Abstract
Vegetables are critical for human health as they are a source of multiple vitamins including vitamin E (VTE). In plants, the synthesis of VTE compounds, tocopherol and tocotrienol, derives from precursors of the shikimate and methylerythritol phosphate pathways. Quantitative trait loci (QTL) for α-tocopherol content in ripe fruit have previously been determined in an Solanum pennellii tomato introgression line population. In this work, variations of tocopherol isoforms (α, β, γ, and δ) in ripe fruits of these lines were studied. In parallel all tomato genes structurally associated with VTE biosynthesis were identified and mapped. Previously identified VTE QTL on chromosomes 6 and 9 were confirmed whilst novel ones were identified on chromosomes 7 and 8. Integrated analysis at the metabolic, genetic and genomic levels allowed us to propose 16 candidate loci putatively affecting tocopherol content in tomato. A comparative analysis revealed polymorphisms at nucleotide and amino acid levels between Solanum lycopersicum and S. pennellii candidate alleles. Moreover, evolutionary analyses showed the presence of codons evolving under both neutral and positive selection, which may explain the phenotypic differences between species. These data represent an important step in understanding the genetic determinants of VTE natural variation in tomato fruit and as such in the ability to improve the content of this important nutriceutical.
Keywords: Fruit metabolism, Solanum pennellii, tocopherol, tomato, vitamin E
Introduction
Vegetables are critical for human health as they are a source of multiple vitamins and other essential compounds. In particular, tomato fruits are an important dietary source of antioxidants for humans due both to the fact that have a high intrinsic content of these compounds and the elevated consumption of this crop by the western population. The main non-enzymatic antioxidants found in tomato fruits are ascorbic acid (VTC), lycopene and carotenoids, phenolics, and vitamin E (VTE) (Abushita et al., 1997; Frusciante et al., 2007). Recent studies have reinforced the hypothesis of beneficial effects of VTE on human health, mainly in the prevention of coronary heart disease, breast cancer, and protection against nicotine-induced oxidative stress in the brain (Das et al., 2009; Ros 2009; Zhang et al., 2009). Although its function in plants remains somewhat undefined, several reports link VTE to the protection of pigments, proteins, and polyunsaturated fatty acids of the photosynthetic apparatus against reactive oxygen species (ROS) generated during photosynthesis (Semchuk et al., 2009). It has additionally been proposed that VTE interacts with other antioxidant mechanisms in order to maintain cellular redox homeostasis (Foyer and Noctor, 2005).
The synthesis of VTE occurs in photosynthetic organisms and its major constituents are a group of amphipathic molecules containing a polar chromanol head group derived from homogentisate and a polyprenyl lipophilic side chain, products of the shikimate (SK) and methylerythritol phosphate (MEP) pathways, respectively. VTE compounds, collectively termed tocochromanols, can be classified into two groups on the basis of the degree of saturation of their hydrophilic tails. Tocopherols, which are the most abundant in plants, have saturated tails derived from phytyl 2P, whereas tocotrienols have an unsaturated tail derived from geranylgeranyl 2P. The VTE biosynthesis pathway proceeding from the reduction of hydroxyphenylpyruvate to homogentisate is considered the ‘VTE core pathway’ and comprises seven enzymes: 4-hydroxyphenylpyruvate dioxygenase (HPPD, EC 1.13.11.27), homogentisic acid geranylgeranyl transferase (HGGT/HST, EC 2.5.1.-), homogentisate phytyl transferase (VTE2, EC 2.5.1.-), dimethyl-phytylquinol methyl transferase (VTE3, EC 2.1.1.-), tocopherol cyclase (VTE1, EC 5.3.-.-), γ-tocopherol C-methyl transferase (VTE4, EC 2.1.1.95), and phytol kinase (VTE5, EC 2.7.-.-). There are four naturally occurring forms of tocopherols and tocotrienols (α, β, γ, and δ), which differ in the position and number of methyl groups on the chromanol ring (Munné-Bosch and Alegre, 2002). Although all VTE isoforms are potent antioxidants in vitro, α-tocopherol is the most active in terms of vitamin activity, partly because it is retained in the human body in preference to other tocopherols and tocotrienols (Traber and Sies, 1996). In plants, tocochromanol has been found exclusively in plastids. Since it has not been proved thus far that any isoform can be transported within the plant, and the enzymes of the core pathway have been found in plastids (Sun et al., 2009), it is assumed that the biosynthesis also occurs in this compartment.
Although the VTE biosynthetic pathway was elucidated in 1979 (Soll and Schultz, 1979), the identification of the genes involved is much more recent. In the last decade, via the use of genetic and genomics-based methods, the genes encoding the enzymes for most of the steps of the VTE core biosynthesis pathway have been identified and cloned. However, as yet, this is confined to the model organisms Arabidopsis thaliana and Synechocystis sp. PCC6803 (Li et al., 2008). Indeed, the characterization of VTE mutants and transgenic lines has provided considerable insight into the regulatory network of tocochromanol biosynthesis (for review see Mène-Saffrané and DellaPenna, 2009; Falk and Munné-Bosch, 2010). These combined studies have additionally suggested roles for VTE compounds beyond their antioxidant function including their participation in diverse physiological processes including germination, photoassimilate partitioning, growth, leaf senescence, and plant responses to abiotic stress (Falk and Munné-Bosch, 2010). Moreover, several studies have demonstrated a close interaction between VTE and other metabolic pathways. Tomato fruits overexpressing phytoene synthase (PSY), a key enzyme in carotenoid biosynthesis, displayed increased levels of tocopherol (Fraser et al., 2007). Moreover, tocochromanol content is additionally affected when the post-chorismate pathway is manipulated. Arabidopsis transgenic plants overexpressing the bacterial bi-functional chorismate mutase (CM)/prephenate dehydratase (PDT), displayed significantly higher levels of phenylalanine, as well as γ-tocopherol and γ-tocotrienol, besides other secondary metabolites (Tzin et al., 2009). Plant tocochromanol biosynthesis is furthermore subjected to control by both environmental and endogenous signals. In agreement with this statement, the silencing of the light response factor DE-ETIOLATED1 resulted in tomato fruits with enhanced levels of antioxidants, including carotenoids, flavonoids, and tocopherol (Davuluri et al., 2005; Enfissi et al., 2010).
Cultivated tomato (Solanum lycopersicum) is the most consumed vegetable globally. The fact that its wild relatives display tremendous variation in metabolite content in both leaves and fruits (Schauer et al., 2005), renders wild germplasm an important source for metabolic gene discovery focused on aiding efforts to improve the nutritional and industrial quality of crop species (Zamir, 2001; Fernie et al., 2006; Tohge and Fernie, 2010). Utilizing this approach, Schauer et al. (2006) reported a detailed metabolite profile of 76 tomato introgression lines (ILs) containing chromosome segments of the wild species Solanum pennellii in the genetic background of the cultivated S. lycopersicum (cv M82; Eshed and Zamir, 1995). Following the quantification of 74 metabolites of known chemical structure, they were able to identify 889 quantitative fruit metabolic loci for variations in the content of amino and organic acids, sugars, alcohols, fatty acids, VTC, and VTE. Two of these quantitative trait loci (QTL), explaining variation in the α-tocopherol fruit content, were located on chromosomes 6 and 9. Independent experiments available at the Tomato Functional Genomics Database (Fei et al., 2006; http://ted.bti.cornell.edu/) have also revealed differences in tocopherol content associated with the exact same genomic regions. However, the mechanisms explaining these variations are currently not understood, partially due to a lack of knowledge of the complete VTE biosynthetic pathway in tomato.
The aim of the current report is to provide a framework for associating gene sequence with fruit tocopherol content phenotypes by (i) characterizing and mapping all genes involved in the VTE biosynthesis pathway in tomato, (ii) identifying QTL for the content of the vitamers of VTE and their candidate genes, (iii) cloning and sequencing these genes from S. pennellii, and (iv) examining evolutionary patterns of candidates genes by comparing orthologues from S. pennelli, S. lycopersicum, and A. thaliana. The combined results of this study will be discussed in the context of the fundamental understanding of the accumulation of VTE opening further stages for functional analyses.
Materials and methods
Plant material
Tomato seeds from S. lycopersicum L. (cv M82) and S. pennellii introgressed lines were obtained from Tomato Genetic Resource Center (http://tgrc.ucdavis.edu). Tomato plants were grown in 20-l pots under greenhouse conditions: 16/8 h photoperiod, 24±3 °C, 60% humidity, and 140±40 μmol m−2 s−1 incident photo-irradiance. Cloning was carried out from fully expanded source leaves and mature fruits (60 d after flowering). For tocopherol quantification six ripe fruits were taken from six individual plants of ILs 6-1, 6-2, 7-4, 7-4-1, 7-5, 8-2, 8-2-1, 9-1, 9-2-6. Tissue was collected, immediately frozen into liquid nitrogen, and stored at –80 °C until use.
Survey of tocopherol biosynthesis enzymes, genome mapping, and expression analyses
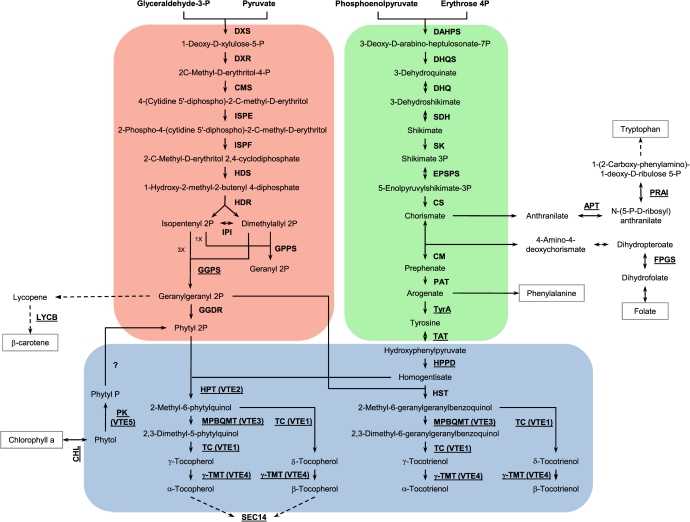
The VTE pathway presented in Fig. 1 was outlined by combining data reported for enzymatic steps involved in SK, MEP, and VTE biosynthesis available on KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/) and related scientific literature. Arabidopsis loci were obtained from the KEGG database and these sequences were used to perform TBLASTN (Altschul et al., 1990) searches against tomato expressed sequences from the Lycopersicon Combined Build # 3 unigene database housed by the Solanaceae Genomics Network (http://solgenomic.net). The criteria used to determine orthology were ≥40% identity at the amino acid level and ≥65% coverage of the Arabidopsis protein. For uncompleted unigenes, the coverage cut-off was set at ≥30%. Based on BLASTN results, all unigene sequences were used as queries to identify the corresponding genomic sequences from the S. lycopersicum genome assembly—version 2.31 (3,230 sequences; 781,381,961 total letters)—The International Tomato Genome Sequencing Project at the Solanaceae Genomics Network (http://solgenomic.net). In silico prediction of the subcellular localization of tomato unigenes was performed using TargetP (http://www.cbs.dtu.dk/services/TargetP) and ChloroP program (http://www.cbs.dtu.dk/services/ChloroP) based on their deduced amino acid sequences.
Fig. 1.
