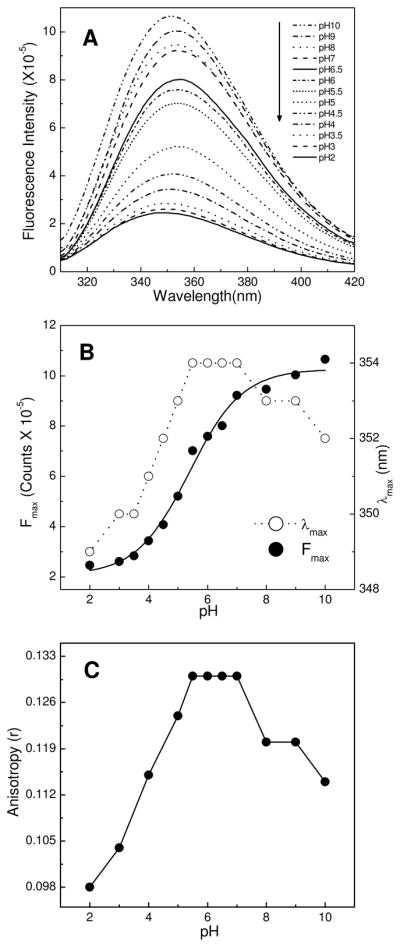
Effect of pH on intrinsic tryptophan fluorescence of HET-C2. A) Emission spectra of HET-C2 acquired at indicated pH values; B) Fluorescence emission intensity maxima and wavelength maxima (λmax) plotted as a function of pH. C) Fluorescence anisotropy values of HET-C2 at various pH values. Protein concentration was 1 μM. Fluorescence excitation occurred at 295 nm and emission measurements were performed as detailed in the Experimental Procedures.
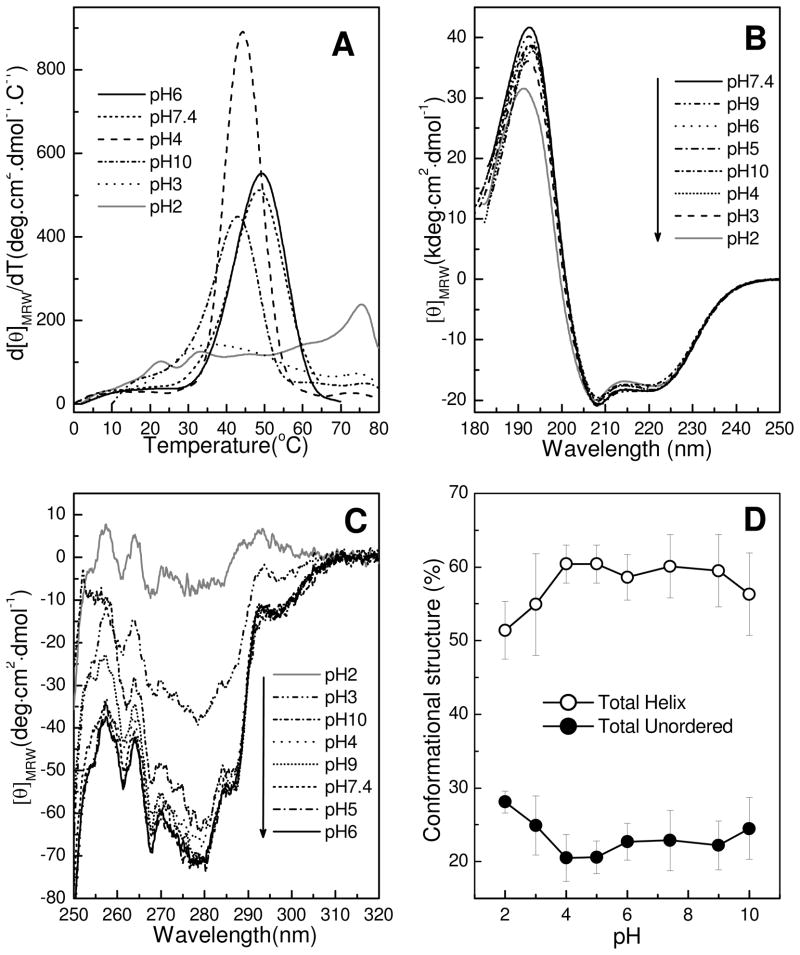
To gain additional insights into HET-C2 conformational stability at various pHs, we used far-UV CD spectroscopy to monitor the first derivative of the CD signal at 222 nm as a function of temperature (37). The choice of wavelength was dictated by the well-established property of α-helical proteins having a large CD signal at 222 nm that dramatically decreases upon unfolding. The heat-induced denaturation profiles in Figure 3A show that HET-C2 undergoes highly cooperative unfolding at pH 7.4 between 25 and 55°C, with a transition midpoint of ~49°C. The relatively low temperature midpoint for unfolding is consistent with a lack of stabilization by intramolecular disulfides. At pH 6, the thermal unfolding profile of HET-C2 remained nearly unaltered. At pH 10, the transition temperature midpoint decreased to 44°C and the unfolding cooperativity decreased, consistent with moderate destabilization of HET-C2. Similar destabilization was indicated at pH 4 by the 43°C midpoint of the unfolding transition, but there was significant enhancement in the unfolding cooperativity. At pH 3, the unfolding transition temperature midpoint was strongly decreased (Tm ~36°C) and the unfolding transition was extremely broad, occurring over the range of 15°C to 68°C (Fig. 3A). A smaller magnitude transition peak also became evident near 75°C. At pH 2, the 75°C transition peak was significantly enhanced while only two minor peaks (Tm ~23°C and ~34°C) were evident at lower temperature indicating a highly unfolded structure. The heat-induced unfolding transitions of HET-C2 were found to be ~85–90% reversible at pHs 2 and 3, but were only 20% reversible at pH 7.4, upon heating 20–25 °C above the midpoint of the unfolding transition temperature. This finding is consistent with diminished HET-C2 solubility near its isoelectric point, which is estimated to be 6.81 from amino acid composition. Inclusion of dithiothreitol did not improve reversibility at neutral pH for GLTP, which responds similarly at low and neutral pH (Kamlekar, Kenoth, & Brown, unpublished observation). Thus, the thermodynamic parameters attributable to the HET-C2 unfolding transition could not be reliably ascertained at physiological pH.
