Abstract
Cdc42 is a Ras-related small G-protein, and functions as a molecular switch in signal transduction pathways linked with cell growth and differentiation. It is controlled by cycling between GTP-bound (active) and GDP-bound (inactive) forms. Nucleotide binding and hydrolysis are modulated by interactions with effectors and/or regulatory proteins. These interactions are centralized in two relatively flexible “Switch” regions as characterized by internal dynamics on multiple timescales (Loh et al., (2001) Biochemistry 40, 4590–4600), and this flexibility may be essential for protein interactions. In the Switch I region, Thr35 seems critical for function, as it is completely invariant in Ras-related proteins. To investigate the importance of conformational flexibility in Switch I of Cdc42, we mutated threonine to alanine, determined the solution structure and characterized the backbone dynamics of the single-point mutant protein, Cdc42(T35A). Backbone dynamics data suggests that the mutation changes the timescale of the internal motions of several residues, with several resonances appearing not discernable in Cdc42 wild type (Adams and Oswald (2007) Biomolecular NMR Assignments 1, 225–227). The mutation does not appear to affect the thermal stability of Cdc42, and chymotrypsin digestion data further suggests that changes in conformational flexibility in Switch I slow proteolytic cleavage relative to wild type. In-vitro binding assays show reduced binding of Cdc42(T35A), relative to wild type, to a GTPase binding protein that inhibits GTP hydrolysis in Cdc42. These results suggest that the mutation of T35 leads to the loss of conformational freedom in Switch I that could affect effector/regulatory protein interactions.
Keywords: Ras GTPase, Signal transduction, Cdc42, Threonine, Alanine, Switch 1 mutant, conformational flexibility, backbone dynamics
The success of target-based approaches to combat Ras-related signal transduction events leading to diseases such as cancer will depend upon understanding the correlations among structural biology, protein-protein interactions and protein function. Indeed, binding events and the activation of protein receptors may play a role in abnormal cell-signaling activities. Understanding such activities will require approaches that elucidate changes in both protein conformation and protein dynamics that lead to particular molecular events and behaviors. Cdc42 is a member of the Rho subfamily of Ras proteins and are closely related to small GTPases. These proteins are ubiquitously expressed, evolutionarily conserved, and among the first human oncogenic proteins to have been identified (1, 2). Mutations or abnormal expression patterns of Ras proteins have been found in up to 30% of cancer cell types (1, 2). Ras proteins act as molecular switches that couple extracellular signals to a variety of cellular responses. Ras protein activities are controlled via cycling between a biologically active GTP form and an inactive GDP-bound form (1, 3). The function of Cdc42, as well as other Ras proteins, is largely mediated through protein-protein interactions. Therefore, understanding the specificity governing these interactions, and associated conformational changes is vital for understanding molecular mechanisms underlying the onset of diseases such as cancer. Different signals can activate guanine nucleotide exchange factors (GEFs) that induce the small G-protein to switch from the inactive, GDP-bound state to the active, GTP-bound state (4, 5). In the active state, the GTPase binds downstream effector and the effector in turn can mediate diverse biological responses. These different signals are terminated when Cdc42 is deactivated, either through its own ability to hydrolyze GTP, or by means of a reaction catalyzed by GTPase-activating proteins (GAPs) (6). Alternatively, interactions with regulatory proteins that block GDP dissociation, such as GDP dissociation inhibitors (GDIs), stabilize the GDP-bound inactive state, and prevent effective GTP-GDP cycling (7). Mechanisms by which Cdc42, as well as most Ras proteins participate in cellular proliferation and/or transformation include mutations that alter the GTPase activity (1, 8–11) and mutations that alter the interactions with effectors or regulators (12). The best understood mutations have been those that result in cellular transformation by inhibiting GTPase activity (1, 8). In addition to this, Cdc42 mutations have also been shown to facilitate cell transformation by increasing the rate of cycling between active and inactive forms of the protein (13 199). Structural studies of the mutant Cdc42(F28L) provided evidence that local environmental changes affect the stability of the nucleotide-binding site leading to aberrant cell-signaling behavior (14).
The binding of effector molecules to Cdc42 essentially involves two regions: Switch I (residues 28–40) and Switch II (residues 57–74), which can undergo rapid conformational changes. In fact, it has been reported that the Switch I region in H-Ras possesses two distinct conformational states, bound to effectors, and unbound (15, 16). Indeed, Switch I is the principal interacting surface for Cdc42 with downstream effectors (11, 17). For Cdc42 in solution, several resonances in the Switch regions cannot be observed due to increased flexibility in these regions (18). The binding of effectors to Cdc42 results in the loss of backbone mobility of the Switch I region, suggesting that the inherent dynamics may allow Cdc42 to bind to different effector proteins (19). It has been widely thought that the conformational freedom of the Switch I region may provide the basis for nucleotide-dependent signal propagation of Cdc42, and that the conformational flexibility of this region is central to the engagement of downstream effectors (17, 20, 21). This premise agrees with biological studies (22). Structural studies of Ras have implicated Thr35 in the Switch I region as being critical for the conformational changes as it is completely invariant in Ras related proteins. 31P-NMR studies with H-Ras(T35A) and Cdc42(T35A) suggested that this mutation causes a signaling-defective state (11, 16). The 31P-NMR spectra of both protein in their GTP-bound state revealed only small changes, suggesting that the nucleotide-binding site of the mutant was similar to wild type. However, binding peptide effectors that target the Switch I region produced significant shifts of 31P resonances in the wild type Ras peptide complexes but not in the T35A-effector complexes, suggesting a weaker interaction between effector peptide derivatives and the mutant protein (11, 16).
To investigate the importance of the conformational flexibility in Switch I of Cdc42, we have determined the solution structure, examined the backbone dynamics, and characterized the biochemical activity of Cdc42(T35A) (2kb0.pdb). Our results suggest that the mutation of Thr35 leads to the loss of conformational freedom in Switch I relative to the rest of the protein. Our results furthermore provide molecular details on why the conformational flexibility of this region is inherently important for the recognition of the Ras-related effectors. Recent studies have implied that innovative strategies to target cancer treatment in the future should target specific mutant Ras proteins and directly block Ras protein interactions with oncogenic potential, for example using antibody fragments (23). Taken together, the structure and dynamics results for Cdc42(T35A) suggest that, in addition to strategies to block Ras-protein interactions, strategies to restrict the conformational flexibility of important binding regions of Ras leading to the inhibition of effector interactions that facilitate aberrant signaling activity may also have therapeutic potential.
Materials and Methods
Protein Expression and Purification
Cdc42(T35A) was cloned, expressed and purified as described previously for Cdc42 (14, 24). Briefly, the Cdc42(T35A) construct was cloned into a pET-15b vector and subsequently over expressed in E. coli. Five-mL cultures were initially grown to saturation and used to seed 1 or 2-L cultures grown in LB broth for unlabelled samples, and in minimal media containing 15N-NH4Cl for 15N-labelled samples, and 15N-NH4Cl and 13C-glucose for 15N,13C-labeled samples. Cell cultures were grown at 37°C to an OD560 reading of 0.4–0.6, and expression induced by adding 1 mM IPTG (isopropyl β-D-thiogalactopyranoside) for 7–12 h. (His)6-Cdc42(T35A) and wild type were expressed, and purified by column chromatography. MALDI mass spectroscopy was used to verify that the molecular weight of the protein was correct, and gel electrophoresis was used to validate the purity of the protein samples.