VTE biosysnthesis pathway. The MEP, SK, and tocopherol core pathways are highlighted in red, green, and blue, respectively. Candidate genes of the VTE-related pathways (carotenoid, chlorophyll, tryptophan and folate metabolism, and the SEC14 protein) are not highlighted. Enzymes are named according to their abbreviations in Table 1. Genes for which wild alleles were cloned, sequenced, and analysed are underlined.
The identified tomato genes were mapped onto the Tomato-EXPEN 2000 genetic map available at the Solanaceae Genomics Network. The genetic positions were obtained by BLASTN (Altschul et al., 1990) searches of the identified unigenes and/or their corresponding genomic sequences against the entire Tomato-EXPEN 2000 map marker sequences database (http://solgenomics.net/index.pl). Map Chart software 2.2 (Voorrips, 2000) was used to construct the graphical representation of the genetic map.
Expression data for the set of genes analysed here were extracted from a TOM1 microarray experiment previously published (Carrari et al., 2006). This experiment comprises transcript analyses from tomato fruits harvested along development and ripening stages (10, 15, 20, 21, 35, 49, 56, and 70 d after anthesis).
Tocopherol quantification by HPLC and QTL mapping
Tocopherol extraction was performed as described by Fraser et al. (2000) with the following modifications: tomato fruit were ground to a fine powder in liquid nitrogen and 500 mg of material was extracted with 1.5 ml of methanol and, after vortex-mixing, 1 ml of chloroform was added. Following 5 min of sonication, 1 ml of Tris buffer (50 mM Tris pH 7.5/1 M NaCl) was added. The chloroform phase was recovered and the methanol phase (remaining pellet) was re-extracted with chloroform (2 ml). Chloroform extracts were pooled and adjusted to a final volume of 4 ml. Two millilitres were dried under nitrogen gas and re-suspended in 0.2 ml of 99.5:0.5 hexane/isopropanol. The tocopherol content was determined using a Hewlett-Packard series 1100 HPLC system coupled with a fluorescence detector (Agilent Technologies series 1200). Separation was carried out on a normal-phase column Metasil Si (250 mm×4.6 mm, 5 μm, Varian; Metachem, Torrance, CA, USA) maintained at room temperature using an isocratic solvent system (mobile phase) consisting of 99.5:0.5 hexane/isopropanol with a flow rate of 1 ml min−1. Eluting compounds were detected and quantified by fluorescence with excitation at 296 nm and emission at 340 nm. Identification and quantification of tocopherol compounds was achieved by comparison with the retention times and peak areas of standards purchased from Merck (tocopherol set; Calbiochem #613424). A daily calibration curve was carried out using a tocopherol solution with a concentration range between 0.31 μg ml−1 and 5 μg ml−1 for each isoform. Data of tocopherol isoforms or total tocopherol content were statistically analysed according to Sokal and Rohlf (1981). When the data pull presented homoscedasticity, with or without data transformation using ln or square root, an ANOVA followed by a Dunnett test (P<0.05) was used to compare tocopherol content between ILs and M82 control. Due to lack of homoscedasticity, a non-parametric comparison was also performed by Kruskal–Wallis test (P<0.05 and P<0.1). Statistical tests were performed with Bioestat 5.0 (Ayres et al., 2007) and InfoStat v. 2009 (Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina).
The position of tocopherol QTL on the Tomato-EXPEN 2000 map was determined according to the flanking markers of the S. pennellii introgression fragments in the analysed ILs (Eshed and Zamir, 1995) and mapping of unigenes performed as described in Kamenetzky et al. (2010).
Identification of VTE-related pathway candidate genes
Candidate genes were surveyed along the genomic regions spanned by the identified VTE QTL as described in Bermúdez et al. (2008). All molecular markers mapped onto the selected genomic regions were identified in the comparison merging the Tomato-EXPEN 2000, the Tomato-EXPEN 1992, and the Tomato IL maps by using the comparative map web interface of the Solanaceae Genomics Network (Mueller et al., 2008). All marker sequences were used as query to identify the corresponding unigenes in the Solanaceae Genomics Network database. Gene product functions were determined according to homology to a previously characterized protein, whose function had been experimentally demonstrated in other related plant species, by using the BLASTX algorithm (Altschul et al., 1990) against the NCBI non-redundant (nr) protein database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The cut-off criteria were: ≥40% identity at amino acid level and ≥65% coverage of the orthologous Arabidopsis protein. For uncompleted unigenes the coverage cut-off was set at ≥30%.
Amplification, cloning, and sequencing
Total RNA from 100 mg of source leaf or fruit tissue was isolated using Trizol reagent (Invitrogen, #15596-026) and 1 μg from each sample was treated with amplification grade DNase I (Invitrogen, #18068-015) to remove potential contamination of genomic DNA. The treated RNA was reverse-transcribed to first-strand cDNA and primed with oligo(dT) using Superscript First-Strand (Invitrogen, #18080-044) following the manufacturer's protocol.
Primers were designed using the software Oligo Analyzer 3.1 (http://www.idtdna.com) on the basis of unigene sequences including the sequence surrounding the putative ATG start and stop codons. The primer sequences are available in Table S1 in Supplementary data available at JXB online.
Full-length cDNA fragments were generated by PCR using Taq Platinum pfx DNA polymerase (Invitrogen, #11708-013). The PCR reactions were conducted in a total volume of 50 μl containing 0.3 mM each dNTPs, 0.4 μM each primer, 1.5× reaction buffer, 1 mM MgSO4, ∼150 ng of cDNA, and 2.5 IU of enzyme. The amplification conditions were: 94 °C for 3 min; 35 cycles of 94 °C for 15 s; primer-specific annealing temperature for 30 s; and then 68 °C for 2 min. Amplification products were purified with GFX purification Kit (Amersham Biosciences, #289034-70) and cloned into a pCR-Blunt II TOPO vector using a TOPO-Zero Blunt cloning kit (Invitrogen, #45-0245). Plasmid DNA was isolated using a Qiagen Miniprep Kit (#27106) and inserts were sequenced with BigDye Terminator (Applied Biosystems, #4336919) on an ABI3700 automated sequencer (Applied Biosystems). Sequence data from this article have been deposited in the GenBank Data Libraries under accession number HQ014366–HQ014383 and HQ219713–HQ219716. Polymorphisms were detected at nucleotide and amino acid levels by aligning S. pennellii and S. lycopersicum sequenced alleles (excluding primer regions) using the MULTALIN program (http://www-archbac.u-psud.fr/genomics/multalin.html; Corpet, 1988).
Evolutionary analyses
The coding region alignments of A. thaliana, S. lycopersicum, and S. pennellii were performed with BioEdit Sequence Alignment Editor (Hall, 1999) using the ClustalW package (v1.81 Thompson et al., 1994) and were manually curated according to amino acid alignment. Non-synonymous (dN) and synonymous (dS) distances and their SE values were estimated with MEGA 4.1 software (Tamura et al., 2007) using the Nei–Gojobori method (p-distance). In order to preserve the reading frames, the alignment gaps were deleted prior to estimation of dS and dN. Codon bias was determined by the effective number of codons (Nc) value computed in the CodonW program (mobyle.pasteur.fr/cgi-bin/portal.py?form=codonw). Nc varies between 21 for maximum codon bias, when only one codon is used per amino acid, and 61 for minimum codon bias, when synonymous codons for each amino acid are used at similar frequencies. One-way ANOVA with Tukey's post-hoc test in the InfoStat software was performed to evaluate significant differences in codon usage.
In order to compare codon evolution models to determine selective constraint, three models were fitted using the CODEML program of the PAML suite (Yang, 2007). The first model, M0, assumes that all codons across the sequences have the same level of dN and dS and estimates these values and the dN/dS ratio (ω). ω is a signal of the selection at protein level thus, 0<ω<1 indicates purifying selection, ω=1 neutral selection, and ω>1 points to the presence of positive selection. The model M1a proposes the existence of two classes of codon, a proportion with 0<ω<1 and the remainder of codons with ω=1. Finally, model M2a divides codons into three classes: those with 0<ω<1, ω=1, and ω≥1. The fit of model M0 versus M1a or M1a versus M2a is evaluated by a likelihood ratio test comparing twice the difference in log likelihoods with a χ2 distribution (Yang, 2007).
Results
Identification, mapping, and expression analyses of tomato genes involved in VTE biosynthesis
VTE compounds, tocopherols, and tocotrienols, are products of the convergence of the plastidial MEP and SK pathways. With the aim of identifying every metabolic step in tocochromanol biosynthesis in tomato, the two routes from their primary metabolism precursors were linked to the VTE core pathway (Fig. 1) based on data available in the KEGG database. After an in-depth search of the unigene database deposited in the Solanaceae Genomics Network (http://solgenomics.net), using Arabidopsis loci as reference sequences, all the putative tomato enzyme encoding genes were identified (Table 1). Twenty-nine biochemical reactions constitute the VTE biosynthesis pathway and these are catalyzed by 28 enzymes, for which a total of 41 different tomato encoding loci were described. For eleven of the enzymes, the number of surveyed tomato genes differs from those described for Arabidopsis, according to the criteria adopted here. Only one point in the pathway shown in Fig. 1 remains obscure in plant metabolism, which is the phosphorylation of phytyl P to provide phytyl 2P from phytol via VTE5, as an alternative to the MEP pathway (Valentin et al., 2006).
Table 1.
Identification of tomato genes involved in VTE biosynthesis and QTL- associated candidate genes. A. thaliana loci used for evolutionary analyses are underlined.
