Abstract
Nova onconeural antigens are neuron-specific RNA-binding proteins implicated in paraneoplastic opsoclonus-myoclonus-ataxia (POMA) syndrome. Nova harbors three K-homology (KH) motifs implicated in alternate splicing regulation of genes involved in inhibitory synaptic transmission. We report the crystal structure of the first two KH domains (KH1/2) of Nova-1 bound to an in vitro selected RNA hairpin, containing a UCAG-UCAC high-affinity binding site. Sequence-specific intermolecular contacts in the complex involve KH1 and the second UCAC repeat, with the RNA scaffold buttressed by interactions between repeats. While the canonical RNA-binding surface of KH2 in the above complex engages in protein-protein interactions in the crystalline state, the individual KH2 domain can sequence-specifically target the UCAC RNA element in solution. The observed anti-parallel alignment of KH1 and KH2 domains in the crystal structure of the complex generates a scaffold that could facilitate target pre-mRNA looping upon Nova binding, thereby potentially explaining Nova’s functional role in splicing regulation.
The generation of a versatile repertoire of functionally diverse proteins is critically dependent on alternate splicing of pre-mRNAs. Alternate splicing of the subunits of all the main neurotransmitter receptors can influence their localization, ligand-binding, signal-transducing and electrophysiological properties. Aberrant splicing of mRNAs in highly specialized cells, such as neurons, can impact proteins critical for neuronal function, leading to the onset of neurological disease (Dredge and Darnell, 2003; Dredge et al., 2001; Licatalosi and Darnell, 2006). The proteins of the Nova (neuro-oncological ventral antigen) family, which are exclusively expressed in central nervous system (CNS) neurons, are important regulators of neuronal RNA metabolism. The Nova proteins are target antigens in the autoimmune disorder paraneoplastic opsoclonus-myoclonus ataxia (POMA) (Musunuru and Darnell, 2001). POMA is a neurodegenerative syndrome that originates when systemic malignant tumors express proteins normally sequestered in the central nervous system (Albert and Darnell, 2004). The immune system recognizes these antigens to be non-self, and the ensuing response results in neuronal degeneration. Homozygous Nova-1 knockout mice appear to exhibit normal gestation and development, but the resulting litters show apoptosis of motor neurons in the brain stem and spinal cord and become incapacitated by progressive motor system failure (Jensen et al., 2000a).
K-homology (KH) domains are RNA-binding elements (Grishin, 2001; Siomi et al., 1993), identified in hnRNP proteins involved in RNA stabilization and translational control (Burd and Dreyfuss, 1994; Ostareck-Lederer et al., 1998; Valverde et al., 2008). Nova-1 and Nova-2 proteins each contain three proteolytically stable KH domains (Figure 1A), connected by flexible linkers. The crystallographic structures of Nova KH3 domains exhibit a three-stranded anti-parallel β-sheet backed by three α-helices (Lewis et al., 1999). RNA targets for the Nova KH3 domain (and adjacent C-terminal residues) have been identified by in vitro selection experiments (Jensen et al., 2000b). The optimal RNA target exhibits a YCAY tetranucleotide element (Y is a pyrimidine) as part of an accessible 12-residue loop segment within the context of a RNA hairpin. The crystal structure of this complex has been determined at 2.4 Å resolution, with the majority of the intermolecular contacts restricted to the extended UCAC segment of the RNA loop, which is pinioned between the invariant Gly-X-X-Gly motif and a variable loop of the KH domain (Lewis et al., 2000). This CA segment, which is most intolerant to mutations, is recognized by Watson-Crick-like hydrogen bonding patterns from protein residues, while the flanking nucleotides are restricted to pyrimidine residues.
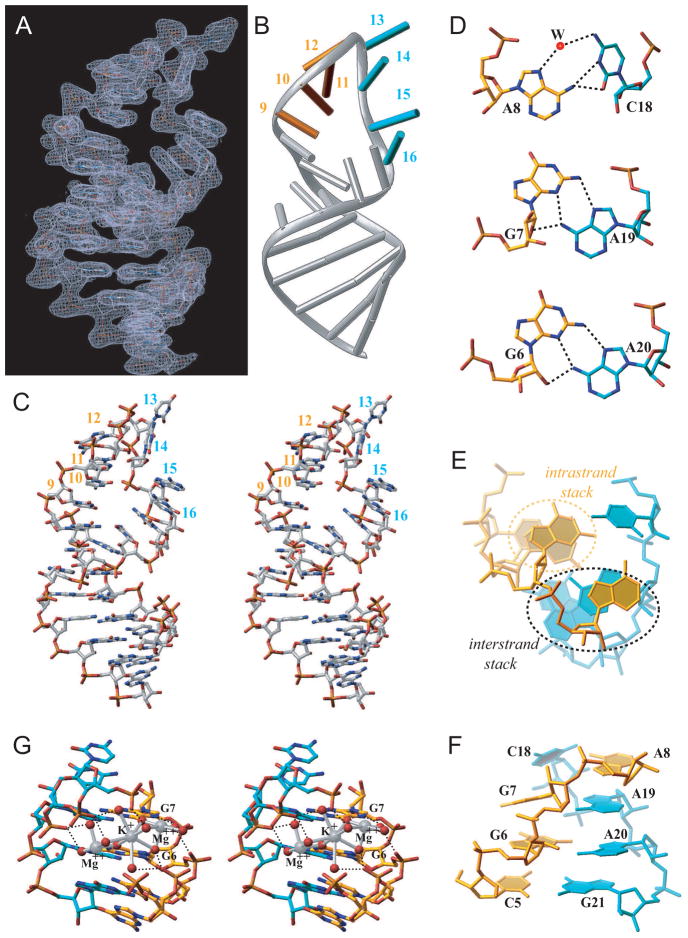
Figure 1. Sequences of Nova KH Domains and RNA Hairpin Target and the Structure of the Nova-1 KH1/2-RNA Hairpin Complex in the crystal.
(A) Schematic of three KH domains and intervening linkers in Nova proteins. Red-colored numbers indicate the length of linker segments in Nova-1.
(B) The Nova-1 KH1/2 construct used in the current project.
(C) The sequence of the in vitro selected 25-mer RNA hairpin. The tandem UCAN sites are colored in gold (U9 to G12) and cyan (U13 to C16). Structural studies were also undertaken on a sequence containing a 5BrU2•A24 pair.
(D) Sequence and secondary structure (Lewis et al, 1999) alignments of Nova KH1, KH2 and KH3 domains, with conserved residues shown in red. The GXXG motif and so-called variable loop, are denoted as I and V, respectively.
(E) Ribbon representation of the structure of the Nova-1 KH1/2-RNA hairpin complex in the crystal. The stoichiometry is KH1/2:RNA hairpin of 2:2. The color codes are RNA in green, KH1 in gold and KH2 in cyan. The UCAC motif interacting with KH1 is colored in blue.
See also Table S1, Figures S1, S2 and S3.
Full-length Nova proteins containing three KH domains have been shown to target and regulate alternative splicing events within neuronal pre-mRNAs which contain repeats of the (YCAY) binding element (Buckanovich and Darnell, 1997; Dredge and Darnell, 2003; Dredge et al., 2005; Licatalosi et al., 2008; Ule et al., 2006; Zhang et al., 2010). RNA aptamers generated against full length Nova protein adopt a RNA hairpin loop scaffold containing a conserved [YCAY-(N)0–2]3 sequence in the loop segment (Buckanovich and Darnell, 1997; Yang et al., 1998). The published structure of the Nova KH3-RNA hairpin complex (Lewis et al., 2000) does not address how multiple KH domains of Nova proteins might cooperate in recognizing tandem YCAY repeat modules. To address this issue in a systematic manner, we set out to determine the structure of a Nova complex containing adjacent KH1 and KH2 domains (designated KH1/2, Figure 1B) targeted to a pair of tandem YCAY RNA sites. Such an opportunity was available since in vitro selection experiments had identified an RNA hairpin architecture (Figure 1C), containing a potential 15-mer loop within which is nestled an UCAG-UCAC segment, which binds the Nova-1 KH1/2 construct (Figure 1B) with high affinity (Musunuru and Darnell, 2004). Mutagenic analysis established that both CA steps within the UCAG-UCAC segment are most intolerant to mutations and therefore represent key residues involved either in protein recognition, and/or in buttressing the recognition interface.
We now report the crystal structure of the Nova-1 KH1/2 construct bound to the UCAG-UCAC segment-containing RNA hairpin. Strikingly, the RNA hairpin is targeted solely by the KH1 domain, with both loop UCAG and UCAC tetranucleotide sites contributing to recognition. Both the crystal structure and binding studies establish that the KH2 domain does not interact with the UCAG-UCAC-containing RNA hairpin. In the crystal, the RNA-binding surface of the KH2 domain instead engages in protein-protein interactions with symmetry-related KH2 domain. By contrast, NMR chemical shift perturbations, gel-shift, and ITC binding studies demonstrate that the KH2 domain does target UCAY tetranucleotide-containing RNAs in a sequence specific manner in solution. Nova’s functional role in splicing regulation could be explained by the observed anti-parallel alignment of KH1 and KH2 domains in the crystal structure of the complex, thereby providing an ideal scaffold to induce target pre-mRNA looping upon Nova binding.