NMR Sample Conditions, Data Collection and Processing
All protein samples were prepared in an NMR sample buffer solution containing 25 mM NaCl, 10 mM NaH2PO4, 5 mM MgCl2, and 1 mM NaN3 with 8% D2O at pH 5.5 (not adjusted for isotope effect) at Cdc42(T35A) concentrations of 0.2–0.5 mM in volumes of 500–550 ml for structural studies. The concentrations of Cdc42 wild type and T35A used for relaxation data collection were 1 mM. NMR spectra for structural studies were obtained using Varian Inova 500 MHz, 600, and 800 MHz spectrometers. All spectrometers were equipped with triple-resonance pulsed-field gradient probes. All NMR spectra were acquired at 25°C in States-TPPI mode for quadrature detection (25). Carrier frequencies for 1H and 15N were 4.77 and 115.9 ppm, respectively. 2D 1H,15N HSQC spectra were acquired on Cdc42(T35A) homogeneously labeled with 15N and resonance assignments were made and deposited (BMRB accession # 15424). Hydrogendeuterium exchange was assessed by lyophilizing Cdc42 wild type or (T35A) and resuspending in 100% D2O. A series of 1H-15N HSQC experiments were performed, and the decrease in the volume and eventual disappearance of 1H-15N cross peaks were used to determine the extent of exchange of deuterons for protons. 3D 1H,15N-TOCSY-HSQC experiments were acquired with saturating delays of 50 and 70 ms. 3D 1H,15N-NOESY-HSQC spectra were acquired using mixing time of 125 ms. The pulse sequences used included a water suppression by a selective pulse on the water resonance followed by dephasing using a gradient in the mixing period, as well as by a gradient applied while the magnetization was spin-ordered during the final INEPT transfer step. NMR spectra were processed using NMRPipe, version 1.6 (26). The spectra were zero-filled, then apodized using a Gaussian window function prior to Fourier transformation. After Fourier transformation, a baseline correction was applied. Spectra were visualized and analyzed using SPARKY (27).
Structure Calculation
The structure of Cdc42(T35A) was modeled and refined using SPARKY (27) and CNS (28). A combination of 1H,15N NOESYHSQC and 1H,13C NOESYHSQC experiments were used to derive distance constraints. In the spectra, the peak volumes of several well dispersed and isolated sequential dαN and dαN(i,i+1) NOE peaks within and in between parallel β-strands, respectively, were compared to the distances expected for ideal β-sheets (2.2 Å and 3 Å, respectively). Approximately 1057 NOESY constraints were classified according to their relative strength in the NOESY spectrum (< 2.4, < 3.4, < 4.0, and < 5.5 Å). Also, 71 hydrogen bond constraints were also input into the structure calculations based on the rate of deuterium exchange, the chemical shift index, and expected NOE patterns indicative of hydrogen bonding. For each hydrogen bond, the constraint between the amide proton to the carbonyl oxygen distance for the residue was held between 1.8–2.3 Å, and for the amide nitrogen to the carbonyl oxygen between 2.5–3.3 Å (18). Dihedral restraints (Total number: ϕ = 101; ψ = 100) were included for residues in α-helices and β-sheets, and the constraints were held to favorable regions of the ϕ,ψ space via: α-helix, ϕ = −80° ± 50°, ψ = −20° ± 50° ; β-sheet, ϕ = −105° ± 65°, ψ = 145° ± 45° using methods developed by Wishart, Sykes and Richardson (29, 30) as done in the solution structure determination of other Cdc42 constructs (14, 18). Residual dipolar coupling restraints were generated using a sample of Cdc42(T35A)-GDP that was partially aligned with a nonionic liquid crystalline medium composed of 4% C12E6 [n-dodecyl hexa(ethylene glycol)] with hexanol as has been done previously (14). RDCs were measured from partially aligned and unaligned samples using TROSY experiments with the phase cycling modified to observe the nitrogen components individually. RDC restraints were developed using PALES software (31), with optimal values of Da and R being −9.8, and 0.64, respectively. All restraints were used to calculate an ensemble of structures for the protein using CNS. Twelve “best” structures were selected based on their overall low energies and minimal constraint violations, and these structures have been deposited into the RCSB Protein Data Bank (2kb0.pdb). Table 1 gives the experimental statistics of the structure calculations. The structures were visualized using Swiss PDBviewer (version 3.7) (32), Chimera (33), analyzed using AQUA (34), PROCHECK-NMR (35), and the NMR ensemble program, OLDERADO (36).
Table 1.
Structural Statistics and Restraint Data for the Structure calculation of Cdc42(T35A)
| ensemble of 12 structures | most representative structure | |
|---|---|---|
| RMSD from experimental constraints | ||
| NOE distances (Å) | 0.018 ± 0.004 | 0.018 |
| dihedral angles (deg) | 0.37 ± 0.05 | 0.38 |
| rmsd from secondary structure | ||
| bond (Å) | 0.0015 ± 0.0001 | 0.0015 |
| angles (deg) | 0.33 ± 0.02 | 0.33 |
| impropers (deg) | 0.35 ± 0.02 | 0.34 |
| Restraint Data | |
|---|---|
| long-range NOEs (|i – j| > 4) | 174 |
| medium-range NOEs (4 > |i – j| > 1) | 883 |
| total NOEs | 1057 |
| intra-residue | 457 |
| sequential | 475 |
| medium-range | 169 |
| long-range | 246 |
| hydrogen bond | 142 |
| Ψ | 101 |
| Φ | 100 |
| NH backbone RDC restraints | 74 |
*From the average structure for backbone non-hydrogen atoms: 0.859 ± 0.01
Dynamics Data Analysis
For relaxation studies, T1, T2, and 1H-15N NOE experiments were acquired on both wild type and T35A samples at 500 and 800 MHz (37, 38). The relaxation delays for T1 experiments were 0, 150, 300, 450, 600, 750, 900, and 1050 milliseconds with triplicate experiments recorded for 0 and 450 millisecond delays at 500 MHz. Delays of 0, 150, 500, 750, 1000, 1250, 1500, and 1750 milliseconds with triplicate experiments for 0 and 1000 millisecond delays were collected at 800 MHz. T2 experiments used 5, 15, 25, 40, 55, 70, 85, and 100 millisecond delays triplicates at 5 and 40 millisecond at 500 MHz. Experiments acquired at 800 MHz used 5, 15, 25, 40, 55, 70, and 85 millisecond delays with 5 and 40 millisecond delay experiments repeated. Heteronuclear NOE experiments utilized a 5 second saturation period and 5 second recycle delay. Spectra collected with a 10 second recycle delay were used as references. All NOE experiments were collected in triplicate. T1 and T2 values were obtained by fitting the peak intensities to a single exponential using NMRPipe (26). Heteronuclear NOEs were measured as the ratio of peak intensity from the spectra collected with saturation divided by the volume of the same peak in the reference spectra. The relaxation data sets were subjected to model-free analysis using the software TENSOR (39) and Modelfree4 (40). The diffusion tensors of Cdc42 wild type and T35A were determined using the three-dimensional coordinate files 1aje.pdb and 2kb0.pdb, respectively.