| Enzymea | A. thaliana locus (no. amino acids) | Curated localization b | Tomato unigenec | Signal peptided | Genomic ide | Linked marker f | Chromosome positiong(cM) |
| 1-Deoxy-D-xylulose-5-P synthase (DXS, EC 2.2.1.7) | At4g15560 (717) | Chloroplast | U567647 (1) | Chloroplast | SL2.31sc05941 | T1704 | 1 (39 cM) |
| U316204 (2) | nd | SL2.31sc03748 | TG523 | 11 (29 cM) | |||
| 2-C-methyl-D-erythritol 4-phosphate synthase (DXR, EC 1.1.1.267) | At5g62790 (477) | Chloroplast | U585813 | Chloroplast | SL2.31sc03701 | C2_At5g23060 | 3 (102.5 cM) |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (CMS, EC 2.7.7.60) | At2g02500 (302) | Chloroplast | U566797 | Chloroplast | SL2.31sc04323 | TG528 | 1 (127.5 cM) |
| 4-(Cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase (ISPE, EC 2.7.1.148) | At2g26930 (383) | Chloroplast | U583224 | Chloroplast | SL2.31sc04133 | cTOC-4-C7 | 1 (19 cM) |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (ISPF, EC 4.6.1.12) | At1g63970 (231) | Chloroplast | U568497 | Chloroplast | SL2.31sc03923 | C2_At3g27530 | 8 (78 cM) |
| 4-Hydroxy-3-methylbut-2-enyl- diphosphate synthase (HDS, EC 1.17.7.1) | At5g60600 (717) | Chloroplast | U567167 | Chloroplast | SL2.31sc03876 | C2_At5g60600 | 11 (79 cM) |
| 4-Hydroxy-3-methylbut-2-enyl- diphosphate reductase (HDR, EC 1.17.1.2) | At4g34350 (466) | Chloroplast | U580658 | Chloroplast | SL2.31sc04323 | C2_At4g34350 | 1 (154 cM) |
| Isopentenyl diphosphate δ-isomerase (IPI, EC 5.3.3.2) | At3g02780 (284) | Mitochondria | U577516 (1) | Chloroplast | SL2.31sc06101 | U49812 | 4 (64 cM) |
| At5g16440 (291) | Chloroplast (Phillips et al., 2008) | U569721 (2) | Chloroplast | SL2.31sc03902 | C2_At5g04270 | 5 (112.7 cM) | |
| Geranyl pyrophosphate synthase (GPPS, EC 2.5.1.1) | At2g34630 (422) | Chloroplast (Bouvier et al.,2000) | U573523 | Mitochondria | SL2.31sc03835 | C2_At1g30360 | 8 (21.3 cM) |
| Geranylgeranyl pyrophosphate synthase (GGPS, EC 2.5.1.29) | At4g36810(371) | Chloroplast | U574849 (1) | Chloroplast | SL2.31sc03748 | C2_At5g16710 | 11 (31.4 cM) |
| U571085 (2) | Chloroplast | SL2.31sc04135 | C2_At1g19340 | 4 (112 cM) | |||
| At4g38460(326) | Chloroplast | U573348 (3) | nd | SL2.31sc03665 | CT232 | 2 (90.1 cM) | |
| U575882 (4) | Chloroplast | SL2.31sc03771 | T0532 | 9 (30 cM) | |||
| Geranylgeranyl reductase (GGDR, EC 1.3.1-) | At1g74470 (467) | Chloroplast | U564571 | Chloroplast | SL2.31sc03701 | C2_At1G74470 | 3 (122 cM) |
| 3-Deoxy-D-arabino-heptulosonate-7-P synthase (DAHPS, EC 2.5.1.54) | At1g22410 (527) | Chloroplast | U581552 (1) | Chloroplast | SL2.31sc03748 | T0408 | 11 (26 cM) |
| At4g33510 (432) | Chloroplast | ||||||
| At4g39980 (525 ) | Chloroplast (Entus et al.,2002) | U566921 (2) | nd | SL2.31sc04135 | T1560 | 4 (83.5 cM) | |
| 3-Dehydroquinate synthase (DHQS, EC 4.2.3.4) | At5g66120 (442) | Chloroplast | U568781 | Chloroplast | SL2.31sc03665 | C2_At3g01160 | 2 (83.4 cM) |
| Shikimate dehydrogenase (SDH, EC 1.1.1.25) / 3-Dehydroquinate dehydratase (DHQ, EC 4.2.1.10) | At3g06350 (603) | Chloroplast | U570855(1) | nd | SL2.31sc05941 | T1704 | 1 (39 cM) |
| U570070(2) | nd | SL2.31sc03622 | TG221 | 6 (101 cM) | |||
| Shikimate kinase (SK, EC 2.7.1.71) | At2g21940 (276) | nd | U582040 | Chloroplast | SL2.31sc06101 | C2_At3g62940 | 4 (56 cM) |
| At4g39540 (300) | Chloroplast | ||||||
| 5-Enolpyruvylshikimate-3-P synthase (EPSPS, EC 2.5.1.19) | At1g48860 (521) | Chloroplast | U577580 | Chloroplast | SL2.31sc04323 | C2_At2g45240 | 1 (57 cM) |
| At2g45300 (520) | Chloroplast | ||||||
| Chorismate synthase (CS, EC 4.2.3.5) | At1g48850 (380) | Chloroplast | U563165(1) | Chloroplast | SL2.31sc06101 | C2_At3g07950 | 4 (56.7 cM) |
| U563163(2) | Chloroplast | SL2.31sc03604 | TG370 | 4 (21.5 cM) | |||
| Chorismate mutase (CM, EC 5.4.99.5) | At1g69370 (316) | Chloroplast (Mobley et al.,1999) | SL2.31sc03665 | T1480 | 2 (106 cM) | ||
| U575627(1) | nd | ||||||
| At5g10870 (340) | Chloroplast (Eberhard et al., 1996) | ||||||
| SL2.31sc03748 | TG147 | 11 (45 cM) | |||||
| At3g29200 (265) | Cytosol (Eberhard et al., 1996) | U585231(2) | nd | ||||
| Prephenate aminotransferase (PAT, EC 2.6.1.57) | At2g22250 (475) | Chloroplast | U567172 | Chloroplast | SL2.31sc06101 | C2_At4g39830 | 4 (61 cM) |
| Arogenate dehydrogenase (TyrA, EC 1.3.1.78) | At1g15710 (358) | Chloroplast | U567861 (1) | Chloroplast | SL2.31sc03731 | C2_At5g34850 | 7 (0.4 cM) |
| At5g34930 (640) | Chloroplast (Rippert et al., 2009) | U570951 (2) | Chloroplast | SL2.31sc03771 | T1212 | 9 (48 cM) | |
| Tyrosine aminotransferase (TAT, EC 2.6.1.5) | At5g53970(414) | nd | U577103(1) | nd | SL2.31sc05925 | C2_At1g53000 | 10 (7.5 cM) |
| U563404 (2) | nd | SL2.31sc03685 | C2_At1g03820 | 7 (43 cM) | |||
| 4-Hydroxyphenylpyruvate dioxygenase (HPPD, EC 1.13.11.27) | At1g06570(473) | Cytosol (Garcia et al.,1999) | U580457(1) | nd | SL2.31sc03685 | TG584 | 7 (36.5 cM) |
| U578997(2) | Chloroplast | SL2.31sc03902 | CLET-6-I4 | 5 (70 cM) | |||
| Homogentisate geranylgeranyl transferase/ homogentisate solanesyl transferase (HGGT/HST, EC 2.5.1.-) | At3g11945.2 (393) | Chloroplast | U585005 | Chloroplast | SL2.31sc06725 | C2_At3g58490 | 3 (72.6cM) |
| Homogentisate phytyl transferase [HPT (VTE2), EC 2.5.1.-] | At2g18950(393) | Chloroplast | U327540(5’) U576207(3’) | Chloroplast | SL2.31sc03731 | cTOA-13-K15 | 7 (17 cM) |
| Dimethyl-phytylquinol methyl transferase [MPBQMT (VTE3), EC 2.1.1.-] | At3g63410(338) | Chloroplast | U578249(1) | Chloroplast | SL2.31sc04777 | T0565 | 9 (52 cM) |
| U581492(2) | Chloroplast | SL2.31sc04439 | TG324 | 3 (4.6 cM) | |||
| Tocopherol cyclase [TC (VTE1), EC 5.3.-.-] | At4g32770(488) | Chloroplast | U570602 | Chloroplast | SL2.31sc04948 | C2_At4g32770 | 8 (34 cM) |
| γ-Tocopherol C-methyl transferase [γ-TMT (VTE4), EC 2.1.1.95] | At1g64970 (348) | Chloroplast (Ferro et al., 2010) | U584511 | Chloroplast | SL2.31sc03923 | TG282 | 8 (41.8 cM) |
| Phytol kinase [PK (VTE5), EC 2.7.-.-] | At5g04490(304) | Chloroplast | U583081 | Chloroplast | SL2.31sc03771 | C2_At5g58240 | 9 (50.5 cM) |
| Anthranilate phosphoribosyltransferase (APT, EC 2.4.2.18) | At5g17990(444) | Chloroplast | U566340 | Chloroplast | SL2.31sc05054 | C2_At5g17990 | 6 (59 cM) |
| Phosphoribosylanthranilate isomerase (PRAI, EC 5.3.1.24) | At1g07780 (275) | Chloroplast | U564371 | Chloroplast | SL2.31sc05732 | cLET-1-I13 | 6 (24 cM) |
| At1g29410 (244) | Chloroplast (Zhao and Last, 1995) | ||||||
| At5g05590 (275) | Chloroplast | ||||||
| Folylpolyglutamate synthase (FPGS, EC 6.3.2.17) | At5g41480(530) | Mitochondria (Ravanel et al., 2001) | U581922h | Mitochondria | SL2.31sc05732 | At5g41480 | 6 (26 cM) |
| Phospholipid transporter SEC14 | At3g51670(409) | Membrane (Peterman et al., 2004) | U583419 | Chloroplast | SL2.31sc03771 | T1095 | 9 (50.5 cM) |
| Chlorophyllase (CHL, EC 3.1.1.14) | At5g43860 (318) | Cytosol (Schenk et al., 2007) | U574853 | nd | SL2.31sc05732 | T0834 | 6 (32 cM) |
| At1g19670 (324) | |||||||
| Lycopeno β-cyclase LYCB | At3g10230(369) | Chloroplast(Ferro et al., 2010) | U570109 | Chloroplast | SL2.31sc05054 | cLET-19-J2 | 6 (74 cM) |
Enzyme name, number according to KEGG (www.genome.jp/kegg/) and abbreviation.
A. thaliana enzyme localization according to The Plant Proteome Database (ppdb.tc.cornell.edu, Sun et al., 2009) or specific reference.
Tomato unigene number according to SGN (solgenomics.net).
Sub-cellular localization prediction according to TargetP 1.1 software (www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2007) and ChloroP (www.cbs.dtu.dk/services/ChloroP).
S. lycopersicum scaffold spanning the corresponding gene (version 2.31).
Closest markers within scaffold.
Chromosome and genetic position in Tomato-EXPEN 2000 v52.
5’ incomplete unigene
Nd, not determined
The subcellular localization of the protein products of previously identified S. lycopersicum loci were predicted by the TargetP and ChloroP softwares. Enzymes that did not contain a predicted targeting peptide were considered cytosolic. This in silico prediction was in general agreement with the experimental evidence reported for the Arabidopsis orthologues (Table 1). For two proteins, however, predictions revealed unexpected results. The tomato geranyl pyrophosphate synthase (GPPS) enzyme was predicted to be targeted to the mitochondria, while for the bi-functional shikimate dehydrogenase/3-dehydroquinate dehydratase (SDH/DHQ) no signal peptides were detected for any of the identified loci. Moreover, none of the tomato CMs presented a predicted plastid signal peptide. The fact that the last enzyme of the post-chorismate portion of the SK pathway, tyrosine aminotransferase (TAT), and HPPD appeared to be localized in the cytosol in Arabidopsis cells provides intrigue regarding the transport of homogentisate across the chloroplast envelope (Joyard et al., 2009). These predictions, with regard to the tomato proteins, also failed to propose a subcellular localization for TAT, even though one of the HPPD unigenes exhibited a chloroplast signal peptide prediction.
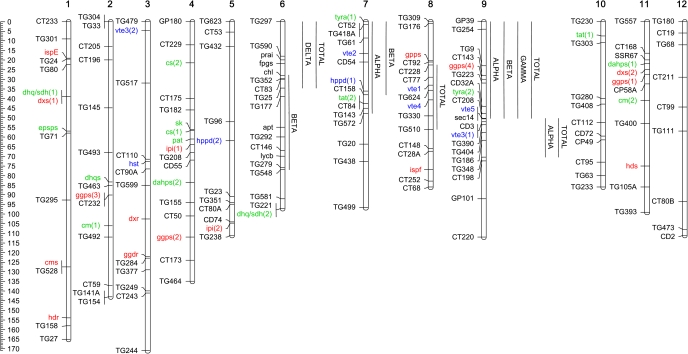
As a second step in the characterization of the genetic basis of tocochromanol biosynthesis, the 41 tomato loci involved in MEP, SK, and VTE core pathways were localized onto the tomato genetic map (Tomato-EXPEN 2000) (Figs 1, 2, Table 1). Mapping was based on physical linkage between mapped markers and the unigenes and/or their corresponding genomic sequences. With the exception of chromosome 12, all other tomato chromosomes harbour at least one of the identified loci. Interestingly, most of the tocochromanol core pathway enzyme-encoding loci were localized on chromosomes 7, 8, and 9, except for hst and vte3(2), mapping to chromosome 3, and hppd(2), mapping to chromosome 5. Remarkably, the four genes mapped on chromosome 9, vte3(1), vte5, arogenate dehydrogenase [tyra(2)], and geranylgeranyl pyrophosphate synthase [ggps(4)] all co-localize with a previously reported QTL for α-tocopherol content described by Schauer et al. (2006).
