Abstract
Total glucose-6-phosphate dehydrogenase (G6PDH) activity, protein abundance, and transcript levels of G6PDH isoforms were measured in response to exogenous abscisic acid (ABA) supply to barley (Hordeum vulgare cv Nure) hydroponic culture. Total G6PDH activity increased by 50% in roots treated for 12 h with exogenous 0.1 mM ABA. In roots, a considerable increase (35%) in plastidial P2-G6PDH transcript levels was observed during the first 3 h of ABA treatment. Similar protein variations were observed in immunoblotting analyses. In leaves, a 2-fold increase in total G6PDH activity was observed after ABA treatment, probably related to an increase in the mRNA level (increased by 50%) and amount of protein (increased by 85%) of P2-G6PDH. Together these results suggest that the plastidial P2-isoform plays an important role in ABA-treated barley plants.
Keywords: ABA, barley, G6PDH, OPPP, stress
Introduction
The oxidative pentose phosphate pathway (OPPP) is the main pathway of production of NADPH for biosyntheses (Bowsher et al., 1989; Esposito et al., 2003; Hutchings et al., 2005), and for redox balance of plant cells (Kruger and von Schaewen, 2003; Scheibe, 2004). The main regulatory step of the OPPP is catalysed by glucose-6-phosphate dehydrogenase (G6PDH; EC 1.1.1.49), a homotetramer with subunits of 50–60 kDa. A recent Arabidopsis genome-wide analysis indicated the presence of two cytosolic (Cy-G6PDH) and four plastidial G6PDH isoforms, named P1-G6PDH and P2-G6PDH (Wakao and Benning, 2005). All these isoforms have been described in a number of higher plants, but the regulatory properties of each G6PDH type are still poorly characterized (Nemoto and Sasakuma, 2000; Wendt et al., 2000; Esposito et al., 2001a, b, 2005; Huang et al., 2002; Hauschild and von Schaewen, 2003). The plastidial enzymes exhibit a complex redox regulation, mostly via the NADPH/NADP+ ratio, and via their cysteine oxidation state (Wenderoth et al., 1997; Esposito et al., 2001a; Kruger and von Schaewen, 2003). Indeed, plastidial G6PDH activity is modulated through dithiol–disulphide exchanges via the ferredoxin/thioredoxin system (Buchanan, 1991; Née et al., 2009), whereas the redox sensitivity of cytosolic isoforms is still controversial (Graeve et al., 1994).
In terms of expression, P1-G6PDH transcripts are detectable in most of the photosynthetic and non-photosynthetic tissues under light conditions (von Schaewen et al., 1995), whereas P2-G6PDH transcripts are expressed throughout the plant, but at higher levels in stems and roots (Knight et al., 2001).
The roles and functions of the three G6PDH classes in plants have also been explored in the last few years (Kruger and von Schaewen, 2003; Hauschild and von Schaewen, 2003; Esposito et al., 2005; Wakao et al., 2008).
The plastidial isoforms are involved in the supply of reducing power for nutrient assimilation, P2-G6PDH protein is induced in roots by nitrogen (Esposito et al., 2001b; Bowsher et al., 2007) while P1-G6PDH protein, which is absent in roots, is increased in leaves in response to both nitrogen supply (Esposito et al., 2005) and inhibition of photosynthesis by dark or paraquat (Hauschild and von Schaewen, 2003).
Regarding cytosolic isoforms, the Arabidopsis thaliana knock-out mutant for the two Cy-G6PDHs produces seeds with a higher oil content, which suggested that G6PDH activity is crucial for the metabolism of developing seeds by increasing carbon substrates for synthesis of storage compounds (Wakao et al., 2008). Other studies have shown that there is an association in plants between the OPPP and the response to different stresses, namely nutrient starvation, drought, salinity, and pathogens (Nemoto and Sasakuma, 2000; Esposito et al., 2003, 2005; Valderrama et al., 2006; Scharte et al., 2009). As an example, Cy-G6PDH isoforms are essential in providing NADPH which is required for a timely defence response as demonstrated in the pathosystem Nicotiana tabacum–Phytophthora nicotianae (Scharte et al., 2009). In leaves of olive plants, the activity of antioxidant enzymes including NADPH-recycling enzymes such as G6PDH is induced in response to salt stress which was shown to increase the levels of reactive oxygen species (ROS) (Valderrama et al., 2006).
Phytohormones such as abscisic acid (ABA), jasmonic acid, ethylene, and salicylic acid appear to be critical components of the complex signalling networks established during the stress response (Zhu, 2002).
ABA plays an important role in a number of physiological processes such as seed maturation and dormancy, stomatal closure, and growth and developmental regulation, especially by regulating gene expression (Zeevaart and Creelman, 1988; Nambara and Marion-Poll, 2005). At the same time it has a pivotal role in the adaptive response to changing environmental conditions. Indeed, application of exogenous ABA is known to regulate a set of different genes, suggesting that ABA is involved in the response to both abiotic (cold, drought, and salinity) (Leung and Giraudat, 1998; Finkelstein et al., 2002; Zhu, 2002; Fujita et al., 2006) and biotic stresses (Mantyla et al., 1995; Li et al., 2004; Fujita et al., 2006; Fan et al., 2009). The ABA-mediated gene regulation occurs through the presence of conserved ABA-responsive elements (ABREs) in gene promoters, usually accompanied by coupling elements. ABREs contain ACGT as a core nucleotide sequence, which acts as a binding site for bZIP family transcription factors (Hatorri et al., 2002). Such an ABRE has been found in both monocotyledonous and dicotyledonous plants, for example in Zea mays and A. thaliana (Lenka et al., 2009), as well as in the promoter of rice P2-G6PDH (Hou et al., 2006), the orthologue of Hordeum vulgare P2-G6PDH (AM398980).
The aim of this work was to investigate the role(s) of the plastidial G6PDH isoform(s) upon exogenous ABA supply to barley plants grown in hydroponic culture. In addition, the importance of the plastidial P2-G6PDH in both roots and leaves is specifically discussed.
Materials and methods
Sequence analysis
The protein sequence of the root barley (H. vulgare) plastidial P2-G6PDH was retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov) and used to search against other genomes using BlastP or tblastn. All the sequences are available at the following websites: for A. thaliana (http://www.arabidopsis.org/), Oryza sativa (http://rice.plantbiology.msu.edu/), Sorghum bicolor (http://genome.jgipsf.org/Sorbi1/Sorbi1.home.html), N. tabacum (http://www.ncbi.nlm.nih.gov), Populus trichocarpa (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), Solanum tuberosum (http://www.ncbi.nlm.nih.gov), Triticum aestivum (http://www.ncbi.nlm.nih.gov), and Spinacia oleracea (http://www.ncbi.nlm.nih.gov). All protein accession numbers used in this article can be found in these databases.