Results
Crystallization and Structure Determination of Complex
A Nova-1 KH1/2-expression construct (Figure 1B) was engineered lacking a 24-amino acid segment between the KH1 and KH2 domains (dashed line in Figure 1A), which is encoded by an alternatively spliced exon. The protein was expressed and purified as described in Supplemental Experimental Procedures. RNA in vitro selection carried out previously against this Nova-1 KH1/2 construct identified a RNA hairpin composed of a 5-base pair stem and a 15-nucleotide loop containing conserved eight-base UCAG-UCAC sequence (Figure 1C) (Musunuru and Darnell, 2004).
We crystallized complexes of Nova-1 KH1/2 protein with two 25-nucleotide RNA aptamer hairpins, which differed at the 2•24 base pair position in the stem segment. One of the RNA hairpins (type I) contained a G2•C24 pair (Figures 1C and S1A), while the other (type II) contained a 5BrU2 A24 pair (Figure S1B). The complexes of Nova-1 KH1/2 with type I and type II RNA hairpins were both crystallized in space group C2, with similar unit cell dimensions, except for a 10.5% difference in the unit cell c dimension (Table S1). Structures were determined via molecular replacement (MR) using the published structure of a KH domain from the Nova-2 KH3-RNA aptamer hairpin complex (Lewis et al., 2000) as a search model (see Supplemental Experimental Procedures). The KH1 and KH2 domains in the Nova-1 KH1/2-RNA hairpin complex adopt the same βααββα topology (Figure 1D), reported earlier for the KH3 domain of Nova-1 and Nova-2 in the free state (Lewis et al., 1999) and in the Nova-2 KH3-RNA hairpin complex (Lewis et al., 2000). The superimposed Nova-1 KH1 and KH2 domains in the RNA bound state exhibit a pair-wise Cα r.m.s.d. of 1.1 Å for residues 6–79 (excluding residues 44–52 of the variable loop). The asymmetric unit contains one KH1/2 molecule and one RNA molecule, with similar overall packing arrangement for both complexes. The differences in the structures of type I and type II complexes are outlined in the Supplemental Information and Figure S1.
Stoichiometry of Complex in Crystal and in Solution
The crystal structure of two Nova-1 KH1/2-RNA hairpin complex molecules related by two-fold crystallographic symmetry is shown in Figure 1E. The KH1 (gold) and KH2 (cyan) domains are aligned in an anti-parallel orientation. The UCAG-UCAC-containing RNA hairpin (green) is targeted solely by the KH1 domain, while the canonical RNA-binding surface of KH2 engages in protein-protein interactions with symmetry-related KH2. The packing arrangement of KH1/2-RNA hairpin complexes in the crystal and the details of KH2-KH2 interface are outlined in the Supplemental Information and Figure S2.
In contrast, analytical ultracentrifugation measurements indicate that KH1/2 exists as a monomer in solution at 10 to 200 μM concentration range, and the KH1/2-RNA complex sediments as a 1:1 complex under these conditions (for details see Supplemental Information and Figure S3).
RNA Hairpin Architecture in the Bound State
The entire UCAG-UCAC-containing RNA hairpin (Figure 2A) can be traced in the crystal structure of the type II complex, while the 5′-C1-G2 segment is disordered in the crystal structure of the type I complex. The simulated annealing 2Fo−Fc omit map of the UCAC RNA site at 1σ level for the type I complex is shown in Figure S1C. The conformation of the UCAG-UCAC is of greatest interest, due to the potential of each tetranucleotide element serving as a KH domain binding site. All four-hairpin residues within the gold-colored U9 to G12 segment (Figure 2B) form a stacked array. By contrast, residues U13 and C14 loop out, while residues A15 and C16 form a stacked pair within the cyan-colored U13 to C16 segment (Figure 2B). The conformation of the bound RNA hairpin is shown in stereo in Figure 2C and establishes extensive stacking interactions that propagates from the paired stem segment into both arms of the hairpin, with the stacking discontinuities restricted to the G12 to A15 segment.
Figure 2. RNA Hairpin Architecture, including Hydrogen-bonding, Stacking, and Cation Coordination, in the KH1/2-RNA Hairpin Complex.
(A) The RNA hairpin portion of the final |2Fo−Fc| electron density map at 1σ level.
(B) The same view of the bound RNA hairpin in a ribbons-and-stick representation. Gold and cyan sticks correspond to U9 to G12 and U13 to C16 segments.
(C) Stereo view of the bound RNA hairpin in the complex. Note unusual positioning of one phosphate group (C14-A15 step).
(D) Pairing alignments of non-canonical A8•C18, G7•A19 and G6•A20 pairs.
(E) Base stack overlap patterns looking down the helical stem axis, highlighting intra-strand and inter-strand overlap patterns.
(F) Cross-strand stacking between A8 and A19 within the zippered-up stem in the complex.
See also Figure S1.
(G) Stereo pair highlighting hydrated Mg2+ and K+ cations positioned within the major groove of the zippered up stem segment. The cations are shown as large silver spheres while the water oxygens and O6 groups of G6 and G7 are shown as small red spheres. Hydrogen bonds are shown as black dashed lines, while ion coordination are shown as silver bonds.
The Watson-Crick paired stem of the RNA hairpin zippers up through formation of G6•A20, G7•A19 and A8•C18 non-canonical pairs (Figure 2D). The two non-canonical G•A pairs are of the sheared type, while the A8•C18 non-canonical pair is buckled and involves bifurcated and water-mediated hydrogen bonds. The small twist angle at the G6–G7 step and the large twist angle at the G7-A8 step, results in a pronounced cross-strand stack between residues A8 and A19 (Figures 2E and 2F). This results in a continuous helical stack that spans G25 to A19 and A8 to G12. The other continuous helical stack spans C1 to G7 and C18 to A15. The zippered-up helical segment involving the three non-canonical pairs, is further stabilized by a pair of hydrated divalent Mg2+ ions and a monovalent K+ ion (with partial occupancies), as shown in stereo in Figure 2G.
Structure of Interface between Nova KH1 and KH2 domains in Complex
The KH1 and KH2 domains align in an anti-parallel orientation across the KH1/2 interface, with interdomain contacts involving segments of α3 and β1 within each domain (colored pink, Figure 3A). The three-stranded anti-parallel β-sheets are on the same side of the protein, thereby forming a continuous six-stranded β-sheet (Figure 3B). The β1 strands are highly twisted relative to each other and the resultant β-sheet between them is stabilized by NH••O main chain hydrogen bonds involving Ile11(KH2) ••Lys9(KH1) and Leu11(KH1) ••Lys9(KH2) (Figure 3C). The KH1/2 interface buries 1,720 Å2 of protein surface, with the interface dominated by van der Waals contacts between conserved hydrophobic and aromatic residues, plus a salt bridge between the side chains of Glu53(KH1) and Arg4(KH2).
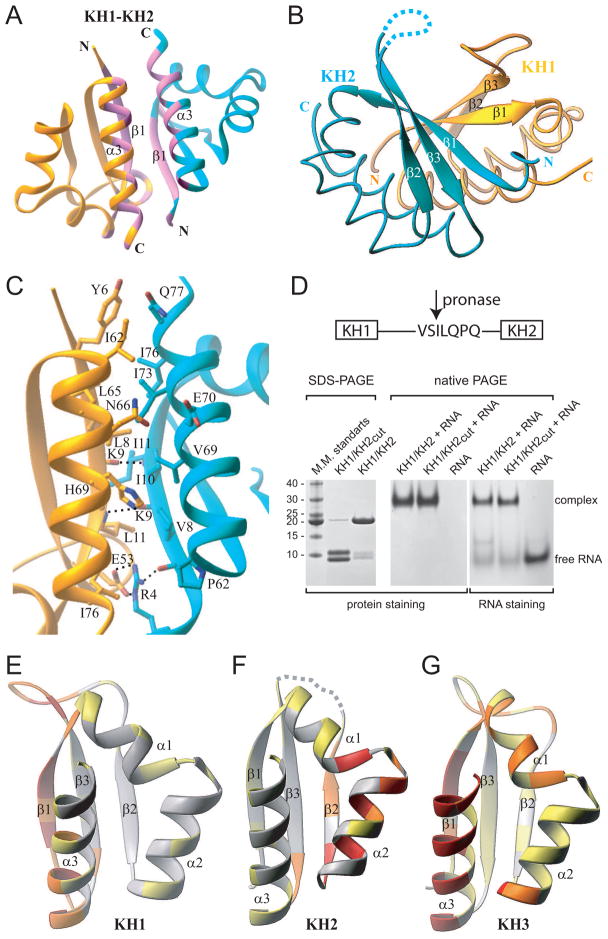
Figure 3. Ribbon Representations of KH1/2 Interface in the KH1/2-RNA Hairpin Complex and Distribution of the Protein-Protein Interaction Propensities over the Surface of Nova KH Domains.
(A) A global overview of the KH1(gold)/KH2(cyan) interface, with the segments constituting the interface, colored in pink.