In-Vitro Binding Assays
For in-vitro pull down assays, a peptide derivative of p21-acivated serine/threonine kinase (PAK) that binds to wild type Cdc42, PBD46 (H2N-GSKERPEISLPSDFEHTIHVGFDAVTGEFTGIPEQWARLLQTSNIT-COO−), was expressed and purified (17), as was GST (control). Five-mg aliquots of GST-PBD46 (or GST alone), were mixed with 5 mg aliquots of partner proteins, Cdc42 wild type or Cdc42(T35A), according to quantification, and 0.5 mM of GMPPCP was added in the mixtures. The total mixture volume was brought up to 2 ml by adding PBS buffer. After incubating the mixture for 4hrs at 4°C, 50 μl of Glutathione Sepharose beads (GE Healthcare) were added to precipitate GST-PBD46 (or GST) incubating for 1 hour then the supernatant was discarded, and the beads were washed by PBS buffer at least 3 times. Fifty ml of 10mM Glutathione in 25mM Tris pH8 was used to elute protein on the sepharose beads. 10 μl of each sample of elution was run on 15% SDS-PAGE, and band intensities were measured to calculate the relative amount of complex formed. In all gels, there was an equal density representing a doublet of GSTPBD46 indicating that equal amounts were pulled down by glutathione beads in both mixture of Cdc42 wild type:PBD46 and Cdc42(T35A):PBD46 as has been seen in other in-vitro binding assays using PBD with Cdc42 bound to various other nucleotides (11). The corresponding intensity of the 20-kDa band on a 15% SDS-PAGE gel was measured using a UVP Biospectrum Imaging System. As a negative control, Cdc42 wild type and Cdc42(T35A) were incubated with GST alone, as mentioned above, and the incubated mixture did not show a band for wild type or T35A in the eluted sample indicating that there was specific binding with PBD46 (Data not shown).
Proteolytic Digestion Assay
Chymotrypsin (Sigma-Aldrich), Trichloroacetic acid (TCA), NaCl (BDH), sodium phosphate, and urea (JT Baker) were of analytical or ultrapure grade. The proteolytic experiments on Cdc42 wild type and Cdc42(T35A) were carried out at 25°C using chymotrypsin in a buffer containing 100mM NaCl, 10mM NaH2PO4 at pH = 8.0). Proteolytic digestions were performed in triplicate at an enzyme /substrate (Cdc42 wild type or T35A) ratio of 10:1 (enzyme:protein). The protease activity was stopped after incremental time intervals (20, 40, 50, 60, 70 80, 90, 100, and 110 minutes) by the addition of 100% TCA. After TCA precipitation, 8M urea was added to the protein sample, and the degree of proteolytic cleavage was measured from the intensity of the 20-kDa band on a 15% SDS-PAGE gel corresponding to the uncleaved protein (time = 0) (Cdc42 wild type or T35A), using the UN-SCAN-IT gel 6.1 software (Silk Scientific, Utah, USA). The control band was taken to be 100% to calculate the percentage of cleavage that occurred.
Differential Scanning Calorimetry
Temperature-induced unfolding of Cdc42 wild type vs. Cdc42(T35A) was performed using a Nano-DSC III Differential Scanning Calorimeter (Calorimetry Sciences Corp). One mg/ml solution of Cdc42 (wild type and T35A) was dialyzed extensively against 1× PBS with 0.5mM Mg2+ ??at pH −7.5 at 4° C. After dialysis, each protein sample was degassed before it was heated from 20 – 80°C using a scan rate of 1.0°C/minute The cells containing each protein sample were equilibrated for approximately 10 minutes before each run, and buffer samples without protein were run prior to each experiment to obtain a baseline and subtracted from the protein scan. Each protein sample was scanned in triplicate. The program CpCalc (Calorimetry Sciences Corp.) was used to construct the melting curve and to determine the Tm value.
Results and Discussion
Overall Structure and Backbone Dynamics of Cdc42(T35A)
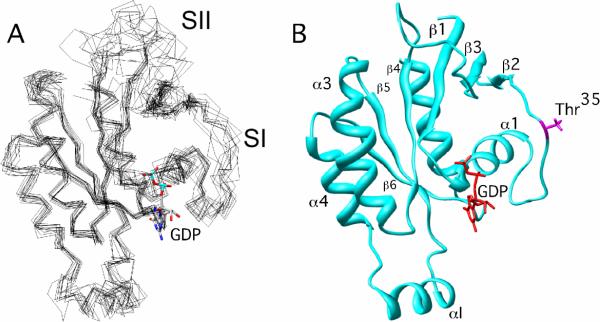
The solution structure of Cdc42(T35A) was characterized to examine conformational differences imparted by the mutation in the Switch I region when the side-chain OH group of residue 35 is removed. An ensemble of the 12 lowest energy structures of Cdc42(T35A) (2kb0.pdb) calculated from NMR-determined distance, hydrogen bonding, dihedral and residual dipolar coupling restraints is shown (Figure 1A). The core of Cdc42(T35A) is comprised of residues 3–11, 15–24, 28–29, 39–46, 48–58, 75–83, 84–103, 109–117, 123–133, 136–150, and 152–177. The core region is similar to Cdc42 wild type (18) and to other Cdc42 mutants, such as Cdc42(F28L) (14). There were no changes observed for the secondary structured regions in Cdc42(T35A) relative to wild type, as Cdc42(T35A) consists of a six-stranded β-sheet structure around its C-terminal α-helix just as in wild type (Figure 1B). As is seen with wild type, and other mutants of Cdc42 (14, 18), the Switch I and II regions, and to a lesser extent, the Insert region (αI), show structural disorder relative to the rest of the protein. The nucleotide-binding site seemed to be unaffected by the mutation, as there were no noticeable differences observed for residues that comprise the nucleotide-binding pocket of Cdc42(T35A) relative to wild type in 15N-HSQC spectra (24). This is in agreement with 31P-NMR studies of H-Ras(T35A) and Cdc42(T35A) that showed very small, if any, changes in the 31P-NMR spectra (11). Therefore, the mutation in Cdc42(T35A) does not seem to alter the overall folding of Cdc42 and the globular structure of Cdc42(T35A) is largely similar to that of wild type Cdc42.
Figure 1.
Structure of Cdc42(T35A)-GDP. The 12 lowest energy structures were superimposed relative to the average structure generated using Xplor-NIH. They were visualized using Chimera (33). The GDP nucleotide was superimposed onto the structure(s) using coordinates from the X-Ray crystal structure of Cdc42 wild-type (1AN0.pdb). (A) All 12 twelve structures overlaid with each other, and (B) The most representative structure, as defined by OLDERADO (36).
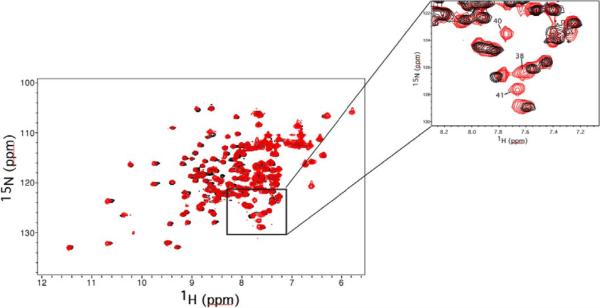
The backbone dynamics for Cdc42(T35A) were characterized from 15N relaxation parameters. Plots of the longitudinal (T1) and transverse relaxation (T2) rates and steady state heteronuclear NOE values are shown in Figure 2. The primary structure of Cdc42(T35A) consists of 166 non-proline residues, and ~72% of the backbone amide resonances for both wild type (119 residues) and T35A (120 residues) were observable. One notable difference was that resonances for residues 40 and 41 were broadened and unobservable in the wild type spectra. This broadening is less significant in the Cdc42(T35A) spectra enabling analysis of NH bond vector internal dynamics for these residues. Due to resonance overlap in both wild type and Cdc42(T35A), there were a number of resonances excluded from the analysis, including residues 35–39 in the Switch I region. However, in contrast to the wild type, the Cdc42(T35A) spectra contained resonances from residues 38, 40, and 41 in Switch I and residues 57, 59, 61, 63, 71, and 74 in Switch II for which internal dynamics parameters could be obtained. The NH resonances from residues 35 and 36 were also observed for Cdc42(T35A); however, they suffered from resonance overlap making confident analysis of relaxation data impossible. Overall, relaxation data from regions with well defined secondary structure of both Cdc42 constructs correspond very well to the previously published data on Cdc42 wild type and Cdc42(F28L) (19, 41). Since the Switch I (residues 28–41) and Switch II (residues 57–74) regions participate in the interaction of Cdc42 with effector proteins, the internal motions of these regions are of particular interest. However, direct comparisons for these regions are not possible due to the resonance broadening and signal overlap in the NMR spectra of wild type Cdc42.