Figure 3.
pH Induced conformational changes of HET-C2 at different pH values. Far-UV and near-UV CD spectra of 50 μM HET-C2 were acquired at 10°C using a J-810 spectropolarimeter as detailed in the Experimental Procedures. A) Temperature dependence of molar ellipticity at 222 nm (derivative plot). Unfolding of HET-C2 is shifted to lower temperature at pH 3 and unfolding cooperativity is completely lacking at pH 2. B) Far-UV CD spectra at different pH values presented in units of molar ellipticity per residue. The negative bands at ~208 nm and positive bands at ~192 nm are characteristic of high helical content, with large negative n-π* transitions at 222 nm and π-π* transitions split into two transitions because of exciton coupling. HET-C2 secondary structure is nearly identical at all pHs tested. C) Near-UV CD spectra. The strong signal response from Tyr and Trp in HET-C2 at moderate pH is moderately disrupted at pH 3 and is almost completely lost at pH 2, consistent with diminished tertiary structure. D) Change in HET-C2 secondary structure induced by pH. The plotted values illustrate calculations of HET-C2 secondary structure, presented as part of Supplementary Table S1 and obtained from the far-UV CD spectra.
Does Extreme pH Induce a Molten Globule-like State for HET-C2?
Partial unfolding to the molten-globule (MG) state is known to occur for many proteins at extreme pHs and has been especially well-studied at low pH (38,39). To determine whether HET-C2 could transition to a MG-like state at low or high pH, secondary and tertiary structure were analyzed by CD spectroscopy. Previously, we showed that the secondary structure of HET-C2 at pH 7.4 is dominated by high helical content, i.e., 56.6% helix, 8.8% β-structure, 12.1% β-turns, and 24.1% random, as assessed by far-UV CD (18). Figure 3B shows far-UV CD spectra acquired at various pHs between 2 and 10. The helical content of HET-C2 remains mostly unaltered over the entire pH range, except for minor changes observed at pH 2. Figure 3D illustrates the stability of HET-C2 helical content and the increase in unordered structure induced by low pH. The data are based on secondary structural calculations (Supplementary Table S1) derived by CDPro software analysis from the far-UV CD spectra (18,25).
Near-UV CD spectra (Fig. 3C), which assess tertiary structure, showed significant environmentally-induced optical activity arising from the four Tyr, two Trp, and 12 Phe residues of HET-C2 in the pH range between 5 and 9. Signals in the 270–290 nm region typically originate from Tyr; those in the 280–300 nm region, from Trp; while those in the 250–270 nm region generally are attributed to Phe (40). In HET-C2, the peak signals at ~257, ~262, and ~271 nm likely belong to the 12 Phe residues, whereas the prominent negative signal near 280 along with the shoulder peaks at 287 and 293 nm could originate from Tyr or Trp. In any case, the near-UV signal response shows little variation between pH 4 and 9. At more basic pH (pH 10), only marginally decreased signal intensity is evident in the spectra. In contrast, at pH 3, pronounced change occurs in the near-UV CD signal suggesting a considerable loss of tertiary folding, while nearly complete loss of tertiary folding is evident at pH 2. Notably, residual signals near 257 nm and 262 nm, originating from the protein interior core Phe residues, persist at extreme pH. Together, the large reduction in tertiary structure while maintaining secondary structure and the dramatic loss of thermally-induced unfolding cooperativity when pH < 4 provide strong evidence for HET-C2 transitioning to a molten globule-like state at low pH.