Fig. 2.
Genomic localization of tocopherol biosynthesis and candidate genes. All genes were localized in the Tomato-EXPEN 2000 genetic map available at the Solanaceae Genomics Network (http://solgenomics.net/index.pl). Markers and genes are indicated on the left side of the chromosomes. Tocopherol QTL are indicated on the right side of the chromosomes. Gene colour code is in accordance with Fig. 1.
Of the 41 identified genes, the expression patterns of 27 of them could be evaluated across fruit development and ripening, by retrieving data from a previously published microarray experiment (Carrari et al., 2006). This analysis revealed that all 27 genes were expressed in tomato fruits in at least one time point. The *omeSOM model of neural clustering recently developed (Milone et al., 2010) revealed six groups. Whilst these clusters grouped genes from all three evaluated pathways, no specific pathway patterns were identified (data not shown).
QTL for fruit tocopherol content and identification of candidate genes
As mentioned above, these mapping results revealed that the majority of the VTE core pathway enzyme-encoding genes are grouped within chromosomes 7, 8, and 9. Previously, using GC-MS analysis, Schauer et al. (2006) reported two QTL for fruit α-tocopherol content localized to chromosomes 6 and 9. In order to investigate further the presence of other QTL associated with the genomic regions that harbour the tocopherol core biosynthesis genes, and as such to obtain a precise and detailed quantification, an HPLC protocol for measuring all four tocopherol isoforms was applied.
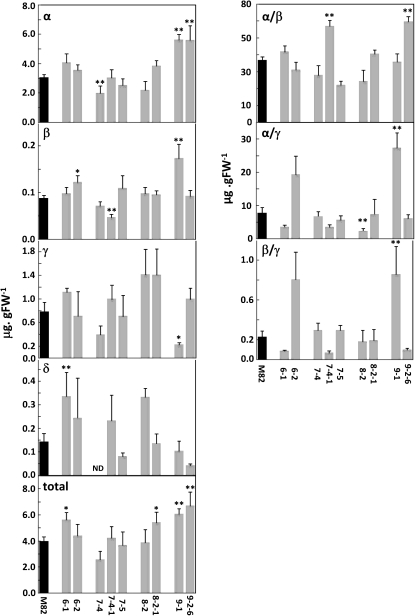
The content of α-, β-, γ-, δ-, and total tocopherol was determined from ripe tomato fruits from the ILs 6-1, 6-2, 7-4, 7-4-1, 7-5, 8-2, 8-2-1, 9-1, 9-2-6 as well as fruits from the corresponding S. lycopersicum control (cv M82). The amount of each isoform and their ratios are presented in Fig. 3, whereas the identified QTL are positioned on the genetic map presented in Fig. 2. On chromosome 9, two QTL were identified for α- and total tocopherol (ILs 9-1 and 9-2-6), one for β-tocopherol (IL 9-1), and one for γ-tocopherol (IL 9-1). Both QTL for total tocopherol are in agreement with the results reported in Schauer et al. (2006). The QTL on IL 9-2-6 co-localize with two VTE core pathway encoding genes: vte3(1) and vte5 (Fig. 2), whilst the QTL on IL 9-1 spans the genomic region containing tyra(2) and ggps(4). Measurements performed in fruits from ILs with S. pennellii introgressions on chromosome 6 showed significant differences from M82 fruits in the levels of β-tocopherol (IL 6-2), δ-, and total tocopherol (IL 6-1). Moreover, QTL were also identified on chromosome 7 (ILs 7-4 and 7-4-1 for α- and β-tocopherol, respectively) and 8 (IL 8-2-1 for total tocopherol) also co-localizing with the genes encoding the VTE core pathway enzymes: vte2 and hppd(1) on chromosome 7 and vte1 and vte4 on chromosome 8 (Fig. 2). Besides the mentioned genes, the presence of tyra(1) at 0.4 cM and tat(2) at 43 cM on chromosome 7 could also be responsible for the α- and β-tocopherol QTL mapped onto these regions.
Fig. 3.
Tocopherol content. Tocopherol content was determined by HPLC. Grey bars indicate means of six biological replicates. Significant differences compared with the M82 control cultivar (black bars) according to Dunnett test (P<0.05) and/or Kruskal–Wallis test (P<0.05 ** and P<0.1*) are indicated.
By surveying the genomic regions spanning the identified QTL, six novel candidate genes belonging to VTE-related pathways were found. On chromosome 6, at 32 cM, a chlorophyllase encoding gene (CHL, EC 3.1.1.14) was identified. It has been proposed that the first step in the degradation of chlorophyll during senescence and fruit ripening is catalysed by this enzyme (Hörtensteiner, 2006) through the synthesis of phytol, providing phytyl 2P to the VTE pathway via VTE5. Moreover, three enzyme-encoding genes for the two branching pathways from chorismate to tryptophan and folate were also identified: a phosphoribosylanthranilate isomerase (PRAI, EC 5.3.1.24), a folylpolyglutamate synthase (FPGS, EC 6.3.2.17), and an anthranilate phosphoribosyltransferase (APT, EC 2.4.2.18) mapping at 24, 26, and 59 cM, respectively. Finally, the lycopene β-cyclase (LYCB) encoding gene was found at 74 cM of chromosome 6. This enzyme, which catalyses the last step in β-carotene biosynthesis, has been deeply characterized after a positional cloning strategy by Ronen et al. (2000) and constitutes a strong candidate for tocopherol content due to the common precursor geranylgeranyl 2P. Another putative candidate, the SEC14 protein-encoding gene, was found on chromosome 9. Several studies have demonstrated the involvement of this protein in tocopherol transport in mammalian cells and lipid traffic in plants (Saito et al., 2007; Bankaitis et al., 2009). With the exception of the chl these novel candidates are expressed in tomato fruits at least one time point of the developmental analysis performed by Carrari et al. (2006).
Taken together the results obtained from tomato gene identification, mapping, tocopherol quantification, and QTL localization, 16 candidate loci putatively affecting tocopherol content in tomato can be proposed: prai, fpgs, chl, apt, and lycb on chromosome 6; tyra(1), vte2, hppd(1), and tat(2) located on chromosome 7; vte1 and vte4 on chromosome 8; and ggps(4), tyra(2), vte5, sec14, and vte3(1) on chromosome 9 (Figs 1–3).
Allele characterization of QTL-associated candidates
The identification of 16 QTL-associated candidate genes prompted us to unearth the allelic differences between S. lycopersicum and S. pennellii (underlined genes in Fig. 1). The coding regions of wild alleles were thus subsequently cloned from the corresponding ILs using primers annealing to the initial and stop codons. Although only minor size differences were observed between the two alleles for most of the genes, TyrA(2)-, VTE4-, PRAI-, and CHL-encoding genes exhibited different amplicon length. These results indicated that neither vast allelic polymorphisms, nor large genomic rearrangements, span the chromosomal region encompassing the analysed genes. This comparison revealed that all analysed candidate genes present at least one non-synonymous polymorphism (Table 2). The most divergent alleles are those encoding the PRAI enzyme for which the S. pennelli allele encodes a protein 26 amino acids shorter in comparison with the S. lycopersicum allele.
Table 2.
Comparison of S. lycopersicum and S. pennellii alleles of cDNA encoding VTE candidate genes. LYC, S. lycopersicum; PEN, S. pennellii.
| Gene | Unigene | Coding sequence (nt) |
Predicted protein (no. amino acids) |
No. of polymorphic nucleotidesa | No. of polymorphic amino acidsa | ||
| LYC | PEN | LYC | PEN | ||||
| ggps(4) | U575882 | 1005 | 1005 | 334 | 334 | 11 | 3 |
| tat(2) | U563404 | 1269 | 1269 | 422 | 422 | 10 | 6 |
| tyra(1) | U567861 | 1134 | 1134 | 377 | 377 | 7 | 1 |
| tyra(2) | U570951 | 1173 | 1182 | 390 | 393 | 24+9insertions | 9+3insertions |
| vte4 | U584511 | 1089 | 1086 | 362 | 361 | 9+3deletions | 2+1deletion |
| vte1 | U570602 | 1497 | 1497 | 498 | 498 | 12 | 5 |
| vte3(1) | U578249 | 1020 | 1020 | 339 | 339 | 7 | 1 |
| vte2 | U327540 (covers 5’ end)U576207 (covers 3’ end) | 1209 | 1209 | 402 | 402 | 8 | 3 |
| vte5 | U583081 | 882 | 882 | 293 | 293 | 10 | 4 |
| hppd(1) | U580457 | 1263 | 1263 b | 420 | 420 b | 16 | 7 |
| apt | U566340 c | 1097 c | 1097 c | 365 | 365 | 8 | 4 |
| prai | U564371 | 906 | 828 | 301 | 275 | 12+78deletions | 3+26deletions |
| fpgs | U581922 c | 1457 c | 1457 c | 485 | 485 | 13 | 4 |
| chl | U574853 | 939 | 948 | 312 | 315 | 22+9insertions | 9+3insertions |
| sec14 | U583419 | 1275 | 1275 | 424 | 424 | 11 | 4 |
| lycb | U570109 | 1497 | 1497 | 498 | 498 | 19 | 9 |
Nucleotide or amino acid insertions or deletions in S. pennellii sequences.
S. pennellii does not present a stop codon along the analysed region.
Probably lacking 3' end.
VTE biosynthesis genes: evolutionary analyses of cultivated and wild tomato alleles
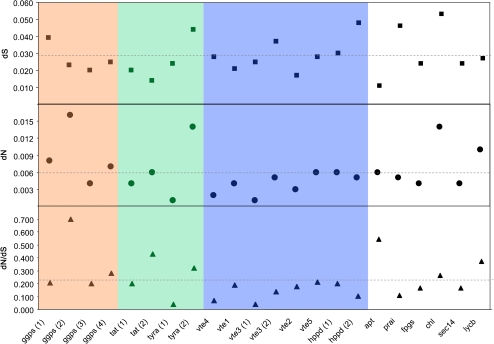
The fate of cellular metabolic networks generally depends on the products of many loci. The inter-relationships between loci at the phenotypic level raise the question of whether they evolved independently. In this work QTL for tocopherol content using S. pennelli ILs were mapped, and 16 candidate loci linked to those QTL were identified and the wild species alleles cloned. Furthermore, in order to study how the structure of the VTE metabolic pathway could have influenced protein evolution rates, the evolutionary pattern among the candidate genes and their paralogues was also investigated, estimating the pairwise synonymous (dS), non-synonymous (dN), and dN/dS divergence between S. lycopersicum and S. pennellii (Fig. 4). The dS and dN values varied greatly between genes, ranging from 4.8 to 16 times for dS and dN, respectively. Four genes, ggps(2), tyra(2), chl, and lycb, displayed particularly high values of dN, above the mean. The dN/dS also varied remarkably among the 22 loci analysed with the highest value being 17.4 times the lowest. The genes of MEP, post-chorismate SK, and tocopherol biosynthesis core pathways displayed values similar to or lower than the average with the exception of those presenting more than one locus. Among candidate genes of related pathways apt, chl, and lycb displayed higher dN/dS values than the mean. Interestingly, the five genes for which more than one locus was identified displayed variations of dN/dS values between paralogues. The dN/dS values for the different ggps, tat, tyra, vte3 and hppd varied by 3.5, 2.1, 7.6, 3.4, and 1.9 times, respectively. Differences in dN/dS values between the paralogues might be due to high dN caused by a weak selection at non-synonymous sites, or related to the intensity of natural selection on synonymous codon usage. To investigate the existence of codon usage bias, the effective number of codons (Nc) was calculated for each gene and species (S. pennellii and S. lycopersicum). No statistically significant differences (ANOVA, P>0.01) in Nc were observed between ggps, tat, tyra, and vte3 paralogues, suggesting that the dN/dS differences are most probably due to a constraint relaxation for one of the gene copies rather than codon bias. In contrast, for the hppd pair a significant Nc bias was observed (ANOVA, P<0.01). This can be also visualized in Fig. 4 comparing dN and dS values, suggesting that the dN/dS rate differences between hppd paralogues might not be explained by constraint relaxation.