Figure 2.
Relaxation data for Cdc42 wild type (black circles) Cdc42(T35A) (Red circles) measured at 800 MHz. The blue shaded bars indicate residues 28–41 (Switch 1), and the pink shaded bars highlight residues 57–74 (Switch 2). (A) T1, (B) T2, and (C) NOE.
Relaxation data were subjected to a model-free formalism (42), in order to extract parameters describing the internal motion of the individual NH bond vectors. We compared the relaxation data collected at both 500 and 800 MHz for both Cdc42 wild type and Cdc42(T35A) and found that the trends in the plots of the data as a function of residue number agreed very well for both proteins. However, to minimize effects that can arise in the amplification of the CSA term at high field strengths, only the data collected at 500 MHz were used in the dynamics analysis. The τC for Cdc42 wild type was 10.9 +/− 0.3 ns and 11.0 +/− 0.2 ns for Cdc42(T35A). An axially symmetric diffusion tensor fit best, with (Dpar/Dper) for Cdc42 wild type equal to 0.84 +/− 0.02 and 0.71 +/− 0.02 for Cdc42(T35A). Most of the core internal dynamics could be described by models 1 and 2 (S2 or S2, τe) (Figure S1 in supplemental material). S2 order parameter values, which are used to characterize the mobility of backbone N-H bonds, calculated from both TENSOR and Modelfree4 were in agreement for both Cdc42 wild type and Cdc42(T35A). The average per residue magnitude of the difference of S2 between TENSOR and Modelfree4 was 0.031 for wild type and 0.039 for Cdc42(T35A), both being on the same order of the error of S2 determination. S2 values were consistently larger in Cdc42(T35A) relative to the corresponding S2 values determined for wild type (Figure 3). Thus it appeared that some regions in Cdc42(T35A) were more conformationally constrained than wild type with the largest differences occurring in the Switch regions. As listed in Table 2, the overall average S2 for both proteins was ~0.89. However, the range of S2 for the Switch I region was 0.78–0.86 in Cdc42(T35A) compared to 0.60–0.79 for wild type. These results indicated that that the Switch I region of T35A showed some differences in the amplitudes of motion, and/or less conformational freedom, relative to the corresponding region in wild type. NMR studies of Cdc42 in complex with PBD46 (a peptide derivative of p21-activated serine/threonine kinase) also revealed chemical shift changes that were largest within Switch I and II, and the P-Loop (Phosphate-binding loop) (17). These results may account for the inhibition of GTP hydrolysis by PBD46 (17).
Figure 3.
Residue-specific S2 values extracted for Cdc42 wild type (black circles) and Cdc42(T35A) (red circles) from the extended Lipari-Szabo modelfree formalism. The Switch I and II regions are highlighted with a light-green block around the regions. The data shown is from analysis of the relaxation data using TENSOR.
Table 2.
Average values of overall order parameters (S2), and deviations from the average overall S2 for the Switch I and II regions for Cdc42 wild type, and Cdc42(T35A). Both proteins were in the GDP-bound form. Residue ranges of each region is given in the parentheses.
| Cdc42(WT) | Cdc42(T35A) | |
|---|---|---|
| Average S2 protein | 0.89 ± 0.04 | 0.89 ± 0.04 |
| Average Switch I S2(28–41) | 0.73 ± 0.10 | 0.81 ± 0.03 |
| Average Switch II S2(57–74) | 0.85 ± 0.091 | 0.88 ± 0.05 |
| %ΔS2 (Switch I) | 18.5% | 8.6% |
| %ΔS2 (Switch II) | 4.6% | 1.5% |
Where %ΔS2 = |(avgS2protein – avgS2region)/avgS2protein|×100.
Structure and Backbone Dynamics of Switch Regions
Switch I
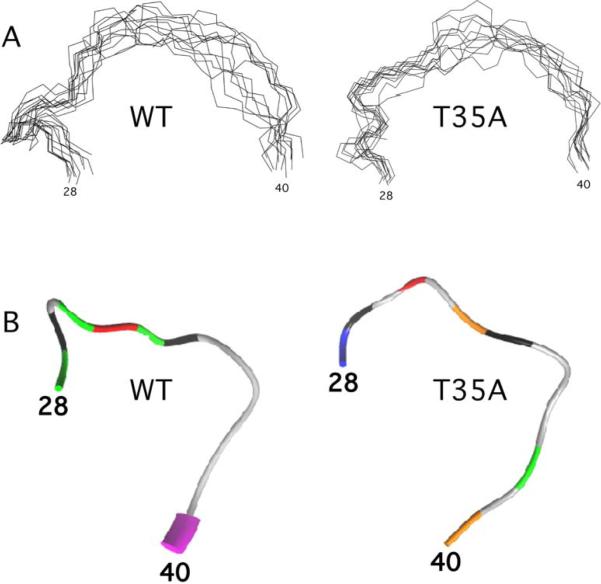
The observation of additional backbone NH resonances suggests that the Switch I region in Cdc42(T35A), while still poorly structured, behaves differently than the corresponding region in wild type (Figure 4). Thr35 in Ras proteins is crucial for binding to effector proteins; however, it may not be directly involved in effector interactions (16). This implies that other residues in the Switch I region may be affected by the T35A mutation, as is observed in the NMR spectra. Shima et al. (43) have shown from X-Ray structures that two conformational states for the Switch I region of H-Ras may account for its ability to interact freely with effector proteins. We were able to observe approximately 1.5 times as many backbone NH NOE restraints in the Switch I region for Cdc42(T35A) (58 total, 23 inter-residue, 8 long-range) relative to wild type (39 total, 20 inter-residue, 3 long-range). Figure 5A depicts only the backbone of the ensemble of structures from Cdc42 wild type (1aje.pdb) and Cdc42(T35A) (2kb0.pdb). A comparison of the backbone RMSD values revealed that Switch I BBRMSDCdc42(T35A) = 1.48 ± 0.065, whereas Switch I BBRMSDCdc42 wild type = 2.95 ± 0.115. As can be seen, the backbone of Switch I from the ensemble of structures, while still random coil in both proteins, highlights a reduced conformational freedom, particularly at the C-terminal end of Switch I (Residues 38–41) for Cdc42(T35A). In addition, we observed new medium to weak NOEs between residues near the C-terminal end of Switch I with residues in and just adjacent to the Switch II region in T35A, for example, one observes with Cdc42(T35A) — but not with wild type — medium and weak backbone NOE's involving 39NH-59-HB, 40HN-55-NH, and a side chain NOE between 39HD-56HZ (aromatic proton). Mutagenesis studies and NMR measurements with Cdc42 and PAK (p21-activated serine/threonine kinase) furthermore have shown that residues 38 and 40 of Switch I are vital to the binding and maximal interaction of the two proteins (7, 44). Both the Switch I region and the β2 region (residues 40–46) following Switch I have been implicated as the interface for effector protein binding with Cdc42 (17). As residues in these regions are not observable on the NMR timescale in Cdc42 wild type, it seems plausible that the fast timescales of motions may result from multiple, interconverting conformations of Switch I made possible by the presence of the −OH group of Thr35 in Switch I, as has been examined previously (24). This could, in turn, lead to the notion that interactions with effectors, such as PAK, are made possible by conformational selectivity (45) for a particular conformation, facilitating the strong binding interactions that occur between effector proteins with wild type Cdc42 (17). Nonetheless, our data highlight an apparent reduced flexibility that may reduce the number of possible conformations for the Switch I region in Cdc42(T35A) when the −OH group of Thr35 is not present. A study of the molecular details of the complex of Cdc42(T35A) and effectors, such as PAK, or GTPase-binding derivatives of PAK, would be useful to confirm these possibilities.
