SUMMARY
TET-family enzymes convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in DNA. Here we show that Tet1 and Tet2 are Oct4-regulated enzymes that together sustain 5hmC in mouse embryonic stem (ES) cells, and are induced concomitantly with 5hmC during reprogramming of fibroblasts to induced pluripotent stem cells. ES cells depleted of Tet1 by RNAi show diminished expression of the Nodal antagonist Lefty1, and display hyperactive Nodal signalling and skewed differentiation into the endoderm-mesoderm lineage in embryoid bodies in vitro. In Fgf4- and heparin-supplemented culture conditions, Tet1-depleted ES cells activate the trophoblast stem cell lineage determinant Elf5 and can colonize the placenta in mid-gestation embryo chimeras. Consistent with these findings, Tet1-depleted ES cells form aggressive hemorrhagic teratomas with increased endoderm, reduced neuroectoderm and ectopic appearance of trophoblastic giant cells. Thus 5hmC is a novel epigenetic modification associated with the pluripotent state, and Tet1 functions to regulate the lineage differentiation potential of ES cells.
INTRODUCTION
The methylation status of DNA influences many biological processes during mammalian development and is known to be highly aberrant in cancer (Gal-Yam et al., 2008; Ooi et al., 2009). DNA methylation is a powerful mechanism of genome defence against transposons and other parasitic DNA (Goll and Bestor, 2005); in addition, promoter methylation in mammals has long been considered suppressive for gene expression (Klose and Bird, 2006). Recent whole-genome analyses have provided insights into the complexity of methylation patterns in plant and animal species (Farthing et al., 2008; Feng et al., 2010a; Lister et al., 2009; Meissner et al., 2008; Zemach et al., 2010). DNA methylation occurs primarily at CpG dinucleotides in mammals: CpG methylation marks that are lost on newly-replicated DNA strands are faithfully restored by the maintenance DNA methyltransferase Dnmt1. In embryonic stem (ES) cells, however, a substantial fraction of cytosine methylation occurs in non-CpG contexts, where it has been attributed to the activity of the de novo methyltransferases Dnmt3a and 3b (Lister et al., 2009; Ramsahoye et al., 2000).
Dynamic changes in DNA methylation occur during early embryogenesis (reviewed in (Feng et al., 2010b)). Shortly after fertilization, the paternal genome loses the methylcytosine mark prior to DNA replication, whereas the maternal genome loses the mark passively in early cell cycles before blastulation. De novo methylation by Dnmt3 occurs around the time of blastocyst implantation, to a greater extent in the inner cell mass (ICM), which remains pluripotent and gives rise to all cell types of the embryo proper, than in the outer trophectoderm (TE) layer, which is restricted to an extraembryonic fate and gives rise to the placenta (Reik et al., 2001). During the formation of primordial germ cells in the mouse, a second wave of genome-wide demethylation occurs during which imprinted marks are erased; they are subsequently reset in the developing gametes through de novo DNA methylation. The tight regulation of DNA methylation and demethylation is developmentally of crucial importance, since Dnmt-deficient (and therefore hypomethylated) ES cells and embryos lose lineage restriction and show transdifferentiation to the extraembryonic trophoblast lineage (Jackson et al., 2004; Ng et al., 2008).
We recently identified the TET proteins TET1, TET2 and TET3 as a new family of enzymes that alter the methylation status of DNA (Iyer et al., 2009; Tahiliani et al., 2009). TET proteins are 2-oxoglutarate (2OG)- and Fe(II)-dependent dioxygenases that catalyse the hydroxylation of 5-methylcytosine to 5-hydroxymethylcytosine (5hmC) in DNA. TET proteins and 5hmC have been reported in many different tissues and both 5hmC and Tet expression/activity are tightly regulated during ES cell differentiation (Ito et al., 2010; Ko et al., 2010; Kriaucionis and Heintz, 2009; Szwagierczak et al., 2010; Tahiliani et al., 2009). TET1 and TET2 are both implicated in cancer: TET1 is an MLL partner in rare cases of acute myeloid (AML) and lymphoid (ALL) leukemias, and loss-of-function of TET2 is strongly associated with AML as well as a variety of myelodysplastic syndromes and myeloproliferative disorders (see references in (Ko et al., 2010)). Together these data suggest that dysregulation of DNA methylation via TET proteins and hmC may have a role in ES cell pluripotency, oncogenic transformation (especially of hematopoietic stem cells towards the myeloid lineage) and neuronal function.
Here we describe the function of Tet proteins (and, by inference, 5hmC) in mouse ES cells. ES cell lines are culture explants from the inner cell mass (ICM) of the developing blastocyst. They can be maintained in the proliferative, undifferentiated state in culture by using the cytokine leukemia inhibitory factor (LIF) to activate STAT3 and the serum component bone morphogenetic protein (BMP) to induce inhibitor-of-differentiation proteins; when given the appropriate cellular signals, they can differentiate into cellular derivatives of the three primary germ layers - ectoderm, mesoderm and endoderm. Withdrawal of LIF from serum-containing media allows BMP to switch from supporting self-renewal to inducing mesodermal and endodermal differentiation while blocking entry into neural lineages; when grown in the absence of both LIF signals and serum, ES cells are predisposed to convert to a neuronal fate (Ying et al., 2003a; Ying et al., 2003b). These features of self-renewal and ability to differentiate, characteristic of a pluripotent state, require a high degree of epigenetic plasticity. Genes crucial for pluripotency are kept active by a self-organizing network of transcription factors and are rapidly silenced by histone modifications and DNA methylation during differentiation, whereas genes that are required later in cellular differentiation are held in a transiently repressed state by chromatin modifications that are easily reversed. Because Tet proteins modify DNA methylation status, it was conceivable that they might influence the expression and functions of either or both classes of genes.
RESULTS
Tet1 and Tet2 regulate 5hmC levels in mouse ES cells and are associated with the pluripotent state
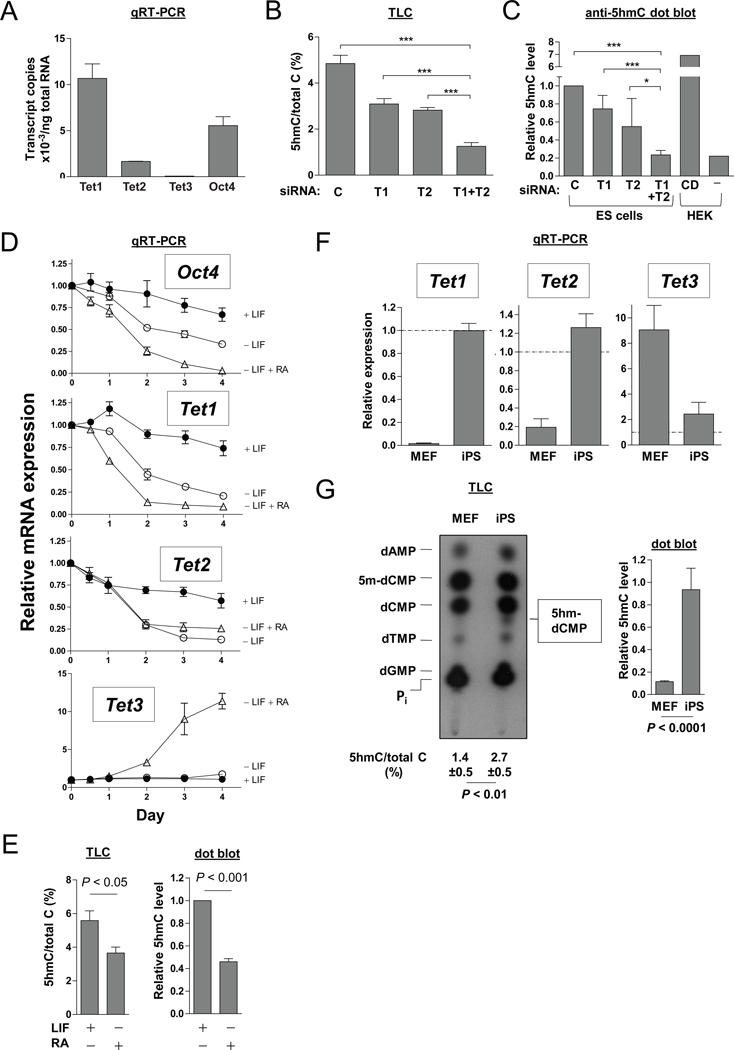
In culture conditions containing LIF and serum, Tet1 transcripts are present at high copy numbers in mouse ES cells, comparable to those of the pluripotency factor Oct4; Tet2 transcripts are about 5-fold less abundant than Tet1 but still well expressed; and Tet3 transcript levels are very low (Figure 1A). Individual depletion of Tet1 or Tet2 mRNAs with SMARTpool siRNA duplexes (Figure S1A) resulted in a moderate decrease in 5hmC, whereas combined depletion of both enzymes reduced 5hmC levels by 75–80% (Figures 1B–C, S1B–C). Thus Tet1 and Tet2 together are responsible for the bulk of 5hmC production in mouse ES cells cultured in the presence of LIF. In contrast to a previous report (Ito et al., 2010), we did not observe major changes in ES cell morphology upon siRNA-mediated depletion of either Tet1 alone or both Tet1 and Tet2 (Figure S1D).