Fig. 4.
Evolutionary rates at synonymous (dS) and non-synonymous (dN) sites of candidate genes linked to tocopherol QTL. Genes belonging to MEP, SK, and tocopherol core pathways are highlighted in red, green, and blue, respectively. VTE-related pathway candidates are not highlighted. Lines indicated estimated means.
Although dN/dS is a useful indicator of selective pressure, there is some oversimplification in its application since it may be that only certain codons in a gene can change in a way that enhances fitness whereas all others cannot accept substitutions without cost to fitness. Thus, to better explore patterns of sequence variation, the orthologous sequences from A. thaliana, S. lycopersicum, and S. pennellii for all 22 genes under study were aligned and a Likelihood Ratio Test (LRT) with three models of molecular evolution was applied. The first, M0, assumes that all positions across the sequences have the same level of dN and dS. The second, M1a, proposes that a proportion of codons is under purifying selection while the remainder have neutral evolution. Finally, M2a divides codons into three classes, those with purifying selection, those with neutral evolution pattern, and the remainder with positive selection. In order to avoid data misinterpretation, the Nc was estimated and the comparison between species did not show statistically significant differences (P>0.01), indicating that there is no codon bias usage. Comparison of the three models showed that for 19 genes M1a displayed the best fit, indicating that, even when a proportion of the codons for every gene are evolving neutrally, there is no support for positive selection. Nevertheless, for ggps(2), ggps(4), and vte1, M2a presented a better fit than M1a, with different levels of significance, showing that these genes exhibited signs of positive selection. Interestingly, while ggps(2) and vte1 showed a higher proportion of codons evolving under positive selection (∼25%) than ggps(4) (0.8%), they displayed lower significance levels. This can be explained by the high ω value of ggps(4) indicating that there is no variation in the synonymous sites along the codons under positive natural selection (Table 3).
Table 3.
Parameter estimates and tests of selection.
| Gene | M0 |
M1a |
M2a |
|||||||
| lnLa | ωb | lnLa | ω0c | ρ0d,e | lnLa | ω0f | ρ0g | ω2h | ρ2i,j | |
| ggps(1) | –2640.1 | 0.041 | –2598.9 | 0.012 | 0.734** | –2598.9 | 0.012 | 0.743 | – | 0.000 |
| ggps(2) | –2594.4 | 0.096 | –2551.5 | 0.028 | 0.730** | –2549.8 | 0.032 | 0.746 | 2.474 | 0.254* |
| ggps(3) | –2444.5 | 0.014 | –2411.5 | 0.018 | 0.812** | –2411.5 | 0.018 | 0.813 | 1.134 | 0.187 |
| ggps(4) | –2233.7 | 0.061 | –2200.8 | 0.019 | 0.811** | –2196.7 | 0.019 | 0.811 | 999.000 | 0.008** |
| tat(1) | –2718.5 | 0.101 | –2690.3 | 0.044 | 0.792** | –2690.3 | 0.044 | 0.792 | – | 0.000 |
| tat(2) | –2857.8 | 0.077 | –2841.7 | 0.046 | 0.817** | –2841.7 | 0.046 | 0.817 | – | 0.000 |
| tyra(1) | –2487.3 | 0.002 | –2461.0 | 0.024 | 0.740** | –2461.0 | 0.024 | 0.740 | – | 0.000 |
| tyra(2) | –2657.4 | 0.108 | –2615.1 | 0.024 | 0.612** | –2615.1 | 0.024 | 0.612 | – | 0.000 |
| vte4 | –2425.8 | 0.062 | –2391.8 | 0.028 | 0.765** | –2391.8 | 0.028 | 0.765 | – | 0.000 |
| vte1 | –3430.5 | 0.072 | –3373.4 | 0.028 | 0.730** | –3371.9 | 0.039 | 0.742 | 2.165 | 0.257* |
| vte3(1) | –2138.5 | 0.051 | –2124.3 | 0.031 | 0.900** | –2124.3 | 0.031 | 0.900 | – | 0.000 |
| vte3(2) | –2256.5 | 0.057 | –2231.5 | 0.022 | 0.842** | –2231.5 | 0.022 | 0.842 | – | 0.000 |
| vte2 | –2693.7 | 0.096 | –2653.3 | 0.030 | 0.739** | –2653.3 | 0.030 | 0.739 | – | 0.000 |
| vte5 | –2302.1 | 0.079 | –2287.6 | 0.023 | 0.541** | –2287.6 | 0.023 | 0.541 | – | 0.000 |
| hppd(1) | –2586.5 | 0.064 | –2552.8 | 0.024 | 0.818** | –2552.8 | 0.024 | 0.820 | 2.258 | 0.016 |
| hppd(2) | –2617.9 | 0.037 | –2579.5 | 0.020 | 0.805** | –2579.5 | 0.020 | 0.805 | – | 0.000 |
| apt | –2447.7 | 0.099 | –2400.6 | 0.024 | 0.770** | –2399.8 | 0.032 | 0.799 | 2.629 | 0.201 |
| prai | -1556.9 | 0.144 | -1551.3 | 0.104 | 0.732** | -1551.9 | 0.104 | 0.732 | – | 0.000 |
| fpgs | -3435.8 | 0.100 | -3410.5 | 0.055 | 0.742** | -3410.5 | 0.055 | 0.742 | – | 0.000 |
| chl | -2198.3 | 0.127 | -2186.0 | 0.078 | 0.846** | -2185.9 | 0.080 | 0.853 | 1.124 | 0.146 |
| sec14 | -2547.0 | 0.030 | -2527.3 | 0.025 | 0.894** | -2527.2 | 0.025 | 0.894 | – | 0.000 |
| lycb | -2628.0 | 0.084 | -2608.2 | 0.046 | 0.772** | -2608.2 | 0.046 | 0.772 | – | 0.000 |
Log likehood of model.
Parameter estimate assuming a single dN/dS ratio per gene.
Estimated dN/dS for proportion of codons (ρ0) under purifying selection; the rest of codons assumed to be evolving neutrally.
Estimated proportion of codons under purifying selection.
Test of M1a versus M0. ** χ2 test using P<0.01.
Estimated dN/dS for proportion of codons (ρ0) under purifying selection.
Estimated proportion of codons under purifying selection.
Estimated dN/dS for proportion of codons (ρ2) under positive selection.
Estimated proportion of codons under positive selection.
Test of M2a versus M1a. * χ2 test using P<0.10; **χ2 test using P<0.01.
–, not available.
Discussion
The two pathways feeding the main precursors of VTE biosynthesis, MEP and SK, as well as the tocopherol core route, were surveyed across the tomato genome with the aim of identifying regulatory steps of the VTE fruit biosynthesis. The metabolic reconstruction of VTE metabolism resulted in the identification of 29 reactions catalysed by the protein products of 28 genes, for which 41 different S. lycopersicum loci were identified (Fig. 1, Table 1). In the course of reconstructing these routes, certain assumptions concerning the presence or absence of reactions involved in VTE metabolism had to be made. First, in the MEP pathway, GPPS is the enzyme leading to monoterpene biosynthesis, while GGPS is responsible for the production of geranylgeranyl 2P by the sequential coupling of three isopentenyl 2P (IPP) molecules to dimethylallyl 2P (DMAPP). Geranylgeranyl 2P serves as a precursor for carotenoid, tocochromanol, gibberelin, and chlorophyll biosynthesis (Aharoni et al., 2005; Joyard et al., 2009). Hence, the classical trend is to assume that GPPS would not be involved in tocochromanol production. Tomato plants silenced for GPPS even displayed a dwarfed phenotype and reduced gibberellin levels, and did not alter carotenoid and chlorophyll content (Van Schie et al., 2007). These results thus indicated that pigments are originated from a geranyl 2P-independent geranylgeranyl 2P pool and suggested that GPPS might not influence VTE biosynthesis. However, further functional experiments that validate this hypothesis are clearly needed. Secondly, HGGT, which condenses homogentisate with geranylgeranyl 2P during tocotrienol synthesis, was identified in grass species in which tocotrienols are the most abundant tocochromanol forms (Cahoon et al., 2003). Neither in Arabidopsis nor in tomato was HGGT identified. However, tocotrienol traces have been detected in both tomato and tobacco (Chun et al., 2006). This might be explained by the promiscuous activity in substrate acceptance of other prenyl transferases such as homogentisate solanesyl transferase (HST) and/or the homogentisate phytyl transferase (VTE2). Flux changes through the SK, MEP, and/or tocopherol pathway could conceivably shift the substrate preference of both these enzymes (Herbers, 2003; Falk and Munné-Bosch, 2010). This hypothesis is supported by the fact that under certain conditions tobacco plants, which do not have a canonical HGGT, and co-express the Arabidopsis HPPD and the yeast prephenate dehydrogenase, exhibit higher levels of tocotrienols (Rippert et al., 2004). Therefore, the enhanced supply of homogentisate may affect the substrate specificity of prenyl transferases leaking through tocotrienol synthesis (Falk and Munné-Bosch, 2010). In addition, A. thaliana HST, which is essential for plastoquinone-9 biosynthesis—catalysing the condensation of solanesyl 2P (SDP) and HGA—also accepts farnesyl 2P and geranylgeranyl 2P as prenyl donors (Sadre et al., 2006; Tian et al., 2007), providing further evidence in support of tocotrienol production in the absence of a specific HGGT.
Prephenate aminotransferase (PAT, EC 2.6.1.57), which converts prephenate intermediate into arogenate, has been characterized in bacteria. Although its activity had also been detected in plants, no associated loci were identified with this enzymatic function (Tzin et al., 2009). Recently, two independent reports performed biochemical and functional characterization of this plant enzyme thus, completing the identification of the genes involved in phenylalanine and tyrosine biosynthesis (Graindorge et al., 2010; Maeda et al., 2010).
One missing links still remains within the metabolic network under study, which is the absence of a phytol-P kinase that could provide phytyl 2P as an alternative to the MEP pathway (Ischebeck et al., 2006; Valentin et al., 2006).