(B) The three-stranded β-sheets of the KH1 and KH2 domains form a continuous twisted six-stranded β-sheet.
(C) Details of the KH1/2 interface, highlighting interactions between side chains of residues involved in inter-facial contacts.
(D) SDS-PAGE electrophoresis (left panel) showing pronase cleavage resulting in two protein fragments (cleavage at Ser90-Ile91 step) with molecular mass of approximately 9 and 10 kDa. Native PAGE (middle and right panels) establishing complex formation between KH1/KH2 and RNA, regardless of cleavage of the linker.
(E to G) Distribution of the protein-protein interaction propensities over the protein surface as calculated by the Optimal Docking Area (ODA) approach for three different KH domains. Ribbons representation of KH1 (panel E, present study) and KH2 domains of the Nova-1 KH1/2 protein (panel F, present study) and Nova-2 KH3 domain (panel G, Lewis et al., 2000). Structures are colored by the absolute magnitude of the ODA signal from the strongest in red, through medium in orange and weak in yellow, to the weakest in white.
See also Figure S5.
The linker segment (residues 81–99) connecting KH1 and KH2 is disordered in the crystal structure of the KH1/2-RNA complex, consistent with its sensitivity to proteases (Lewis et al., 1999). Pronase cleaves the linker at the Ser90-Ile91 step permitting comparative gel shift studies of complex formation for intact and cleaved KH1/2 with the UCAG-UCAC-containing RNA hairpin (Figure 3D). These studies establish that complex formation is independent of the cleavage status of the linker connecting KH1 and KH2, suggestive of linker-independent association across the KH1/2 interface.
Surface Interaction Propensities of Nova KH Domains
To independently evaluate the interaction propensity of the surface regions of Nova KH domains (Figure 3E–G), we applied the “Optimal Docking Area” (ODA) algorithm (Fernandez-Recio et al., 2005). The calculated energy changes reflect the effect of replacing the water environment for a lower dielectric (but still polar) medium, such as the surface of another protein. The highest protein-protein interaction propensities span adjoining β1/α3 elements for Nova KH1 (Figure 3E) and KH3 (Figure 3G) domains. These results are consistent with the β1/α3 elements of Nova KH1 forming contacts with KH2 (Figures 3A) in the Nova KH1/2-RNA hairpin complex, and with the β1/α3 elements of Nova KH3 forming contacts with symmetry-related KH3 in the crystal (Lewis et al., 2000). By contrast, the highest surface interaction propensities span adjoining β2/α2 elements, the loop between α1- and α2-helices, and the C-terminus of α1-helix for Nova KH2 (Figure 3F). These elements are located on the canonical RNA-binding surface of KH domains and it is this surface of KH2 that participates in protein-protein interactions with symmetry-related KH2 in the crystal (Figure S2B).
Recognition between KH1 Domain of Nova KH1/2 and RNA Hairpin
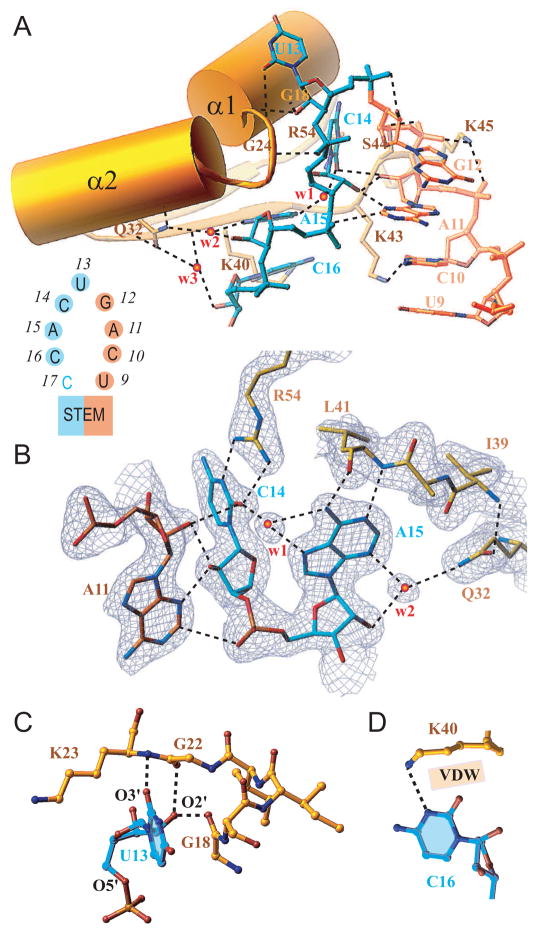
Protein-RNA recognition in the Nova-1 KH1/2-RNA hairpin complex involves only one of the two KH domains and both UCAG-UCAC tetranucleotide elements within the RNA hairpin. Specifically, the KH1 domain primarily targets the U13 to C16 segment, and the C10 base of the preceding U9 to G12 segment (Figure 4A), with C14 and A11 buttressing each other through hydrogen bonding interactions (Figure 4B). The U13 base loops out of the hairpin and interacts with the KH1 domain through both its base and sugar residues (Figure 4A). The U13 base stacks over the Gly18-Ser19 peptide bond, with O4 of U13 forming van der Waals contact with Cβ of Ser19 and O2 of U13 forming a weak hydrogen bond with main chain NH of Lys23 (Figure 4C). In addition, the 2′-OH of the U13 sugar forms a bifurcated hydrogen bond with the main chain O atoms of Gly18 and Gly22. The C14 base forms two hydrogen bonds through its Watson-Crick edge with the guanidinium group of Arg54, with C14 buttressed in position through a network of base-sugar/sugar-base-phosphate hydrogen bonds with A11 of the adjacent U9 to G12 segment (Figure 4B). The next residue, A15 forms a pair of hydrogen bonds to the main chain NH and O atoms of Leu41, and is further buttressed in place by a bifurcated water-mediated hydrogen bond from its base (N3) and sugar (2′-OH) atoms to the side chain of Gln32. The network of hydrogen bonds and water-mediated bridges (w1 and w2) that direct the observed intermolecular recognition and buttressing events, are specific for cytosine at position 14 and adenine residues at positions 15 and 11 in the complex (Figure 4B). The Lys40 and Lys43 side chains are directed towards the C16 and C10 bases, with the specific intermolecular contacts involving these lysine side chains differing between the type I and type II complexes (Figure 4A). The Lys43 ε-amino group forms a hydrogen bond with either N3 atom of C16 or O2 atom of C10. When Lys43 is occupied in the interaction with C10, the N3 atom of C16 is hydrogen bonded to the Lys40 ε-amino group, with the rest of the Lys40 side chain forming van der Waals contacts with O2 atom of C16 (Figure 4D).
Figure 4. KH1-RNA Interactions in Complex.
(A) Overview of interactions between RNA hairpin loop (U9 to G12 in gold and the U13 to C16 in cyan, see insert) and the N-terminal half of the KH1 domain (Tyr15 to Lys45 in gold, encompassing α1, loop I, α2 and β2 segments). Bridging water molecules at the interface are labeled w1, w2 and w3.
(B) Details of intermolecular contacts involving the C14-A15 segment of the RNA, and amino acids Arg54, Lys41 and Gln32 of the KH1 domain. Note buttressing interactions between the base and sugar of A11 and C14. The side chain of Lys40 is removed for clarity.
(C) Details of intermolecular contacts involving U13 base and sugar hydroxyl group of the RNA, and backbone atoms of amino acids Gly18, Gly22 and Lys23 of the KH1 domain.
(D) Intermolecular contacts between the Watson-Crick edge of C16 and the side chain amine (hydrogen bonding) and methylene groups (van der Waals) of Lys40.
Binding of Nova KH1/2 domains to UCAG-UCAC-containing RNA Hairpin
We have used filter-binding assays to measure the binding affinities of the individual KH2 and KH3 domains, KH1/2 dual domains, and full-length KH1/2/3 Nova protein to a panel of three RNA hairpin targets. These include the KH3-binding RNA aptamer containing a single UCAC site (Figure 5A), the KH1/2-binding RNA aptamer containing a UCAG-UCAC site (Figure 5B), and the KH1/2/3-binding RNA aptamer containing a (YCAY-N)3 site (Figure 5C). Thus, while the intact KH1/2/3 Nova protein (open circles, Figure 5D) and KH1/2 dual domains (black circles, Figure 5D) bind the UCAG-UCAC-containing RNA aptamer target (Figure 5B) with μM affinity, the KH3 domain (green circles, Figure 5D) exhibits reduced binding affinity, while the KH2 domain (red circles, Figure 5D) exhibits no measurable binding affinity.