Figure 1. Tet1 and Tet2 regulate 5hmC levels in mouse ES cells and are associated with the pluripotent state.
(A) Normalized transcript copy numbers of Tet1, Tet2, Tet3 and Oct4 in v6.5 mouse ES cells, determined using absolute standard curves of plasmid templates. Gapdh transcript levels were used to estimate RNA content.
(B) Densitometric measurement of 5’-hydroxymethyl-dCMP (5hm-dCMP) spot intensities detected on thin layer chromatography (TLC) analysis of enriched CpG sites in the genome of ES cells after 5 days of siRNA transfection to deplete Tet1 and/or Tet2. Values are depicted as percentages of total dCMP species comprising 5-methyl-dCMP (5m-dCMP), dCMP and hm-dCMP. Error bars indicate SD of 7 replicates from 3 independent experiments. siRNA treatments are denoted: C, control; T1, Tet1 SMARTpool; T2, Tet1 SMARTpool; T1+T2, combined Tet1+Tet2 SMARTpool. A representative TLC autoradiogram is shown in Figure S1B. For ES cell morphology and knockdown efficiency of Tet1 and Tet2, see Figure S1.
(C) Measurement of 5hmC levels in genomic DNA from transfected ES cells and HEK293T based on dot blot analysis. Values are expressed relative to levels in control-transfected ES cells. HEK 293T cells were transfected with either TET1-catalytic domain (CD) or mock-transfected (−). Error bars indicate SD of 5 replicates from 2 experiments. In (B) and (C), P values derived from ANOVA with Bonferroni’s multiple comparison test are denoted: *, P < 0.05; ***, P < 0.001.
(D) Quantitative RT-PCR measurement of Oct4, Tet1, Tet2 and Tet3 transcript levels in mouse ES cells cultured for 1–4 days on gelatinized (feeder-free) plates in the presence of LIF, or differentiated by LIF withdrawal, or LIF withdrawal plus addition of 1 µM all-trans retinoic acid (RA). Data are represented as mean ± SEM from 3 independent experiments.
(E) Measurement of hmC levels by TLC (left) and dot blot (right) in genomic DNA extracted from ES cells after 4 days of LIF withdrawal and treatment with RA. Error bars indicate SD of 3–5 replicates.
(F) Quantitative RT-PCR of Tet1, Tet2 and Tet3 transcript levels expressed relative to levels in ES cells (marked as dotted lines), during reprogramming of MEFs by viral transduction of Oct4, Sox2, Klf4 and c-Myc into iPS cell clones Error bars indicate SD (n=3).
(G)TLC (left) and hmC dot blot (right) analyses showing 5hmC detected in iPS cells but not in fibroblasts. hmC levels from dot blots are expressed relative to levels in ES cells. Error bars indicate SD of 3–5 replicates. P values in (E) and (G) are derived from t-tests.
When plated on gelatin in the presence of LIF, ES cells largely retained expression of Tet1, Tet2 and Oct4 over 4–5 days. Within 3 days of LIF withdrawal, Tet1 and Tet2 mRNA levels declined to 25–30% of starting levels, with a time-course that paralleled the decline of Oct4 mRNA (Figure 1D), and differentiated epithelial-like cells were observed in 4–5 days. When the ES cells were treated with retinoic acid (RA; a potent stimulus for cellular differentiation) at the same time that LIF was withdrawn, Tet1, Tet2 and Oct4 expression declined more rapidly (Figure 1D), and epithelial-like morphology was apparent earlier, by day 3. Tet3 mRNA levels increased more than 10-fold under these conditions (Figure 1D). Under both conditions, 5hmC levels declined significantly, to 40–60% of control (Tahiliani et al., 2009; Figure 1E); the moderate change (compared to that seen in Figures 1B–C) likely reflects both the incomplete loss of Tet1 and Tet2 under conditions of LIF withdrawal and the upregulation of Tet3 in response to RA (Figure 1D).
We examined Tet expression and activity during reprogramming of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem (iPS) cells by transduction with the four reprogramming transcription factors Oct4, Sox2, Klf4 and c-Myc (Takahashi and Yamanaka, 2006). The starting population of fibroblasts expressed almost no Tet1 mRNA and only a basal level of Tet2 mRNA, but fully-reprogrammed iPS cells that had reactivated an endogenous Oct4-GFP reporter (Yeom et al., 1996) displayed levels of Tet1 and Tet2 mRNA comparable to those in ES cells; Tet3 transcripts also decreased, approaching the low level observed in ES cells (Figure 1F). In parallel, 5hmC levels increased, both globally and at MspI sites, from almost undetectable in fibroblasts to levels typical of ES cells in iPS cells (Figure 1G). Similar results were obtained during reprogramming of mouse adult tail-tip fibroblasts into iPS cells (data not shown). Collectively, these data point to a strong association of Tet1, Tet2 and 5hmC with the pluripotent state in both ES and iPS cells, and a contrasting association of Tet3 with the differentiated state.
Oct4 regulates Tet mRNA levels
We measured Tet mRNA levels during ES cell differentiation induced by RNAi-mediated depletion of the key pluripotency factors Oct4, Sox2 and Nanog. ES cells treated with SMARTpool siRNA duplexes targeting Oct4 differentiated rapidly within 3 days. Differentiation induced by Sox2 RNAi was slower, requiring ~5 days, but alkaline phosphatase-positive colonies were still present in ES cells treated with Nanog RNAi for 5 days (Figure S2A). We confirmed that each SMARTpool depleted expression of its target pluripotency factor (Figure S2B), although as expected, depletion of each pluripotency factor in ES cells also downregulated expression of the others due to known cross-regulatory and cooperative interactions (Chew et al., 2005; Loh et al., 2006).
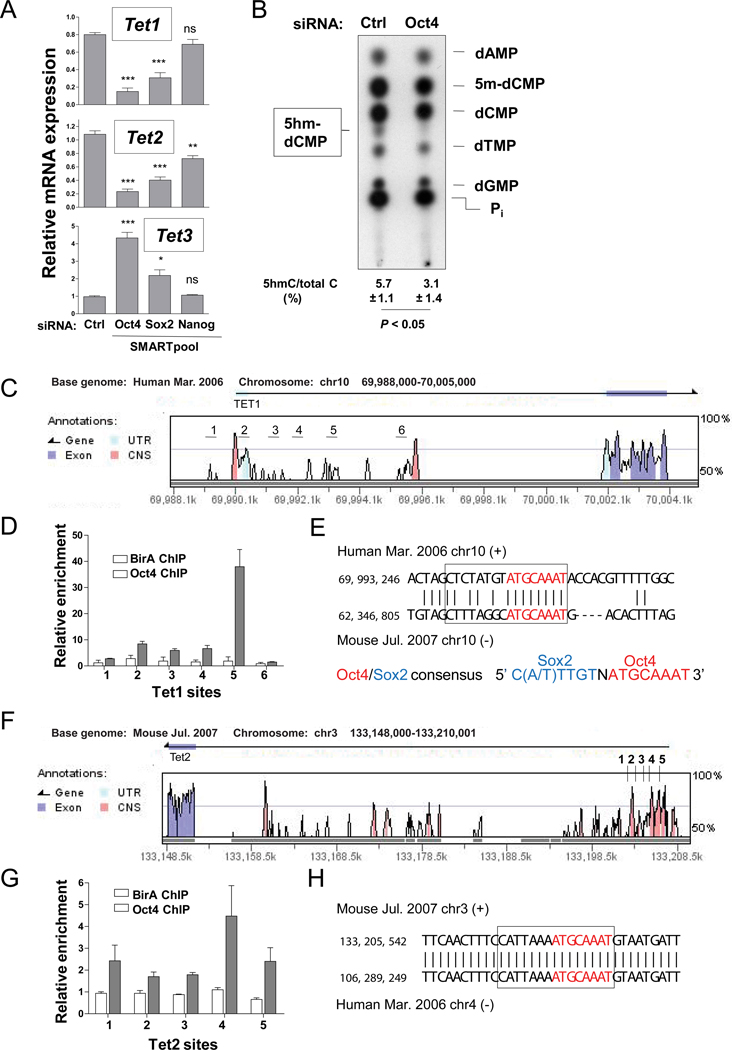
Oct4 and Sox2 RNAi resulted in potent repression of Tet1 and Tet2 mRNA, to <20% and ~30% of control levels respectively; Tet3 mRNA was upregulated by ~4-fold and ~2-fold (Figure 2A). Nanog RNAi had almost no effect on Tet1 and Tet3 while reducing Tet2 expression moderately, to ~60% of control. TLC analysis at day 5 showed a marked loss of 5hmC at MspI sites only in cells treated with Oct4 siRNA (Figure 2B).