Figure 4.
2D 15N-HSQC of Cdc42wt (black) overlaid with Cdc42T35A (red). The inset (upper right corner of figure) highlights a region of the spectrum containing resonances from three residues on the C-terminal portion of the Switch 1 region of Cdc42T35A (Residues 38, 40 and 41). These resonances are well defined in the Cdc42T35A spectrum, but could not be detected in the HSQC spectrum of Cdc42wt due to excessive line broadening (18).
Figure 5.
A) Stereoview of backbone ensemble of Switch I region of Cdc42 wild type and T35A, and B) Backbone of Switch I region colored based on best-fit dynamics model assignments from dynamics calculations. Model assignments are: white, unassigned: blue, model 1 (S2 only); green, model 2 (S2 and τe); model 3, orange (S2 and Rex); model 4, red (S2, Rex, and τe); model 5, yellow (Sf2, Ss2, and τe).
Spoerner at al. (16) suggested from kinetics and 31P-NMR studies there are two dominant conformational states of Ras, one in the absence and another in the presence of effectors; however, in Ras(T35A) this was not the case. The arguments for two conformational states of the Switch I region were strengthened by X-Ray studies of M-, and H-Ras (43). The increased number of observable resonances in Switch I in Cdc42(T35A) indicates that the conformational flexibility of Switch I in T35A may be more restricted than in Cdc42 wild type. This result could be due to a change in the transition rate and/or populations of the possible conformational states that have been suggested for H- and M-Ras by Shima et al (43).
The “Switch” binding regions in wild type Cdc42 are unstructured and flexible, yet crucial for interactions of Cdc42 with effectors that inhibit important regulatory control mechanisms, such as GTP hydrolysis (17), a property seen also with other Ras proteins (46, 47). Cdc42 behaves similarly to H-, K, and M-Ras, as well as Rap in that, in these Ras proteins, millisecond time scale motions are seen for the Switch I region (15, 48). We characterized the backbone dynamics of Cdc42(T35A) to discover which regions of the protein might be most influenced by the T35A mutation. Figure 5B depicts the models of motion, extracted for the observable residues of Switch I for wild type relative to Cdc42(T35A). Even though the average overall order parameter, S2, is 0.89 for both proteins (Table 2), a residue-by-residue comparison leads to an average S2 difference of −0.06 +/− 0.04 between the two forms, (Cdc42(T35A)-Cdc42 wild type). There were furthermore regions where backbone order parameters differed significantly. As seen in Figure 3, the S2 values for residues observable in both wild type and Cdc42(T35A) in the Switch regions are consistently larger in the mutant. For Cdc42(T35A), a number of observable residues in Switch I have S2 that is near the average value for the entire protein. For wild type Cdc42, some values of S2 are significantly lower than the average S2 value calculated for the protein, particularly NH bond vectors of residues 31 and 32 (32 not measured for T35A) which have values of S2 < 0.65 (Figure 3), with the internal dynamics of NH bond of residue 32 being characterized by model 5 (Ss2, Sf2 and τe). This suggests that the internal dynamics of NH bond vectors of residues following Y32 were probably complicated and contributed to resonance broadening in the NMR spectra of the wild type. The Switch I region shows no residues with an S2 value less than 0.7 in T35A.
To understand the extent of the changes in the backbone dynamics, we calculated %ΔS2 for the observable residues of Switch I relative to the average S2 for each entire protein (Table 2). We observed that S2 was 18.5% lower for the Switch I residues in wild type, but only 8.6% lower for T35A. Previous studies have implicated that Thr mutants in the Switch I region of Ras proteins show a reduced affinity for effector proteins, but yet, Thr35 is not directly involved in protein interactions in this region (16). Our results clearly show that the T35A mutation affects the backbone mobility of the Switch I region of Cdc42. As previous studies suggested, when Ser is present at position 35, two conformational states and a two-state binding model have been implied for Ras wild type and Ras(T35S), but not seen in Ras(T35A) (16). Therefore, the backbone flexibility in Switch I seems likely important for effector interactions for Cdc42 and other Ras proteins, and is lost in Cdc42(T35A).
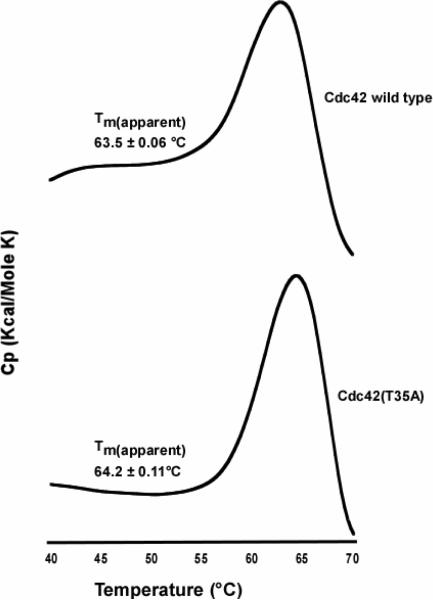
Single point mutations in Switch I involving residues 35, 37 and 40 markedly reduce the association of the Ras protein with effector proteins (49). To test whether the binding of a GTP hydrolysis inhibitory protein is altered for Cdc42(T35A) as postulated from other studies, we performed an in-vitro binding assay of PBD46 with Cdc42 wild type and Cdc42(T35A) by measuring the intensity change of the pull-down band of Cdc42 relative to Cdc42(T35A) in the presence of equilmolar amounts of PBD46 and compared the change in the intensity (ΔI) (Figure 6). The ΔI from wildtype:PBD46 compared to T35A:PBD46 was 4.2±0.59, consistent with previous observations of Cdc42(T35A) showing decreased binding to effector proteins (50). We also examined the overall stability of Cdc42 wild type relative to the Switch I mutant protein using differential scanning calorimetry (Figure 7). The apparent Tm is seemingly unaffected by the mutation as Cdc42(wt)Tm = 63.5±0.06°C and Cdc42(T35A)Tm = 64.2±0.11°C, suggesting no change in the overall conformational stability or folding of the constructs. The finding of similar overall folding and stability, yet reduced conformational flexibility for Switch I in Cdc42(T35A) may help to explain some of the biological consequences of mutations in the Switch I region (49).
Figure 6.
Figure 7.
Representative differential scanning calorimetry thermograms showing the unfolding of Cdc42 wild type (Top panel), and Cdc42(T35A) (Bottom panel). The Tm values are calculated from experiments done in triplicate for both proteins. The protein concentration for each protein was 1.0 mg/mL in buffer as described in the Methods section. Both thermograms were corrected for background noise, and each of the experiments (run in triplicate) were performed at a scan rate of 1.0 °C/min.