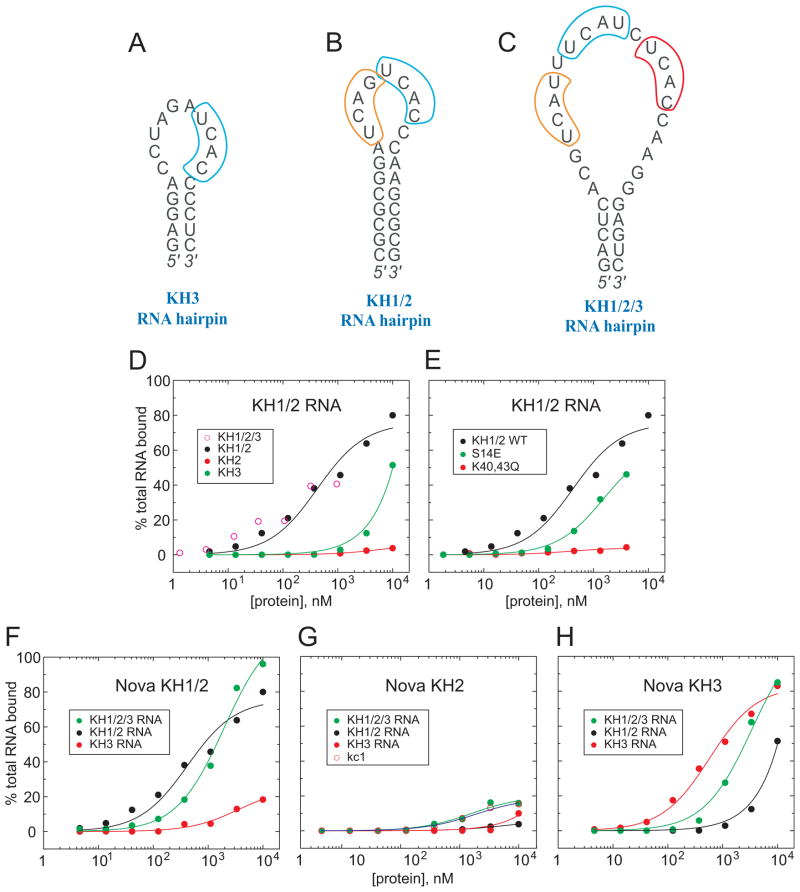
Figure 5. Filter Binding Assays for Nova KH Domain-RNA Complexes.
(A) The sequence of the Nova-2 KH3-binding 20-nt RNA hairpin containing a single UCAN site (Lewis et al., 2000).
(B) The sequence of the Nova-1 KH1/2-binding 25-nt RNA hairpin containing a (UCAN)2 site (Musunuru and Darnell, 2004) that was used in this work.
(C) The sequence of the Nova-1 KH1/2/3-binding 32-nt RNA hairpin containing a (UCAN-N)3 site (Buckanovich and Darnell, 1997).
(D) Filter-binding assays for complex formation of Nova KH1/2/3 (open circles), KH1/2 (black circles), KH2 (red circles), and KH3 (green circles) with KH1/2 RNA hairpin containing a (UCAN)2 site (panel B).
(E) Filter-binding assays for complex formation of wild-type Nova KH1/2 (black circles), Ser14Glu mutant (green circles), and Lys40Gln, Lys43Gln dual mutant (red circles) with KH1/2 RNA hairpin containing a (UCAN)2 site (panel B).
(F) Filter-binding assays for complex formation of Nova KH1/2 with RNA hairpins containing a single UCAN site (panel A) (red circles), (UCAN)2 site (panel B) (black circles), and (UCAN-N)3 site (panel C) (green circles).
(G) Filter-binding assays for complex formation of Nova KH2 with RNA hairpins containing a single UCAN site (panel A) (red circles), (UCAN)2 site (panel B) (black circles), (UCAN-N)3 site (panel C) (green circles), and a RNA hairpin specific for FMRP KH2 domain (open circles).
(H) Filter-binding assays for complex formation of Nova KH3 with RNA hairpins containing a single UCAN site (panel A) (red circles), (UCAN)2 site (panel B) (black circles), and (UCAN-N)3 site (panel C) (green circles).
We have used filter-binding assays to compare the binding affinities of wild-type (KD of 0.42 ± 0.11 μM; black circles, Figure 5E) and mutant KH1/2 dual domains for the KH1/2-binding UCAG-UCAC-containing RNA hairpin (Figure 5B). Replacement of Ser14 in KH1 by its Glu counterpart observed at this position in KH3, within the context of the KH1/2 construct, resulted in an approximate 4-fold decrease in RNA hairpin binding affinity to 1.6 ± 0.2 μM (green circles, Figure 5E). Replacement of Lys40 and Lys43 in KH1 by their Gln counterparts observed at these positions in KH2, within the context of the KH1/2 construct, resulted in no measurable RNA hairpin binding affinity (red circles, Figure 5E).
Binding of Nova KH Domains to (YCAY)n-containing RNA Hairpins
Filter-binding assays have also been used to measure the binding affinities of the KH2 and KH3 individual domains (KH1 is unstable as an individual domain) and KH1/2 dual domains to RNA aptamer hairpin targets containing (YCAY)n sites, where n=1,2,3 (Figure 5A–C). The KH1/2 dual domains bind most tightly to the RNA aptamers containing UCAG-UCAC (black circles, Figure 5F; KD = 0.42 ± 0.11 μM) sites and (YCAY-N)3 (green circles, Figure 5F; KD = 1.9 ± 0.3 μM), with weaker binding affinity to the RNA aptamer containing a single UCAC site (red circles, Figure 5F).
The KH2 domain binds very weakly to all three RNA aptamers (Figure 5G), with the residual binding observed for the RNA aptamer containing a (YCAY-N)3 site (green circles, Figure 5G) being non-specific, since a similar binding curve is observed for an unrelated FMRP KH2 domain-binding RNA aptamer, kc1 (open circles, Figure 5G).
Finally, the KH3 domain binds most tightly to its own RNA aptamer containing a single UCAC site (red circles, Figure 5H; KD = 0.58 ± 0.09 μM), less tightly to the RNA aptamer containing a (YCAY-N)3 site (green circles, Figure 5H; KD = 3.4 ± 0.6 μM) and weakest to the RNA aptamer containing a UCAG-UCAC site (black circles, Figure 5H).
NMR Chemical Shift Mapping of RNA-binding Surface of Nova KH2 Domain
Structural similarities between all three Nova-1 KH domains, as well as the conservation of a key recognition residue Arg54, suggests that the KH2 domain could recognize the YCAY sequence in a manner similar to that observed for KH1 (this study) and KH3 (Lewis et al., 2000) domains in their respective crystal structures. Nevertheless, we detect no binding of the KH2 domain to (YCAY)n-containing RNA hairpins in either crystal (Figure 1E) or solution (Figure 5G) states.
To elucidate whether Nova-1 KH2 is capable of YCAY-containing single-stranded RNA binding, we performed chemical shift perturbation studies of the KH2 domain in the free state and bound to the UCAC RNA tetranucleotide by NMR spectroscopy. We could assign 96% of backbone residues (excluding prolines) in a 1H-15N-HSQC spectrum of the complex, with slow exchange observed between free and bound states (Figure 6A). The largest weighted 1H/15N chemical shift changes on complex formation map to the β2 and β3 strands and their connecting loop, as well as to the residues located on the α1 and α2 helices near the GXXG loop (Figure 6B). These changes when mapped onto the backbone structure of the KH2 domain (Figure 6C), show that KH2 residues Arg54, Leu41, Lys40, Gly22, Ser19 and Gly18, whose KH1 equivalents mediate intermolecular contacts with 13-UCAC-16 motif in the crystal structures of KH1/2-RNA complexes (Figure 4), are located either within or adjacent to these regions (Figure 6C).
Figure 6. NMR-monitored Complexation Shifts in Nova-1 KH2 Domain on Addition of UCAC RNA Tetranucleotide.
(A) Selected regions of 1H,15N-HSQC of Nova-1_KH2 in presence of various amount of UCAC RNA tetranucleotide (1:0 left; 1:4 middle; 1:36 right). Cross peaks of V28 in free (V28f) and RNA-bound state (V28c) are shown.
(B) Histogram outlining the magnitude of the average chemical shift perturbation of the 15N and 1H backbone amide resonances of Nova-1 KH2 on complex formation with UCAC RNA tetranucleotide. The average chemical shift difference Δδave between the free and RNA-bound forms of KH2 was calculated using a correlation: , where ΔδH is the chemical shift of amide proton and ΔδN is the chemical shift of amide nitrogen. Protein secondary structure elements are indicated on the top.
(C) Comparison of the residues involved in KH2-UCAC interactions indicated by the average amide chemical shift perturbations (left panel) with the residues involved in direct KH1-UCAC interactions observed in the crystal structure of KH1/2-RNA hairpin complex (right panel). The RNA is shown in green, the KH1 residues directly contacting U13-C14-A15-C16 segment are shown in red on the KH1 domain backbone (right panel), with the magnitude of the KH2 complexation shifts on RNA binding color-coded on the KH2 domain backbone with red representing the largest and grey the smallest shifts (left panel).
See also Figure S4.