Figure 2. Tet mRNA levels are regulated by Oct4.
(A) Quantitative RT-PCR of Tet1, Tet2 and Tet3 transcript levels in ES cells transfected for 5 days with Oct4, Sox2 and Nanog SMARTpool siRNA. Normalized transcript levels are expressed relative to levels at the start of transfection. Data are mean ± SEM from 3 experiments. P values derived from ANOVA with Bonferroni’s multiple comparison test are denoted: *, P < 0.05; **, P < 0.01; ***, P < 0.001. For ES cell morphology and knockdown efficiency of Oct4, Sox2 and Nanog, see Figure S2.
(B) TLC analysis of genomic DNA purified from mouse ES cells differentiated by Oct4 RNAi. Values of 5hmC levels are mean ± SD of 3 replicates from 2 independent experiments.
(C) Vista plot of sequence conservation between the human and mouse Tet1 gene loci upstream of the first coding exon, depicting CNS regions in the vicinity of the Tet1 transcription start site and coding exon 1. Regions numbered 1 to 6 spaced at 1 kb intervals were probed by ChIP PCR. For a diagram of all the CNS regions in the Tet1 locus, see Figure S2C.
(D) Oct4 binding was detected as amplification from Oct4 biotin-mediated ChIP samples from ES cells at the target sites depicted in (C). Control ChIP was performed in cells expressing only the biotin ligase (BirA). For a complete analysis of Oct4 binding to all CNS regions in the Tet1 locus, see Figure S2D.
(E) The mouse-human sequence alignment at the Tet1 site 5 showing a conserved Oct4-Sox2 consensus element (boxed). The Oct4 consensus sequence is highlight in red.
(F) Sequence conservation between the human and mouse Tet2 gene loci upstream of the first coding exon. Regions numbered 1 to 5 spaced at 1 kb intervals were probed.
(G) Oct4 binding was detected as amplification from Oct4 biotin-mediated ChIP samples from ES cells at the target sites depicted in (F).
(H) The mouse-human sequence alignment at the Tet2 site 4 showing a conserved Oct4-Sox2 consensus element (boxed).
Chromatin immunoprecipitation of biotin-tagged Oct4 from ES cells stably expressing the BirA biotin ligase (Kim et al., 2008) showed that Oct4 bound to sites located within conserved non-coding sequence (CNS) regions of both the Tet1 and Tet2 genes (Figure 2C–H). In both cases, the sites resembled consensus Oct4-Sox2 composite sites (Loh et al., 2006) and especially the Oct4 portion of the site was strongly conserved between human and mouse (Figures 2E, H). Oct4 binding sites were not detected in other CNS regions of the Tet1 locus (Figure S2C–D), or at two other predicted Oct4-Sox2 binding elements in CNS regions at ~ −140 kb and −200 kb 5’ of the Tet2 transcription start site. Although we have not tested formally whether these conserved Oct4-Sox2 composite sites function as transcriptional regulatory elements, the combined data suggest strongly that Tet1 and Tet2 are regulated by the Oct4-Sox2 complex.
Tet1 depletion skews ES cell differentiation
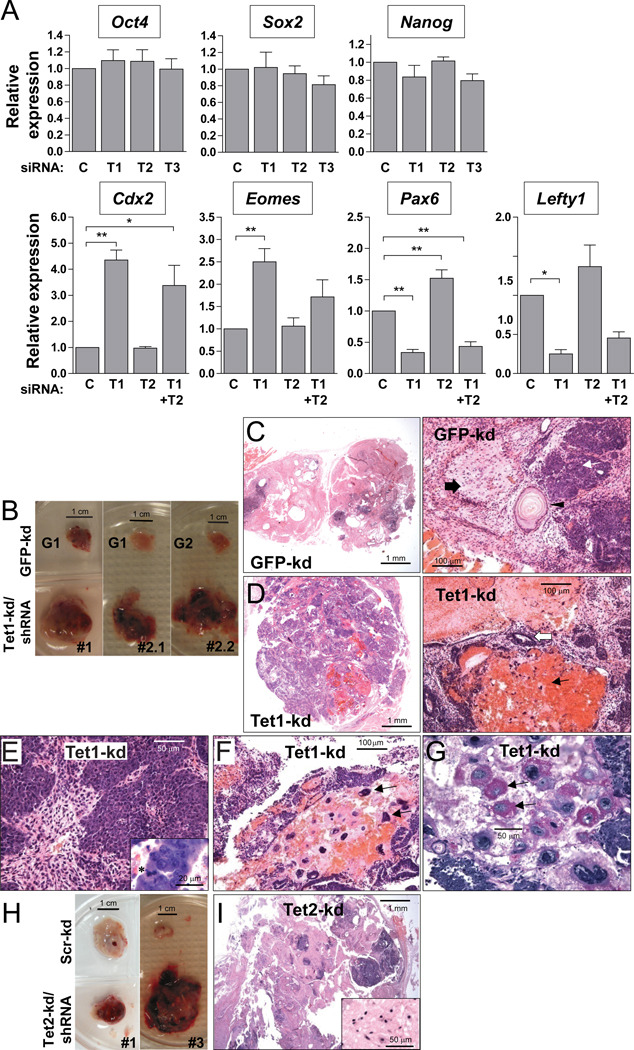
We characterized gene expression in ES cells after siRNA-mediated depletion of each of the three Tet proteins by quantitative RT-PCR. Tet mRNAs were maximally depleted by 3 days of transfection. Tet1 depletion had no effect on Tet2 mRNA expression and vice versa (Figure S1A). In contrast to a previous report that Tet1 depletion led to diminished Nanog mRNA and protein (Ito et al., 2010), Tet depletion did not affect expression of the key pluripotency factors Oct4, Sox2 and Nanog under our conditions for up to 5 days (Figure 3A), nor was there a marked change in the undifferentiated appearance of ES cells maintained in LIF (Figure S1D). Rather, Tet1 depletion resulted in reproducible changes in expression of a panel of lineage-specific markers within 3–5 days: there was a reproducible increase in expression of mRNAs encoding the trophectoderm markers Cdx2, Eomes and Hand1, and a consistent decrease in expression of the neuroectoderm markers Pax6 and Neurod1 and the Nodal antagonists Lefty1 and Lefty2 (Figures 3A, S3A and data not shown). Tet2 depletion had no effect on trophectoderm, endoderm and mesoderm markers, but consistently caused a small increase in expression of Pax6, Neurod1, Lefty1 and Lefty2 (Figures 3A, S3A), whereas Tet3 knockdown caused a 50% repression of Lefty2 but otherwise had no effect on all other targets tested (Figure S3A and data not shown). Combined depletion of Tet1 and Tet2, shown above to decrease genomic 5hmC levels almost to baseline (Figure 1B–C), had a similar but less striking effect compared to Tet1 depletion alone (Figure 3A), suggesting that Tet2 antagonizes the dominant effect of Tet1 at specific target genes.
Figure 3. Tet depletion selectively affects cell lineage markers and skews ES cell differentiation.
(A) Effect of Tet1 (T1), Tet2 (T2), Tet3 (T3) or combined Tet1+Tet2 (T1+T2) SMARTpool siRNAs on the expression of pluripotency and selected lineage marker genes, assessed after 5 days of transfection. C, non-targeting control siRNA. Data are represented as mean ± SEM, n=3–4. *, P < 0.05; **, P < 0.01; P < 0.001 by ANOVA with a post-hoc test for comparison to control. See also Fig S3A.
(B) Enhanced growth and extensive hemorrhagy in Tet1-kd tumors compared to GFP-kd controls. Refer to Fig S3 for knockdown efficiency in stable Tet-kd ES cell clones.
(C) Gross histology of teratomas formed after injection of control clones, revealed through low power images of hematoxylin/eosin (H&E)-stained sections (left), showing high contribution of differentiated neuronal tissue (pink). Higher power image (right) shows mature epithelium with squamous differentiation (arrowhead), terminally differentiated neuronal tissue (block arrow) and immature glandular tissue (white arrow).
(D) Low power image of a Tet1-kd teratoma (from Tet1-kd/shRNA#2.1), showing predominance of immature glandular tissue (purple). Higher power image (right) shows necrotic tissue with blood and scattered giant cells (black arrow) and glandular tissue (white arrow).
(E) Tet1-kd teratoma showing highly proliferative glandular tissues. Inset: a cell undergoing mitosis is marked with asterisk.