The amino acid alignments were performed using ClustalW (www.ebi.ac.uk/Tools/clustalw2/index.html) and the phylogenetic tree was constructed using the Neighbor–Joining tree algorithm in MEGA version 4 (Tamura et al., 2007).
Supplemental data files available at JXB online contain all the protein sequences indexed in this study including the accession numbers.
Plant culture
Barley seeds (H. vulgare, Istituto Sperimentale di Cerealicoltura, Fiorenzuola D'Arda, Italy) were germinated for 5 d in the dark on wet paper. The young seedlings were transferred in hydroculture, according to Esposito et al. (2005), and grown for 3 d in the absence of any external nitrogen source, under a photoperiod of 16 h light/8 h dark and then 5 mM ammonium phosphate was supplied as the sole nitrogen source. After 7 d of growth (‘0’ experimental time), 0.1 mM ABA was added to the nutrient medium. Plants were harvested at different times (3, 6, 9, 12, 24, and 48 h) of exposure to ABA, and G6PDH activity was measured as described in Esposito et al. (2001a). Data shown in figures or tables are representative of five separate experiments.
Preparation of crude extracts for enzyme activities
Plants were collected 2 h after the beginning of the light period (16 h), thus all plant groups were under illumination. Each replicate is done with a group of 10–40 plants to generate at least 2 g (usually 3–5 g) of roots or leaves. Plant material was quickly frozen in liquid nitrogen, and powdered in a mortar with a pestle, and proteins were extracted in 100 mM TRIS-HCl pH 7.9, 10 mM MgCl2, 4 mM EDTA, 1 mM phenylmethylsulphonyl fluoride (PMSF), 10% glycerol, 15 μM NADP+. The homogenate was then filtered through four layers of muslin and centrifuged at 20 000 g for 20 min at 4 °C. The supernatant (the fraction designated as the crude extract) was used for G6PDH assays.
G6PDH activity assay
G6PDH activity was assayed by monitoring NADP+ reduction at 340 nm. The assay mixture contained: 50 mM TRIS-HCl pH 8.0, 50 mM MgCl2, 1.5 mM NADP+, 30 mM glucose-6-phosphate (G6P), and extract (10–100 μl; 3–60 μg of protein). For enzyme activity measurements against a blank (without G6P), three different replicates were performed. The activity was expressed as nmol NADP+ reduced min−1 mg−1 protein.
Western blot analysis
The electrophoresis and western blotting analyses were carried out using crude extracts from roots and leaves at the given experimental times. A total of three separate experiments were performed, and data shown in the figures are representative of the general, similar behaviour. The proteins (15 μg or 50 μg for root and leaf extracts, respectively) were resolved by 10% SDS–PAGE, according to Esposito et al. (2005). Gels were run for 120 min at 40 mA, 180 V and the separated polypeptides were transferred on a Hybond membrane (GE Healthcare). After the transfer (2 h at 25 V, 300 mA), the membrane was incubated with primary G6PDH antibody from potato for P1-, P2-, and Cy-G6PDH isoforms (Wendt et al., 2000). These antibodies have been proven to react with and discriminate the different barley G6PDH isoforms in previous studies (Esposito et al., 2001b, 2003, 2005). After washing, the membranes were incubated with secondary antibodies coupled to alkaline phosphatase.
To assess equal loading of protein in each lane, western blots were carried out on control gels loaded with the same amounts of extracts, and β-tubulin was detected using specific antibodies.
Semi-quantitative RT-PCR assay
Semi-quantitative reverse transcrption-PCR (RT-PCR) experiments were used to estimate the expression of G6PDH transcripts in leaves and roots. Total RNAs were extracted from 100 mg of barley roots and leaves using Trizol Reagent (Invitrogen) as described by the manufacturer. The final RNA pellet was dissolved in diethylpyrocarbonate (DEPC)-treated water and quantified with UV spectrophotometry. cDNA synthesis was carried out using ThermoScript RT-PCR System (Invitrogen). PCR amplification was performed with 10 ng μl−1 of cDNA as PCR template and 2.5 pmol of the primers in a final volume of 25 μl. cDNA synthesis and PCR amplification were carried out with a thermal cycler (GeneAmp PCR System 2700, Applied Biosystems). The program used was as follows: 3 min denaturation at 94 °C, and 40 cycles of 1 min denaturation at 94 °C, 30 s annealing at 58 °C, 30 s extension at 72 °C; extension in the last cycle was prolonged for 10 min. Primers were designed for P2-G6PDH (forward, 5′-GGGAAAGGAGCTGGTGGAGAAC-3′; reverse, 5′-TATTCTCAGAAGACTTTGGCAC-3′) and for Cy-G6PDH (forward, 5′-ATACGAGCGCCTCATTTTGG-3′; reverse, 5′-ACAACATCGACGCTGGCAA-3′).
As an internal control, the constitutively expressed ribosomal 18S gene was amplified from various samples to generate a 600 bp fragment. The PCR conditions for amplifying the 18S gene were a pre-denaturation of 5 min at 94 °C; 35 cycles of 30 s at 94 °C, 45 s at 50 °C, 30 s at 72 °C; and an extension for 10 min at 72 °C.
The sequences of the primers for the 18S gene are as follows (forward, 5′- GGAGAAGTCGTAACAAGGTTTCCG 3′; reverse, 5′ -TTCGCTCGCCGTTACTAAGGG 3′).
As a control, PCR analyses were made on extracts of roots and leaves from untreated plants at the same given times; in these samples the expression levels of both Cy-G6PDH and P2-G6PDH remained constant throughout the experiments (not shown).
Analysis of PCR products
The amplified products were resolved on a 1.5% agarose gel, and the DNA was visualized by ethidium bromide, using an UV transilluminator, and images were acquired by a camera. Each gel photo was processed using Image J (NIH) to obtain a threshold black and white pixel map. The spots were quantified as arbitrary units (pixel) in an Excel worksheet, and used to construct a graph using the Sigmaplot (Jandel) software. The experiment shown is representative of three experiments which showed the same general behaviour; the average values and the standard errors are given in the chart below the gel photos in the figures.