HET-C2 Unfolding assessed by ANS Fluorescence Analysis
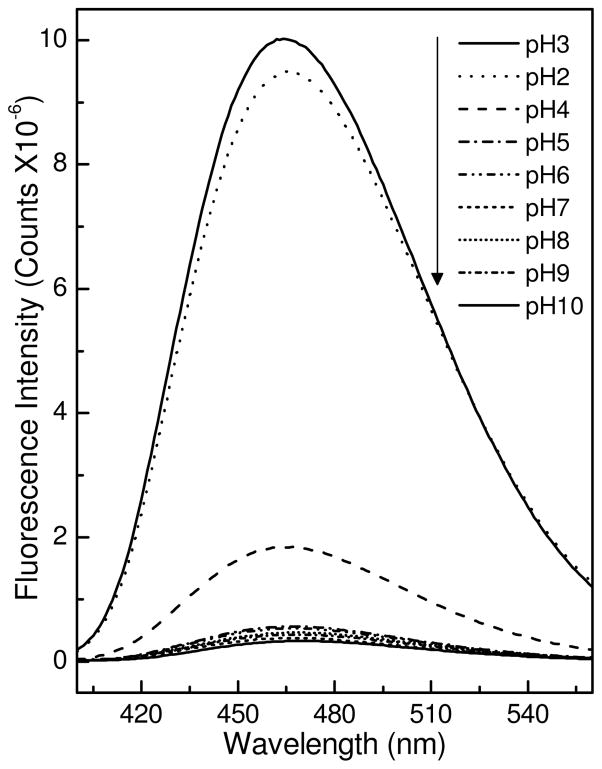
Changes in ANS fluorescence often are used to monitor formation of folding intermediates during protein unfolding and refolding (28,41,42). The presence of large solvent-exposed hydrophobic patches is a general property of a partially-folded protein. ANS interaction with exposed hydrophobic sites on a protein is accompanied by considerable increase in dye fluorescence intensity and a substantially blue shifted emission maximum (41–43). Accordingly, we used ANS fluorescence to study pH-induced conformational changes in HET-C2. Figure 4 shows that ANS emission intensity and λ max values remain nearly unchanged in the presence of HET-C2 over the pH range from 5 to 9. At pH 4, the intensity is slightly elevated. However, at pH 2 and pH 3, dramatic increases in intensity are observed along with substantial λ max blue shifting, consistent with the appearance of solvent-exposed hydrophobic surfaces. The findings support the CD data showing that low pH transforms HET-C2 to a partially-folded intermediate state with molten globule-like properties.
Figure 4.
ANS Fluorescence at Different pHs. HET-C2 (2 μM) was incubated with ANS (10 μM) for 10 min at the indicated pH values and the fluorescence spectra were recorded. Samples were excited at 375 nm and emission was measured from 400–560 nm.
Is Glycolipid Transfer Activity by HET-C2 Helped or Hindered by the pH-Induced Molten Globule-like State?
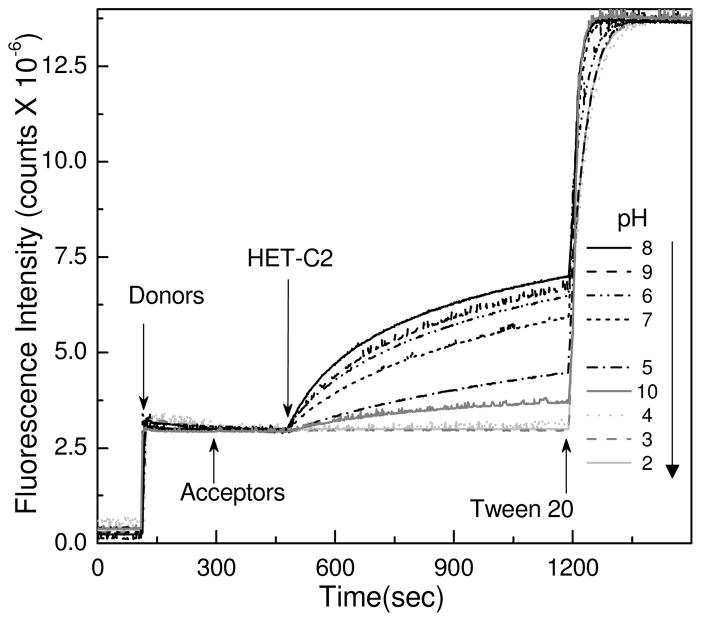
Figure 5 shows how pH affects the glycolipid transfer activity of HET-C2. After pre-incubation for 10 min at the indicated pH, glycolipid transfer activity was monitored using a well-established Förster resonance energy transfer (FRET) assay that enables real time kinetic monitoring of fluorescent glycolipid transfer between membranes (17). Maximum transfer activity was found between pH 6 and 9 (Table 1). Activity was strongly diminished (~90%) at pH 10 and ~75% diminished at pH 5. More acidic pH (e.g. pH 4) resulted in almost complete loss of activity. At pH 3 or 2, glycolipid transfer by HET-C2 ceased. Having membrane vesicles present during HET-C2 incubation at the indicated pH (prior to assay initiation) did not change the outcome. These findings suggest that formation of an acid-induced, molten globule-like state strongly inhibits the ability of HET-C2 to transfer glycolipid between membranes.
Figure 5.
pH-Induced Change in Glycolipid Transfer Activity of HET-C2. Förster energy transfer involving fluorescently-labeled glycolipid (AV-GalCer) and phospholipid (Perylenoyl-PC) was used to assess glycolipid intervesicular transfer by HET-C2. Stock protein at pH 8 was diluted ~10-fold into buffer of the specified pH and incubated at room temperature for 10 min. Assays were initiated by ~1000-fold dilution of HET-C2 into indicated pH. AV-GalCer was excited at 370 nm and emission was monitored at 425 nm. Additional details are provided in the Experimental Procedures.
Table 1.