(F) Tet1-kd teratoma showing a cluster of trophoblastic giant cells (arrows).
(G) Periodic acid-Schiff staining of a serial section from the Tet1-kd tumor shown in (G), showing glycogen-rich granules (purple stain) in giant cells (arrows).
(H) Hemorrhagy in Tet2-kd tumors compared to scrambled-kd controls.
(I) Low power image of H&E stained Tet2-kd teratoma (from Tet2-kd/shRNA#3), showing areas of fully differentiated neuronal tissue (pink) and glandular tissue (purple). Inset: neuronal tissue.
Histology of Tet1-kd shown is representative of tumors from Tet1-kd/shRNA#2 (each clone #2.1 and #2.2 injected into 2 mice per experiment) from 3 independent experiments performed with 2 different strains of immunodeficient mice. Each Tet2-kd clone was injected in 2 mice per experiment in 2 independent experiments.
To explore the effect of prolonged depletion of Tet1 on ES cell developmental potential, we generated ES cell clones stably expressing shRNAs against Tet1 and Tet2. Two Tet1-depleted clones, generated using Tet1 shRNA#2, showed >80% decrease in Tet1 mRNA levels (Tet1-kd/shRNA#2.1 and #2.2; Figure S3B, left); likewise two Tet2-depleted clones, generated using Tet2 siRNA sequences #1 and #3, showed ~55% and 75% decrease in Tet2 mRNA levels respectively (Tet2-kd/shRNA#1 and #3; Figure S3B, right). Control clones expressed an irrelevant shRNA (scramble (Scr)-kd) or shRNA directed against GFP (GFP-kd). The clones could be propagated serially on feeder cells in the absence of further selection and were morphologically indistinguishable from parental and control clones (Figure S3C). Growth proliferation rates of the selected Tet-kd clones were similar to or slightly enhanced compared to control clones (Figure S3D). Whole-genome transcriptome analysis of stable Tet1-kd versus control ES cell clones largely confirmed the results from transient RNAi transfections; gene ontology (GO) analysis of differentially expressed genes yielded many terms related to embryonic development and cell cycle regulation (Table S1). Possibly because of incomplete knockdown, most gene expression changes in Tet1-kd ES cells were fairly modest (~2 to 5 fold), and the cells retained a genome-wide molecular signature typical of normal ES cells.
We injected control shRNA and Tet-kd ES clones intramuscularly into immunodeficient mice and observed teratoma formation. Within 4–7 weeks, control ES cell lines formed well-differentiated benign teratomas containing cells representative of all three embryonic germ layers, whereas Tet1-kd clones formed large aggressive tumors with massive internal hemorrhage (Figures 3B–D). Histologically, all three primary germ layer lineages could be found in Tet1-kd teratomas, but the relative contributions of each lineage appeared altered compared to controls (Figures 3C–D). Tet1-kd teratomas contained predominantly immature glandular tissue with surrounding stromal cells, indicative of definitive endoderm and mesoderm respectively; many of the glandular cells contained nuclei in mitotic phases, suggestive of a highly proliferative state (Figure 3E). There was considerably less neuroectoderm in the teratoma and many regions with necrotic tissue and blood (Figure 3D). A striking feature was the presence of many giant cells with large nuclei, found especially within and near the necrotic regions but also forming distinct clusters (Figure 3F); many of these cells contained glycogen-rich inclusion bodies, indicative of trophoblastic giant cells of the extra-embryonic lineage (Figure 3G). These histological features were independent of tumor size, since sized-matched control teratomas grown to full size were typically not hemorrhagic, contained more neural tissue and rarely contained any trophoblastic giant cells. Moreover, smaller Tet1-kd teratomas obtained with injection of fewer cells (105 instead of 106) still formed hemorrhagic tumors containing many giant cells (Figure S3E–F).
Like Tet1-kd clones, Tet2-kd clones also formed large hemorrhagic teratomas that grew more aggressively than controls (Figure 3H). Both Tet2-kd clones, generated by stable expression of independent shRNA hairpins, displayed a similar phenotype of hemorrhagy, although the phenotype was stronger in Tet2-kd/shRNA#3-derived teratomas, correlating with stronger constitutive Tet2 knockdown (Figure S3A, right). Despite the hemorrhagic appearance, there was more neuroectoderm contribution in Tet2-kd teratomas, such that apart from the appearance of regions with necrotic tissue, most cellular features still resembled those of control teratomas (Figure 3I). Trophoblastic giant cells were also less apparent in Tet2-kd compared to Tet1-kd teratomas, appearing in clusters in only one oversized tumor harvested but otherwise barely represented in all other Tet2-kd tumors (data not shown). We conclude that Tet1 loss-of-function in ES cells results in developmental skewing towards endoderm/mesoderm and trophoblast lineages, whereas Tet2 loss-of-function maintains a tendency towards neuroectoderm.
Altered developmental potential of Tet1-depleted ES cells: skewing to trophectoderm
The upregulation of transcripts encoding the trophectodermal transcription factors Cdx2 and Eomes (Figure 3A), and the appearance of trophoblastic giant cells in Tet1-kd tumors (Figure 3F–I), suggested that Tet1 deficiency might attenuate the normal restriction of ES cells to embryonic tissues and permit their transdifferentiation into extra-embryonic trophoblast derivatives. Indeed, a previous report showed that when Tet1 siRNA was injected together with a marker gene into mouse embryos at the two-cell stage, the marked (presumably Tet1-depleted) cells were moderately excluded from the inner cell mass (ICM) and enriched in the trophectoderm (Ito et al., 2010).
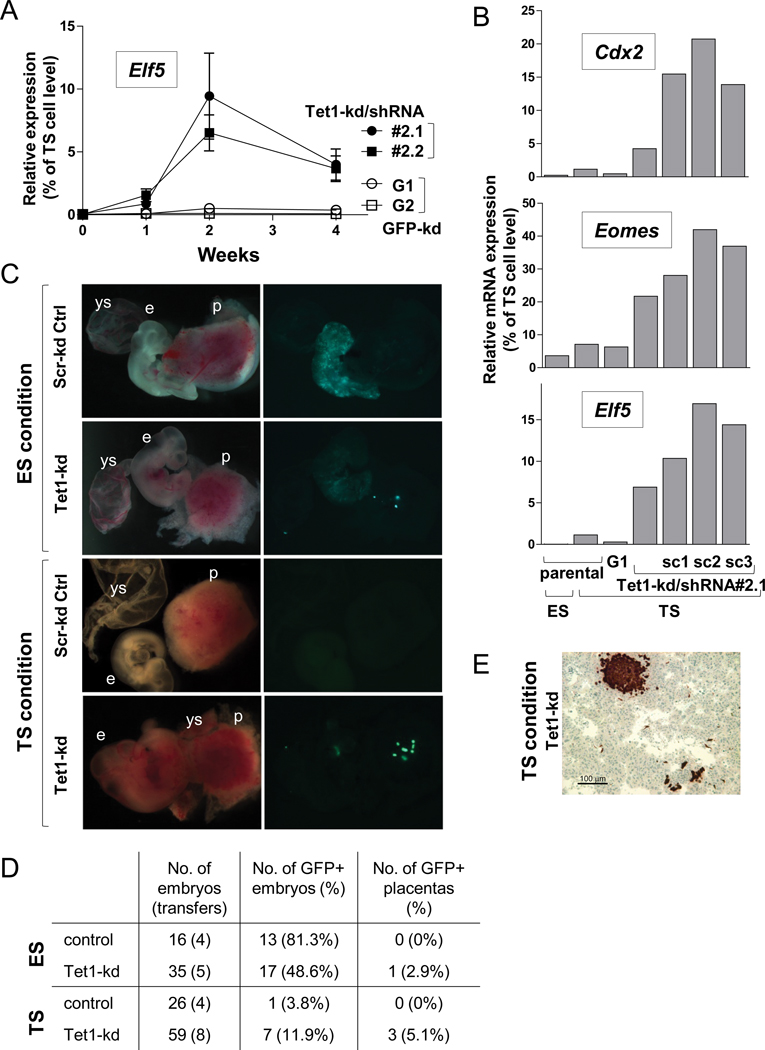
To explore this phenotype further, we cultured control and Tet1-kd clones on feeders in the presence of FGF4 and heparin but without exogenous LIF, a culture condition previously described to favor the derivation of trophoblast stem (TS) cells from the trophectoderm of blastocysts (Tanaka et al 1998). In these alternative “TS” culture conditions, Tet1 depletion did not result in obvious morphological changes: both control (scramble shRNA) and Tet1-kd ES cells formed dense undifferentiated colonies which tended to be flatter with jagged edges, thus showing some resemblance to true TS cells which are flat with a ridge-like periphery (Figure S4A). After 2 weeks in TS cell culture conditions, we observed a robust and reproducible induction of Elf5 transcripts in Tet1-kd clones (50–200 fold increase in Elf5 mRNA over the low background expression seen in control clones, approaching ~10–20% of the levels observed in TS cells; Figure 4A). Elf5 lies downstream of the early trophoblast lineage determinants Cdx2 and Eomes, and was recently described as a commitment marker for the trophoblastic fate (Ng et al., 2008). Moreover, whole-genome gene set enrichment analysis of Tet1-kd clones compared to control clones in TS conditions revealed significant enrichment (both FDR q-value and FWER p-value of 0.02) of a core set of genes defining trophectodermal cell differentiation, including Cdx2, Eomes and Tead4 (Figure S4B). Expression of intermediate trophoblast (Tpbpa) or differentiated giant cell markers (PL1) in Tet1-kd clones was not observed during the course of TS cell culture, suggesting that the cells were being sustained in a TS-like state without overt differentiation into trophoblasts. To further investigate trophectoderm skewing, we cultured the Tet1-kd/shRNA#2.1 ES cell clone for 2 weeks in TS culture conditions, picked three subclones, Tet1-kd/shRNA#2.1-sc1, .2, and .3 based on flattened TS-like morphology, and propagated them in TS culture conditions. Quantitative RT-PCR analysis of these subclones showed a dramatic induction of Cdx2, Eomes and Elf5 expression compared to control shRNA and parental cell lines (Figure 4B); again, however, expression of these TE markers in the subclones was only a fraction of levels in TS cells, suggesting that the cells are propagating as an intermediate cell type between the ES and TS cell states.