Statistical analysis
Data obtained on leaf and root length, G6PDH total activity, and PCR were statistically analysed using a one-way analysis of variance (ANOVA) test; Jandel Sigma Plot 11 software. When differences in the mean values among the treatment groups were greater than would be expected by chance and a statistically significant difference (P <0.001) was observed, data were compared by using a multiple evaluation procedure.
Results and Discussion
Although barley is a diploid inbreeding species with a genome of 5 Gbp, not presently suited to whole-genome sequencing (because 80% of its sequence is composed of repetitive DNA), it is a useful model to study cereals due to its smaller genome compared with all other Triticeae species (Bennett and Smith, 1976; Sreenivasulu et al., 2008). Thus, the current genome sequence available represents an important tool for research in cereal crops, particularly for functional genomics applications developed by the international barley sequencing consortium (http://barleygenome.org). These data have been completed by the generation of ∼435 000 expressed sequence tags (ESTs) covering different cDNA libraries from various stages of barley plant development and tissues challenged with abiotic and biotic stresses (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). The alignment of these ESTs led to the identification of a set of 50 435 unigenes with 23 176 tentative consensus and 27 094 singletons (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=barley), representing ∼75% of the barley genes (Sreenivasulu et al., 2008).
Phylogenetic analysis and G6PDH characteristics
G6PDHs are tetrameric proteins formed by monomers of ∼50–60 kDa, which are present in all photosynthetic and non-photosynthetic organisms except the Archaea (Wendt et al., 1999).
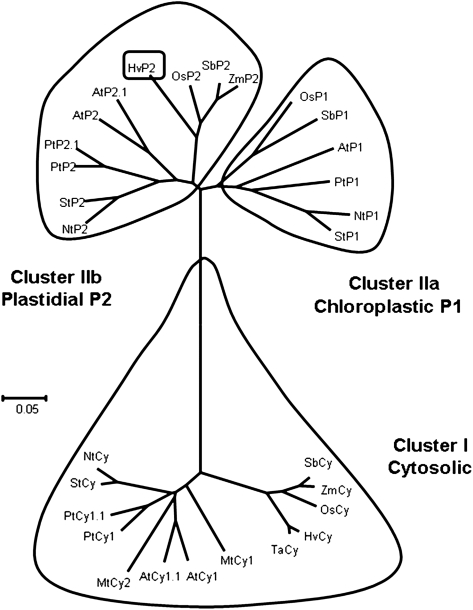
In order to study the evolution and distribution of the G6PDH protein family, a phylogenetic tree was constructed (Fig. 1) using G6PDH amino acid sequences (cytosolic and plastidial) from several higher plants: A. thaliana, O. sativa, N. tabacum, S. bicolor, P. trichocarpa, M. truncatula, S. tuberosum, S. oleracea, Z. mays, T. aestivum, and H. vulgare. Searches for sequences from barley and other non-annotated genomes were essentially performed by a BlastP or tblastn analysis on protein and genomic sequences using various predicted sequences from monocotyledons and dicotyledons. The primary structure analysis and the predicted subcellular localization allow the classification of G6PDHs of higher plants in two different clusters that here are called cluster I and cluster II (subdivided into IIa and IIb).
Fig. 1.
Phylogenetic tree of higher plant G6PDHs. The organism codes are as follow: St, Solanum tuberosum; Nt, Nicotiana tabacum; Pt, Populus trichocarpa; At, Arabidopsis thaliana; Sb, Sorghum bicolor; Os, Oryza sativa; Ta, Triticum aestivum; Mt, Medicago truncatula; and Zm, Zea mays. The corresponding accession numbers are available in the Supplementary data file at JXB online containing all the G6PDH sequences used in this study.
Cluster I represents Cy-G6PDHs, proteins of ∼50 kDa and 500 amino acids. This branch contains two subgroups representing the monocotyledons and dicotyledons. The monocotyledon subgroup includes species of the Poacea family (S. bicolor, Z. mays, T. aestivum, and H. vulgare). The sequences in this subgroup present a high similarity, from 87% to 97% identity. The identity between sequences present in the dicotyledon subgroup (A. thaliana, P. trichocarpa, N. tabacum, S. tuberosum, and M. truncatula) ranges from 78% to 95%. It should be noted that P. trichocarpa and A. thaliana possess two cytosolic isoforms, most probably arising from specific duplication events. The similarity between all sequences of cluster I is quite high and it ranges between 72% and 97% (data not shown). All of them display the strictly conserved active site motif DHYLGKE.
The phylogenetic analysis revealed that the second cluster is split into two distinct subgroups representing the two known plastidial isoforms, P1 (cluster IIa) and P2 (cluster IIb). In each cluster, the monocotyledon and dicotyledon G6PDH sequences form separate classes. These proteins are composed of ∼580 amino acids with a predicted mol. wt of 66 kDa. It is important to stress that these proteins exhibit N-terminal extensions of ∼80 amino acids corresponding to putative plastidial targeting sequences. As they are generally cleaved during protein import, the size of the mature proteins is very close to that of cytosolic proteins. The chloroplastic localization of the P1 and P2 isoforms has been confirmed by green fluorescent protein (GFP) fusion experiments (Wendt et al., 2000).
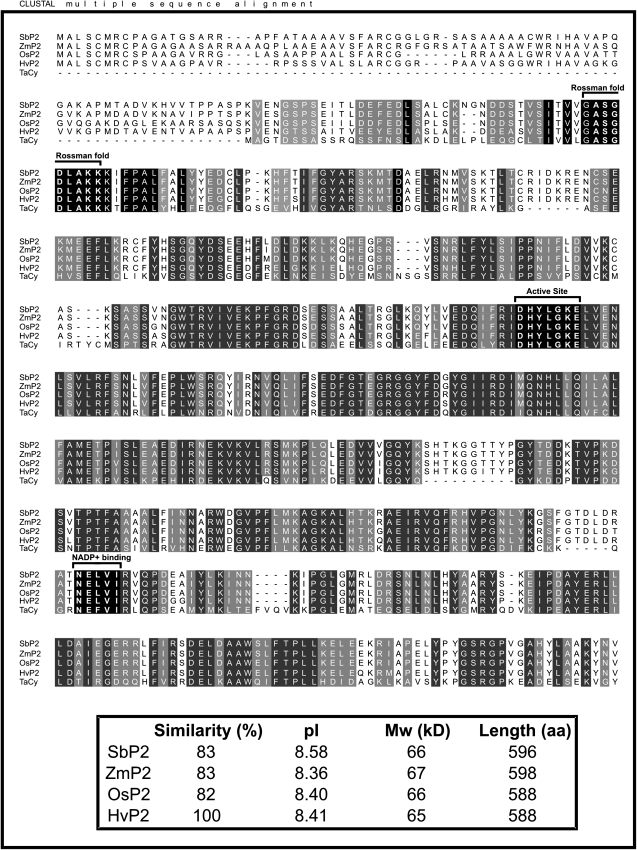
The barley (H. vulgare) P2-G6PDH coding sequence has recently been isolated from roots (accession no. AM398980). It shows 82% identity with the rice P2-G6PDH (LOC_Os07g22350). The two isoforms share a strict identity in the Rossman-fold motif (ASGDLAKK) and in the active site (DHYLGKE), and a good conservation of the NADP+-binding site (NELVI sequence) (Fig. 2).