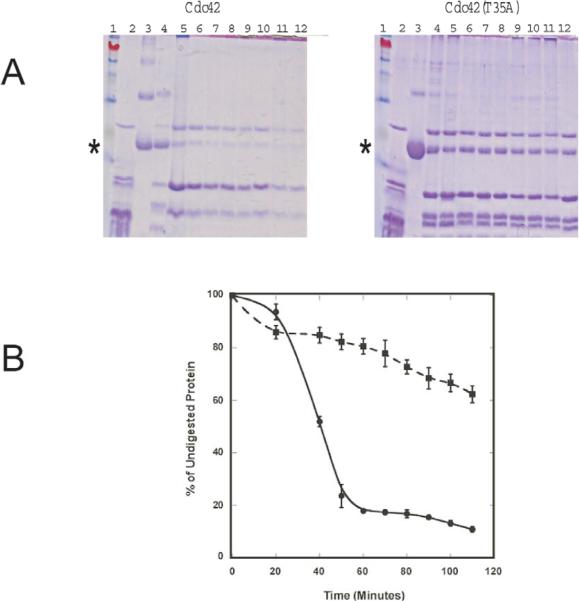
We also examined whether the Switch I region of Cdc42(T35A) would exhibit altered sensitivity to chymotrypsin as 4 of the 13 residues in Switch I are aromatic. We observed that about 90% of the parent band for Cdc42 wild type, but only ~38% of Cdc42(T35A), is cleaved during a 110-minute exposure to chymotrypsin (Figure 8), which suggests that the Cdc42(T35A) construct is somewhat stabilized against chymotrypsin cleavage. This finding is consistent with the structural and dynamics results that Switch I is more conformationally restricted in Cdc42(T35A). In summary, our results point to reduced conformational flexibility for Cdc42(T35A). The reduced flexibility in turn may lead to less efficient interactions with effector proteins.
Figure 8.
A) chymotrypsin digestion products of Cdc42 wild type and Cdc42(T35A). The intensity of the parent Cdc42 protein bands in lane 3 in both gels that were not treated with chymotrypsin (Incubation time with enzyme = 0) and assumed to 100% protein for each gel. Initial concentrations used for each protein were the same. In each gel, corresponding Cdc42 (wild type) or (T35A) bands were normalized to the parent band (Lane 3) at ~20 kDa (indicated by asterisk). Lanes 4–12 in each gel represents the chymotrypsin digestion products obtained after incubation with either Cdc42 or Cdc42(T35A) for 20, 40, 50, 60, 70, 80, 90, 100, and 110 minutes respectively. In each gel, lane 1 represents the MW marker, and lane 2 represents that of chymotrypsin alone to show corresponding bands from the autolysis of chymotrypsin. B) A graphical depiction of the % of undigested Cdc42 wild type (Circles) or Cdc42(T35A) (Squares) incubated with chymotrypsin as a function of time. Error bars represent the standard deviation of triplicate measurements.
Switch II
Switch II, as well as Switch I shows increased conformational flexibility in wild type Cdc42 (19). The Switch II region in Cdc42 is also believed to serve as a part of the binding pocket for effector proteins, such as the Ser/Thr PAK kinase that inhibits GTP hydrolysis. Indeed, chemical shifts could be observed for some residues of Switch II upon binding of effector proteins to Cdc42 (17). The structure and the backbone dynamics of this region are also changed by the T35A mutation in Cdc42. Indeed, several NH backbone resonances can be identified relative to the Switch II region in wild type. In Cdc42(T35A), only one non-proline residue (R68) is unobserved, whereas the wild type is missing resonances for 59, 60, 66, 67, 68, 71, 72, and 74. The NOE data for Switch II resulted in more than three times as many observed backbone NH NOEs in Cdc42(T35A) (76 total, 45 inter-residue, 15 long-range) relative to wild type (24 total, 13 inter-residue, 2 long-range). The region is random coil, however, unlike Switch I in T35A, the backbone residue of Switch II display similar conformational freedom as the region in wild type, as most of the NOEs are sequential and there are no NOEs seen in Switch II of Cdc42(T35A) that are more than 2 residues apart in the primary sequence.
The dynamics of Switch II of Cdc42(T35A) (See Figure 3) reveal comparisons of S2 values for wild type and Cdc42(T35A). Thirteen residues in Switch II of wild type have either broad, barely visible, or unassigned peaks; compared to only nine in T35A. As was true to Switch I, many Switch II residues have S2 values that are higher in Cdc42(T35A) than in wild type. In Switch II, many of the NH bond vectors are best described by model 3 (S2 and Rex), with contributions of motions on the μs-ms time scale. The S2 values for observable residues in Switch II for both proteins (Table 2) show very little change relative to the overall S2 values, as there is only a 4.6% change in wild type, and a 1.5% change in Cdc42(T35A). The small changes suggest that, while important for regulatory and effector interactions in Ras-related proteins, Switch II may serve more of a complementary role in effector binding, whereas the functionally significant binding region is likely Switch I (16, 17, 43). Solution structures studies have shown that when the PAK derivative, PBD46, binds to Cdc42, the Switch II region shows some chemical shift changes, but there are no NOE constraints between the effector peptide and Switch II (17, 21). X-Ray studies have suggested that the primary importance of Switch II is direct interaction with the phosphate-binding or P-Loop, as residues 59–61 show contacts with the γ-PO4 in Ras and Cdc42 (10, 51). These studies also argued that GTP hydrolysis results in the loss of stabilizing interactions with residues 59–61 when the γ-PO4 group leaves, suggesting that Switch II may provide stability to the nucleotide cavity (51, 52). In addition, structural studies using other transforming mutants have implicated a series of congruent hydrophobic interactions between Switch II and the nucleotide (53). Recent 31P-NMR studies showed significant chemical shift changes upon PBD46 binding for wild type (γ–31P shift 7.473 free vs. 6.740 bound), whereas for Cdc42(T35A) the chemical shift difference was negligible (γ–31P shift 7.808 free vs. 7.844 bound) (11). Our data suggests that, for Cdc42(T35A), the dynamics and conformational changes observed for Switch I might facilitate the weakened binding of PBD46, and thus the Switch II–γPO4 interaction in Cdc42(T35A) is minimally disturbed.
X-Ray studies of transforming Switch II mutants (such as Q61) of Ras have suggested a more ordered Switch II region than that in wild type Ras (53, 54). The studies also suggested that, in each Switch II mutant studied, Switch I and II come closer together forming a more hydrophobic pocket around the nucleotide. In-vitro binding studies using the Q61 mutant of Cdc42 with two Ras effector proteins (IQGAP1 and 2) suggested that both proteins could bind at various concentrations to this Cdc42 mutant, but that at least two Switch I mutations (Y32K and T35A) lead to no appreciable binding (50). Moreover, structural studies of Cdc42 with Cdc42GAPs have highlighted several Switch II residues (residues 61–64) that are oriented toward Switch I residues, and also make direct contacts with GAPs (55). Therefore, while Switch II may not be critical for the optimal binding of all Ras-related effectors, Switch II is nevertheless likely to be important in facilitating interactions with Switch I that indirectly support the binding of some effectors. The conformational freedom exhibited by both of these regions likely helps in effector identification for the wild type protein. We are not able to compare through-space distances between Switch I and II in wild type Cdc42 relative to Cdc42(T35A), primarily due to the lack of observable resonances for some Switch residues in wild type. However, it seems plausible, in part due to the presence of long-range NOEs between portions of the two regions, that the slowed dynamics caused by the T35A mutation in Switch I facilitates interactions between portions of Switch I and Switch II that do not allow optimal interaction with effector proteins, in part because of some lost entropic component in the mutant construct.
The backbone dynamics in the Switch II region in Cdc42 wild type and T35A are similar to that of Cdc42 as a whole. However, our studies imply that the formation of the binding pocket involving the backbone residues of Switch I and II is altered, and perhaps in a more packed environment in T35A than for wild type. The major change again is the reduced conformational flexibility of Switch I in this mutant. The minimal changes seen in the backbone dynamics of Switch II suggest a minimal or indirect role for this region in terms of effector interactions. A more systematic investigation of Switch II, via site-directed mutagenesis, in Cdc42 with PBD46, should help confirm this hypothesis.