Glycolipid transfer activity (initial transfer rate) by HET-C2 as a function of pH.
| pH | Initial transfer rate of AV-GalCer (pmol/min)* |
|---|---|
| 2 | 0.75 ± 0.39 |
| 3 | 0.25 ± 0.25 |
| 4 | 1.30 ± 0.08 |
| 5 | 7.78 ± 0.35 |
| 6 | 22.07 ± 0.25 |
| 7 | 16.39 ± 1.46 |
| 8 | 33.70 ± 0.70 |
| 9 | 27.79 ± 1.45 |
| 10 | 3.46 ± 1.04 |
Determined from Förster resonance energy transfer data shown in Figure 5
DISCUSSION
The present study provides fundamental insights into the stability and folding of HET-C2, a fungal GLTP-like protein with a core architecture that strongly resembles the human GLTP-fold. HET-C2 contains a α-helically-dominated, two-layer ‘sandwich’ topology characteristic of the GLTP-fold in which the α1, α2, α6 and α7 helices make up one layer and the α3, α4, α5 and α8 helices make up the other layer (18). HET-C2 contains two Cys residues, Cys118 in helix 4 and Cys162 in the α6–7 loop, that are too far apart (15.6 Å) to stabilize conformation via disulfide bridging (Fig. S2A). An important consequence is moderate temperature stability at neutral pH with an unfolding transition temperature midpoint of ~49°C (18). The 4–5°C lower unfolding transition temperature midpoint of HET-C2 compared to human GLTP could reflect adaptation for growth conditions typical for Podospora anserina, a saprophytic fungus inhabiting the dung of herbivores. In any case, the lack of Cys residue conservation (Fig. S2) and of intra-molecular disulfide bridging in HET-C2 and GLTP clearly distinguishes the GLTP-fold from other lipid binding/transfer protein folds that also are dominated by α-helices, i.e. saposins and nonspecific plant lipid transfer proteins (nsLTP-folds) (22). The saposin motif is characterized by a set of five helices, sometimes arranged in two-layers, but always connected by a conserved pattern of three intramolecular disulphide bonds that impart heat stability (44–49). In nsLTP-folds, the core tertiary superstructure consists of multiple helices, bundled to form a large central, tunnel-like hydrophobic cavity (50–55). The spatial arrangement of eight cysteines, interspersed to form four interhelical disulfide bridges, stabilizes the bundle-like structure and is thought to be important for maintaining tertiary structure when the large central hydrophobic tunnel is devoid of lipid ligand (52,55). A consequence of the multiple disulfide bridging is remarkable heat stability with unfolding transition temperatures in the 90–100°C range (50).
What Fluorescence and CD reveal about HET-C2 Stability and Functionality
Despite the moderate temperature stability, the HET-C2 GLTP-fold remains relatively stable over a wide pH range extending from 5 to 10, as revealed by intrinsic signal changes obtained from fluorescence (e.g. Trp) and CD spectroscopic analyses. The indole rings for both Trp residues in HET-C2 are solvent-exposed (Fig. S3) resulting in a more highly red-shifted emission λ max (~355 nm) compared to GLTP (λ max≈348 nm) in which the solvent accessibility of one of three Trps is relatively restricted. The responsiveness of HET-C2 Trp emission to pH change appears to reflect altered inter-actions with neighboring His residues. In the case of Trp208, the indole ring stacks against the His101 imidazole ring (~3.2Ǻ). Water-bridged hydrogen bonding of His101 with Glu105 helps keep the imidazole ring favorably aligned for stacking against the Trp208 indole ring. The His101/Trp208 π-stack and associated hydrogen-bonding interactions have the net effect of altering the conformation of the C-terminal region of HET-C2 compared to GLTP (18). With Trp109, interaction with His146 is indirect and occurs by networking through Arg112 as shown in Figure 1 and detailed earlier. Because of the involvement of the His residues, which change their ionization state at relatively moderate pH (56), the responsiveness of the two Trp residues is not surprising. Quenching by increased levels of protonated histidine probably contributes to the diminished Trp emission intensity accompanying the stepwise pH decreases between 6 and 4. Considering the surface location of the His101/Trp208 stack and the aqueous accessibility of the Trp109/Arg112/His146 network, the net pK value of ~5.5 that can be approximated from the Trp fluorescence data is reasonable (56,57). It is noteworthy that other residues known to quench Trp emission, i.e. Cys and Tyr, are not sufficiently close to be effective quenchers (58). Both Cys residues (pKa ~8.6) are located >17Ǻ from either Trp residue. All four Tyr residues (pKa ~9.8) are >20Ǻ away except for Tyr139 which is 8–10Ǻ from Trp109. The closest Lys residues (Lys73, Lys152, Lys207) (pKa ~10.4) are 8–11Ǻ away, diminishing their ability to quench by H+ transfer (58). In contrast, the close proximity of His and the established ability of protonated histidine to quench Trp fluorescence are consistent with a major role for His in the regulation of HET-C2 Trp emission intensity (57–62).