Fig. 2.
Amino acid sequence alignment of P2-G6PDHs from Hordeum vulgare (Hv, CAL44728), Oryza sativa (Os, LOC_Os07g22350), Zea mays (Zm, ACG29334), Sorghum bicolor (Sb, estExt_Genewise1Plus.C_chr_60876), and Triticum aestivum (Ta, BAA97663). The alignment was performed with ClustalW. Strictly conserved amino acids are on a black background, while functionally conserved amino acids are on a grey background. The active site (DHYLGKEL), the Rossman-fold motif (ASGDLAKK), and the core of the NADP+-binding site (NELVI) are indicated by brackets. In the lower box, the percentage similarity, deduced isoelectric points (pI), molecular weights (MW), and protein lengths in amino acids (aa) of the different sequences are shown.
Effects of ABA supply on barley plants
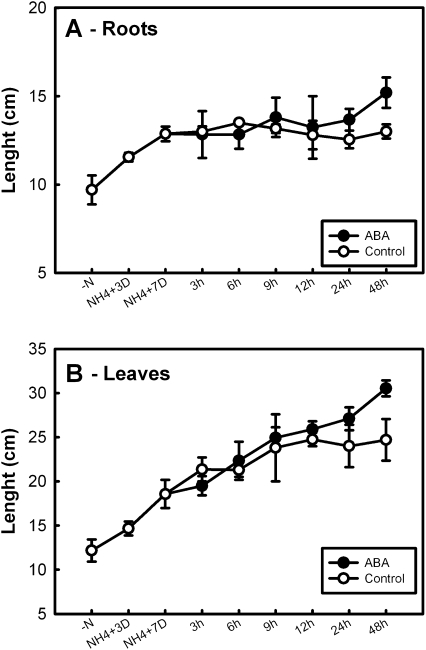
Application of ABA to the barley seedlings during the first 24 h of experimental treatment had no visible effect on the root length (Fig. 3A), and slightly increased the length of the leaves (Fig. 3B), as previously reported in maize (Sharp, 2002). After 48 h of ABA treatment, a 23% increase in leaf (P <0.001) and a 17% increase in root (P <0.001) length were observed with respect to control plants. These effects could be linked to the ABA-induced restriction of ethylene production (Sharp, 2002). Images of ABA-treated plants and control plants at different times during the experiments are shown in Supplementary Fig. S1 at JXB online.
Fig. 3.
Relative growth rate of barley roots (A) and leaves (B) after addition of 0.1 mM ABA. Barley seedlings were grown for 3 d in hydroculture without any nitrogen source, then grown under 5 mM ammonium phosphate for 7 d; seedlings were then supplied with 0.1 mM ABA and representative samples were collected at the given times. Measurements are the average from three different plants ±SE; a statistical one-way ANOVA was performed using Jandel SigmaPlot 11.0 Software; other details are given in the text.
Total G6PDH activity in roots and leaves upon ABA treatment
It has been shown earlier that ABA levels directly affect several basic metabolism enzymes. On the other hand, the role of OPPP in the response and/or tolerance to drought, salinity, and nutrient starvation in plants has been established (Esposito et al., 2001b, 2005; Valderrama et al., 2006). In particular, a central role has been described for G6PDHs both in the ABA signalling pathway and in the salt stress response (Kempa et al., 2008). For instance, an increase in total G6PDH activity has been observed in plants upon nitrogen starvation (Esposito et al., 2001b, 2005), under biotic (Sindelar et al., 1999) and abiotic (Bredemeijer and Esselink, 1995) stresses.
Considering the presence of an ABRE in the rice P2-G6PDH promoter and its high degree of similarity to the barley P2-G6PDH sequence, it was hypothesized that ABA could have a direct effect on P2-G6PDH expression which would open up new perspectives in the study of the regulation of the adaptation of the whole plant to abiotic stresses. For this purpose, total enzymatic activity, protein occurrence, and transcript levels of the plastidial G6PDH(s) have been investigated during ABA treatment, keeping in mind that ABA also plays pivotal roles in various aspects of plant growth and development (Hou et al., 2006; Lenka et al., 2009).
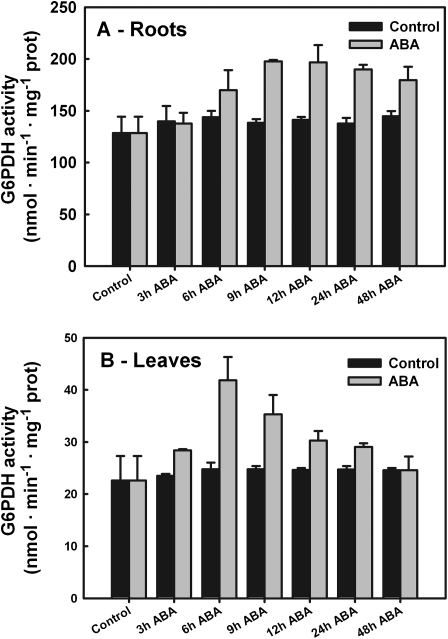
To study the effect of ABA on G6PDH activity, 0.1 mM ABA was applied exogenously to seedlings (15 d old) and the total G6PDH enzymatic rates were measured in root (Fig. 4A) and leaf (Fig. 4B) extracts. ABA (0.1 mM) caused a rapid increase (+85%) in G6PDH activity in leaves within 6 h (P <0.001). Thereafter enzyme activity rates decreased constantly until they reached the control values within 48 h (Fig. 4A). Control plants (no ABA supply) showed no appreciable variation (<9%; P <0.215) in G6PDH activity throughout the experiments (Fig. 4A).
Fig. 4.