All the MEP enzyme-encoding genes surveyed here are in accordance with sequences previously reported for tomato (Lois et al., 2000; Rohdich et al., 2000; Rodríguez-Concepción et al., 2001, 2003; Botella-Pavía et al., 2004; Ament et al., 2006; Paetzold et al., 2010; and GenBank database direct submission for IPI -GQ169536 and EU253957-), with the exception of the 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (CMS)- and geranylgeranyl reductase (GGDR)-encoding genes, which have not been reported before in this species. Moreover, two additional loci for GGPS were identified here. For the SK pathway, enzyme-encoding genes involved in reactions upstream of the prephenate intermediate have already been reported in tomato (Gasser et al., 1988; Schmid et al., 1992; Görlach et al., 1993, 1995; Eberhard et al., 1996; Bischoff et al., 1996, 2001). Moreover, novel loci for SDH/DHQ and CM were identified, whilst the loci encoding PAT, TyrA, and TAT were first described in tomato.
Protein localization data are highly valuable information in order to elucidate gene function. The MEP and SK pathways are well accepted to operate in the chloroplast (Rippert et al., 2009). Even so, the recent identification of some cytosolic isoforms points to the existence of an extraplastidial SK pathway (Ding et al., 2007). The results presented here for the in silico prediction of the subcellular localization of the tomato deduced protein sequences indicated that the reactions of VTE metabolism occur mostly in chloroplasts. In contrast, for the bi-functional SDH/DHQ no signal peptide was detected, most probably due to the failure to detect true targeting, since reported analysis of subcellular fractions indicated that tomato SDH/DHQ is localized in the chloroplast (Bischoff et al., 2001). However, Ding et al. (2007) functionally characterized a cytosolic SDH/DHQ isoform in tobacco and identified the tomato orthologue (BF096277/SGN-U578253). This locus was not included in this study due to its low identity to Arabidopsis protein according to the Pipeline criteria adopted and the absence of functional support so far. Unsuccessful subcellular prediction is not an unusual scenario, since in silico, signal peptides are occasionally misidentified. However, these predictions indicated mitochondrial targeting for the A. thaliana GPPS (data not shown), whilst experimental data revealed that this enzyme is directed to the plastid (Bouvier et al., 2000). It is important to point out the existence of some ambiguous reports on actual GPPS localization where experimental evidence supports its localization either in plastid or in the cytosol depending on the species (Nagegowda, 2010). Moreover, for the TAT and CM enzymes, the plastidial localization could not be confirmed by in silico analysis in agreement with the undetermined subcellular occurrence of the post-chorismate portion of the SK pathway.
Following the identification of the tomato genes encoding all enzymes for the tocochromanol synthesis pathway, the 41 loci were further mapped. Furthermore, a detailed profile for all tocopherol isoforms was performed in fruits from ILs harbouring introgressed regions spanning the VTE biosynthesis core pathway genes (ILs 7-4, 7-4-1, 8-2, 8-2-1, 9-1, and 9-2-6) along with those in chromosome 6 (ILs 6-1 and 6-2) for which α-tocopherol QTL have been previously reported (Schauer et al., 2006). Although VTE content has been demonstrated to be a highly environmentally affected trait (Schauer et al., 2008), the results reported here indicate that at least those QTL mapped on chromosomes 6 and 9 show a relatively high heritability level as they are in accordance with previous experiments reported by Schauer et al. (2006). Moreover it is worth noting that, as opposed to the previous study, which was carried out on field-grown plants, the data reported here were obtained from greenhouse plants.
Integrated analysis at metabolic, genomic, and genetic levels allowed us to propose 16 candidate loci putatively affecting tocopherol content in tomato: prai, fpgs, chl, apt, and lycb on chromosome 6; tyra(1), vte2, hppd(1), and tat(2) located on chromosome 7; vte1 and vte4 on chromosome 8; and ggps(4), tyra(2), vte5, sec14, and vte3(1) on chromosome 9. In plants, several QTL controlling tocopherol content have been identified in soybean, maize, oilseed rape, and Arabidopsis. As revealed in this report, only some of those QTL localized to areas of the genome where tocopherol core pathway genes occur (Marwede et al., 2005; Gilliland et al., 2006; Chander et al., 2008; Li et al., 2010).
Detailed analysis of the identified QTL together with the co-localizing genes raised interesting features concerning VTE content regulation. None of chromosome 6 QTL co-localize with any of the MEP, SK, or tocopherol core pathway genes, thus suggesting that tocopherol content variation observed in ILs 6-1 and 6-2 might be determined by the effect of genes belonging to VTE-related pathways. IL 6-1 showed elevated δ- and total tocopherol content in comparison with S. lycopersicum (M82) control. This line harbours the S. pennellii alleles of the prai, fpgs, and chl genes. The first two could be regulating hydroxyphenylpyruvate fluxes into the tocopherol pathway by the deviation of chorismate from tryptophan and folate biosynthetic routes. On the other point of the pathway drawn in Fig. 1, the hypothesis that chlorophyll degradation-derived phytol serves as an important intermediate for tocopherol synthesis has been demonstrated by characterization of the Arabidopsis vte5 mutant (Valentin et al., 2006). In this sense, the presence of S. pennellii chl allele in IL 6-1 might be raising the phytyl 2P input into tocopherol synthesis. On the same chromosome, IL 6-2 displayed significantly higher levels of β-tocopherol than the control. This IL carries S. pennellii alleles of chl, apt, and lycb. Even when these genes are linked to intermediate metabolites of the tocopherol pathway (chorismate and geranylgeranyl 2P) no evident links can be specifically associated with the β-isoform. The candidature of genes not directly involved in the VTE structural pathway is supported by the regulatory network acting on branching points. The CM acting on the SK pathway is allosterically feedback-inhibited in plants by phenylalanine and tyrosine and induced by tryptophan (Tzin and Galili, 2010). Allelic variation in prai and apt could be modifying tryptophan synthesis, altering influx through the SK pathway and then, increasing homogentisate precursor, finally resulting in the tocopherol content variation observed in ILs 6-1 and 6-2.
Two QTL have been detected on chromosome 7; while IL 7-4 displayed lower levels of α-tocopherol, IL 7-4-1 showed reduced amounts of the β-isoform. The introgressed wild genome fragments in these ILs harbour S. pennellii alleles of tyra(1), vte2, and hppd(1). IL 7-4 also spans the wild allele of tat(2). These four candidates can alter total precursor influx to tocopherol biosynthesis. Nevertheless, the way that these genes could differentially modify the amounts of tocopherol isoforms is currently unclear.
On chromosome 8, the IL 8-2 displayed a significantly lower α/γ-tocopherol ratio when compared with control (Fig. 3). Interestingly, this IL bears S. pennellii alleles of vte1 and vte4 genes whose protein product activities synthesize these two tocopherol isoforms. Therefore, the low α/γ-tocopherol ratio could be caused by lower VTE1 and/or higher VTE4 activity of wild alleles. Intriguingly, IL 8-2-1 also harbours the wild alleles of vte1 and vte4. Even though no significant alteration in α/γ-tocopherol ratio was observed, a significant increase in total tocopherol was detected in the fruits.
IL 9-1 exhibits increased levels of α-, β-, and total tocopherol most probably due to differential activity levels of the enzymes encoded by the S. pennellii allele of the tyra(2) and ggps(4) loci that could lead to higher input of hydroxyphenylpyruvate and phytyl 2P to the tocopherol core pathway. Elevated α/γ and β/γ ratios support this hypothesis (Fig. 3). Interesting to note is the fact that tyra(2) shows one of the highest dN values, indicating protein divergences between S. pennellii and S. lycopersicum (Fig. 4). In this sense, the results presented here reinforce the hypothesis of Rippert et al. (2004) that hydroxyphenylpyruvate is a key step in the accumulation of VTE in plants. IL 9-2-6 exhibits increased levels of α- and total tocopherol and harbours the S. pennellii allele of the vte5 gene, which could improve the input of phytyl 2P via the phytol alternative pathway (Valentin et al., 2006). However, levels of β-tocopherol are not increased in IL 9-2-6, which could be explained by the more efficient activity of the S. pennellii vte3 allele. This is also reflected in the significant differences found in the α/β ratio (Fig. 3).
Although there is a reliable link between the identified candidate genes and fruit tocopherol content, the effect of unidentified loci within the QTL cannot be discarded and, due to the considerable size of the S. pennellii fragments in the ILs, the differences observed in tocopherol accumulation could also be caused by undetected genes located within them.
Candidate gene approach has proved to be extremely powerful for studying the genetic architecture of complex traits (Zhu and Zao, 2007). By revealing the pattern of molecular genetic variation, the evolutionary analyses offer complementary data for strengthening gene candidature (Moyle and Muir, 2010). In this sense, the pattern of selection of different genes within a metabolic pathway allows the determination of whether they are subject to equivalent evolutionary forces underlying trait phenotypic variation. Comparing S. lycopersicum and S. pennellii alleles, out of the 22 genes studied, those encoding MEP, post-chorismate SK, and VTE core pathway enzymes presented dN/dS ratios below the mean, excluding those with more than one loci. In contrast, out of the six analysed candidate genes of related pathways, apt, chl, and lycb displayed dN/dS ratios values above the mean (Fig. 4). This constraint relaxation cannot be related to a higher region-specific mutation rate on chromosome 6 because two other genes mapped also on chromosome 6, prai and fpgs, presented low dN/dS ratios. Therefore, these results suggest a strong purifying selection for tocopherol central biosynthesis genes while there is a more relaxed constraint for those genes of related pathways. When the selection constraint is evaluated codon by codon applying a likelihood ratio test, new insights about the evolutionary history of the genes are revealed. Nineteen genes exhibit purifying selection associated with neutral evolving codons (Table 3) in agreement with the dN/dS analysis and previous reports that concluded that significant heterogeneity of evolutionary rates in metabolic pathway genes is mainly ascribed to differential constraint relaxation rather than to positive selection (Livingston and Anderson, 2009; Yang et al., 2009). Even so, three loci exhibited patterns consistent with positive selection evolving codons. In tomato, loci showing positive selection have been identified associated with biotic and abiotic stresses (Jiménez-Gomez and Maloof, 2009). It would be unexpected to envisage signs of positive selection in loci of major biosynthetic pathways that feed multiple metabolic routes such as MEP and SK. However, the signs of diversifying selection found for ggps(2) and ggps(4) could be explained by the existence of two other paralogues evolving under a more conservative evolutionary pattern. In the case of vte1, the reduction in protein negative selective pressure might indicate that this is not a committed step in tocopherol production.
Studies of evolutionary rates of genes in the plant anthocyanin (Lu and Rausher, 2003) and carotenoid (Livingstone and Anderson, 2009) pathways have demonstrated that upstream genes in the pathway evolved more slowly than downstream genes. However, this seems not to be a constant trend. Downstream genes in the gibberellin pathway did not exhibit elevated substitution rates and instead, genes encoding either the branch point enzyme or those catalysing multiple steps in the pathway showed the lowest evolutionary rates due to strong purifying selection (Yang et al., 2009). This observation is in close agreement with the theory of pathway fluxes, which indicates that natural selection would target enzymes controlling metabolic fluxes between converging pathways. Consequently, these branch points are usually targets of selection, experiencing higher evolutionary constraints (Flowers et al., 2007). In this sense, the lowest value of dN/dS was observed for the tyra(1) gene whose protein product shares its substrate with phenylalanine/tyrosine biosynthesis resulting in branching points, whilst the tyra(2) paralogue displays a relaxed evolutionary constraint, indicative of a functional divergence.