Figure 4. Tet1 depletion in ES cells predisposes cells to differentiate along a trophoblastic lineage.
(A) Time-course of expression of Elf5 in control and Tet1-kd clones grown in TS conditions for 1–4 weeks. Data represent mean ± SEM of 4 independent experiments.
(B) Expression of trophectoderm markers Cdx2, Eomes and Elf5 in the Tet1-kd/shRNA#2 line and subclones cultured in TS cell conditions for 4 weeks compared to parental cells in TS or ES cell conditions. Normalized transcript levels in (B–C) are expressed relative to levels in TS cells (set as 100).
(C) Mid-gestation embryo chimerism of GFP-labeled control (scrambled shRNA) or Tet1-knockdown (Tet1-kd/shRNA#2) cells injected into blastocysts after culture in ES or TS conditions. Brightfield (left) and wholemount GFP fluorescence (right) images were taken at E10.5. Abbreviations: e, embryo; ys, yolk sac and p, placenta.
(D) Scoring of GFP presence in embryos and placentas at midgestation.
(E) GFP antibody staining of placental section showing chimerism of a Tet1-kd subclone (sc3) from TS cell culture.
The association of Tet1 knockdown with Cdx2, Eomes and Elf5 activation suggested that Tet1 might function to repress trophectoderm development during early embryogenesis. To test this hypothesis, we injected GFP-labeled Tet1-kd ES cell lines, cultured either in ES or TS cell conditions, into mouse blastocysts, and observed chimerism in mid-gestation (E10.5) embryos by GFP fluorescence. Typically, ES cells injected into the ICM of blastocysts contribute only to the developing embryo and not to placental tissues (Beddington and Robertson, 1989), and this was indeed observed with ES cells expressing a scrambled control shRNA (Figure 4C). Tet1-kd ES cells from ES cell cultures also chimerized the developing embryo, consistent with our data from teratomas that differentiation into the three primary germ layers is not completely blocked; however, the contribution to embryos appeared reduced and in rare cases, GFP+ cells could even be detected in placental tissue (manifested as spots of extraembryonic GFP+ cells in one embryo, or as faint GFP+ cells in another 2/35 embryos obtained from 5 uterine transfers) (Figure 4D). When the same GFP-labelled ES cells were cultured for 4 weeks in TS cell conditions, there was a marked reduction in the ability of both control and Tet1-kd clones to chimerize the embryos based on GFP fluorescence; this in part reflects a technical shortcoming due to silencing of GFP observed in prolonged TS culture conditions. However, injection of a Tet1-kd clone or subclone from TS cell culture occasionally produced embryos with bright aggregates of GFP-positive cells in the placenta (Figure 4C). The presence of GFP+ cells in the placenta was confirmed by immunohistochemical staining for GFP (Figure 4E). In contrast, none of the control ES cells expressing control shRNA gave rise to any bright GFP-fluorescent cells in the placenta, whether cultured under ES or TS conditions. Together these data suggest that a small subset of Tet1-kd ES cells cultured in either ES or TS conditions are able to cross an embryonic restriction barrier to colonize the placenta.
Altered developmental potential of Tet1-depleted ES cells: skewing to mesendoderm
We asked whether the observed increase in the representation of cells of the mesoderm and endoderm lineages in teratomas formed from Tet1-kd ES cells (Figure 3D) could reflect decreased expression of the Nodal antagonist Lefty (Figure 3A). Nodal and Lefty are both members of the TGFβ superfamily (Schier, 2009; Shen, 2007; Tabibzadeh and Hemmati-Brivanlou, 2006). Nodal signals act as morphogens and are essential for the induction of mesoderm and definitive endoderm in the gastrulation-stage embryo when uncommitted epiblast cells move through the primitive streak, a structure marked by expression of the transcription factor Brachyury (Bry) (Tam and Loebel, 2007). Mesoderm is induced from the posterior primitive streak in response to Wnt or low levels of TGFβ/Nodal/Activin signaling, whereas definitive endoderm arises in response to high, sustained Nodal/Activin signals from “mesendoderm” progenitors in the anterior posterior streak that are marked by expression of Foxa2 and Goosecoid. We postulated that Tet1 depletion, by decreasing Lefty expression, would increase Nodal signals and result in the mesoderm/endoderm skewing observed in our teratoma assays.
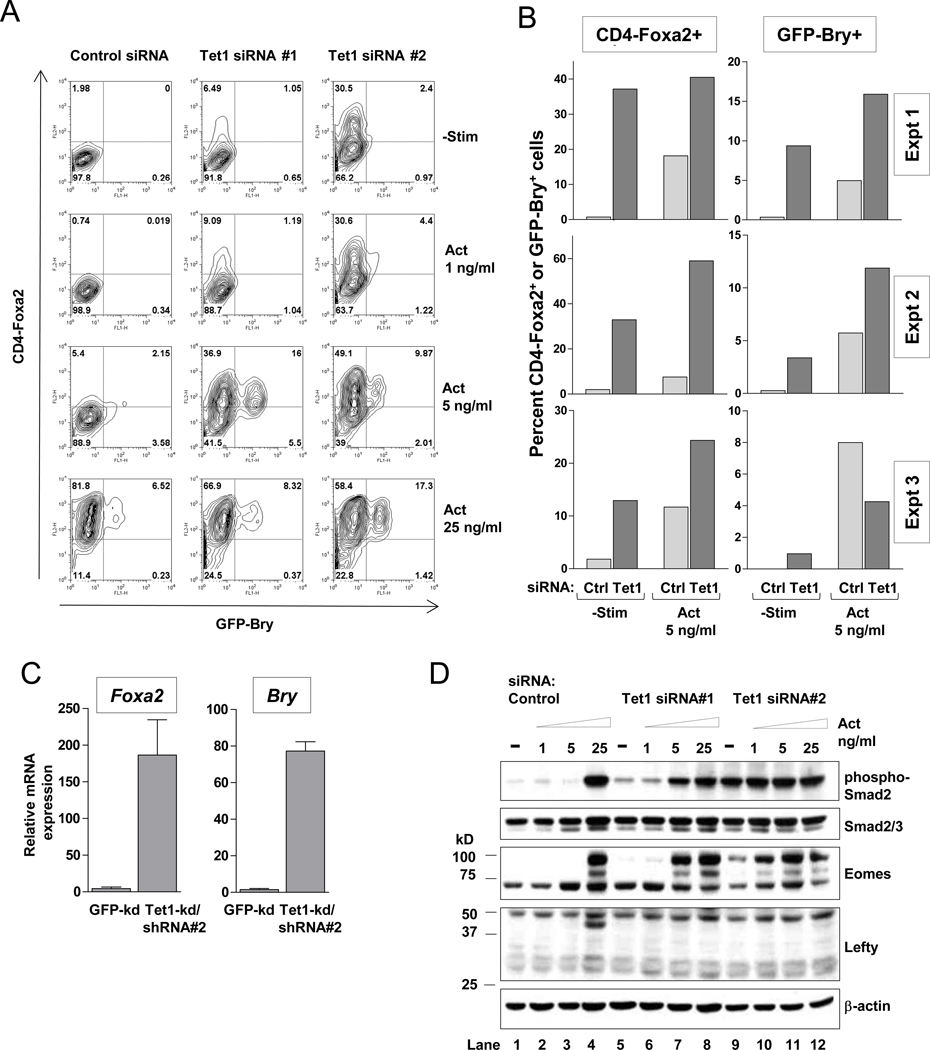
To test this hypothesis, we used the CD4-Foxa2/GFP-Bry ES cell line (E14.1, 129/Ola) in which Brachyury and Foxa2 expression are read out as expression of GFP and a cell-surface receptor, human CD4, respectively (Gadue et al., 2006). If Tet1 depletion in this cell line indeed led to mesoderm and/or endoderm skewing, this would be apparent in ES cell in vitro differentiation assays as increased expression of Brachyury and/or Foxa2 respectively. We depleted Tet1 in CD4-Foxa2/GFP-Bry ES cells using 2 independent Tet1 siRNAs and then allowed the cells to differentiate into embryoid bodies (EB) for 4 days. During the last 2 days of differentiation, we supplemented some cultures with graded concentrations of TGFβ family member Activin A (1, 5 or 25 ng/ml) as a positive control to stimulate mesoderm and endoderm formation. Unlike Nodal, Activin is not inhibited by Lefty (Chen and Shen, 2004). As expected, Tet1 transcripts declined to ~50% of control levels by Day 2 of EB formation (Figure 1D), but siRNA treatment decreased Tet1 mRNA expression even further (data not shown). Control siRNA-transfected ES cells remained CD4- and GFP-negative during EB differentiation, but treatment with Tet1 siRNA (especially siRNA#2) led to the emergence of subpopulations expressing CD4 and GFP indicating strong expression of Foxa2 and low expression of Brachyury respectively (Figure 5A, top panels; Figure 5B). GFP-Bry and CD4-Foxa2 expression were increased in Tet1 siRNA-treated cells that were also exposed to low concentrations of activin (5 ng/ml) (Figure 5A, middle panels; Figure 5B). Similarly, when stable Tet1-kd ES cell clones (v6.5 cell line) were subjected to in vitro EB differentiation, we again observed induction of Foxa2 and Brachyury as measured by qRT-PCR (Figure 5C).