Total G6PDH activity in barley roots (A) and leaves (B). Barley seedlings were grown for 3 d in hydroculture without any nitrogen source, then with 5 mM ammonium phosphate for 7 d. The seedlings were then supplied with 0.1 mM ABA, samples were collected at the given times, and the crude extracts of leaves and roots were assayed for G6PDH activity. Data shown are the average ±SE of four different determinations measured at least in duplicate. A statistical one-way ANOVA was performed using Jandel SigmaPlot 11.0 Software; other details are given in the text.
Application of ABA caused a gradual increase in total G6PDH activity between 3 h and 12 h in roots (+54%; P <0.001) compared with the activity detected under the experimental starting conditions (plants grown for 7 d in hydroponic culture). Over the following days, G6PDH activity remained at the same high values (Fig. 4B). In control plants (without ABA supply), enzyme activity did not change appreciably (±10%; P <0.059) throughout the experiments (Fig. 4B).
Effect of ABA on plastidial G6PDHs
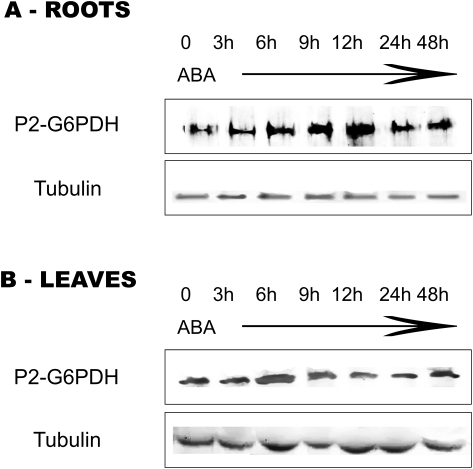
The abundance of G6PDH proteins was investigated by western blotting using antibodies able to discriminate the plastidial P2-isoform (Wendt et al., 2000; Esposito et al., 2005).
In roots, supply of exogenous ABA progressively increased the abundance of P2-G6PDH with respect to control plants, the densitometry analysis indicating an increase of 70% after 3–12 h and up to 2.8-fold at 12 h. Therefore, changes in abundance of P2-G6PDH (Fig. 5A) suggest that there is a gradual and consistent increase of P2-G6PDH protein levels in roots, mimicking the situation observed for total activity (Fig. 4A).
Fig. 5.
Western blots of P2-G6PDH from crude extracts of roots (A) and leaves (B) of barley plants subjected to ABA treatment. The seedlings were grown on a medium supplied with 0.1 mM ABA and samples were collected at the given times. Detection was carried out using antibodies raised against potato plastidial P2-G6PDH enzyme (Wendt et al., 2000). Tubulin: western analysis of the same samples using tubulin antibodies to ensure equal protein loading.
In leaf crude extracts, the amount of P2-G6PDH strongly increased (1.9-fold) within 6 h following the ABA treatment, and then it returned to levels comparable with those of control plants in the following hours/days (Fig. 5B). It should be noted that nitrogen starvation and nitrogen supply did not change the levels of the P2-G6PDH isoform in leaves (Esposito et al., 2005).
In roots, the Cy-G6PDH protein content showed a slight increase within 12–24 h following ABA treatment (Supplementary Fig. S2A at JXB online). The Cy-G6PDH protein content in leaves did not appear to change upon ABA treatment (Supplementary Fig. S2B). These patterns are similar to that observed for Cy-G6PDH in barley roots after nitrogen supply (Esposito et al., 1998). A similar increase and decrease in accumulation related to ABA levels was observed in non-dormant Arabidopsis with respect to dormant seeds for two isoforms of cytosolic glyceraldehyde-3-P dehydrogenase (Chibani et al., 2006).
Regarding P1-G6PDH, the absence of signal for the plastidial P1-G6PDH in barley roots confirmed that it is not or only very weakly synthesized in non-photosynthetic tissues, making the plastidial P2-type isoform the only G6PDH representative for this compartment in roots. The amount of P1-G6PDH decreased rapidly in leaves 3 h after ABA treatment and this decreased level remained constant, whereas it is not detected in roots (Supplementary Fig. S2B).
Altogether, these results indicate that the increase in G6PDH activity observed in crude leaf and root extracts correlates with changes in P2-G6PDH abundance. This increase in P2-G6PDH protein content following ABA supply strongly supports the involvement of this isoform in the response to abiotic stress in plants. This result further demonstrates that changes in enzymes of primary metabolism can modify the stress response. In this respect, a recent study clearly demonstrates the central role played by Cy-G6PDH for the provision of reductants to NADPH oxidases during the oxidative burst occurring upon Phytophthora infection of tobacco plants (Scharte et al., 2009).
Expression of the P2-G6PDH gene in ABA-treated plants
To understand the role of ABA concerning transcriptional regulation of P2-G6PDH, semi-quantitative RT-PCRs were performed on roots and leaves of ABA-treated barley plants.
The P2-G6PDH-encoding gene was amplified from H. vulgare root and leaf cDNAs with a specific pair of primers. Although P1-G6PDH is expressed in leaves, the absence of nucleotidic sequence for this gene unfortunately prevented the design of specific primers and its detection in RT-PCR measurements.
Semi-quantitative RT-PCR analysis indicates that P2-G6PDHs are expressed both in roots and in leaves.
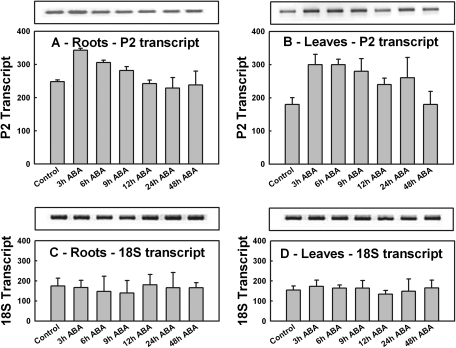
A 40% increase in P2-G6PDH transcript levels was observed in roots within 3 h of ABA supply as attested by the densitometry analysis (P <0.05). Afterwards, the transcript levels slowly declined until they reached the initial levels at 12 h, then remained unchanged at 24 h and 48 h (Fig. 6A). Thus, the progressive increase in total G6PDH activity measured in roots in the first 12 h in response to ABA could be due at least in part to an increase in transcription and synthesis of P2-G6PDH protein (Fig. 5A).
Fig. 6.