Although quenching of Trp by protonated His is likely to increase over the pH range from 6 to 4, the diminished Trp anisotropy also suggests accompanying Trp conformational change. The local nature of the conformational change is supported by the nearly unaltered far-UV and near-UV CD spectra at pH 4, 5, 6, and 7.4 as well as by only slightly altered thermal midpoints of unfolding. What is noteworthy with respect HET-C2 GLTP-fold functionality is the robust glycolipid transfer activity over the pH range of 6 to 9, but the precipitous drop in glycolipid transfer activity at moderately acidic pHs of 5 and 4, when native global folding still remains largely unaltered. Thus, the local interactions between Trp and His appear to play a crucial regulatory role in controlling the glycolipid transfer activity of HET-C2.
Below pH 4, the fluorescence data for Trp and ANS, and the near-UV CD spectra indicate global alterations in HET-C2 conformation that result in loss of glycolipid transfer activity. The low-pH induced changes are likely to alter the two interchain cation-pi interactions involving His101 and His146 as well as disrupt one or more of the numerous salt bridges involving Asp and Glu residues (pKa 3.9 & 4.3, respectively) (Table 2). Among the salt bridges, two are complex or networked bridges (Glu46-Arg75-Glu72 and Asp90-Arg93-Glu205) along with half dozen single bridges that are interchain rather than intrahelical in nature (63). While the effect of pH change on GLTP has been reported by West et al. (34), their focus was exclusively on transfer activity over the pH range of 4 to 10 without accompanying conformational insights.
Table 2.
Inter-Residue Interactions of the HET-C2 GLTP-fold
| Salt Bridges (<4.0Å) | Nature of Interaction |
|---|---|
| E17 ↔ K178 (~3.6Å) | (η1↔α8) - interchain |
| *E46 ↔ R75 (~2.8Å) | (α1↔α2) - interchain |
| D53 ↔ K64 (~3.6Å) | (α1↔α2) - interchain |
| *E72 ↔ R75 (~2.8Å) | (α2↔α2) – intrahelix |
| E84 ↔ R77 (~3.5Å) | (α2–3 coil↔α2) - interchain |
| *D90 ↔ R93 (~2.8Å) | (α3↔α3) – intrahelix |
| *E205 ↔ R93 (~2.9Å) | (η6↔α3) - interchain |
| E105 ↔ H101 (~3.7Å) | (α4↔α3–4 coil) - interchain |
| E115 ↔ K198 (~3.4Å) | (α4↔α8) - interchain |
| K123 ↔ E129 (~3.9Å) | (α4↔α4–5 coil) - interchain |
| D133 ↔ R136 (~3.1Å) | (α5↔α5) - intrahelix |
| Cation-Pi (<5.0Å) | |
| W96 ↔ R112 (~3.4Å) | (α4↔α4) - intrahelix |
| ** W208 ↔ H101 (~3.5Å) | (COOH↔α4–5 coil) - interchain |
| F149 ↔ K152 (~5.1Å) | (α6↔α6) - intrahelix |
| ** H146 ↔ R112 (~3.4Å) | (η5↔α4) - interchain |
| H2O-bridging (<3.0Å) | |
| E105↔ H2O ↔ H101 (2.6Å - 2.6Å) | (α4↔α3–4 coil) - interchain |
| H147 ↔ H2O ↔ D66 (2.8Å - 2.5Å) | (η5↔α2) - interchain |
| E105↔ H2O↔ W96 (~2.8Å - 2.4Å) | (α4↔ α4) - intrachain |
The distances between residue side-chains are derived from our HET-C2 X-ray structure (1.9 Å; PDB 3kv0) (18) which also identifies residue location within secondary structural elements (see Fig S2, lower panel). Salt bridges are indicated by oppositely charged residues within 4 Å of each other (e.g., 63). The asterisk indicates networked salt bridges involving Arg75 and Arg93. The cation-π interaction between Arg112 and Trp109 is predicted as energetically favored by the on-line CaPTURE program (http://capture.caltech.edu) (35). His146 and Arg112 π-stacking and
Functional Consequences of the Low pH Induced MG-like State in HET-C2
MG-like states reportedly help many amphitropic peripheral membrane proteins perform their biological functions by promoting association with membranes (24). Lactalbumin (8), retinol binding protein (38), colicin A (7), diphtheria toxin T-domain (5), StAR lipid binding domains (10), and barley nsLTP (12) are a few such proteins. The GLTP-fold represents both a novel motif among lipid binding/transfer proteins and a novel membrane interaction motif among amphitropic peripheral proteins that translocate to membranes during function (25). For this reason, the existence and potential role of a MG-like state for the HET-C2 GLTP-fold was of interest. Such states typically are characterized by maintenance of or increase in secondary structure, a reduction/loss of tertiary structure, and loss of cooperativity of melting (1–4). In this respect, it is noteworthy that our CD spectra reveal maintenance of HET-C2 regular secondary structure and partial loss of tertiary structure at pH 3, in contrast to the complete loss of tertiary structure observed at pH 2. Also, considerable lowering of the unfolding transition temperature and loss of unfolding cooperativity are observed at pH 3 but complete loss occurs at pH 2. Dramatic enhancement of ANS fluorescence intensity is evident upon binding by HET-C2 at pH 3 and 2. Thus, the experimental data are consistent with the existence of an MG-like state at low pH. However, it is clear from the activity measurements that glycolipid transfer activity by HET-C2 is completely lost at pH 3, indicating that the pH-induced, MG-like state does not promote glycolipid transfer by the HET-C2 GLTP-fold.