Figure 5. RNAi depletion of Tet1 in ES cells skews differentiation towards the endoderm-mesoderm lineages in vitro.
(A) Expression of CD4 and GFP in CD4-Foxa2/GFP-Bry ES cells transfected with Tet1 siRNA and differentiated in serum-free media for 4 days to form EB. EB cultures were reaggregated and treated with Activin A (Act) at the indicated concentrations at Day 2. Numbers in quadrants denote percentage of gated cell populations.
(B) Percentages of CD4-high and GFP-high cell populations in Day 4 EB after siRNA treatment. Three independent experiments are shown comparing control and Tet1 siRNA#2 treatment.
(C) Quantitative RT-PCR analysis of Foxa2 and Brachyury in Day 4 EB differentiated from Tet1-kd/shRNA#2 stable clones. Data are mean ± SEM of 3 clones in each group, representative of 2 independent experiments.
(D) Western blot analysis of phosphor-Smad2, total Smad2/3, Eomes and Lefty in whole-cell lysates of Day 4 EB.
We examined Nodal/Activin signaling in whole cell lysates of control and Tet1-depleted EB at Day 4 by Western blotting (Figure 5D). In control-siRNA treated cells, high doses of Activin A (25 ng/ml) stimulated Smad2 phosphorylation, upregulation of Eomes, and upregulation of Lefty (Figure 5D, lanes 4; the Lefty antiserum recognizes both Lefty 1 and Lefty 2; note the particularly strong upregulation of high molecular weight forms of Eomes.) These results are consistent with previous reports that Eomes is a downstream target of Nodal/Activin signaling (Arnold et al., 2008; Brennan et al., 2001), and that both Nodal itself and the inhibitor Lefty are induced by Nodal/Activin signals in positive and negative auto-regulatory feedback loops (Besser, 2004; Saijoh et al., 2000). Notably, Tet1-depleted ES cells also showed increased Smad2 phosphorylation and increased Eomes expression in the absence of activin (particularly striking in the case of Tet1 siRNA#2, lane 9), suggesting that decreased levels of Tet1 promote increased signaling in the TGFβ pathway. These effects of Tet1 depletion were potentiated by activin treatment (lanes 7, 10, 11). Interestingly, Tet1 depletion abolished the activin-induced increase in Lefty expression.
Tet1 depletion has complex effects on DNA methylation at target gene promoters
Tet enzymes regulate DNA methylation by modifying 5mC, and have been proposed to promote DNA demethylation in multiple ways (Tahiliani et al., 2009). By converting 5mC to 5hmC, Tet proteins diminish DNA methylation. Moreover, because 5hmC is not recognized by Dnmt1, its presence would promote passive demethylation. Finally, 5hmC might be actively removed by a DNA repair system and replaced by unmodified cytosine. Consistent with these possibilities, the Nanog promoter has been reported to become hypermethylated in ES cells depleted of Tet1 (Ito et al., 2010). In contrast, however, we have shown that Tet2 loss-of-function in myeloid tumours results in global hypomethylation rather than localized hypermethylation at CpG dinucleotides in the genome (Ko et al., 2010).
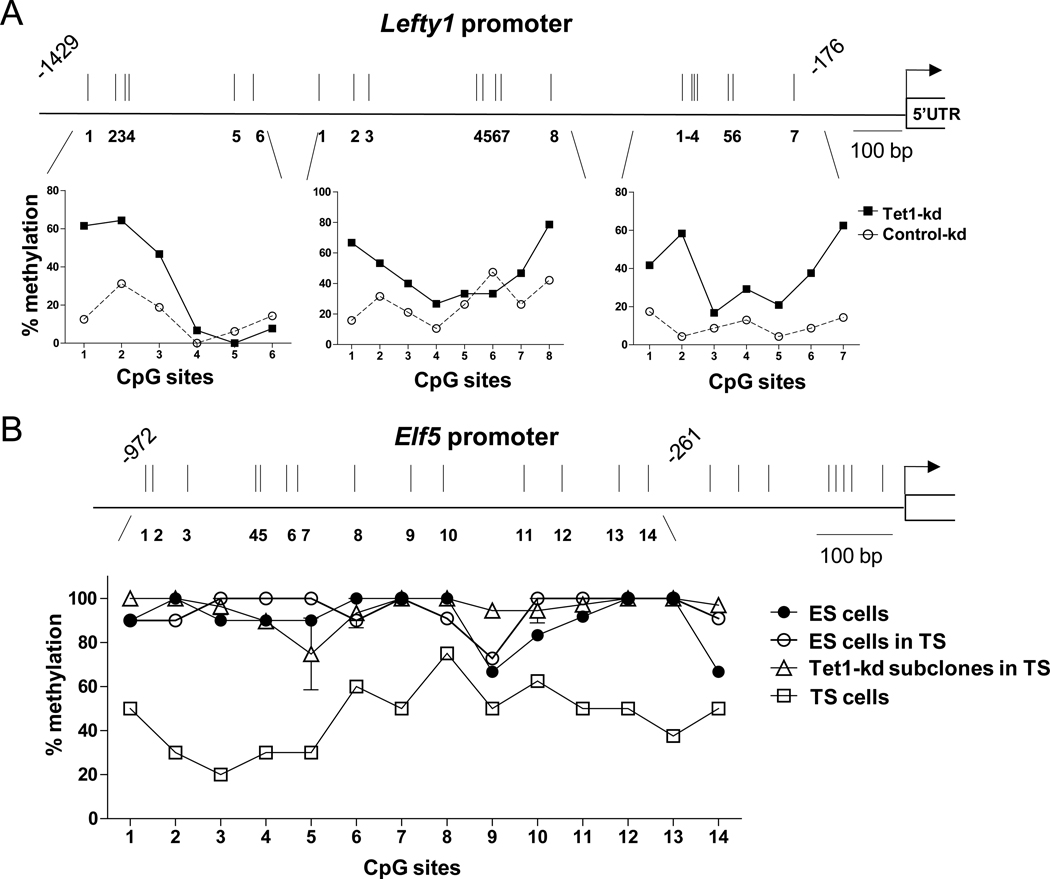
To investigate the relation of Tet1 depletion to changes in DNA methylation, we examined the promoters of two Tet1-regulated genes, Lefty1 and Elf5. The Lefty1 promoter is hypomethylated in stem cells and hypermethylated in differentiated cells (Farthing et al., 2008), whereas the Elf5 promoter is hypermethylated (and repressed) in ES compared to TS cells (Ng et al., 2008). We first examined DNA “methylation” at the Lefty1 promoter in ES cells depleted of Tet1 by RNAi, using the bisulfite sequencing technique which does not distinguish 5mC and 5hmC (Huang et al., 2010). Compared to control-treated cells in which the locus was hypomethylated, Tet1-depleted ES cells showed an increase in CpG “methylation” levels at specific regions of the 1.4 kb Lefty1 promoter region (Figure 6A), consistent with the notion that Tet1 directly or indirectly regulates Lefty1 expression by facilitating DNA demethylation. In contrast, the Elf5 promoter was as highly “methylated” in Tet1-kd ES cell subclones as in the parental ES cells (Figure 6B), despite the fact that Elf5 transcripts were more highly expressed (Figure 4A, B).