P2-G6PDH transcript expression profiles after ABA treatment. Semi-quantitative RT-PCRs were performed with RNA extracted from roots (left) and leaves (right) of samples collected at the given times from seedlings supplied with 0.1 mM ABA. (A and B) P2-G6PDH transcript levels; (C and D) ribosomal 18S transcript levels used as control for roots (A, C) and leaves (B, D), respectively. The graphs show the quantification of transcript obtained using Image J software (NIH, USA) indicated by bars. Data shown are the average ±SE of five different determinations. A statistical one-way ANOVA was performed using Jandel SigmaPlot 11.0 Software; other details are given in the text.
Cy-G6PDH transcript levels in both roots and leaves did not change appreciably after ABA supply (no significant difference—see Supplementary Fig. S3A, B at JXB online) as previously observed in wheat (Nemoto and Sasakuma, 2000) and rice (Hou et al., 2006). However, this is in contrast to the 2-fold increase described upon salt stress (Nemoto and Sasakuma, 2000).
Curiously, in A. thaliana, Cy-G6PDH transcript levels were down-regulated upon ABA treatment (Wang et al., 2008).
In barley leaves, ABA supply caused a rapid and significant increase (+66%) in the P2-G6PDH transcript abundance within 3 h (P <0.001), after which the levels remained higher than the initial levels until 48 h, when they returned to the initial value (Fig. 6B).
In silico analysis of barley P2-G6PDH gene expression
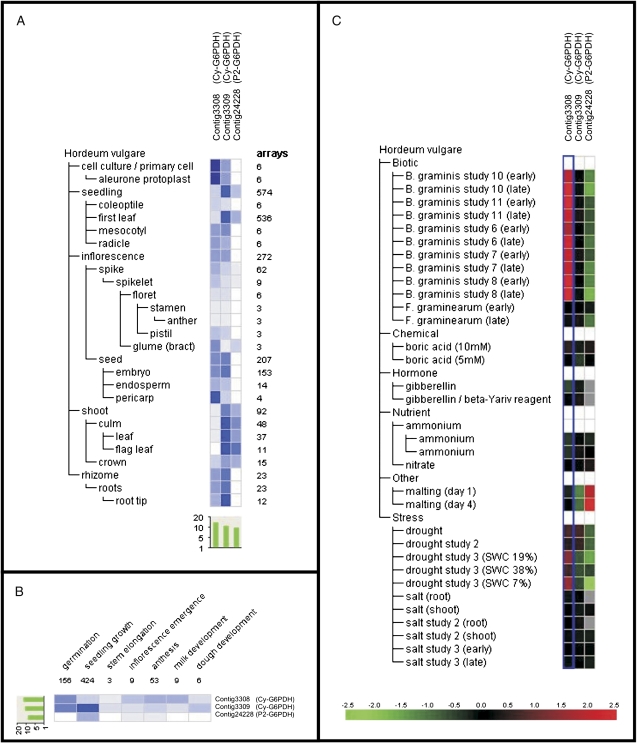
The recent release of barley transcriptomic data in the genevestigator website (https://www.genevestigator.com) allowed the regulation of the plastidial barley P2-G6PDH-encoding gene (contig24228) to be studied in more detail in the different plant organs and under stress conditions (Zimmermann et al., 2008).
This P2-G6PDH is relatively highly expressed in shoots (leaf and flag leaf) compared with the other organs (Fig. 7A) during seedling growth (Fig. 7B), a result in accordance with semi-quantitative RT-PCRs in barley leaves and roots presented in Fig. 6A. In contrast, in N. tabacum and A. thaliana, P2-G6PDH was expressed in all tissues, including roots, leaves, stem, and flowers (Knight et al., 2001; Wakao and Benning, 2005). In S. tuberosum, P2-G6PDH transcripts are prominent in roots and growing tissues, whereas both P1- and P2- isoforms could be detected in leaves (Wendt et al., 2000).
Fig. 7.
Transcript level analysis of barley P2-G6PDH (contig24228) and of two different Cy-G6PDHs (contig3308 and contig3309), using the meta-profile analysis tool in Genevestigator V3 (www.genevestigator.ethz.ch). Transcript levels have been analysed under different abiotic stimuli (A), in different barley organs (B), and at different developmental stages of barley plants (C). Data are log2. In A, the red colour marks up-regulation and the green color marks down-regulation (see reference bar).
Regarding the stress response, the transcript levels of barley P2-G6PDH are markedly down-regulated by drought stress and up-regulated during malting (Fig. 7C). The latter process is a consequence of release of gibberellic acid (GA)—an antagonist of ABA. In the experiments shown here, the P2-G6PDH isoform is up-regulated by ABA treatment in mature barley plants. This apparent discrepancy could be explained by the existence of several (but as yet unidentified in barley) P2-G6PDH isoforms with a distinct expression pattern, as in A. thaliana or P. trichocarpa. Alternatively, it could be the same isoform which is differentially regulated in germinating seeds compared with leaves.
The Genevestigator database contains at least two more contigs probably corresponding to Cy-G6PDH isoforms. Contig3308 is highly expressed in seed pericarp and cell cultures, whereas contig3309 is preferentially expressed in leaves and roots of barley (Fig. 7A).
It should be noted that the two different cytosolic isoforms described in A. thaliana, AtG6PDH5 and AtG6PDH6 (Wakao and Benning, 2005), are also differentially distributed, the former being expressed mainly in roots, and the latter constitutively expressed in leaves (Wakao and Benning, 2005; Wakao et al., 2008).
Regarding environmental constraints, contig3308 is strongly up-regulated by biotic stress, in particular during infection by Blumeria graminis, and is slightly up-regulated by drought (Fig. 7C). In contrast, contig3309 is not regulated under any stress conditions tested except for a slight down-regulation during malting and drought stress (Fig. 7C).
Conclusions
In the experiments presented here, ABA was supplied to the roots of barley plants in order to define its role(s) in the expression and activity of plastidial P2-G6PDH isoforms. First of all, it should be mentioned that the Cy-G6PDH isoform(s) accounts for 78–92% of the total measurable activity in plant tissues (Debnam and Emes, 1999; Esposito et al., 2001b, 2005), whereas the plastidial activity, which probably represents the remaining 8–22% of G6PDH activity, is undetectable upon nitrogen starvation, but induced by both ammonium (Esposito et al., 2001b) and nitrate (Esposito et al., 2005).