Genes responsible for adaptive morphological and physiological differences between species carry signatures of positive selection (Aguileta et al., 2010). In this sense, regarding the variation in VTE content observed in the S. pennellii introgressed lines in comparison with that of S. lycopersicum, the presence of neutral and/or positive evolving codons could result in novel protein features being a source of new functional profiles. Even when coding sequences are relevant to phenotype, they might not be the location at which key evolutionary changes occur. Analyses across coding sequences do not reveal allelic differences in regulatory sequences that could also be determining the observed phenotypic variations. In fact, a co-response analysis of 32 of the genes identified here revealed an intricate network suggesting these pathways to be finely regulated at the level of gene expression (data not shown).
This report describes a comprehensive survey of the genes encoding VTE biosynthesis pathway enzymes in tomato, and the methods adopted allowed the identification of novel tocopherol QTL. By an integrated analysis of the genome sequence data together with a well-characterized biosynthetic pathway, like that for VTE in Arabidopsis model species, this genetic/genomic approach described loci and allelic variations that probably impact antioxidant content in tomato fruit. The identified candidate genes support cross-talk between the MEP, SK, and tocopherol core pathways through the control of VTE accumulation in tomato fruit. In addition, the VTE-related pathway genes might contribute to regulation of the supply of intermediates for plastid tocopherol biosynthesis. The data produced provide a platform for functional studies that will contribute to elucidation of the biosynthesis and catabolism of tocochromanols, and their role in plant physiology.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1 lists the gene primers used.
Acknowledgments
The authors thank Gregorio Ceccantini for assistance with statistical analyses. This work was partially supported by grants from FAPESP, CNPq, and USP (Brazil); Max Planck Society (Germany); INTA, CONICET, and ANPCyT (Argentina); and under the auspices of the EU SOL Integrated Project FOOD-CT-2006-016214. J.A. was a recipient of a CAPES (Brazil). L.B., F.G., and N.S. were recipients of a FAPESP (Brazil) fellowships. L.Q. was a recipient of an ANPCyT (Argentina) fellowship. R.A. and F.C. are members of CONICET (Argentina). This work was carried out in compliance with current laws governing genetic experimentation in Brazil and in Argentina.
Glossary
Abbreviations
- IL
introgression line
- LRT
likelihood ratio test
- MEP
methylerythritol phosphate
- QTL
quantitative trait loci
- ROS
reactive oxygen species
- SK
shikimate
- VTC
vitamin C
- VTE
vitamin E
References
- Abushita AA, Hebshi EA, Daood HG, Biacs PA. Determination of antioxidant vitamins in tomatoes. Food Chemistry. 1997;60:207–212. [Google Scholar]
- Aguileta G, Lengelle J, Marthey S, et al. Finding candidate genes under positive selection in non-model species: examples of genes involved in host specialization in pathogens. Molecular Ecology. 2010;19:292–306. doi: 10.1111/j.1365-294X.2009.04454.x. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Jongsma MA, Bouwmeester HJ. Volatile science? Metabolic engineering of terpenoids in plants. Trends in Plant Science. 2005;10:594–602. doi: 10.1016/j.tplants.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ament K, Van Schie CC, Bouwmeester HJ, Haring MA, Schuurink RC. Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E, E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta. 2006;224:1197–1208. doi: 10.1007/s00425-006-0301-5. [DOI] [PubMed] [Google Scholar]
- Ayres M, Ayres Júnior M, Ayres DL, Santos AA. Bioestat – aplicações estatísticas nas áreas das ciências bio-médicas. Sociedade Civil Mamirauá/MCT CNPq, Belém, PA. 2007 Brasil. [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends in Biochemical Sciences. 2009;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez L, Urias U, Milstein D, Kamenetzky L, Asis R, Fernie AR, Van Sluys MA, Carrari F, Rossi M. A candidate gene survey of quantitative trait loci affecting chemical composition in tomato fruit. Journal of Experimental Botany. 2008;59:2875–2890. doi: 10.1093/jxb/ern146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M, Rösler J, Raesecke HR, Görlach J, Amrhein N, Schmid J. Cloning of a cDNA encoding 3-dehydroquinate synthase from a higher plant, and analysis of the organ-specific and elicitor-induced expression of the corresponding gene. Plant Molecular Biology. 1996;31:69–76. doi: 10.1007/BF00020607. [DOI] [PubMed] [Google Scholar]
- Bischoff M, Schaller A, Bieri F, Kessler F, Amrhein N, Schmid J. Molecular characterization of tomato 3-dehydroquinate dehydratase-shikimate:NADP oxidoreductase. Plant Physiology. 2001;125:1891–1900. doi: 10.1104/pp.125.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella-Pavía P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodriguez-Concepcion M. Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. The Plant Journal. 2004;40:188–199. doi: 10.1111/j.1365-313X.2004.02198.x. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Suire C, d'Harlingue A, Backhaus RA, Camara B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. The Plant Journal. 2000;24:241–252. doi: 10.1046/j.1365-313x.2000.00875.x. [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ. Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nature Biotechnology. 2003;21:1082–1087. doi: 10.1038/nbt853. [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, et al. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behaviour. Plant Physiology. 2006;142:1380–1396. doi: 10.1104/pp.106.088534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander S, Guo YQ, Yang XH, Yan JB, Zhang YR, Song TM, Li JS. Genetic dissection of tocopherol content and composition in maize grain using quantitative trait loci analysis and the candidate gene approach. Molecular Breeding. 2008;22:353–365. [Google Scholar]
- Chun J, Lee J, Ye L, Exler J, Eitenmiller RR. Tocopherol and tocotrienol contents of raw and processed fruits and vegetables in the United States diet. Journal of Food Composition and Analysis. 2006;19:196–204. [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Research. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Gautam N, Dey SK, Maiti T, Roy S. Oxidative stress in the brain of nicotine-induced toxicity: protective role of Andrographis paniculata Nees and vitamin E. Applied Physiology, Nutrition, and Metabolism. 2009;34:124–135. doi: 10.1139/H08-147. [DOI] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Fraser PD, et al. Fruit-specific RNAi-mediated suppression of DET1 enhances tomato nutritional quality. Nature Biotechnology. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Hofius D, Hajirezaei MR, Fernie AR, Börnke F, Sonnewald U. Functional analysis of the essential bifunctional tobacco enzyme 3-dehydroquinate dehydratase/shikimate dehydrogenase in transgenic tobacco plants. Journal of Experimental Botany. 2007;58:2053–2067. doi: 10.1093/jxb/erm059. [DOI] [PubMed] [Google Scholar]
- Eberhard J, Bischoff M, Raesecke HR, Amrhein N, Schmid J. Isolation of a cDNA from tomato coding for an unregulated, cytosolic chorismate mutase. Plant Molecular Biology. 1996;31:917–922. doi: 10.1007/BF00019479. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijine G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Enfissi EM, Barneche F, Ahmed I, et al. Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. The Plant Cell. 2010;22:1190–1215. doi: 10.1105/tpc.110.073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entus R, Poling M, Herrmann KM. Redox regulation of Arabidopsis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Plant Physiology. 2002;129:1866–1871. doi: 10.1104/pp.002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics. 1995;141:1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J, Munné-Bosch S. Tocochromanol functions in plants: antioxidation and beyond. Journal of Experimental Botany. 2010;61:1549–1566. doi: 10.1093/jxb/erq030. [DOI] [PubMed] [Google Scholar]
- Fei Z, Tang X, Alba R, Giovannoni J. Tomato Expression Database (TED): a suite of data presentation and analysis tools. Nucleic Acids Research. 2006;34:D766–D770. doi: 10.1093/nar/gkj110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Yaakov T, Zamir D. Natural genetic variation for improving crop quality. Current Opinion in Plant Biology. 2006;9:196–202. doi: 10.1016/j.pbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Ferro M, Brugiere S, Salvi D, et al. AT_CHLORO: a comprehensive chloroplast proteome database with sub-plastidial localization and information for functional genomics using quantitative label-free analyses. Molecular and Celllular Proteomics. 2010;9:1063–1084. doi: 10.1074/mcp.M900325-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Sezgin E, Kumagai S, Duvernell DD, Matzkin LM, Schmidt PS, Eanes WF. Adaptive evolution of metabolic pathways in. Drosophila. Molecular Biology and Evolution. 2007;24:1347–1354. doi: 10.1093/molbev/msm057. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. The Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EM, Halket JH, Truesdale MR, Yu DM, Gerrish C, Bramley PM. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids and intermediary metabolism. The Plant Cell. 2007;19:3194–3211. doi: 10.1105/tpc.106.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Pinto ME, Holloway DE, Bramley PM. Application of high-performance liquid chromatography with photodiode array detection to the metabolite profiling of plant isoprenoids. The Plant Journal. 2000;24:551–558. doi: 10.1046/j.1365-313x.2000.00896.x. [DOI] [PubMed] [Google Scholar]
- Frusciante L, Carli P, Ercolano MR, Pernice R, Di Matteo A, Fogliano V, Pellegrini N. Antioxidant nutritional quality of tomato. Molecular Nutrition and Food Research. 2007;51:609–617. doi: 10.1002/mnfr.200600158. [DOI] [PubMed] [Google Scholar]
- Garcia I, Rodgers M, Pepin R, Hssich T, Matringe M. Characterization and subcellular compartmentation of recombinant 4-hydroxyphenylpyruvate dioxygenase from Arabidopsis in transgenic tobacco. Plant Physiology. 1999;119:1507–1516. doi: 10.1104/pp.119.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser CS, Winter JA, Hironaka CM, Shah DM. Structure, expression, and evolution of the 5-enolpyruvylshikimate-3-phosphate synthase genes of petunia and tomato. Journal of Biological Chemistry. 1988;263:4280–4287. [PubMed] [Google Scholar]
- Gilliland LU, Magallanes-Lundback M, Hemming C, Supplee A, Koornneef M, Bentsink L, Dellapenna D. Genetic basis for natural variation in seed vitamin E levels in. Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2006;103:18834–18841. doi: 10.1073/pnas.0606221103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach J, Beck A, Henstrand JM, Handa AK, Herrmann KM, Schmid J, Amrhein N. Differential expression of tomato (Lycopersicon esculentum L.) genes encoding shikimate pathway isoenzymes: I. 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Plant Molecular Biology. 1993;23:697–706. doi: 10.1007/BF00021525. [DOI] [PubMed] [Google Scholar]
- Görlach J, Raesecke HR, Abel G, Wehrli R, Amrhein N, Schmid J. Organ-specific differences in the ratio of alternatively spliced chorismate synthase (LeCS2) transcripts in tomato. The Plant Journal. 1995;8:451–456. doi: 10.1046/j.1365-313x.1995.08030451.x. [DOI] [PubMed] [Google Scholar]