Figure 6. Tet1 depletion has different effects on DNA methylation at target gene promoters.
(A) Bisulphite sequencing analysis of CpG methylation status at the Lefty1 promoter in ES cells transfected with control or Tet1 SMARTpool siRNA. The average % methylation at each CpG site is derived from sequencing of 20–24 clones.
(B) Bisulphite sequencing analysis of the Elf5 promoter in ES cells, TS cells and various Tet1-kd ES clones in TS culture condition. The % methylation at each CpG site is derived from sequencing of 10–12 clones. Three Tet1-kd/shRNA#2.1 subclones were analyzed and errors bars are mean ± SEM.
DISCUSSION
In this study we report the functional roles of Tet proteins, a newly-discovered family of DNA-modifying enzymes, in mouse ES and iPS cells. We show that Tet1 and Tet2 are the key enzymes responsible for the presence of 5hmC in mouse ES and iPS cells, that their expression is regulated by Oct4, and that their activity (as judged by 5hmC levels) correlates closely with the pluripotent state. In contrast to a previous report (Ito et al., 2010), acute RNAi-mediated depletion of Tet1 alone, or both Tet1 and Tet2, did not in our hands cause overt ES cell differentiation, diminish ES cell proliferation, or affect expression of the key pluripotency factors Oct4, Sox2 and Nanog. Rather, Tet1 depletion correlated with decreased expression of the Nodal antagonists Lefty1 and Lefty 2, increased expression of the mesoderm/endoderm transcription factors Brachyury and Foxa2, and increased expression of the trophoectoderm transcription factors Cdx2, Eomes and Elf5. While we have not yet established whether these genes are direct or indirect targets of Tet1, it is notable that each lies at a “crossroads” of cell fate determination during embryonic development (Ng et al., 2008; Tabibzadeh and Hemmati-Brivanlou, 2006).
Tet enzymes are downstream targets of the transcription factor network that maintains ES cell pluripotency. Oct4 depletion led to rapid ES cell differentiation, a parallel strong decrease in Tet1 and Tet2 mRNA expression, and an increase in Tet3 mRNA expression. Sox2 RNAi had a similar but less dramatic effect. BIO-ChIP assays showed Oct4 binding to both the Tet1 and Tet2 loci at composite Oct4/Sox2 sites, suggesting strongly that Tet1 and Tet2 are directly regulated by the cooperative Oct4/Sox2 complex. A previous genome-wide ChIP-seq analysis showed Oct4 binding to the Tet2 locus (Chen et al., 2008); however neither this nor earlier studies (Boyer et al., 2005; Kim et al., 2008; Loh et al., 2006) identified Tet1 or Tet3 as Oct4 target genes, possibly because the signals did not reach statistical significance. We note that the mild effect of Nanog depletion on Tet2 gene expression may be indirect, through the ability of Nanog to regulate Oct4 and Sox2 (Loh et al., 2006).
Our studies highlight a strong correlation between Tet1 and Tet2 expression and the pluripotent state. Stimuli that induced ES cell differentiation -- LIF withdrawal, RA addition and Oct4 RNAi -- caused loss of Tet1 and Tet2 expression and a parallel loss of genomic 5hmC. A previous kinetic analysis of gene expression in ES cells undergoing RA-induced differentiation also identified Tet1 (D10Ertd17e) as one of 65 rapidly-downregulated candidate genes (Ivanova et al., 2006); however, Tet1 was not characterized further in this study, because – as also observed in our hands -- shRNA-mediated knockdown of Tet1 did not dramatically affect morphology or alkaline phosphatase activity in ES cells. Conversely, reprogramming of fibroblasts to iPS cells was associated with increases in Tet1, Tet2 and 5hmC, and combined depletion of both Tet1 and Tet2 during early reprogramming by doxycycline-inducible RNAi reduced the number of iPS cell colonies by about 50% (KPK, CAS, GM, AR, unpubl.). A formal demonstration of the role of Tet enzymes in iPS cell reprogramming will require the use of robust overexpression systems as well as fibroblasts from Tet-deficient mice.
At several genes examined (e.g. Cdx2, Eomes, Lefty, Pax6), the effects of Tet1 depletion were dominant over an often mildly opposing effect of Tet2 depletion. Since both enzymes catalyze the conversion of 5mC to 5hmC, it is likely that they are recruited to disparate (possibly overlapping) sets of target genes and in turn recruit distinct transcriptional regulatory complexes through their divergent N-terminal regions. Notably, TET2 and 5hmC are also expressed, albeit at lower levels, in other tissues and cell types including haematopoietic stem cells (HSC) (Delhommeau et al., 2009; Ito et al., 2010; Ko et al., 2010); thus TET2 may regulate cell fate decisions in adult stem /precursor cells as well. Indeed, RNAi-mediated depletion of Tet2 in early haematopoietic precursors resulted in skewed differentiation, with an enhanced propensity to commit to the myeloid lineage in response to appropriate cytokines in vitro (Ko et al., 2010).
The gene expression changes observed in ES cells in response to acute Tet1 depletion were consistent with the developmental effects observed in teratoma assays in vivo and embryoid body formation in vitro. Teratomas formed by Tet1 and Tet2-kd ES cells contained cells from all three germ layers, albeit with altered relative contributions compared to control ES cells, indicating that Tet1 and Tet2-kd ES cells retained pluripotency. Tet1-kd teratomas contained a high proportion of glandular tissue interspersed with stromal cells, indicative of definitive endoderm and mesoderm lineages respectively; cells of the neuroectoderm lineage were considerably fewer. Tet2-kd clones also formed large hemorrhagic teratomas, but with greater contribution from neuroectoderm. These features are consistent with the role of Lefty proteins (positively regulated by Tet1, mildly repressed by Tet2) as inhibitors of the TGF-β family member Nodal. Downstream of Activin/Nodal receptors, moderate and strong inductive signals mediated by Smad2/3 support differentiation into mesoderm and definitive endoderm lineages marked by Brachyury and Foxa2 expression respectively (Gadue et al., 2006; Kubo et al., 2004). Thus, diminished Lefty expression associated with Tet1 depletion would be expected to increase Smad signaling under conditions where the Nodal pathway was active; increase expression of the downstream target of Nodal signaling, Eomes; increase Brachyury and Foxa2 expression in differentiation assays in vitro; and skew development towards mesoderm/endoderm lineages in vivo, exactly as actually observed. Reciprocally, the slight increase in Lefty expression caused by Tet2 depletion would be expected to decrease Smad signaling and decrease the constraint on neuroectoderm gene expression. Although downregulation of Pax6 due to Tet1 depletion can also skew differentiation of mesendoderm by causing loss of neural progenitors, we did not observe any perceptible loss of Pax6 and NeuroD1 proteins when Tet1-depleted ES cells were differentiated for 4 days into embryoid bodies (data not shown). Thus small changes in gene expression in Tet1-kd ES cells can be amplified into major changes in the strength of Nodal/Activin signalling, resulting in pronounced mesendoderm skewing during ES cell differentiation.
Tet1-kd teratomas also showed a marked increase in the number of trophoblastic giant cells, especially amidst hemorrhagic and necrotic tissue. Furthermore, Tet1-kd ES cells chimerized placental tissue ectopically in mid-gestation stage embryos following blastocyst injection, albeit at low frequency. Again, this tendency was also apparent in vitro: Tet1-kd ES cells showed only a small increase in expression of the trophectoderm markers Cdx2 and Eomes and did not express Elf5, but increased the expression of all three genes upon switching to TS culture conditions that promote derivation of trophoblast stem cells. Thus, an induction signal for differentiation (LIF withdrawal and supplementation with Fgf4 and heparin) accentuates the effect of Tet1 deficiency on lineage commitment markers.