Here it is demonstrated that ABA supply causes an increase in total G6PDH activity both in roots and in leaves, with the plastidial P2-isoform probably being an important player in barley plants. In roots, since the P1-G6PDH protein is not detectable, and the abundance of Cy-G6PDH only varied slightly, the increase in total G6PDH activity upon ABA supply can most probably be attributed to the plastidial P2-G6PDH isoform(s) whose protein content gradually increased after 6–12 h of ABA treatment. This increase in P2 protein level correlates well with a preceding increase in the corresponding transcript levels, ∼3–9 h after treatment. These results suggest that a considerable part of the response to ABA in roots involves the plastidial P2-G6PDH. Similarly, the 2-fold increase in total G6PDH activities observed at 6 h is likely to be related to an increase in P2-G6PDH activity in leaves.
Briefly, this paper supports a direct effect of ABA on P2-G6PDH gene expression, possibly due to the presence of an ABRE in the promoter, which is subsequently translated into an increase in protein abundance and activity.
Supplementary data
Supplementary data are available at JXB online
In addition to the figures below, the sequences, accession numbers, and relative databases used are given in the Supplementary data.
Figure S1. Relative growth rate of barley seedlings upon trreatment with 0.1 mM ABA. Barley seedlings were grown for 3 d in hydroculture without any nitrogen source, then grown under 5 mM ammonium phosphate for 7 d; seedlings were then supplied with 0.1 mM ABA and representative samples were collected at the given times. The appearance of control plants is shown above in comparison with ABA-treated plants (below).
Figure S2. Western blots of P1- and Cy-G6PDH isoforms from crude extracts of roots (A) and leaves (B) of barley plants subjected to ABA treatment. The seedlings were grown on a medium supplied with 0.1 mM ABA and samples were collected at the given times. Detection of Cy- and P1-G6PDH isoforms was performed using antibodies raised against potato proteins (Wendt et al., 2000).
Figure S3. G6PDH transcript expression profiles after ABA treatment. Semi-quantitative RT-PCRs were performed with RNA extracted from roots (A) and leaves (B) of samples collected at the given times from seedlings supplied with 0.1 mM ABA. The graphs show the quantification of transcripts obtained using Image J software (NIH, USA) indicated by bars. Data shown are the average±SE of five different determinations. A statistical one-way ANOVA was performed using Jandel SigmaPlot 11.0 Software; other details are given in the text.
Acknowledgments
The authors wish to dedicate this paper to Mauro Cardi. We acknowledge the Istituto Sperimentale per la Cerealicoltura, Fiorenzuola D'Arda (PC, Italy) for the gift of barley seeds. We thank Antje von Schaewen (University of Münster, Germany) for potato G6PDH antibodies. The authors are grateful to PhD student Daniela Castiglia (DBSF, University of Naples ‘Federico II’) for cDNA preparation, assistance, and suggestions. Thanks to Dr Giulia Maisto (DBSF, University of Naples ‘Federico II’) for her help and assistance in statistical analysis of the results. The financial support for MC from the Egide programme (dossier no. 672700K) is acknowledged.
Glossary
Abbreviations
- Cy
cytosolic
- G6PDH
glucose-6-phosphate dehydrogenase
- OPPP
oxidative pentose phosphate pathway
- P
plastidial
References
- Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society B: Biological Sciences. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bowsher CG, Hucklesby DP, Emes MJ. Nitrite reduction and carbohydrate metabolism in plastids purified from roots of Pisum sativum. Planta. 1989;177:359–366. doi: 10.1007/BF00403594. [DOI] [PubMed] [Google Scholar]
- Bowsher CG, Lacey AE, Hanke GT, Clarkson DT, Saker LR, Stulen I, Emes JM. The effect of Glc6P uptake and its subsequent oxidation within pea root plastids on nitrite reduction and glutamate synthesis. Journal of Experimental Botany. 2007;58:1109–1118. doi: 10.1093/jxb/erl269. [DOI] [PubMed] [Google Scholar]
- Bredemeijer GMM, Esselink G. Glucose 6-phosphate dehydrogenase during cold-hardening in Lolium perenne. Journal of Plant Physiology. 1995;145:565–569. [Google Scholar]
- Buchanan BB. Regulation of CO, assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Archives of Biochemistry and Biophysics. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiology. 2006;142:1493–1510. doi: 10.1104/pp.106.087452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnam PM, Emes MJ. Subcellular distribution of enzymes of the oxidative pentose phosphate pathway in root and leaf tissues. Journal of Experimental Botany. 1999;50:1653–1661. [Google Scholar]
- Esposito S, Carfagna S, Massaro G, Vona V, Di Martino Rigano V. Glucose-6P dehydrogenase in barley roots: kinetic properties and localisation of the isoforms. Planta. 2001a;212:627–634. doi: 10.1007/s004250000443. [DOI] [PubMed] [Google Scholar]
- Esposito S, Carillo P, Carfagna S. Ammonium metabolism stimulation of glucose-6P dehydrogenase and phosphoenol-pyruvate carboxylase in young barley roots. Journal of Plant Physiology. 1998;153:61–66. [Google Scholar]
- Esposito S, Guerriero G, Vona V, Di Martino Rigano V, Carfagna S, Rigano C. Glutamate synthase activities and protein changes in relation to nitrogen nutrition in barley: the dependence on different plastidial glucose-6P dehydrogenase isoforms. Journal of Experimental Botany. 2005;56:55–64. doi: 10.1093/jxb/eri006. [DOI] [PubMed] [Google Scholar]
- Esposito S, Massaro G, Vona V, Di Martino Rigano V, Carfagna S, Rigano C. Ammonium induction of a novel isoform of glucose-6Pdehydrogenase in barley roots. Physiologia Plantarum. 2001b;113:469–476. [Google Scholar]
- Esposito S, Massaro G, Vona V, Di Martino Rigano V, Carfagna S. Glutamate synthesis in barley roots: the role of the plastidial glucose-6-phosphate dehydrogenase. Planta. 2003;216:639–647. doi: 10.1007/s00425-002-0892-4. [DOI] [PubMed] [Google Scholar]
- Fan J, Hill, Crooks C, Doerner P, Lamb C L. Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiology. 2009;150:1750–1761. doi: 10.1104/pp.109.137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signalling in seeds and seedlings. The Plant Cell. 2002;14:15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Graeve K, von Schaewen A, Scheibe R. Purification, characterisation, and cDNA sequence of glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.) The Plant Journal. 1994;5:353–361. doi: 10.1111/j.1365-313x.1994.00353.x. [DOI] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant and Cell Physiology. 2002;43:136–140. doi: 10.1093/pcp/pcf014. [DOI] [PubMed] [Google Scholar]