Clearly the Switch regions have important features necessary for proper effector binding, and Switch I has been shown to be largely responsible for the regulation of effector binding that could potentially influence cell signaling. Cdc42 mutations that alter the intrinsic GTPase activity (9), facilitate antiapoptotic events (56), or alter interactions with regulatory proteins (12), are all involved in aberrant signaling activity. These findings have their foundation in the impact of the Switch regions on the effector interactions of Ras proteins. Therefore, Cdc42 has been and will continue to be an excellent model to probe new aspects of structure-function relationships with regard to cell signaling processes. Our characterization of the solution structure and dynamics of Cdc42(T35A) have outlined molecular details of the Switch regions, particularly Switch I, that likely account for the importance of this region in faciliatating interactions with a number of Ras-related effectors.
The Ras GTPases are promising drug targets for anti-cancer therapy, presumably due to the high abundance of abnormal cell-signaling activities produced by mutation. Progress is evident in the development of approaches to target aberrantly functioning Ras proteins. For example, small molecule inhibitors have shown promise in impacting Ras-related cell death by inhibiting transformation events by preventing Ras proteins from attaching to the cell membrane; however, these drugs have also shown differences in selectivity for individual Ras proteins (57, 58). Also, progress has been made in the design of small molecule inhibitors to block interactions of Rho GTPases with GEFs via screening of large databases (59, 60). Recently, it was shown that Ras signaling in transgenic mouse models could be altered by hindering RAS-protein interactions using an antibody fragment, and that this blocked Ras protein interaction resulted in the regression of tumor growth (61). Characterization of human papilloma virus protein complexes also has suggested that alternative mechanisms to block HPV-related infections should include the development of ways to inhibit protein interactions (62). These findings highlight the importance of understanding Ras-related protein interactions that contribute to abnormal cell signaling activities that lead to proliferation and transformation. New approaches to inhibiting the transformation capability of Ras proteins should involve considerations of dynamics as well as structure. The results of our study of the solution structure and backbone dynamics of Cdc42(T35A) suggest that, in addition to strategies to block Ras-protein interactions with small molecules, the development of strategies to restrict the conformational flexibility of regions of Ras that interact with effectors may be a beneficial way to target Ras-related abnormal cell signaling activity. One such approach could involve the use of molecules to catalyze the formation of covalent bonds to “tie down” important interacting regions. Efforts are underway in our laboratory to develop approaches to restrict the conformational freedom of Switch I of Cdc42 without altering its overall globular conformation. This study foreshadows the importance of understanding how aberrant Ras-related protein interactions might be controlled via restriction of conformational flexibility of the Switch region(s).
Supplementary Material
Acknowledgements
We thank Drs. Roger Koeppe II, Colin Heyes, and Robert Oswald for critical review of the manuscript.
The authors thank the National Institutes of Health Grants 1K-01-CA113753 and ARRA/3K01CA113753 to P.D.A., NCRR COBRE Grant 1P30RR031154-01, and the Arkansas Biosciences Institute for financial support. OS was supported by an Undergraduate Research Fellowship from the State of Arkansas.
Abbreviations
- Cdc42
Cell division cycle 42
- T35A
Threonine 35 to Alanine
- C12E6
[n-dodecyl hexa(ethylene glycol)]
- D2O
Deuterium oxide
- PAK
P(21)-activated kinase
- PBD46
PAK Binding Domain 46
- IPTG
Isopropyl β-D-thiogalactopyranoside
- BMRB
Biological Magnetic Resonance Data Bank
- HSQC
Heteronuclear Single Quantum Coherence
- TOCSY
Total Correlation Spectroscopy
- NOE
Nuclear Overhauser Effect
- INEPT
Insensitive Nuclei Enhanced by Polarization
- RDCs
Residual Dipolar couplings
- TROSY
Transverse Relaxation Optimized Spectroscopy
- GMPPCP
Guanosine-5'-[(α, β)-methyleno]triphosphate
Footnotes
SUPPORTING INFORMATION AVAILABLE. Figure S1. Best-fit dynamics model assignment as a function of residue for Cdc42 wild type and Cdc42(T35A). Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
REFERENCES
- 1.Barbacid M. ras Genes. Ann. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 3.Bourne HR, A. SD, F. M. The GTPase superfamily: Conserved Structure and Molecular Mechanism. Nature. 1991:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 4.Hart M, Eva A, Evans T, Aaronson S, Cerione R. Catalysis of Guanine Nucleotide Exchange on the Cdc42Hs Protein by the dbl Oncogene Product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 5.Horii Y, Beeler JF, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. Embo J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart M, Shinjo K, Hall A, Evans T, Cerione RA. Identification of the Human Platelet GTPase Activating Protein for the CDC42Hs Protein. J. Biol. Chem. 1991;266:20840–20848. [PubMed] [Google Scholar]
- 7.Leonard D, Hart MJ, Platko JV, Eva A, Henzel W, Evans T, Cerione RA. The identification and characterization of a GDP-dissociation inhibitor (GDI) for the CDC42Hs protein. J Biol Chem. 1992;267:22860–22868. [PubMed] [Google Scholar]
- 8.Bos JL, Toksoz D, Marshall CJ, Verlaan-de Vries M, Veeneman GH, van der Eb AJ, van Boom JH, Janssen JW, Steenvoorden AC. Amino-acid substitutions at codon 13 of the N-ras oncogene in human acute myeloid leukaemia. Nature. 1985;315:726–730. doi: 10.1038/315726a0. [DOI] [PubMed] [Google Scholar]
- 9.Miller A-F, Halkides C, Redfield A. An NMR Comparison of the Changes Produced by Different Guanosine 5'-Triphosphate Analogs in Wildtype and Oncogenic Mutant p21ras. Biochemistry. 1993;32:7367–7376. doi: 10.1021/bi00080a006. [DOI] [PubMed] [Google Scholar]
- 10.Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of h-ras p21 at 1.35 Å resolution: implication for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips MJ, Calero G, Chan B, Ramachandran S, Cerione RA. Effector proteins exert an important influence on the signaling-active state of the small GTPase Cdc42. J Biol Chem. 2008;283:14153–14164. doi: 10.1074/jbc.M706271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 13.Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- 14.Adams PD, Oswald RE. Solution structure of an oncogenic mutant of Cdc42Hs. Biochemistry. 2006;45:2577–2583. doi: 10.1021/bi051686g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geyer M, Schweins T, Herrmann C, Prisner T, Wittinghofer A, Kalbitzer HR. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996;35:10308–10320. doi: 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]
- 16.Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci U S A. 2001;98:4944–4949. doi: 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo W, Sutcliffe MJ, Cerione RA, Oswald RE. Identification of the binding surface on Cdc42Hs for p21-activated kinase. Biochemistry. 1998;37:14030–14037. doi: 10.1021/bi981352+. [DOI] [PubMed] [Google Scholar]
- 18.Feltham JL, Dotsch V, Raza S, Manor D, Cerione RA, Sutcliffe MJ, Wagner G, Oswald RE. Definition of the switch surface in the solution structure of Cdc42Hs. Biochemistry. 1997;36:8755–8766. doi: 10.1021/bi970694x. [DOI] [PubMed] [Google Scholar]