Our data suggest a complex relation between Tet proteins and DNA methylation. Tet1 depletion resulted in increased DNA methylation at the Lefty1 promoter in parallel with decreased expression of Lefty1 mRNA and protein. These data are consistent with the possibility that Tet1 promotes hydroxymethylation of the Lefty1 promoter, facilitating demethylation and hence promoting Lefty1 transcription. This hypothesis is clearly inadequate in the case of Elf5, however. The Elf5 promoter is normally silenced in ES cells by DNA methylation and its demethylation and activation are required for ES cells to differentiate into trophoblast derivatives (Ng et al., 2008). Thus our finding that Tet1 depletion correlated with increased Elf5 expression and trophectoderm skewing is not consistent with the fact that ES cells, ES cells cultured under TS conditions, and Tet1-kd ES cells cultured under TS conditions show similar hypermethylation at the Elf5 promoter, in contrast to the hypomethylation observed in TS cells. Since conventional bisulfite sequencing does not distinguish 5mC and 5hmC (Huang et al., 2010; Jin et al., 2010; Nestor et al., 2010), we have not formally ruled out the possibility that 5hmC is present at a subset of CpG sites at the Elf5 promoter. Although further studies are required to ascertain whether Tet1 binds directly to Lefty, Elf5 or other target genes, it is clear that the effect of Tet1 on DNA methylation and gene expression in ES cells cannot be explained by the simple postulate that 5hmC is an intermediate in a DNA demethylation pathway. Because Elf5 is located downstream of the trophoblast differentiation cascade and is induced by the early trophoblast lineage determinants Cdx2 and Eomes, one possibility is that Tet1 depletion increases Elf5 expression indirectly, through upregulation of Cdx2 and Eomes.
In summary, our studies identify Tet proteins as key regulators of early embryonic differentiation. Our data suggest that these enzymes do not act alone, but rather operate in coordination with developmental signals to regulate lineage determination at decision points that are critical for early lineage commitment. We propose that Tet1 functions downstream of Oct4 in the first lineage split between inner cell mass and trophectoderm to constrain Elf5 expression in the inner cell mass; later in development, when the epiblast differentiates into the three somatic germ layers, Tet1 coordinates the canalization of developmental pathways by regulating Lefty (Figure S5). An understanding of the roles of Tet proteins and the novel epigenetic mark, 5hmC, in ES cell function and embryonic development will require analysis of Tet-disrupted mice and the genome-wide localization of 5hmC.
EXPERIMENTAL PROCEDURES
Oct4 binding site bioinformatics and chromatin immunoprecipitation
Alignments between the mouse and human Tet1 gene loci were performed on the Vista Browser (http://pipeline.lbl.gov/cgi-bin/gateway2?bg=mm9&selector=vista) to include all intervening untranscribed region between neighboring genes. Putative Oct4-Sox2 consensus sites were detected by the EMBOSS program (http://emboss.sourceforge.net/docs/adminguide/node6.html) using the fuzznuc command line to detect the pattern 'C[AT]TTGTN(0,5)ATGCAAAT' tolerating a mismatch of 2 bases. Predicted sites within CNS regions were manually assessed. PCR primers were designed to probe CNS regions at 1-kb intervals and shown in Supplementary Information. ChIP reactions were performed as previously described (Rao et al., 2010). Samples were analyzed under similar conditions and fold enrichments were calculated by comparing ChIP samples to genomic DNA controls after normalizing to GAPDH.
Teratoma formation
Tumors and teratomas were obtained by intramuscular injection of 1×106 ES cells into the each quadripcep of either NOD/SCID (Charles River Laboratories, Wilmington, MA) or Rag2−/−;γC−/− mice with similar results. Specimens were collected when tumors exceeded 2.0 cm in diameter and fixed overnight in 4% paraformaldehyde. Paraffin sections and staining were performed by the Specialized Rodent Histopathology Services at Brigham and Women’s Hospital. Animal handling and maintenance were performed in accordance with institutional guidelines.
Chimera generation
15 ES or ES-TS cells labeled with GFP by lentiviral vectors were injected into mouse blastocysts and implanted by uterus transfer into pseudopregnant foster mothers using standard methods. Pregnant mice were sacrificed at day E10.5 and whole embryos were photographed with an inverted fluorescent microscope. Specimens were fixed overnight in 10% formalin, mounted in HistoGel (Richard-Allan Scientific, MI) and stored in 80% ethanol. Paraffin sections and anti-GFP staining were performed at the Specialized Rodent Histopathology Services, Brigham and Women’s Hospital.
ES cell serum-free embryoid body (EB) differentiation
CD4-Foxa2/GFP-Bry ES cells were maintained in serum-containing media with feeder cells before adaptation on feeder-free gelatin-coated wells in serum-free culture media (Invitrogen. Carlsbad, CA unless otherwise indicated) consisting of Knockout DMEM/F12 supplemented with 0.5× of both N2 and B27, penicillin/streptomycin, 0.05% BSA, LIF (1% conditional medium), human BMP4 (10 ng/ml; R&D Systems, Minneapolis, MN) and 1.5 × 10−4 M 1-thioglycerol (Sigma, St. Louis, MO) as previously described (Gadue et al., 2006). ES cells were adapted for 2 passages in serum/feeder-free culture and transfected at 1.5 × 105 cells/ml for 2 days with 50 nM siRNA using Lipofectamine RNAiMAX according to manufacturer’s instruction. To induce EB differentiation (day 0), cells were dissociated by TrypLE Express and cultured at 1 × 105 cells/ ml in serum-free differentiation media consisting of 75% Iscove’s modified Dulbecco’s medium and 25% Ham’s F12 media supplemented with 0.5× of both N2 and B27 (without retinoic acid), 0.05% BSA, 50 µg/ml ascorbic acid (Sigma) and 4.5 × 10−4 M 1-thioglycerol. At day 2, the EBs were dissociated with TrypLE Express and reaggregated in serum-free differentiation media with the addition of human activin A (R & D systems). Re-transfections of siRNA were performed at days 0 and 2 of EB aggregation. At day 4, EBs generated were dissociated by incubation with trypsin and stained with anti-human CD4-phycoerythrin (BioLegend, San Diego, CA). The cells were analysed by a FacsCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) and data processed with FlowJo software (TreeS Star, Ashland, OR). Note that Tet1 siRNA#1 and #2 correspond to Tet1 siRNA 1.1 and 1.2 respectively in Fig S1A. For unknown reasons Tet1 siRNA#1 was less effective than siRNA#2 in CD4-Foxa2/GFP-Bry ES cells, although in v6.5 ES cells the two siRNAs were equivalent (see Figure S1A).
Supplementary Material
ACKNOWLEDGEMENTS
We thank P. Gadue and G. Keller for CD4-Foxa2/GFP-Bry ES cells, C. Unitt and staff at the Specialized Rodent Histopathology Services, Brigham and Women’s Hospital for mouse histology and GFP staining, P. Junni at the Finnish Microarray and Sequencing Centre, Turku Centre for Biotechnology for sample processing and array hybridization, and K. E. Waraska at the Biopolymers facility, Harvard Medical School for low-density array-based q-PCR. This work was funded by NIH grants AI44432, CA42471 and RC1-DA028422 (to A.R.), RO1-DK70055, RO1-DK59279, UO1-HL100001, and special funds from the ARRA stimulus package- RC2-HL102815 (to G.Q.D), the Roche Foundation for Anemia Research, Alex’s Lemonade Stand, and the Harvard Stem Cell Institute (to G.Q.D), an American Heart Association postdoctoral fellowship (to K.P.K) and grants from The Academy of Finland and The Sigrid Juselius Foundation (to R.L.). GQD is an affiliate member of the Broad Institute, a recipient of Clinical Scientist Awards in Translational Research from the Burroughs Wellcome Fund and the Leukemia and Lymphoma Society, and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. The Journal of biological chemistry. 2004;279:45076–45084. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RSP, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Chen C, Shen MM. Two modes by which Lefty proteins inhibit nodal signaling. Curr Biol. 2004;14:618–624. doi: 10.1016/j.cub.2004.02.042. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS genetics. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010a;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010b;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-Î2 signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annual review of biochemistry. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe Global DNA Hypomethylation Blocks Differentiation and Induces Histone Hyperacetylation in Embryonic Stem Cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic acids research. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48:317–319. doi: 10.2144/000113403. [DOI] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SKT, O'Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Shao Z, Roumiantsev S, McDonald LT, Yuan GC, Orkin SH. Differential Roles of Sall4 Isoforms in Embryonic Stem Cell Pluripotency. Mol Cell Biol. 2010;30:5364–5380. doi: 10.1128/MCB.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Sakuma R, Yeo CY, Yashiro K, Watanabe M, Hashiguchi H, Mochida K, Ohishi S, Kawabata M, et al. Left-right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal morphogens. Cold Spring Harbor perspectives in biology. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabibzadeh S, Hemmati-Brivanlou A. Lefty at the crossroads of "stemness" and differentiative events. Stem Cells. 2006;24:1998–2006. doi: 10.1634/stemcells.2006-0075. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Ying Q-L, Nichols J, Chambers I, Smith A. BMP Induction of Id Proteins Suppresses Differentiation and Sustains Embryonic Stem Cell Self-Renewal in Collaboration with STAT3. Cell. 2003a;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying Q-L, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotech. 2003b;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.