- Graindorge M, Giustini C, Jacomin AC, Kraut A, Curien G, Matringe M. Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: the last homeless enzyme of aromatic amino acids biosynthesis. FEBS Letters. 2010;584:4357–4360. doi: 10.1016/j.febslet.2010.09.037. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Herbers K. Vitamin production in transgenic plants. Journal of Plant Physiology. 2003;160:821–829. doi: 10.1078/0176-1617-01024. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. Chlorophyll degradation during senescence. Annual Review of Plant Biology. 2006;57:55–77. doi: 10.1146/annurev.arplant.57.032905.105212. [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Zbierzak AM, Kanwischer M, Dormann P. A salvage pathway for phytol metabolism in Arabidopsis. Journal of Biological Chemistry. 2006;281:2470–2477. doi: 10.1074/jbc.M509222200. [DOI] [PubMed] [Google Scholar]
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N. Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Molecular Plant. 2009;2:1154–1180. doi: 10.1093/mp/ssp088. [DOI] [PubMed] [Google Scholar]
- Jiménez-Gomez JM, Maloof JN. Sequence diversity in three tomato species: SNPs, markers, and molecular evolution. BMC Plant Biology. 2009;9:85. doi: 10.1186/1471-2229-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetzky L, Asís R, Bassi S, et al. Genomic analysis of wild tomato introgressions determining metabolic- and yield-associated traits. Plant Physiology. 2010;152:1772–1786. doi: 10.1104/pp.109.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Z, Sun X, Tang K. The current opinions on the functions of tocopherol based on the genetic manipulation of tocopherol biosynthesis in plants. Journal of Integrative Plant Biology. 2008;50:1057–1069. doi: 10.1111/j.1744-7909.2008.00689.x. [DOI] [PubMed] [Google Scholar]
- Li H, Liu H, Han Y, Wu X, Teng W, Liu G, Li W. Identification of QTL underlying vitamin E contents in soybean seed among multiple environments. TAG Theoretical and Applied Genetics. 2010;120:1405–1413. doi: 10.1007/s00122-010-1264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone K, Anderson S. Patterns of variation in the evolution of carotenoid biosynthetic pathway enzymes of higher plants. Journal of Heredity. 2009;100:754–761. doi: 10.1093/jhered/esp026. [DOI] [PubMed] [Google Scholar]
- Lois LM, Rodríguez-Concepción M, Gallego F, Campos N, Boronat A. Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-d-xylulose 5-phosphate synthase. The Plant Journal. 2000;22:503–513. doi: 10.1046/j.1365-313x.2000.00764.x. [DOI] [PubMed] [Google Scholar]
- Lu Y, Rausher MD. Evolutionary rate variation in anthocyanin pathway genes. Molecular Biology and Evolution. 2003;20:1844–1853. doi: 10.1093/molbev/msg197. [DOI] [PubMed] [Google Scholar]
- Maeda H, Yoo H, Dudareva N. Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nature Chemical Biology. 2010;7:19–21. doi: 10.1038/nchembio.485. [DOI] [PubMed] [Google Scholar]
- Marwede V, Gul MK, Becker HC, Ecke W. Mapping of QTL controlling tocopherol content in winter oilseed rape. Plant Breeding. 2005;124:20–26. [Google Scholar]
- Mène-Saffrané L, DellaPenna D. Biosynthesis, regulation and functions of tocochromanols in plants. Plant Physiology and Biochemistry. 2009;48:301–309. doi: 10.1016/j.plaphy.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Milone D, Stegmayer G, Kamenetzky L, Lopez M, Lee J-M, Giovannoni JJ, Carrari F. *omeSOM: a software for clustering and visualization of transcriptional and metabolite data mined from interspecific crosses of crop plants. BMC Bioinformatics. 2010;11:438–447. doi: 10.1186/1471-2105-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley EM, Kunkel BN, Keith B. Identification, characterization and comparative analysis of a novel chorismate mutase gene in. Arabidopsis thaliana. Gene. 1999;240:115–123. doi: 10.1016/s0378-1119(99)00423-0. [DOI] [PubMed] [Google Scholar]
- Moyle L, Muir CD. Reciprocal insights into adaptation from agricultural and evolutionary studies in tomato. Evolutionary Applications. 2010;3:409–421. doi: 10.1111/j.1752-4571.2010.00143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Mills A, Skwarecki B, Buels R, Menda N, Tanksley SD. The SGN Comparative Map Viewer. Bioinformatics. 2008;24:422–423. doi: 10.1093/bioinformatics/btm597. [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S, Alegre L. The function of tocopherols and tocotrienols in plants. Critical Reviews in Plant Sciences. 2002;21:31–57. [Google Scholar]
- Nagegowda DA. Plant volatile terpenoid metabolism: biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Letters. 2010;584:2965–2973. doi: 10.1016/j.febslet.2010.05.045. [DOI] [PubMed] [Google Scholar]
- Paetzold H, Garms S, Bartram S, Wieczorek J, Uros-Gracia EM, Rodriguez-Concepción M, Boland W, Strack D, Hause B, Walter MH. The isogene 1-deoxy-D-xylulose 5-phosphate synthase II controls isoprenoid profiles, precursor pathway allocation and density of tomato trichomes. Molecular Plant. 2010;3:904–916. doi: 10.1093/mp/ssq032. [DOI] [PubMed] [Google Scholar]
- Peterman TK, Ohol YM, McReynolds LJ, Luna EJ. Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiology. 2004;136:3080–3094. doi: 10.1104/pp.104.045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MA, D'Auria JC, Gershenzon J, Pichersky E. The Arabidopsis thaliana type I isopentenyl diphosphate isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. The Plant Cell. 2008;20:677–696. doi: 10.1105/tpc.107.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Cherest H, Jabrin S, Grunwald D, Surdin-Kerjan Y, Douce R, Rebeille F. Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2001;98:1523–1535. doi: 10.1073/pnas.261585098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippert P, Scimemi C, Dubald M, Matringe M. Engineering plant shikimate pathway for production of tocotrienol and improving herbicide resistance. Plant Physiology. 2004;134:92–100. doi: 10.1104/pp.103.032441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippert P, Puyaubert J, Grisollet D, Derrier L, Matringe M. Tyrosine and phenylalanine are synthesized within the plastids in. Arabidopsis. Plant Physiology. 2009;149:1251–1260. doi: 10.1104/pp.108.130070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Ahumada I, Diez-Juez E, Sauret-Güeto S, Lois LM, Gallego F, Carretero-Paulet L, Campos N, Boronat A. 1-Deoxy-D-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. The Plant Journal. 2001;27:213–222. doi: 10.1046/j.1365-313x.2001.01089.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Querol J, Lois LM, Imperial S, Boronat A. Bioinformatic and molecular analysis of hydroxymethylbutenyl diphosphate synthase (GCPE) gene expression during carotenoid accumulation in ripening tomato fruit. Planta. 2003;217:476–482. doi: 10.1007/s00425-003-1008-5. [DOI] [PubMed] [Google Scholar]
- Rohdich F, Wungsintaweekul J, Lüttgen H, Fischer M, Eisenreich W, Schuhr CA, Fellermeier M, Schramek N, Zenk MH, Bacher A. Biosynthesis of terpenoids: 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase from tomato. Proceedings of the National Academy of Sciences, USA. 2000;97:8251–8256. doi: 10.1073/pnas.140209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proceedings of the National Academy of Sciences, USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros E. Nuts and novel biomarkers of cardiovascular disease. American Journal of Clinical Nutrition. 2009;89:1649S–1656S. doi: 10.3945/ajcn.2009.26736R. [DOI] [PubMed] [Google Scholar]
- Sadre R, Gruber J, Frentzen M. Characterization of homogentisate prenyltransferases involved in plastoquinone-9 and tocochromanol biosynthesis. FEBS Letters. 2006;580:5357–5362. doi: 10.1016/j.febslet.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Saito K, Tautz L, Mustelin T. The lipid-binding SEC14 domain. Biochimica et Biophysica Acta. 2007;1771:719–726. doi: 10.1016/j.bbalip.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. Mode of inheritance of primary metabolic traits in tomato. The Plant Cell. 2008;20:509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nature Biotechnology. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- Schauer N, Zamir D, Fernie AR. Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicon complex. Journal of Experimental Botany. 2005;56:297–307. doi: 10.1093/jxb/eri057. [DOI] [PubMed] [Google Scholar]
- Schmid J, Schaller A, Leibinger U, Boll W, Amrhein N. The in-vitro synthesized tomato shikimate kinase precursor is enzymatically active and is imported and processed to the mature enzyme by chloroplasts. The Plant Journal. 1992;2:375–383. [PubMed] [Google Scholar]
- Schenk N, Schelbert S, Kanwischer M, Goldschmidt EE, Dörmann P, Hörtensteiner S. The chlorophyllases At CLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in. Arabidopsis thaliana. FEBS Letters. 2007;581:5517–5525. doi: 10.1016/j.febslet.2007.10.060. [DOI] [PubMed] [Google Scholar]
- Semchuk NM, Lushchak OV, Falk J, Krupinska K, Lushchak VI. Inactivation of genes, encoding tocopherol biosynthetic pathway enzymes, results in oxidative stress in outdoor grown. Arabidopsis thaliana. Plant Physiology and Biochemistry. 2009;47:384–390. doi: 10.1016/j.plaphy.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. Second Edition. 1981. Freeman, New York. [Google Scholar]
- Soll J, Schultz G. Comparison of geranylgeranyl and phytyl substituted methylquinols in the tocopherol synthesis of spinach chloroplasts. Biochemical and Biophysical Research Communications. 1979;91:715–720. doi: 10.1016/0006-291x(79)91939-9. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PDB, van Wijk KJ. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Research. 2009;37:969–974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, DellaPenna D, Dixon RA. The pds2 mutation is a lesion in the Arabidopsis homogentisate solanesyltransferase gene involved in plastoquinone biosynthesis. Planta. 2007;226:1067–1073. doi: 10.1007/s00425-007-0564-5. [DOI] [PubMed] [Google Scholar]
- Tohge T, Fernie AR. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nature Protocols. 2010;5:1210–1227. doi: 10.1038/nprot.2010.82. [DOI] [PubMed] [Google Scholar]
- Traber MG, Sies H. Vitamin E in humans: demand and delivery. Annual Review of Nutrition. 1996;16:321–347. doi: 10.1146/annurev.nu.16.070196.001541. [DOI] [PubMed] [Google Scholar]
- Tzin V, Malitsky S, Aharoni A, Galili G. Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in. Arabidopsis. The Plant Journal. 2009;60:156–167. doi: 10.1111/j.1365-313X.2009.03945.x. [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Molecular Plant. 2010;3:956–972. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- Valentin HE, Lincoln K, Moshiri F, et al. The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. The Plant Cell. 2006;18:212–224. doi: 10.1105/tpc.105.037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schie CCN, Ament K, Schmidt A, Lange T, Haring MA, Schuurink RC. Geranyl diphosphate synthase is required for biosynthesis of gibberellins. The Plant Journal. 2007;52:752–762. doi: 10.1111/j.1365-313X.2007.03273.x. [DOI] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang F, Ge S. Evolutionary rate patterns of the gibberellin pathway genes. BMC Evolutionary Biology. 2009;9:206–217. doi: 10.1186/1471-2148-9-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir D. Improving plant breeding with exotic genetic libraries. Nature Reviews Genetics. 2001;2:983–989. doi: 10.1038/35103590. [DOI] [PubMed] [Google Scholar]
- Zhang CX, Ho SC, Chen YM, Fu JH, Cheng SZ, Lin FY. Greater vegetable and fruit intake is associated with a lower risk of breast cancer among Chinese women. International Journal of Cancer. 2009;125:181–188. doi: 10.1002/ijc.24358. [DOI] [PubMed] [Google Scholar]
- Zhao J, Last RL. Immunological characterization and chloroplast localization of the tryptophan biosynthetic enzymes of the flowering plant Arabidopsis thaliana. Journal of Biological Chemistry. 1995;270:6081–6087. doi: 10.1074/jbc.270.11.6081. [DOI] [PubMed] [Google Scholar]
- Zhu M, Zhao S. Candidate gene identification approach: progress and challenges. International Journal of Biological Sciences. 2007;3:420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.