Nova Tandem and Single KH domains Bind to [UCAU(N)2]3 motif
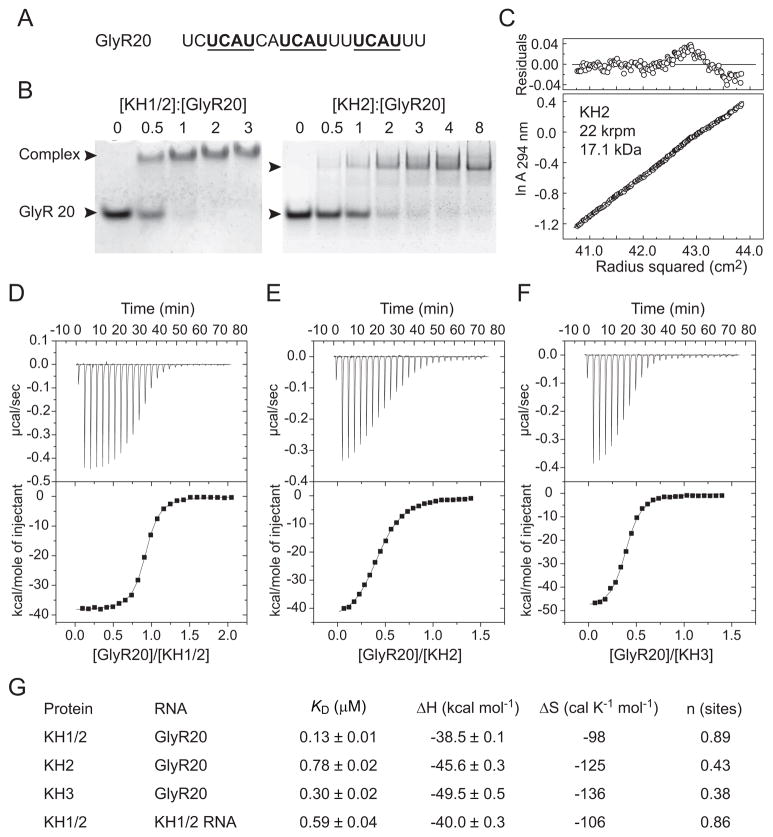
In view of the observation that the KH2 domain is able to specifically recognize an isolated UCAC tetranucleotide RNA sequence, we next studied binding of tandem KH1/2 and single KH2 to in vivo Nova-1 target – an intronic UCAU repeat element of glycine receptor α2 (GlyRα2) pre-mRNA (Figure 7A). Previous studies have identified that Nova-1 binds to the GlyRα2 50-nt sequence in vitro with approximately 20 nM affinity via the [UCAU(N)2]3 motif (Buckanovich and Darnell, 1997), and enhances utilization of an adjacent 3′-splice site of an alternatively spliced exon in vivo (Jensen et al., 2000a). KH1/2 binds GlyRα2 20-nt RNA element (designated GlyR20, Figure 7A) with 1:1 stoichiometry, while KH2 binds this RNA with 2:1 stoichiometry, as determined by electrophoretic mobility shift (gel-shift) assay (Figure 7B). According to analytical ultracentrifugation measurements over a ten-fold range of protein concentrations, the apparent molecular mass of KH2 in the free state (10 to 100μM concentration range) is 17.1 kDa (Figure 7C), indicating that KH2 forms a dimer in solution. Consistently, the gel-filtration column elution volume of KH2 also indicates that it is dimeric in solution (data not shown). Single RNA band shifts observed upon KH1/2 binding at 1:1 protein:RNA molar ratio and KH2 binding at 2:1 protein:RNA molar ratio (Figure 7B) suggest similar modes of KH1/KH2 intramolecular pseudodimer and of KH2/KH2 homodimer binding to GlyR20, with each KH domain of the dimer bound to one of three UCAU motifs of the RNA. We have monitored the binding affinities of Nova-1 KH1/2, KH2 or KH3 to GluR20 by isothermal titration calorimetry (ITC). The dissociation constant (KD) values are 0.13 μM for the KH1/2 (Figures 7D,G), 0.78 μM for the KH2 (Figures 7E,G) and 0.30 μM for the KH3 (Figures 7F,G) complexes with GlyR20. The n value (a number of binding sites per protein monomer) estimated from ITC curves using a single site binding model is 0.89 for the KH1/2-RNA complex, whereas n = 0.43 for KH2-RNA complex, consistent with 1:1 and 2:1 protein:RNA stoichiometries of these respective complexes (Figures 7D,E and G). The n value of 0.38 estimated from ITC curve (Figures 7F,G) for the KH3-RNA complex indicates that stoichiometry of KH3-GlyR20 complex is either 2:1 or 3:1. These experiments demonstrate that individual Nova KH2 and KH3 domains bind [UCAU(N)2]3 motif of single stranded RNA with comparable (2.6 fold difference) binding affinity, while dual KH1/2 domain binds this ssRNA with 6-fold higher affinity compared to a single KH2 domain.
Figure 7. Binding of Nova-1 tandem KH1/2 and single KH2 and KH3 domains to UCAU repeat element of GlyRα2 pre-mRNA.
(A) RNA sequence of an intronic 20-nt UCAU repeat element of GlyRα2 pre-mRNA used in binding experiments. Three non-overlaping UCAU repeats (underlined) separated by 2 nucleotides represent potential binding sites for individual Nova-1 KH domains.
(B) Electrophoretic mobility gel shift data for binding of KH1/2 to GlyR20 establishing 1:1 stoichiometry of the complex (left panel); and of KH2 to GlyR20 establishing 2:1 stoichiometry of the complex (right panel).
(C) Analytical ultracentrifugation data of a 30 μM sample of KH2 at 20°C and 22,000 rpm in 25 mM Tris-HCl (pH 7.5) and 500 mM KCl.
(D–F) Isothermal titration calorimetry (ITC) binding curves of GlyR20 binding to KH1/2 (panel D), KH2 (panel E) and KH3 (panel F) domains. A thermogram as a result of titration is shown in the top panel, and a plot of the total heat released as a function of the molar RNA/protein ratio is shown in the bottom panel. Solid lines indicate non-linear least-squares fit to a theoretical titration curve using Microcal software, with ΔH (binding enthalpy kcal mol−1), KD (association constant), and n (number of binding sites per monomer) as variable parameters.
(G) Summary of ITC-based energetic parameters for tandem and single KH-RNA complexes.
We have also monitored binding of KH1/2 to GlyR20 RNA by NMR spectroscopy. We observed significant chemical shift perturbations between free and RNA-bound states (Figure S4A). The number of KH1/2 resonances that experienced chemical shift changes is at least twice the number observed for individual KH2 (Figure 6B), suggesting that both KH2 and KH1 domains are bound to GlyR20 in the complex. For example, in addition to Gly22 and Gly25 of KH2 domain, there are two other cross-peaks in the same region of the KH1/2 1H-15N-HSQC spectrum that undergo significant chemical shift changes upon RNA binding (compare the selected regions of the spectra of KH2 and KH1/2 in Figures S4B and S4C, respectively).
Discussion
KH domains, which constitute one of the most abundant RNA-binding motifs (Messias and Sattler, 2004), are of particular interest because of their role in regulating gene expression at both transcriptional and translational levels, in alternate splicing, and in the maturation of mRNA (Valverde et al., 2008). Eukaryotic KH domains bind their mRNA targets with sequence specificity and affinities spanning the μM to nM range, and target tetranucleotide sequences where the first and last bases are pyrimidines, with structures of complexes available where the third base is an adenine or cytosine. Our current understanding of the principles underlying KH-RNA recognition emerged following determination of the crystal structure of Nova-2 KH3 domain bound to a UCAC element within the hairpin segment of a RNA stem-loop (Lewis et al., 2000), the NMR structure of the STAR (signal transduction and activation of RNA) member KH domain augmented by a QUA1 (quaker homology 1) domain of SF1 (splicing factor 1) bound to a UAAC segment in a single-stranded RNA context (Liu et al., 2001), the complex between NusA, a key regulator of bacterial antitermination, and RNAs derived from the antitermination region (Beuth et al., 2005), and the crystal structures of KH1 domain of poly(C)-binding protein 2 (PCBP2) bound to C-rich telomeric DNA and RNA (Du et al., 2007). In each case, the RNA tetranucleotide is gripped in a molecular vise composed of a hydrophobic α-helix/β-strand platform, an invariant GXXG motif and a variable loop, thereby aligning the Watson-Crick edge of the adenine at the third position for hydrogen-bonding with the peptide backbone.
The current contribution reports the crystal structure of dual eukaryotic KH domains of Nova-1 targeted to tandem RNA tetranucleotide repeats, where KH1 and KH2 domains form an intramolecular pseudodimer, with KH1 solely involved in RNA recognition and KH2 involved in protein-protein contacts. These results highlight the ability of KH domains to participate in protein-RNA and protein-protein recognition and outline how RNA-RNA buttressing interactions expand on our current understanding of KH-RNA tetranucleotide recognition events.
Comparison of Nova KH1/2 interface with other KH-KH interfaces
Our structure of the complex establishes that KH1 and KH2 have extensive inter-domain interactions, forming an intramolecular pseudodimer. Two recently reported structures revealed a similar arrangement of tandem KH domains in the RNA-free state: the NMR structure of KH1/2 domains of human PCBP2 (Figure S5A) (Du et al., 2008), and the crystal structure of IMP1 mRNA-Binding Protein 1 (also called ZBP1) KH3/4 domains (Figure S5B), whereas three other structures of tandem KH domains in the free state revealed different modes of arrangement of their respective KH domains (Figure S5C–E) (Diaz-Moreno et al., 2010; Gopal et al., 2001; Valverde et al., 2007; Worbs et al., 2001). A detailed comparison of the Nova KH1/2 interface with these KH-KH interfaces is given in the Supplemental Information.