- Hauschild R, von Schaewen A. Differential regulation of glucose-6-phosphate dehydrogenase isoenzyme activities in potato. Plant Physiology. 2003;133:47–62. doi: 10.1104/pp.103.025676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F-Y, Hueng J, Lu J-F, Wang Z-F, Zhang H- S. Isolation and expression analysis of plastidial glucose-6-phosphate dehydrogenase gene from rice (Oryza sativa L.) Acta Genetica Sinica. 2006;33:441–448. doi: 10.1016/S0379-4172(06)60071-1. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang JF, Zhang HS, Cao YJ, Lin CF, Wang D, Yang JS. In silico cloning of glucose-6-phosphate dehydrogenase cDNA from rice (Oryza sativa L.) Acta Genetica Sinica. 2002;29:1012–1016. [PubMed] [Google Scholar]
- Hutchings D, Rawsthorne S, Emes MJ. Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus) Journal of Experimental Botany. 2005;56:577–585. doi: 10.1093/jxb/eri046. [DOI] [PubMed] [Google Scholar]
- Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C. A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS ONE. 2008;3:e3935. doi: 10.1371/journal.pone.0003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Emes MJ, Debnam PM. Isolation and characterisation of a full-length genomic clone encoding a plastidial glucose 6-phosphate dehydrogenase from Nicotiana tabacum. Planta. 2001;212:499–507. doi: 10.1007/s004250000419. [DOI] [PubMed] [Google Scholar]
- Kruger NJ, von Schaewen A. The pentose phosphate pathway: structure and organisation. Current Opinion in Plant Biology. 2003;6:236–246. doi: 10.1016/s1369-5266(03)00039-6. [DOI] [PubMed] [Google Scholar]
- Lenka SK, Lohia B, Kumar A, Chinnusamy V, Bansal KC. Genome-wide targeted prediction of ABA responsive genes in rice based on over-represented cis-motif in coexpressed genes. Plant Molecular Biology. 2009;69:261–271. doi: 10.1007/s11103-008-9423-4. [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Li C, Yin C, Liu S. Different responses of two contrasting Populus davidiana populations to exogenous abscisic acid application. Environmental and Experimental Botany. 2004;51:237–246. [Google Scholar]
- Mantyla E, Lang V, Palva ET. Role of abscissic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LTI and RAB18 proteins in Arabidopsis thaliana. Plant Physiology. 1995;l07:141–148. doi: 10.1104/pp.107.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Née G, Zaffagnini M, Trost P, Issakidis-Bourguet E. Redox regulation of chloroplastic glucose-6-phosphate dehydrogenase: a new role for f-type thioredoxin. FEBS Letters. 2009;17:2827–2832. doi: 10.1016/j.febslet.2009.07.035. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Sasakuma T. Specific expression of glucose-6-phosphate dehydrogenase (G6PDH) gene by salt stress in wheat (Triticum aestivum L.) Plant Science. 2000;158:53–60. doi: 10.1016/s0168-9452(00)00305-8. [DOI] [PubMed] [Google Scholar]
- Scharte J, Schön H, Tjaden Z, Weis E, von Schaewen A. Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proceedings of the National Academy of Sciences, USA. 2009;106:8061–8066. doi: 10.1073/pnas.0812902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R. Malate valves to balance cellular energy supply. Physiologia Plantarum. 2004;120:21–26. doi: 10.1111/j.0031-9317.2004.0222.x. [DOI] [PubMed] [Google Scholar]
- Sharp RE. Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant, Cell and Environment. 2002;25:211–222. doi: 10.1046/j.1365-3040.2002.00798.x. [DOI] [PubMed] [Google Scholar]
- Sindelar L, Sindelarova M, Burketova L. Changes in activity of glucose-6-phosphate and 6-phosphogluconate dehydrogenase isozymes upon potato virus Y infection in tobacco leaf tissue and protoplasts. Plant Physiology and Biochemistry. 1999;37:195–201. [Google Scholar]
- Sreenivasulu N, Graner A, Wobus U. Barley genomics: an overview. International Journal of Plant Genomics. 2008;2008:486258. doi: 10.1155/2008/486258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Valderrama R, Corpas FJ, Carreras A, Gomez-Rodriguez MV, Chaki M, Pedrajas JR, Fernandez-Ocana A, Del rio LA, Barroso JB. The dehydrogenase-mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plants. Plant, Cell and Environment. 2006;29:1449–1459. doi: 10.1111/j.1365-3040.2006.01530.x. [DOI] [PubMed] [Google Scholar]
- von Schaewen A, Langenkamper G, Graeve K, Wenderoth I, Scheibe R. Molecular characterisation of the plastidial glucose-6-phosphate dehydrogenase from potato in comparison to its cytosolic counterpart. Plant Physiology. 1995;109:1327–1335. doi: 10.1104/pp.109.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S, Andre C, Benning C. Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis. Plant Physiology. 2008;146:277–288. doi: 10.1104/pp.107.108423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S, Benning C. Genome-wide analysis of glucose-6-phosphate dehydrogenase in Arabidopsis. The Plant Journal. 2005;41:243–256. doi: 10.1111/j.1365-313X.2004.02293.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Ma Y, Huang C, Wan Q, Li N, Bi Y. Glucose-6-phosphate dehydrogenase plays a central role in modulating reduced glutathione levels in reed callus under salt stress. Planta. 2008;227:611–623. doi: 10.1007/s00425-007-0643-7. [DOI] [PubMed] [Google Scholar]
- Wenderoth I, Scheibe R, von Schaewen A. Identification of the cysteine residues involved in redox modification of plant plastidial glucose-6-phosphate dehydrogenase. Journal of Biological Chemistry. 1997;272:26985–26990. doi: 10.1074/jbc.272.43.26985. [DOI] [PubMed] [Google Scholar]
- Wendt UK, Hauschild R, Lange C, Pietersma M, Wenderoth I, von Schaewen A. Evidence for functional convergence of redox regulation in G6PDH isoforms of cyanobacteria and higher plants. Plant Molecular Biology. 1999;40:487–494. doi: 10.1023/a:1006257230779. [DOI] [PubMed] [Google Scholar]
- Wendt UK, Wenderoth I, Tegeler A, von Schaewen A. Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.) The Plant Journal. 2000;23:723–733. doi: 10.1046/j.1365-313x.2000.00840.x. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annual Review of Plant Physiology and Plant Molecular Biology. 1988;39:439–473. [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W. Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Molecular Plant. 2008;1:851–857. doi: 10.1093/mp/ssn048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.