- 19.Loh AP, Guo W, Nicholson LK, Oswald RE. Backbone dynamics of inactive, active, and effector-bound Cdc42Hs from measurements of (15)N relaxation parameters at multiple field strengths. Biochemistry. 1999;38:12547–12557. doi: 10.1021/bi9913707. [DOI] [PubMed] [Google Scholar]
- 20.Mott HR, Owen D, Nietlispach D, Lowe PN, Manser E, Lim L, Laue ED. Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature. 1999;399:384–388. doi: 10.1038/20732. [DOI] [PubMed] [Google Scholar]
- 21.Rittinger K, Walker PA, Eccleston JF, Nurmahomed K, Owen D, Laue E, Gamblin SJ, Smerdon SJ. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 22.Gladfelter AS, Moskow JJ, Zyla TR, Lew DJ. Isolation and characterization of effector-loop mutants of CDC42 in yeast. Mol Biol Cell. 2001;12:1239–1255. doi: 10.1091/mbc.12.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T, Rabbitts TH. Interfering with RAS-effector protein interactions prevent RAS-dependent tumour initiation and causes stop-start control of cancer growth. Oncogene. 2010;29:6064–6070. doi: 10.1038/onc.2010.346. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Oswald RE. NMR assignment of Cdc42(T35A), an active Switch I mutant of Cdc42. Biomolecular NMR Assignments. 2007;1:225–227. doi: 10.1007/s12104-007-9062-8. [DOI] [PubMed] [Google Scholar]
- 25.States D, Haberkorn R, Ruben D. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn. Reson. 1982;48:282–292. [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Dunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: A New Software suite for Macromolecular Structure Determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 31.Zweckstetter M, Bax A. Prediction of Sterically Induced Alignment in a Dilute Liquid Crystalline Phase: Aid to Protein Structure Determination by NMR. Journal of the American Chemical Society. 2000;122:3791–3792. [Google Scholar]
- 32.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 35.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 36.Kelley LA, Sutcliffe MJ. OLDERADO: On-Line Database of Ensemble Representatives And DOmains. Protein Sci. 1997;6:2628–2630. doi: 10.1002/pro.5560061215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kay L, Nicholson L, Delaglio F, Bax A, Torchia D. Pulse sequences for removal of the effects of cross-correlation between dipolar and chemical-shift anisotropy relaxation mechanism on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 1992;97:359–375. [Google Scholar]
- 38.Kay L, Torchia D, Bax A. Backbone Dynamics of Proteins as Studied by 15N Inverse Detected Heteronuclear NMR Spectroscopy: Application to Staphylococcal Nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 39.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 40.Palmer A. ModelFree. v. 4.0. 1998. http://cpmcnet.columbia.edu/dept/gsas/biochem/labs/palmer. [Google Scholar]
- 41.Adams PD, Loh AP, Oswald RE. Backbone dynamics of an oncogenic mutant of Cdc42Hs shows increased flexibility at the nucleotide-binding site. Biochemistry. 2004;43:9968–9977. doi: 10.1021/bi0490901. [DOI] [PubMed] [Google Scholar]
- 42.Lipari G, Szabo A. Model-free Approach to the Interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 1. Theory and Range of Validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 43.Shima F, Ijiri Y, Muraoka S, Liao J, Ye M, Araki M, Matsumoto K, Yamamoto N, Sugimoto T, Yoshikawa Y, Kumasaka T, Yamamoto M, Tamura A, Kataoka T. Structural basis for conformational dynamics of GTP-bound Ras protein. J Biol Chem. 2010;285:22696–22705. doi: 10.1074/jbc.M110.125161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenstroem P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce action polymerization and G1 cell cycle progression independently of p65 and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 45.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc Natl Acad Sci U S A. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, Rosen MK. Structure of Cdc42 in complex with the GTPase-binding domain of the `Wiskott-Aldrich syndrome' protein. Nature. 1999;399:379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 47.Gizachew D, Guo W, Chohan KK, Sutcliffe MJ, Oswald RE. Structure of the complex of Cdc42Hs with a peptide derived from P-21 activated kinase. Biochemistry. 2000;39:3963–3971. doi: 10.1021/bi992646d. [DOI] [PubMed] [Google Scholar]
- 48.Liao J, Shima F, Araki M, Ye M, Muraoka S, Sugimoto T, Kawamura M, Yamamoto N, Tamura A, Kataoka T. Two conformational states of Ras GTPase exhibit differential GTP-binding kinetics. Biochem Biophys Res Commun. 2008;369:327–332. doi: 10.1016/j.bbrc.2008.01.169. [DOI] [PubMed] [Google Scholar]
- 49.Fiordalisi JJ, Holly SP, Johnson RL, 2nd, Parise LV, Cox AD. A distinct class of dominant negative Ras mutants: cytosolic GTP-bound Ras effector domain mutants that inhibit Ras signaling and transformation and enhance cell adhesion. J Biol Chem. 2002;277:10813–10823. doi: 10.1074/jbc.M107684200. [DOI] [PubMed] [Google Scholar]
- 50.McCallum SJ, Wu WJ, Cerione RA. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- 51.Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine nucleotide binding domain of the Ha-Ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 52.Stouten PF, Sander C, Wittinghofer A, Valencia A. How does the switch II region of G-domains work? FEBS Lett. 1993;320:1–6. doi: 10.1016/0014-5793(93)81644-f. [DOI] [PubMed] [Google Scholar]
- 53.Buhrman G, Wink G, Mattos C. Transformation efficiency of RasQ61 mutants linked to structural features of the switch regions in the presence of Raf. Structure. 2007;15:1618–1629. doi: 10.1016/j.str.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krengel U, Schlichting L, Scherer A, Schumann R, Frech M, John J, Kabsch W, Pai E, Wittinghofer A. Three-Dimensional Structures of H-Ras p21 Mutants: Molecular Basis for their Inability to Function as Sifnal Switch Molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 55.Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- 56.Tu SS, Wu WJ, Yang W, Nolbant P, Hahn K, Cerione RA. Antiapoptotic Cdc42 mutants are potent activators of cellular transformation. Biochemistry. 2002;41:12350–12358. doi: 10.1021/bi026167h. [DOI] [PubMed] [Google Scholar]
- 57.Lebowitz PF, Prendergast GC. Non-Ras targets of farnesyltransferase inhibitors: focus on Rho. Oncogene. 1998;17:1439–1445. doi: 10.1038/sj.onc.1202175. [DOI] [PubMed] [Google Scholar]
- 58.Schmidmaier R, Baumann P, Simsek M, Dayyani F, Emmerich B, Meinhardt G. The HMG-CoA reductase inhibitor simvastatin overcomes cell adhesion-mediated drug resistance in multiple myeloma by geranylgeranylation of Rho protein and activation of Rho kinase. Blood. 2004;104:1825–1832. doi: 10.1182/blood-2003-12-4218. [DOI] [PubMed] [Google Scholar]
- 59.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nassar N, Cancelas J, Zheng J, Williams DA, Zheng Y. Structure-function based design of small molecule inhibitors targeting Rho family GTPases. Curr Top Med Chem. 2006;6:1109–1116. doi: 10.2174/156802606777812095. [DOI] [PubMed] [Google Scholar]
- 61.Yagi R, Tanaka M, Sasaki K, Kamata R, Nakanishi Y, Kanai Y, Sakai R. ARAP3 inhibits peritoneal dissemination of scirrhous gastric carcinoma cells by regulating cell adhesion and invasion. Oncogene. 2010 doi: 10.1038/onc.2010.522. [DOI] [PubMed] [Google Scholar]
- 62.Chi CN, Bach A, Engstrom A, Stromgaard K, Lundstrom P, Ferguson N, Jemth P. Biophysical Characterization of the Complex between Human Papillomavirus E6 Protein and Synapse-associated Protein 97. J Biol Chem. 2011;286:3597–3606. doi: 10.1074/jbc.M110.190264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.