Nova KH1/2-RNA recognition involving tandem YCAY Repeats
The three KH domains of Nova proteins have been shown to target [YCAY-(N)2]3 sites in natural α-2 glycine receptor subunit, GABAA and Nova-1 pre-mRNAs (Buckanovich and Darnell, 1997; Dredge and Darnell, 2003; Dredge et al., 2005) and [YCAY-(N)0–2]3 sites in RNA hairpin aptamers identified through in vitro selection (Buckanovich and Darnell, 1997). Nevertheless, it was not clear whether individual KH domains target one or more tandem YCAY sites or whether one or more KH domains target a single YCAY site. The structure of the Nova-1 KH1/2-RNA hairpin complex reported in the present study addresses this issue by revealing a novel recognition mode, where the KH1 domain differentially recognizes adjacent tandem UCAG and UCAC sites, mediated by specific buttressing interactions between the two sites. The intermolecular interactions are primarily between the KH1 domain and all four nucleotides of the second UCAC tetranucleotide (U13 to C16) site, with a few additional interactions with C10 and its phosphate of the first UCAG tetranucleotide (U9 to G12) site in the complex (Figure 4A). Both UCAG and UCAC sites are involved in sculpting the RNA scaffold due to the extensive base-sugar and base-phosphate interactions associated with the mutual buttressing of A11 of the first tetranucleotide repeat and C14 of the second tetranucleotide repeat in the complex (Figure 4B).
The recognition elements between the KH1 domain and the second UCAC tetranucleotide (U13 to C16) site in the Nova-1 KH1/2-RNA hairpin complex reported in this study are very similar to those reported previously between the KH3 domain and the single UCAC site in the Nova-2 KH3-RNA hairpin complex (Lewis et al., 2000). In both complexes, the sequential uracil, cytosine and adenine residues of the UCAC segment are positioned atop an α/β platform and pinioned in place by interactions with the invariant Gly-X-X-Gly motif and the variable loop. The cytosine pairs with R54 using Watson-Crick-like hydrogen bonding alignments and the adenine pairs with the backbone using Watson-Crick-like hydrogen bonding alignments (Figure 4B).
We highlight below a common intermolecular recognition feature that has been observed in the eukaryotic Nova-2 KH3-RNA hairpin (Lewis et al., 2000) and Nova-1 KH1/2-RNA hairpin (this study) complexes, as well as in the splicing factor 1-intron branch site RNA complex (Liu et al., 2001) and the bacterial NusA KH1/2 complex with a GAACUCAAUAG RNA sequence (Beuth et al., 2005). All these complexes have one key element in common, namely that the main chain of the β2-strand forms two hydrogen bonds with the Watson-Crick edge of the adenine base (Figure 4B). A more detailed comparison of Nova KH1/2-RNA hairpin and NusA KH1/2-ssRNA complexes is provided in the Supplemental Information.
Comparison of RNA Hairpin Topologies in Nova KH1/2 and KH3 Complexes
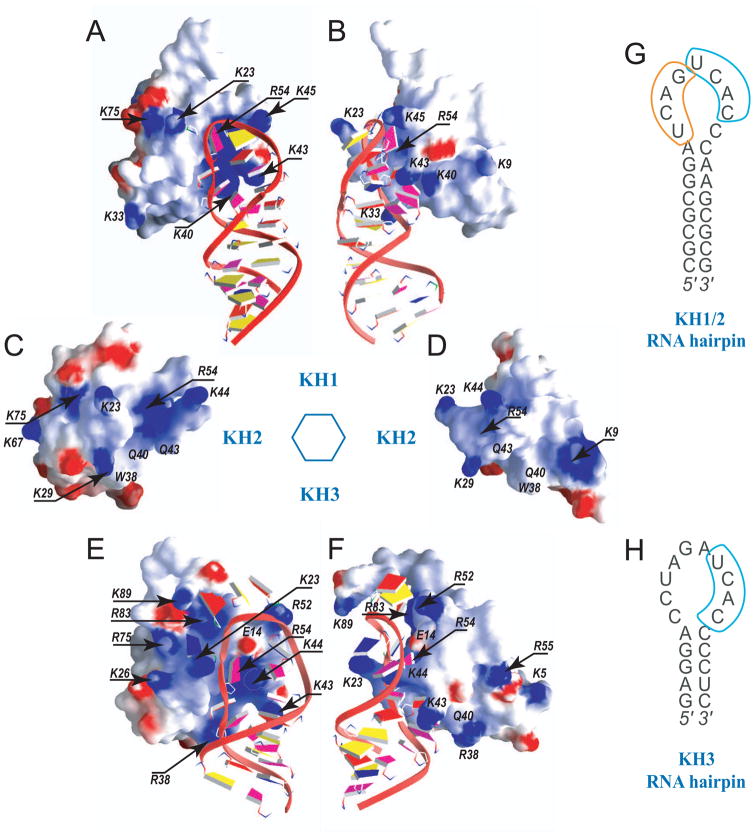
The Nova KH1/2 and KH3 domains bind preferentially their respective RNA hairpin aptamers, while they bind each other’s aptamers significantly weaker (Figure 6F,H), implying that not only the presence of primary UCAY recognition site but also the RNA hairpin loop structure determine binding affinity and specificity of individual Nova KH domains. The charge distributions are somewhat different on the canonical RNA-binding surfaces of the three Nova KH domains (Figures 8). The electrostatics of the RNA-binding KH1 surface in the KH1/2-RNA hairpin complex (Figure 8A,B) appears to be designed to favor binding by a compact RNA loop (contains nine bases), as reflected in a sharp turn facilitated by the Lys45 side chain. The conformation of the primary U13-C14-A15-C16 target site (cyan in Figure 4A), together with the sharp turn, results in the mutual buttressing of C14 by A11 of the neighboring site (Figure 4B). At the same time, two appropriately located positively charged residues Lys40 and Lys43, which adopt finger-like conformations, recognize cytosine bases C16 and C10, respectively (Figure 4A). This network of intermolecular and buttressing interactions results in both the primary UCAC target site and the CA step of the preceding U9-C10-A11-G12 site participating in KH1 domain recognition and stabilization.
Figure 8. Two Perpendicular Views of the Electrostatic Surface of Nova KH1 (with bound RNA), KH2 and KH3 (with bound RNA) Domains.
(A, B) Two alternate views of KH1 domain of KH1/2 complexed with RNA hairpin containing tandem UCAN-sites within the loop segment.
(C, D) Two alternate views of KH2 domain of KH1/2, that does not form a complex with RNA hairpin.
(E, F) Two alternate views of KH3 domain complexed with RNA hairpin containing single UCAC site (Lewis et al., 2000; PDB entry 1EC6).
Blue and red patches are associated with positively- and negatively-charged KH surface segments. The RNA backbone is shown in a ribbon and the bases in a slab representation. The slabs are colored as follows: U, blue; C, magenta; A, red; G, yellow.
(G) The sequence of the Nova-1 KH1/2-binding 25-nt RNA hairpin containing a (UCAN)2 site (Musunuru and Darnell, 2004) that was used in this work.
(H) The sequence of the Nova-2 KH3-binding 20-nt RNA hairpin containing a single UCAN site (Lewis et al., 2000).
The electrostatics of the RNA-binding KH3 surface in the KH3-RNA hairpin complex (Lewis et al., 2000) appears to be designed to favor a less-restrictive RNA loop (contains ten bases) topology, as reflected in no sharp turns in the loop trajectory (Figure 8E). Two important differences between the electrostatics of the RNA-binding surfaces of KH1 (Figure 8A) and KH3 (Figure 8E) are the presence of Glu14 and Lys44 in the latter complex. This distribution of charged amino acids appears to contribute to the separation of the 5′- and 3′-halves of the loop in the KH3 complex, such that the loop topology adopts a more gradual turn trajectory (Figure 8E). The positively charged residues Arg83 and Arg52 together with the negatively charged Glu14 side chain direct the RNA backbone towards the C-terminus of the KH3 domain (Figure 8E). No buttressing interactions are observed across the loop in the KH3-RNA hairpin complex, due to the increased separation between the opposite sides of the loop.
The importance of the electrostatics of the RNA interacting surface of the Nova KH domains to recognition is reinforced from the observed reduction in binding affinities associated with the KH1/2-RNA hairpin complex for the KH1 Ser14Glu single mutant and the KH1 Lys40,43Gln dual mutant (Figure 5E). In the single mutant, neutral serine in KH1 is replaced by negatively-charged glutamate observed at this position in KH3, while in the dual mutant, positively-charged lysines observed at these positions in KH1 are replaced by neutral glutamines observed at these positions in KH2.