It is worthwhile to recall that HET-C2 carries out important functional roles that may occur independently of its glycolipid binding/transfer ability. Het genes exhibit extensive polymorphism and generally encode proteins carrying a HET domain. In Podospora anserina, the process of protoplasmic incompatibility involves cell destruction that can be triggered by interaction of het-C alleles, (e.g. het-c2 encoding GLTP-like HET-C2) and het-E, encoding a HET domain and a WD repeat domain involved in recognition (64). The structural similarity of HET-E with mammalian APAF-1, a protein controlling cytochrome c-induced apoptosis, suggests parallels between cell death by incompatibility and by programmed cell death mechanisms in higher eukaryotes. It has been proposed that cytochrome c binds to the APAF-1 in the cytoplasm to form a complex that initiates enzymatic cascades leading to apoptosis. Cytochrome c is thought to be released from mitochondria in the MG state during apoptosis. Within this context, the current observation of HET-C2 transition to the MG-like state at low pH may be significant. Modifications of the vacuolar compartment are frequently observed during cell death reactions. Vacuole membrane permeabilization or rupture followed by liberation of lytic enzymes and acidification of the cytoplasm is proposed to be involved in cell death by incompatibility in filamentous fungi. In this acidic environment, it is tempting to speculate that HET-C2 may achieve a partially unfolded conformation of functional importance to the fungal incompatibility cell death response. Clearly additional cell biological experiments will be needed to further evaluate these ideas.
Supplementary Material
Acknowledgments
We thank Helen Pike for expressing and purifying the HET-C2 protein and the Resource for Biocomputing, Visualization, and Informatics of UCSF (NIH P41 RR-01081) for the use of Chimera.
Abbreviations
- HET-C2
Heterokaryon incompatability C2 protein
- GLTP
glycolipid transfer protein
- MG
molten globule
- ANS
1-anilinonaphthalene-8-sulfonic acid
- CD
circular dichroism
- FRET
Förster resonance energy transfer
- UV
ultraviolet
- AV
anthyrlvinyl
- Per
3-perylenoyl
- PC
1,2-acyl-sn-glycero-3-phosphocholine
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
Footnotes
We are grateful for financial support by NIH/NIGMS GM45928 & GM34847, NIH/NCI CA121493, Spanish Ministerio de Ciencia e Innovacion (MICINN BFU2010–17711), Russian Foundation for Basic Research #09–04–00313, the Abby Rockefeller Mauzé Trust, and the Dewitt Wallace, Maloris, Mayo, and Hormel Foundations.
One table summarizing HET-C2 secondary structure, as computed from the far-UV CD spectra, and three images of HET-C2 conformation are provided as Supplemental Materials and may be accessed free of charge online at http://pubs.acs.org
References
- 1.Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov SY, Ptitsyn OB. Alpha-lactalbumin: Compact state with fluctuating tertiary structure. FEBS Lett. 1981;136:311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- 2.Dobson CM. Unfolded proteins, compact states and molten globules. Curr Opin Struct Biol. 1992;2:6–12. [Google Scholar]
- 3.Dobson CM. Protein folding: Solid evidence for molten globules. Curr Biol. 1994;4:636–640. doi: 10.1016/s0960-9822(00)00141-x. [DOI] [PubMed] [Google Scholar]
- 4.Ptitsyn OB, Bychkova VE, Uversky VN. Kinetic and equilibrium folding intermediates. Phil Trans R Soc Lond B. 1995;348:35–41. doi: 10.1098/rstb.1995.0043. [DOI] [PubMed] [Google Scholar]
- 5.Ren J, Kachel K, Kim H, Malenbaum SE, Collier RJ, London E. Interaction of diphtheria toxin T domain with molten globule-like proteins and its implications for translocation. Science. 1999;284:955–957. doi: 10.1126/science.284.5416.955. [DOI] [PubMed] [Google Scholar]
- 6.Bychkova VE, Pain RH, Ptitsyn OB. The ‘molten globule’ state is involved in the translocation of proteins across membranes? FEBS Lett. 1988;238:231–234. doi: 10.1016/0014-5793(88)80485-x. [DOI] [PubMed] [Google Scholar]
- 7.van der Goot FG, Gonzalez-Manas JM, Lakey JH, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 8.Bañuelos S, Muga A. Binding of molten globule-like conformations to lipid bilayers. J Biol Chem. 1995;270:29910–29915. doi: 10.1074/jbc.270.50.29910. [DOI] [PubMed] [Google Scholar]
- 9.Bose HS, Whittal RM, Baldwin MA, Miller WL. The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule. Proc Natl Acad Sci USA. 1999;96:7250–7255. doi: 10.1073/pnas.96.13.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song M, Shao H, Mujeeb A, James TL, Miller WL. Molten-globule structure and membrane binding of the N-terminal protease-resistant domain (63–193) of the steroidogenic acute regulatory protein (StAR) Biochem J. 2001;356:151–158. doi: 10.1042/0264-6021:3560151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene LH, Wijesinha-Bettoni R, Redfield C. Characterization of the molten globule of human serum retinol-binding protein using NMR spectroscopy. Biochemistry. 2006;45:9475–9484. doi: 10.1021/bi060229c. [DOI] [PubMed] [Google Scholar]