The KH2 domain does not participate in RNA recognition in the Nova-1 KH1/2-RNA hairpin complex (Figure 1E), and it does not appear to bind either UCAG-UCAC- or UCAC-containing RNA hairpin aptamers with high affinity and binds very weakly to a RNA hairpin containing (YCAY-N)3 sites (Figure 5G). The charge distributions are somewhat different on the canonical RNA-binding face of the KH2 domain (Figures 8C,D), due to Gln40 and Gln43 replacing their lysine counterparts in the KH1 domain (Figures 8A,B). We attempt to explain why the canonical RNA-binding surface of the KH1 domain as part of a KH1/2 construct (black circles, Figure 5D), but not the KH2 domain (red circles, Figure 5D), targets the Nova-1 KH1/2-RNA hairpin. We put forward two potential contributors to such discriminative recognition following a detailed structural analysis of protein-RNA contacts in the complex, as outlined in the Supplemental Information.
All Nova KH Domains Bind UCAY Repeat Single-Stranded RNA
We demonstrated that all three Nova-1 KH domains bind [UCAU(N)2]3 repeats of a single-stranded RNA, with comparable binding affinities between individual KH2 and KH3 domains (Figure 7). NMR chemical shift perturbation studies (Figure 6A,B) establish that Nova KH2 binds the UCAC RNA tetranucleotide in slow exchange, a feature characteristic of a tightly bound complex in solution. These results on a single-stranded RNA that spans the KH domain binding pocket contrasts with no measurable binding when the same tetranucleotide element, or its tandem repeats, is part of the loop segment of RNA hairpins (Figure 5G). The chemical shift changes in the KH2 domain on UCAC binding are mainly distributed within the nucleic acid-binding channel (Figure 6C), and their magnitude is in the range reported for Nova KH3 binding to ssRNA pentanucleotide sequences containing UCAN (N is either pyrimidine or purine) elements (Beuth et al., 2007).
The ability of KH2 to recognize UCAC motif is fully expected based on structural similarity of its RNA-binding surface to that of the other two Nova KH domains, as well as the conservation of a key recognition residue Arg54. A 2.6-fold lower binding affinity of KH2 than KH3 for [UCAU(N)2]3 repeat ssRNA is consistent with less positively charged RNA binding surface of KH2 domain compared to that of KH3 domain (Figure 8C–F). We have established that KH2 domain in an isolated state forms a dimer in solution. Since hydrophobic interactions dominate the KH1/KH2 interface of the dual KH1/2 domain, the KH2 domain in the absence of KH1 binding partner is likely forms the KH2/KH2 homodimer through the same hydrophobic β1/α3 surface. Such very similar dimeric interfaces have been previously observed between KH1 domains of PCBP2 in the crystal structures of its complexes with poly(C) nucleic acids (Du et al., 2007) and between the tandem KH1 and KH2 domains of the protein (Du et al., 2008) (Figure S5A). Therefore, it is possible that KH2/KH2 homodimer and KH1/KH2 intramolecular pseudodimer bind RNA in a similar way, such that each KH domain of a dimer bind the two terminal UCAU motifs of GlyR20, consistent with 1:1 (RNA:dimer) stoichiometry determined for each complex. The NMR chemical shift perturbations observed for the intramolecular KH1/2 pseudodimer upon binding GlyR20 (Figure S4) are also consistent with the involvement of both KH domains in the interaction with the RNA. This model is stereochemically possible as the 8 nucleotide intervening sequence is of sufficient length to connect the two terminal UCAU motifs simultaneously bound by the two KH domains of a dimer.
Nova KH Domain Surface Mimicry Associated with Protein and RNA Recognition
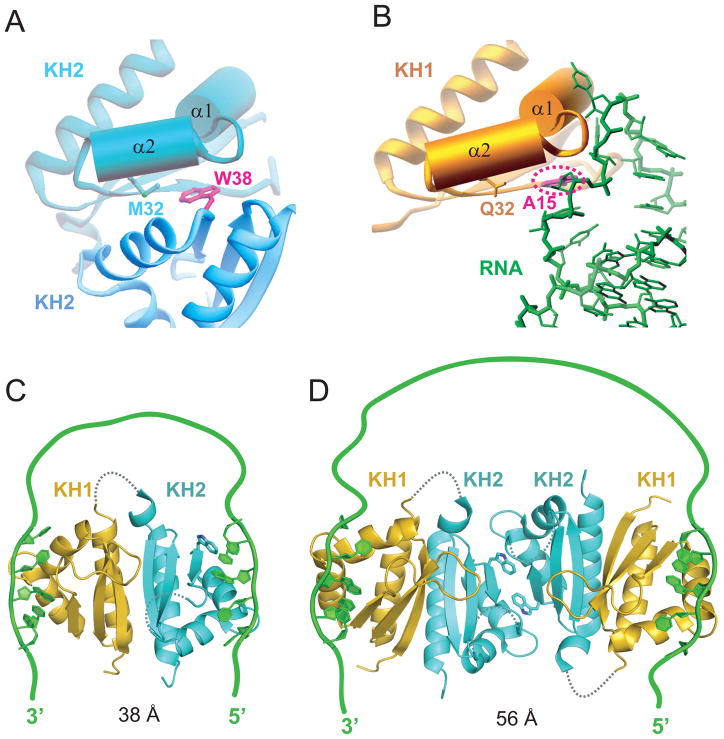
The same canonical RNA-binding surface of the KH domain participates in protein-protein interactions in the crystal (KH2 domain, top left panel of Figure 9A) or in RNA recognition (KH1 domain, top left panel of Figure 9B) in the Nova-1 KH1/2-RNA hairpin complex. A competing molecular recognition duality has also been reported for the Y14 protein, whose canonical RNA-binding surface, is also involved in protein-protein interactions with Mago, with both Y14 and Mago being components of the exon junction complex (Fribourg et al., 2003; Lau et al., 2003; Shi and Xu, 2003).
Figure 9. Models for Interaction of Nova KH1/2 Domains with Wild-type RNA Targets through a RNA Looping Mechanism.
(A) The KH2-KH2 interaction between symmetry-related molecules in the crystal of the KH1/2-RNA hairpin complex. Note the juxtaposition of Met32 and Trp38 within van der Waals contact across the interface.
(B) The KH1 (gold) - RNA (green) interface in the KH1/2-RNA hairpin complex. Note the juxtaposition of Gln32 on KH1 and A15 on the RNA within van der Waals contact across the intermolecular interface.
(C) Model where Nova-1 KH1/2 monomer (based on ultracentrifugation data in solution) is targeted by a RNA containing a pair of sequence elements capable of targeting KH1 (with high affinity) and KH2 (most likely with lower affinity) and separated by a linker segment of sufficient length. The stoichiometry of this complex is 1:1 Nova-1 KH1/2:RNA.
(D) Model of Nova-1 KH1/2 based on the x-ray structure of the complex (Figure 2A), where the RNA-binding surfaces of the KH2 domains pack against each other in the crystal lattice. The target RNA contains a pair of sequence elements capable of targeting KH1 with high affinity and separated by a longer linker segment of sufficient length. The stoichiometry of this complex is 2:1 Nova-1 KH1/2:RNA.
Equally striking is the extension of the structural mimicry to an element of the interacting partner. Thus, the indole ring of a Trp residue mediates the protein-protein contacts involving KH2 domains in the crystal (Figure 9A), while the purine ring of the key adenine base occupies the same interfacial position in the KH1-RNA interaction (Figure 9B) in the Nova-1 KH1/2-RNA hairpin complex.
Alternate Models involving RNA Looping on the Surface of Nova-1 KH1/2
Our ultracentrifugation studies imply that Nova-1 KH1/2 forms a monomer in the free state (Figure S3A) and when bound to its RNA aptamer target (Figure S3B) in dilute solution. The KH1 and KH2 domains interact with each other through an extensive interface (Figures 3A and 3C), thereby positioning their canonical RNA-binding surfaces at opposite ends of the molecule and accessible for recognition. Thus, an RNA containing a pair of sequence elements capable of targeting KH1 (with high affinity) and KH2 (most likely with lower affinity) and separated by a linker segment of sufficient length, should be capable of complex formation through a RNA looping mechanism (Figure 9C), as initially proposed for splicing regulation from the structure of tandem RNA-binding domains RBD3/4 of polypyrimidine tract-binding protein bound to RNA (Oberstrass et al., 2005). An RNA looping mechanism has also been proposed for RNA recognition based on the structure of tandem zinc-finger domains of Muscleblind-like protein bound to RNA (Teplova and Patel, 2008), KH1/2 domains of polyC-binding protein in the free state (Du et al., 2008) and KH3/4 domains of IMP1 protein in the free state (Chao et al., 2010).
An alternate possibility for an RNA looping mechanism is based on the x-ray structure of the complex (Figure 1E), where the RNA-binding surfaces of the KH2 domains pack against each other in the crystal lattice. Thus, an RNA containing a pair of sequence elements capable of targeting KH1 and separated by a linker segment of sufficient length, could also be capable of complex formation through an RNA looping mechanism (Figure 9D).
We have no definitive evidence differentiating between models of RNA looping between the 1:1 KH1/2-RNA complex shown in Figure 9C and the 2:1 KH1/2-RNA complex shown in Figure 9D, especially in the context of full-length Nova protein containing KH1/2/3 domains and under cellular conditions, where additional binding proteins could modulate the recognition process. These models are proposed in the spirit of stimulating further research in this area.
Biological Implications
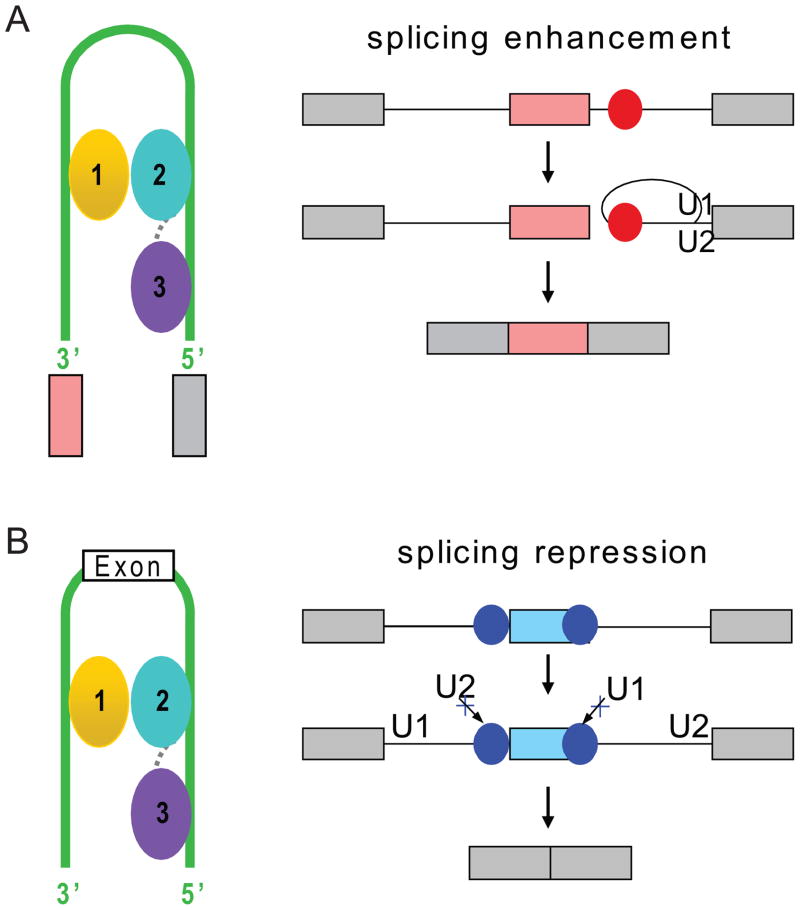
Nova proteins can either enhance or inhibit exon inclusion depending on the position of its binding sites relative to the regulated exon (Ule et al., 2006). If the KH domains of Nova bind intronic YCAY clusters, it could enhance exon inclusion, whereas if it binds YCAY clusters located within the exon or within intronic region immediately adjacent to an alternative exon, it could inhibit exon inclusion. Our structural models of target pre-mRNA looping induced by Nova binding (Figure 9C,D) could provide one explanation of Nova’s function in splicing regulation. Looping of the intron induced by Nova binding might help to bring 5′ and 3′ splice cites in close proximity and thereby enhance spliceosome assembly and exon inclusion (Figure 10A). By contrast, looping out an entire exon or adjacent regions could prevent assembly of the spliceosome components on pre-mRNA, subsequently resulting in exon skipping (Figure 10B).
Figure 10. Models of RNA looping induced by Nova KH1–3 domains binding to splicing enhancers and splicing silencers predicted by Nova RNA map of splicing regulation (Ule et al., 2006).
(A) Model based on the examples of Nova upregulation of exon inclusion by binding to intronic splicing enhancer elements (red circles) to enhance spliceosome assembly.
(B) Model based on the examples of Nova inhibition of exon inclusion by binding to exonic splicing silencing element (blue circle) and blocking U1 snRNP (U1) assembly on the pre-mRNA, and Nova inhibition of exon inclusion by binding to intronic splicing silencing element immediately upstream of alternative exon by blocking recognition of the 3′ splice site by U2 snRNP (U2). KH1, KH2 and KH3 are colored gold, cyan and purple, respectively. The linkers between KH1 and KH2, and between KH2 and KH3 are represented by the dotted gray lines. The green line represents RNA loop.
The current structural data provides a satisfying mechanistic explanation for the functional interaction maps generated that describe Nova actions on target pre-mRNAs (Licatalosi and Darnell, 2010; Licatalosi et al., 2008; Ule et al., 2006). The looping model discussed above not only suggests explanations for these position-dependent interactions, but may be extended in several additional directions. Hints of a Nova-dependent positional map in which regulation of alternative polyadenylation (Licatalosi et al., 2008) may have analogous explanations in the looping model described. Moreover, we have found that Nova has functional interactions with other RNABPs. These include inhibitory interactions with PTBP2 (Polydorides et al., 2000), suggesting that some protein-protein interactions may inhibit Nova-RNA-looping bridges from forming. In contrast, interactions with the Fox1/2 RNA binding protein (Zhang et al., 2010) are agonistic, suggesting that dimer/heterodimer formation may indeed play an important role providing sufficient RNA binding affinities to allow loop formation.
EXPERIMENTAL PROCEDURES
Detailed procedures for protein and RNA preparation, ITC measurements, crystallization and data collection, structure determination and refinement, NMR sample preparation, NMR spectroscopy and chemical shift assignments, NMR relaxation measurements, gel electrophoretic mobility shift binding assays, and filter binding assays are listed under Experimental Procedures in the Supplemental Information.
Supplementary Material
Acknowledgments
DJP received support from NIH grant CA49982, JCD from NIH grant HD40647, RBD from NIH grant NS34389, and K.M., S.K.B., and R.B.D. from NIH grant NS40955 and the Howard Hughes Medical Institute. K.M. received support from the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD program and NIH MSTP grant GM07739.
Footnotes
Coordinates Deposition
Atomic coordinates and structure factors for the type I and type II Nova-1 KH1/KH2-RNA hairpin complexes have been deposited in the Protein Data Bank (www.rutgers.edu/pub) under accession codes 2ANN and 2ANR, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert ML, Darnell RB. Paraneoplastic neurological degenerations: keys to tumour immunity. Nat Rev Cancer. 2004;4:36–44. doi: 10.1038/nrc1255. [DOI] [PubMed] [Google Scholar]
- Beuth B, Garcia-Mayoral MF, Taylor IA, Ramos A. Scaffold-independent analysis of RNA-protein interactions: the Nova-1 KH3-RNA complex. J Am Chem Soc. 2007;129:10205–10210. doi: 10.1021/ja072365q. [DOI] [PubMed] [Google Scholar]
- Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–3587. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Moreno I, Hollingworth D, Kelly G, Martin S, Garcia-Mayoral M, Briata P, Gherzi R, Ramos A. Orientation of the central domains of KSRP and its implications for the interaction with the RNA targets. Nucleic Acids Res. 2010;38:5193–5205. doi: 10.1093/nar/gkq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Darnell RB. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol Cell Biol. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Polydorides AD, Darnell RB. The splice of life: alternative splicing and neurological disease. Nat Rev Neurosci. 2001;2:43–50. doi: 10.1038/35049061. [DOI] [PubMed] [Google Scholar]
- Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Fenn S, Tjhen R, James TL. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J Biol Chem. 2008;283:28757–28766. doi: 10.1074/jbc.M803046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Lee JK, Fenn S, Tjhen R, Stroud RM, James TL. X-ray crystallographic and NMR studies of protein-protein and protein-nucleic acid interactions involving the KH domains from human poly(C)-binding protein-2. RNA. 2007;13:1043–1051. doi: 10.1261/rna.410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Recio J, Totrov M, Skorodumov C, Abagyan R. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins. 2005;58:134–143. doi: 10.1002/prot.20285. [DOI] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- Gopal B, Haire LF, Gamblin SJ, Dodson EJ, Lane AN, Papavinasasundaram KG, Colston MJ, Dodson G. Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution. J Mol Biol. 2001;314:1087–1095. doi: 10.1006/jmbi.2000.5144. [DOI] [PubMed] [Google Scholar]
- Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000a;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci U S A. 2000b;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. Structure of the Y14-Magoh core of the exon junction complex. Curr Biol. 2003;13:933–941. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, Burley SK. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure. 1999;7:191–203. doi: 10.1016/S0969-2126(99)80025-2. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- Messias AC, Sattler M. Structural basis of single-stranded RNA recognition. Acc Chem Res. 2004;37:279–287. doi: 10.1021/ar030034m. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Darnell RB. Paraneoplastic neurologic disease antigens: RNA-binding proteins and signaling proteins in neuronal degeneration. Annu Rev Neurosci. 2001;24:239–262. doi: 10.1146/annurev.neuro.24.1.239. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Darnell RB. Determination and augmentation of RNA sequence specificity of the Nova K-homology domains. Nucleic Acids Res. 2004;32:4852–4861. doi: 10.1093/nar/gkh799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Hentze MW. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci U S A. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xu RM. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev. 2003;17:971–976. doi: 10.1101/gad.260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- Valverde R, Pozdnyakova I, Kajander T, Venkatraman J, Regan L. Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure. 2007;15:1090–1098. doi: 10.1016/j.str.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- Yang YY, Yin GL, Darnell RB. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci U S A. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.