- 12.Mills ENC, Gao C, Wilde PJ, Rigby NM, Wijesinha-Bettoni R, Johnson VE, Smith LJ, Mackie AR. Partially-folded forms of barley lipid transfer protein are more surface active. Biochemistry. 2009;48:12081–12088. doi: 10.1021/bi901328f. [DOI] [PubMed] [Google Scholar]
- 13.Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous Ascomycetes. Microbiol Mol Biol Rev. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell. 2003;2:1–8. doi: 10.1128/EC.2.1.1-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Federova ND, Badger JH, Robson GD, Wortman JR, Nierman WC. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics. 2005;6:177. doi: 10.1186/1471-2164-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paoletti M, Saupe SJ, Clave C. Genesis of a fungal non-self recognition repertoire. PLoS ONE. 2007;2:e283. doi: 10.1371/journal.pone.0000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattjus P, Turcq B, Pike HM, Molotkovsky JG, Brown RE. Glycolipid intermembrane transfer is accelerated by HET-C2, a filamentous fungus gene product involved in the cell-cell incompatibility response. Biochemistry. 2003;42:535–542. doi: 10.1021/bi026896x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenoth R, Simanshu DK, Kamlekar RK, Pike HM, Molotkovsky JG, Benson LM, Bergen HR, III, Prendergast FG, Malinina L, Venyaminov SY, Patel DJ, Brown RE. Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold. J Biol Chem. 2010;285:13066–13078. doi: 10.1074/jbc.M109.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malinina L, Malakhova ML, Kanack AT, Lu M, Abagyan R, Brown RE, Patel DJ. The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure. PLoS Biol. 2006;4:e362. doi: 10.1371/journal.pbio.0040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Airenne TT, Kidron H, Nymalm Y, Nylund M, West GP, Mattjus P, Salminen TA. Structural evidence for adaptive ligand binding of glycolipid transfer protein. J Mol Biol. 2006;355:224–236. doi: 10.1016/j.jmb.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 22.Brown RE, Mattjus P. Glycolipid transfer proteins. Biochim Biophys Acta. 2007;1771:746–760. doi: 10.1016/j.bbalip.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JE, Cornell RB. Amphitropic proteins: regulation by reversible membrane interactions. Mol Membr Biol. 1999;16:217–235. doi: 10.1080/096876899294544. [DOI] [PubMed] [Google Scholar]
- 24.Halskau O, Arturo Muga A, Martínez A. Linking new paradigms in protein chemistry to reversible membrane-protein interactions. Cur Protein Peptide Sci. 2009;10:339–359. doi: 10.2174/138920309788922199. [DOI] [PubMed] [Google Scholar]
- 25.Kamlekar RK, Gao Y, Kenoth R, Pike HM, Molotkovsky JG, Prendergast FG, Malinina L, Patel DJ, Wessels W, Venyaminov SY, Brown RE. Human GLTP: Three distinct functions for the three tryptophans in a novel peripheral amphitropic fold. Biophys J. 2010;90:2626–2635. doi: 10.1016/j.bpj.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nylund M, Mattjus P. Protein mediated glycolipid transfer is inhibited FROM sphingomyelin membranes but enhanced TO sphingomyelin containing raft like membranes. Biochim Biophys Acta. 2005;1669:87–94. doi: 10.1016/j.bbamem.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Malakhova ML, Malinina L, Pike HM, Kanack AT, Patel DJ, Brown RE. Point mutational analysis of the liganding site in human glycolipid transfer protein. Functionality of the complex. J Biol Chem. 2005;280:26312–26320. doi: 10.1074/jbc.M500481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li XM, Malakhova ML, Lin X, Pike HM, Chung T, Molotkovsky JG, Brown RE. Human glycolipid transfer protein: Probing conformation using fluorescence spectroscopy. Biochemistry. 2004;43:10285–10294. doi: 10.1021/bi0495432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhai X, Malakhova ML, Pike HM, Benson LM, Bergen HR, III, Sugar IP, Malinina L, Patel DJ, Brown RE. Glycolipid acquisition by human glycolipid transfer protein dramatically alters intrinsic tryptophan fluorescence: insights into glycolipid liganding affinity. J Biol Chem. 2009;284:13620–13628. doi: 10.1074/jbc.M809089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mihaly EJ. Numerical values of the absorbances of the aromatic amino acids in acid, neutral and alkaline solutions. Chem Eng Data. 1968;13:179–182. [Google Scholar]
- 31.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 32.Mach H, Middaugh CR, Lewis RV. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal Biochem. 1992;200:74–80. doi: 10.1016/0003-2697(92)90279-g. [DOI] [PubMed] [Google Scholar]
- 33.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West G, Nylund M, Slotte JP, Mattjus P. Membrane interaction and activity of the glycolipid transfer protein. Biochim Biophys Acta. 2006;1758:1732–1742. doi: 10.1016/j.bbamem.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 35.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heyda J, Mason PE, Jungwirth P. Attractive interactions between side chains of histidine-histidine and histidine-arginine-based cationic dipeptides in water. J Phys Chem B. 2010;114:8744–8749. doi: 10.1021/jp101031v. [DOI] [PubMed] [Google Scholar]
- 37.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Bychkova VE, Berni R, Rossi GL, Kutyshenko VP, Ptitsyn OB. Retinol-binding protein is in the molten globule state at low pH. Biochemistry. 1992;31:7566–7571. doi: 10.1021/bi00148a018. [DOI] [PubMed] [Google Scholar]
- 39.Redfield C, Smith RAG, Dobson CM. Structural characterization of a highly structured ‘molten globule’ at low pH. Nature Struct Biol. 1994;1:23–29. doi: 10.1038/nsb0194-23. [DOI] [PubMed] [Google Scholar]
- 40.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nature Protocols. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Cripaś AF, Gilmanshin RI. Study of the “Molten Globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 42.Rosen CG, Weber G. Dimer formation from 1-anilino-8-naphthalenesulfonate catalyzed by bovine serum albumin. Fluorescent molecule with exceptional binding properties. Biochemistry. 1969;8:3915–39. doi: 10.1021/bi00838a006. [DOI] [PubMed] [Google Scholar]
- 43.Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res. 2008;25:1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kishimoto Y, Hiraiwa M, O’Brien JS. Saposins: structure, function, distribution, and molecular genetics. J Lipid Res. 1992;33:1255–1267. [PubMed] [Google Scholar]
- 45.Liepinsh E, Andersson M, Ruysschaert JM, Otting G. Saposin fold revealed by the NMR structure of NK-lysin. Nature Struct Biol. 1997;4:793–795. doi: 10.1038/nsb1097-793. [DOI] [PubMed] [Google Scholar]
- 46.Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Privé GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci USA. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahn VE, Leyko P, Alattia J-R, Chen L, Privé GG. Crystal structures of saposins A and C. Protein Sci. 2006;15:1849–1857. doi: 10.1110/ps.062256606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.John M, Wendeler M, Heller M, Sandhoff K, Kessler H. Characterization of human saposins by NMR spectroscopy. Biochemistry. 2006;45:5206–5216. doi: 10.1021/bi051944+. [DOI] [PubMed] [Google Scholar]
- 49.Popovic K, Privé GG. Structures of the human ceramide activator protein saposin D. Acta Cryst. 2008;D64:589–594. doi: 10.1107/S0907444908003120. [DOI] [PubMed] [Google Scholar]
- 50.Samuel D, Liu YJ, Cheng CS, Lyu PC. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa) J Biol Chem. 2002;277:35267–35273. doi: 10.1074/jbc.M203113200. [DOI] [PubMed] [Google Scholar]
- 51.Pons JL, de Lamotte F, Gautier M-F, Delsuc MA. Refined solution structure of a liganded type 2 wheat nonspecific lipid transfer protein. J Biol Chem. 2003;278:14249–14256. doi: 10.1074/jbc.M211683200. [DOI] [PubMed] [Google Scholar]
- 52.José-Estanyol M, Gomis-Rüth FX, Puigdomènech P. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem. 2004;42:355–365. doi: 10.1016/j.plaphy.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 53.Hoh F, Pons J-L, Gautier M-F, de Lamotte F, Dumas C. Structure of a liganded type 2 non-specific lipid transfer protein from wheat and the molecular basis of lipid binding. Acta Cryst. 2005;D61:397–406. doi: 10.1107/S0907444905000417. [DOI] [PubMed] [Google Scholar]
- 54.de Oliveira Carvalho A, Gomes VM. Role of plant lipid transfer proteins in plant cell physiology—A concise review. Peptides. 2007;28:1144–1153. doi: 10.1016/j.peptides.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Yeats TH, Rose JKC. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs) Protein Sci. 2008;17:191–198. doi: 10.1110/ps.073300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18:247–51. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okada A, Miura T, Takeuchi H. Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from Influenza A virus. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, Barkley MD. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 59.Shinitzky M, Goldman R. Fluorometric detection of histidine-tryptophan complexes in peptides and proteins. Eur J Biochem. 1967;3:139–144. doi: 10.1111/j.1432-1033.1967.tb19508.x. [DOI] [PubMed] [Google Scholar]
- 60.Vos R, Engelborghs Y. A fluorescence study of tryptophan-histidine interactions in the peptide anantin and in solution. Photochem Photobiol. 1994;60:24–32. doi: 10.1111/j.1751-1097.1994.tb03938.x. [DOI] [PubMed] [Google Scholar]
- 61.Otomo K, Toyama A, Miura T, Takeuchi H. Interactions between histidine and tryptophan residues in the BM2 proton channel from Influenza B virus. J Biochem. 2009;145:543–554. doi: 10.1093/jb/mvp009. [DOI] [PubMed] [Google Scholar]
- 62.Smirnova I, Kasho V, Sugihara J, Kaback HR. Probing of the rates of alternating access in LacY with Trp fluorescence. Proc Natl Acad Sci USA. 2009;106:21561–21566. doi: 10.1073/pnas.0911434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donald JE, Kulp DW, DeGrado WF. Salt bridges: Geometrically specific, designable interactions. Proteins. 2011;79:898–915. doi: 10.1002/prot.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paoletti M, Clave C. The fungus-specific HET domain mediates programmed cell death in Podospora anserine. Eukaryot Cell. 2007;6:2001–2008. doi: 10.1128/EC.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.